Pangenome-Guided Reverse Vaccinology and Immunoinformatics Approach for Rational Design of a Multi-Epitope Subunit Vaccine Candidate Against the Multidrug-Resistant Pathogen Chromobacterium violaceum: A Computational Immunopharmacology Perspective
Abstract
1. Introduction
2. Results
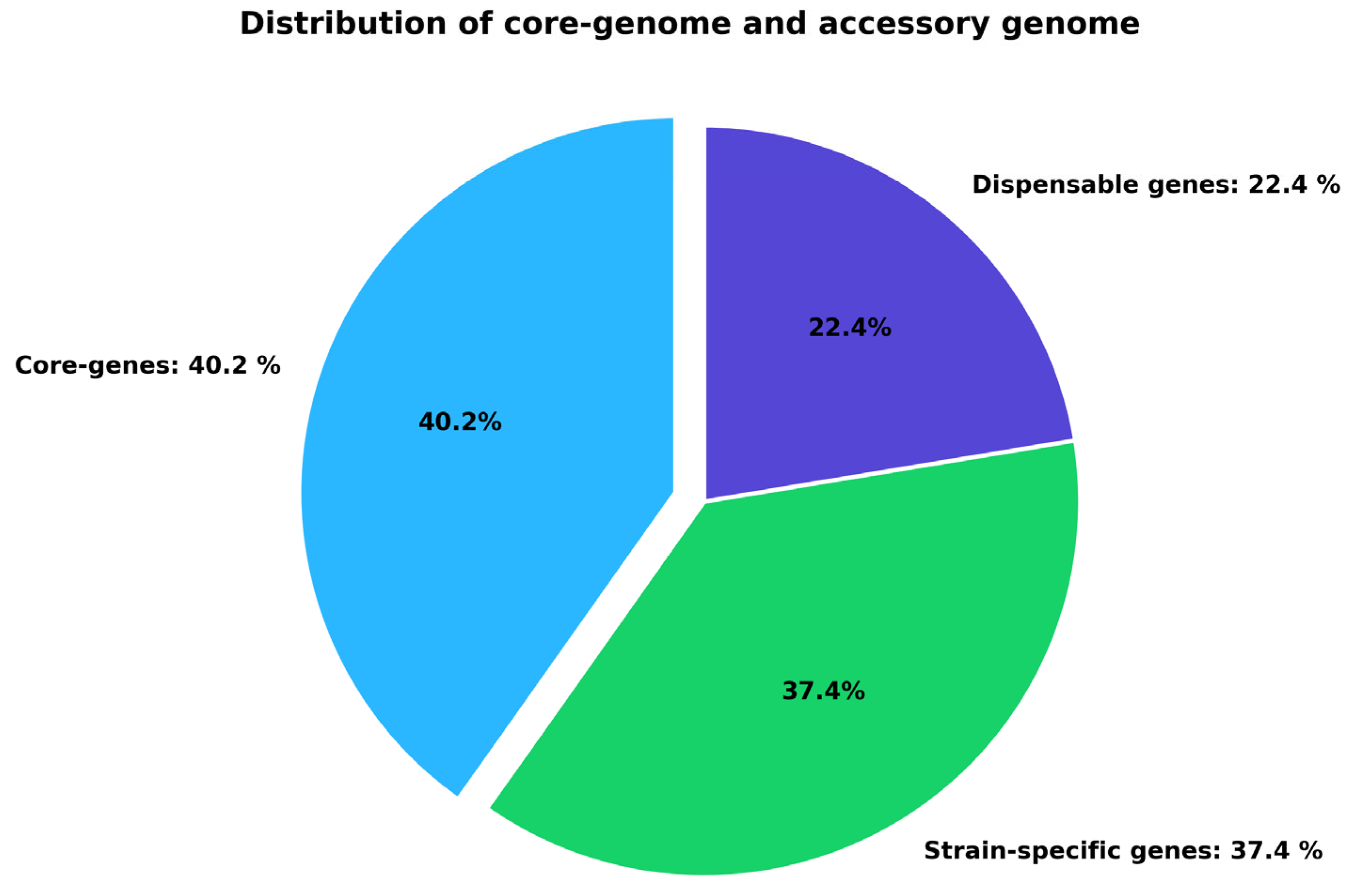
2.1. Pangenome Analysis of C. violaceum
2.2. Subtractive Proteomics
2.3. Epitope Prediction
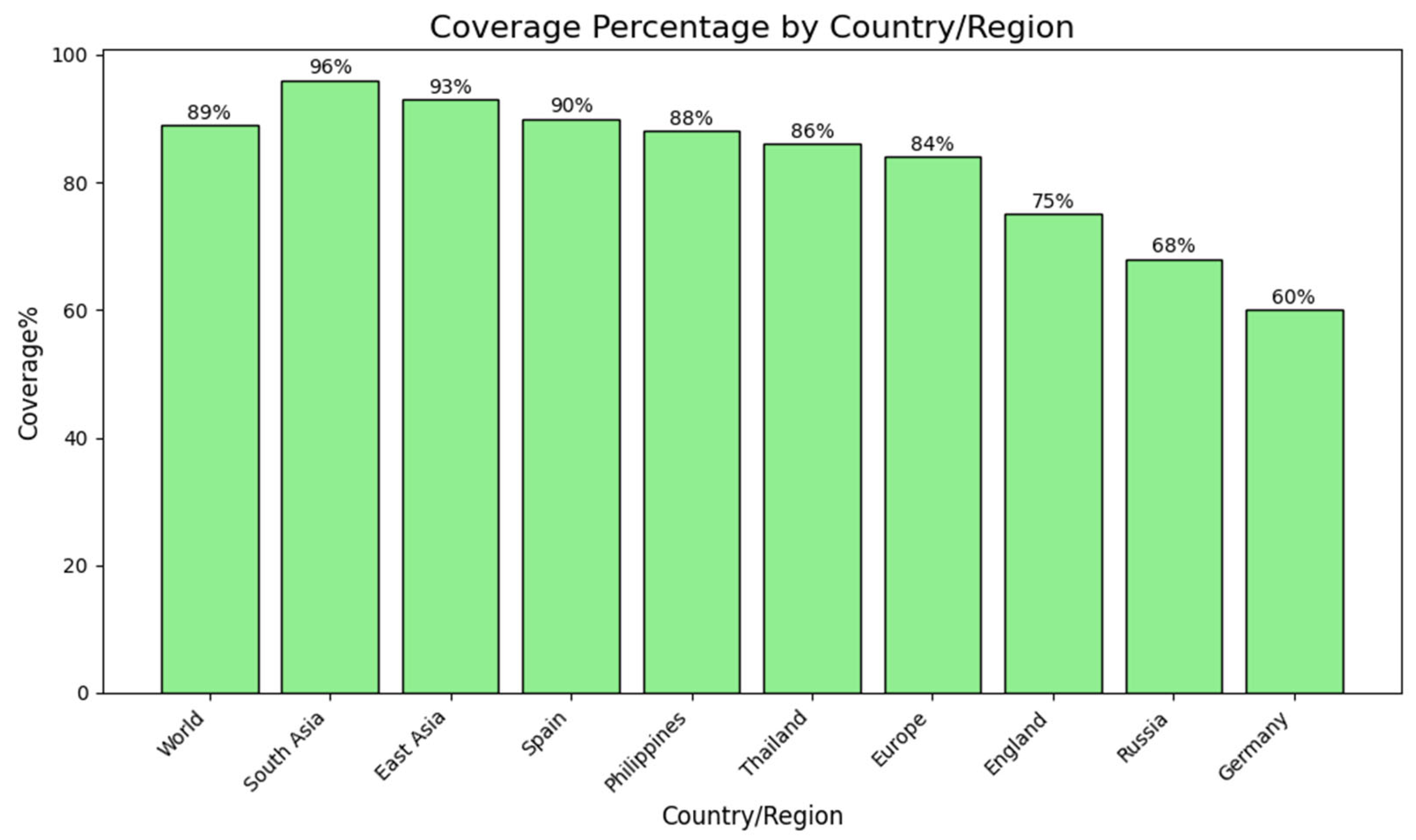
2.4. Population Coverage Analysis
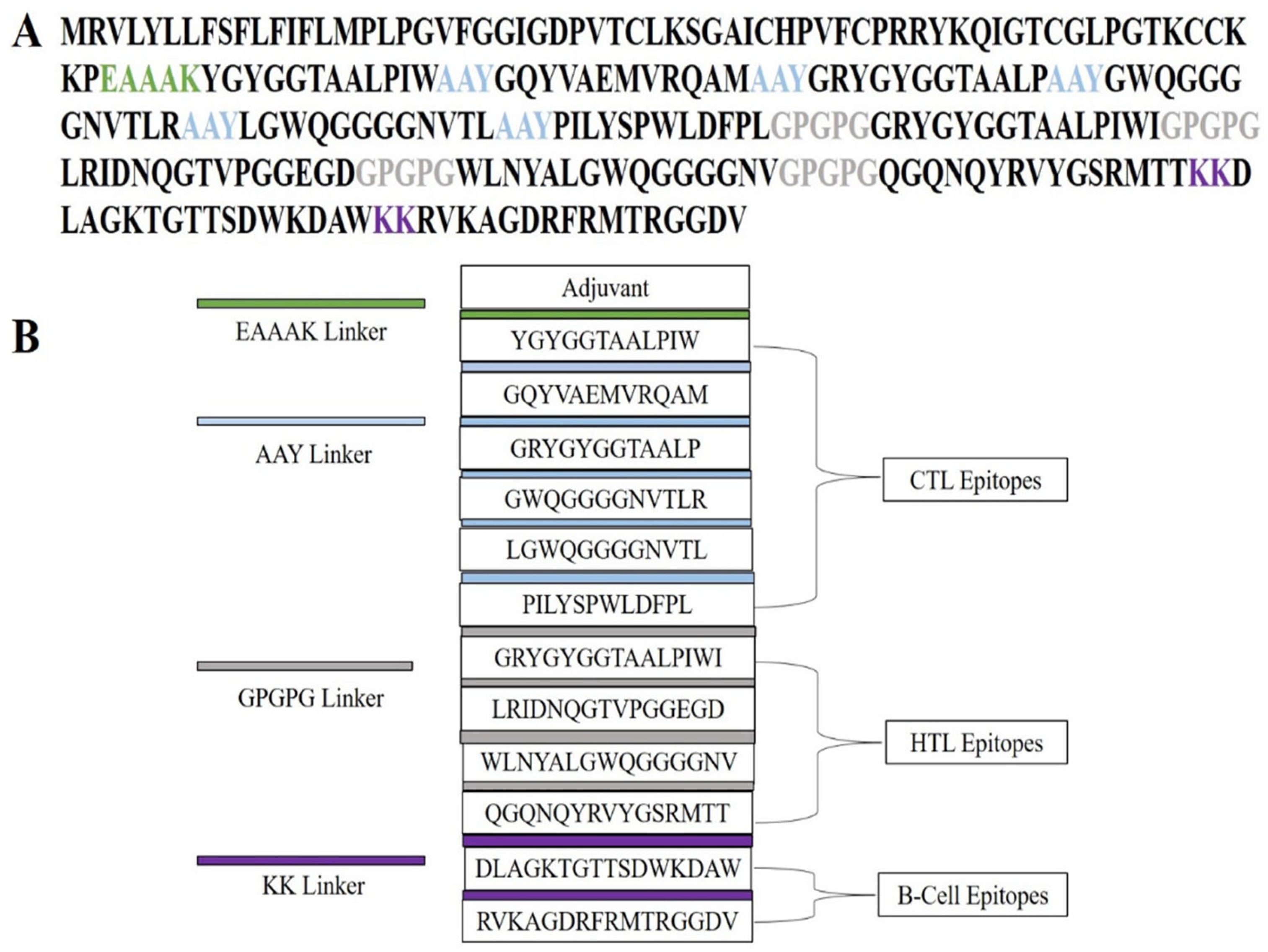
2.5. Multi-Epitope Vaccine Construction: Integration of Adjuvant and Linkers for Enhanced Immunogenicity
2.6. Post-Translational and Physicochemical Characterization of the Vaccine Construct
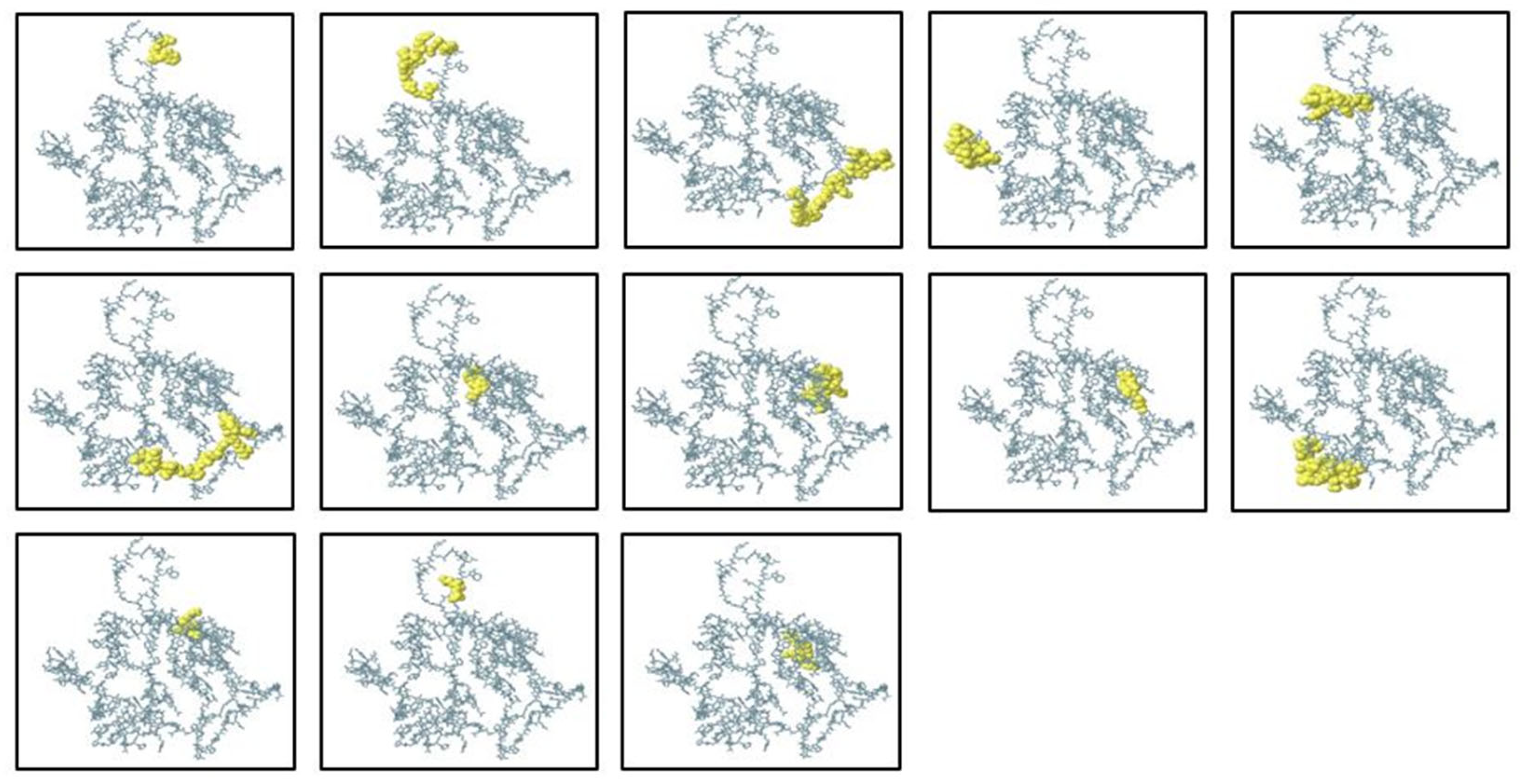
2.7. Structural Conformity Analysis of B-Cell Epitopes
2.8. Prediction and Selection of B-Cell Epitopes
2.9. Molecular Docking Analysis with Host Immune Receptor
2.10. Normal Mode Analysis of the MEV–TLR4 Complex
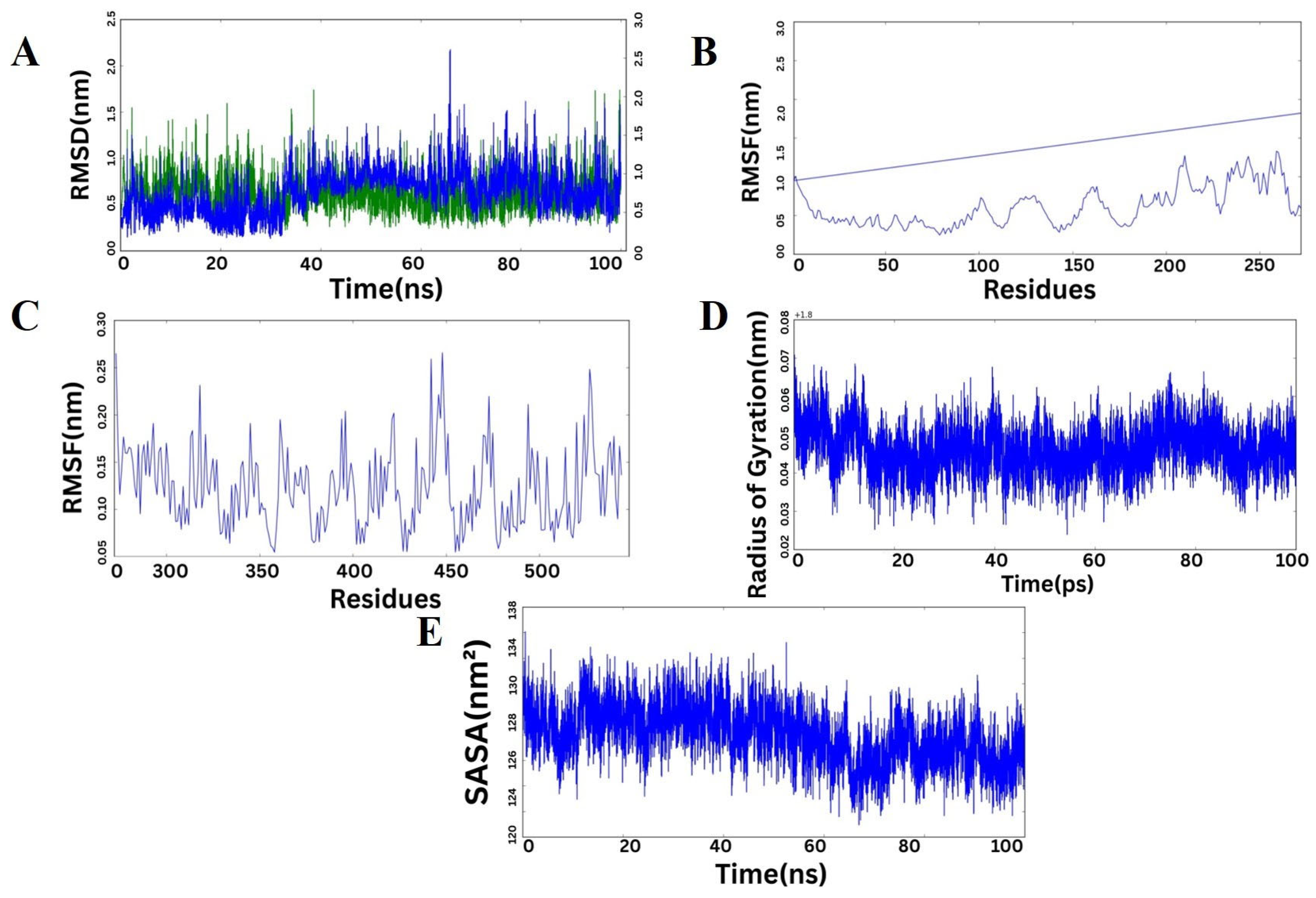
2.11. Molecular Dynamics Simulation of the Protein Complex
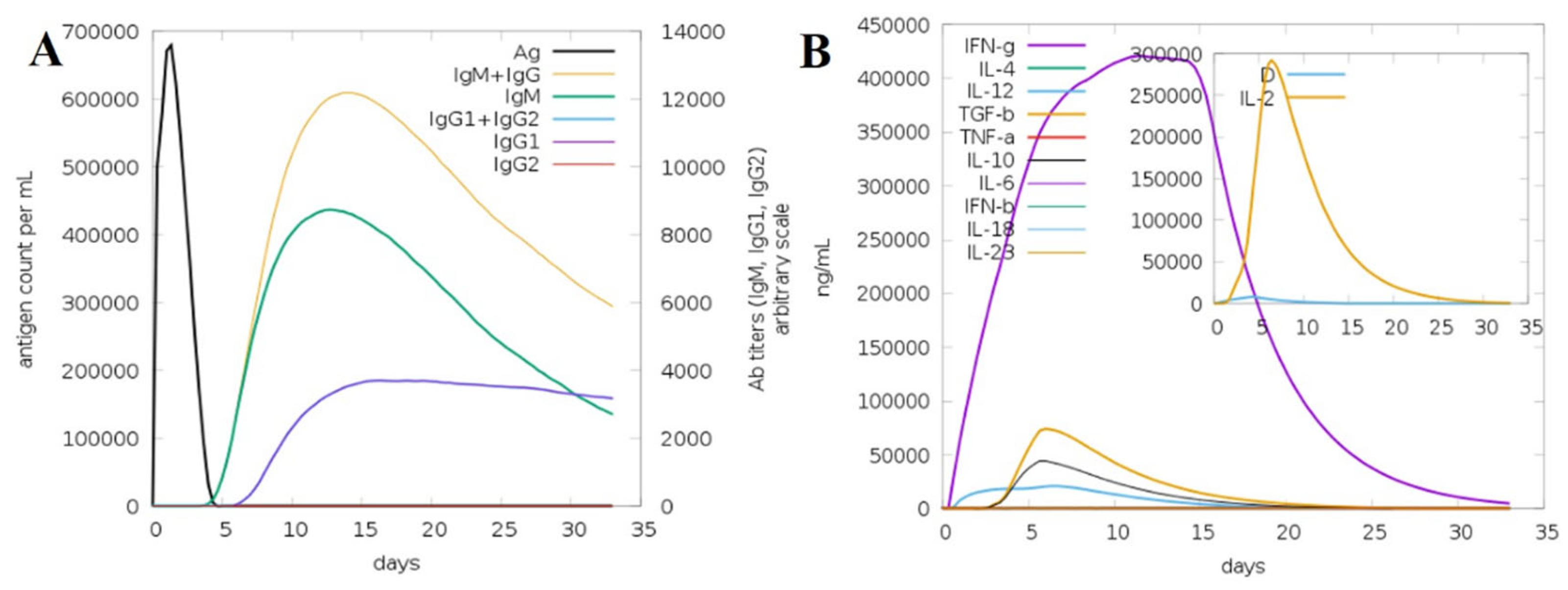
2.12. Immune Simulation Analysis
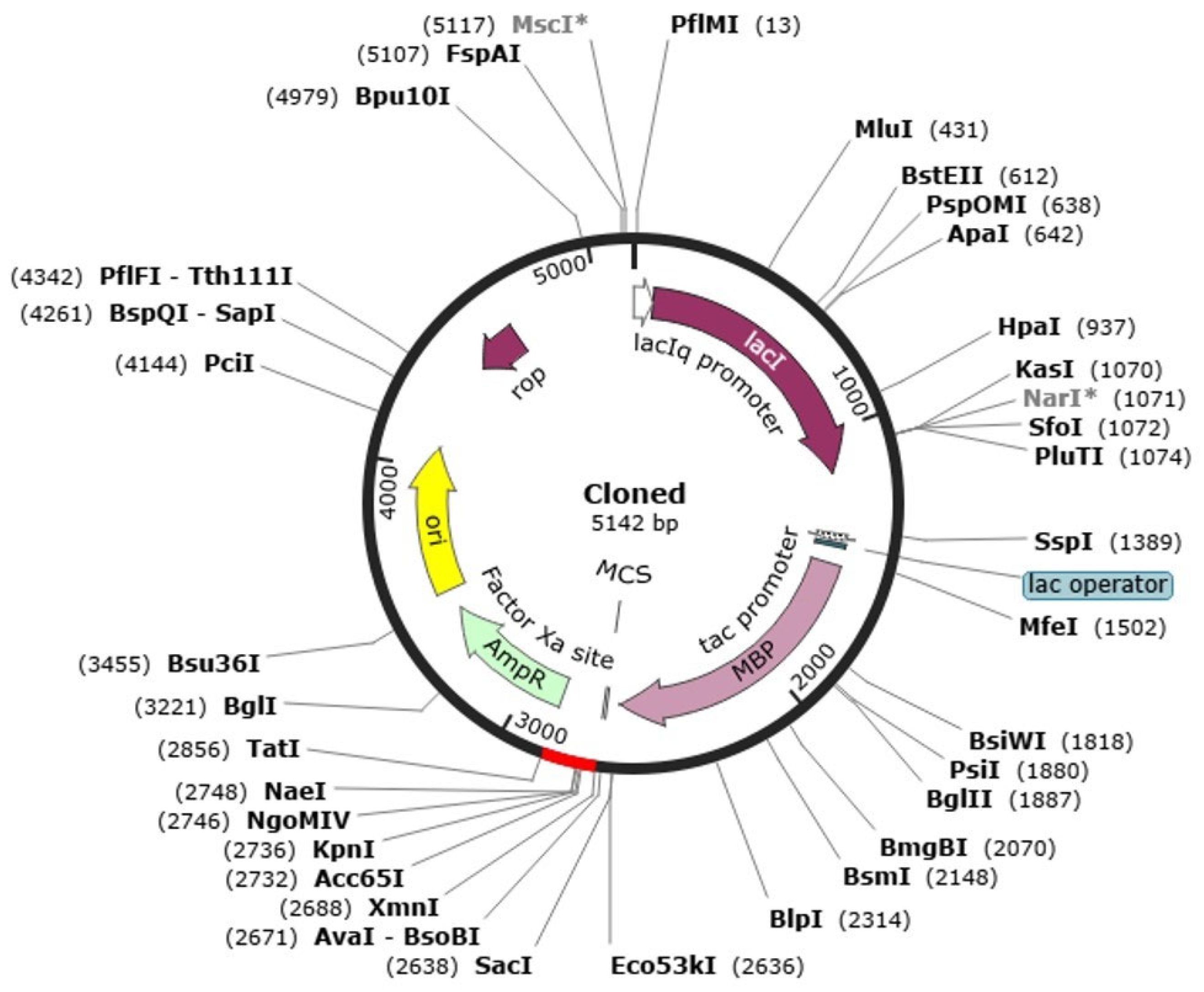
2.13. Codon Optimization and In Silico Cloning
3. Discussion
4. Materials and Methods
4.1. Genome Retrieval and Dataset Preparation for Pangenome Analysis
4.2. Subtractive Proteomics Analysis
4.3. Epitope Prediction and Screening
4.3.1. CTL Epitope Selection and Assessment
4.3.2. HTL Epitope Selection and Analysis
4.3.3. LBL Epitope Identification and Analysis
4.4. Population Coverage Analysis
4.5. Designing of the Vaccine Construct
4.6. Structural Analysis
4.7. Refinement, Confirmation and Prediction of Tertiary Structure
4.8. B-Cell Epitope Screening
4.9. Molecular Docking Analysis
4.10. Normal Mode Analysis of MEV–TLR4 Docked Complex
4.11. Molecular Dynamics (MD) Simulations
4.12. Immune Simulation
4.13. Reverse Translation, Codon Optimization, and In Silico Cloning
5. Conclusions
Supplementary Materials
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| MEV | Multi-Epitope Vaccine |
| CTL | Cytotoxic T Lymphocyte |
| HTL | Helper T Lymphocyte |
| LBL | Linear B Lymphocyte |
| CTB | Cholera Toxin B Subunit |
| NCBI | National Center for Biotechnology Information |
| TLR4 | Toll-Like Receptor 4 |
| MHC | Major Histocompatibility Complex |
| MD | Molecular Dynamics |
| NMA | Normal Mode Analysis |
| OMV | Outer Membrane Vesicle |
| LPS | Lipopolysaccharide |
| RMSD | Root Mean Square Deviation |
| RMSF | Root Mean Square Fluctuation |
| SASA | Solvent Accessible Surface Area |
References
- Kumar, M. Chromobacterium violaceum: A rare bacterium isolated from a wound over the scalp. Int. J. App. Basic. Med. Res. 2012, 2, 70. [Google Scholar] [CrossRef] [PubMed]
- Barnes, P.; Gonzales, J.; Hammond, D. Chromobacterium violaceum: A rare opportunistic pathogen and clue for pediatric chronic granulomatous disease. Pediatr. Dermatol. 2023, 40, 396–397. [Google Scholar] [CrossRef] [PubMed]
- Siqueira, C.D.; Dias, J.; Ruf, H.; Ramos, E.A.G.; Maciel, E.A.P.; Rolim, A.; Jabur, L.; Vasconcelos, L.; Silvany, C. Chromobacterium violaceum in Siblings, Brazil. Emerg. Infect. Dis. 2005, 11, 1443–1445. [Google Scholar] [CrossRef] [PubMed]
- Klenk, C.; Schnieders, M.; Heinemann, M.; Wiegard, C.; Büttner, H.; Ramharter, M.; Jordan, S.; Mackroth, M.S. Severe Systemic Chromobacterium violaceum Infection: A Case Study of a German Long-Term Resident in French Guyana. TropicalMed 2024, 9, 242. [Google Scholar] [CrossRef] [PubMed]
- Meher-Homji, Z.; Mangalore, R.P.; DR Johnson, P.; YL Chua, K. Chromobacterium violaceum infection in chronic granulomatous disease: A case report and review of the literature. JMM Case Rep. 2017, 4, e005084. [Google Scholar] [CrossRef]
- Alisjahbana, B.; Debora, J.; Susandi, E.; Darmawan, G. Chromobacterium violaceum: A Review of an Unexpected Scourge. Int. J. Gen. Med. 2021, 14, 3259–3270. [Google Scholar] [CrossRef] [PubMed]
- Batista, J.H.; Da Silva Neto, J.F. Chromobacterium violaceum Pathogenicity: Updates and Insights from Genome Sequencing of Novel Chromobacterium Species. Front. Microbiol. 2017, 8, 2213. [Google Scholar] [CrossRef]
- Sadique, T.; Khan, A.R.; Ahmed, D.; Kabir, F.; Chowdhury, A.; Islam, N.; Halim, F.; Akter, N.; Islam, S.B. A Case of Severe Septicemia With Chromobacterium violaceum in Bangladesh. Clin. Case Rep. 2025, 13, e70603. [Google Scholar] [CrossRef]
- Venkatramanan, M.; Nalini, E. Regulation of virulence in Chromobacterium violaceum and strategies to combat it. Front. Microbiol. 2024, 15, 1303595. [Google Scholar] [CrossRef]
- Choi, S.Y.; Yoon, K.; Lee, J.I.; Mitchell, R.J. Violacein: Properties and Production of a Versatile Bacterial Pigment. BioMed Res. Int. 2015, 2015, 465056. [Google Scholar] [CrossRef]
- Dimitrova, P.D.; Damyanova, T.; Paunova-Krasteva, T. Chromobacterium violaceum: A Model for Evaluating the Anti-Quorum Sensing Activities of Plant Substances. Sci. Pharm. 2023, 91, 33. [Google Scholar] [CrossRef]
- Alves, J.A.; Leal, F.C.; Previato-Mello, M.; Da Silva Neto, J.F. A Quorum Sensing-Regulated Type VI Secretion System Containing Multiple Nonredundant VgrG Proteins Is Required for Interbacterial Competition in Chromobacterium violaceum. Microbiol. Spectr. 2022, 10, e01576-22. [Google Scholar] [CrossRef] [PubMed]
- Pena, R.T.; Blasco, L.; Ambroa, A.; González-Pedrajo, B.; Fernández-García, L.; López, M.; Bleriot, I.; Bou, G.; García-Contreras, R.; Wood, T.K.; et al. Relationship Between Quorum Sensing and Secretion Systems. Front. Microbiol. 2019, 10, 1100. [Google Scholar] [CrossRef] [PubMed]
- Ciprandi, A.; Da Silva, W.M.; Santos, A.V.; De Castro Pimenta, A.M.; Carepo, M.S.P.; Schneider, M.P.C.; Azevedo, V.; Silva, A. Chromobacterium violaceum: Important Insights for Virulence and Biotechnological Potential by Exoproteomic Studies. Curr. Microbiol. 2013, 67, 100–106. [Google Scholar] [CrossRef]
- De Brito, C.F.A.; Carvalho, C.B.; Santos, F.; Gazzinelli, R.T.; Oliveira, S.C.; Azevedo, V.; Teixeira, S.M.R. Chromobacterium violaceum genome: Molecular mechanisms associated with pathogenicity. Genet. Mol. Res. 2004, 3, 148–161. [Google Scholar]
- Miki, T.; Iguchi, M.; Akiba, K.; Hosono, M.; Sobue, T.; Danbara, H.; Okada, N. Chromobacterium pathogenicity island 1 type III secretion system is a major virulence determinant for Chromobacterium violaceum -induced cell death in hepatocytes. Mol. Microbiol. 2010, 77, 855–872. [Google Scholar] [CrossRef]
- Ahmed, S.K.; Hussein, S.; Qurbani, K.; Ibrahim, R.H.; Fareeq, A.; Mahmood, K.A.; Mohamed, M.G. Antimicrobial resistance: Impacts, challenges, and future prospects. J. Med. Surg. Public Health 2024, 2, 100081. [Google Scholar] [CrossRef]
- Mirghani, R.; Saba, T.; Khaliq, H.; Mitchell, J.; Do, L.; Chambi, L.; Diaz, K.; Kennedy, T.; Alkassab, K.; Huynh, T.; et al. Biofilms: Formation, drug resistance and alternatives to conventional approaches. AIMS Microbiol. 2022, 8, 239–277. [Google Scholar] [CrossRef]
- Yahya, M.F.Z.R.; Jalil, M.T.M.; Jamil, N.M.; Nor, N.H.M.; Alhajj, N.; Siburian, R.; Majid, N.A. Biofilms and multidrug resistance: An emerging crisis and the need for multidisciplinary interventions. Front. Bioeng. Biotechnol. 2025, 13, 1625356. [Google Scholar] [CrossRef]
- Ghattas, M.; Dwivedi, G.; Lavertu, M.; Alameh, M.-G. Vaccine Technologies and Platforms for Infectious Diseases: Current Progress, Challenges, and Opportunities. Vaccines 2021, 9, 1490. [Google Scholar] [CrossRef]
- Ghaffar, S.A.; Tahir, H.; Muhammad, S.; Shahid, M.; Naqqash, T.; Faisal, M.; Albekairi, T.H.; Alshammari, A.; Albekairi, N.A.; Manzoor, I. Designing of a multi-epitopes based vaccine against Haemophilius parainfluenzae and its validation through integrated computational approaches. Front. Immunol. 2024, 15, 1380732. [Google Scholar] [CrossRef]
- Dingding, H.; Muhammad, S.; Manzoor, I.; Ghaffar, S.A.; Alodaini, H.A.; Moubayed, N.M.; Hatamleh, A.A.; Songxiao, X. Subtractive proteomics and reverse-vaccinology approaches for novel drug targets and designing a chimeric vaccine against Ruminococcus gnavus strain RJX1120. Front. Immunol. 2025, 16, 1555741. [Google Scholar] [CrossRef] [PubMed]
- Aslam, S.; Ashfaq, U.A.; Zia, T.; Aslam, N.; Alrumaihi, F.; Shahid, F.; Noor, F.; Qasim, M. Proteome based mapping and reverse vaccinology techniques to contrive multi-epitope based subunit vaccine (MEBSV) against Streptococcus pyogenes. Infect. Genet. Evol. 2022, 100, 105259. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Chen, M.; Bian, Y.; Zheng, X.; Tong, R.; Sun, X. Advanced subunit vaccine delivery technologies: From vaccine cascade obstacles to design strategies. Acta Pharm. Sin. B 2023, 13, 3321–3338. [Google Scholar] [CrossRef] [PubMed]
- Allemailem, K.S. A Comprehensive Computer Aided Vaccine Design Approach to Propose a Multi-Epitopes Subunit Vaccine against Genus Klebsiella Using Pan-Genomics, Reverse Vaccinology, and Biophysical Techniques. Vaccines 2021, 9, 1087. [Google Scholar] [CrossRef] [PubMed]
- Allemailem, K.S. In Silico Development of a Chimeric Multi-Epitope Vaccine Targeting Helcococcus kunzii: Coupling Subtractive Proteomics and Reverse Vaccinology for Vaccine Target Discovery. Pharmaceuticals 2025, 18, 1258. [Google Scholar] [CrossRef]
- Li, J.; Ju, Y.; Jiang, M.; Li, S.; Yang, X.-Y. Epitope-Based Vaccines: The Next Generation of Promising Vaccines Against Bacterial Infection. Vaccines 2025, 13, 248. [Google Scholar] [CrossRef]
- Moradkasani, S.; Esmaeili, S.; Asadi Karam, M.R.; Mostafavi, E.; Shahbazi, B.; Salek Farrokhi, A.; Chiani, M.; Badmasti, F. Development of a multi-epitope vaccine from outer membrane proteins and identification of novel drug targets against Francisella tularensis: An In Silico approach. Front. Immunol. 2025, 16, 1479862. [Google Scholar] [CrossRef]
- Khan, K.; Jalal, K.; Uddin, R. Pangenome diversification and resistance gene characterization in Salmonella typhi prioritized RfaJ as a significant therapeutic marker. J. Genet. Eng. Biotechnol. 2023, 21, 125. [Google Scholar] [CrossRef]
- Du, Y.; Qian, C.; Li, X.; Zheng, X.; Huang, S.; Yin, Z.; Chen, T.; Pan, L. Unveiling intraspecific diversity and evolutionary dynamics of the foodborne pathogen Bacillus paranthracis through high-quality pan-genome analysis. Curr. Res. Food Sci. 2024, 9, 100867. [Google Scholar] [CrossRef]
- González-Cruz, M.; Reyes-Gastellou, A.; Castelán-Vega, J.A.; Monterrubio-López, G.P.; Jiménez-Alberto, A.; Aparicio-Ozores, G.; Ribas-Aparicio, R.M. In silico development of a broad-spectrum vaccine against ESKAPE pathogens. J. Mol. Graph. Model. 2025, 140, 109120. [Google Scholar] [CrossRef]
- Dolley, A.; Goswami, H.B.; Dowerah, D.; Dey, U.; Kumar, A.; Hmuaka, V.; Mukhopadhyay, R.; Kundu, D.; Varghese, G.M.; Doley, R.; et al. Reverse vaccinology and immunoinformatics approach to design a chimeric epitope vaccine against Orientia tsutsugamushi. Heliyon 2024, 10, e23616. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.; Ali, S.L.; Alamri, A.; Khatrawi, E.M.; Baiduissenova, A.; Suleimenova, F.; Mishra, V.K.; Khan, A.; Dusmagambetov, M.; Askarova, G. Multi-epitope-based vaccine models prioritization against Astrovirus MLB1 using immunoinformatics and reverse vaccinology approaches. J. Genet. Eng. Biotechnol. 2025, 23, 100451. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.; Ali, S.S.; Khan, A.; Zahid, M.A.; Alshabrmi, F.M.; Waheed, Y.; Agouni, A. Structural proteomics guided annotation of vaccine targets and designing of multi-epitopes vaccine to instigate adaptive immune response against Francisella tularensis. Microb. Pathog. 2024, 194, 106777. [Google Scholar] [CrossRef] [PubMed]
- Rahman, S.; Chiou, C.-C.; Ahmad, S.; Islam, Z.U.; Tanaka, T.; Alouffi, A.; Chen, C.-C.; Almutairi, M.M.; Ali, A. Subtractive Proteomics and Reverse-Vaccinology Approaches for Novel Drug Target Identification and Chimeric Vaccine Development against Bartonella henselae Strain Houston-1. Bioengineering 2024, 11, 505. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, M.Z.; Rao, T.; Mutahir, Z.; Ahmed, S.; Ullah, N.; Ojha, S.C. Immunoinformatic-driven design and evaluation of multi-epitope mRNA vaccine targeting HIV-1 gp120. Front. Immunol. 2025, 16, 1480025. [Google Scholar] [CrossRef]
- Raj, K.H.; Hossain, E.; Zahin, H.; Noman, A.A.; Saba, A.A.; Sayem, M.; Yasmin, T.; Nabi, A.H.M.N. A robust comprehensive immunoinformatics approach for designing a potential multi-epitope based vaccine against a reiterated monkeypox virus. Biochem. Biophys. Rep. 2025, 43, 102075. [Google Scholar] [CrossRef]
- Barh, D.; Tiwari, S.; Jain, N.; Ali, A.; Santos, A.R.; Misra, A.N.; Azevedo, V.; Kumar, A. In silico subtractive genomics for target identification in human bacterial pathogens. Drug Dev. Res. 2011, 72, 162–177. [Google Scholar] [CrossRef]
- Gupta, S.; Kapoor, P.; Chaudhary, K.; Gautam, A.; Kumar, R. Open Source Drug Discovery Consortium; Raghava, G.P.S. In Silico Approach for Predicting Toxicity of Peptides and Proteins. PLoS ONE 2013, 8, e73957. [Google Scholar] [CrossRef]
- Lin, Y.D.; Majumdar, S.S.; Hennessy, J.; Baird, R.W. The Spectrum of Chromobacterium violaceum Infections from a Single Geographic Location. Am. Soc. Trop. Med. Hyg. 2016, 94, 710–716. [Google Scholar] [CrossRef]
- Fantinatti-Garboggini, F.; Rd, A.; Barbosa, T.A.; Trevilato, P.B.; Neto, C.E.; Coêlho, R.D.; Silva, D.W.; Bartoleti, L.A.; Hanna, E.S.; Brocchi, M.; et al. Drug resistance in Chromobacterium violaceum. Genet. Mol. Res. 2004, 3, 134–147. [Google Scholar]
- Shovon, M.H.J.; Khan, D.A.; Tareq, M.M.I.; Imtiaz, M.; Zilani, M.N.H.; Hasan, M.N. A comprehensive assessment of VCAN transcriptional expression and evaluation as an effective prognostic biomarker against breast cancer: In silico study. Bull. Natl. Res. Cent. 2023, 47, 83. [Google Scholar] [CrossRef]
- Jiang, F.; Han, Y.; Liu, Y.; Xue, Y.; Cheng, P.; Xiao, L.; Gong, W. A comprehensive approach to developing a multi-epitope vaccine against Mycobacterium tuberculosis: From in silico design to in vitro immunization evaluation. Front. Immunol. 2023, 14, 1280299. [Google Scholar] [CrossRef] [PubMed]
- Nayak, S.S.; Sethi, G.; Ramadas, K. Design of multi-epitope based vaccine against Mycobacterium tuberculosis: A subtractive proteomics and reverse vaccinology based immunoinformatics approach. J. Biomol. Struct. Dyn. 2023, 41, 14116–14134. [Google Scholar] [CrossRef] [PubMed]
- Liao, J.; Zhang, X.; Zeng, X.; Zhao, Z.; Sun, T.; Xia, Z.; Jing, H.; Yuan, Y.; Chen, Z.; Gou, Q.; et al. A rational designed multi-epitope vaccine elicited robust protective efficacy against Klebsiella pneumoniae lung infection. Biomed. Pharmacother. 2024, 174, 116611. [Google Scholar] [CrossRef] [PubMed]
- Goodarzi, N.N.; Barzi, S.M.; Ajdary, S.; Chiani, M.; Yekaninejad, M.S.; Badmasti, F.; Pourmand, M.R. Immunogenic evaluation of LptD + LtgC as a bivalent vaccine candidate against Neisseria gonorrhoeae. J. Transl. Med. 2025, 23, 261. [Google Scholar] [CrossRef]
- Peele, K.A.; Srihansa, T.; Krupanidhi, S.; Ayyagari, V.S.; Venkateswarulu, T.C. Design of multi-epitope vaccine candidate against SARS-CoV-2: A in-silico study. J. Biomol. Struct. Dyn. 2021, 39, 3793–3801. [Google Scholar] [CrossRef]
- Caputo, A.; Fournier, P.-E.; Raoult, D. Genome and pan-genome analysis to classify emerging bacteria. Biol. Direct 2019, 14, 5. [Google Scholar] [CrossRef]
- Alghamdi, M.; Al-Judaibi, E.; Al-Rashede, M.; Al-Judaibi, A. Comparative De Novo and Pan-Genome Analysis of MDR Nosocomial Bacteria Isolated from Hospitals in Jeddah, Saudi Arabia. Microorganisms 2023, 11, 2432. [Google Scholar] [CrossRef]
- Khan, A.; Khanzada, M.H.; Khan, K.; Jalal, K.; Uddin, R. Integrating core subtractive proteomics and reverse vaccinology for multi-epitope vaccine design against Rickettsia prowazekii endemic typhus. Immunol. Res. 2024, 72, 82–95. [Google Scholar] [CrossRef]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.G.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef]
- Khan, M.T.; Mahmud, A.; Islam, M.M.; Sumaia, M.S.N.; Rahim, Z.; Islam, K.; Iqbal, A. Multi-epitope vaccine against drug-resistant strains of Mycobacterium tuberculosis: A proteome-wide subtraction and immunoinformatics approach. Genom. Inf. 2023, 21, e42. [Google Scholar] [CrossRef] [PubMed]
- Maldonado, R.F.; Sá-Correia, I.; Valvano, M.A. Lipopolysaccharide modification in Gram-negative bacteria during chronic infection. FEMS Microbiol. Rev. 2016, 40, 480–493. [Google Scholar] [CrossRef] [PubMed]
- Botos, I.; Majdalani, N.; Mayclin, S.J.; McCarthy, J.G.; Lundquist, K.; Wojtowicz, D.; Barnard, T.J.; Gumbart, J.C.; Buchanan, S.K. Structural and Functional Characterization of the LPS Transporter LptDE from Gram-Negative Pathogens. Structure 2016, 24, 965–976. [Google Scholar] [CrossRef]
- Roumia, A.F.; Tsirigos, K.D.; Theodoropoulou, M.C.; Tamposis, I.A.; Hamodrakas, S.J.; Bagos, P.G. OMPdb: A Global Hub of Beta-Barrel Outer Membrane Proteins. Front. Bioinform. 2021, 1, 646581. [Google Scholar] [CrossRef] [PubMed]
- Malinverni, J.C.; Silhavy, T.J. Assembly of Outer Membrane β-Barrel Proteins: The Bam Complex. EcoSal Plus 2011, 4, 2. [Google Scholar] [CrossRef] [PubMed]
- Scheffers, D.-J.; Pinho, M.G. Bacterial Cell Wall Synthesis: New Insights from Localization Studies. Microbiol. Mol. Biol. Rev. 2005, 69, 585–607. [Google Scholar] [CrossRef]
- Silhavy, T.J.; Kahne, D.; Walker, S. The Bacterial Cell Envelope. Cold Spring Harb. Perspect. Biol. 2010, 2, a000414. [Google Scholar] [CrossRef]
- Murphy, S.G.; Murtha, A.N.; Zhao, Z.; Alvarez, L.; Diebold, P.; Shin, J.-H.; VanNieuwenhze, M.S.; Cava, F.; Dörr, T. Class A Penicillin-Binding Protein-Mediated Cell Wall Synthesis Promotes Structural Integrity during Peptidoglycan Endopeptidase Insufficiency in Vibrio cholerae. mBio 2021, 12, e03596-20. [Google Scholar] [CrossRef]
- Montaner, M.; Lopez-Argüello, S.; Oliver, A.; Moya, B. PBP Target Profiling by β-Lactam and β-Lactamase Inhibitors in Intact Pseudomonas aeruginosa: Effects of the Intrinsic and Acquired Resistance Determinants on the Periplasmic Drug Availability. Microbiol. Spectr. 2023, 11, e03038-22. [Google Scholar] [CrossRef]
- Munita, J.M.; Arias, C.A. Mechanisms of Antibiotic Resistance. Microbiol. Spectr. 2016, 4, 481–511. [Google Scholar] [CrossRef]
- Sethuvel, D.P.M.; Bakthavatchalam, Y.D.; Karthik, M.; Irulappan, M.; Shrivastava, R.; Periasamy, H.; Veeraraghavan, B. β-Lactam Resistance in ESKAPE Pathogens Mediated Through Modifications in Penicillin-Binding Proteins: An Overview. Infect. Dis. Ther. 2023, 12, 829–841. [Google Scholar] [CrossRef] [PubMed]
- Yao, J.; Zou, P.; Cui, Y.; Quan, L.; Gao, C.; Li, Z.; Gong, W.; Yang, M. Recent Advances in Strategies to Combat Bacterial Drug Resistance: Antimicrobial Materials and Drug Delivery Systems. Pharmaceutics 2023, 15, 1188. [Google Scholar] [CrossRef] [PubMed]
- Parvizpour, S.; Pourseif, M.M.; Razmara, J.; Rafi, M.A.; Omidi, Y. Epitope-based vaccine design: A comprehensive overview of bioinformatics approaches. Drug Discov. Today 2020, 25, 1034–1042. [Google Scholar] [CrossRef] [PubMed]
- Mir, S.A.; Alaidarous, M.; Alshehri, B.; Bin Dukhyil, A.A.; Banawas, S.; Madkhali, Y.; Alsagaby, S.A.; Al Othaim, A. Immunoinformatics-Based Identification of B and T Cell Epitopes in RNA-Dependent RNA Polymerase of SARS-CoV-2. Vaccines 2022, 10, 1660. [Google Scholar] [CrossRef]
- Zhuang, L.; Ali, A.; Yang, L.; Ye, Z.; Li, L.; Ni, R.; An, Y.; Ali, S.L.; Gong, W. Leveraging computer-aided design and artificial intelligence to develop a next-generation multi-epitope tuberculosis vaccine candidate. Infect. Med. 2024, 3, 100148. [Google Scholar] [CrossRef]
- Naorem, R.S.; Pangabam, B.D.; Bora, S.S.; Fekete, C.; Teli, A.B. Immunoinformatics Design of a Multiepitope Vaccine (MEV) Targeting Streptococcus mutans: A Novel Computational Approach. Pathogens 2024, 13, 916. [Google Scholar] [CrossRef]
- Stratmann, T. Cholera Toxin Subunit B as Adjuvant—An Accelerator in Protective Immunity and a Break in Autoimmunity. Vaccines 2015, 3, 579–596. [Google Scholar] [CrossRef]
- Wlodawer, A. Stereochemistry and Validation of Macromolecular Structures. In Protein Crystallography; Methods in Molecular Biology; Wlodawer, A., Dauter, Z., Jaskolski, M., Eds.; Springer: New York, NY, USA, 2017; Volume 1607, pp. 595–610. ISBN 978-1-4939-6998-2. [Google Scholar]
- AlChalabi, R.; Al-Rahim, A.; Omer, D.; Suleiman, A.A. Immunoinformatics design of multi-epitope peptide-based vaccine against Haemophilus influenzae strain using cell division protein. Netw. Model. Anal. Health Inf. Bioinform. 2022, 12, 1. [Google Scholar] [CrossRef]
- Ayyagari, V.S.; Venkateswarulu, T.C.; Karlapudi, A.P.; Srirama, K. Design of a Multi-Epitope-Based Vaccine Targeting M-Protein of SARS-CoV2: An Immunoinformatics Approach. J. Biomol. Struct. Dyn. 2022, 40, 2963–2977. [Google Scholar] [CrossRef]
- Dowling, J.K.; Mansell, A. Toll-like receptors: The swiss army knife of immunity and vaccine development. Clin. Trans. Immunol. 2016, 5, e85. [Google Scholar] [CrossRef]
- Kang, S.-M.; Compans, R.W. Host Responses from Innate to Adaptive Immunity after Vaccination: Molecular and Cellular Events. Mol. Cells 2009, 27, 5–14. [Google Scholar] [CrossRef]
- Rasool, D.; Jan, S.A.; Khan, S.U.; Nahid, N.; Ashfaq, U.A.; Umar, A.; Qasim, M.; Noor, F.; Rehman, A.; Shahzadi, K.; et al. Subtractive proteomics-based vaccine targets annotation and reverse vaccinology approaches to identify multiepitope vaccine against Plesiomonas shigelloides. Heliyon 2024, 10, e31304. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, M.; Sharma, A.R.; Patra, P.; Ghosh, P.; Sharma, G.; Patra, B.C.; Saha, R.P.; Lee, S.-S.; Chakraborty, C. A SARS-CoV-2 vaccine candidate: In-silico cloning and validation. Inform. Med. Unlocked 2020, 20, 100394. [Google Scholar] [CrossRef]
- Kitts, P.A.; Church, D.M.; Thibaud-Nissen, F.; Choi, J.; Hem, V.; Sapojnikov, V.; Smith, R.G.; Tatusova, T.; Xiang, C.; Zherikov, A.; et al. Assembly: A resource for assembled genomes at NCBI. Nucleic Acids Res. 2016, 44, D73–D80. [Google Scholar] [CrossRef]
- Mazumder, R.; Abdullah, A.; Hussain, A.; Ahmed, D.; Mondal, D. Draft Genome Sequence of Chromobacterium violaceum RDN09, Isolated from a Patient with a Wound Infection in Bangladesh. Microbiol. Resour. Announc. 2020, 9, e00957-20. [Google Scholar] [CrossRef] [PubMed]
- Dereeper, A.; Summo, M.; Meyer, D.F. PanExplorer: A web-based tool for exploratory analysis and visualization of bacterial pan-genomes. Bioinformatics 2022, 38, 4412–4414. [Google Scholar] [CrossRef] [PubMed]
- Navasca, A.; Singh, J.; Rivera-Varas, V.; Gill, U.; Secor, G.; Baldwin, T. Dispensable genome and segmental duplications drive the genome plasticity in Fusarium solani. Front. Fungal Biol. 2025, 6, 1432339. [Google Scholar] [CrossRef]
- O’Leary, N.A.; Wright, M.W.; Brister, J.R.; Ciufo, S.; Haddad, D.; McVeigh, R.; Rajput, B.; Robbertse, B.; Smith-White, B.; Ako-Adjei, D.; et al. Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016, 44, D733–D745. [Google Scholar] [CrossRef]
- Cock, P.J.A.; Antao, T.; Chang, J.T.; Chapman, B.A.; Cox, C.J.; Dalke, A.; Friedberg, I.; Hamelryck, T.; Kauff, F.; Wilczynski, B.; et al. Biopython: Freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 2009, 25, 1422–1423. [Google Scholar] [CrossRef]
- Acar, M.B.; Ayaz-Güner, Ş.; Güner, H.; Dinç, G.; Ulu Kılıç, A.; Doğanay, M.; Özcan, S. A subtractive proteomics approach for the identification of immunodominant Acinetobacter baumannii vaccine candidate proteins. Front. Immunol. 2022, 13, 1001633. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Ning, L.-W.; Ye, Y.-N.; Guo, F.-B. Geptop: A Gene Essentiality Prediction Tool for Sequenced Bacterial Genomes Based on Orthology and Phylogeny. PLoS ONE 2013, 8, e72343. [Google Scholar] [CrossRef] [PubMed]
- Wen, Q.-F.; Liu, S.; Dong, C.; Guo, H.-X.; Gao, Y.-Z.; Guo, F.-B. Geptop 2.0: An Updated, More Precise, and Faster Geptop Server for Identification of Prokaryotic Essential Genes. Front. Microbiol. 2019, 10, 1236. [Google Scholar] [CrossRef] [PubMed]
- Yu, N.Y.; Wagner, J.R.; Laird, M.R.; Melli, G.; Rey, S.; Lo, R.; Dao, P.; Sahinalp, S.C.; Ester, M.; Foster, L.J.; et al. PSORTb 3.0: Improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 2010, 26, 1608–1615. [Google Scholar] [CrossRef]
- Krogh, A.; Larsson, B.; Von Heijne, G.; Sonnhammer, E.L.L. Predicting transmembrane protein topology with a hidden markov model: Application to complete genomes11Edited by F. Cohen. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef]
- Ong, E.; Cooke, M.F.; Huffman, A.; Xiang, Z.; Wong, M.U.; Wang, H.; Seetharaman, M.; Valdez, N.; He, Y. Vaxign2: The second generation of the first Web-based vaccine design program using reverse vaccinology and machine learning. Nucleic Acids Res. 2021, 49, W671–W678. [Google Scholar] [CrossRef]
- Vita, R.; Mahajan, S.; Overton, J.A.; Dhanda, S.K.; Martini, S.; Cantrell, J.R.; Wheeler, D.K.; Sette, A.; Peters, B. The Immune Epitope Database (IEDB): 2018 update. Nucleic Acids Res. 2019, 47, D339–D343. [Google Scholar] [CrossRef]
- Kim, Y.; Ponomarenko, J.; Zhu, Z.; Tamang, D.; Wang, P.; Greenbaum, J.; Lundegaard, C.; Sette, A.; Lund, O.; Bourne, P.E.; et al. Immune epitope database analysis resource. Nucleic Acids Res. 2012, 40, W525–W530. [Google Scholar] [CrossRef]
- Calis, J.J.A.; Maybeno, M.; Greenbaum, J.A.; Weiskopf, D.; De Silva, A.D.; Sette, A.; Keşmir, C.; Peters, B. Properties of MHC Class I Presented Peptides That Enhance Immunogenicity. PLoS Comput. Biol. 2013, 9, e1003266. [Google Scholar] [CrossRef]
- Doytchinova, I.A.; Flower, D.R. VaxiJen: A server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinform. 2007, 8, 4. [Google Scholar] [CrossRef]
- Dimitrov, I.; Bangov, I.; Flower, D.R.; Doytchinova, I. AllerTOP v.2—A server for in silico prediction of allergens. J. Mol. Model. 2014, 20, 2278. [Google Scholar] [CrossRef] [PubMed]
- Paul, S.; Weiskopf, D.; Angelo, M.A.; Sidney, J.; Peters, B.; Sette, A. HLA Class I Alleles Are Associated with Peptide-Binding Repertoires of Different Size, Affinity, and Immunogenicity. J. Immunol. 2013, 191, 5831–5839. [Google Scholar] [CrossRef] [PubMed]
- Saha, S.; Raghava, G.P.S. Prediction of continuous B-cell epitopes in an antigen using recurrent neural network. Proteins Struct. Funct. Bioinform. 2006, 65, 40–48. [Google Scholar] [CrossRef] [PubMed]
- Bui, H.-H.; Sidney, J.; Dinh, K.; Southwood, S.; Newman, M.J.; Sette, A. Predicting population coverage of T-cell epitope-based diagnostics and vaccines. BMC Bioinform. 2006, 7, 153. [Google Scholar] [CrossRef]
- Greenbaum, J.; Sidney, J.; Chung, J.; Brander, C.; Peters, B.; Sette, A. Functional classification of class II human leukocyte antigen (HLA) molecules reveals seven different supertypes and a surprising degree of repertoire sharing across supertypes. Immunogenetics 2011, 63, 325–335. [Google Scholar] [CrossRef]
- Khatoon, N.; Pandey, R.K.; Prajapati, V.K. Exploring Leishmania secretory proteins to design B and T cell multi-epitope subunit vaccine using immunoinformatics approach. Sci. Rep. 2017, 7, 8285. [Google Scholar] [CrossRef]
- Kruppa, J.; Jo, W.K.; Van Der Vries, E.; Ludlow, M.; Osterhaus, A.; Baumgaertner, W.; Jung, K. Virus detection in high-throughput sequencing data without a reference genome of the host. Infect. Genet. Evol. 2018, 66, 180–187. [Google Scholar] [CrossRef]
- Semple, F.; Dorin, J.R. β-Defensins: Multifunctional Modulators of Infection, Inflammation and More? J. Innate Immun. 2012, 4, 337–348. [Google Scholar] [CrossRef]
- Funderburg, N.; Lederman, M.M.; Feng, Z.; Drage, M.G.; Jadlowsky, J.; Harding, C.V.; Weinberg, A.; Sieg, S.F. Human β-defensin-3 activates professional antigen-presenting cells via Toll-like receptors 1 and 2. Proc. Natl. Acad. Sci. USA 2007, 104, 18631–18635. [Google Scholar] [CrossRef]
- Pandey, R.K.; Bhatt, T.K.; Prajapati, V.K. Novel Immunoinformatics Approaches to Design Multi-epitope Subunit Vaccine for Malaria by Investigating Anopheles Salivary Protein. Sci. Rep. 2018, 8, 1125. [Google Scholar] [CrossRef]
- Galanis, K.A.; Nastou, K.C.; Papandreou, N.C.; Petichakis, G.N.; Pigis, D.G.; Iconomidou, V.A. Linear B-Cell Epitope Prediction for In Silico Vaccine Design: A Performance Review of Methods Available via Command-Line Interface. Int. J. Gen. Med. 2021, 22, 3210. [Google Scholar] [CrossRef]
- Panda, A.; Kapoor, J.; Hareramadas, B.; Naqvi, I.; Ganta, S.; Chhabra, R.; Kumar, S.; Bandyopadhyay, A. Design of a Multi-Epitope Vaccine using β-barrel Outer Membrane Proteins Identified in Chlamydia trachomatis. J. Membr. Biol. 2025, 6, 473–496. [Google Scholar] [CrossRef] [PubMed]
- Garg, V.K.; Avashthi, H.; Tiwari, A.; Jain, P.A.; Ramkete, P.W.R.; Kayastha, A.M.; Singh, V.K. MFPPI—Multi FASTA ProtParam Interface. Bioinformation 2016, 12, 74–77. [Google Scholar] [CrossRef]
- Geourjon, C.; Deléage, G. SOPMA: Significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Bioinformatics 1995, 11, 681–684. [Google Scholar] [CrossRef] [PubMed]
- Bertoline, L.M.F.; Lima, A.N.; Krieger, J.E.; Teixeira, S.K. Before and after AlphaFold2: An overview of protein structure prediction. Front. Bioinform. 2023, 3, 1120370. [Google Scholar] [CrossRef] [PubMed]
- Heo, L.; Park, H.; Seok, C. GalaxyRefine: Protein structure refinement driven by side-chain repacking. Nucleic Acids Res. 2013, 41, W384–W388. [Google Scholar] [CrossRef]
- Wang, W.; Xia, M.; Chen, J.; Deng, F.; Yuan, R.; Zhang, X.; Shen, F. Data set for phylogenetic tree and RAMPAGE Ramachandran plot analysis of SODs in Gossypium raimondii and G. arboreum. Data Brief 2016, 9, 345–348. [Google Scholar] [CrossRef]
- Binbay, F.A.; Rathod, D.C.; George, A.A.P.; Imhof, D. Quality Assessment of Selected Protein Structures Derived from Homology Modeling and AlphaFold. Pharmaceuticals 2023, 16, 1662. [Google Scholar] [CrossRef]
- Ponomarenko, J.; Bui, H.-H.; Li, W.; Fusseder, N.; Bourne, P.E.; Sette, A.; Peters, B. ElliPro: A new structure-based tool for the prediction of antibody epitopes. BMC Bioinform. 2008, 9, 514. [Google Scholar] [CrossRef]
- Ain, Q.U.; Batool, M.; Choi, S. TLR4-Targeting Therapeutics: Structural Basis and Computer-Aided Drug Discovery Approaches. Molecules 2020, 25, 627. [Google Scholar] [CrossRef]
- Kozakov, D.; Hall, D.R.; Xia, B.; Porter, K.A.; Padhorny, D.; Yueh, C.; Beglov, D.; Vajda, S. The ClusPro web server for protein–protein docking. Nat. Protoc. 2017, 12, 255–278. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; Jabłońska, J.; Pravda, L.; Vařeková, R.S.; Thornton, J.M. PDBsum: Structural summaries of PDB entries. Protein Sci. 2018, 27, 129–134. [Google Scholar] [CrossRef] [PubMed]
- López-Blanco, J.R.; Aliaga, J.I.; Quintana-Ortí, E.S.; Chacón, P. iMODS: Internal coordinates normal mode analysis server. Nucleic Acids Res. 2014, 42, W271–W276. [Google Scholar] [CrossRef] [PubMed]
- Bahar, I.; Lezon, T.R.; Bakan, A.; Shrivastava, I.H. Normal Mode Analysis of Biomolecular Structures: Functional Mechanisms of Membrane Proteins. Chem. Rev. 2010, 110, 1463–1497. [Google Scholar] [CrossRef] [PubMed]
- Abraham, M.J.; Gready, J.E. Optimization of parameters for molecular dynamics simulation using smooth particle-mesh Ewald in GROMACS 4.5. J. Comput. Chem. 2011, 32, 2031–2040. [Google Scholar] [CrossRef]
- Giese, T.J.; Panteva, M.T.; Chen, H.; York, D.M. Multipolar Ewald Methods, 1: Theory, Accuracy, and Performance. J. Chem. Theory Comput. 2015, 11, 436–450. [Google Scholar] [CrossRef]
- Lemkul, J.A. Introductory Tutorials for Simulating Protein Dynamics with GROMACS. J. Phys. Chem. B 2024, 128, 9418–9435. [Google Scholar] [CrossRef]
- Hunt-Isaak, I.; Russell, J.; Hekstra, D. mpl-interactions: A Python Package for InteractiveMatplotlib Figures. J. Open Source Softw. 2024, 9, 5651. [Google Scholar] [CrossRef]
- AlMalki, F. In Silico Subtractive Proteome Analysis to Design Multi-Epitope-Based Subunit Vaccine against Eikenella corrodens. J. Microbiol. Biotechnol. 2024, 35, e2410015. [Google Scholar] [CrossRef]
- Choudhury, A.; Sen Gupta, P.S.; Panda, S.K.; Rana, M.K.; Mukherjee, S. Designing AbhiSCoVac—A single potential vaccine for all ‘corona culprits’: Immunoinformatics and immune simulation approaches. J. Mol. Liq. 2022, 351, 118633. [Google Scholar] [CrossRef]
- Grote, A.; Hiller, K.; Scheer, M.; Munch, R.; Nortemann, B.; Hempel, D.C.; Jahn, D. JCat: A novel tool to adapt codon usage of a target gene to its potential expression host. Nucleic Acids Res. 2005, 33, W526–W531. [Google Scholar] [CrossRef]




| Strain Name | Country | Continent | Organism |
|---|---|---|---|
| Chromobacterium violaceum ATCC 12472 | – | – | C. violaceum ATCC 12472 |
| Chromobacterium violaceum CV1192 | Brazil | South America | C. violaceum |
| Chromobacterium violaceum CV1197 | – | – | C. violaceum |
| Chromobacterium violaceum CV20 | – | – | C. violaceum |
| Chromobacterium violaceum FDAARGOS_1273 | USA | North America | C. violaceum |
| Chromobacterium violaceum FDAARGOS_1274 | USA | North America | C. violaceum |
| Chromobacterium violaceum FDAARGOS_635 | USA | North America | C. violaceum |
| Chromobacterium violaceum NCTC9695_assembly | – | – | C. violaceum |
| Accession No. | Protein | Antigenicity | Allergenicity | Toxicity |
|---|---|---|---|---|
| WP_232514932.1 | penicillin-binding protein 1A | 0.4664 | Non-allergen | Non-toxin |
| VEB45604.1 | Organic solvent tolerance protein | 0.6948 | Non-allergen | Non-toxin |
| Epitope | Protein | Allele | Position | Antigenicity | Immunogenicity |
|---|---|---|---|---|---|
| YGYGGTAALPIW | penicillin-binding protein 1A | HLA-C*03:03 HLA-C*12:03 HLA-B*58:01 | 270–281 | 1.2146 | 0.23786 |
| GQYVAEMVRQAM | penicillin-binding protein 1A | HLA-C*14:02 | 246–357 | 0.9856 | 0.01896 |
| GRYGYGGTAALP | penicillin-binding protein 1A | HLA-C*14:02 HLA-C*03:03 HLA-B*48:01 | 668–679 | 0.9568 | 0.19647 |
| GWQGGGGNVTLR | Organic solvent tolerance protein | HLA-B*38:01 HLA-B*48:01 | 312–323 | 2.4988 | 0.15434 |
| LGWQGGGGNVTL | Organic solvent tolerance protein | HLA-B*38:01 HLA-B*48:01 | 311–322 | 1.9613 | 0.13877 |
| PILYSPWLDFPL | Organic solvent tolerance protein | HLA-A*24:02 HLA-E*01:01 | 165–176 | 1.8126 | 0.13888 |
| Epitope | Protein | Allele | Position | Antigenicity | Immunogenicity |
|---|---|---|---|---|---|
| GRYGYGGTAALPIWI | penicillin-binding protein 1A | HLA-DRB1*07:03 | 668–682 | 1.5906 | 0.4298 |
| LRIDNQGTVPGGEGD | penicillin-binding protein 1A | HLA-DRB1*11:07 HLA-DRB1*03:09 HLA-DRB1*03:05 | 729–743 | 1.5661 | 0.21582 |
| WLNYALGWQGGGGNV | Organic solvent tolerance protein | HLA-DRB1*08:01 | 306–320 | 1.3753 | 0.2801 |
| QGQNQYRVYGSRMTT | Organic solvent tolerance protein | HLA-DRB1*15:02 HLA-DRB1*15:01 | 114–128 | 1.0514 | −0.27841 |
| Epitope | Protein | Score | Position | Antigenicity | Immunogenicity |
|---|---|---|---|---|---|
| DLAGKTGTTSDWKDAW | penicillin-binding protein 1A | 0.85 | 674 | 1.2111 | −0.10202 |
| RVKAGDRFRMTRGGDV | Organic solvent tolerance protein | 0.81 | 87 | 1.6479 | 0.22943 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Allemailem, K.S. Pangenome-Guided Reverse Vaccinology and Immunoinformatics Approach for Rational Design of a Multi-Epitope Subunit Vaccine Candidate Against the Multidrug-Resistant Pathogen Chromobacterium violaceum: A Computational Immunopharmacology Perspective. Pharmaceuticals 2026, 19, 29. https://doi.org/10.3390/ph19010029
Allemailem KS. Pangenome-Guided Reverse Vaccinology and Immunoinformatics Approach for Rational Design of a Multi-Epitope Subunit Vaccine Candidate Against the Multidrug-Resistant Pathogen Chromobacterium violaceum: A Computational Immunopharmacology Perspective. Pharmaceuticals. 2026; 19(1):29. https://doi.org/10.3390/ph19010029
Chicago/Turabian StyleAllemailem, Khaled S. 2026. "Pangenome-Guided Reverse Vaccinology and Immunoinformatics Approach for Rational Design of a Multi-Epitope Subunit Vaccine Candidate Against the Multidrug-Resistant Pathogen Chromobacterium violaceum: A Computational Immunopharmacology Perspective" Pharmaceuticals 19, no. 1: 29. https://doi.org/10.3390/ph19010029
APA StyleAllemailem, K. S. (2026). Pangenome-Guided Reverse Vaccinology and Immunoinformatics Approach for Rational Design of a Multi-Epitope Subunit Vaccine Candidate Against the Multidrug-Resistant Pathogen Chromobacterium violaceum: A Computational Immunopharmacology Perspective. Pharmaceuticals, 19(1), 29. https://doi.org/10.3390/ph19010029








