Biocontrol Potential and Catabolic Profile of Endophytic Diaporthe eres Strain 1420S from Prunus domestica L. in Poland—A Preliminary Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Fungal Isolation
2.2. DNA Extraction, PCR Amplification, and Sequencing
2.3. Antagonistic Activity of Endophytic Diaporthe Strain Based on Dual Culture Assay
- I% = inhibition of radial mycelial growth;
- RC = colony radial growth of the pathogen in control plate;
- RT = colony radial growth of the pathogen in the presence of endophyte.
2.4. Application of Biolog FF Plates for Catabolic Profile of Diaporthe Isolate
3. Results
3.1. Identification of Endophytic Diaporthe Strain
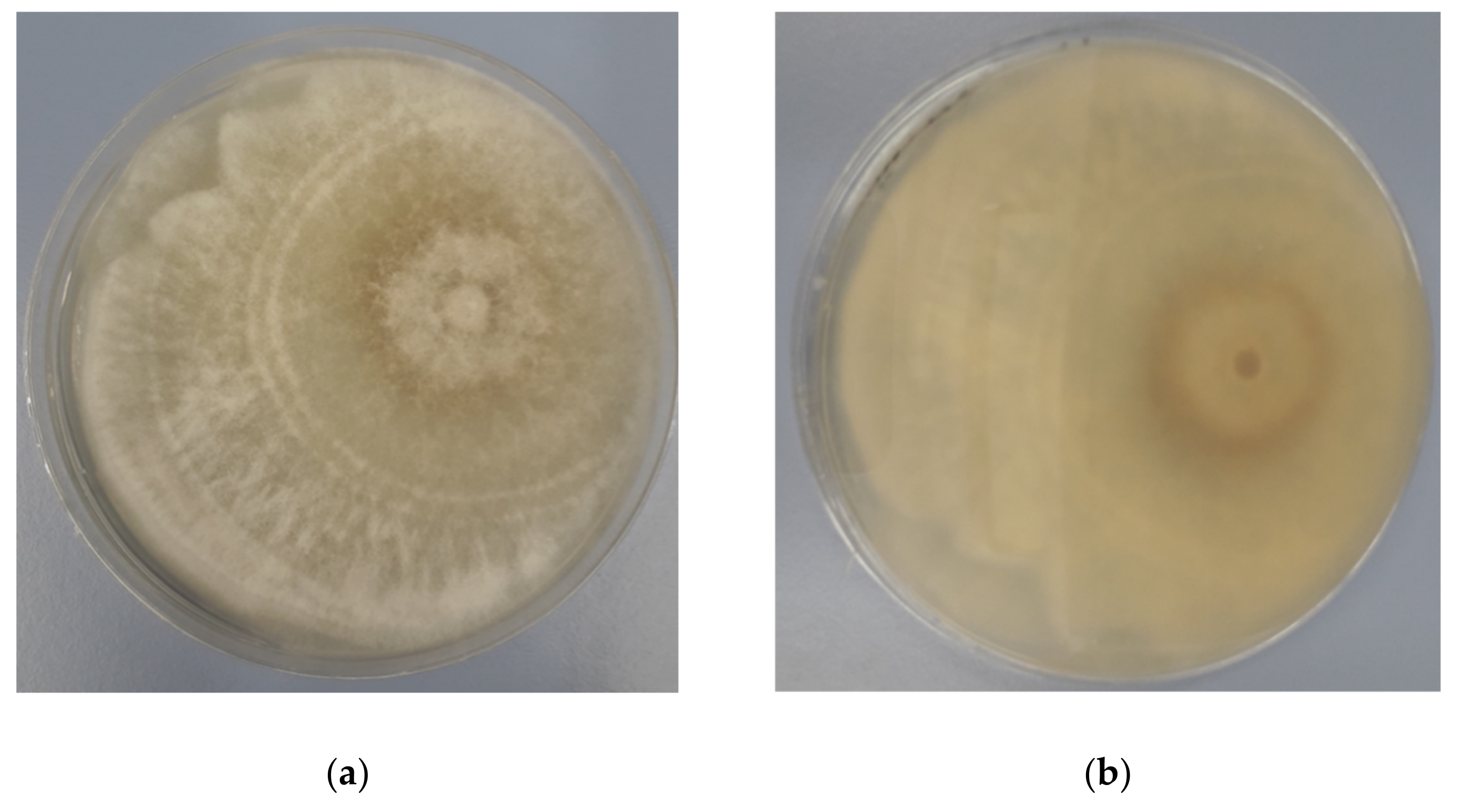
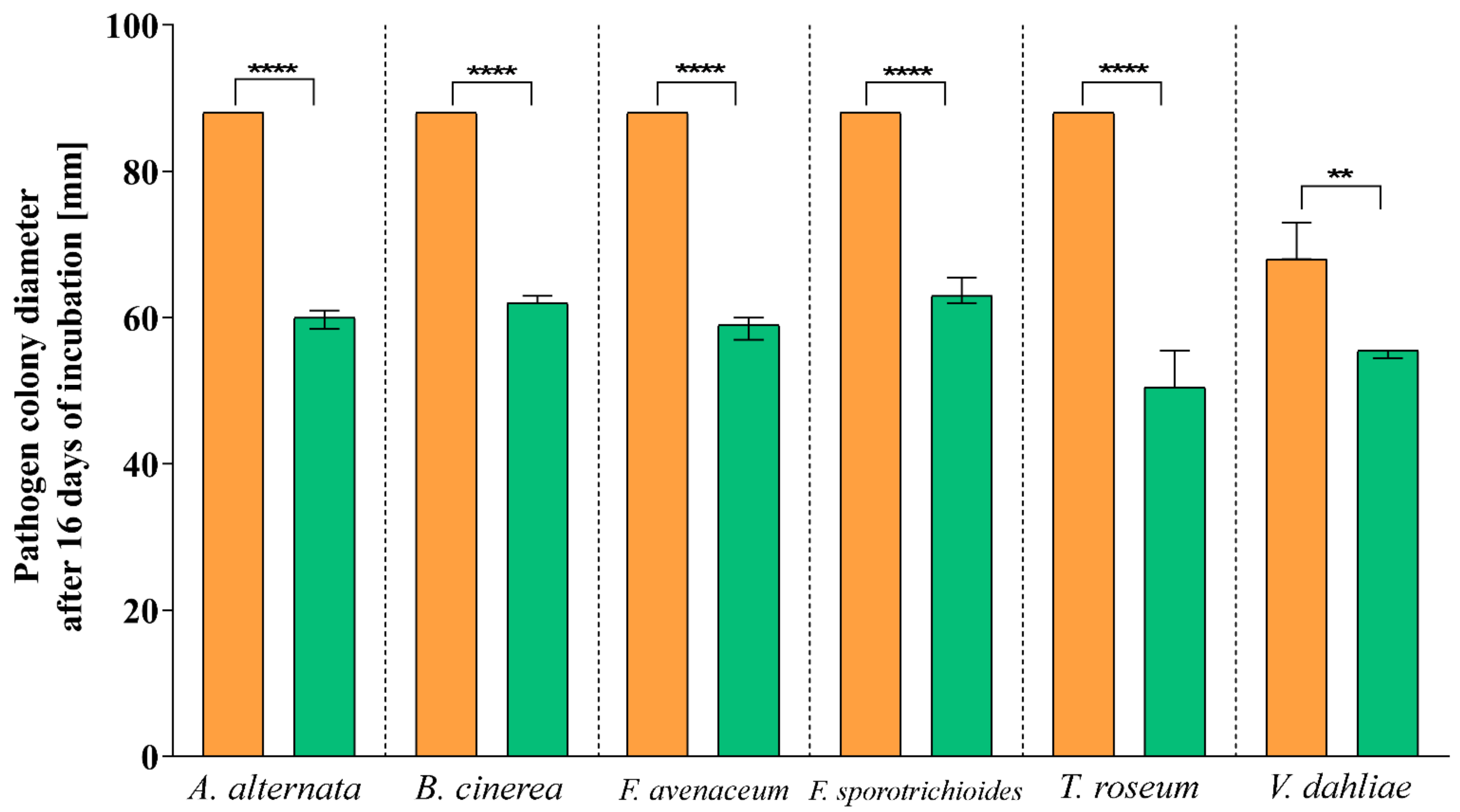
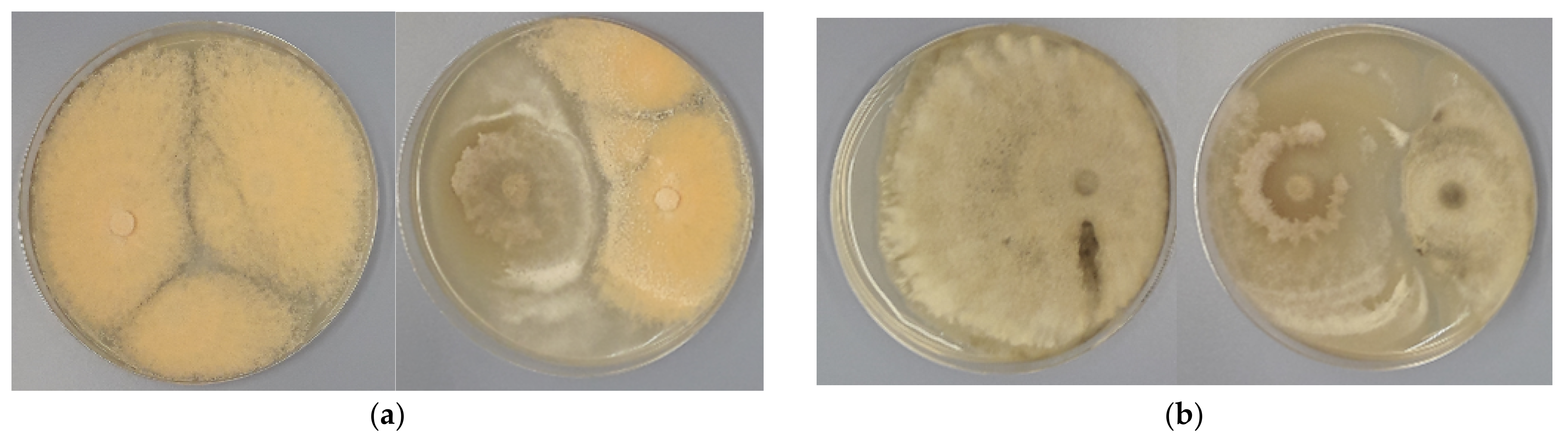
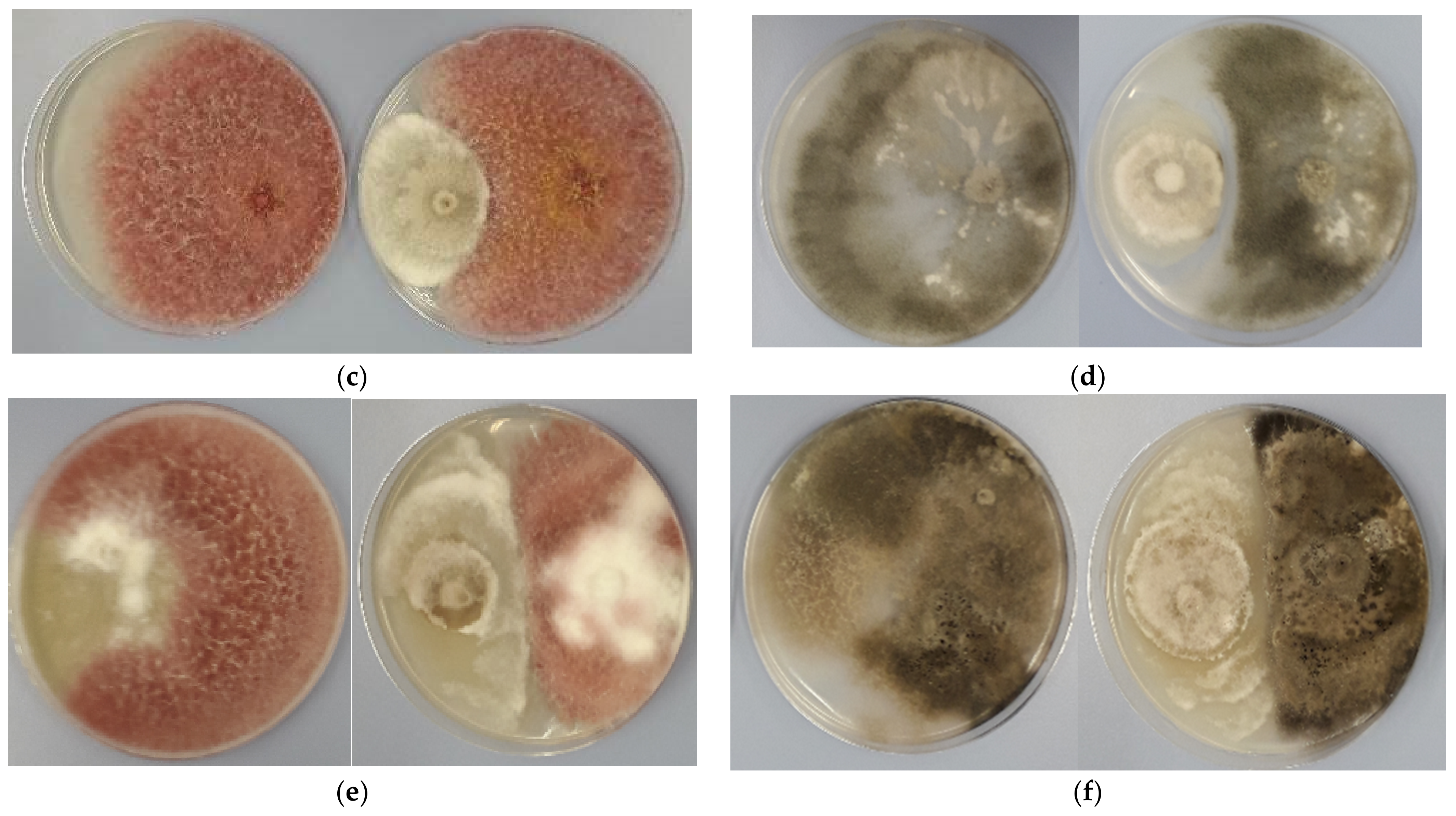
3.2. Antagonistic Activity
3.3. BIOLOG Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Arnold, A.E.; Mejia, L.C.; Kyllo, D.; Rojas, E.I.; Maynard, Z.; Robbins, N.; Herre, E.A. Fungal endophytes limit pathogen damage in a tropical tree. Proc. Natl. Acad. Sci. USA 2003, 100, 15649–15654. [Google Scholar] [CrossRef]
- Doty, L.S. Growth-promoting endophytic fungi of forest trees. In Endophytes of Forest Trees: Biology and Applications, Forestry Sciences 80; Pirttilä, A.M., Frank, A.C., Eds.; Springer Science, Business Media: Berlin/Heidelberg, Germany, 2011; pp. 151–156. [Google Scholar]
- Rakshith, D.; Santosh, P.; Satish, S. Isolation and characterization of antimicrobial metabolite producing endophytic Phomopsis sp. from Ficus pumila Linn. (Moraceae). Int. J. Chem. Anal. Sci. 2013, 4, 156–160. [Google Scholar] [CrossRef]
- Khan, A.L.; Hussain, J.; Al-Harrasi, A.; Al-Rawahi, A.; Lee, I.J. Endophytic fungi: Resource for gibberellins and crop abiotic stress resistance. Crit. Rev. Biotechnol. 2015, 35, 62–74. [Google Scholar] [CrossRef] [PubMed]
- Dutta, D.; Puzari, K.C.; Gogoi, R.; Dutta, P. Endophytes: Exploitation as a Tool in Plant Protection. Braz. Arch. Biol. Technol. 2014, 57, 621–629. [Google Scholar] [CrossRef]
- Santos, C.M.; da Silva, R.A.; Garcia, A.; Polli, A.D.; Polonio, J.C.; Azevedo, J.L.; Pamphile, J.A. Enzymatic and antagonist activity of endophytic fungi from Sapindus saponaria L. (Sapindaceae). Acta Biol. Colomb. 2019, 24, 322. [Google Scholar] [CrossRef]
- Petrini, O.; Sieber, T.N.; Toti, L.; Viret, O. Ecology, metabolic production, and substrate utilization in endophytic fungi. Nat. Toxins 1992, 1, 185–196. [Google Scholar] [CrossRef]
- Hyde, K.D.; Soytong, K. The fungal endophyte dilemma. Fungal Divers. 2008, 33, 163–173. [Google Scholar]
- De Silva, N.I.; Brooks, S.; Lumyong, S.; Hyde, K.D. Use of endophytes as biocontrol agents. Fungal Biol. Rev. 2019, 33, 133–148. [Google Scholar] [CrossRef]
- Chepkirui, C.; Stadler, M. The genus Diaporthe: A rich source of diverse and bioactive metabolites. Mycol. Prog. 2017, 16, 477–494. [Google Scholar] [CrossRef]
- Xu, T.-C.; Lu, Y.-H.; Wang, J.-F.; Song, Z.-Q.; Hou, Y.-G.; Liu, S.-S.; Liu, C.-S.; Wu, S.-H. Bioactive Secondary Metabolites of the Genus Diaporthe and Anamorph Phomopsis from Terrestrial and Marine Habitats and Endophytes: 2010–2019. Microorganisms 2021, 9, 217. [Google Scholar] [CrossRef]
- Udayanga, D.; Liu, X.; McKenzie, E.H.C.; Chukeatirote, E.; Bahkali, A.H.A.; Hyde, K.D. The genus Phomopsis: Biology, applications, species concepts and names of common phytopathogens. Fungal Divers. 2011, 50, 189–225. [Google Scholar] [CrossRef]
- Gomes, R.R.; Glienke, C.; Videira, S.I.R.; Lombard, L.; Groenewald, J.Z.; Crous, P.W. Diaporthe: A genus of endophytic, saprobic and plant pathogenic fungi. Persoonia 2013, 31, 1–41. [Google Scholar] [CrossRef]
- Król, E.D.; Abramczyk, B.A.; Zalewska, E.D.; Zimowska, B. Fungi inhabiting fruit tree shoots with special reference to the Diaporthe (Phomopsis) genus. Acta Sci. Pol. Hortorum Cultus 2017, 16, 113–126. [Google Scholar] [CrossRef]
- Santos, L.; Alves, A.; Alves, R. Evaluating multi-locus phylogenies for species boundaries determination in the genus Diaporthe. PeerJ 2017, 5, e3120. [Google Scholar] [CrossRef]
- Abramczyk, B.A.; Król, E.D.; Zalewska, E.D.; Zimowska, B. Influence of temperature and fungal community on growth and sporulation of Diaporthe from fruit plants. Acta Sci. Pol. Hortorum Cultus 2020, 19, 71–79. [Google Scholar] [CrossRef]
- Sun, W.; Huang, S.; Xia, J.; Zhang, X.; Li, Z. Morphological and molecular identification of Diaporthe species in south-western China, with description of eight new species. MycoKeys 2021, 77, 65–95. [Google Scholar] [CrossRef] [PubMed]
- Baumgartner, K.; Fujiyoshi, P.T.; Travadon, R.; Castlebury, L.A.; Wilcox, W.F.; Rolshausen, P.E. Characterization of species of Diaporthe from wood cankers of grape in eastern North American vineyards. Plant Dis. 2013, 97, 912–920. [Google Scholar] [CrossRef]
- Huang, F.; Hou, X.; Dewdney, M.M.; Fu, Y.; Chen, G.Q.; Hyde, K.D.; Li, H. Diaporthe species occurring on Citrus in China. Fungal Divers. 2013, 61, 237–250. [Google Scholar] [CrossRef]
- Huang, F.; Udayanga, D.; Wang, X.H.; Hou, X.; Mei, X.F.; Fu, Y.S.; Hyde, K.D.; Li, H.Y. Endophytic Diaporthe associated with Citrus: A phylogenetic reassessment with seven new species from China. Fungal Biol. 2015, 119, 331–347. [Google Scholar] [CrossRef]
- Tan, Y.P.; Edwards, J.; Grice, K.R.E.; Shivas, R.G. Molecular phylogenetic analysis reveals six new Diaporthe species from Australia. Fungal Divers. 2013, 61, 251–260. [Google Scholar] [CrossRef]
- Udayanga, D.; Castlebury, L.A.; Rossman, A.Y.; Chukeatirote, E.; Hyde, K.D. Insights into the genus Diaporthe: Phylogenetic species delimitation in the D. eres species complex. Fungal Divers. 2014, 67, 203–229. [Google Scholar] [CrossRef]
- Udayanga, D.; Castlebury, L.A.; Rossman, A.Y.; Hyde, K.D. Species limits in Diaporthe: Molecular reassessment of D. citri, D. cytosporella, D. foeniculina and D. rudis. Persoonia 2014, 32, 83–101. [Google Scholar] [CrossRef] [PubMed]
- Udayanga, D.; Castlebury, L.A.; Rossman, A.Y.; Chukeatirote, E.; Hyde, K.D. The Diaporthe sojae species complex: Phylogenetic reassessment of pathogens associated with soybean, cucurbits and other field crops. Fungal Biol. 2015, 119, 383–407. [Google Scholar] [CrossRef]
- Gao, Y.H.; Liu, F.; Duan, W.; Crous, P.W.; Cai, L. Diaporthe is paraphyletic. IMA Fungus 2017, 8, 153–187. [Google Scholar] [CrossRef] [PubMed]
- Guarnaccia, V.; Groenewald, J.Z.; Woodhall, J.; Armengol, J.; Cinelli, T.; Eichmeier, A.; Ezra, D.; Fontaine, F.; Gramaje, D.; Gutierrez-Aguirregabiria, A. Diaporthe diversity and pathogenicity revealed from a broad survey of grapevine diseases in Europe. Persoonia 2018, 40, 135–153. [Google Scholar] [CrossRef]
- Guo, Y.S.; Crous, P.W.; Bai, Q.; Fu, M.; Yang, M.M.; Wang, X.H.; Du, Y.M.; Hong, N.; Xu, W.X.; Wang, G.P. High diversity of Diaporthe species associated with pear shoot canker in China. Persoonia 2020, 45, 132–162. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Jiang, N.; Tian, C.M. Three new Diaporthe species from Shaanxi Province, China. MycoKeys 2020, 67, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Zapata, M.; Palma, M.A.; Aninat, M.J.; Piontelli, E. Polyphasic studies of new species of Diaporthe from native forest in Chile, with descriptions of Diaporthe araucanorum sp. nov., Diaporthe foikelawen sp. nov. and Diaporthe patagonica sp. nov. Int. J. Syst. Evol. Microbiol. 2020, 70, 3379–3390. [Google Scholar] [CrossRef] [PubMed]
- Dvořák, M.; Palovčíková, D.; Jankovský, L. The occurrence of endophytic fungus Phomopsis oblonga on elms in the area of southern Bohemia. J. For. Sci. 2006, 52, 531–535. [Google Scholar] [CrossRef]
- Murali, T.S.; Suryanarayanan, T.S.; Geeta, R. Endophytic Phomopsis species: Host range and implications for diversity estimates. Can. J. Microbiol. 2006, 52, 673–680. [Google Scholar] [CrossRef]
- Ferreira, M.C.; Vieira, M.L.A.; Zani, C.L.; Alves, T.M.A.; Sales, J.P.A.; Murta, S.M.F.; Romanha, A.J.; Gil, L.H.V.G.; Carvalho, A.G.O.; Zilli, J.E.; et al. Molecular phylogeny, diversity, symbiosis and discover of bioactive compounds of endophytic fungi associated with the medicinal Amazonian plant Carapa guianensis Aublet (Meliaceae). Biochem. Syst. Ecol. 2015, 59, 36–44. [Google Scholar] [CrossRef]
- Martins, F.; Pereira, J.A.; Bota, P.; Bento, A.; Baptista, P. Fungal endophyte communities in above-and belowground olive tree organs and the effect of season and geographic location on their structures. Fungal Ecol. 2016, 20, 193–201. [Google Scholar] [CrossRef]
- Hanácková, Z.; Havrdová, L.; Cerný, L.; Zahradník, D.; Koukol, O. Fungal Endophytes in Ash Shoots—Diversity and Inhibition of Hymenoscyphus fraxineus. Balt. For. 2017, 23, 89–106. [Google Scholar]
- Ignjatović, J.; Maljurić, N.; Golubović, J.; Ravnikar, M.; Petković, M.; Savodnik, N.; Štrukelj, B.; Otašević, B. Characterization of Biomolecules with Antibiotic Activity from Endophytic Fungi Phomopsis Species. Acta Chim. Slov. 2020, 67, 445–461. [Google Scholar] [CrossRef]
- Król, E. Grzyby zasiedlające zdrowe łozy winorośli (Vitis spp.) w wybranych szkółkach. Acta Agrobot. 2006, 59, 163–173. [Google Scholar] [CrossRef]
- Castillo-Pando, M.S.; Nair, N.G.; Emmett, R.W.; Wicks, T.J. Inhibition in pycnidial viability of Phomopsis viticola on canes in situ as an aid to reducing inoculum potential of cane and leaf blight disease of grapevines. Australas. Plant Pathol. 1997, 26, 21–25. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, J.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- Glass, N.L.; Donaldson, G. Development of primer sets designed for use with PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microbiol. 1995, 61, 1323–1330. [Google Scholar] [CrossRef]
- Carbone, I.; Kohn, L.M. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 1999, 91, 553–556. [Google Scholar] [CrossRef]
- Katoch, M.; Pull, S. Endophytic fungi associated with Monarda citriodora, an aromatic and medicinal plant and their biocontrol potential. Pharm. Biol. 2017, 55, 1528–1535. [Google Scholar] [CrossRef]
- Hajieghrari, B.; Torabi-Giglou, M.; Mohammadi, M.R.; Davari, M. Biological potential of some Iranian Trichoderma isolates in the control of soil borne plant pathogenic fungi. Afr. J. Biotechnol. 2008, 7, 967–972. [Google Scholar]
- Kaur, T. Fungal Endophyte-Host Plant Interactions: Role in Sustainable Agriculture. In Sustainable Crop Production; IntechOpen: London, UK, 2020. [Google Scholar]
- Kowalski, T.; Kehr, R.D. Endophytic fungi colonization of branch bases in several forest tree species. Sydowia 1992, 44, 137–168. [Google Scholar]
- Sieber, T.; Hugentobler, C. Endophytische Pilze in Blattern und Asten gesunder und geschadigter Buchen (Fagus sylvatica L.). Eur. J. For. Pathol. 1987, 17, 411–425. [Google Scholar] [CrossRef]
- Sieber, T.N. Endophytic fungi in forest trees: Are they mutualists? Fungal Biol. Rev. 2007, 21, 75–89. [Google Scholar] [CrossRef]
- Sieber, T.N.; Dorworth, C.E. An ecological study about assemblages of endophytic fungi in Acer macrophyllum in British Columbia—In search of candidate mycoherbicides. Can. J. Bot. 1994, 72, 1397–1402. [Google Scholar] [CrossRef]
- Liu, K.; Ding, X.; Deng, B.; Chen, W. Isolation and characterization of endophytic taxol-producing fungi from Taxus chinensis. J. Ind. Microbiol. Biotechnol. 2009, 36, 1171–1177. [Google Scholar] [CrossRef] [PubMed]
- Moricca, S.; Ginetti, B.; Ragazzi, B. Species and organ-specificity in endophytes colonizing healthy and declining Mediterranean oaks. Phytopathol. Mediterr. 2012, 51, 587–598. [Google Scholar]
- Kim, C.K.; Eo, J.K.; Eom, A.H. Diversity and seasonal variation of endophytic fungi isolated from three conifers in Mt. Taehwa, Korea. Mycobiology 2014, 41, 82–85. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, A.D.S.; Polonio, J.C.; Costa, A.T.; Dos Santos, C.M.; Rhoden, S.A.; Azevedo, J.L.; Pamphile, J.A. Bioprospection of culturable endophytic fungi associated with the ornamental plant Pachystachys lutea. Curr. Microbiol. 2018, 75, 588–596. [Google Scholar] [CrossRef] [PubMed]
- Alabouvette, C.; Olivain, C.; Steinberg, C. Biological control of plant diseases: The European situation. Eur. J. Plant Pathol. 2006, 114, 329–341. [Google Scholar] [CrossRef]
- Landum, M.; Felix, M.; Alho, J.; Garcia, R.; Cabrita, M.J.; Rei, F.; Varanda, C.M. Antagonistic activity of fungi of Olea europaea L. against Colletotrichum acutatum. Microbiol. Res. 2016, 183, 100–108. [Google Scholar] [CrossRef]
- Jaber, L.R.; Ownley, B.H. Can we use entomopathogenic fungi as endophytes for dual biological control of insect pests and plant pathogens? Biol. Control 2018, 116, 36–45. [Google Scholar] [CrossRef]
- Santos, P.J.; Savi, D.C.; Gomes, R.R.; Goulin, E.H.; Da Costa Senkiv, C.; Tanaka, F.A.; Almeida, Á.M.; Galli-Terasawa, L.; Kava, V.; Glienke, C. Diaporthe endophytica and D. terebinthifolii from medicinal plants for biological control of Phyllosticta citricarpa. Microbiol. Res. 2016, 186–187, 153–160. [Google Scholar] [CrossRef] [PubMed]
- Polonio, J.C.; Almeida, T.; Garcia, A.; Mariucci, G.; Azevedo, J.; Rhoden, S.; Pamphile, J.A. Biotechnological prospecting of foliar endophytic fungi of guaco (Mikania glomerata Spreng.) with antibacterial and antagonistic activity against phytopathogens. Genet. Mol. Res. 2015, 14, 7297–7309. [Google Scholar] [CrossRef]
- Prada, H.; Avila, L.; Sierra, R.; Bernal, A.; Restrepo, S. Caracterizacion morfologica y molecular del antagonismo entre el endofito Diaporthe sp. aislado de frailejon (Espeletia sp.) y el fitopatogeno Phytophthora infestans. (Morphological and molecular characterization of the antagonistic interaction between the endophyte Diaporthe sp. isolated from frailejon (Espeletia sp.) and the plant pathogen Phytophthora infestans). Rev. Iberoam. Micol. 2009, 26, 198–201. [Google Scholar] [CrossRef] [PubMed]
- Cheong, S.L.; Cheow, Y.L.; Ting, A.S. Characterizing antagonistic activities and host compatibility (via simple endophyte-calli test) of endophytes as biocontrol agents of Ganoderma boninense. Biol. Control 2017, 105, 86–92. [Google Scholar] [CrossRef]
- Li, G.; Kusari, S.; Kusari, P.; Kayser, O.; Spiteller, M. Endophytic Diaporthe sp. LG23 produces a potent antibacterial tetracyclic triterpenoid. J. Nat. Prod. 2015, 78, 2128–2132. [Google Scholar] [CrossRef] [PubMed]
- Tanney, J.B.; McMullin, D.R.; Blake, D.; Green, B.D.; Miller, J.D.; Seifert, K.A. Production of antifungal and antiinsectan metabolites by the Picea endophyte Diaporthe maritima sp. nov. Fungal Biol. 2016, 120, 1448–1457. [Google Scholar] [CrossRef]
- Singh, G.; Katoch, A.; Razak, M.; Kitchlu, S.; Goswami, A.; Katoch, M. Bioactive and biocontrol potential of endophytic fungi associated with Brugmansia aurea Lagerh. FEMS Microbiol. Lett. 2017, 364, fnx194. [Google Scholar] [CrossRef]
- Fu, J.; Zhou, Y.; Li, H.F.; Ye, Y.H.; Guo, J.H. Antifungal metabolites from Phomopsis sp. By254, an endophytic fungus in Gossypium hirsutum. Afr. J. Microbiol. Res. 2011, 5, 1231–1263. Available online: https://www.academicjournals.org/ajmr (accessed on 4 December 2021). [CrossRef]
- Webber, J.F.; Gibbs, J.N. Colonization of elm bark by Phomopsis oblonga. Trans. Br. Mycol. Soc. 1984, 82, 348–352. [Google Scholar] [CrossRef]
- Pertile, G.; Panek, J.; Oszust, K.; Siczek, A.; Frac, M. Intraspecific functional and genetic diversity of Petriella setifera. PeerJ 2018, 6, e4420. [Google Scholar] [CrossRef]
- Mchunu, N.P.; Permaul, K.; Alam, M.; Singh, S. Carbon utilization profile of a thermophilic fungus, Thermomyces lanuginosus using phenotypic microarray. Adv. Biosci. Biotechnol. 2013, 4, 24–32. [Google Scholar] [CrossRef]
- Barreraa, V.A.; Martin, M.E.; Aulicino, M.; Martínez, S.; Chiessa, G.; Saparrat, M.C.N.; Gasoni, A.L. Carbon-substrate utilization profiles by Cladorrhinum (Ascomycota) Rev. Argent. Microbiol. 2019, 51, 302–306. [Google Scholar]
- Oszust, K.; Cybulska, J.; Frąc, M. How Do Trichoderma Genus Fungi Win a Nutritional Competition Battle against Soft Fruit Pathogens? A Report on Niche Overlap Nutritional Potentiates. Int. J. Mol. Sci. 2020, 21, 4235. [Google Scholar] [CrossRef]
- Dela Cruz, T.E.E.; Schulz, B.E.; Kubicek, C.P.; Druzhinina, I.S. Carbon source utilization by the marine Dendryphiella species D. arenaria and D. salina. FEMS Microbiol. Ecol. 2006, 58, 343–353. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.P. Application of Biolog FF MicroPlate for substrate utilization and metabolite profiling of closely related fungi. J. Microbiol. Methods 2009, 77, 102–108. [Google Scholar] [CrossRef]
- Tang, E.; Hill, C.B.; Hartman, G.L. Carbon utilization profiles of Fusarium virguliforme isolates. Can. J. Microbiol. 2010, 56, 979–986. [Google Scholar]
- Hothersall, J.S.; Ahmed, A. Metabolic fate of the increased yeast amino acid uptake subsequent to catabolite derepression. J. Amino Acids 2013, 2013, 461901. [Google Scholar] [CrossRef] [PubMed]
- Rossman, A.Y.; Farr, D.F.; Castlebury, L.A. A review of the phylogeny and biology of the Diaporthales. Mycoscience 2007, 48, 135–144. [Google Scholar] [CrossRef]
- Garcia, A.; Rhoden, S.A.; Filho, C.J.R.; Nakamura, C.V.; Pamphile, J.A. Diversity of foliar endophytic fungi from the medicinal plant Sapindus saponaria L. and their localization by scanning electron microscopy. Biol. Res. 2012, 45, 139–148. [Google Scholar] [CrossRef][Green Version]
- Sessa, L.; Abreo, E.; Lupo, S. Diversity of fungal latent pathogens and true endophytes associated with fruit trees in Uruguay. J. Phytopathol. 2018, 166, 633–647. [Google Scholar] [CrossRef]
- Abramczyk, B.A.; Król, E.D.; Zalewska, E.D.; Zimowska, B. Morphological characteristics and pathogenicity of Diaporthe eres isolates to the fruit tree shoots. Acta Sci. Pol. Hortorum Cultus 2018, 17, 125–133. [Google Scholar] [CrossRef]
- Dong, Z.; Manawasinghe, I.S.; Huang, Y.; Shu, Y.; Phillips, A.J.L.; Dissanayake, A.J.; Hyde, K.D.; Xiang, M.; Luo, M. Endophytic Diaporthe associated with Citrus grandis cv. Tomentosa in China. Front. Microbiol. 2021, 11, 609387. [Google Scholar] [CrossRef]
- Guarnaccia, V.; Crous, P.W. Emerging citrus diseases in Europe caused by Diaporthe spp. IMA Fungus 2017, 8, 317–334. [Google Scholar] [CrossRef]
- Thompson, S.M.; Grams, R.A.; Neate, S.M.; Shivas, R.G.; Ryley, M.J.; Tan, Y.P.; Aitken, E.; Wright, G.C.; O’Connor, D.J. First reports of Diaporthe kongii, D. masirevicii, and D. ueckerae associated with stem and peg dieback on peanut in Australia. Plant Dis. 2018, 102, 1459. [Google Scholar] [CrossRef]
- Naveen, J.; Navya, H.M.; Hithamani, G.; Niranjana, S.R.; Hariprasad, P. First report of Diaporthe masirevicii causing leaf blight of Gloriosa superba in India. New Dis. Rep. 2018, 37, 13. [Google Scholar] [CrossRef]
- Pazdiora, P.C.; Hoffmann, J.F.; Chaves, F.C.; Dallagnol, L.J. First report of fruit rot caused by Diaporthe masirevicii on Physalis peruviana in Brazil. Plant Dis. 2018, 102, 441–442. [Google Scholar] [CrossRef]
- Lim, L.; Mohd, M.H.; Zakaria, L. Identification and pathogenicity of Diaporthe species associated with stem-end rot of mango (Mangifera indica L.). Eur. J. Plant Pathol. 2019, 155, 687–696. [Google Scholar] [CrossRef]
- Manawasinghe, I.S.; Dissanayake, A.J.; Li, X.; Liu, M.; Wanasinghe, D.; Xu, J.; Zhao, W.; Zhang, W.; Zhou, Y.; Hyde, K.D.; et al. High genetic diversity and species complexity of diaporthe associated with grapevine dieback in China. Front. Microbiol. 2019, 10, 1936. [Google Scholar] [CrossRef] [PubMed]
- Brader, G.; Compant, S.; Vescio, K.; Mitter, B.; Trognitz, F.; Ma, L.J.; Sessitsch, A. Ecology and genomic insights into plant-pathogenic and plant-nonpathogenic endophytes. Annu. Rev. Phytopathol. 2017, 55, 61–83. [Google Scholar] [CrossRef] [PubMed]
- Hardoim, P.R.; Van Overbeek, L.S.; Berg, G.; Pirttilä, A.M.; Compant, S.; Campisano, A.; Döring, M.; Sessitsch, A. The hidden world within plants: Ecological and evolutionary considerations for defining functioning of microbial endophytesfining functioning of microbial endophytes. Microbiol. Mol. Biol. Rev. 2015, 79, 293–320. [Google Scholar] [CrossRef]
- Qi, F.; Jing, T.; Zhan, Y. Characterization of endophytic fungi from Acer ginnala Maxim. in an artificial plantation: Media effect and tissue-dependent variation. PLoS ONE 2012, 7, e46785. [Google Scholar] [CrossRef][Green Version]
- Aly, A.H.; Debbab, A.; Proksch, P. Fungal endophytes: Unique plant inhabitants with great promises. Appl. Microbiol. Biotechnol. 2011, 90, 1829–1845. [Google Scholar] [CrossRef]
- Kuo, H.C.; Hui, S.; Choi, J.; Asiegbu, F.O.; Valkonen, J.P.T.; Lee, Y. Secret lifestyles of Neurospora crassa. Sci. Rep. 2014, 4, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Mengistu, A.A. Endophytes: Colonization, Behaviour, and Their Role in Defense Mechanism. Int. J. Microbiol. 2020, 2020, 6927219. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Xu, X.; Mao, L.; Lao, J.; Lin, F.; Yuan, Z.; Zhang, C. Endophytic Diaporthe from Southeast China are genetically diverse based on multi-locus phylogeny analyses. World J. Microbiol. Biotechnol. 2014, 30, 237–243. [Google Scholar] [CrossRef] [PubMed]


| LOCUS | GenBank Accession No. | Closest Related Species | Similarity [%] | Coverage [%] |
|---|---|---|---|---|
| ITS | MW664034 | D. eres EU571099 | 100 | 99.8 |
| TEF | OK506723 | D. eres JF461475 | 100 | 100 |
| TUB | OK506724 | D. eres MG281207 | 100 | 100 |
| CAL | OK490500 | D. eres MH051294 | 100 | 100 |
| Pathogen | Inhibition Rate (I%) |
|---|---|
| A. alternata | 32.04 |
| B. cinerea | 29.20 |
| F. avenaceum | 33.40 |
| F. sporotrichioides | 27.84 |
| T. roseum | 40.68 |
| V. dahliae | 20.68 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abramczyk, B.; Marzec-Grządziel, A.; Grządziel, J.; Król, E.; Gałązka, A.; Oleszek, W. Biocontrol Potential and Catabolic Profile of Endophytic Diaporthe eres Strain 1420S from Prunus domestica L. in Poland—A Preliminary Study. Agronomy 2022, 12, 165. https://doi.org/10.3390/agronomy12010165
Abramczyk B, Marzec-Grządziel A, Grządziel J, Król E, Gałązka A, Oleszek W. Biocontrol Potential and Catabolic Profile of Endophytic Diaporthe eres Strain 1420S from Prunus domestica L. in Poland—A Preliminary Study. Agronomy. 2022; 12(1):165. https://doi.org/10.3390/agronomy12010165
Chicago/Turabian StyleAbramczyk, Barbara, Anna Marzec-Grządziel, Jarosław Grządziel, Ewa Król, Anna Gałązka, and Wiesław Oleszek. 2022. "Biocontrol Potential and Catabolic Profile of Endophytic Diaporthe eres Strain 1420S from Prunus domestica L. in Poland—A Preliminary Study" Agronomy 12, no. 1: 165. https://doi.org/10.3390/agronomy12010165
APA StyleAbramczyk, B., Marzec-Grządziel, A., Grządziel, J., Król, E., Gałązka, A., & Oleszek, W. (2022). Biocontrol Potential and Catabolic Profile of Endophytic Diaporthe eres Strain 1420S from Prunus domestica L. in Poland—A Preliminary Study. Agronomy, 12(1), 165. https://doi.org/10.3390/agronomy12010165








