Identification of Aly1 and Aly2 as Modulators of Cytoplasmic pH in Saccharomyces cerevisiae
Abstract
1. Introduction
2. Materials and Methods
2.1. Yeast Strains
2.2. Gene Deletion Assay
2.3. Suppressor Screening Assay
2.4. SNARF-AM Staining Assay
3. Results
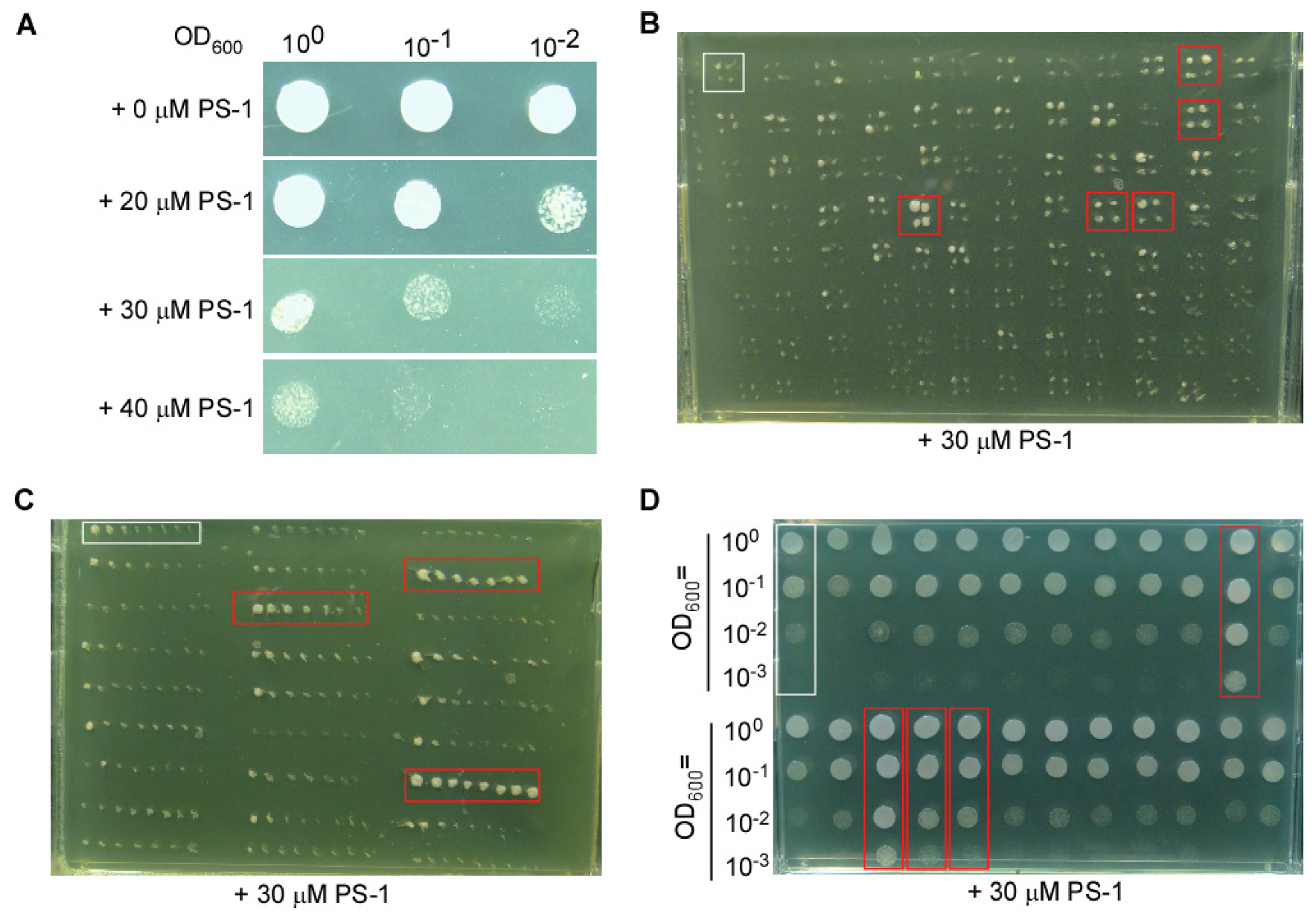
3.1. Screening for New Components Possibly Involved in Cytosolic pH Homeostasis
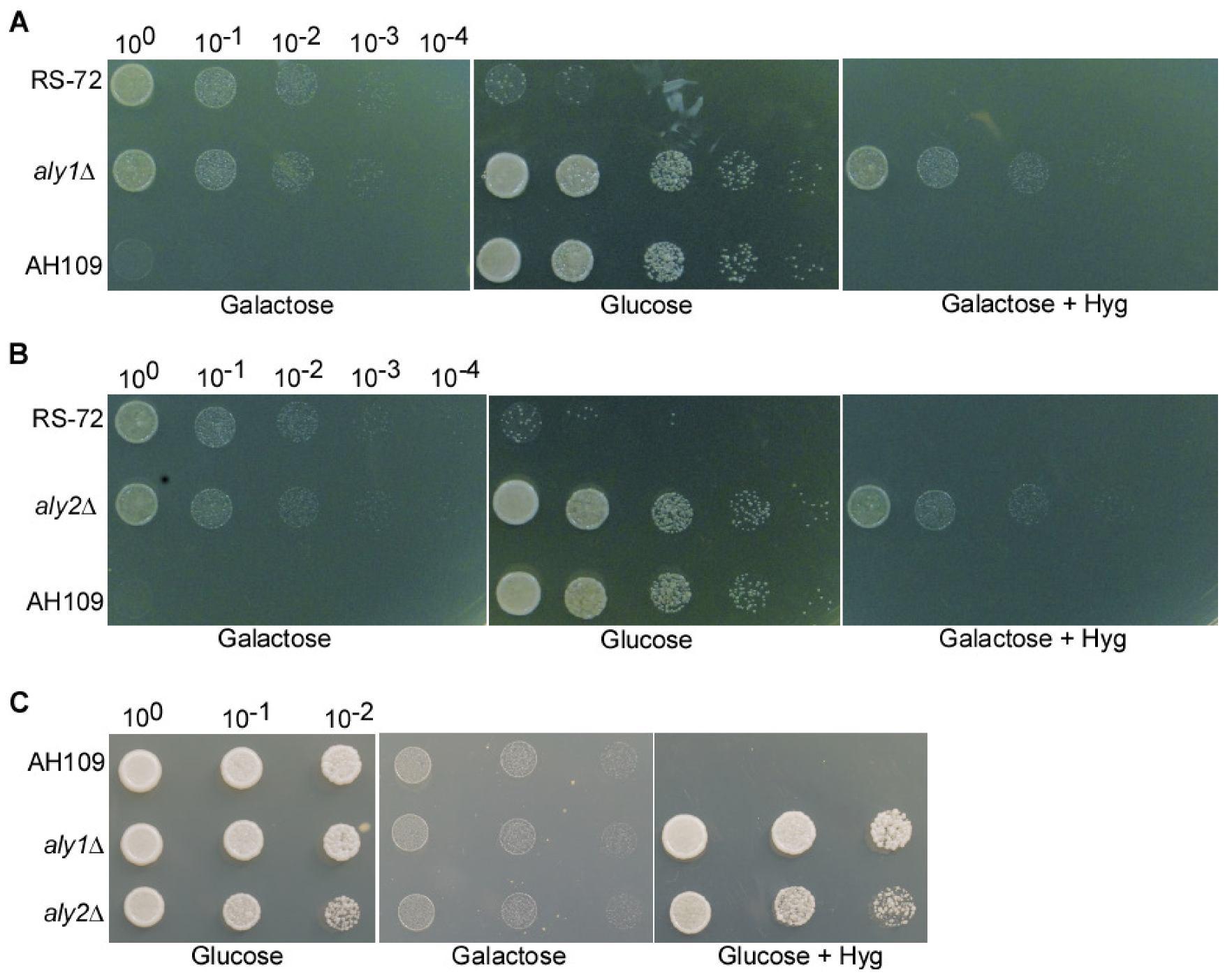
3.2. The Loss of Aly1 and Aly2 Function Can Rescue Growth of Yeast Cells with Reduced PMA1 Activity
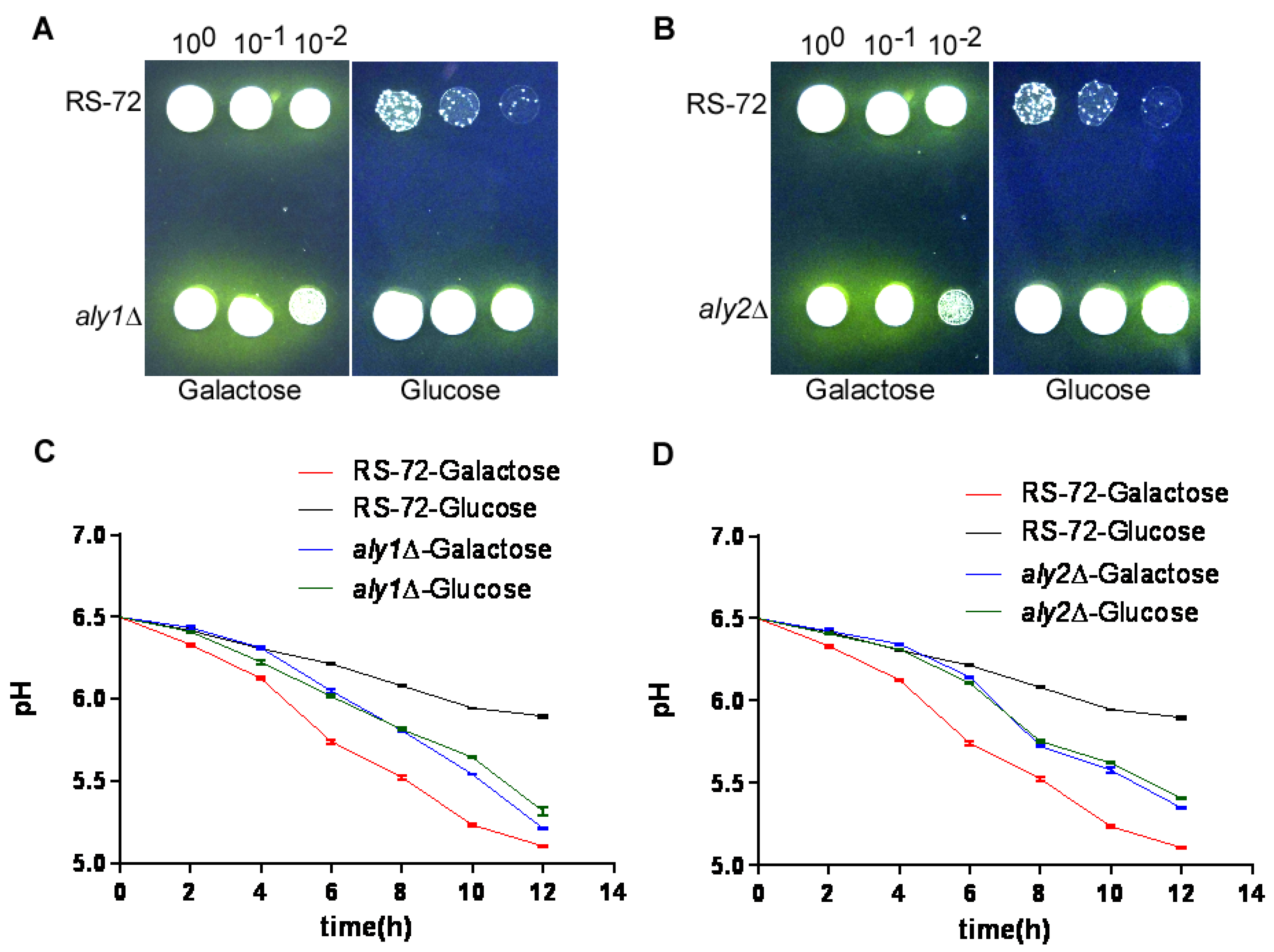
3.3. The Loss of Aly1 or Aly2 Function Promotes H+ Extrusion in Yeast Cells with Reduced PMA1 Activity
3.4. The Loss of Aly1 or Aly2 Function Recovers Cytosolic pH in Yeast Cells with Reduced PMA1 Activity
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Deprez, M.A.; Eskes, E.; Wilms, T.; Ludovico, P.; Winderickx, J. pH homeostasis links the nutrient sensing PKA/TORC1/Sch9 ménage-à-trois to stress tolerance and longevity. Microb. Cell 2018, 5, 119–136. [Google Scholar] [CrossRef] [PubMed]
- Isom, D.G.; Page, S.C.; Collins, L.B.; Kapolka, N.J.; Taghon, G.J.; Dohlman, H.G. Coordinated regulation of intracellular pH by two glucose-sensing pathways in yeast. J. Biol. Chem. 2018, 293, 2318–2329. [Google Scholar] [CrossRef] [PubMed]
- Ramos, S.; Balbín, M.; Raposo, M.; Valle, E.; Pardo, L.A. The mechanism of intracellular acidification induced by glucose in Saccharomyces cerevisiae. J. Gen. Microbiol. 1989, 135, 2413–2422. [Google Scholar] [CrossRef] [PubMed]
- Eskes, E.; Deprez, M.A.; Wilms, T.; Winderickx, J. pH homeostasis in yeast; the phosphate perspective. Curr. Genet. 2018, 64, 155–161. [Google Scholar] [CrossRef]
- Huang, C.; Chang, A. pH-dependent cargo sorting from the Golgi. J. Biol. Chem. 2011, 286, 10058–10065. [Google Scholar] [CrossRef]
- Kane, P.M. Proton Transport and pH Control in Fungi. Adv. Exp. Med. Biol. 2016, 892, 33–68. [Google Scholar]
- Serrano, R. In vivo glucose activation of the yeast plasma membrane ATPase. FEBS Lett. 1983, 156, 11–14. [Google Scholar] [CrossRef]
- Ma, T.Y.; Deprez, M.A.; Callewaert, G.; Winderickx, J. Coordinated glucose-induced Ca2+ and pH responses in yeast Saccharomyces cerevisiae. Cell Calcium 2021, 100, 102479. [Google Scholar] [CrossRef]
- Lecchi, S.; Nelson, C.J.; Allen, K.E.; Swaney, D.L.; Thompson, K.L.; Coon, J.J.; Sussman, M.R.; Slayman, C.W. Tandem phosphorylation of Ser-911 and Thr-912 at the C terminus of yeast plasma membrane H+-ATPase leads to glucose-dependent activation. J. Biol. Chem. 2007, 282, 35471–35481. [Google Scholar] [CrossRef]
- Saliba, E.; Evangelinos, M.; Gournas, C.; Corrillon, F.; Georis, I.; André, B. The yeast H+-ATPase Pma1 promotes Rag/Gtr-dependent TORC1 activation in response to H+-coupled nutrient uptake. eLife 2018, 7, e31981. [Google Scholar] [CrossRef]
- Tomashevsky, A.A.; Petrov, V.V. Point mutations in the different domains of the Saccharomyces cerevisiae plasma membrane PMA1 ATPase cause redistribution among fractions of inorganic polyphosphates. J. Biomol. Struct. Dyn. 2022, 40, 635–647. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Nasution, O.; Lee, Y.M.; Kim, E.; Choi, W.; Kim, W. Overexpression of PMA1 enhances tolerance to various types of stress and constitutively activates the SAPK pathways in Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2017, 101, 229–239. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.Y.; Jang, E.; Ko, N.; Kim, M.; Kim, S.Y.; Moon, Y.; Nam, J.S.; Lee, S.; Jun, Y. A genome-wide screen reveals that endocytic genes are important for Pma1p asymmetry during cell division in Saccharomyces cerevisiae. Int. J. Mol. Sci. 2022, 23, 2364. [Google Scholar] [CrossRef] [PubMed]
- Tung, T.T.; Nielsen, J. Drug discovery and development on Pma1, where are we now? A critical review from 1995 to 2022. ChemMedChem 2022, 17, e202200356. [Google Scholar] [CrossRef] [PubMed]
- Wilms, T.; Swinnen, E.; Eskes, E.; Dolz-Edo, L.; Uwineza, A.; Van Essche, R.; Rosseels, J.; Zabrocki, P.; Cameroni, E.; Franssens, V.; et al. The yeast protein kinase Sch9 adjusts V-ATPase assembly/disassembly to control pH homeostasis and longevity in response to glucose availability. PLoS Genet. 2017, 13, e1006835. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Muñoz, G.A.; Kane, P. Vacuolar and plasma membrane proton pumps collaborate to achieve cytosolic pH homeostasis in yeast. J. Biol. Chem. 2008, 283, 20309–20319. [Google Scholar] [CrossRef] [PubMed]
- Smardon, A.M.; Kane, P.M. Loss of vacuolar H+-ATPase activity in organelles signals ubiquitination and endocytosis of the yeast plasma membrane proton pump Pma1p. J. Biol. Chem. 2014, 289, 32316–32326. [Google Scholar] [CrossRef] [PubMed]
- Velivela, S.D.; Kane, P.M. Compensatory Internalization of Pma1 in V-ATPase Mutants in Saccharomyces cerevisiae Requires Calcium- and Glucose-Sensitive Phosphatases. Genetics 2018, 208, 655–672. [Google Scholar] [CrossRef]
- Tarsio, M.; Zheng, H.; Smardon, A.M.; Martínez-Muñoz, G.A.; Kane, P.M. Consequences of loss of Vph1 protein-containing vacuolar ATPases (V-ATPases) for overall cellular pH homeostasis. J. Biol. Chem. 2011, 286, 28089–28096. [Google Scholar] [CrossRef]
- Cid, A.; Perona, R.; Serrano, R. Replacement of the promoter of the yeast plasma membrane ATPase gene by a galactose-dependent promoter and its physiological consequences. Curr. Genet. 1987, 12, 105–110. [Google Scholar] [CrossRef]
- Yang, Y.; Liu, X.; Guo, W.; Liu, W.; Shao, W.; Zhao, J.; Li, J.; Dong, Q.; Ma, L.; He, Q.; et al. Testing the polar auxin transport model with a selective plasma membrane H+-ATPase inhibitor. J. Integr. Plant Biol. 2022, 64, 1229–1245. [Google Scholar] [CrossRef] [PubMed]
- Palmgren, M.G.; Christensen, G. Functional comparisons between plant plasma membrane H+-ATPase isoforms expressed in yeast. J. Biol. Chem. 1994, 269, 3027–3033. [Google Scholar] [CrossRef] [PubMed]
- Hatakeyama, R.; Kamiya, M.; Takahara, T.; Maeda, T. Endocytosis of the aspartic acid/glutamic acid transporter Dip5 is triggered by substrate-dependent recruitment of the Rsp5 ubiquitin ligase via the arrestin-like protein Aly2. Mol. Cell Biol. 2010, 30, 5598–5607. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, A.F.; Huang, L.; Thorner, J.; Cyert, M.S. A calcineurin-dependent switch controls the trafficking function of α-arrestin Aly1/Art6. J. Biol. Chem. 2013, 288, 24063–24080. [Google Scholar] [CrossRef] [PubMed]
- Kahlhofer, J.; Leon, S.; Teis, D.; Schmidt, O. The α-arrestin family of ubiquitin ligase adaptors links metabolism with selective endocytosis. Biol. Cell 2021, 113, 183–219. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, A.F.; Apffel, A.; Gardner, R.G.; Cyert, M.S. Alpha-arrestins Aly1 and Aly2 regulate intracellular trafficking in response to nutrient signaling. Mol. Biol. Cell 2010, 21, 3552–3566. [Google Scholar] [CrossRef] [PubMed]
- Becuwe, M.; Léon, S. Integrated control of transporter endocytosis and recycling by the arrestin-related protein Rod1 and the ubiquitin ligase Rsp5. eLife 2014, 3, e03307. [Google Scholar] [CrossRef][Green Version]
- Ivashov, V.; Zimmer, J.; Schwabl, S.; Kahlhofer, J.; Weys, S.; Gstir, R.; Jakschitz, T.; Kremser, L.; Bonn, G.K.; Lindner, H.; et al. Complementary α-arrestin-ubiquitin ligase complexes control nutrient transporter endocytosis in response to amino acids. eLife 2020, 9, e58246. [Google Scholar] [CrossRef]
- Wawrzycka, D.; Sadlak, J.; Maciaszczyk-Dziubinska, E.; Wysocki, R. Rsp5-dependent endocytosis and degradation of the arsenite transporter Acr3 requires its N-terminal acidic tail as an endocytic sorting signal and arrestin-related ubiquitin-ligase adaptors. Biochim. Biophys. Acta Biomembr. 2019, 1861, 916–925. [Google Scholar] [CrossRef]
- Hager, N.A.; Krasowski, C.J.; Mackie, T.D.; Kolb, A.R.; Needham, P.G.; Augustine, A.A.; Dempsey, A.; Szent-Gyorgyi, C.; Bruchez, M.P.; Bain, D.J.; et al. Select α-arrestins control cell-surface abundance of the mammalian Kir2.1 potassium channel in a yeast model. J. Biol. Chem. 2018, 293, 11006–11021. [Google Scholar] [CrossRef]
- Young, B.P.; Shin, J.J.; Orij, R.; Chao, J.T.; Li, S.C.; Guan, X.L.; Khong, A.; Jan, E.; Wenk, M.R.; Prinz, W.A.; et al. Phosphatidic acid is a pH biosensor that links membrane biogenesis to metabolism. Science 2010, 329, 1085–1088. [Google Scholar] [CrossRef] [PubMed]
- Robinson, B.P.; Hawbaker, S.; Chiang, A.; Jordahl, E.M.; Anaokar, S.; Nikiforov, A.; Bowman, R.W., 2nd; Ziegler, P.; McAtee, C.K.; Patton-Vogt, J.; et al. Alpha-arrestins Aly1/Art6 and Aly2/Art3 regulate trafficking of the glycerophosphoinositol transporter Git1 and impact phospholipid homeostasis. Biol. Cell 2022, 114, 3–31. [Google Scholar] [CrossRef] [PubMed]
- Li, S.C.; Diakov, T.T.; Xu, T.; Tarsio, M.; Zhu, W.; Couoh-Cardel, S.; Weisman, L.S.; Kane, P.M. The signaling lipid PI(3,5)P₂ stabilizes V₁-V(o) sector interactions and activates the V-ATPase. Mol. Biol. Cell 2014, 25, 1251–1262. [Google Scholar] [CrossRef] [PubMed]

| NO. | Ver4.0 Position | Systemic ID | Ortholog in Budding Yeast | Gene Description |
|---|---|---|---|---|
| 1 | V4-P02-12 (A12) | SPAC12G12.13c | YNL299W| YOL115W | poly(A) polymerase Cid14 |
| 2 | V4-P02-54 (E6) | SPAC13G7.11 | YBR185C | mitochondrial respiratory complex assembly protein (predicted) |
| 3 | V4-P04-33 (C9) | SPAC17G8.13c | YBL052C | histone acetyltransferase Mst2 |
| 4 | V4-P04-46 (D10) | SPAC1805.01c | YAL017W| YOL045W | serine/threonine protein kinase Ppk6 (predicted) |
| 5 | V4-P06-75 (G3) | SPAC22H10.09 | NONE | sequence orphan |
| 6 | V4-P07-62 (F2) | SPAC25B8.18 | YKR049C | mitochondrial electron carrier (predicted) |
| 7 | V4-P12-04 (A4) | SPAC6G10.08 | YDL066W|YLR174W|YNL009W | isocitrate dehydrogenase Idp1 (predicted) |
| 8 | V4-P13-47 (D11) | SPACUNK4.12c | YLR389C | metallopeptidase (predicted) |
| 9 | V4-P17-74 (G2) | SPBC18H10.06c | YKL018W | Set1C complex subunit Swd2.1 |
| 10 | V4-P18-15 (B3) | SPBC19G7.04 | NONE | HMG box protein |
| 11 | V4-P19-85 (H1) | SPBC2D10.04 | YJL084C| YKR021W | arrestin Aly1 related, implicated in endocytosis |
| 12 | V4-P22-41 (D5) | SPBC609.02 | YNL128W | phosphatidylinositol-3,4,5-trisphosphate3-phosphatasePtn1 |
| 13 | V4-P25-06 (A6) | SPCC1235.09 | YBR103W | Set3 complex subunit Hif2 |
| 14 | V4-P26-63 (F3) | SPCC1906.02c | YGL110C | CUE domain protein Cue3 (predicted) |
| 15 | V4-P27-17 (B5) | SPCC31H12.05c | YER133W | serine/threonine protein phosphatase Sds21 |
| 16 | V4-P28-81 (G9) | SPCC794.10 | YHL012W| YKL035W | UTP-glucose-1-phosphate uridylyltransferase (predicted) |
| 17 | V4-P30-05 (A5) | SPAC186.02c | NONE | hydroxyacid dehydrogenase (predicted) |
| 18 | V4-P33-74 (G2) | SPBC29A3.21 | NONE | sequence orphan |
| 19 | V4-P34-95 (H11) | SPBPB10D8.07c | YPL092W | membrane transporter (predicted) |
| 20 | V4-P35-39 (D3) | SPCC162.11c | YNR012W | uridine kinase/uracil phosphoribosyltransferase (predicted) |
| 21 | V4-P35-50 (E2) | SPCC16C4.17 | NONE | meiotically upregulated gene Mug123 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, G.; Han, X.; Yu, X.; Wang, Y.; Ma, J.; Yang, Y. Identification of Aly1 and Aly2 as Modulators of Cytoplasmic pH in Saccharomyces cerevisiae. Curr. Issues Mol. Biol. 2024, 46, 171-182. https://doi.org/10.3390/cimb46010013
Liu G, Han X, Yu X, Wang Y, Ma J, Yang Y. Identification of Aly1 and Aly2 as Modulators of Cytoplasmic pH in Saccharomyces cerevisiae. Current Issues in Molecular Biology. 2024; 46(1):171-182. https://doi.org/10.3390/cimb46010013
Chicago/Turabian StyleLiu, Guoyong, Xiuli Han, Xiang Yu, Yu Wang, Jinbiao Ma, and Yongqing Yang. 2024. "Identification of Aly1 and Aly2 as Modulators of Cytoplasmic pH in Saccharomyces cerevisiae" Current Issues in Molecular Biology 46, no. 1: 171-182. https://doi.org/10.3390/cimb46010013
APA StyleLiu, G., Han, X., Yu, X., Wang, Y., Ma, J., & Yang, Y. (2024). Identification of Aly1 and Aly2 as Modulators of Cytoplasmic pH in Saccharomyces cerevisiae. Current Issues in Molecular Biology, 46(1), 171-182. https://doi.org/10.3390/cimb46010013





