The Search for Cancer Biomarkers: Assessing the Distribution of INDEL Markers in Different Genetic Ancestries
Abstract
1. Introduction
2. Materials and Methods
2.1. DNA Extraction and Quantification
2.2. Genotyping of Investigated Polymorphisms
2.3. Data Analyses
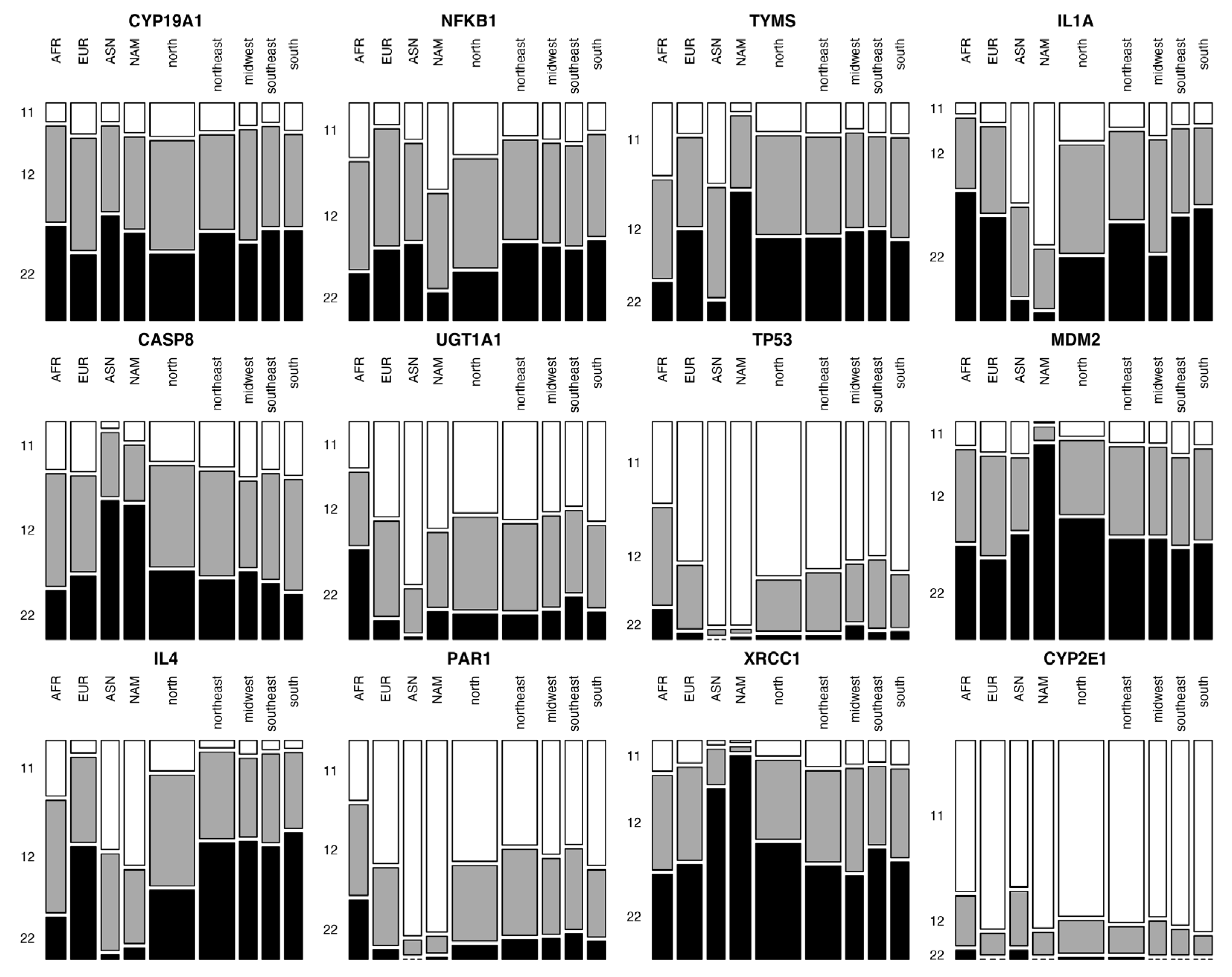
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Globocan Cancer Today. Available online: http://gco.iarc.fr/today/home (accessed on 6 March 2019).
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Giampazolias, E.; Tait, S.W.G. Mitochondria and the Hallmarks of Cancer. FEBS J. 2016, 283, 803–814. [Google Scholar] [CrossRef] [PubMed]
- INCA. Estimativa 2020: Incidência de Câncer No Brasil|INCA—Instituto Nacional de Câncer. Available online: https://www.inca.gov.br/publicacoes/livros/estimativa-2020-incidencia-de-cancer-no-brasil (accessed on 17 October 2020).
- Amador, M.A.T.; Cavalcante, G.C.; Santos, N.P.C.; Gusmão, L.; Guerreiro, J.F.; Ribeiro-dos-Santos, Â.; Santos, S. Distribution of Allelic and Genotypic Frequencies of IL1A, IL4, NFKB1 and PAR1 Variants in Native American, African, European and Brazilian Populations. BMC Res. Notes 2016, 9, 101. [Google Scholar] [CrossRef] [PubMed]
- Suarez-Kurtz, G. Pharmacogenetics in the Brazilian Population. Front. Pharm. 2010, 1, 118. [Google Scholar] [CrossRef]
- Yuan, F.; Sun, R.; Li, L.; Jin, B.; Wang, Y.; Liang, Y.; Che, G.; Gao, L.; Zhang, L. A Functional Variant Rs353292 in the Flanking Region of MiR-143/145 Contributes to the Risk of Colorectal Cancer. Sci. Rep. 2016, 6, 30195. [Google Scholar] [CrossRef]
- Wang, C.; Zhao, H.; Zhao, X.; Wan, J.; Wang, D.; Bi, W.; Jiang, X.; Gao, Y. Association between an Insertion/Deletion Polymorphism within 3′UTR of SGSM3 and Risk of Hepatocellular Carcinoma. Tumour Biol. 2014, 35, 295–301. [Google Scholar] [CrossRef]
- 1000 Genomes Project Consortium; Auton, A.; Brooks, L.D.; Durbin, R.M.; Garrison, E.P.; Kang, H.M.; Korbel, J.O.; Marchini, J.L.; McCarthy, S.; McVean, G.A.; et al. A Global Reference for Human Genetic Variation. Nature 2015, 526, 68–74. [Google Scholar] [CrossRef]
- Ahirwar, D.; Kesarwani, P.; Manchanda, P.K.; Mandhani, A.; Mittal, R.D. Anti- and Proinflammatory Cytokine Gene Polymorphism and Genetic Predisposition: Association with Smoking, Tumor Stage and Grade, and Bacillus Calmette-Guérin Immunotherapy in Bladder Cancer. Cancer Genet. Cytogenet. 2008, 184, 1–8. [Google Scholar] [CrossRef]
- Yang, C.-M.; Chen, H.-C.; Hou, Y.-Y.; Lee, M.-C.; Liou, H.-H.; Huang, S.-J.; Yen, L.-M.; Eng, D.-M.; Hsieh, Y.-D.; Ger, L.-P. A High IL-4 Production Diplotype Is Associated with an Increased Risk but Better Prognosis of Oral and Pharyngeal Carcinomas. Arch. Oral Biol. 2014, 59, 35–46. [Google Scholar] [CrossRef]
- Dong, D.; Gao, X.; Zhu, Z.; Yu, Q.; Bian, S.; Gao, Y. A 40-Bp Insertion/Deletion Polymorphism in the Constitutive Promoter of MDM2 Confers Risk for Hepatocellular Carcinoma in a Chinese Population. Gene 2012, 497, 66–70. [Google Scholar] [CrossRef]
- Zhang, Y.J.; Zhong, X.P.; Chen, Y.; Liu, S.R.; Wu, G.; Liu, Y.F. Association between CASP-8 Gene Polymorphisms and Cancer Risk in Some Asian Population Based on a HuGE Review and Meta-Analysis. Genet. Mol. Res. 2013, 12, 6466–6476. [Google Scholar] [CrossRef] [PubMed]
- Ramalhinho, A.C.M.; Fonseca-Moutinho, J.A.; Breitenfeld Granadeiro, L.A.T.G. Positive Association of Polymorphisms in Estrogen Biosynthesis Gene, CYP19A1, and Metabolism, GST, in Breast Cancer Susceptibility. DNA Cell Biol. 2012, 31, 1100–1106. [Google Scholar] [CrossRef] [PubMed]
- Kuhlmann, J.D.; Bankfalvi, A.; Schmid, K.W.; Callies, R.; Kimmig, R.; Wimberger, P.; Siffert, W.; Bachmann, H.S. Prognostic Relevance of Caspase 8 -652 6N InsDel and Asp302His Polymorphisms for Breast Cancer. BMC Cancer 2016, 16, 618. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Karakosta, M.; Kalotychou, V.; Kostakis, A.; Pantelias, G.; Rombos, I.; Kouraklis, G.; Manola, K.N. UGT1A1*28 Polymorphism in Chronic Lymphocytic Leukemia: The First Investigation of the Polymorphism in Disease Susceptibility and Its Specific Cytogenetic Abnormalities. Acta Haematol. 2014, 132, 59–67. [Google Scholar] [CrossRef]
- Bajro, M.H.; Josifovski, T.; Panovski, M.; Jankulovski, N.; Nestorovska, A.K.; Matevska, N.; Petrusevska, N.; Dimovski, A.J. Promoter Length Polymorphism in UGT1A1 and the Risk of Sporadic Colorectal Cancer. Cancer Genet. 2012, 205, 163–167. [Google Scholar] [CrossRef]
- Jiang, O.; Zhou, R.; Wu, D.; Liu, Y.; Wu, W.; Cheng, N. CYP2E1 Polymorphisms and Colorectal Cancer Risk: A HuGE Systematic Review and Meta-Analysis. Tumor Biol. 2013, 34, 1215–1224. [Google Scholar] [CrossRef]
- Jirásková, A.; Novotný, J.; Novotný, L.; Vodicka, P.; Pardini, B.; Naccarati, A.; Schwertner, H.A.; Hubácek, J.A.; Puncochárová, L.; Šmerhovský, Z.; et al. Association of Serum Bilirubin and Promoter Variations in HMOX1 and UGT1A1 Genes with Sporadic Colorectal Cancer. Int. J. Cancer 2012, 131, 1549–1555. [Google Scholar] [CrossRef]
- Morita, M.; Le Marchand, L.; Kono, S.; Yin, G.; Toyomura, K.; Nagano, J.; Mizoue, T.; Mibu, R.; Tanaka, M.; Kakeji, Y.; et al. Genetic Polymorphisms of CYP2E1 and Risk of Colorectal Cancer: The Fukuoka Colorectal Cancer Study. Cancer Epidemiol. Biomark. Prev. 2009, 18, 235–241. [Google Scholar] [CrossRef]
- Silva, T.D.; Felipe, A.V.; Pimenta, C.A.; Barão, K.; Forones, N.M. CYP2E1 RsaI and 96-Bp Insertion Genetic Polymorphisms Associated with Risk for Colorectal Cancer. Genet. Mol. Res. 2012, 11, 3138–3145. [Google Scholar] [CrossRef]
- Santoro, A.B.; Vargens, D.D.; Barros Filho Mde, C.; Bulzico, D.A.; Kowalski, L.P.; Meirelles, R.M.R.; Paula, D.P.; Neves, R.R.S.; Pessoa, C.N.; Struchine, C.J.; et al. Effect of UGT1A1, UGT1A3, DIO1 and DIO2 Polymorphisms on L-Thyroxine Doses Required for TSH Suppression in Patients with Differentiated Thyroid Cancer. Br. J. Clin. Pharm. 2014, 78, 1067–1075. [Google Scholar] [CrossRef]
- Fujimoto, D.; Hirono, Y.; Goi, T.; Katayama, K.; Matsukawa, S.; Yamaguchi, A. The Activation of Proteinase-Activated Receptor-1 (PAR1) Promotes Gastric Cancer Cell Alteration of Cellular Morphology Related to Cell Motility and Invasion. Int. J. Oncol. 2013, 42, 565–573. [Google Scholar] [CrossRef] [PubMed]
- Qiao, W.; Wang, T.; Zhang, L.; Tang, Q.; Wang, D.; Sun, H. Association Study of Single Nucleotide Polymorphisms in XRCC1 Gene with the Risk of Gastric Cancer in Chinese Population. Int. J. Biol. Sci. 2013, 9, 753–758. [Google Scholar] [CrossRef] [PubMed]
- Santos, N.P.C.; Ribeiro-Rodrigues, E.M.; Ribeiro-dos-Santos, Â.K.C.; Pereira, R.; Gusmão, L.; Amorim, A.; Guerreiro, J.F.; Zago, M.A.; Matte, C.; Hutz, M.H.; et al. Assessing Individual Interethnic Admixture and Population Substructure Using a 48-Insertion-Deletion (INSEL) Ancestry-Informative Marker (AIM) Panel. Hum. Mutat. 2010, 31, 184–190. [Google Scholar] [CrossRef] [PubMed]
- Ramos, B.R.D.A.; Mendes, N.D.; Tanikawa, A.A.; Amador, M.A.T.; dos Santos, N.P.C.; dos Santos, S.E.B.; Castelli, E.C.; Witkin, S.S.; da Silva, M.G. Ancestry Informative Markers and Selected Single Nucleotide Polymorphisms in Immunoregulatory Genes on Preterm Labor and Preterm Premature Rupture of Membranes: A Case Control Study. BMC Pregnancy Childbirth 2016, 16, 30. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro-Rodrigues, E.M.; Palha, T.D.J.B.F.; Bittencourt, E.A.; Ribeiro-Dos-Santos, A.; Santos, S. Extensive Survey of 12 X-STRs Reveals Genetic Heterogeneity among Brazilian Populations. Int. J. Leg. Med 2011, 125, 445–452. [Google Scholar] [CrossRef] [PubMed]
- Silva, W.A.; Bortolini, M.C.; Schneider, M.P.C.; Marrero, A.; Elion, J.; Krishnamoorthy, R.; Zago, M.A. MtDNA Haplogroup Analysis of Black Brazilian and Sub-Saharan Populations: Implications for the Atlantic Slave Trade. Hum. Biol. 2006, 78, 29–41. [Google Scholar] [CrossRef] [PubMed]
- Palha, T.; Gusmão, L.; Ribeiro-Rodrigues, E.; Guerreiro, J.F.; Ribeiro-dos-Santos, Â.; Santos, S. Disclosing the Genetic Structure of Brazil through Analysis of Male Lineages with Highly Discriminating Haplotypes. PLoS ONE 2012, 7, e40007. [Google Scholar] [CrossRef]
- Andrade, R.B.; Amador, M.A.T.; Cavalcante, G.C.; Leitão, L.P.C.; Fernandes, M.R.; Modesto, A.A.C.; Moreira, F.C.; Khayat, A.S.; Assumpção, P.P.; Ribeiro-dos-Santos, Â.; et al. Estimating Asian Contribution to the Brazilian Population: A New Application of a Validated Set of 61 Ancestry Informative Markers. G3 Genes Genomes Genet. 2018, 8, 3577–3582. [Google Scholar] [CrossRef]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: Long Island, NY, USA, 1989. [Google Scholar]
- Cavalcante, G.C.; Amador, M.A.; Ribeiro Dos Santos, A.M.; Carvalho, D.C.; Andrade, R.B.; Pereira, E.E.; Fernandes, M.R.; Costa, D.F.; Santos, N.P.; Assumpção, P.P.; et al. Analysis of 12 Variants in the Development of Gastric and Colorectal Cancers. World J. Gastroenterol. 2017, 23, 8533–8543. [Google Scholar] [CrossRef]
- Excoffier, L.; Lischer, H.E.L. Arlequin Suite Ver 3.5: A New Series of Programs to Perform Population Genetics Analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Core Team: Vienna, Austria, 2014. [Google Scholar]
- GTEx Consortium. The Genotype-Tissue Expression (GTEx) Project. Nat. Genet. 2013, 45, 580–585. [Google Scholar] [CrossRef] [PubMed]
- Fritsche, E.; Pittman, G.S.; Bell, D.A. Localization, Sequence Analysis, and Ethnic Distribution of a 96-Bp Insertion in the Promoter of the Human CYP2E1 Gene. Mutat. Res. 2000, 432, 1–5. [Google Scholar] [CrossRef]
- Shin, H.J.; Kim, J.Y.; Cheong, H.S.; Na, H.S.; Shin, H.D.; Chung, M.W. Functional Study of Haplotypes in UGT1A1 Promoter to Find a Novel Genetic Variant Leading to Reduced Gene Expression. Drug Monit. 2015, 37, 369–374. [Google Scholar] [CrossRef] [PubMed]
- Cavalcante, G.C.; Ribeiro-Dos-Santos, A.M.; Carvalho, D.C.D.; Silva, E.M.D.; Assumpção, P.P.D.; Ribeiro-Dos-Santos, Â.; Santos, S. Investigation of Potentially Deleterious Alleles for Response to Cancer Treatment with 5-Fluorouracil. Anticancer Res. 2015, 7, 6971–6977. [Google Scholar]
- Gansmo, L.B.; Vatten, L.; Romundstad, P.; Hveem, K.; Ryan, B.M.; Harris, C.C.; Knappskog, S.; Lønning, P.E. Associations between the MDM2 Promoter P1 Polymorphism Del1518 (Rs3730485) and Incidence of Cancer of the Breast, Lung, Colon and Prostate. Oncotarget 2016, 7, 28637–28646. [Google Scholar] [CrossRef] [PubMed]
- Castro-Rojas, C.A.; Esparza-Mota, A.R.; Hernandez-Cabrera, F.; Romero-Diaz, V.J.; Gonzalez-Guerrero, J.F.; Maldonado-Garza, H.; Garcia-Gonzalez, I.S.; Buenaventura-Cisneros, S.; Sanchez-Lopez, J.Y.; Ortiz-Lopez, R.; et al. Thymidylate Synthase Gene Variants as Predictors of Clinical Response and Toxicity to Fluoropyrimidine-Based Chemotherapy for Colorectal Cancer. Drug Metab. Pers. 2017, 32, 209–218. [Google Scholar] [CrossRef]
- Pastorakova, A.; Chandogova, D.; Chandoga, J.; Luha, J.; Bohmer, D.; Malova, J.; Braxatorisova, T.; Juhosova, M.; Reznakova, S.; Petrovic, R. Distribution of the Most Common Polymorphisms in TYMS Gene in Slavic Population of Central Europe. Neoplasma 2017, 64, 962–970. [Google Scholar] [CrossRef]
- Summers, C.M.; Cucchiara, A.J.; Nackos, E.; Hammons, A.L.; Mohr, E.; Whitehead, A.S.; Von Feldt, J.M. Functional Polymorphisms of Folate-Metabolizing Enzymes in Relation to Homocysteine Concentrations in Systemic Lupus Erythematosus. J. Rheumatol. 2008, 35, 2179–2186. [Google Scholar] [CrossRef]
- Pardini, B.; Verderio, P.; Pizzamiglio, S.; Nici, C.; Maiorana, M.V.; Naccarati, A.; Vodickova, L.; Vymetalkova, V.; Veneroni, S.; Daidone, M.G.; et al. Association between CASP8 –652 6N Del Polymorphism (Rs3834129) and Colorectal Cancer Risk: Results from a Multi-Centric Study. PLoS ONE 2014, 9, e85538. [Google Scholar] [CrossRef]
- Pittman, A.M.; Broderick, P.; Sullivan, K.; Fielding, S.; Webb, E.; Penegar, S.; Tomlinson, I.; Houlston, R.S. CASP8 Variants D302H and −652 6N Ins/Del Do Not Influence the Risk of Colorectal Cancer in the United Kingdom Population. Br. J. Cancer 2008, 98, 1434–1436. [Google Scholar] [CrossRef][Green Version]
- Brown, K.L.; Seale, K.B.; El Khoury, L.Y.; Posthumus, M.; Ribbans, W.J.; Raleigh, S.M.; Collins, M.; September, A.V. Polymorphisms within the COL5A1 Gene and Regulators of the Extracellular Matrix Modify the Risk of Achilles Tendon Pathology in a British Case-Control Study. J. Sports Sci. 2017, 35, 1475–1483. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, K.; Williamson, A.-L.; Hoffman, M.; Dandara, C. CASP8 Promoter Polymorphism Is Associated with High-Risk HPV Types and Abnormal Cytology but Not with Cervical Cancer. J. Med. Virol. 2011, 83, 630–636. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, D.C.; Wanderley, A.V.; Amador, M.A.T.; Fernandes, M.R.; Cavalcante, G.C.; Pantoja, K.B.C.C.; Mello, F.A.R.; de Assumpção, P.P.; Khayat, A.S.; Ribeiro-dos-Santos, Â.; et al. Amerindian Genetic Ancestry and INDEL Polymorphisms Associated with Susceptibility of Childhood B-Cell Leukemia in an Admixed Population from the Brazilian Amazon. Leuk. Res. 2015, 39, 1239–1245. [Google Scholar] [CrossRef]
- Sanjari Moghaddam, A.; Nazarzadeh, M.; Sanjari Moghaddam, H.; Bidel, Z.; Keramatinia, A.; Darvish, H.; Mosavi-Jarrahi, A. XRCC1 Gene Polymorphisms and Breast Cancer Risk: A Systematic Review and Meta- Analysis Study. Asian Pac. J. Cancer Prev. 2016, 17, 323–330. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fehringer, G.; Kraft, P.; Pharoah, P.D.; Eeles, R.A.; Chatterjee, N.; Schumacher, F.R.; Schildkraut, J.M.; Lindström, S.; Brennan, P.; Bickeböller, H.; et al. Cross-Cancer Genome-Wide Analysis of Lung, Ovary, Breast, Prostate, and Colorectal Cancer Reveals Novel Pleiotropic Associations. Cancer Res. 2016, 76, 5103–5114. [Google Scholar] [CrossRef] [PubMed]
- Hashemi, M.; Aftabi, S.; Moazeni-Roodi, A.; Sarani, H.; Wiechec, E.; Ghavami, S. Association of CASP8 Polymorphisms and Cancer Susceptibility: A Meta-Analysis. Eur. J. Pharm. 2020, 881, 173201. [Google Scholar] [CrossRef]
- Marques, D.; Ferreira-Costa, L.R.; Ferreira-Costa, L.L.; da Silva Correa, R.; Borges, A.M.P.; Ito, F.R.; de Oliveira Ramos, C.C.; Bortolin, R.H.; Luchessi, A.D.; Ribeiro-Dos-Santos, Â.; et al. Association of Insertion-Deletions Polymorphisms with Colorectal Cancer Risk and Clinical Features. World J. Gastroenterol. 2017, 23, 6854–6867. [Google Scholar] [CrossRef]
- Cătană, A.; Pop, M.; Hincu, B.D.; Pop, I.V.; Petrişor, F.M.; Porojan, M.D.; Popp, R.A. The XRCC1 Arg194Trp Polymorphism Is Significantly Associated with Lung Adenocarcinoma: A Case-Control Study in an Eastern European Caucasian Group. Onco Targets 2015, 8, 3533–3538. [Google Scholar] [CrossRef]
- Bu, H.; Rosdahl, I.; Sun, X.-F.; Zhang, H. Importance of Polymorphisms in NF-KappaB1 and NF-KappaBIalpha Genes for Melanoma Risk, Clinicopathological Features and Tumor Progression in Swedish Melanoma Patients. J. Cancer Res. Clin. Oncol. 2007, 133, 859–866. [Google Scholar] [CrossRef]
- Escobar, G.F.; Arraes, J.A.A.; Bakos, L.; Ashton-Prolla, P.; Giugliani, R.; Callegari-Jacques, S.M.; Santos, S.; Bakos, R.M. Polymorphisms in CYP19A1 and NFKB1 Genes Are Associated with Cutaneous Melanoma Risk in Southern Brazilian Patients. Melanoma Res. 2016, 26, 348–353. [Google Scholar] [CrossRef]
- Chojnacka, K.; Bilinska, B.; Mruk, D.D. Interleukin 1alpha-Induced Disruption of the Sertoli Cell Cytoskeleton Affects Gap Junctional Communication. Cell. Signal. 2016, 28, 469–480. [Google Scholar] [CrossRef] [PubMed]
- Griswold, M.D. The Central Role of Sertoli Cells in Spermatogenesis. Semin. Cell Dev. Biol. 1998, 9, 411–416. [Google Scholar] [CrossRef] [PubMed]
- Gutti, R.K.; Tsai-Morris, C.-H.; Dufau, M.L. Gonadotropin-Regulated Testicular Helicase (DDX25), an Essential Regulator of Spermatogenesis, Prevents Testicular Germ Cell Apoptosis. J. Biol. Chem. 2008, 283, 17055–17064. [Google Scholar] [CrossRef]
- O’Bryan, M.K.; Hedger, M.P. Inflammatory Networks in the Control of Spermatogenesis: Chronic Inflammation in an Immunologically Privileged Tissue? Adv. Exp. Med. Biol. 2008, 636, 92–114. [Google Scholar] [CrossRef] [PubMed]
- Singh, V.; Kumar Mohanty, S.; Verma, P.; Chakraborty, A.; Trivedi, S.; Rajender, S.; Singh, K. XRCC1 Deficiency Correlates with Increased DNA Damage and Male Infertility. Mutat. Res. Toxicol. Environ. Mutagen. 2019, 839, 1–8. [Google Scholar] [CrossRef]
- Walter, C.A.; Trolian, D.A.; McFarland, M.B.; Street, K.A.; Gurram, G.R.; McCarrey, J.R. Xrcc-1 Expression during Male Meiosis in the Mouse. Biol. Reprod. 1996, 55, 630–635. [Google Scholar] [CrossRef]
| Gene | ID | Size (bp) | Primers | Amplicon (bp) |
|---|---|---|---|---|
| CASP8 | rs3834129 | 6 | F-5′CTCTTCAATGCTTCCTTGAGGT3′ R-5′CTGCATGCCAGGAGCTAAGTAT3′ | 249–255 |
| CYP2E1 | - | 96 | F-5′TGTCCCAATACAGTCACCTCTTT3′ R-5′GGCTTTTATTTGTTTTGCATCTG3′ | 397–493 |
| CYP19A1 | rs28892005 | 3 | F-5′TGCATGAGAAAGGCATCATATT3′ R-5′AAAAGGCACATTCATAGACAAAAA3′ | 122–125 |
| IL1A | rs3783553 | 4 | F-5′TGGTCCAAGTTGTGCTTATCC3′ R-5′ACAGTGGTCTCATGGTTGTCA3′ | 230–234 |
| IL4 | rs79071878 | 70 (1–3 repeats) | F-5′AGGGTCAGTCTGGCTACTGTGT3′ R-5′CAAATCTGTTCACCTCAACTGC3′ | 147/217/287 |
| MDM2 | rs3730485 | 40 | F-5′GGAAGTTTCCTTTCTGGTAGGC3′ R-5′TTTGATGCGGTCTCATAAATTG3′ | 192–232 |
| NFKB1 | rs28362491 | 4 | F-5′TATGGACCGCATGACTCTATCA3′ R-5′GGCTCTGGCATCCTAGCAG3′ | 366–370 |
| PAR1 | rs11267092 | 13 | F-5′AAAACTGAACTTTGCCGGTGT3′ R-5′GGGCCTAGAAGTCCAAATGAG | 265–277 |
| TP53 | rs17878362 | 16 | F-5′GGGACTGACTTTCTGCTCTTGT3′ R-5′GGGACTGTAGATGGGTGAAAAG3′ | 135–141 |
| TYMS | rs151264360 | 6 | F-5′ATCCAAACCAGAATACAGCACA3′ R-5′CTCAAATCTGAGGGAGCTGAGT3′ | 148–164 |
| UGT1A1 | rs8175347 | 2 (5–8 repeats) | F-5′CTCTGAAAGTGAACTCCCTGCT3′ R-5′AGAGGTTCGCCCTCTCCTAT3′ | 133/135/137/139 |
| XRCC1 | rs3213239 | 4 | F-5′GAACCAGAATCCAAAAGTGACC3′ R-5′AGGGGAAGAGAGAGAAGGAGAG3′ | 243–247 |
| Frequencies | δ | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Markers | EUR | ASN | AFR | NAM | EUR/ASN | AFR/ASN | NAM/ASN | EUR/AFR | EUR/NAM | AFR/NAM |
| IL1A | 0.3 | 0.69 | 0.22 | 0.82 | 0.39 | 0.47 | 0.13 | 0.08 | 0.52 | 0.6 |
| IL4 | 0.26 | 0.69 | 0.58 | 0.77 | 0.43 | 0.11 | 0.08 | 0.32 | 0.51 | 0.19 |
| NFKB1 | 0.38 | 0.40 | 0.52 | 0.64 | 0.02 | 0.12 | 0.24 | 0.14 | 0.26 | 0.12 |
| PAR1 | 0.77 | 0.96 | 0.5 | 0.95 | 0.19 | 0.46 | 0.01 | 0.27 | 0.18 | 0.45 |
| UGT1A1 | 0.68 | 0.89 | 0.45 | 0.70 | 0.21 | 0.44 | 0.19 | 0.23 | 0.02 | 0.25 |
| CYP19A1 | 0.42 | 0.3 | 0.32 | 0.37 | 0.12 | 0.02 | 0.07 | 0.1 | 0.06 | 0.05 |
| CYP2E1 | 0.94 | 0.8 | 0.83 | 0.95 | 0.14 | 0.03 | 0.15 | 0.11 | 0.01 | 0.12 |
| CASP8 | 0.47 | 0.18 | 0.5 | 0.23 | 0.29 | 0.32 | 0.05 | 0.03 | 0.24 | 0.27 |
| TYMS | 0.36 | 0.65 | 0.58 | 0.21 | 0.29 | 0.07 | 0.44 | 0.22 | 0.15 | 0.37 |
| XRCC1 | 0.33 | 0.1 | 0.83 | 0.02 | 0.23 | 0.73 | 0.08 | 0.5 | 0.31 | 0.81 |
| MDM2 | 0.38 | 0.33 | 0.33 | 0.04 | 0.05 | 0.00 | 0.29 | 0.05 | 0.34 | 0.29 |
| TP53 | 0.82 | 0.99 | 0.62 | 0.98 | 0.17 | 0.37 | 0.01 | 0.2 | 0.16 | 0.36 |
| Average | 0.21 | 0.26 | 0.14 | 0.19 | 0.23 | 0.32 | ||||
| Gene | Variant ID | p-Value | NES | Tissue |
|---|---|---|---|---|
| CASP8 | rs3834129 | 4.9 × 10−13 | 0.28 | Cells-Cultured fibroblasts |
| 1.1 × 10−9 | 0.18 | Esophagus-Mucosa | ||
| 1.5 × 10−7 | −0.27 | Pituitary gland | ||
| 2.6 × 10−7 | −0.50 | Brain-Cerebellum | ||
| 6.9 × 10−7 | 0.16 | Adipose-Visceral (Omentum) | ||
| 7.3 × 10−7 | −0.48 | Brain-Frontal Cortex (BA9) | ||
| 0.0000019 | −0.16 | Thyroid | ||
| 0.0000030 | 0.18 | Skin-Sun Exposed (Lower leg) | ||
| 0.0000035 | 0.22 | Breast-Mammary Tissue | ||
| 0.000011 | 0.24 | Spleen | ||
| 0.000024 | −0.38 | Brain-Cortex | ||
| 0.000055 | 0.14 | Adipose-Subcutaneous | ||
| 0.00012 | 0.17 | Skin-Not Sun Exposed (Suprapubic) | ||
| 0.00012 | 0.12 | Muscle-Skeletal | ||
| 0.00023 | −0.12 | Nerve-Tibial | ||
| CYP19A1 | rs28892005 | 1.1 × 10−9 | 0.20 | Skin-Sun Exposed (Lower leg) |
| 0.00020 | 0.14 | Skin-Not Sun Exposed (Suprapubic) | ||
| IL1A | rs3783553 | 3.9 × 10−14 | 0.35 | Skin-Not Sun Exposed (Suprapubic) |
| 2.6 × 10−11 | 0.30 | Skin-Sun Exposed (Lower leg) | ||
| 2.0 × 10−7 | 0.37 | Spleen | ||
| 0.0000018 | 0.19 | Testis | ||
| 0.000028 | 0.16 | Esophagus-Mucosa | ||
| 0.00016 | 0.19 | Thyroid | ||
| NFKB1 | rs28362491 | 9.9 × 10−14 | 0.15 | Muscle-Skeletal |
| 6.0 × 10−8 | 0.12 | Cells-Cultured fibroblasts | ||
| 2.4 × 10−7 | 82 | Whole Blood | ||
| 0.0000018 | −0.12 | Testis | ||
| 0.000099 | −0.12 | Heart-Atrial Appendage | ||
| 0.00021 | −77 | Skin-Not Sun Exposed (Suprapubic) | ||
| TYMS | rs151264360 | 6.6 × 10−34 | 0.66 | Esophagus-Muscularis |
| 1.7 × 10−22 | −0.28 | Testis | ||
| 8.1 × 10−17 | 0.53 | Esophagus-Gastroesophageal Junction | ||
| XRCC1 | rs3213239 | 4.4 × 10−45 | −0.43 | Thyroid |
| 5.4 × 10−42 | −0.53 | Pancreas | ||
| 2.0 × 10−26 | −0.49 | Testis | ||
| 2.0 × 10−24 | −0.58 | Adrenal Gland | ||
| 3.8 × 10−19 | −0.23 | Muscle-Skeletal | ||
| 6.3 × 10−15 | −0.32 | Pituitary | ||
| 4.1 × 10−13 | 0.20 | Nerve-Tibial | ||
| 4.2 × 10−13 | −0.46 | Brain-Hypothalamus | ||
| 8.7 × 10−11 | −0.23 | Colon-Sigmoid | ||
| 1.5 × 10−10 | −0.54 | Brain-Anterior cingulate cortex (BA24) | ||
| 4.8 × 10−10 | −0.38 | Brain-Caudate (basal ganglia) | ||
| 6.8 × 10−10 | −0.23 | Stomach | ||
| 1.1 × 10−9 | −0.34 | Ovary | ||
| 1.7 × 10−9 | −0.24 | Heart-Left Ventricle | ||
| 1.2 × 10−8 | −0.41 | Brain-Hippocampus | ||
| 2.0 × 10−8 | −0.33 | Brain-Nucleus accumbens (basal ganglia) | ||
| 3.7 × 10−8 | −0.28 | Liver | ||
| 3.8 × 10−8 | −0.14 | Colon-Transverse | ||
| 1.7 × 10−7 | −0.29 | Brain-Frontal Cortex (BA9) | ||
| 1.9 × 10−7 | −0.29 | Brain-Cortex | ||
| 2.0 × 10−7 | −0.48 | Brain-Amygdala | ||
| 4.5 × 10−7 | −0.15 | Esophagus-Mucosa | ||
| 0.0000022 | −0.40 | Minor Salivary Gland | ||
| 0.0000028 | −0.55 | Brain-Substantia nigra | ||
| 0.0000045 | −0.30 | Brain-Putamen (basal ganglia) | ||
| 0.000011 | −84 | Whole Blood | ||
| 0.000018 | −78 | Cells-Cultured fibroblasts | ||
| 0.000031 | −0.24 | Prostate | ||
| 0.000056 | −0.15 | Heart-Atrial Appendage | ||
| 0.00012 | −94 | Artery-Tibial |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Andrade, R.B.; Cavalcante, G.C.; Amador, M.A.T.; Moreira, F.C.; Khayat, A.S.; Assumpção, P.P.; Ribeiro-dos-Santos, Â.; Santos, N.P.C.; Santos, S. The Search for Cancer Biomarkers: Assessing the Distribution of INDEL Markers in Different Genetic Ancestries. Curr. Issues Mol. Biol. 2022, 44, 2275-2286. https://doi.org/10.3390/cimb44050154
Andrade RB, Cavalcante GC, Amador MAT, Moreira FC, Khayat AS, Assumpção PP, Ribeiro-dos-Santos Â, Santos NPC, Santos S. The Search for Cancer Biomarkers: Assessing the Distribution of INDEL Markers in Different Genetic Ancestries. Current Issues in Molecular Biology. 2022; 44(5):2275-2286. https://doi.org/10.3390/cimb44050154
Chicago/Turabian StyleAndrade, Roberta B., Giovanna C. Cavalcante, Marcos A. T. Amador, Fabiano Cordeiro Moreira, André S. Khayat, Paulo P. Assumpção, Ândrea Ribeiro-dos-Santos, Ney P. C. Santos, and Sidney Santos. 2022. "The Search for Cancer Biomarkers: Assessing the Distribution of INDEL Markers in Different Genetic Ancestries" Current Issues in Molecular Biology 44, no. 5: 2275-2286. https://doi.org/10.3390/cimb44050154
APA StyleAndrade, R. B., Cavalcante, G. C., Amador, M. A. T., Moreira, F. C., Khayat, A. S., Assumpção, P. P., Ribeiro-dos-Santos, Â., Santos, N. P. C., & Santos, S. (2022). The Search for Cancer Biomarkers: Assessing the Distribution of INDEL Markers in Different Genetic Ancestries. Current Issues in Molecular Biology, 44(5), 2275-2286. https://doi.org/10.3390/cimb44050154








