Plant Metabolomics: Current Initiatives and Future Prospects
Abstract
:1. Introduction
2. Metabolomic Platforms and Large-Scale Metabolite Databases
3. Role of Metabolomics in Crop Improvement
4. Metabolomics and Its Regulations in Abiotic Stresses
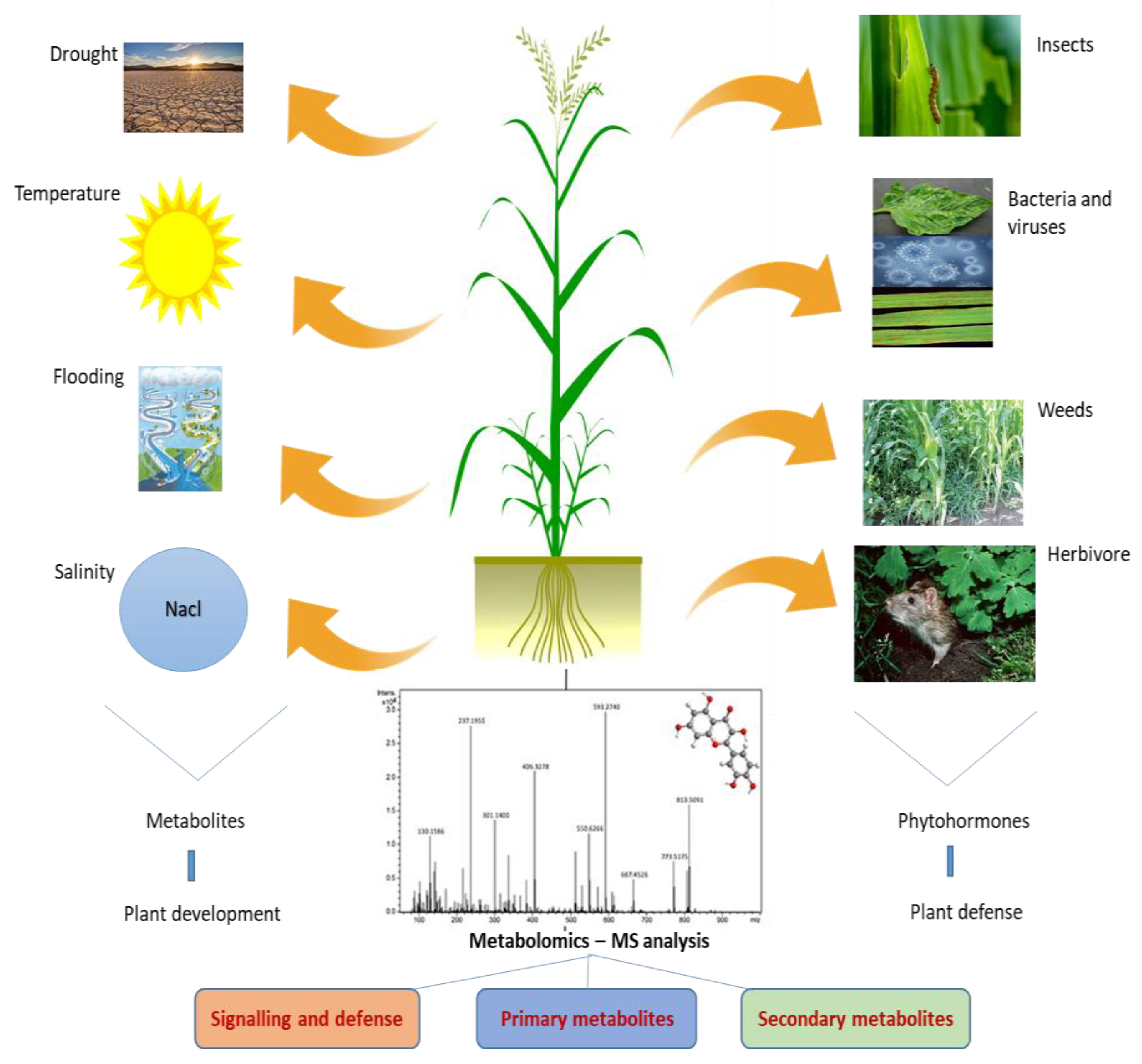
5. Metabolite Accumulation in Biotic Stresses
6. Metabolomics in Assessing the Nutrients
7. Metabolomics in Discovering Biomarkers
8. Metabolomics-Assisted Breeding
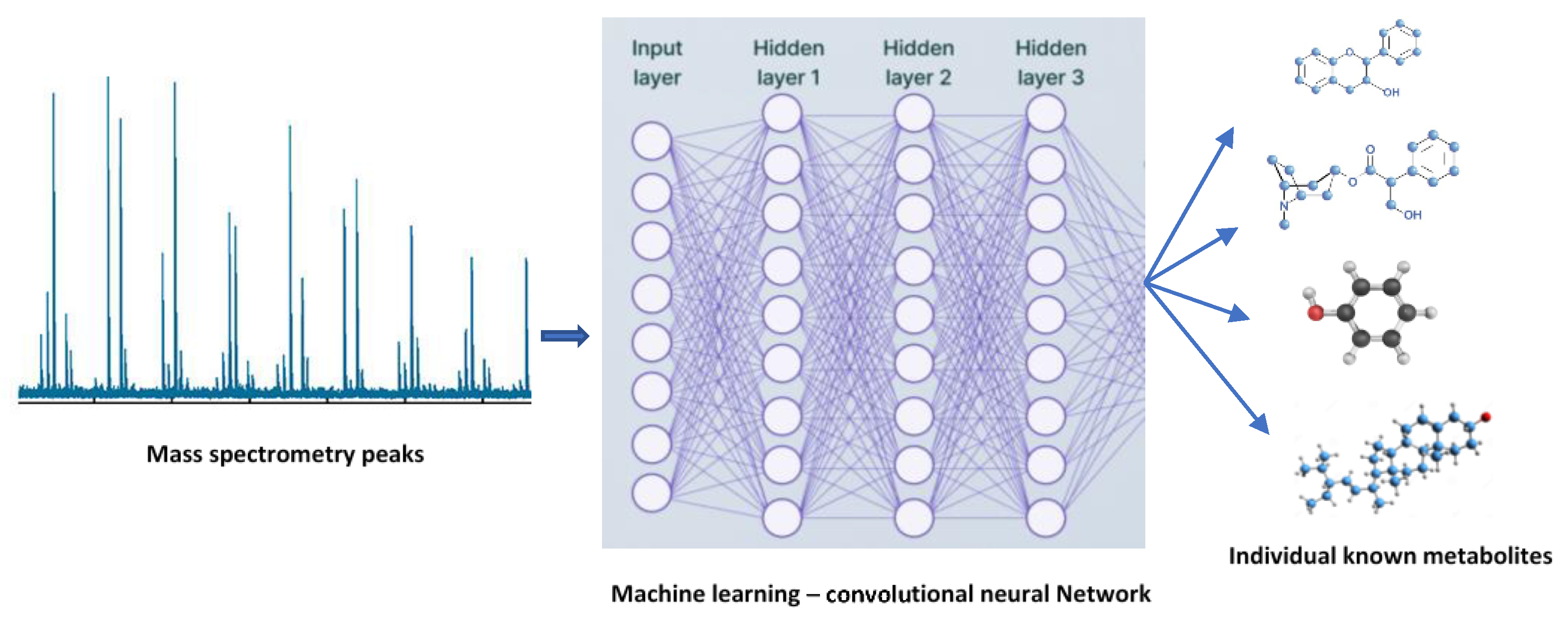
9. Implications of Data Science in Plant Metabolomics
10. Metabolomics for Plant-Microbe Interactions Research
11. Interrelationship between Different Omics

12. Concluding Remarks and Future Prospectives
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Issaq, H.J.; Van, Q.N.; Waybright, T.J.; Muschik, G.M.; Veenstra, T.D. Analytical and statistical approaches to metabolomics research. J. Sep. Sci. 2009, 32, 2183–2199. [Google Scholar] [CrossRef] [PubMed]
- Ralston-Hooper, K.J.; Adamec, J.; Jannash, A.; Mollenhauer, R.; Ochoa-Acuña, H.; Sepúlveda, M.S. Use of GC × GC/TOF-MS and LC/TOF-MS for metabolomic analysis of Hyalella azteca chronically exposed to atrazine and its primary metabolite, desethylatrazine. J. Appl. Toxicol. 2011, 31, 399–410. [Google Scholar] [CrossRef] [PubMed]
- Turner, M.F.; Heuberger, A.L.; Kirkwood, J.S.; Collins, C.C.; Wolfrum, E.J.; Broeckling, C.D.; Prenni, J.E.; Jahn, C.E. Non-targeted metabolomics in diverse sorghum breeding lines indicates primary and secondary metabolite profiles are associated with plant biomass accumulation and photosynthesis. Front. Plant Sci. 2016, 7, 953. [Google Scholar] [CrossRef] [PubMed]
- Ramautar, R.; de Jong, G.J. Recent developments in liquid-phase separation techniques for metabolomics. Bioanalysis 2014, 6, 1011–1026. [Google Scholar] [CrossRef] [PubMed]
- Zrodnikov, Y.; Davis, C.E. The highs and lows of FAIMS: Predictions and future trends for high field asymmetric waveform ion mobility spectrometry. J. Nanomed. Nanotechnol. 2012, 3, 109e. [Google Scholar] [CrossRef]
- Ma, X.; Xia, H.; Liu, Y.; Wei, H.; Zheng, X.; Song, C.; Chen, L.; Liu, H.; Luo, L. Transcriptomic and metabolomic studies disclose key metabolism pathways contributing to well-maintained photosynthesis under the drought and the consequent drought-tolerance in rice. Front. Plant Sci. 2016, 7, 1886. [Google Scholar] [CrossRef]
- Misra, B.B.; van der Hooft, J.J. Updates in metabolomics tools and resources: 2014–2015. Electrophoresis 2016, 37, 86–110. [Google Scholar] [CrossRef]
- Perez de Souza, L.; Naake, T.; Tohge, T.; Fernie, A.R. From chromatogram to analyte to metabolite. How to pick horses for courses from the massive web resources for mass spectral plant metabolomics. Gigascience 2017, 6, gix037. [Google Scholar] [CrossRef]
- Johnson, C.H.; Ivanisevic, J.; Benton, H.P.; Siuzdak, G. Bioinformatics: The next frontier of metabolomics. Anal. Chem. 2015, 87, 147–156. [Google Scholar] [CrossRef]
- Tsugawa, H.; Cajka, T.; Kind, T.; Ma, Y.; Higgins, B.; Ikeda, K.; Kanazawa, M.; VanderGheynst, J.; Fiehn, O.; Arita, M. MS-DIAL: Data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat. Methods 2015, 12, 523–526. [Google Scholar] [CrossRef]
- Xie, W.; Lv, X.; Ye, L.; Zhou, P.; Yu, H. Construction of lycopene-overproducing Saccharomyces cerevisiae by combining directed evolution and metabolic engineering. Metab. Eng. 2015, 30, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Hein, J.A.; Sherrard, M.E.; Manfredi, K.P.; Abebe, T. The fifth leaf and spike organs of barley (Hordeum vulgare L.) display different physiological and metabolic responses to drought stress. BMC Plant Biol. 2016, 16, 248. [Google Scholar] [CrossRef] [PubMed]
- García-Cristobal, J.; García-Villaraco, A.; Ramos, B.; Gutierrez-Mañero, J.; Lucas, J. Priming of pathogenesis related-proteins and enzymes related to oxidative stress by plant growth promoting rhizobacteria on rice plants upon abiotic and biotic stress challenge. J. Plant Physiol. 2015, 188, 72–79. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.Y.; Li, A.X.; Li, F.; Zhao, M.R.; Wang, W. Characterization of a wheat (Triticum aestivum L.) expansin gene, TaEXPB23, involved in the abiotic stress response and phytohormone regulation. Plant Physiol. Biochem. 2012, 54, 49–58. [Google Scholar] [CrossRef]
- Mhlongo, M.I.; Piater, L.A.; Madala, N.E.; Labuschagne, N.; Dubery, I.A. The chemistry of plant–microbe interactions in the rhizosphere and the potential for metabolomics to reveal signaling related to defense priming and induced systemic resistance. Front. Plant Sci. 2018, 9, 112. [Google Scholar] [CrossRef]
- Guo, R.; Shi, L.; Jiao, Y.; Li, M.; Zhong, X.; Gu, F.; Liu, Q.; Xia, X.; Li, H. Metabolic responses to drought stress in the tissues of drought-tolerant and drought-sensitive wheat genotype seedlings. AoB Plants 2018, 10, ply016. [Google Scholar] [CrossRef]
- Kang, Z.; Babar, M.A.; Khan, N.; Guo, J.; Khan, J.; Islam, S.; Shrestha, S.; Shahi, D. Comparative metabolomic profiling in the roots and leaves in contrasting genotypes reveals complex mechanisms involved in post-anthesis drought tolerance in wheat. PLoS ONE 2019, 14, e0213502. [Google Scholar] [CrossRef]
- Khan, F.; Fuentes, D.; Threthowan, R.; Mohammad, F.; Ahmad, M. Comparative metabolite profiling of two wheat genotypes as affected by nitrogen stress at seedling stage. JAPS J. Anim. Plant Sci. 2019, 29, 260–268. [Google Scholar]
- Blum, A. Osmotic adjustment is a prime drought stress adaptive engine in support of plant production. Plant Cell Environ. 2017, 40, 4–10. [Google Scholar] [CrossRef]
- Krasensky, J.; Jonak, C. Drought, salt, and temperature stress-induced metabolic rearrangements and regulatory networks. J. Exp. Bot. 2012, 63, 1593–1608. [Google Scholar] [CrossRef]
- Yadav, A.K.; Carroll, A.J.; Estavillo, G.M.; Rebetzke, G.J.; Pogson, B.J. Wheat drought tolerance in the field is predicted by amino acid responses to glasshouse-imposed drought. J. Exp. Bot. 2019, 70, 4931–4948. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Fountain, J.C.; Ji, P.; Ni, X.; Chen, S.; Lee, R.D.; Kemerait, R.C.; Guo, B. Deciphering drought-induced metabolic responses and regulation in developing maize kernels. Plant Biotechnol. J. 2018, 16, 1616–1628. [Google Scholar] [CrossRef] [PubMed]
- Skirycz, A.; Inzé, D. More from less: Plant growth under limited water. Curr. Opin. Biotechnol. 2010, 21, 197–203. [Google Scholar] [CrossRef] [PubMed]
- Nam, K.-H.; Kim, D.Y.; Kim, H.J.; Pack, I.-S.; Kim, H.J.; Chung, Y.S.; Kim, S.Y.; Kim, C.-G. Global metabolite profiling based on GC–MS and LC–MS/MS analyses in ABF3-overexpressing soybean with enhanced drought tolerance. Appl. Biol. Chem. 2019, 62, 15. [Google Scholar] [CrossRef]
- Gayen, D.; Barua, P.; Lande, N.V.; Varshney, S.; Sengupta, S.; Chakraborty, S.; Chakraborty, N. Dehydration-responsive alterations in the chloroplast proteome and cell metabolomic profile of rice reveals key stress adaptation responses. Environ. Exp. Bot. 2019, 160, 12–24. [Google Scholar] [CrossRef]
- Gupta, P.; De, B. Metabolomics analysis of rice responses to salinity stress revealed elevation of serotonin, and gentisic acid levels in leaves of tolerant varieties. Plant Signal. Behav. 2017, 12, e1335845. [Google Scholar] [CrossRef]
- Shelden, M.C.; Dias, D.A.; Jayasinghe, N.S.; Bacic, A.; Roessner, U. Root spatial metabolite profiling of two genotypes of barley (Hordeum vulgare L.) reveals differences in response to short-term salt stress. J. Exp. Bot. 2016, 67, 3731–3745. [Google Scholar] [CrossRef]
- Chang, J.; Cheong, B.E.; Natera, S.; Roessner, U. Morphological and metabolic responses to salt stress of rice (Oryza sativa L.) cultivars which differ in salinity tolerance. Plant Physiol. Biochem. 2019, 144, 427–435. [Google Scholar] [CrossRef]
- Kurotani, K.-I.; Hayashi, K.; Hatanaka, S.; Toda, Y.; Ogawa, D.; Ichikawa, H.; Ishimaru, Y.; Tashita, R.; Suzuki, T.; Ueda, M. Elevated levels of CYP94 family gene expression alleviate the jasmonate response and enhance salt tolerance in rice. Plant Cell Physiol. 2015, 56, 779–789. [Google Scholar] [CrossRef]
- Zörb, C.; Geilfus, C.-M.; Mühling, K.H.; Ludwig-Müller, J. The influence of salt stress on ABA and auxin concentrations in two maize cultivars differing in salt resistance. J. Plant Physiol. 2013, 170, 220–224. [Google Scholar] [CrossRef]
- Borrelli, G.M.; Fragasso, M.; Nigro, F.; Platani, C.; Papa, R.; Beleggia, R.; Trono, D. Analysis of metabolic and mineral changes in response to salt stress in durum wheat (Triticum turgidum ssp. durum) genotypes, which differ in salinity tolerance. Plant Physiol. Biochem. 2018, 133, 57–70. [Google Scholar] [CrossRef] [PubMed]
- Rouphael, Y.; Raimondi, G.; Lucini, L.; Carillo, P.; Kyriacou, M.C.; Colla, G.; Cirillo, V.; Pannico, A.; El-Nakhel, C.; De Pascale, S. Physiological and metabolic responses triggered by omeprazole improve tomato plant tolerance to NaCl stress. Front. Plant Sci. 2018, 9, 249. [Google Scholar] [CrossRef] [PubMed]
- Thomason, K.; Babar, M.A.; Erickson, J.E.; Mulvaney, M.; Beecher, C.; MacDonald, G. Comparative physiological and metabolomics analysis of wheat (Triticum aestivum L.) following post-anthesis heat stress. PLoS ONE 2018, 13, e0197919. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Hou, L.; Lu, Y.; Wu, B.; Gong, X.; Liu, M.; Wang, J.; Sun, Q.; Vierling, E.; Xu, S. Metabolic adaptation of wheat grain contributes to a stable filling rate under heat stress. J. Exp. Bot. 2018, 69, 5531–5545. [Google Scholar] [CrossRef] [PubMed]
- Chebrolu, K.K.; Fritschi, F.B.; Ye, S.; Krishnan, H.B.; Smith, J.R.; Gillman, J.D. Impact of heat stress during seed development on soybean seed metabolome. Metabolomics 2016, 12, 28. [Google Scholar] [CrossRef]
- Sun, C.; Gao, X.; Li, M.; Fu, J.; Zhang, Y. Plastic responses in the metabolome and functional traits of maize plants to temperature variations. Plant Biol. 2016, 18, 249–261. [Google Scholar] [CrossRef]
- Paupière, M.J.; Müller, F.; Li, H.; Rieu, I.; Tikunov, Y.M.; Visser, R.G.; Bovy, A.G. Untargeted metabolomic analysis of tomato pollen development and heat stress response. Plant Reprod. 2017, 30, 81–94. [Google Scholar] [CrossRef]
- Qi, X.; Xu, W.; Zhang, J.; Guo, R.; Zhao, M.; Hu, L.; Wang, H.; Dong, H.; Li, Y. Physiological characteristics and metabolomics of transgenic wheat containing the maize C4 phosphoenolpyruvate carboxylase (PEPC) gene under high temperature stress. Protoplasma 2017, 254, 1017–1030. [Google Scholar] [CrossRef]
- Sung, J.; Lee, S.; Lee, Y.; Ha, S.; Song, B.; Kim, T.; Waters, B.M.; Krishnan, H.B. Metabolomic profiling from leaves and roots of tomato (Solanum lycopersicum L.) plants grown under nitrogen, phosphorus or potassium-deficient condition. Plant Sci. 2015, 241, 55–64. [Google Scholar] [CrossRef]
- Ghosson, H.; Schwarzenberg, A.; Jamois, F.; Yvin, J.-C. Simultaneous untargeted and targeted metabolomics profiling of underivatized primary metabolites in sulfur-deficient barley by ultra-high performance liquid chromatography-quadrupole/time-of-flight mass spectrometry. Plant Methods 2018, 14, 62. [Google Scholar] [CrossRef]
- Huang, C.Y.; Roessner, U.; Eickmeier, I.; Genc, Y.; Callahan, D.L.; Shirley, N.; Langridge, P.; Bacic, A. Metabolite profiling reveals distinct changes in carbon and nitrogen metabolism in phosphate-deficient barley plants (Hordeum vulgare L.). Plant Cell Physiol. 2008, 49, 691–703. [Google Scholar] [CrossRef] [PubMed]
- Hernández, G.; Valdés-López, O.; Ramírez, M.; Goffard, N.; Weiller, G.; Aparicio-Fabre, R.; Fuentes, S.I.; Erban, A.; Kopka, J.; Udvardi, M.K. Global changes in the transcript and metabolic profiles during symbiotic nitrogen fixation in phosphorus-stressed common bean plants. Plant Physiol. 2009, 151, 1221–1238. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Ma, X.-M.; Wang, X.-C.; Liu, J.-H.; Huang, B.-Y.; Guo, X.-Y.; Xiong, S.-P.; La, G.-X. UPLC-QTOF analysis reveals metabolomic changes in the flag leaf of wheat (Triticum aestivum L.) under low-nitrogen stress. Plant Physiol. Biochem. 2017, 111, 30–38. [Google Scholar] [CrossRef] [PubMed]
- Allwood, J.W.; Ellis, D.I.; Goodacre, R. Metabolomic technologies and their application to the study of plants and plant–host interactions. Physiol. Plant. 2008, 132, 117–135. [Google Scholar] [CrossRef] [PubMed]
- Tenenboim, H.; Brotman, Y. Omic relief for the biotically stressed: Metabolomics of plant biotic interactions. Trends Plant Sci. 2016, 21, 781–791. [Google Scholar] [CrossRef] [PubMed]
- Balmer, D.; Flors, V.; Glauser, G.; Mauch-Mani, B. Metabolomics of cereals under biotic stress: Current knowledge and techniques. Front. Plant Sci. 2013, 4, 82. [Google Scholar] [CrossRef]
- Niculaes, C.; Abramov, A.; Hannemann, L.; Frey, M. Plant protection by benzoxazinoids—Recent insights into biosynthesis and function. Agronomy 2018, 8, 143. [Google Scholar] [CrossRef]
- López-Gresa, M.P.; Maltese, F.; Bellés, J.M.; Conejero, V.; Kim, H.K.; Choi, Y.H.; Verpoorte, R. Metabolic response of tomato leaves upon different plant–pathogen interactions. Phytochem. Anal. Int. J. Plant Chem. Biochem. Tech. 2010, 21, 89–94. [Google Scholar] [CrossRef]
- Suharti, W.S.; Nose, A.; Zheng, S.-H. Metabolomic study of two rice lines infected by Rhizoctonia solani in negative ion mode by CE/TOF-MS. J. Plant Physiol. 2016, 206, 13–24. [Google Scholar] [CrossRef]
- Rajaprakasam, S.; Shanmugavel, P.; Chockalingam, V.; Jegadeesan, S.; Latha, T.K.S.; Ananthan, S.N.; Muthurajan, R.; Kanagarajan, S. Comparative Metabolomic Profiling of Horse Gram (Macrotyloma uniflorum (Lam.) Verdc.) Genotypes for Horse Gram Yellow Mosaic Virus Resistance. Metabolites 2023, 13, 165. [Google Scholar] [CrossRef]
- Sudhagar, R.; Priyanka, S.; Chockalingam, V.; Sendhilvel, V.; Souframanien, J.; Raja, K.; Kanagarajan, S. Development and metabolic characterization of horse gram (Macrotyloma uniflorum lam.(Verdc.)) mutants for powdery mildew resistance. Agronomy 2022, 12, 800. [Google Scholar] [CrossRef]
- Agarrwal, R.; Bentur, J.S.; Nair, S. Gas chromatography mass spectrometry based metabolic profiling reveals biomarkers involved in rice-gall midge interactions. J. Integr. Plant Biol. 2014, 56, 837–848. [Google Scholar] [CrossRef] [PubMed]
- Sana, T.R.; Fischer, S.; Wohlgemuth, G.; Katrekar, A.; Jung, K.-H.; Ronald, P.C.; Fiehn, O. Metabolomic and transcriptomic analysis of the rice response to the bacterial blight pathogen Xanthomonas oryzae pv. oryzae. Metabolomics 2010, 6, 451–465. [Google Scholar] [CrossRef]
- Zhou, S.; Zhang, Y.K.; Kremling, K.A.; Ding, Y.; Bennett, J.S.; Bae, J.S.; Kim, D.K.; Ackerman, H.H.; Kolomiets, M.V.; Schmelz, E.A. Ethylene signaling regulates natural variation in the abundance of antifungal acetylated diferuloylsucroses and Fusarium graminearum resistance in maize seedling roots. New Phytol. 2019, 221, 2096–2111. [Google Scholar] [CrossRef] [PubMed]
- Vasmatkar, P.; Kaur, K.; Pannu, P.; Kaur, G.; Kaur, H. Unraveling the metabolite signatures of maize genotypes showing differential response towards southern corn leaf blight by 1H-NMR and FTIR spectroscopy. Physiol. Mol. Plant Pathol. 2019, 108, 101441. [Google Scholar] [CrossRef]
- Foito, A.; Byrne, S.L.; Hackett, C.A.; Hancock, R.D.; Stewart, D.; Barth, S. Short-term response in leaf metabolism of perennial ryegrass (Lolium perenne) to alterations in nitrogen supply. Metabolomics 2013, 9, 145–156. [Google Scholar] [CrossRef]
- Comadira, G.; Rasool, B.; Karpinska, B.; Morris, J.; Verrall, S.R.; Hedley, P.E.; Foyer, C.H.; Hancock, R.D. Nitrogen deficiency in barley (Hordeum vulgare) seedlings induces molecular and metabolic adjustments that trigger aphid resistance. J. Exp. Bot. 2015, 66, 3639–3655. [Google Scholar] [CrossRef]
- Rajagopalan, V.R.; Manickam, S.; Muthurajan, R. A comparative metabolomic analysis reveals the nutritional and therapeutic potential of grains of the traditional rice variety Mappillai Samba. Plants 2022, 11, 543. [Google Scholar] [CrossRef]
- Heyneke, E.; Watanabe, M.; Erban, A.; Duan, G.; Buchner, P.; Walther, D.; Kopka, J.; Hawkesford, M.J.; Hoefgen, R. Characterization of the wheat leaf metabolome during grain filling and under varied N-supply. Front. Plant Sci. 2017, 8, 2048. [Google Scholar] [CrossRef]
- Steinfath, M.; Strehmel, N.; Peters, R.; Schauer, N.; Groth, D.; Hummel, J.; Steup, M.; Selbig, J.; Kopka, J.; Geigenberger, P. Discovering plant metabolic biomarkers for phenotype prediction using an untargeted approach. Plant Biotechnol. J. 2010, 8, 900–911. [Google Scholar] [CrossRef]
- Gärtner, T.; Steinfath, M.; Andorf, S.; Lisec, J.; Meyer, R.C.; Altmann, T.; Willmitzer, L.; Selbig, J. Improved heterosis prediction by combining information on DNA-and metabolic markers. PLoS ONE 2009, 4, e5220. [Google Scholar] [CrossRef]
- Fiehn, O.; Kopka, J.; Dörmann, P.; Altmann, T.; Trethewey, R.N.; Willmitzer, L. Metabolite profiling for plant functional genomics. Nat. Biotechnol. 2000, 18, 1157–1161. [Google Scholar] [CrossRef] [PubMed]
- Guijas, C.; Montenegro-Burke, J.R.; Warth, B.; Spilker, M.E.; Siuzdak, G. Metabolomics activity screening for identifying metabolites that modulate phenotype. Nat. Biotechnol. 2018, 36, 316–320. [Google Scholar] [CrossRef] [PubMed]
- Fernie, A.R.; Schauer, N. Metabolomics-assisted breeding: A viable option for crop improvement? Trends Genet. 2009, 25, 39–48. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Gao, Y.; Xie, W.; Gong, L.; Lu, K.; Wang, W.; Li, Y.; Liu, X.; Zhang, H.; Dong, H. Genome-wide association analyses provide genetic and biochemical insights into natural variation in rice metabolism. Nat. Genet. 2014, 46, 714–721. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Buckler, E.S. Genetic association mapping and genome organization of maize. Curr. Opin. Biotechnol. 2006, 17, 155–160. [Google Scholar] [CrossRef] [PubMed]
- Kisiel, A.; Krzemińska, A.; Cembrowska-Lech, D.; Miller, T. Data Science and Plant Metabolomics. Metabolites 2023, 13, 454. [Google Scholar] [CrossRef]
- Piasecka, A.; Kachlicki, P.; Stobiecki, M. Analytical methods for detection of plant metabolomes changes in response to biotic and abiotic stresses. Int. J. Mol. Sci. 2019, 20, 379. [Google Scholar] [CrossRef]
- Hasan, M.M.; Chopin, J.P.; Laga, H.; Miklavcic, S.J. Detection and analysis of wheat spikes using convolutional neural networks. Plant Methods 2018, 14, 100. [Google Scholar] [CrossRef]
- Fuentes, A.; Yoon, S.; Lee, M.H.; Park, D.S. Improving accuracy of tomato plant disease diagnosis based on deep learning with explicit control of hidden classes. Front. Plant Sci. 2021, 12, 682230. [Google Scholar] [CrossRef]
- van de Mortel, J.E.; de Vos, R.C.; Dekkers, E.; Pineda, A.; Guillod, L.; Bouwmeester, K.; van Loon, J.J.; Dicke, M.; Raaijmakers, J.M. Metabolic and transcriptomic changes induced in Arabidopsis by the rhizobacterium Pseudomonas fluorescens SS101. Plant Physiol. 2012, 160, 2173–2188. [Google Scholar] [CrossRef] [PubMed]
- Zhalnina, K.; Louie, K.B.; Hao, Z.; Mansoori, N.; Da Rocha, U.N.; Shi, S.; Cho, H.; Karaoz, U.; Loqué, D.; Bowen, B.P. Dynamic root exudate chemistry and microbial substrate preferences drive patterns in rhizosphere microbial community assembly. Nat. Microbiol. 2018, 3, 470–480. [Google Scholar] [CrossRef] [PubMed]
- Negrel, L.; Halter, D.; Wiedemann-Merdinoglu, S.; Rustenholz, C.; Merdinoglu, D.; Hugueney, P.; Baltenweck, R. Identification of lipid markers of Plasmopara viticola infection in grapevine using a non-targeted metabolomic approach. Front. Plant Sci. 2018, 9, 360. [Google Scholar] [CrossRef] [PubMed]
- Gilard, F.; Brule, L.; Quillere, I.; Gourion, B.; Ratet, P.; de Souza, E.M.; Lea, P.J.; Hirel, B. Metabolic profiling of two maize (Zea mays L.) inbred lines inoculated with the nitrogen fixing plant-interacting bacteria Herbaspirillum seropedicae and Azospirillum brasilense. PLoS ONE 2017, 12, e0174576. [Google Scholar]
- Gupta, S.; Schillaci, M.; Roessner, U. Metabolomics as an emerging tool to study plant–microbe interactions. Emerg. Top. Life Sci. 2022, 6, 175–183. [Google Scholar] [CrossRef]
- Wen, W.; Li, K.; Alseekh, S.; Omranian, N.; Zhao, L.; Zhou, Y.; Xiao, Y.; Jin, M.; Yang, N.; Liu, H. Genetic determinants of the network of primary metabolism and their relationships to plant performance in a maize recombinant inbred line population. Plant Cell 2015, 27, 1839–1856. [Google Scholar] [CrossRef]
- Fernandez, O.; Urrutia, M.; Bernillon, S.; Giauffret, C.; Tardieu, F.; Le Gouis, J.; Langlade, N.; Charcosset, A.; Moing, A.; Gibon, Y. Fortune telling: Metabolic markers of plant performance. Metabolomics 2016, 12, 158. [Google Scholar] [CrossRef]
- Scossa, F.; Brotman, Y.; e Lima, F.d.A.; Willmitzer, L.; Nikoloski, Z.; Tohge, T.; Fernie, A.R. Genomics-based strategies for the use of natural variation in the improvement of crop metabolism. Plant Sci. 2016, 242, 47–64. [Google Scholar] [CrossRef]
- Beleggia, R.; Rau, D.; Laido, G.; Platani, C.; Nigro, F.; Fragasso, M.; De Vita, P.; Scossa, F.; Fernie, A.R.; Nikoloski, Z. Evolutionary metabolomics reveals domestication-associated changes in tetraploid wheat kernels. Mol. Biol. Evol. 2016, 33, 1740–1753. [Google Scholar] [CrossRef]
- Gong, L.; Chen, W.; Gao, Y.; Liu, X.; Zhang, H.; Xu, C.; Yu, S.; Zhang, Q.; Luo, J. Genetic analysis of the metabolome exemplified using a rice population. Proc. Natl. Acad. Sci. USA 2013, 110, 20320–20325. [Google Scholar] [CrossRef]
- Toubiana, D.; Semel, Y.; Tohge, T.; Beleggia, R.; Cattivelli, L.; Rosental, L.; Nikoloski, Z.; Zamir, D.; Fernie, A.R.; Fait, A. Metabolic profiling of a mapping population exposes new insights in the regulation of seed metabolism and seed, fruit, and plant relations. PLoS Genet. 2012, 8, e1002612. [Google Scholar] [CrossRef]
- Carreno-Quintero, N.; Acharjee, A.; Maliepaard, C.; Bachem, C.W.; Mumm, R.; Bouwmeester, H.; Visser, R.G.; Keurentjes, J.J. Untargeted metabolic quantitative trait loci analyses reveal a relationship between primary metabolism and potato tuber quality. Plant Physiol. 2012, 158, 1306–1318. [Google Scholar] [CrossRef]
- Matsuda, F.; Okazaki, Y.; Oikawa, A.; Kusano, M.; Nakabayashi, R.; Kikuchi, J.; Yonemaru, J.I.; Ebana, K.; Yano, M.; Saito, K. Dissection of genotype–phenotype associations in rice grains using metabolome quantitative trait loci analysis. Plant J. 2012, 70, 624–636. [Google Scholar] [CrossRef]
- Piasecka, A.; Sawikowska, A.; Kuczyńska, A.; Ogrodowicz, P.; Mikołajczak, K.; Krystkowiak, K.; Gudyś, K.; Guzy-Wróbelska, J.; Krajewski, P.; Kachlicki, P. Drought-related secondary metabolites of barley (Hordeum vulgare L.) leaves and their metabolomic quantitative trait loci. Plant J. 2017, 89, 898–913. [Google Scholar] [CrossRef]
- Templer, S.E.; Ammon, A.; Pscheidt, D.; Ciobotea, O.; Schuy, C.; McCollum, C.; Sonnewald, U.; Hanemann, A.; Förster, J.; Ordon, F. Metabolite profiling of barley flag leaves under drought and combined heat and drought stress reveals metabolic QTLs for metabolites associated with antioxidant defense. J. Exp. Bot. 2017, 68, 1697–1713. [Google Scholar] [CrossRef]
- Feng, J.; Long, Y.; Shi, L.; Shi, J.; Barker, G.; Meng, J. Characterization of metabolite quantitative trait loci and metabolic networks that control glucosinolate concentration in the seeds and leaves of Brassica napus. New Phytol. 2012, 193, 96–108. [Google Scholar] [CrossRef]
- Alseekh, S.; Tohge, T.; Wendenberg, R.; Scossa, F.; Omranian, N.; Li, J.; Kleessen, S.; Giavalisco, P.; Pleban, T.; Mueller-Roeber, B. Identification and mode of inheritance of quantitative trait loci for secondary metabolite abundance in tomato. Plant Cell 2015, 27, 485–512. [Google Scholar] [CrossRef]
- Alseekh, S.; Tong, H.; Scossa, F.; Brotman, Y.; Vigroux, F.; Tohge, T.; Ofner, I.; Zamir, D.; Nikoloski, Z.; Fernie, A.R. Canalization of tomato fruit metabolism. Plant Cell 2017, 29, 2753–2765. [Google Scholar] [CrossRef]
- Hill, C.B.; Taylor, J.D.; Edwards, J.; Mather, D.; Langridge, P.; Bacic, A.; Roessner, U. Detection of QTL for metabolic and agronomic traits in wheat with adjustments for variation at genetic loci that affect plant phenology. Plant Sci. 2015, 233, 143–154. [Google Scholar] [CrossRef]
- Kazmi, R.H.; Willems, L.A.; Joosen, R.V.; Khan, N.; Ligterink, W.; Hilhorst, H.W. Metabolomic analysis of tomato seed germination. Metabolomics 2017, 13, 145. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Manickam, S.; Rajagopalan, V.R.; Kambale, R.; Rajasekaran, R.; Kanagarajan, S.; Muthurajan, R. Plant Metabolomics: Current Initiatives and Future Prospects. Curr. Issues Mol. Biol. 2023, 45, 8894-8906. https://doi.org/10.3390/cimb45110558
Manickam S, Rajagopalan VR, Kambale R, Rajasekaran R, Kanagarajan S, Muthurajan R. Plant Metabolomics: Current Initiatives and Future Prospects. Current Issues in Molecular Biology. 2023; 45(11):8894-8906. https://doi.org/10.3390/cimb45110558
Chicago/Turabian StyleManickam, Sudha, Veera Ranjani Rajagopalan, Rohit Kambale, Raghu Rajasekaran, Selvaraju Kanagarajan, and Raveendran Muthurajan. 2023. "Plant Metabolomics: Current Initiatives and Future Prospects" Current Issues in Molecular Biology 45, no. 11: 8894-8906. https://doi.org/10.3390/cimb45110558
APA StyleManickam, S., Rajagopalan, V. R., Kambale, R., Rajasekaran, R., Kanagarajan, S., & Muthurajan, R. (2023). Plant Metabolomics: Current Initiatives and Future Prospects. Current Issues in Molecular Biology, 45(11), 8894-8906. https://doi.org/10.3390/cimb45110558





