Genome-Wide Identification of DUF26 Domain-Containing Genes in Dongxiang Wild Rice and Analysis of Their Expression Responses under Submergence
Abstract
:1. Introduction
2. Materials and Methods
2.1. Identification of DUF26 Domain-Containing Genes in DXWR
2.2. Analysis of the Main Characteristics of DUF26 Domain-Containing Genes in DXWR
2.3. Repeat Events and Collinearity Analysis of DUF26 Domain-Containing Genes
2.4. Expression Data of DUF26 Domain-Containing Genes under Submergence
2.5. Correlation Analysis between DUF26 Domain-Containing Genes and Submergence Tolerance Genes
3. Results
3.1. Identification of DUF26 Domain-Containing Genes in DXWR
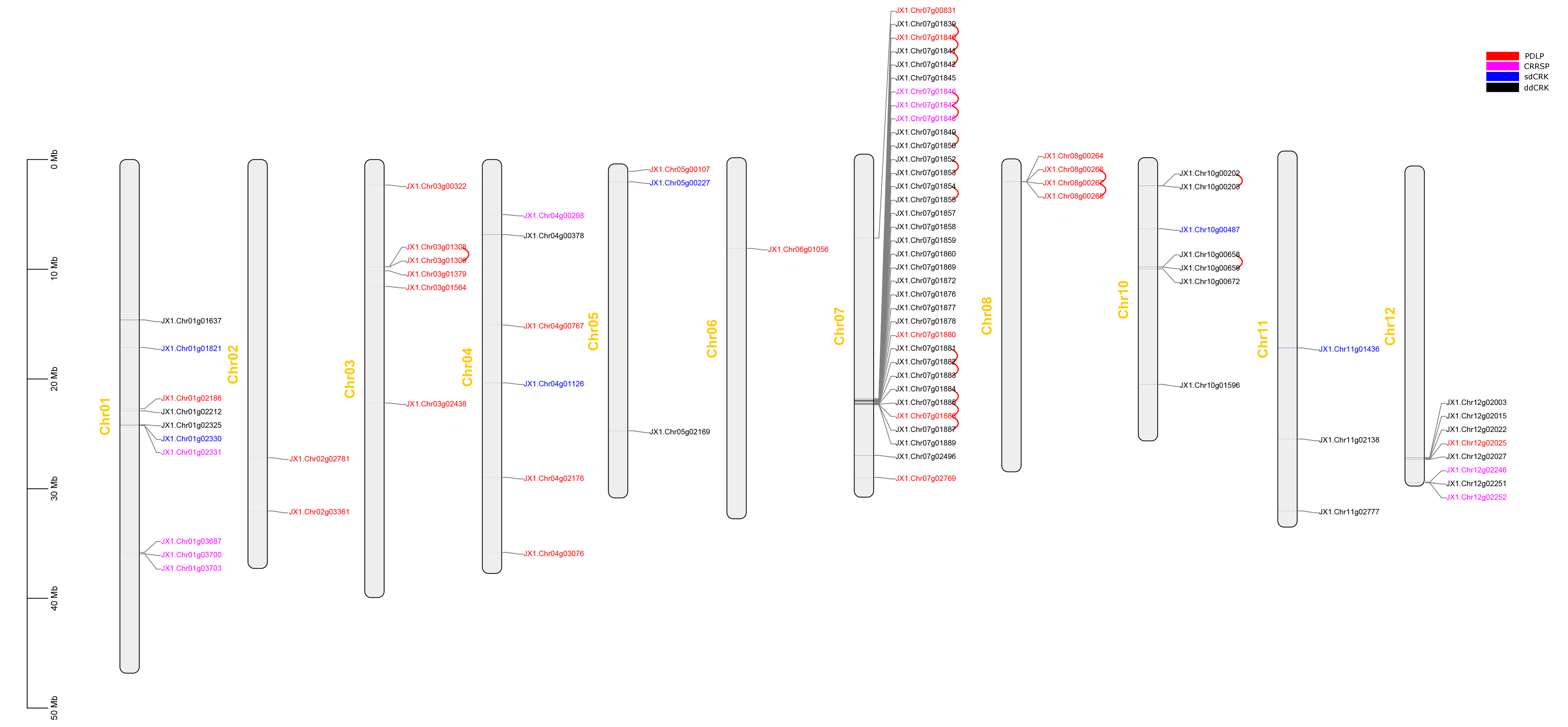
3.2. Chromosomal Localization of DUF26 Domain-Containing Genes in DXWR
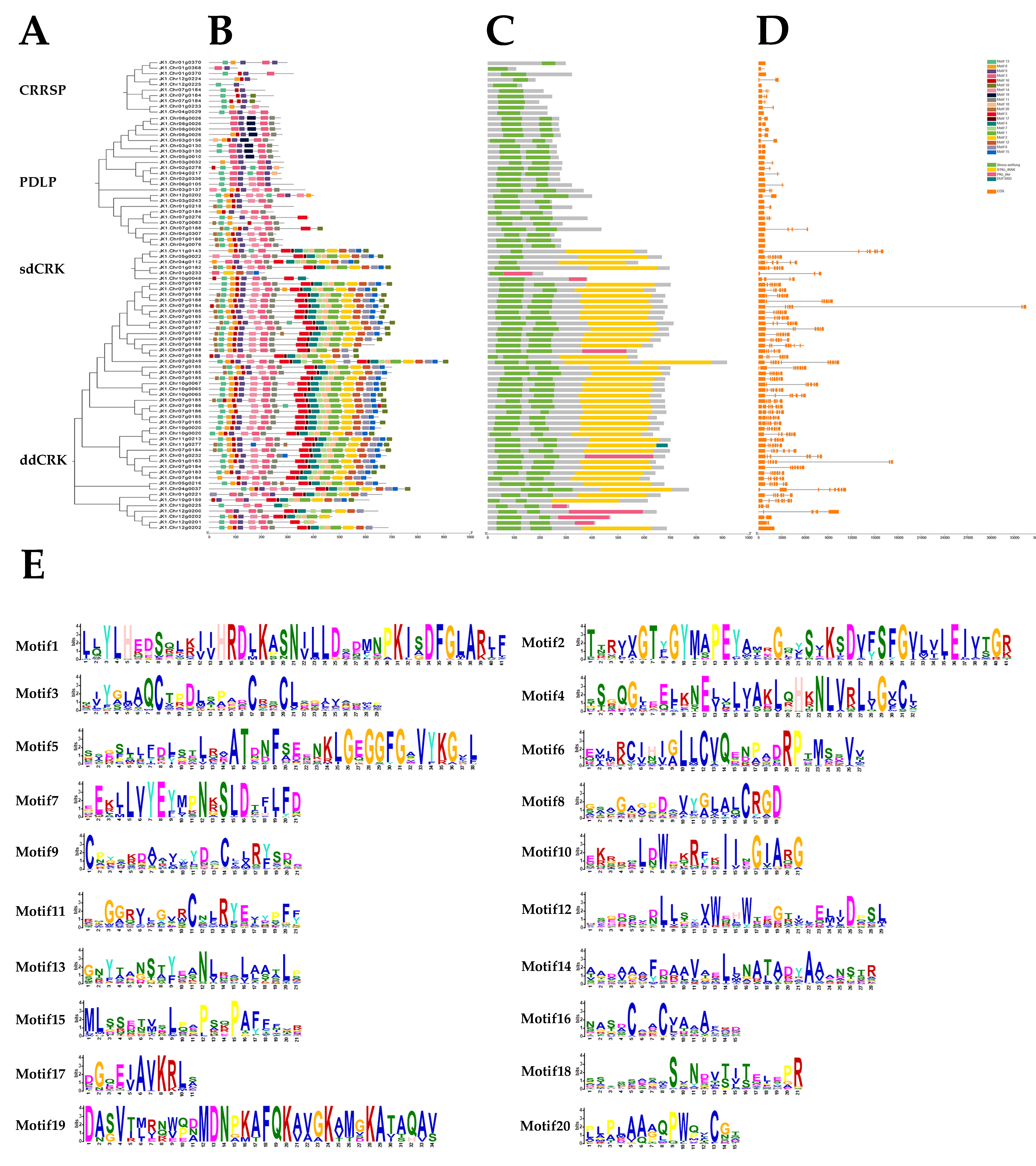
3.3. Motifs and Structural Composition of DUF26 Domain-Containing Genes in DXWR
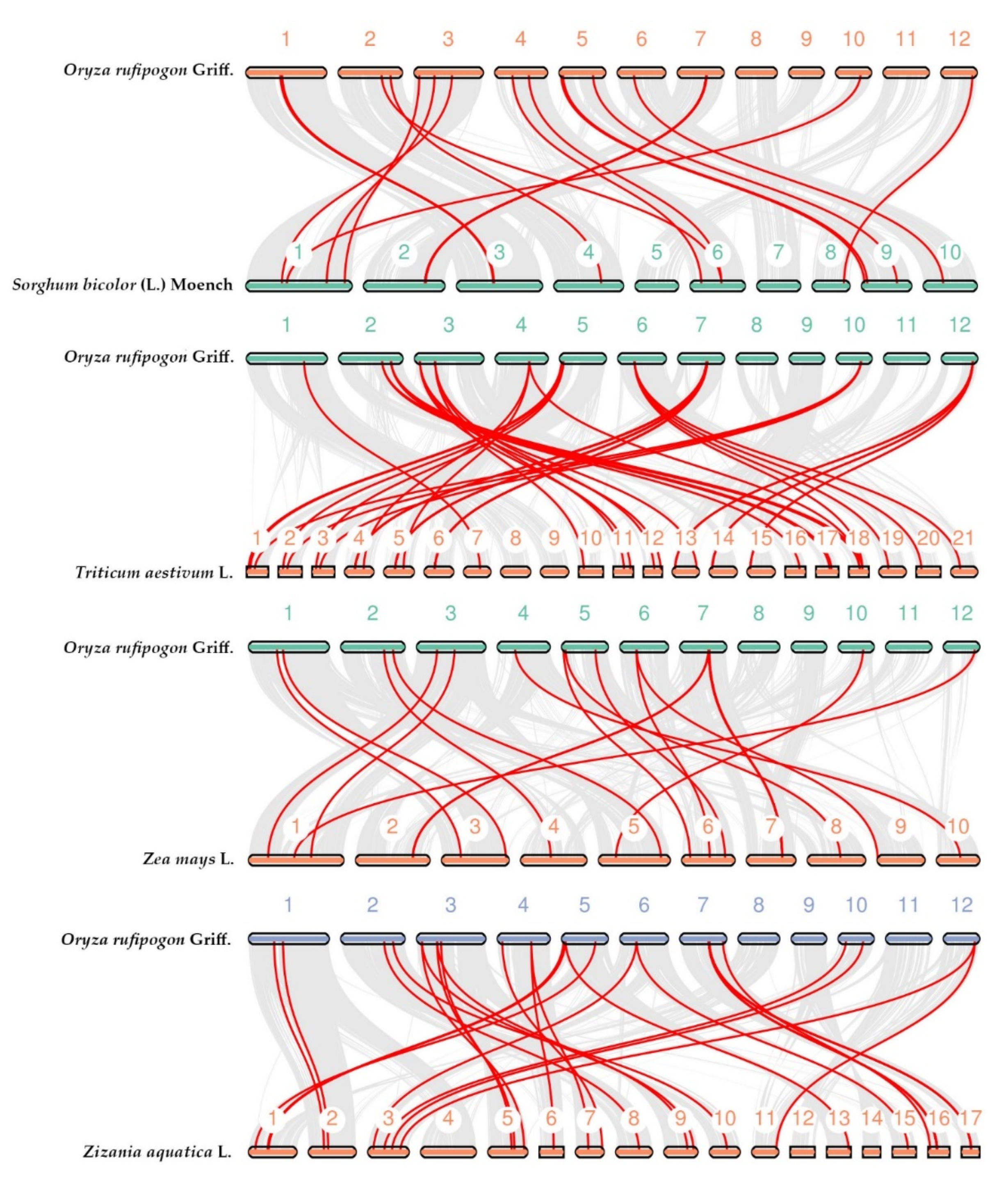
3.4. Collinearity Analysis of DUF26 Domain-Containing Genes in DXWR among Gramineous Species
3.5. Expression Analysis of DUF26 Domain-Containing Genes in DXWR under Submergence
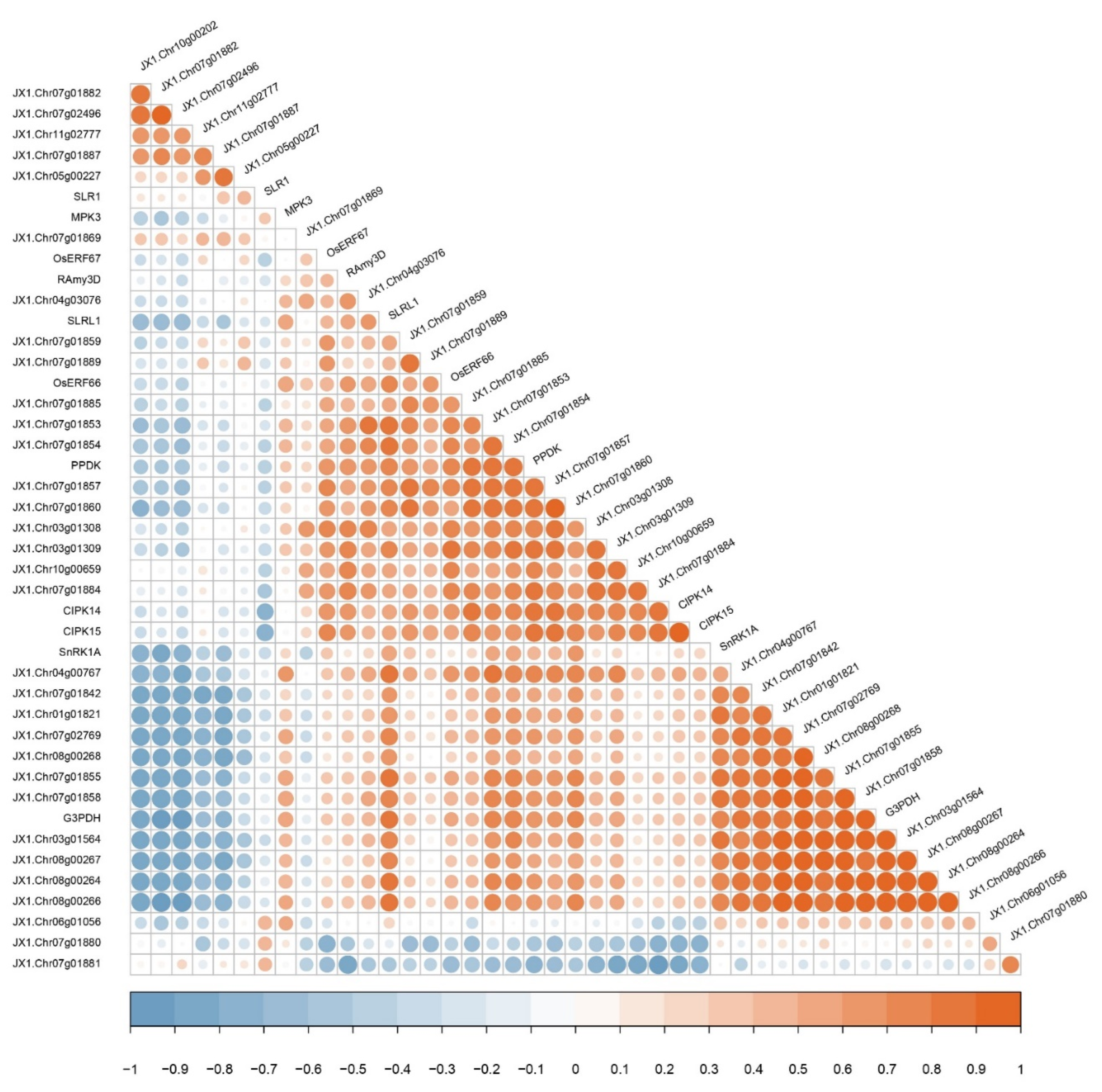
3.6. Correlation Analysis between DUF26 Domain-Containing Genes and Submergence Tolerant Response Genes in DXWR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nakamura, S.; Suzuki, T.; Kawamukai, M.; Nakagawa, T. Expression analysis of Arabidopsis thaliana small secreted protein genes. Biosci. Biotechnol. Biochem. 2012, 76, 436–446. [Google Scholar] [CrossRef] [Green Version]
- Agrawal, G.K.; Jwa, N.S.; Lebrun, M.H.; Job, D.; Rakwal, R. Plant secretome: Unlocking secrets of the secreted proteins. Proteomics 2010, 10, 799–827. [Google Scholar] [CrossRef]
- Tavormina, P.; De Coninck, B.; Nikonorova, N.; De Smet, I.; Cammue, B.P. The Plant Peptidome: An Expanding Repertoire of Structural Features and Biological Functions. Plant Cell 2015, 27, 2095–2118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miyakawa, T.; Miyazono, K.; Sawano, Y.; Hatano, K.; Tanokura, M. Crystal structure of ginkbilobin-2 with homology to the extracellular domain of plant cysteine-rich receptor-like kinases. Proteins 2009, 77, 247–251. [Google Scholar] [CrossRef] [PubMed]
- Miyakawa, T.; Hatano, K.; Miyauchi, Y.; Suwa, Y.; Sawano, Y.; Tanokura, M. A secreted protein with plant-specific cysteine-rich motif functions as a mannose-binding lectin that exhibits antifungal activity. Plant Physiol. 2014, 166, 766–778. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, L.S.; Wang, L.; Trippel, C.; Mendoza-Mendoza, A.; Ullmann, S.; Moretti, M.; Carsten, A.; Kahnt, J.; Reissmann, S.; Zechmann, B.; et al. The Ustilago maydis repetitive effector Rsp3 blocks the antifungal activity of mannose-binding maize proteins. Nat. Commun. 2018, 9, 1711. [Google Scholar] [CrossRef] [Green Version]
- Lee, D.S.; Kim, Y.C.; Kwon, S.J.; Ryu, C.M.; Park, O.K. The Arabidopsis Cysteine-Rich Receptor-Like Kinase CRK36 Regulates Immunity through Interaction with the Cytoplasmic Kinase BIK1. Front. Plant Sci. 2017, 8, 1856. [Google Scholar] [CrossRef] [Green Version]
- Yeh, Y.H.; Chang, Y.H.; Huang, P.Y.; Huang, J.B.; Zimmerli, L. Enhanced Arabidopsis pattern-triggered immunity by overexpression of cysteine-rich receptor-like kinases. Front. Plant Sci. 2015, 6, 322. [Google Scholar] [CrossRef] [Green Version]
- Wrzaczek, M.; Brosche, M.; Salojarvi, J.; Kangasjarvi, S.; Idanheimo, N.; Mersmann, S.; Robatzek, S.; Karpinski, S.; Karpinska, B.; Kangasjarvi, J. Transcriptional regulation of the CRK/DUF26 group of receptor-like protein kinases by ozone and plant hormones in Arabidopsis. BMC Plant Biol. 2010, 10, 95. [Google Scholar] [CrossRef] [Green Version]
- Idanheimo, N.; Gauthier, A.; Salojarvi, J.; Siligato, R.; Brosche, M.; Kollist, H.; Mahonen, A.P.; Kangasjarvi, J.; Wrzaczek, M. The Arabidopsis thaliana cysteine-rich receptor-like kinases CRK6 and CRK7 protect against apoplastic oxidative stress. Biochem. Biophys. Res. Commun. 2014, 445, 457–462. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Fan, B.; Du, L.; Chen, Z. Activation of hypersensitive cell death by pathogen-induced receptor-like protein kinases from Arabidopsis. Plant Mol. Biol. 2004, 56, 271–283. [Google Scholar] [CrossRef]
- Chen, K.; Du, L.; Chen, Z. Sensitization of defense responses and activation of programmed cell death by a pathogen-induced receptor-like protein kinase in Arabidopsis. Plant Mol. Biol. 2003, 53, 61–74. [Google Scholar] [CrossRef]
- Acharya, B.R.; Raina, S.; Maqbool, S.B.; Jagadeeswaran, G.; Mosher, S.L.; Appel, H.M.; Schultz, J.C.; Klessig, D.F.; Raina, R. Overexpression of CRK13, an Arabidopsis cysteine-rich receptor-like kinase, results in enhanced resistance to Pseudomonas syringae. Plant J. 2007, 50, 488–499. [Google Scholar] [CrossRef] [PubMed]
- Caillaud, M.C.; Wirthmueller, L.; Sklenar, J.; Findlay, K.; Piquerez, S.; Jones, A.; Robatzek, S.; Jones, J.; Faulkner, C.; Birch, P.J.P.P. The Plasmodesmal Protein PDLP1 Localises to Haustoria-Associated Membranes during Downy Mildew Infection and Regulates Callose Deposition. PLoS Pathog. 2014, 10, e1004496. [Google Scholar] [CrossRef]
- Lim, G.H.; Shine, M.B.; Delorenzo, L.; Yu, K.; Cui, W.; Navarre, D.; Hunt, A.; Lee, J.Y.; Kachroo, A.; Kachroo, P.J.C.H.; et al. Plasmodesmata Localizing Proteins Regulate Transport and Signaling during Systemic Acquired Immunity in Plants. Cell Host Microbe 2016, 19, 541–549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sasaki, T.; Burr, B. International Rice Genome Sequencing Project: The effort to completely sequence the rice genome. Curr. Opin. Plant Biol. 2000, 3, 138–141. [Google Scholar] [CrossRef]
- Bailey-Serres, J.; Lee, S.C.; Brinton, E. Waterproofing crops: Effective flooding survival strategies. Plant Physiol. 2012, 160, 1698–1709. [Google Scholar] [CrossRef] [Green Version]
- Ma, X.; Han, B.; Tang, J.; Zhang, J.; Cui, D.; Geng, L.; Zhou, H.; Li, M.; Han, L. Construction of chromosome segment substitution lines of Dongxiang common wild rice (Oryza rufipogon Griff.) in the background of the japonica rice cultivar Nipponbare (Oryza sativa L.). Plant Physiol. Biochem. 2019, 144, 274–282. [Google Scholar] [CrossRef] [PubMed]
- Potter, S.C.; Luciani, A.; Eddy, S.R.; Park, Y.; Lopez, R.; Finn, R.D. HMMER web server: 2018 update. Nucleic Acids Res. 2018, 46, W200–W204. [Google Scholar] [CrossRef] [Green Version]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Bryant, S.H. CD-Search: Protein domain annotations on the fly. Nucleic Acids Res. 2004, 32, W327–W331. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Khedkar, S.; Bork, P. SMART: Recent updates, new developments and status in 2020. Nucleic Acids Res. 2021, 49, D458–D460. [Google Scholar] [CrossRef] [PubMed]
- Artimo, P.; Jonnalagedda, M.; Arnold, K.; Baratin, D.; Csardi, G.; de Castro, E.; Duvaud, S.; Flegel, V.; Fortier, A.; Gasteiger, E.; et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012, 40, W597–W603. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [Green Version]
- Tang, H.; Bowers, J.E.; Wang, X.; Ming, R.; Alam, M.; Paterson, A.H. Synteny and collinearity in plant genomes. Science 2008, 320, 486–488. [Google Scholar] [CrossRef] [Green Version]
- Vaattovaara, A.; Brandt, B.; Rajaraman, S.; Safronov, O.; Veidenberg, A.; Luklova, M.; Kangasjarvi, J.; Loytynoja, A.; Hothorn, M.; Salojarvi, J.; et al. Mechanistic insights into the evolution of DUF26-containing proteins in land plants. Commun. Biol. 2019, 2, 56. [Google Scholar] [CrossRef]
- Rogers, R.L.; Shao, L.; Thornton, K.R. Tandem duplications lead to novel expression patterns through exon shuffling in Drosophila yakuba. PLoS Genet. 2017, 13, e1006795. [Google Scholar] [CrossRef] [Green Version]
- Wu, S.; Han, B.; Jiao, Y. Genetic Contribution of Paleopolyploidy to Adaptive Evolution in Angiosperms. Mol. Plant 2020, 13, 59–71. [Google Scholar] [CrossRef] [PubMed]
- Ren, R.; Wang, H.; Guo, C.; Zhang, N.; Zeng, L.; Chen, Y.; Ma, H.; Qi, J. Widespread Whole Genome Duplications Contribute to Genome Complexity and Species Diversity in Angiosperms. Mol. Plant 2018, 11, 414–428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, N.; Yang, T.; Yu, X.T.; Shang, L.G.; Guo, D.P.; Zhang, Y.; Meng, L.; Qi, Q.Q.; Li, Y.L.; Du, Y.M.; et al. Chromosome-level genome assembly of Zizania latifolia provides insights into its seed shattering and phytocassane biosynthesis. Commun. Biol. 2022, 5, 36. [Google Scholar] [CrossRef]
- Dwivedi, S.K.; Kumar, S.; Bhakta, N.; Singh, S.K.; Rao, K.K.; Mishra, J.S.; Singh, A.K. Improvement of submergence tolerance in rice through efficient application of potassium under submergence-prone rainfed ecology of Indo-Gangetic Plain. Funct. Plant Biol. 2017, 44, 907–916. [Google Scholar] [CrossRef]
- Kurusu, T.; Hamada, J.; Nokajima, H.; Kitagawa, Y.; Kiyoduka, M.; Takahashi, A.; Hanamata, S.; Ohno, R.; Hayashi, T.; Okada, K.; et al. Regulation of microbe-associated molecular pattern-induced hypersensitive cell death, phytoalexin production, and defense gene expression by calcineurin B-like protein-interacting protein kinases, OsCIPK14/15, in rice cultured cells. Plant Physiol. 2010, 153, 678–692. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, C.A.; Lin, C.C.; Lee, K.W.; Chen, J.L.; Huang, L.F.; Ho, S.L.; Liu, H.J.; Hsing, Y.I.; Yu, S.M. The SnRK1A protein kinase plays a key role in sugar signaling during germination and seedling growth of rice. Plant Cell 2007, 19, 2484–2499. [Google Scholar] [CrossRef] [Green Version]
- Kollmer, I.; Novak, O.; Strnad, M.; Schmulling, T.; Werner, T. Overexpression of the cytosolic cytokinin oxidase/dehydrogenase (CKX7) from Arabidopsis causes specific changes in root growth and xylem differentiation. Plant J. 2014, 78, 359–371. [Google Scholar] [CrossRef]
- Singh, P.; Sinha, A.K. A Positive Feedback Loop Governed by SUB1A1 Interaction with MITOGEN-ACTIVATED PROTEIN KINASE3 Imparts Submergence Tolerance in Rice. Plant Cell 2016, 28, 1127–1143. [Google Scholar] [CrossRef] [Green Version]
- Moons, A.; Valcke, R.; Van Montagu, M. Low-oxygen stress and water deficit induce cytosolic pyruvate orthophosphate dikinase (PPDK) expression in roots of rice, a C3 plant. Plant J. 1998, 15, 89–98. [Google Scholar] [CrossRef]
- Park, M.; Yim, H.K.; Park, H.G.; Lim, J.; Kim, S.H.; Hwang, Y.S. Interference with oxidative phosphorylation enhances anoxic expression of rice alpha-amylase genes through abolishing sugar regulation. J. Exp. Bot. 2010, 61, 3235–3244. [Google Scholar] [CrossRef] [Green Version]
- Xiong, M.; Yu, J.; Wang, J.; Gao, Q.; Huang, L.; Chen, C.; Zhang, C.; Fan, X.; Zhao, D.; Liu, Q.Q.; et al. Brassinosteroids regulate rice seed germination through the BZR1-RAmy3D transcriptional module. Plant Physiol. 2022, 189, 402–418. [Google Scholar] [CrossRef] [PubMed]
- Toyofuku, K.; Umemura, T.; Yamaguchi, J. Promoter elements required for sugar-repression of the RAmy3D gene for alpha-amylase in rice. FEBS Lett. 1998, 428, 275–280. [Google Scholar] [CrossRef] [Green Version]
- Lin, C.C.; Chao, Y.T.; Chen, W.C.; Ho, H.Y.; Chou, M.Y.; Li, Y.R.; Wu, Y.L.; Yang, H.A.; Hsieh, H.; Lin, C.S.; et al. Regulatory cascade involving transcriptional and N-end rule pathways in rice under submergence. Proc. Natl. Acad. Sci. USA 2019, 116, 3300–3309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ayano, M.; Kani, T.; Kojima, M.; Sakakibara, H.; Kitaoka, T.; Kuroha, T.; Angeles-Shim, R.B.; Kitano, H.; Nagai, K.; Ashikari, M. Gibberellin biosynthesis and signal transduction is essential for internode elongation in deepwater rice. Plant Cell Environ. 2014, 37, 2313–2324. [Google Scholar] [CrossRef] [PubMed]
- Fukao, T.; Bailey-Serres, J. Submergence tolerance conferred by Sub1A is mediated by SLR1 and SLRL1 restriction of gibberellin responses in rice. Proc. Natl. Acad. Sci. USA 2008, 105, 16814–16819. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Itoh, H.; Shimada, A.; Ueguchi-Tanaka, M.; Kamiya, N.; Hasegawa, Y.; Ashikari, M.; Matsuoka, M. Overexpression of a GRAS protein lacking the DELLA domain confers altered gibberellin responses in rice. Plant J. 2005, 44, 669–679. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, C.; Wang, D.; Chen, H.; Deng, W.; Chen, D.; Chen, P.; Wang, J. Genome-Wide Identification of DUF26 Domain-Containing Genes in Dongxiang Wild Rice and Analysis of Their Expression Responses under Submergence. Curr. Issues Mol. Biol. 2022, 44, 3351-3363. https://doi.org/10.3390/cimb44080231
Huang C, Wang D, Chen H, Deng W, Chen D, Chen P, Wang J. Genome-Wide Identification of DUF26 Domain-Containing Genes in Dongxiang Wild Rice and Analysis of Their Expression Responses under Submergence. Current Issues in Molecular Biology. 2022; 44(8):3351-3363. https://doi.org/10.3390/cimb44080231
Chicago/Turabian StyleHuang, Cheng, Dianwen Wang, Hongping Chen, Wei Deng, Dazhou Chen, Ping Chen, and Jilin Wang. 2022. "Genome-Wide Identification of DUF26 Domain-Containing Genes in Dongxiang Wild Rice and Analysis of Their Expression Responses under Submergence" Current Issues in Molecular Biology 44, no. 8: 3351-3363. https://doi.org/10.3390/cimb44080231
APA StyleHuang, C., Wang, D., Chen, H., Deng, W., Chen, D., Chen, P., & Wang, J. (2022). Genome-Wide Identification of DUF26 Domain-Containing Genes in Dongxiang Wild Rice and Analysis of Their Expression Responses under Submergence. Current Issues in Molecular Biology, 44(8), 3351-3363. https://doi.org/10.3390/cimb44080231





