Transcriptomic Establishment of Pig Macrophage Polarization Signatures
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Cell Culture
2.3. RNA-Seq
2.4. Gene Set Enrichment Analysis (GSEA) and Network Construction
3. Results
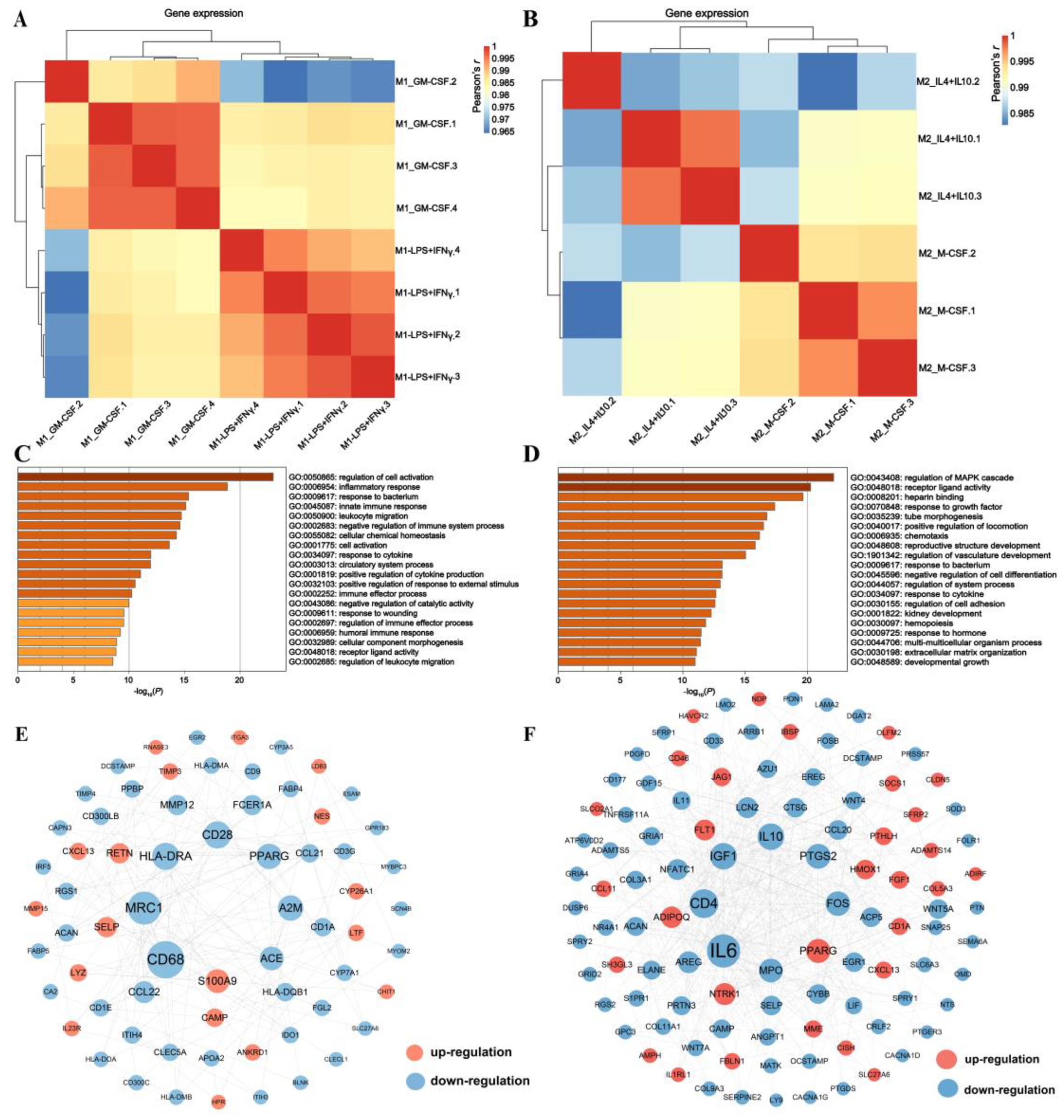
3.1. Comparing M1 with M2 to Generate Porcine Macrophage Gene Signatures
3.2. Revealing Transcriptomic Differences of Macrophages within Phenotypes
3.3. Application of M1_IFNγ + LPS, M1_GM-CSF, M2_IL4 + IL10, and M2_M-CSF Signatures to the Identification of Swine Disease and Its Molecular Mechanism
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Takahashi, K. Development and Differentiation of Macrophages and Related Cells: Historical Review and Current Concepts. J. Clin. Exp. Hematop. 2001, 41, 1–31. [Google Scholar] [CrossRef]
- Murray, P.J.; Wynn, T.A. Protective and Pathogenic Functions of Macrophage Subsets. Nat. Rev. Immunol. 2011, 11, 723–737. [Google Scholar] [CrossRef] [PubMed]
- Sica, A.; Mantovani, A. Plasticity and Polarization. J. Clin. Investig. 2012, 122, 787–795. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Gao, F.; Gu, K.; Chen, D. The Role of Monocytes and Macrophages in Autoimmune Diseases: A Comprehensive Review. Front. Immunol. 2019, 10, 1140. [Google Scholar] [CrossRef]
- Rendra, E.; Riabov, V.; Mossel, D.M.; Sevastyanova, T.; Harmsen, M.C.; Kzhyshkowska, J. Reactive Oxygen Species (ROS) in Macrophage Activation and Function in Diabetes. Immunobiology 2019, 224, 242–253. [Google Scholar] [CrossRef]
- Martinez, F.O.; Gordon, S. The M1 and M2 Paradigm of Macrophage Activation: Time for Reassessment. F1000Prime Rep. 2014, 6, 13. [Google Scholar] [CrossRef]
- Bility, M.T.; Cheng, L.; Zhang, Z.; Luan, Y.; Li, F.; Chi, L.; Zhang, L.; Tu, Z.; Gao, Y.; Fu, Y.-X.; et al. Hepatitis B Virus Infection and Immunopathogenesis in a Humanized Mouse Model: Induction of Human-Specific Liver Fibrosis and M2-Like Macrophages. PLoS Pathog. 2014, 10, e1004032. [Google Scholar] [CrossRef]
- Anderson, N.R.; Minutolo, N.G.; Gill, S.; Klichinsky, M. Macrophage-Based Approaches for Cancer Immunotherapy. Cancer Res. 2021, 81, 1201–1208. [Google Scholar] [CrossRef]
- Lacey, D.C.; Achuthan, A.; Fleetwood, A.J.; Dinh, H.; Roiniotis, J.; Scholz, G.M.; Chang, M.W.; Beckman, S.K.; Cook, A.D.; Hamilton, J.A. Defining GM-CSF– and Macrophage-CSF–Dependent Macrophage Responses by In Vitro Models. J. Immunol. 2012, 188, 5752–5765. [Google Scholar] [CrossRef]
- Martinez, F.O.; Gordon, S.; Locati, M.; Mantovani, A. Transcriptional Profiling of the Human Monocyte-to-Macrophage Differentiation and Polarization: New Molecules and Patterns of Gene Expression. J. Immunol. 2006, 177, 7303–7311. [Google Scholar] [CrossRef]
- Vogel, D.Y.S.; Vereyken, E.J.F.; Glim, J.E.; Heijnen, P.D.A.M.; Moeton, M.; van der Valk, P.; Amor, S.; Teunissen, C.E.; van Horssen, J.; Dijkstra, C.D. Macrophages in Inflammatory Multiple Sclerosis Lesions Have an Intermediate Activation Status. J. Neuroinflamm. 2013, 10, 809. [Google Scholar] [CrossRef] [PubMed]
- Swindle, M.M.; Makin, A.; Herron, A.J.; Clubb, F.J.; Frazier, K.S. Swine as Models in Biomedical Research and Toxicology Testing. Vet. Pathol. 2012, 49, 344–356. [Google Scholar] [CrossRef] [PubMed]
- Sironi, M. Differential Regulation of Chemokine Production by Fc Receptor Engagement in Human Monocytes: Association of CCL1 with a Distinct Form of M2 Monocyte Activation (M2b, Type 2). J. Leukoc. Biol. 2006, 80, 342–349. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, A.; Sozzani, S.; Locati, M.; Allavena, P.; Sica, A. Macrophage Polarization: Tumor-Associated Macrophages as a Paradigm for Polarized M2 Mononuclear Phagocytes. Trends Immunol. 2002, 23, 549–555. [Google Scholar] [CrossRef] [PubMed]
- Morris, S.M. Arginine Metabolism: Boundaries of Our Knowledge. J. Nutr. 2007, 137, 1602S–1609S. [Google Scholar] [CrossRef]
- Perleberg, C.; Kind, A.; Schnieke, A. Genetically Engineered Pigs as Models for Human Disease. DMM Dis. Model. Mech. 2018, 11, dmm030783. [Google Scholar] [CrossRef]
- Pepe, G.; Calderazzi, G.; De Maglie, M.; Villa, A.M.; Vegeto, E. Heterogeneous Induction of Microglia M2a Phenotype by Central Administration of Interleukin-4. J. Neuroinflamm. 2014, 11, 211. [Google Scholar] [CrossRef]
- Giraud, E.; Lestinova, T.; Derrick, T.; Martin, O.; Dillon, R.J.; Volf, P.; Műller, I.; Bates, P.A.; Rogers, M.E. Leishmania Proteophosphoglycans Regurgitated from Infected Sand Flies Accelerate Dermal Wound Repair and Exacerbate Leishmaniasis via Insulin-like Growth Factor 1-Dependent Signalling. PLoS Pathog. 2018, 14, e1006794. [Google Scholar] [CrossRef]
- Leidi, M.; Gotti, E.; Bologna, L.; Miranda, E.; Rimoldi, M.; Sica, A.; Roncalli, M.; Palumbo, G.A.; Introna, M.; Golay, J. M2 Macrophages Phagocytose Rituximab-Opsonized Leukemic Targets More Efficiently than M1 Cells In Vitro. J. Immunol. 2009, 182, 4415–4422. [Google Scholar] [CrossRef]
- Harusato, A.; Geem, D.; Denning, T.L. Macrophage Isolation from the Mouse Small and Large Intestine. Methods Mol. Biol. 2016, 1422, 171–180. [Google Scholar] [CrossRef]
- Truty, M.J.; Smoot, R.L. Animal Models in Pancreatic Surgery: A Plea for Pork. Pancreatology 2008, 8, 546–550. [Google Scholar] [CrossRef] [PubMed]
- Martinez, F.O.; Helming, L.; Milde, R.; Varin, A.; Melgert, B.N.; Draijer, C.; Thomas, B.; Fabbri, M.; Crawshaw, A.; Ho, L.P. Genetic Programs Expressed in Resting and IL-4 Alternatively Activated Mouse and Human Macrophages: Similarities and Differences. Blood J. Am. Soc. Hematol. 2013, 121, e57–e69. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Scheenstra, M.R.; van Dijk, A.; Veldhuizen, E.J.A.; Haagsman, H.P. A New and Efficient Culture Method for Porcine Bone Marrow-Derived M1-and M2-Polarized Macrophages. Vet. Immunol. Immunopathol. 2018, 200, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Raes, G.; Van den Bergh, R.; De Baetselier, P.; Ghassabeh, G.H. Arginase-1 and Ym1 Are Markers for Murine, but Not Human, Alternatively Activated Myeloid Cells. J. Immunol. 2005, 174, 6561–6562. [Google Scholar] [CrossRef]
- Schneemann, M.; Schoeden, G. Macrophage Biology and Immunology: Man Is Not a Mouse. J. Leukoc. Biol. 2007, 81, 579. [Google Scholar] [CrossRef]
- Meurens, F.; Summerfield, A.; Nauwynck, H.; Saif, L.; Gerdts, V. The Pig: A Model for Human Infectious Diseases. Trends Microbiol. 2012, 20, 50–57. [Google Scholar] [CrossRef]
- Mehle, A.; Doudna, J.A. Adaptive Strategies of the Influenza Virus Polymerase for Replication in Humans. Proc. Natl. Acad. Sci. USA 2009, 106, 21312–21316. [Google Scholar] [CrossRef]
- Mclaughlin, T.; Ackerman, S.E.; Shen, L.; Engleman, E. Role of Innate and Adaptive Immunity in Obesity-Associated Metabolic Disease. J. Clin. Investig. 2017, 127, 5–13. [Google Scholar] [CrossRef]
- Björck, P. Isolation and Characterization of Plasmacytoid Dendritic Cells from Flt3 Ligand and Granulocyte-Macrophage Colony-Stimulating Factor-Treated Mice. Blood 2001, 98, 3520–3526. [Google Scholar] [CrossRef]
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-Optimal Probabilistic RNA-Seq Quantification. Nat. Biotechnol. 2016, 34, 525–527. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene Set Enrichment Analysis: A Knowledge-Based Approach for Interpreting Genome-Wide Expression Profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Cirulli, E.T.; Singh, A.; Shianna, K.V.; Ge, D.; Smith, J.P.; Maia, J.M.; Heinzen, E.L.; Goedert, J.J.; Goldstein, D.B. Screening the Human Exome: A Comparison of Whole Genome and Whole Transcriptome Sequencing. Genome Biol. 2010, 11, R57. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.; Ma, C. A Novel Ferroptosis-Associated Gene Signature to Predict Prognosis in Patients with Uveal Melanoma. Diagnostics 2021, 11, 219. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yu, W.; Wollenweber, T.; Lu, X.; Wei, Y.; Beitzke, D.; Wadsak, W.; Kropf, S.; Wester, H.J.; Haug, A.R. [68 Ga] Pentixafor PET/MR Imaging of Chemokine Receptor 4 Expression in the Human Carotid Artery. Eur. J. Nucl. Med. Mol. Imaging 2019, 46, 1616–1625. [Google Scholar] [CrossRef]
- Tang, C.; Lei, X.; Xiong, L.; Hu, Z.; Tang, B. HMGA1B/2 Transcriptionally Activated-POU1F1 Facilitates Gastric Carcinoma Metastasis via CXCL12/CXCR4 Axis-Mediated Macrophage Polarization. Cell Death Dis. 2021, 12, 1–15. [Google Scholar] [CrossRef]
- Emokpae, M.A.; Mrakpor, B.A. Do Sex Differences in Respiratory Burst Enzyme Activities Exist in Human Immunodeficiency Virus-1 Infection? Med. Sci. 2016, 4, 19. [Google Scholar] [CrossRef]
- Wu, W.; Hou, B.; Tang, C.; Liu, F.; Yang, J.; Pan, T.; Si, K.; Lu, D.; Wang, X.; Wang, J. (+)-Usnic Acid Inhibits Migration of c-KIT Positive Cells in Human Colorectal Cancer. Evid.-Based Complement. Altern. Med. 2018, 2018, 5149436. [Google Scholar] [CrossRef]
- Van Rossum, D.; Hilbert, S.; Straßenburg, S.; Hanisch, U.; Brück, W. Myelin-phagocytosing Macrophages in Isolated Sciatic and Optic Nerves Reveal a Unique Reactive Phenotype. Glia 2008, 56, 271–283. [Google Scholar] [CrossRef]
- Cao, W.; Wei, W.; Zhan, Z.; Xie, D.; Xie, Y.; Xiao, Q. Regulation of Drug Resistance and Metastasis of Gastric Cancer Cells via the MicroRNA647-ANK2 Axis. Int. J. Mol. Med. 2018, 41, 1958–1966. [Google Scholar] [CrossRef]
- Auburger, G.; Gispert, S.; Torres-Odio, S.; Jendrach, M.; Brehm, N.; Canet-Pons, J.; Key, J.; Sen, N.-E. SerThr-PhosphoProteome of Brain from Aged PINK1-KO+ A53T-SNCA Mice Reveals PT1928-MAP1B and PS3781-ANK2 Deficits, as Hub between Autophagy and Synapse Changes. Int. J. Mol. Sci. 2019, 20, 3284. [Google Scholar] [CrossRef]
- Wu, X.; Lv, D.; Cai, C.; Zhao, Z.; Wang, M.; Chen, W.; Liu, Y. A TP53-Associated Immune Prognostic Signature for the Prediction of Overall Survival and Therapeutic Responses in Muscle-Invasive Bladder Cancer. Front. Immunol. 2020, 11, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Foss, D.L.; Bennaars, A.M.; Pennell, C.A.; Moody, M.D.; Murtaugh, M.P. Differentiation of Porcine Dendritic Cells by Granulocyte-Macrophage Colony-Stimulating Factor Expressed in Pichia Pastoris. Vet. Immunol. Immunopathol. 2003, 91, 205–215. [Google Scholar] [CrossRef] [PubMed]
- Bézie, S.; Freuchet, A.; Sérazin, C.; Salama, A.; Vimond, N.; Anegon, I.; Guillonneau, C. IL-34 Actions on FOXP3+ Tregs and CD14+ Monocytes Control Human Graft Rejection. Front. Immunol. 2020, 11, 1496. [Google Scholar] [CrossRef] [PubMed]
- Mossel, D.M.; Moganti, K.; Riabov, V.; Weiss, C.; Kopf, S.; Cordero, J.; Dobreva, G.; Rots, M.G.; Klüter, H.; Harmsen, M.C. Epigenetic Regulation of S100A9 and S100A12 Expression in Monocyte-Macrophage System in Hyperglycemic Conditions. Front. Immunol. 2020, 11, 1071. [Google Scholar] [CrossRef] [PubMed]
- Pan, L.; Wei, N.; Jia, H.; Gao, M.; Chen, X.; Wei, R.; Sun, Q.; Gu, S.; Du, B.; Xing, A. Genome-Wide Transcriptional Profiling Identifies Potential Signatures in Discriminating Active Tuberculosis from Latent Infection. Oncotarget 2017, 8, 112907. [Google Scholar] [CrossRef]
- Carroll, R.G.; Riley, J.L.; Levine, B.L.; Feng, Y.; Kaushal, S.; Ritchey, D.W.; Bernstein, W.; Weislow, O.S.; Brown, C.R.; Berger, E.A. Differential Regulation of HIV-1 Fusion Cofactor Expression by CD28 Costimulation of CD4+ T Cells. Science 1997, 276, 273–276. [Google Scholar] [CrossRef]
- Gladow, N.; Hollmann, C.; Ramos, G.; Frantz, S.; Kerkau, T.; Beyersdorf, N.; Hofmann, U. Treatment of Mice with a Ligand Binding Blocking Anti-CD28 Monoclonal Antibody Improves Healing after Myocardial Infarction. PLoS ONE 2020, 15, e0227734. [Google Scholar]
- Vozza, E.G.; Mulcahy, M.E.; McLoughlin, R.M. Making the Most of the Host; Targeting the Autophagy Pathway Facilitates Staphylococcus Aureus Intracellular Survival in Neutrophils. Front. Immunol. 2021, 12, 2304. [Google Scholar] [CrossRef]
- Perry, S.E.; Mostafa, S.M.; Wenstone, R.; Shenkin, A.; McLaughlin, P.J. HLA-DR Regulation and the Influence of GM-CSF on Transcription, Surface Expression and Shedding. Int. J. Med. Sci. 2004, 1, 126. [Google Scholar] [CrossRef]
- Peng, P.; Zhu, H.; Liu, D.; Chen, Z.; Zhang, X.; Guo, Z.; Dong, M.; Wan, L.; Zhang, P.; Liu, G. TGFBI Secreted by Tumor-Associated Macrophages Promotes Glioblastoma Stem Cell-Driven Tumor Growth via Integrin Avβ5-Src-Stat3 Signaling. Theranostics 2022, 12, 4221. [Google Scholar] [CrossRef]
- Pasqualetti, F.; Giampietro, C.; Montemurro, N.; Giannini, N.; Gadducci, G.; Orlandi, P.; Natali, E.; Chiarugi, P.; Gonnelli, A.; Cantarella, M.; et al. Old and New Systemic Immune-Inflammation Indexes Are Associated with Overall Survival of Glioblastoma Patients Treated with Radio-Chemotherapy. Genes 2022, 13, 1054. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Zhang, Q.; Li, L.; Chen, K.; Yang, J.; Dixit, D.; Gimple, R.C.; Ci, S.; Lu, C.; Hu, L.; et al. Β2-Microglobulin Maintains Glioblastoma Stem Cells and Induces M2-like Polarization of Tumor-Associated Macrophages. Cancer Res. 2022, 82, 3321–3334. [Google Scholar] [CrossRef] [PubMed]
- Vikhe, P.P.; Purnell, T.; Brown, S.D.M.; Hood, D.W. Cellular Immune Response against Nontypeable Haemophilus Influenzae Infecting the Preinflamed Middle Ear of the Junbo Mouse. Infect. Immun. 2019, 87, e00689-19. [Google Scholar] [CrossRef] [PubMed]
- Glas, J.; Seiderer, J.; Markus, C.; Pfennig, S.; Wetzke, M.; Paschos, E.; Göke, B.; Ochsenkühn, T.; Müller-Myhsok, B.; Diegelmann, J. Role of PPARG Gene Variants in Inflammatory Bowel Disease. Inflamm. Bowel Dis. 2011, 17, 1057–1058. [Google Scholar] [CrossRef]
- Zahuczky, G.; Kristóf, E.; Majai, G.; Fésüs, L. Differentiation and Glucocorticoid Regulated Apopto-Phagocytic Gene Expression Patterns in Human Macrophages. Role of Mertk in Enhanced Phagocytosis. PLoS ONE 2011, 6, e21349. [Google Scholar] [CrossRef]
- Pajtler, K.W.; Rebmann, V.; Lindemann, M.; Schulte, J.H.; Schulte, S.; Stauder, M.; Leuschner, I.; Schmid, K.; Köhl, U.; Schramm, A. Expression of NTRK1/TrkA Affects Immunogenicity of Neuroblastoma Cells. Int. J. Cancer 2013, 133, 908–919. [Google Scholar] [CrossRef]
- Pereira, G.; Bexiga, R.; Silva, J.C.E.; Silva, E.; Ramé, C.; Dupont, J.; Guo, Y.; Humblot, P.; Lopes-da-Costa, L. Adipokines as Biomarkers of Postpartum Subclinical Endometritis in Dairy Cows. Reproduction 2020, 160, 417–430. [Google Scholar] [CrossRef]
- Otowa, Y.; Moriwaki, K.; Sano, K.; Shirakabe, M.; Yonemura, S.; Shibuya, M.; Rossant, J.; Suda, T.; Kakeji, Y.; Hirashima, M. Flt1/VEGFR1 Heterozygosity Causes Transient Embryonic Edema. Sci. Rep. 2016, 6, 27186. [Google Scholar] [CrossRef]
- Lokki, A.I.; Heikkinen-Eloranta, J.K.; Laivuori, H. The Immunogenetic Conundrum of Preeclampsia. Front. Immunol. 2018, 9, 2630. [Google Scholar] [CrossRef]
- Fernando, M.R.; Giembycz, M.A.; McKay, D.M. Bidirectional Crosstalk via IL-6, PGE2 and PGD2 between Murine Myofibroblasts and Alternatively Activated Macrophages Enhances Anti-Inflammatory Phenotype in Both Cells. Br. J. Pharmacol. 2016, 173, 899–912. [Google Scholar] [CrossRef]
- Keränen, T.; Hömmö, T.; Hämäläinen, M.; Moilanen, E.; Korhonen, R. Anti-Inflammatory Effects of Β2-Receptor Agonists Salbutamol and Terbutaline Are Mediated by MKP-1. PLoS ONE 2016, 11, e0148144. [Google Scholar] [CrossRef]
- Barroso, F.A.L.; de Jesus, L.C.L.; de Castro, C.P.; Batista, V.L.; Ferreira, Ê.; Fernandes, R.S.; de Barros, A.L.B.; Leclerq, S.Y.; Azevedo, V.; Mancha-Agresti, P. Intake of Lactobacillus Delbrueckii (PExu: Hsp65) Prevents the Inflammation and the Disorganization of the Intestinal Mucosa in a Mouse Model of Mucositis. Microorganisms 2021, 9, 107. [Google Scholar] [CrossRef] [PubMed]
- Bergmann, C.B.; Beckmann, N.; Salyer, C.E.; Hanschen, M.; Crisologo, P.A.; Caldwell, C.C. Potential Targets to Mitigate Trauma-or Sepsis-Induced Immune Suppression. Front. Immunol. 2021, 12, 121. [Google Scholar] [CrossRef] [PubMed]
- Fernando, M.R.; Reyes, J.L.; Iannuzzi, J.; Leung, G.; McKay, D.M. The Pro-Inflammatory Cytokine, Interleukin-6, Enhances the Polarization of Alternatively Activated Macrophages. PLoS ONE 2014, 9, e94188. [Google Scholar] [CrossRef] [PubMed]
- Messing, M.; Jan-Abu, S.C.; McNagny, K. Group 2 Innate Lymphoid Cells: Central Players in a Recurring Theme of Repair and Regeneration. Int. J. Mol. Sci. 2020, 21, 1350. [Google Scholar] [CrossRef]
- WERNER, S.; GROSE, R. Regulation of Wound Healing by Growth Factors and Cytokines. Physiol. Rev. 2003, 83, 835–870. [Google Scholar] [CrossRef]
- Acosta, J.R.; Tavira, B.; Douagi, I.; Kulyté, A.; Arner, P.; Rydén, M.; Laurencikiene, J. Human-Specific Function of IL-10 in Adipose Tissue Linked to Insulin Resistance. J. Clin. Endocrinol. Metab. 2019, 104, 4552–4562. [Google Scholar] [CrossRef]
- Al-Rubaie, A.; Wise, A.F.; Sozo, F.; De Matteo, R.; Samuel, C.S.; Harding, R.; Ricardo, S.D. The therapeutic effect of mesenchymal stem cells on pulmonary myeloid cells following neonatal hyperoxic lung injury in mice. Respir. Res. 2018, 19, 1–11. [Google Scholar] [CrossRef]
- Awad, F.; Assrawi, E.; Jumeau, C.; Georgin-Lavialle, S.; Cobret, L.; Duquesnoy, P.; Piterboth, W.; Thomas, L.; Stankovic-Stojanovic, K.; Louvrier, C.; et al. Impact of human monocyte and macrophage polarization on NLR expression and NLRP3 inflammasome activation. PLoS ONE 2017, 12, e0175336. [Google Scholar] [CrossRef]
- Beceiro, S.; Radin, J.; Chatuvedi, R.; Piazuelo, M.; Horvarth, D.; Cortado, H.; Gu, Y.; Dixon, B.; Gu, C.; Lange, I.; et al. TRPM2 ion channels regulate macrophage polarization and gastric inflammation during Helicobacter pylori infection. Mucosal Immunol. 2017, 10, 493–507. [Google Scholar] [CrossRef]
- Binnemars-Postma, K.; Bansal, R.; Storm, G.; Prakash, J. Targeting the Stat6 pathway in tumor-associated macrophages reduces tumor growth and metastatic niche formation in breast cancer. FASEB J. 2018, 32, 969–978. [Google Scholar] [CrossRef] [PubMed]
- Camiolo, G.; Barbato, A.; Giallongo, C.; Vicario, N.; Romano, A.; Parrinello, N.L.; Parenti, R.; Sandoval, J.C.; García-Moreno, D.; Lazzarino, G.; et al. Iron regulates myeloma cell/macrophage interaction and drives resistance to bortezomib. Redox Biol. 2020, 36, 101611. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Perry, T.L.; Chitko-McKown, C.G.; Smith, A.D.; Cheung, L.; Beshah, E.; Urban, J.F.; Dawson, H.D. The regulatory actions of retinoic acid on M2 polarization of porcine macrophages. Dev. Comp. Immunol. 2019, 98, 20–33. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.-C.; Cheng, H.-C.; Wang, J.; Wang, S.-W.; Tai, H.-C.; Lin, C.-W.; Tang, C.-H. Prostate cancer-derived CCN3 induces M2 macrophage infiltration and contributes to angiogenesis in prostate cancer microenvironment. Oncotarget 2014, 5, 1595–1608. [Google Scholar] [CrossRef]
- Cheng, Y.; Si, Y.; Wang, L.; Ding, M.; Yu, S.; Lu, L.; Guo, Y.; Zong, M.; Fan, L. The regulation of macrophage polarization by hypoxia-PADI4 coordination in Rheumatoid arthritis. Int. Immunopharmacol. 2021, 99, 107988. [Google Scholar] [CrossRef]
- Chiba, Y.; Mizoguchi, I.; Furusawa, J.; Hasegawa, H.; Ohashi, M.; Xu, M.; Owaki, T.; Yoshimoto, T. Interleukin-27 exerts its antitumor effects by promoting differentiation of hematopoietic stem cells to M1 macrophages. Cancer Res. 2018, 78, 182–194. [Google Scholar] [CrossRef]
- Cui, Y.; Chen, F.; Gao, J.; Lei, M.; Wang, D.; Jin, X.; Guo, Y.; Shan, L.; Chen, X. Comprehensive landscape of the renin-angiotensin system in Pan-cancer: A potential downstream mediated mechanism of SARS-CoV-2. Int. J. Biol. Sci. 2021, 17, 3795. [Google Scholar] [CrossRef]
- Cui, Z.; Feng, Y.; Li, D.; Li, T.; Gao, P.; Xu, T. Activation of aryl hydrocarbon receptor (AhR) in mesenchymal stem cells modulates macrophage polarization in asthma. J. Immunotoxicol. 2020, 17, 21–30. [Google Scholar] [CrossRef]
- Dong, H.; Xie, C.; Jiang, Y.; Li, K.; Lin, Y.; Pang, X.; Xiong, X.; Zheng, J.; Ke, X.; Chen, Y.; et al. Tumor-Derived Exosomal Protein Tyrosine Phosphatase Receptor Type O Polarizes Macrophage to Suppress Breast Tumor Cell Invasion and Migration. Front. Cell Dev. Biol. 2021, 2731. [Google Scholar] [CrossRef]
- Fang, X.Y.; Zhan, Y.X.; Zhou, X.M.; Wu, L.N.; Lin, J.; Yi, Y.T.; Jiang, C.M.; Wang, J.; Liu, J. CXCL12/CXCR4 Mediates Orthodontic Root Resorption via Regulating the M1/M2 Ratio. J. Dent. Res. 2021, 101, 569–579. [Google Scholar] [CrossRef]
- Guo, Y.; Lin, C.; Xu, P.; Wu, S.; Fu, X.; Xia, W.; Yao, M. AGEs induced autophagy impairs cutaneous wound healing via stimulating macrophage polarization to M1 in diabetes. Sci. Rep. 2016, 6, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Li, Q.; Hu, R.; Li, R.; Yang, Y. Five immune-related genes as diagnostic markers for endometriosis and their correlation with immune infiltration. Front. Endocrinol. 2022, 13, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Inoue, T. M1 macrophage triggered by Mincle leads to a deterioration of acute kidney injury. Kidney Int. 2017, 91, 526–529. [Google Scholar] [CrossRef] [PubMed]
- Jablonski, K.A.; Gaudet, A.D.; Amici, S.A.; Popovich, P.G.; Guerau-De-Arellano, M. Control of the inflammatory macrophage transcriptional signature by miR-155. PLoS ONE 2016, 11, e0159724. [Google Scholar] [CrossRef]
- Joerink, M.; Rindsjö, E.; van Riel, B.; Alm, J.; Papadogiannakis, N. Placental macrophage (Hofbauer cell) polarization is independent of maternal allergen-sensitization and presence of chorioamnionitis. Placenta 2011, 32, 380–385. [Google Scholar] [CrossRef]
- Ganta, V.C.; Choi, M.; Farber, C.R.; Annex, B.H. Antiangiogenic VEGF165b regulates macrophage polarization via S100A8/S100A9 in peripheral artery disease. Circulation 2019, 139, 226–242. [Google Scholar] [CrossRef]
- Le, Y.; Gao, H.; Bleday, R.; Zhu, Z. The homeobox protein VentX reverts immune suppression in the tumor microenvironment. Nat. Commun. 2018, 9, 1–10. [Google Scholar] [CrossRef]
- Liu, M.; Li, F.; Bin Liu, B.; Jian, Y.; Zhang, D.; Zhou, H.; Wang, Y.; Xu, Z. Profiles of immune cell infiltration and immune-related genes in the tumor microenvironment of esophageal squamous cell carcinoma. BMC Med. Genomics 2021, 14, 75. [Google Scholar] [CrossRef]
- Lv, D.; Wu, X.; Chen, X.; Yang, S.; Chen, W.; Wang, M.; Liu, Y.; Gu, D.; Zeng, G. A novel immune-related gene-based prognostic signature to predict biochemical recurrence in patients with prostate cancer after radical prostatectomy. Cancer Immunol. Immunother. 2021, 70, 3587–3602. [Google Scholar] [CrossRef]
- Müller, E.; Christopoulos, P.F.; Halder, S.; Lunde, A.; Beraki, K.; Speth, M.; Øynebråten, I.; Corthay, A. Toll-like receptor ligands and interferon-γ synergize for induction of antitumor M1 macrophages. Front. Immunol. 2017, 8, 1383. [Google Scholar] [CrossRef]
- Oliveira, L.; McClellan, S.; Hansen, P.J. Differentiation of the endometrial macrophage during pregnancy in the cow. PLoS ONE 2010, 5, e13213. [Google Scholar] [CrossRef] [PubMed]
- Pagie, S.; Gérard, N.; Charreau, B. Notch signaling triggered via the ligand DLL4 impedes M2 macrophage differentiation and promotes their apoptosis. Cell Commun. Signal. 2018, 16, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Quan, Q.; Xiong, X.; Wu, S.; Yu, M. Identification of Immune-Related Key Genes in Ovarian Cancer Based on WGCNA. Front. Genet. 2021, 12, 760225. [Google Scholar] [CrossRef] [PubMed]
- Quero, L.; Tiaden, A.N.; Hanser, E.; Roux, J.; Laski, A.; Hall, J.; Kyburz, D. miR-221-3p Drives the Shift of M2-Macrophages to a Pro-Inflammatory Function by Suppressing JAK3/STAT3 Activation. Front. Immunol. 2020, 10, 3087. [Google Scholar] [CrossRef]
- Sanjurjo, L.; Aran, G.; Téllez, É.; Amézaga, N.; Armengol, C.; López, D.; Prats, C.; Sarrias, M.-R. CD5L promotes M2 macrophage polarization through autophagy-mediated upregulation of ID3. Front. Immunol. 2018, 9, 480. [Google Scholar] [CrossRef]
- Sato, K.; Yamashita, T.; Shirai, R.; Shibata, K.; Okano, T.; Yamaguchi, M.; Mori, Y.; Hirano, T.; Watanabe, T. Adropin contributes to anti-atherosclerosis by suppressing monocyte-endothelial cell adhesion and smooth muscle cell proliferation. Int. J. Mol. Sci. 2018, 19, 1293. [Google Scholar] [CrossRef]
- Seneviratne, A.; Cave, L.; Hyde, G.; Moestrup, S.K.; Carling, D.; Mason, J.; O Haskard, D.; Boyle, J.J. Metformin directly suppresses atherosclerosis in normoglycaemic mice via haematopoietic adenosine monophosphate-activated protein kinase. Cardiovasc. Res. 2021, 117, 1295–1308. [Google Scholar] [CrossRef]
- Shapouri-Moghaddam, A.; Mohammadian, S.; Vazini, H.; Taghadosi, M.; Esmaeili, S.-A.; Mardani, F.; Seifi, B.; Mohammadi, A.; Afshari, J.T.; Sahebkar, A. Macrophage plasticity, polarization, and function in health and disease. J. Cell. Physiol. 2018, 233, 6425–6440. [Google Scholar] [CrossRef]
- Song, W.-M.; Agrawal, P.; Von Itter, R.; Fontanals-Cirera, B.; Wang, M.; Zhou, X.; Mahal, L.K.; Hernando, E.; Zhang, B. Network models of primary melanoma microenvironments identify key melanoma regulators underlying prognosis. Nat. Commun. 2021, 12, 1–14. [Google Scholar] [CrossRef]
- Talamonti, E.; Pauter, A.M.; Asadi, A.; Fischer, A.W.; Chiurchiù, V.; Jacobsson, A. Impairment of systemic DHA synthesis affects macrophage plasticity and polarization: Implications for DHA supplementation during inflammation. Cell. Mol. Life Sci. 2017, 74, 2815–2826. [Google Scholar] [CrossRef]
- Tan, Y.; Sun, R.; Liu, L.; Yang, D.; Xiang, Q.; Li, L.; Tang, J.; Qiu, Z.; Peng, W.; Wang, Y.; et al. Tumor suppressor DRD2 facilitates M1 macrophages and restricts NF-κB signaling to trigger pyroptosis in breast cancer. Theranostics 2021, 11, 5214. [Google Scholar] [CrossRef] [PubMed]
- Bossche, J.V.D.; Laoui, D.; Morias, Y.; Movahedi, K.; Raes, G.; De Baetselier, P.; Van Ginderachter, J.A. Claudin-1, Claudin-2 and Claudin-11 Genes Differentially Associate with Distinct Types of Anti-inflammatory Macrophages In vitro and with Parasite- and Tumour-elicited Macrophages In vivo. Scand. J. Immunol. 2012, 75, 588–598. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Liu, X.; Song, X.; Dong, L.; Liu, D. MiR-202-5p promotes M2 polarization in allergic rhinitis by targeting MATN2. Int. Arch. Allergy Immunol. 2019, 178, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Weng, Y.; Feng, Y.; Liang, B.; Wang, H.; Li, L.; Wang, Z. Trem1 Induces Periodontal Inflammation via Regulating M1 Polarization. J. Dent. Res. 2021, 101, 437–447. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.F.; Lin, C.A.; Yuan, T.H.; Yeh, H.Y.; Su, S.F.; Guo, C.L.; Chang, G.C.; Li, K.C.; Ho, C.C.; Chen, H.W. The M1/M2 spectrum and plasticity of malignant pleural effusion-macrophage in advanced lung cancer. Cancer Immunol. Immunother. 2021, 70, 1435–1450. [Google Scholar] [CrossRef]
- Wu, X.; Giobbie-Hurder, A.; Liao, X.; Connelly, C.; Connolly, E.M.; Li, J.; Manos, M.P.; Lawrence, D.; McDermott, D.; Severgnini, M.; et al. Angiopoietin-2 as a Biomarker and Target for Immune Checkpoint Therapy. Cancer Immunol. Res. 2017, 5, 17–28. [Google Scholar] [CrossRef]
- Yan, C.; Zhou, Q.-Y.; Wu, J.; Xu, N.; Du, Y.; Li, J.; Liu, J.-X.; Koda, S.; Zhang, B.-B.; Yu, Q.; et al. Csi-let-7a-5p delivered by extracellular vesicles from a liver fluke activates M1-like macrophages and exacerbates biliary injuries. Proc. Natl. Acad. Sci. USA 2021, 118, e2102206118. [Google Scholar] [CrossRef]
- Yan, J.; Wu, X.; Yu, J.; Zhu, Y.; Cang, S. Prognostic Role of Tumor Mutation Burden Combined With Immune Infiltrates in Skin Cutaneous Melanoma Based on Multi-Omics Analysis. Front. Oncol. 2020, 10, 2403. [Google Scholar] [CrossRef]
- Zhang, A.Z.; Yuan, X.; Liang, W.H.; Zhang, H.J.; Li, Y.; Xie, Y.F.; Li, J.F.; Jiang, C.H.; Li, F.P.; Shen, X.H.; et al. Immune Infiltration in Gastric Cancer Microenvironment and Its Clinical Significance. Front. Cell Dev. Biol. 2021, 9, 762029. [Google Scholar] [CrossRef]


| Abbreviation | Phenotype | Method |
|---|---|---|
| M1_IFNγ + LPS | M1 | 100 ng/mL IFNγ (Proteintech, Rosemount, IL, USA, HZ-1301), and 20 ng/mL LPS (sigma, USA, L4516) |
| M1_GM-CSF | M1 | 20 ng/mL GM-CSF (absin, Shanghai, China, abs04132) |
| M2_IL4 + IL10 | M2 | 10 ng/mL IL10(Proteintech, USA, HZ-1145), and 10 ng/mL IL4 (Proteintech, Rosemount, IL, USA, HZ-1004) |
| M2_M-CSF | M2 | 20 ng/mL M-CSF (absin, Shanghai, China, abs00846) |
| GEO ID | Cohort Description | Experimental Groups |
|---|---|---|
| GSE174494 | Macrophages were obtained by lung perfusion in pigs infected with the PRRSV virus. The harvested infected macrophages were detected by RNA-seq. | Alveolar M ϕ from PRRSV (n = 3) vs. PPRRSV-GFP (n = 3) vs. RPMI-1640 (n = 3). |
| GSE145954 | PAMs infected ASFV isolates (MOI = 1), and transcriptome analysis was performed on infected cells and normal PAM at 0, 6, 12, and 24 h. | Alveolar M ϕ from ASFV-0h (n = 3) vs. ASFV-6h (n = 3) vs. ASFV-12h (n = 3) vs. ASFV-24 h (n = 3) vs. uninfected (n = 3). |
| GSE146715 | PAM-infected with T. gondii Me49 for 15 h. Transcriptome analysis was performed on infected and uninfected PAM. | Alveolar M ϕ from T. gondii Me49-PAM (n = 3) vs. uninfected (n = 3). |
| GSE153330 | PAM-infected with TgRH (type i), TgME49 (type ii), or TgHB1-Toxoplasma for 6, 12, and 24 h. Transcriptome analysis was performed on infected and uninfected PAM. | Alveolar M ϕ from PAM at different infection times for each infection strain (n = 3) vs. uninfected (n = 3). |
| GSE34544 | The piglets were infected with SS2 and HPS4 28 days later. Transcriptome analysis of infected and uninfected PAM. | Alveolar M ϕ from PAM for each infection strain (n = 6) vs. uninfected (n = 6). |
| GSE30696 | ||
| GSE181105 | Infected PAM were obtained 6 and 15 h after infection with Mhp or PRRSV. Microarray analysis of infected and uninfected PAM. | Alveolar M ϕ from PAM for each infection strain (n = 3) vs. uninfected (n = 3). |
| GSE22782 | Infected PAM were obtained after piglets were infected with HP-PRRSV for 5 days. Microarray analysis of infected and uninfected PAM. | Alveolar M ϕ from PAM for each infection strain (n = 6) vs. uninfected (n = 6). |
| GSE30918 | Infected PAM were obtained 48 h after PCV2 infection. Microarray analysis of infected and uninfected PAM. | Alveolar M ϕ from PAM for each infection strain (n = 3) vs. uninfected (n = 3). |
| GSE156504 | Infected PAM were obtained after 21 h of infection with different strains of PRRSV. Transcriptome analysis of infected and uninfected PAM. | Alveolar M ϕ from PAM for each infection strain (n = 3) vs. uninfected (n = 3). |
| GSE49882 | Infected PAM were obtained after 6 and 15 h of infection with Mycoplasma hyopneumoniae and PRRSV. | Alveolar M ϕ from PAM for each infection strain (n = 3) vs. uninfected (n = 3). |
| GSE30956 | BMDM were stimulated with LPS from Salmonella enterica serotype Minnesota Re 595 for 0, 2, 7, and 24 h to obtain macrophages after a reaction. | Alveolar M ϕ from PAM for each infection strain (n = 3) vs. uninfected (n = 3). |
| GSE45145 | BMDM from different breeds of pigs were stimulated with LPS from Salmonella enterica serotype Minnesota Re 595 for 0, 2, 7, and 24 h to obtain macrophages after the reaction. | Alveolar M ϕ from PAM for each infection strain (n = 3) vs. uninfected (n = 3). |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J.; Yuan, T.; Zhang, A.; Yang, P.; He, L.; Long, K.; Tang, C.; Chen, L.; Li, M.; Lu, L. Transcriptomic Establishment of Pig Macrophage Polarization Signatures. Curr. Issues Mol. Biol. 2023, 45, 2338-2350. https://doi.org/10.3390/cimb45030151
Li J, Yuan T, Zhang A, Yang P, He L, Long K, Tang C, Chen L, Li M, Lu L. Transcriptomic Establishment of Pig Macrophage Polarization Signatures. Current Issues in Molecular Biology. 2023; 45(3):2338-2350. https://doi.org/10.3390/cimb45030151
Chicago/Turabian StyleLi, Jing, Teng Yuan, Anjing Zhang, Peidong Yang, Li He, Keren Long, Chuang Tang, Li Chen, Mingzhou Li, and Lu Lu. 2023. "Transcriptomic Establishment of Pig Macrophage Polarization Signatures" Current Issues in Molecular Biology 45, no. 3: 2338-2350. https://doi.org/10.3390/cimb45030151
APA StyleLi, J., Yuan, T., Zhang, A., Yang, P., He, L., Long, K., Tang, C., Chen, L., Li, M., & Lu, L. (2023). Transcriptomic Establishment of Pig Macrophage Polarization Signatures. Current Issues in Molecular Biology, 45(3), 2338-2350. https://doi.org/10.3390/cimb45030151







