Large-Scale Production of Anti-RNase A VHH Expressed in pyrG Auxotrophic Aspergillus oryzae
Abstract
1. Introduction
2. Materials and Methods
2.1. Construction of Auxotrophic A. oryzae Strain by the Deletion of pyrG
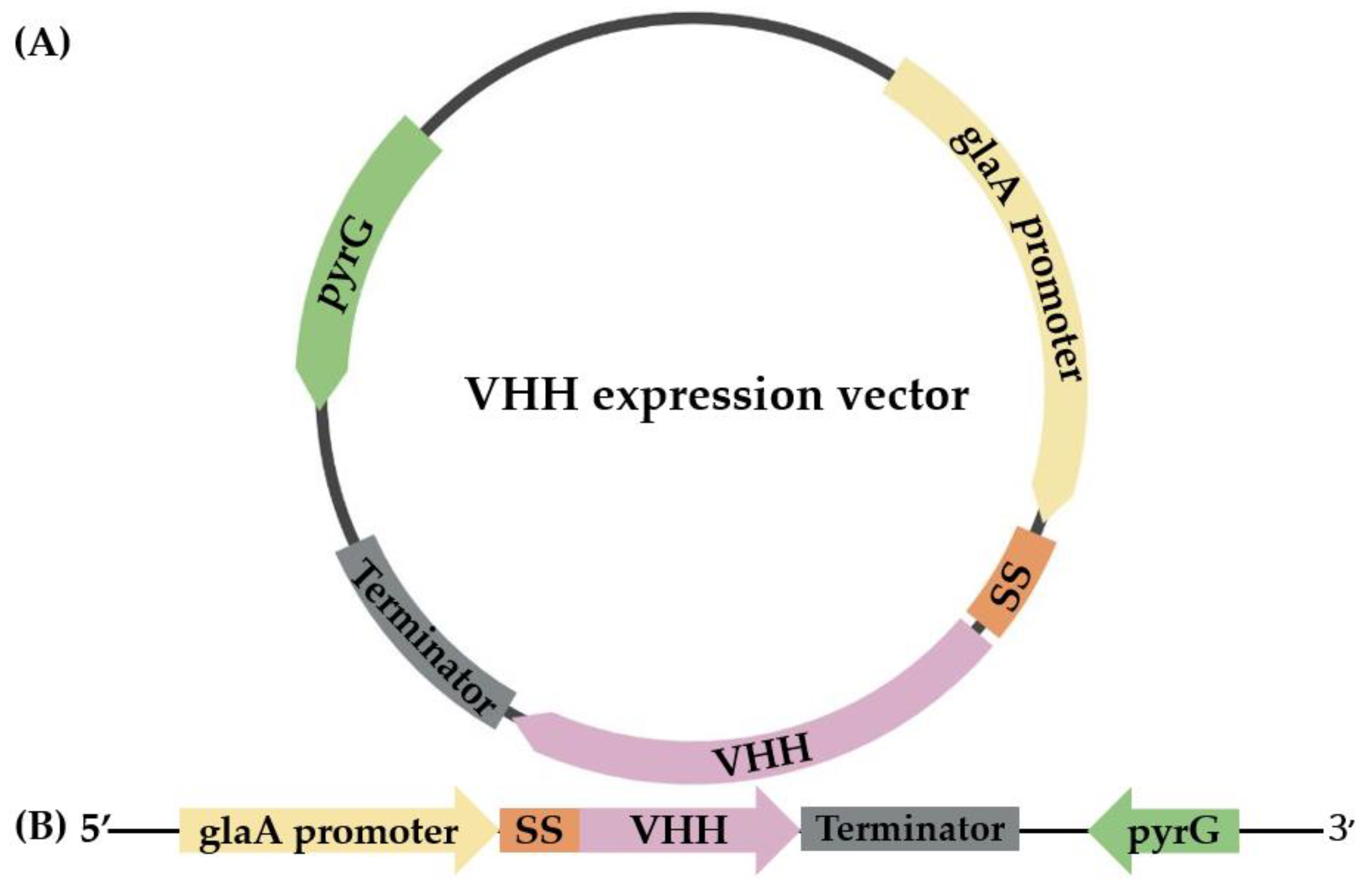
2.2. Cloning and Expression of VHH in pyrG(-) A. oryzae
2.3. Purification of the VHH Expressed in A. oryzae
2.4. Western Blot Analysis of Asp. VHH
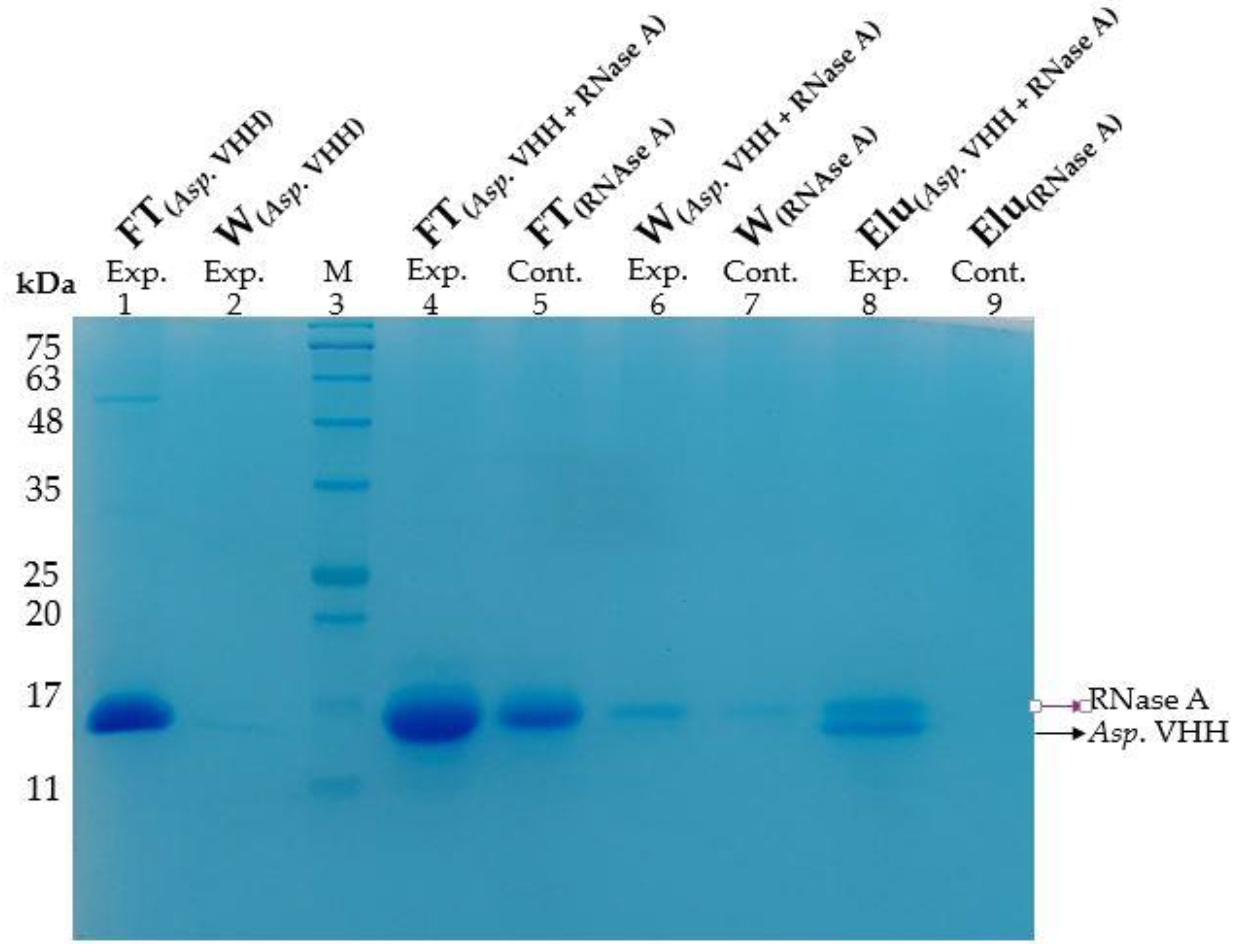
2.5. Pull down Assay
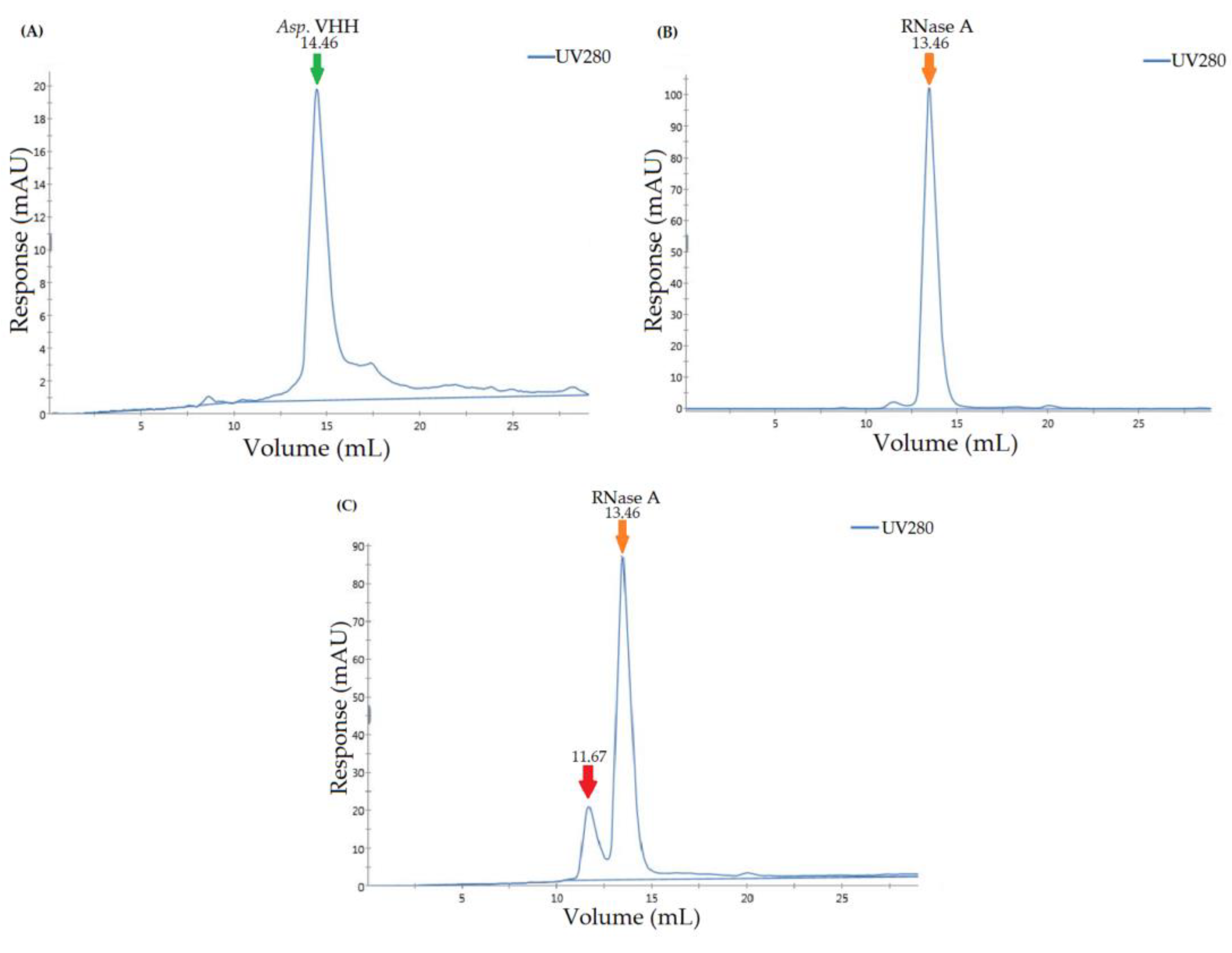
2.6. Size Exclusion Assay for Determining the Specificity of Asp. VHH against RNase A
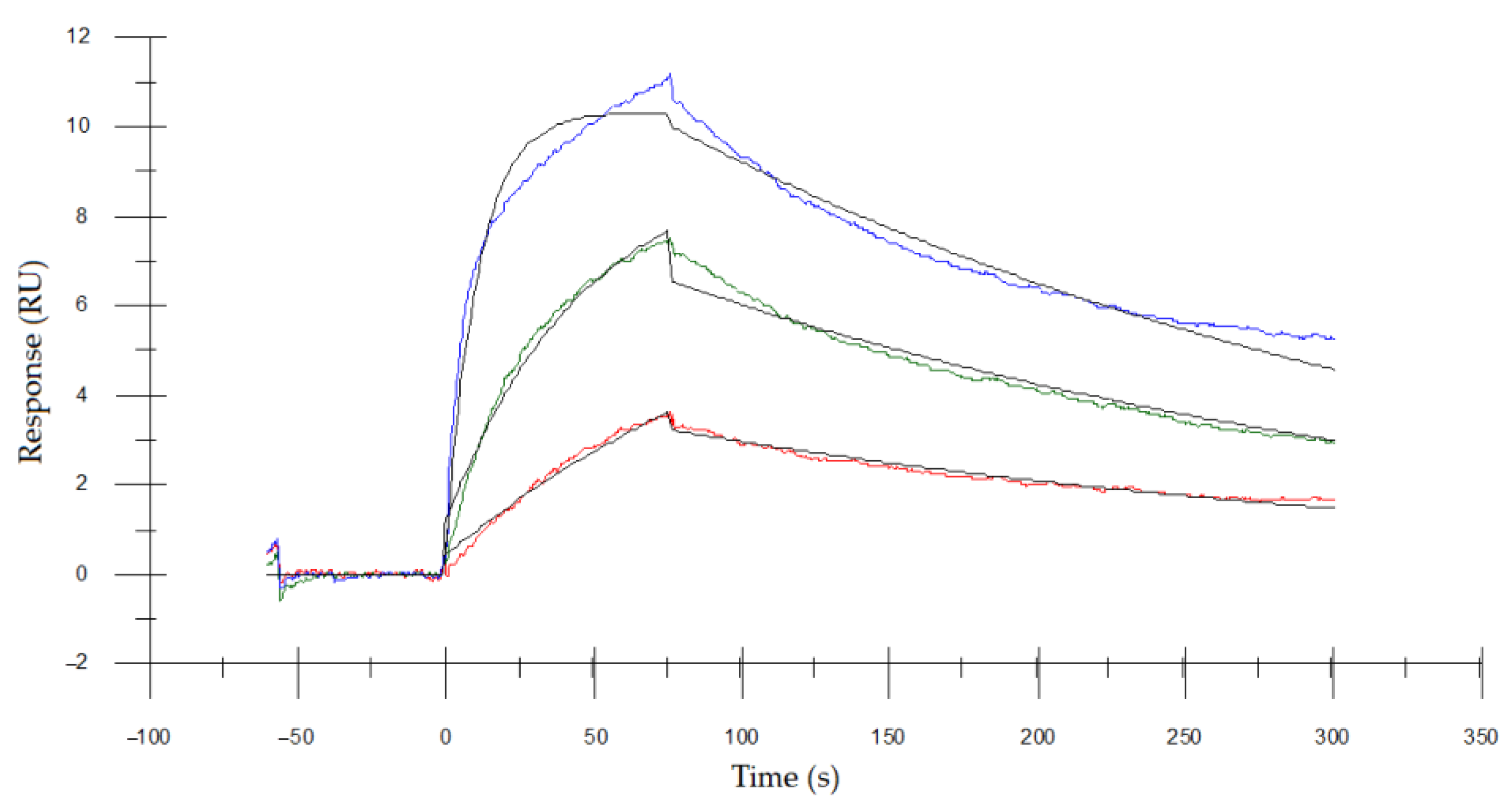
2.7. Surface Plasmon Resonance (SPR) Analysis for Measuring the Binding Affinity of Asp. VHH and RNase A
3. Results
3.1. The pyrG Auxotrophic A. oryzae
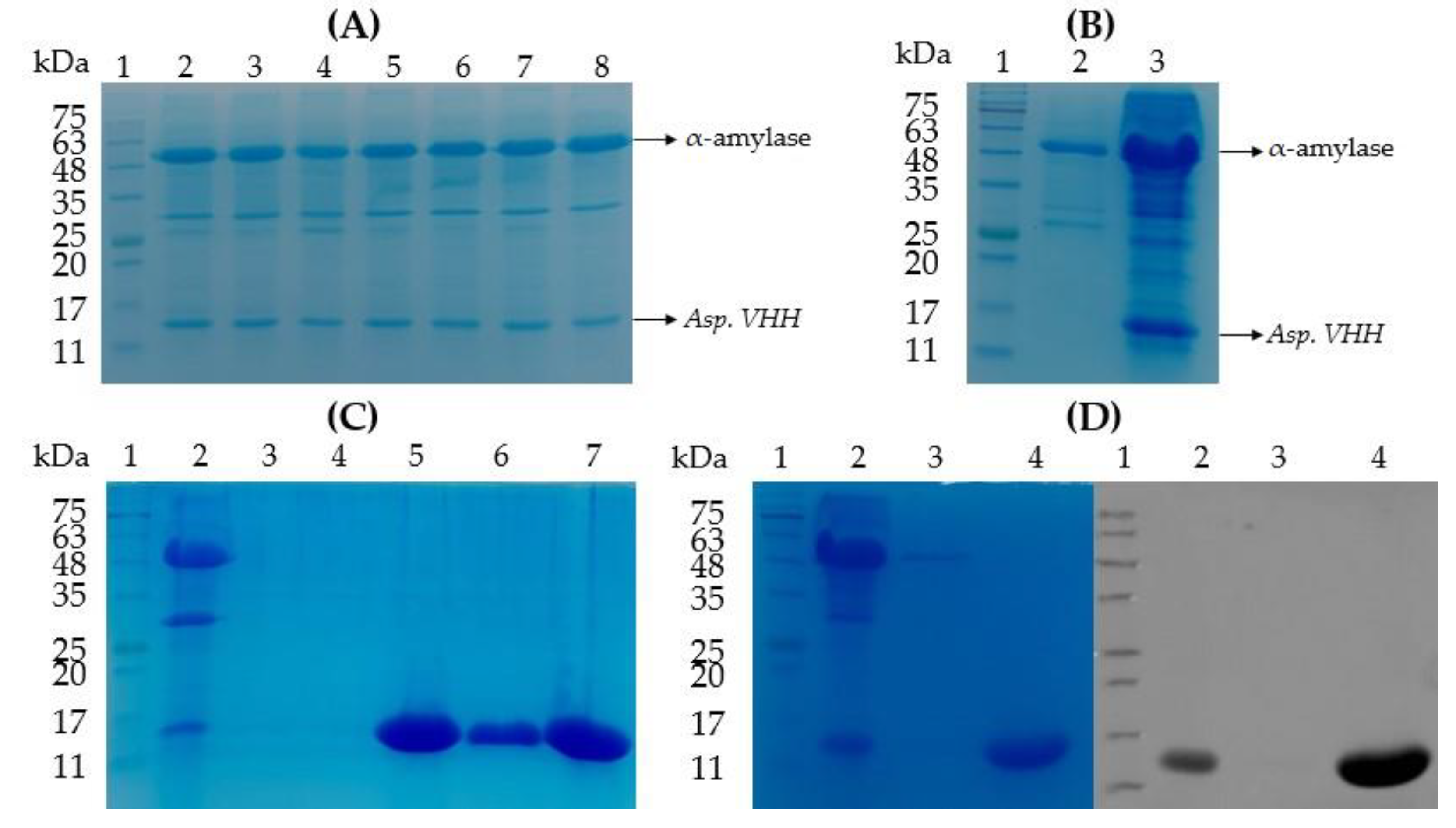
3.2. The Expression of VHH in pyrG(-) A. oryzae
3.3. The Purification of Asp. VHH
3.4. Determining the Specificity of Asp. VHH against RNase A
3.5. Determining the Binding Affinity of Asp. VHH to RNase A
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hamers-Casterman, C.; Atarhouch, T.; Muyldermans, S.; Robinson, G.; Hamers, C.; Bajyana Songa, E.; Bendahman, N.; Hammers, R. Naturally occurring antibodies devoid of light chains. Nature 1993, 363, 446–448. [Google Scholar] [CrossRef] [PubMed]
- Vincke, C.; Muyldermans, S. Introduction to heavy chain antibodies and derived Nanobodies. In Single Domain Antibodies. Methods in Molecular Biology; Saerens, D., Muyldermans, S., Eds.; Humana Press: New York, NY, USA, 2012; Volume 911, pp. 15–26. [Google Scholar] [CrossRef]
- Nguyen, V.K.; Hamers, R.; Wyns, L.; Muyldermans, S. Loss of splice consensus signal is responsible for the removal of the entire CH1 domain of the functional camel IGG2A heavy-chain antibodies. Mol. Immunol. 1999, 36, 515–524. [Google Scholar] [CrossRef] [PubMed]
- Govaert, J.; Pellis, M.; Deschacht, N.; Vincke, C.; Conrath, K.; Muyldermans, S.; Saerens, D. Dual beneficial effect of interloop disulfide bond for single domain antibody fragments. J. Biol. Chem. 2012, 287, 1970–1979. [Google Scholar] [CrossRef]
- Perruchini, C.; Pecorari, F.; Bourgeois, J.P.; Duyckaerts, C.; Rougeon, F.; Lafaye, P. Llama VHH antibody fragments against GFAP: Better diffusion in fixed tissues than classical monoclonal antibodies. Acta Neuropathol. 2009, 118, 685–695. [Google Scholar] [CrossRef] [PubMed]
- De Genst, E.; Silence, K.; Decanniere, K.; Conrath, K.; Loris, R.; Kinne, J.; Muyldermans, S.; Wyns, L. Molecular basis for the preferential cleft recognition by dromedary heavy-chain antibodies. Proc. Natl. Acad. Sci. USA 2006, 103, 4586–4591. [Google Scholar] [CrossRef]
- Muyldermans, S. Nanobodies: Natural single-domain antibodies. Annu. Rev. Biochem. 2013, 82, 775–797. [Google Scholar] [CrossRef]
- van der Linden, R.H.; Frenken, L.G.; de Geus, B.; Harmsen, M.M.; Ruuls, R.C.; Stok, W.; de Ron, L.; Wilson, S.; Davis, P.; Verrips, C.T. Comparison of physical chemical properties of llama VHH antibody fragments and mouse monoclonal antibodies. Biochim. Biophys. Acta (BBA)-Protein Struct. Mol. Enzymol. 1999, 1431, 37–46. [Google Scholar] [CrossRef]
- Dumoulin, M.; Conrath, K.; Van Meirhaeghe, A.; Meersman, F.; Heremans, K.; Frenken, L.G.J.; Muyldermans, S.; Wyns, L.; Matagne, A. Single-domain antibody fragments with high conformational stability. Protein Sci. 2002, 11, 500–515. [Google Scholar] [CrossRef]
- Pérez, J.M.; Renisio, J.G.; Prompers, J.J.; van Platerink, C.J.; Cambillau, C.; Darbon, H.; Frenken, L.G. Thermal unfolding of a llama antibody fragment: A two-state reversible process. Biochemistry 2001, 40, 74–83. [Google Scholar] [CrossRef]
- Lafaye, P.; Li, T. Use of camel single-domain antibodies for the diagnosis and treatment of zoonotic diseases. Comp. Immunol. Microbiol. Infect. Dis. 2018, 60, 17–22. [Google Scholar] [CrossRef]
- Van Audenhove, I.; Gettemans, J. Nanobodies as versatile tools to understand, diagnose, visualize and treat cancer. EBioMedicine 2016, 8, 40–48. [Google Scholar] [CrossRef] [PubMed]
- Salvador, J.-P.; Vilaplana, L.; Marco, M.-P. Nanobody: Outstanding features for diagnostic and therapeutic applications. Anal. Bioanal. Chem. 2019, 411, 1703–1713. [Google Scholar] [CrossRef] [PubMed]
- Jovčevska, I.; Muyldermans, S. The therapeutic potential of nanobodies. BioDrugs 2020, 34, 11–26. [Google Scholar] [CrossRef] [PubMed]
- Koide, S. Engineering of recombinant crystallization chaperones. Curr. Opin. Struct. Biol. 2009, 19, 449–457. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, S.G.; Choi, H.J.; Fung, J.J.; Pardon, E.; Casarosa, P.; Chae, P.S.; Devree, B.T.; Rosenbaum, D.M.; Thian, F.S.; Kobilka, T.S.; et al. Structure of a nanobody-stabilized active state of the β2 adrenoceptor. Nature 2011, 469, 175–180. [Google Scholar] [CrossRef]
- Verheesen, P.; ten Haaft, M.R.; Lindner, N.; Verrips, C.T.; De Haard, J.J. Beneficial properties of single-domain antibody fragments for application in immunoaffinity purification and immuno-perfusion chromatography. Biochim. Biophys. Acta (BBA)-Gen. Subj. 2003, 1624, 21–28. [Google Scholar] [CrossRef]
- Hussack, G.; Arbabi-Ghahroudi, M.; van Faassen, H.; Songer, J.G.; Ng, K.K.-S.; MacKenzie, R.; Tanha, J. Neutralization of Clostridium difficile toxin A with single-domain antibodies targeting the cell receptor binding domain. J. Biol. Chem. 2011, 286, 8961–8976. [Google Scholar] [CrossRef]
- Khirehgesh, M.R.; Sharifi, J.; Safari, F.; Akbari, B. Immunotoxins and nanobody-based immunotoxins: Review and update. J. Drug Target. 2021, 29, 848–862. [Google Scholar] [CrossRef] [PubMed]
- Erreni, M.; Schorn, T.; D’Autilia, F.; Doni, A. Nanobodies as versatile tool for multiscale imaging modalities. Biomolecules 2020, 10, 1695. [Google Scholar] [CrossRef] [PubMed]
- Bastos-Soares, E.A.; Sousa, R.M.O.; Gómez, A.F.; Alfonso, J.; Kayano, A.M.; Zanchi, F.B.; Funes-Huacca, M.E.; Stábelli, R.G.; Soares, A.M.; Pereira, S.S.; et al. Single domain antibodies in the development of immunosensors for diagnostics. Int. J. Biol. Macromol. 2020, 165, 2244–2252. [Google Scholar] [CrossRef]
- Piramoon, M.; Khodadust, F.; Hosseinimehr, S.J. Radiolabeled nanobodies for tumor targeting: From bioengineering to imaging and therapy. Biochim. Biophys. Acta (BBA)-Rev. Cancer 2021, 1875, 188529. [Google Scholar] [CrossRef]
- Kirchhofer, A.; Helma, J.; Schmidthals, K.; Frauer, C.; Cui, S.; Karcher, A.; Pellis, M.; Muyldermans, S.; Casas-Delucchi, C.S.; Cardoso, M.C.; et al. Modulation of protein properties in living cells using nanobodies. Nat. Struct. Mol. Biol. 2010, 17, 133–138. [Google Scholar] [CrossRef] [PubMed]
- Rothbauer, U.; Zolghadr, K.; Tillib, S.; Nowak, D.; Schermelleh, L.; Gahl, A.; Backmann, N.; Conrath, K.; Muyldermans, S.; Cardoso, M.C.; et al. Targeting and tracing antigens in live cells with fluorescent nanobodies. Nat. Methods 2006, 3, 887–889. [Google Scholar] [CrossRef] [PubMed]
- Bao, C.; Gao, Q.; Li, L.L.; Han, L.; Zhang, B.; Ding, Y.; Song, Z.; Zhang, R.; Wu, X.H. The application of nanobody in CAR-T therapy. Biomolecules 2021, 11, 238. [Google Scholar] [CrossRef]
- Kozani, P.S.; Naseri, A.; Mirarefin, S.M.J.; Salem, F.; Nikbakht, M.; Bakhshi, S.E.; Kozani, P.S. Nanobody-based CAR-T cells for cancer immunotherapy. Biomark. Res. 2022, 10, 1–18. [Google Scholar] [CrossRef]
- Behar, G.; Sibéril, S.; Groulet, A.; Chames, P.; Pugniére, M.; Boix, C.; Sautés-Fridman, C.; Teillaud, J.-L.; Baty, D. Isolation and characterization of anti-FcγRIII (CD16) llama single-domain antibodies that activate natural killer cells. Protein Eng. Des. Sel. 2008, 21, 1–10. [Google Scholar] [CrossRef]
- Knoebl, P.; Cataland, S.; Peyvandi, F.; Coppo, P.; Scully, M.; Hovinga, J.A.K.; Metjian, A.; de la Rubia, J.; Pavenski, K.; Edou, J.M.M.; et al. Efficacy and safety of open-label caplacizumab in patients with exacerbations of acquired thrombotic thrombocytopenic purpura in the HERCULES study. J. Thromb. Haemost. 2020, 18, 479–484. [Google Scholar] [CrossRef]
- Sarker, S.A.; Jäkel, M.; Sultana, S.; Alam, N.H.; Bardhan, P.K.; Chisti, M.J.; Salam, M.A.; Theis, W.; Hammarström, L.; Frenken, L.G. Anti-rotavirus protein reduces stool output in infants with diarrhea: A randomized placebo-controlled trial. Gastroenterology 2013, 145, 740–748. [Google Scholar] [CrossRef]
- Ablynx, Identifier NCT01284569. In Study to Assess Safety and Efficacy of Anti-Interleukin 6-Receptor (IL6R) Nanobody in Rheumatoid Arthritis (RA) Patients; National Library of Medicine (US): Bethesda, MD, USA. Available online: https://clinicaltrials.gov/ct2/show/NCT01284569?term=ALX-0061&draw=2&rank=6 (accessed on 14 October 2022).
- Harmsen, M.M.; De Haard, H.J. Properties, production, and applications of camelid single-domain antibody fragments. Appl. Microbiol. Biotechnol. 2007, 77, 13–22. [Google Scholar] [CrossRef]
- Teillaud, J.-L. From whole monoclonal antibodies to single domain antibodies: Think small. In Single Domain Antibodies. Methods and Protocols; Saerens, D., Muyldermans, S., Eds.; Humana Press: New York, NY, USA, 2012; Volume 911, pp. 3–13. [Google Scholar] [CrossRef]
- Arbabi-Ghahroudi, M.; Tanha, J.; MacKenzie, R. Prokaryotic expression of antibodies. Cancer Metastasis Rev. 2005, 24, 501–519. [Google Scholar] [CrossRef]
- Liu, Y.; Huang, H. Expression of single-domain antibody in different systems. Appl. Microbiol. Biotechnol. 2018, 102, 539–551. [Google Scholar] [CrossRef] [PubMed]
- Asaadi, Y.; Jouneghani, F.F.; Janani, S.; Rahbarizadeh, F. A comprehensive comparison between camelid nanobodies and single chain variable fragments. Biomark. Res. 2021, 9, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Rahbarizadeh, F.; Rasaee, M.J.; Forouzandeh-Moghadam, M.; Allameh, A.-A. High expression and purification of the recombinant camelid anti-MUC1 single domain antibodies in Escherichia coli. Protein Expr. Purif. 2005, 44, 32–38. [Google Scholar] [CrossRef] [PubMed]
- Zarschler, K.; Witecy, S.; Kapplusch, F.; Foerster, C.; Stephan, H. High-yield production of functional soluble single-domain antibodies in the cytoplasm of Escherichia coli. Microb. Cell Factories 2013, 12, 1–13. [Google Scholar] [CrossRef]
- Salema, V.; Fernández, L.Á. High yield purification of nanobodies from the periplasm of E. coli as fusions with the maltose binding protein. Protein Expr. Purif. 2013, 91, 42–48. [Google Scholar] [CrossRef]
- Shriver-Lake, L.C.; Goldman, E.R.; Zabetakis, D.; Anderson, G.P. Improved production of single domain antibodies with two disulfide bonds by co-expression of chaperone proteins in the Escherichia coli periplasm. J. Immunol. Methods 2017, 443, 64–67. [Google Scholar] [CrossRef]
- Thomassen, Y.E.; Meijer, W.; Sierkstra, L.; Verrips, C.T. Large-scale production of VHH antibody fragments by Saccharomyces cerevisiae. Enzym. Microb. Technol. 2002, 30, 273–278. [Google Scholar] [CrossRef]
- Gorlani, A.; Hulsik, D.L.; Adams, H.; Vriend, G.; Hermans, P.; Verrips, T. Antibody engineering reveals the important role of J segments in the production efficiency of llama single-domain antibodies in Saccharomyces cerevisiae. Protein Eng. Des. Sel. 2012, 25, 39–46. [Google Scholar] [CrossRef]
- Omidfar, K.; Rasaee, M.J.; Kashanian, S.; Paknejad, M.; Bathaie, Z. Studies of thermostability in Camelus bactrianus (Bactrian camel) single-domain antibody specific for the mutant epidermal-growth-factor receptor expressed by Pichia. Biotechnol. Appl. Biochem. 2007, 46, 41–49. [Google Scholar]
- Ezzine, A.; El Adab, S.M.; Bouhaouala-Zahar, B.; Hmila, I.; Baciou, L.; Marzouki, M.N. Efficient expression of the anti-AahI’scorpion toxin nanobody under a new functional form in a Pichia pastoris system. Biotechnol. Appl. Biochem. 2012, 59, 15–21. [Google Scholar] [CrossRef]
- Barbesgaard, P.; Heldt-Hansen, H.P.; Diderichsen, B. On the safety of Aspergillus oryzae: A review. Appl. Microbiol. Biotechnol. 1992, 36, 569–572. [Google Scholar] [CrossRef] [PubMed]
- He, B.; Tu, Y.; Jiang, C.; Zhang, Z.; Li, Y.; Zeng, B. Functional genomics of Aspergillus oryzae: Strategies and progress. Microorganisms 2019, 7, 103. [Google Scholar] [CrossRef]
- Archer, D.B.; Peberdy, J.F. The molecular biology of secreted enzyme production by fungi. Crit. Rev. Biotechnol. 1997, 17, 273–306. [Google Scholar] [CrossRef] [PubMed]
- Ntana, F.; Mortensen, U.H.; Sarazin, C.; Figge, R. Aspergillus: A powerful protein production platform. Catalysts 2020, 10, 1064. [Google Scholar] [CrossRef]
- Huynh, H.H.; Morita, N.; Sakamoto, T.; Katayama, T.; Miyakawa, T.; Tanokura, M.; Chiba, Y.; Shinkura, R.; Maruyama, J. Functional production of human antibody by the filamentous fungus Aspergillus oryzae. Fungal Biol. Biotechnol. 2020, 7, 1–15. [Google Scholar] [CrossRef]
- Sakekar, A.A.; Gaikwad, S.R.; Punekar, N.S. Protein expression and secretion by filamentous fungi. J. Biosci. 2021, 46, 1–18. [Google Scholar] [CrossRef]
- Meyer, V.; Wu, B.; Ram, A.F.J. Aspergillus as a multi-purpose cell factory: Current status and perspectives. Biotechnol. Lett. 2011, 33, 469–476. [Google Scholar] [CrossRef]
- Decanniere, K.; Desmyter, A.; Lauwereys, M.; Ghahroudi, M.A.; Muyldermans, S.; Wyns, L. A single-domain antibody fragment in complex with RNase A: Non-canonical loop structures and nanomolar affinity using two CDR loops. Structure 1999, 7, 361–370. [Google Scholar] [CrossRef]
- Koide, A.; Tereshko, V.; Uysal, S.; Margalef, K.; Kossiakoff, A.A.; Koide, S. Exploring the capacity of minimalist protein interfaces: Interface energetics and affinity maturation to picomolar KD of a single-domain antibody with a flat paratope. J. Mol. Biol. 2007, 373, 941–953. [Google Scholar] [CrossRef]
- Nguyen, K.T.; Ho, Q.N.; Do, L.T.B.X.; Mai, L.T.D.; Pham, D.-N.; Tran, H.T.T.; Le, D.H.; Nguyen, H.Q.; Tran, V.G. A new and efficient approach for construction of uridine/uracil auxotrophic mutants in the filamentous fungus Aspergillus oryzae using Agrobacterium tumefaciens-mediated transformation. World J. Microbiol. Biotechnol. 2017, 33, 1–11. [Google Scholar] [CrossRef]
- Ling, S.S.; Storms, R.; Zheng, Y.; Rodzi, M.M.; Mahadi, N.M.; Illias, R.M.; Abdul Murad, A.M.; Abu Bakar, F.D. Development of a pyrG mutant of Aspergillus oryzae strain S1 as a host for the production of heterologous proteins. Sci. World J. 2013, 2013, 634317. [Google Scholar] [CrossRef] [PubMed]
- Sakai, K.; Kinoshita, H.; Nihira, T. Heterologous expression system in Aspergillus oryzae for fungal biosynthetic gene clusters of secondary metabolites. Appl. Microbiol. Biotechnol. 2012, 93, 2011–2022. [Google Scholar] [CrossRef] [PubMed]
- Landberg, J.; Wright, N.R.; Wulff, T.; Herrgård, M.J.; Nielsen, A.T. CRISPR interference of nucleotide biosynthesis improves production of a single-domain antibody in Escherichia coli. Biotechnol. Bioeng. 2020, 117, 3835–3848. [Google Scholar] [CrossRef]
- Du, Y.; Xie, G.; Yang, C.; Fang, B.; Chen, H. Construction of brewing-wine Aspergillus oryzae pyrG− mutant by pyrG gene deletion and its application in homology transformation. Acta Biochim. Biophys. Sin. 2014, 46, 477–483. [Google Scholar] [CrossRef] [PubMed]
- Veggiani, G.; de Marco, A. Improved quantitative and qualitative production of single domain intrabodies mediated by the co-expression of Erv1p sulfhydryl oxidase. Protein Expr. Purif. 2011, 79, 111–114. [Google Scholar] [CrossRef]
- Järviluoma, A.; Strandin, T.; Lülf, S.; Bouchet, J.; Mäkelä, A.R.; Geyer, M.; Benichou, S.; Saksela, K. High-affinity target binding engineered via fusion of a single-domain antibody fragment with a ligand-tailored SH3 domain. PLoS ONE 2012, 7, e40331. [Google Scholar] [CrossRef]
- Veggiani, G.; Giabbai, B.; Semrau, M.S.; Medagli, B.; Riccio, V.; Bajc, G.; Storici, P.; de Marco, A. Comparative analysis of fusion tags used to functionalize recombinant antibodies. Protein Expr. Purif. 2020, 166, 105505. [Google Scholar] [CrossRef]
- Bao, X.; Xu, L.; Lu, X.; Jia, L. Optimization of dilution refolding conditions for a camelid single domain antibody against human beta-2-microglobulin. Protein Expr. Purif. 2016, 117, 59–66. [Google Scholar] [CrossRef]
- Xue, X.; Fan, X.; Qu, Q.; Wu, G. Bioscreening and expression of a camel anti-CTGF VHH nanobody and its renaturation by a novel dialysis–dilution method. AMB Express 2016, 6, 1–11. [Google Scholar] [CrossRef]
- Yu, J.; Guo, Y.; Gu, Y.; Fan, X.; Li, F.; Song, H.; Nian, R.; Liu, W. A novel silk fibroin protein–based fusion system for enhancing the expression of nanobodies in Escherichia coli. Appl. Microbiol. Biotechnol. 2022, 106, 1967–1977. [Google Scholar] [CrossRef]
- Thomassen, Y.E.; Verkleij, A.J.; Boonstra, J.; Verrips, C.T. Specific production rate of VHH antibody fragments by Saccharomyces cerevisiae is correlated with growth rate, independent of nutrient limitation. J. Biotechnol. 2005, 118, 270–277. [Google Scholar] [CrossRef] [PubMed]
- Frenken, L.G.; Van Der Linden, R.H.; Hermans, P.W.; Bos, J.W.; Ruuls, R.C.; De Geus, B.; Verrips, C.T. Isolation of antigen specific llama VHH antibody fragments and their high level secretion by Saccharomyces cerevisiae. J. Biotechnol. 2000, 78, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Zhou, Y.; Yu, J.; Liu, W.; Li, F.; Xian, M.; Nian, R.; Song, H.; Feng, D. An efficient constitutive expression system for Anti-CEACAM5 nanobody production in the yeast Pichia pastoris. Protein Expr. Purif. 2019, 155, 43–47. [Google Scholar] [CrossRef]
- Rahbarizadeh, F.; Rasaee, M.J.; Forouzandeh, M.; Allameh, A.A. Over expression of anti-MUC1 single-domain antibody fragments in the yeast Pichia pastoris. Mol. Immunol. 2006, 43, 426–435. [Google Scholar] [CrossRef] [PubMed]
- Maras, M.; Die, I.V.; Contreras, R.; van den Hondel, C.A. Filamentous Fungi as Production Organisms for Glycoproteins of Bio-Medical Interest; Glycotechnology; Springer: Berlin/Heidelberg, Germany, 1999; pp. 19–27. [Google Scholar]
- Joosten, V.; Gouka, R.J.; Van Den Hondel, C.A.; Verrips, C.T.; Lokman, B.C. Expression and production of llama variable heavy-chain antibody fragments (VHHs) by Aspergillus awamori. Appl. Microbiol. Biotechnol. 2005, 66, 384–392. [Google Scholar] [CrossRef]
- Joosten, V.; Roelofs, M.S.; Van den Dries, N.; Goosen, T.; Verrips, C.T.; Van den Hondel, C.A.; Lokman, B.C. Production of bifunctional proteins by Aspergillus awamori: Llama variable heavy chain antibody fragment (VHH) R9 coupled to Arthromyces ramosus peroxidase (ARP). J. Biotechnol. 2005, 120, 347–359. [Google Scholar] [CrossRef]
- Okazaki, F.; Aoki, J.I.; Tabuchi, S.; Tanaka, T.; Ogino, C.; Kondo, A. Efficient heterologous expression and secretion in Aspergillus oryzae of a llama variable heavy-chain antibody fragment VHH against EGFR. Appl. Microbiol. Biotechnol. 2012, 96, 81–88. [Google Scholar] [CrossRef]
- Hisada, H.; Tsutsumi, H.; Ishida, H.; Hata, Y. High production of llama variable heavy-chain antibody fragment (VHH) fused to various reader proteins by Aspergillus oryzae. Appl. Microbiol. Biotechnol. 2013, 97, 761–766. [Google Scholar] [CrossRef]
- Leow, C.H.; Fischer, K.; Leow, C.Y.; Cheng, Q.; Chuah, C.; McCarthy, J. Single domain antibodies as new biomarker detectors. Diagnostics 2017, 7, 52. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karaman, E.; Eyüpoğlu, A.E.; Mahmoudi Azar, L.; Uysal, S. Large-Scale Production of Anti-RNase A VHH Expressed in pyrG Auxotrophic Aspergillus oryzae. Curr. Issues Mol. Biol. 2023, 45, 4778-4795. https://doi.org/10.3390/cimb45060304
Karaman E, Eyüpoğlu AE, Mahmoudi Azar L, Uysal S. Large-Scale Production of Anti-RNase A VHH Expressed in pyrG Auxotrophic Aspergillus oryzae. Current Issues in Molecular Biology. 2023; 45(6):4778-4795. https://doi.org/10.3390/cimb45060304
Chicago/Turabian StyleKaraman, Elif, Alp Ertunga Eyüpoğlu, Lena Mahmoudi Azar, and Serdar Uysal. 2023. "Large-Scale Production of Anti-RNase A VHH Expressed in pyrG Auxotrophic Aspergillus oryzae" Current Issues in Molecular Biology 45, no. 6: 4778-4795. https://doi.org/10.3390/cimb45060304
APA StyleKaraman, E., Eyüpoğlu, A. E., Mahmoudi Azar, L., & Uysal, S. (2023). Large-Scale Production of Anti-RNase A VHH Expressed in pyrG Auxotrophic Aspergillus oryzae. Current Issues in Molecular Biology, 45(6), 4778-4795. https://doi.org/10.3390/cimb45060304





