Profiling the Complexity of Resistance Factors in Cancer Cells Towards Berberine and Its Derivatives
Abstract
1. Introduction
2. Results
2.1. Cytotoxicity of Berberine Chloride as Determined by the Resazurin Reduction Assay
2.2. Drug Resistance Profiling of Berberine Chloride
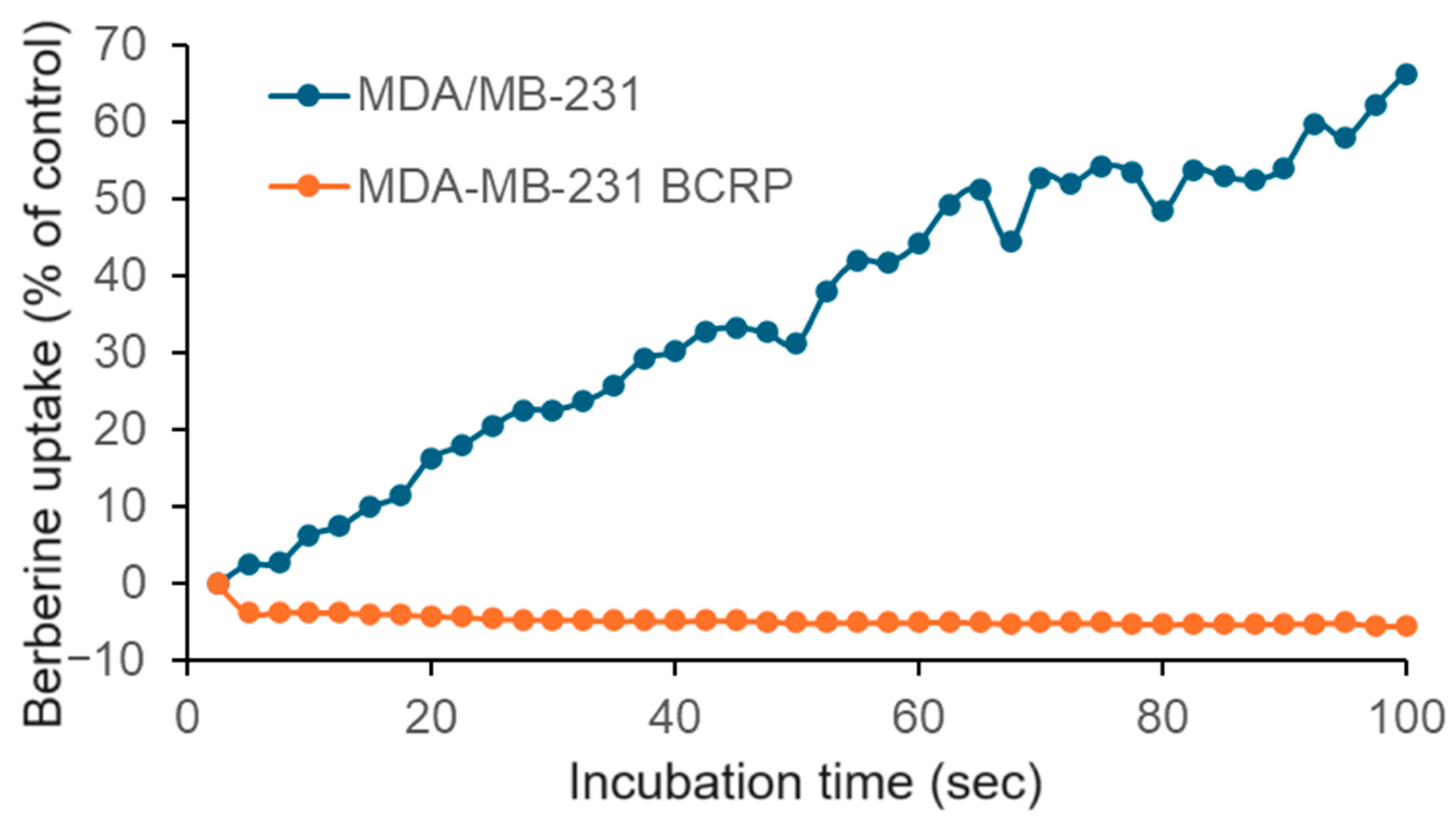
2.3. Berberine Chloride Uptake Measured by Live Cell Microscopy
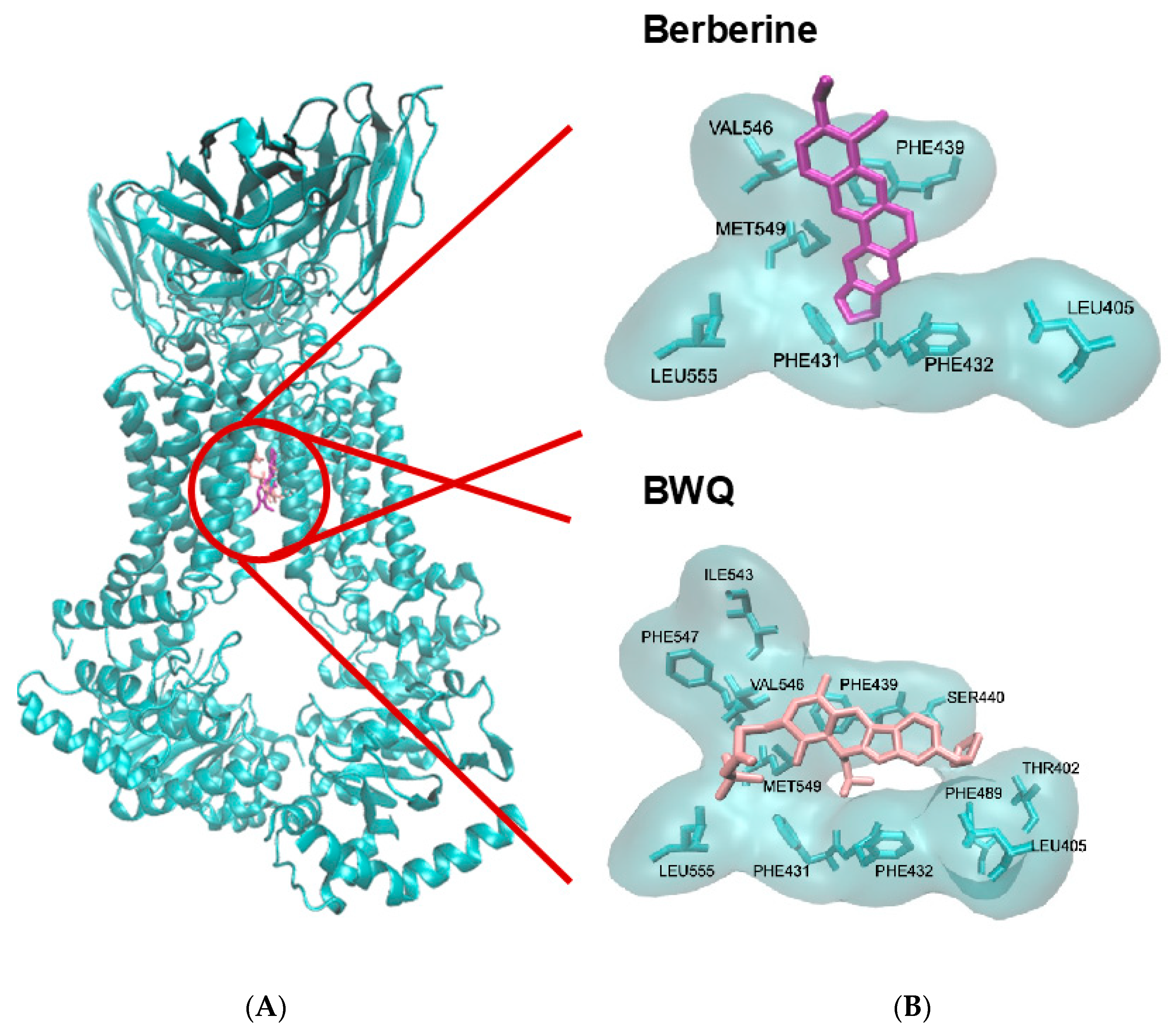
2.4. Molecular Docking of Berberine to BCRP
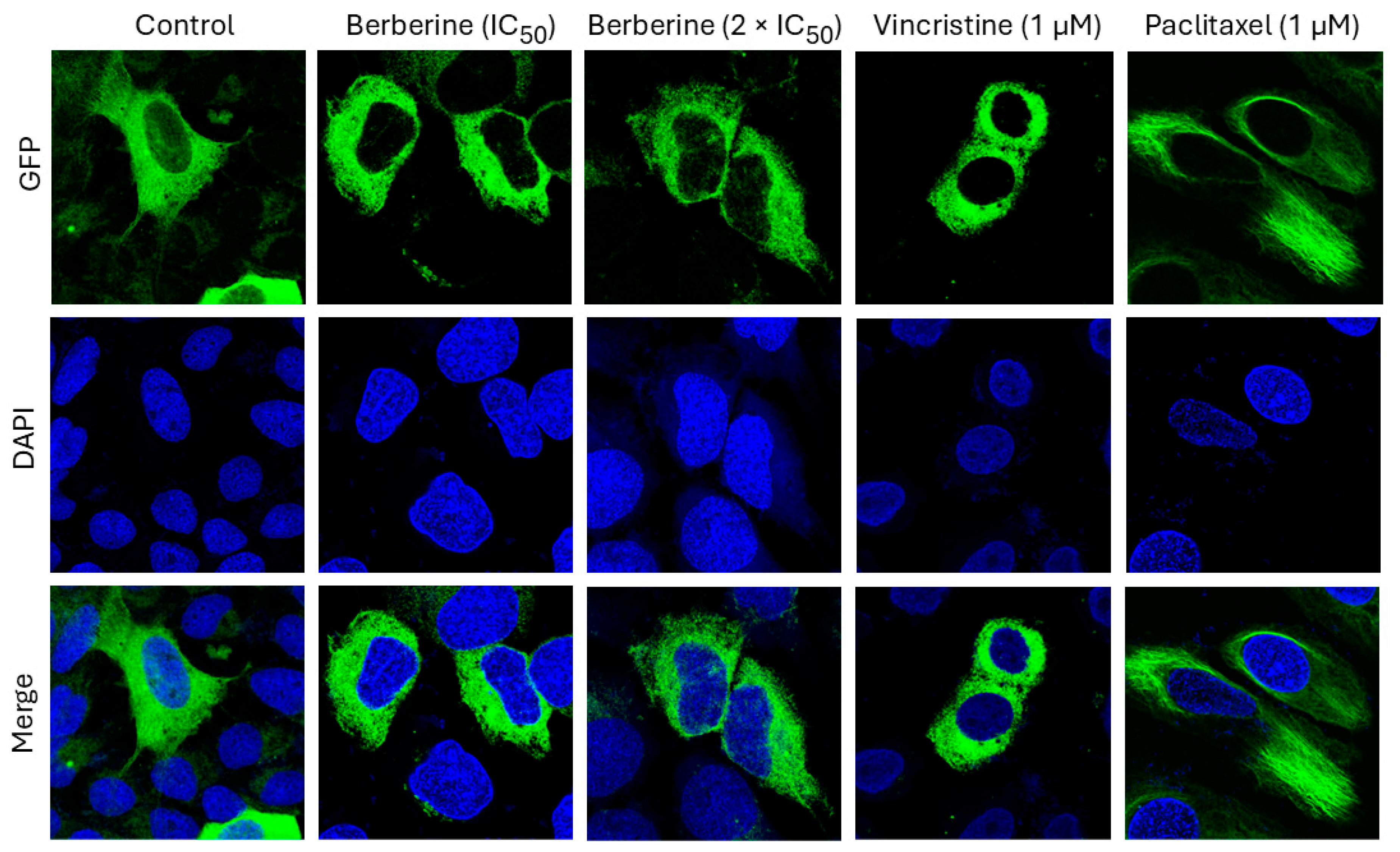
2.5. Effect of Berberine Chloride on Microtubules
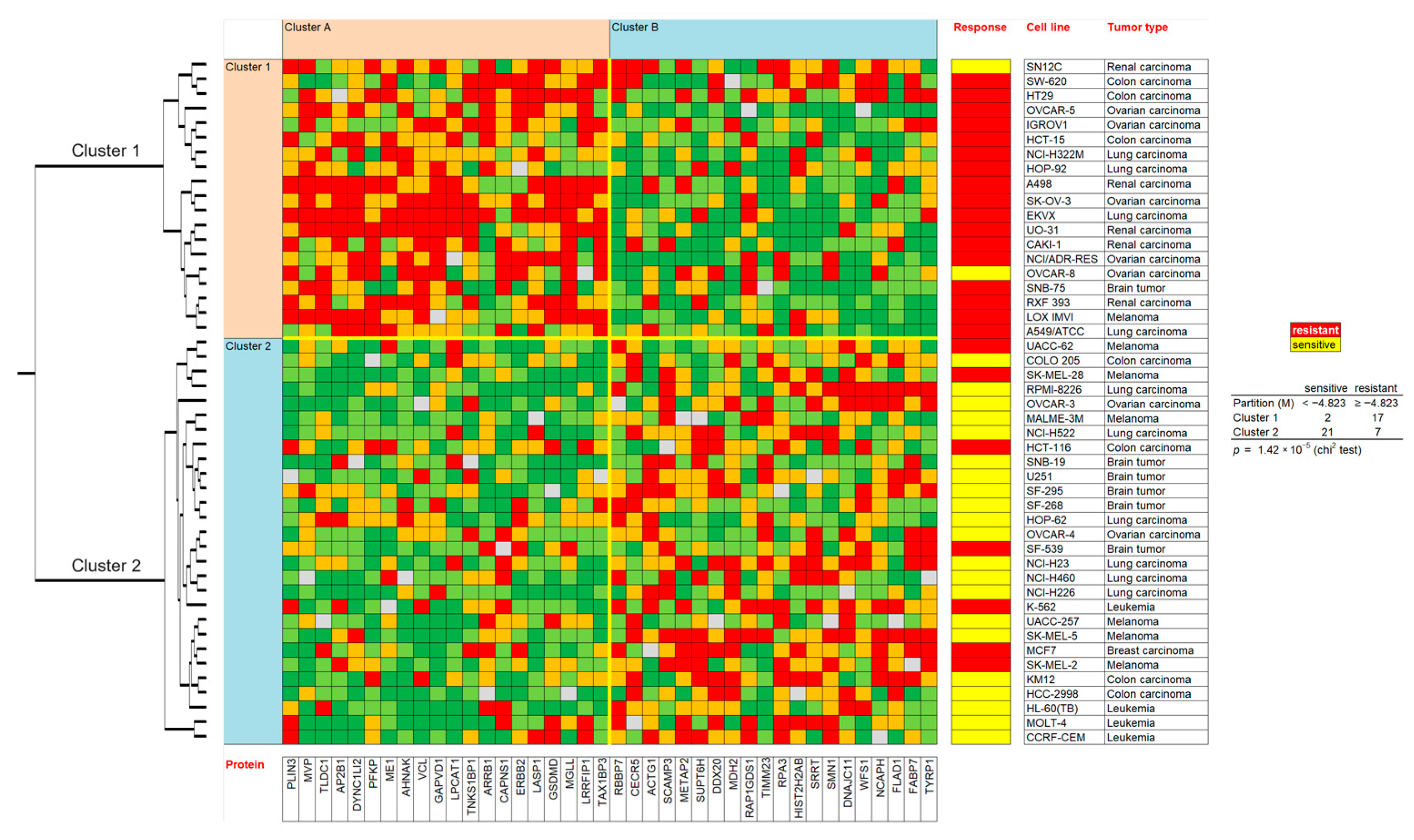
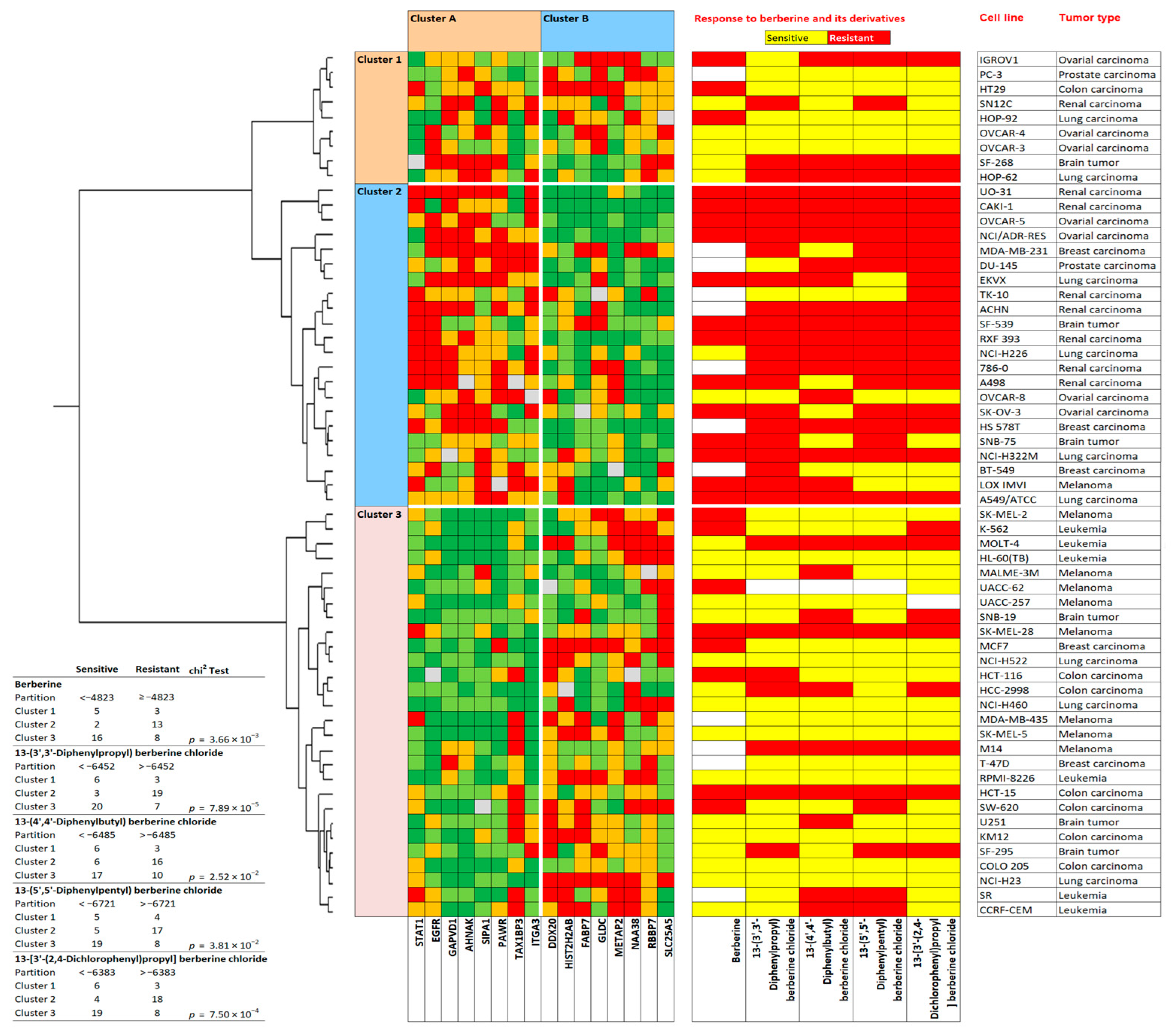
2.6. Prediction of Sensitivity and Resistance to Berberine by Proteomic Profiling
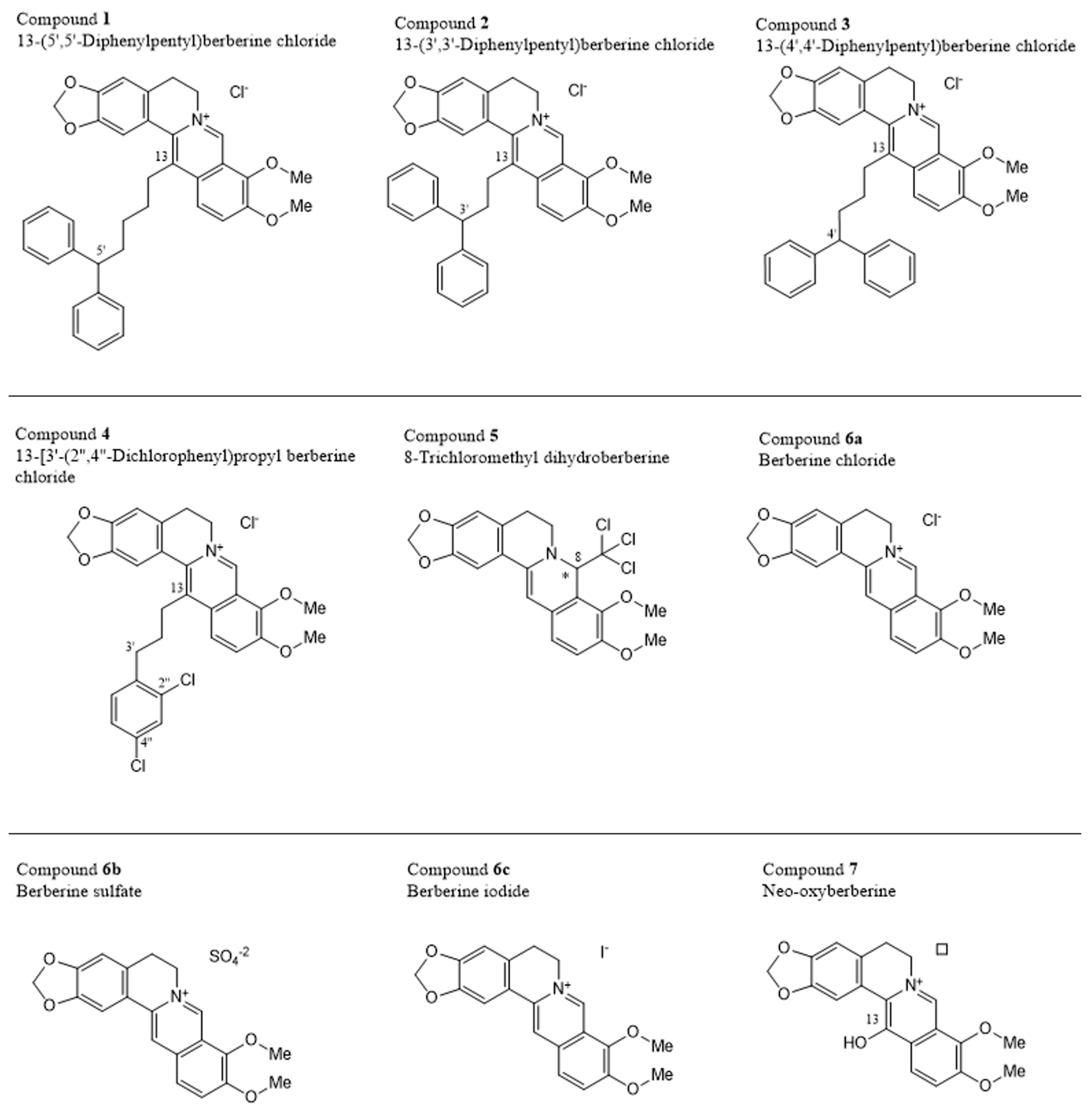
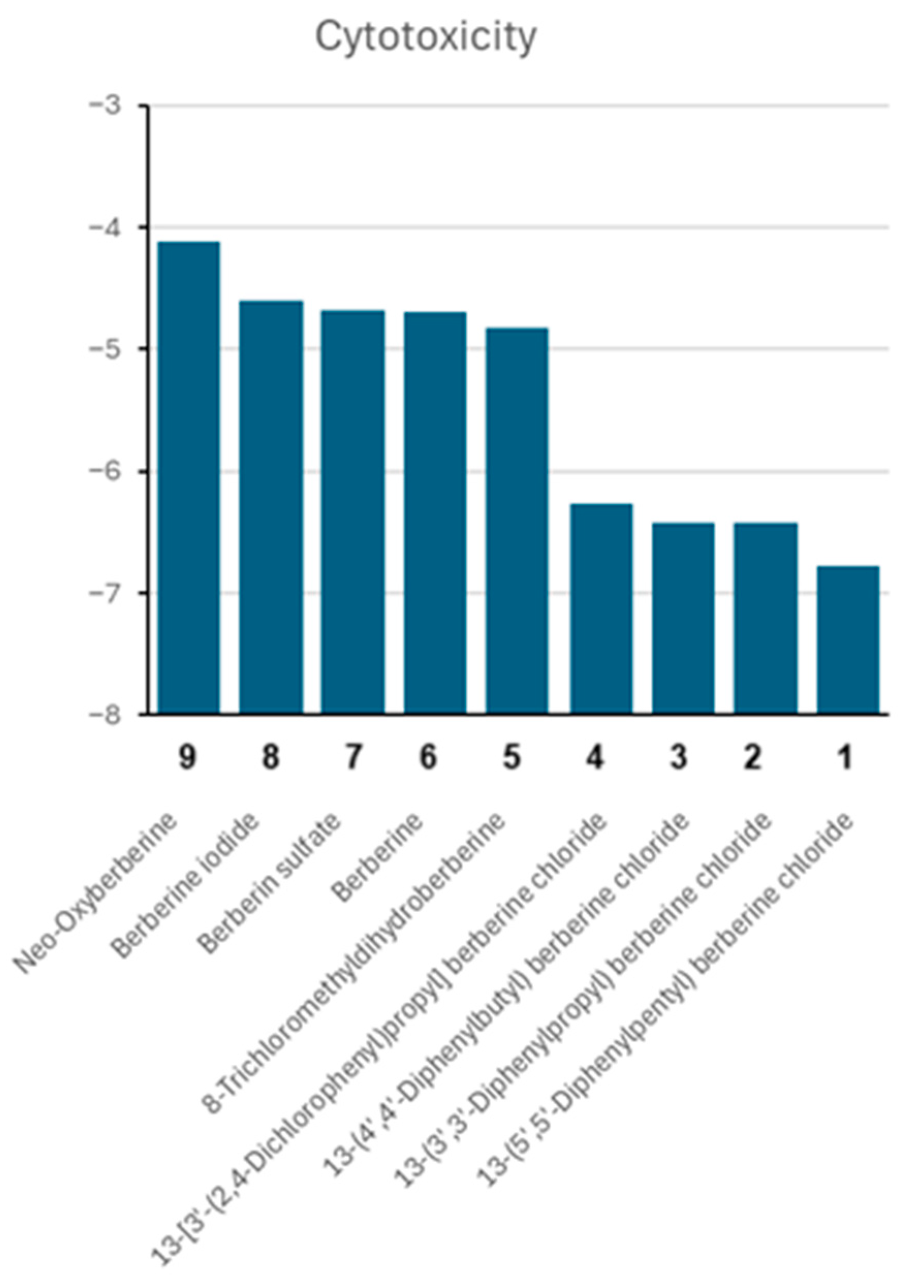
2.7. Cytotoxicity of Berberine Derivatives
2.8. Structure-Activity Relationships of Berberine Derivatives
2.9. Prediction of Sensitivity and Resistance to Berberine Derivatives by Proteomic Profiling
3. Discussion
4. Materials and Methods
4.1. Cell Lines
4.2. Resazurin Reduction Assay
4.3. Hierarchical Cluster Analyses of Proteomic Expression Profiles
4.4. Molecular Docking with AutoDock 1.5.7
4.5. Berberine Uptake by Live-Cell Microscopy
4.6. Imaging of Structure and Dynamics of the Microtubule Cytoskeleton by Fluorescence Microscopy
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ABCG2 | ATP-binding cassette sub-family G member 2 |
| AMPK | AMP-activated protein kinase |
| GPX | Glutathione peroxidase |
| DAPI | 4′,6-Diamidino-2-phenylindole |
| DDT | Dichlorodiphenyltrichloroethane |
| GFP | Green fluorescent protein |
| dlg | Docking log |
| FOXO1 | Forkhead box O1 |
| NCI | National Cancer Institute |
| Nrf2 | Nuclear factor erythroid 2-related factor 2 |
| mTOR | Mammalian target of rapamycin |
| NQOq | NAD(P)H quinone oxidoreductase 1 |
| pKi | Predicted inhibition constant |
| pdb | Protein Data Bank |
| SIRT1 | Sirtuin |
| PIK3CA | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit α |
| TCM | Traditional Chinese medicine |
| TUBA | α-tubulin |
| WT1 | Wilms’ tumor gene |
| VMD | Visual Molecular Dynamics |
References
- World Health Organization (WHO). Available online: https://www.who.int/news-room/fact-sheets/detail/cancer (accessed on 16 March 2025).
- Newman, D.J.; Cragg, G.M. Natural products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019. J. Nat. Prod. 2020, 83, 770–803. [Google Scholar] [CrossRef]
- Cushnie, T.P.T.; Cushnie, B.; Lamb, A.J. Alkaloids: An overview of their antibacterial, antibiotic-enhancing and antivirulence activities. Int. J. Antimicrob. Agents 2014, 44, 377–386. [Google Scholar] [CrossRef]
- Daley, S.K.; Cordell, G.A. Alkaloids in contemporary drug discovery to meet global disease needs. Molecules 2021, 26, 3800. [Google Scholar] [CrossRef]
- Olofinsan, K.; Abrahamse, H.; George, B.P. Therapeutic role of alkaloids and alkaloid derivatives in cancer management. Molecules 2023, 28, 5578. [Google Scholar] [CrossRef]
- Birdsall, T. Berberine: Therapeutic potential of an alkaloid found in several medicinal plants. Altern. Med. Rev. 1997, 2, 94–103. [Google Scholar]
- Yin, J.; Zhang, H.; Ye, J. Traditional Chinese medicine in treatment of metabolic syndrome. Endocr. Metab. Immune Disord. Drug Targets 2008, 8, 99–111. [Google Scholar] [CrossRef]
- Gu, Y.; Zhang, Y.; Shi, X.; Li, X.; Hong, J.; Chen, J.; Gu, W.; Lu, X.; Xu, G.; Ning, G. Effect of traditional Chinese medicine berberine on type 2 diabetes based on comprehensive metabonomics. Talanta 2010, 81, 766–772. [Google Scholar] [CrossRef]
- Kumar, A.; Ekavali Chopra, K.; Mukherjee, M.; Pottabathini, R.; Dhull, D.K. Current knowledge and pharmacological profile of berberine: An update. Eur. J. Pharmacol. 2015, 761, 288–297. [Google Scholar] [CrossRef]
- Battu, S.K.; Repka, M.A.; Maddineni, S.; Chittiboyina, A.G.; Avery, M.A.; Majumdar, S. Physicochemical characterization of berberine chloride: A perspective in the development of a solution dosage form for oral delivery. AAPS PharmSciTech 2010, 11, 1466–1475. [Google Scholar] [CrossRef]
- Ye, Y.; Liu, X.; Wu, N.; Han, Y.; Wang, J.; Yu, Y.; Chen, Q. Efficacy and safety of berberine alone for several metabolic disorders: A systematic review and meta-analysis of randomized clinical trials. Front. Pharmacol. 2021, 12, 653887. [Google Scholar] [CrossRef]
- Fang, S.; Guo, S.; Du, S.; Cao, Z.; Yang, Y.; Su, X.; Wei, W. Efficacy and safety of berberine in preventing recurrence of colorectal adenomas: A systematic review and meta-analysis. J. Ethnopharmacol. 2022, 282, 114617. [Google Scholar] [CrossRef]
- Och, A.; Och, M.; Nowak, R.; Podgórska, D.; Podgórski, R. Berberine, a herbal metabolite in the metabolic syndrome: The risk factors, course, and consequences of the disease. Molecules 2022, 27, 1351. [Google Scholar] [CrossRef]
- Pu, Z.; Wen, H.; Jiang, H.; Hou, Q.; Yan, H. Berberine improves negative symptoms and cognitive function in patients with chronic schizophrenia via anti-inflammatory effect: A randomized clinical trial. Chin. Med. 2023, 18, 41. [Google Scholar] [CrossRef]
- Almatroodi, S.A.; Alsahli, M.A.; Rahmani, A.H. Berberine: An important emphasis on its anticancer effects through modulation of various cell signaling pathways. Molecules 2022, 27, 5889. [Google Scholar] [CrossRef]
- Xiong, R.G.; Huang, S.Y.; Wu, S.X.; Zhou, D.D.; Yang, Z.J.; Saimaiti, A.; Zhao, C.N.; Shang, A.; Zhang, Y.J.; Gan, R.Y.; et al. Anticancer effects and mechanisms of berberine from medicinal herbs: An update review. Molecules 2022, 27, 4523. [Google Scholar] [CrossRef]
- Jackson, S.M.; Manolardis, I.; Kowal, J.; Zechner, M.; Altmann, K.H.; Locher, K.P. Structure of inhibitor-bound ABCG2. Nat. Struct. Mol. Biol. 2018, 25, 333–340. [Google Scholar] [CrossRef]
- Raghav, D.; Ashraf, S.M.; Mohan, L.; Rathinasamy, K. Berberine induces toxicity in hela cells through perturbation of microtubule polymerization by binding to tubulin at a unique site. Biochemistry 2017, 56, 2594–2611. [Google Scholar] [CrossRef]
- Holohan, C.; Van Schaeybroeck, S.; Longley, D.B.; Johnston, P.G. Cancer drug resistance: An evolving paradigm. Nat. Rev. Cancer 2013, 13, 714–726. [Google Scholar] [CrossRef]
- Gottesman, M.M.; Lavi, O.; Hall, M.D.; Gillet, J.P. Toward a better understanding of the complexity of cancer drug resistance. Annu. Rev. Pharmacol. Toxicol. 2016, 56, 85–102. [Google Scholar] [CrossRef]
- Mansoori, B.; Mohammadi, A.; Davudian, S.; Shirjang, S.; Baradaran, B. The different mechanisms of cancer drug resistance: A brief review. Adv. Pharm. Bull. 2017, 7, 339–348. [Google Scholar] [CrossRef]
- Yao, Y.; Chen, H.; Yan, L.; Wang, W.; Wang, D. Berberine alleviates type 2 diabetic symptoms by altering gut microbiota and reducing aromatic amino acids. Biomed. Pharmacother. 2020, 131, 110669. [Google Scholar] [CrossRef]
- Pan, L.; Zhai, X.; Duan, Z.; Xu, K.; Liu, G. Systematic review and meta-analysis of Coptis chinensis Franch.-containing traditional Chinese medicine as an adjunct therapy to metformin in the treatment of type 2 diabetes mellitus. Front. Pharmacol. 2022, 13, 956313. [Google Scholar] [CrossRef]
- Ma, L.; Bai, Y.; Liu, J.; Gong, K.; He, Q.; Zhao, J.; Suo, Y.; Wang, W.; Chen, G.; Lu, Z. The therapeutic effects of traditional Chinese medicine on insulin resistance in obese mice by modulating intestinal functions. Heliyon 2024, 10, e30379. [Google Scholar] [CrossRef]
- Potdar, D.; Hirwani, R.R.; Dhulap, S. Phyto-chemical and pharmacological applications of Berberis aristata. Fitoterapia 2012, 83, 817–830. [Google Scholar] [CrossRef]
- Almeleebia, T.M.; Alsayari, A.; Wahab, S. Pharmacological and clinical efficacy of Picrorhiza kurroa and its secondary metabolites: A comprehensive review. Molecules 2022, 27, 8316. [Google Scholar] [CrossRef]
- Arunachalam, K.; Yang, X.; San, T.T. Tinospora cordifolia (Willd.) Miers: Protection mechanisms and strategies against oxidative stress-related diseases. J. Ethnopharmacol. 2022, 283, 114540. [Google Scholar] [CrossRef]
- Xiang, Y.; Guo, Z.; Zhu, P.; Chen, J.; Huang, Y. Traditional Chinese medicine as a cancer treatment: Modern perspectives of ancient but advanced science. Cancer Med. 2019, 8, 1958–1975. [Google Scholar] [CrossRef]
- Cicero, A.F.; Baggioni, A. Berberine and its role in chronic disease. Adv. Exp. Med. Biol. 2016, 928, 27–45. [Google Scholar]
- Liu, B.; Wang, G.; Yang, J.; Pan, X.; Yang, Z.; Zang, L. Berberine inhibits human hepatoma cell invasion without cytotoxicity in healthy hepatocytes. PLoS ONE 2011, 6, e21416. [Google Scholar] [CrossRef]
- El Khalki, L.; Tilaoui, M.; Jaafari, A.; Ait Mouse, H.; Zyad, A. Studies on the dual cytotoxicity and antioxidant properties of Berberis vulgaris extracts and its main constituent berberine. Adv. Pharmacol. Pharm. Sci. 2018, 2018, 3018498. [Google Scholar] [CrossRef]
- Yang, X.; Huang, N. Berberine induces selective apoptosis through the AMPK mediated mitochondrial/caspase pathway in hepatocellular carcinoma. Mol. Med. Rep. 2013, 8, 505–510. [Google Scholar] [CrossRef]
- Demirgan, R.; Karagöz, A.; Pekmez, M.; Önay-Uçar, E.; Artun, F.T.; Gürer, Ç.; Mat, A. In vitro anticancer activity and cytotoxicity of some papaver alkaloids on cancer and normal cell lines. Afr. J. Tradit. Complement. Altern. Med. 2016, 13, 22–26. [Google Scholar] [CrossRef]
- Imenshahidi, M.; Hosseinzadeh, H. Berberine and barberry (Berberis vulgaris): A clinical review. Phytother. Res. 2019, 33, 504–523. [Google Scholar] [CrossRef]
- Li, Z.; Wang, Y.; Xu, Q.; Ma, J.; Li, X.; Yan, J.; Tian, Y.; Wen, Y.; Chen, T. Berberine and health outcomes: An umbrella review. Phytother. Res. 2023, 37, 2051–2066. [Google Scholar] [CrossRef]
- Collins, M.A.; Neafsey, E.J.; Cheng, B.Y.; Hurley-Gius, K.; Ung-Chhun, N.A.; Pronger, D.A.; Christensen, M.A.; Hurley-Gius, D. Endogenous analogs of N-methyl-4-phenyl-1,2,3,6-tetrahydro-pyridine: Indoleamine derived tetrahydro-ß-carbolines as potential causative factors in Parkinson’s Disease. Adv. Neurol. 1986, 45, 179–182. [Google Scholar]
- Bringmann, G.; God, R.; Feineis, D.; Wesemann, W.; Riederer, P.; Rausch, W.D.; Reichmann, H.; Sontag, K.H. The TaClo concept: 1-trichloromethyl-1,2,3,4-tetrahydro-beta-carboline (TaClo), a new toxin for dopaminergic neurons. J. Neural Transm. Suppl. 1995, 46, 235–244. [Google Scholar]
- Bringmann, G.; God, R.; Feineis, D.; Janetzky, B.; Reichmann, H. TaClo as a neurotoxic lead: Improved synthesis, stereochemical analysis, and inhibition of the mitochondrial respiratory chain. J. Neural Transm. Suppl. 1995, 46, 245–254. [Google Scholar]
- Bringmann, G.; Feineis, D.; God, R.; Peters, K.; Peters, E.M.; Scholz, J.; Riederer, F.; Moser, A. 1-Trichloromethyl-1,2,3,4-tetrahydro-beta-carboline (TaClo) and related derivatives: Chemistry and biochemical effects on catecholamine biosynthesis. Bioorganic Med. Chem. 2002, 10, 2207–2214. [Google Scholar] [CrossRef]
- Efferth, T.; Chen, Z.; Kaina, B.; Wang, G. Molecular determinants of response of tumor cells to berberine. Cancer Genom. Proteom. 2005, 2, 115–124. [Google Scholar]
- Wang, Y.; Liu, Y.; Du, X.; Ma, H.; Yao, J. The anti-cancer mechanisms of berberine: A review. Cancer Manag. Res. 2020, 12, 695–702. [Google Scholar] [CrossRef]
- Goel, A. Current understanding and future prospects on Berberine for anticancer therapy. Chem. Biol. Drug Des. 2023, 102, 177–200. [Google Scholar] [CrossRef] [PubMed]
- Ho, Y.T.; Yang, J.S.; Lu, C.C.; Chiang, J.H.; Li, T.C.; Lin, J.J.; Lai, K.; Liao, C.L.; Lin, J.G.; Chung, J.G. Berberine inhibits human tongue squamous carcinoma cancer tumor growth in a murine xenograft model. Phytomedicine 2009, 16, 887–890. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Wang, Q.; Lin, Z.; Yang, P.; Dou, K.; Zhang, R. Berberine inhibits growth of liver cancer cells by suppressing glutamine uptake. Onco Targets Ther. 2019, 12, 11751–11763. [Google Scholar] [CrossRef]
- Liang, H.; Liu, Y.; Fu, L.; Li, L.; Gong, N. Berberine inhibits the development of endometrial cancer through circ_ZNF608/miR-377-3p/COX2 axis. Autoimmunity 2022, 55, 485–495. [Google Scholar] [CrossRef]
- Xu, M.; Ren, L.; Fan, J.; Huang, L.; Zhou, L.; Li, X.; Ye, X. Berberine inhibits gastric cancer development and progression by regulating the JAK2/STAT3 pathway and downregulating IL-6. Life Sci. 2022, 290, 120266. [Google Scholar] [CrossRef]
- Feng, X.; Wang, K.; Cao, S.; Ding, L.; Qiu, F. Pharmacokinetics and excretion of berberine and its nine metabolites in rats. Front. Pharmacol. 2021, 11, 594852. [Google Scholar] [CrossRef]
- Nie, Q.; Li, M.; Huang, C.; Yuan, Y.; Liang, Q.; Ma, X.; Qiu, T.; Li, J. The clinical efficacy and safety of berberine in the treatment of non-alcoholic fatty liver disease: A meta-analysis and systematic review. J. Transl. Med. 2024, 22, 225. [Google Scholar] [CrossRef]
- Chen, Y.X.; Gao, Q.Y.; Zou, T.H.; Wang, B.M.; Liu, S.D.; Sheng, J.Q.; Ren, J.L.; Zou, X.P.; Liu, Z.J.; Song, Y.Y.; et al. Berberine versus placebo for the prevention of recurrence of colorectal adenoma: A multicentre, double-blinded, randomised controlled study. Lancet Gastroenterol. Hepatol. 2020, 5, 267–275. [Google Scholar] [CrossRef]
- Gansauge, F.; Ramadani, M.; Pressmar, J.; Gansauge, S.; Muehling, B.; Stecker, K.; Cammerer, G.; Leder, G.; Beger, H.G. NSC-631570 (Ukrain) in the palliative treatment of pancreatic cancer. Results of a phase II trial. Langenbecks Arch. Surg. 2002, 386, 570–574. [Google Scholar] [CrossRef]
- Maggini, V.; Lombardi, N.; Crescioli, G.; Gallo, E.; Sivelli, F.; Gensini, G.F.; Vannacci, A.; Firenzuoli, F. Chelidonium majus: Relevant safety aspects of a hepatotoxic plant, trawling the web. Phytother. Res. 2019, 33, 2465–2469. [Google Scholar] [CrossRef] [PubMed]
- Pantano, F.; Mannocchi, G.; Marinelli, E.; Gentili, S.; Graziano, S.; Busardò, F.P.; di Luca, N.M. Hepatotoxicity induced by greater celandine (Chelidonium majus L.): A review of the literature. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 46–52. [Google Scholar] [PubMed]
- Efferth, T.; Kahl, S.; Paulus, K.; Adams, M.; Rauh, R.; Boechzelt, H.; Hao, X.; Kaina, B.; Bauer, R. Phytochemistry and pharmacogenomics of natural products derived from traditional Chinese medicine and Chinese Materia Medica with activity against tumor cells. Mol. Cancer Ther. 2008, 7, 152–161. [Google Scholar] [CrossRef]
- Khalid, S.A.; Dawood, M.; Boulos, J.C.; Wasfi, M.; Drif, A.; Bahramimehr, F.; Shahhamzehei, N.; Shan, L.; Efferth, T. Identification of gedunin from a phytochemical depository as a novel multidrug resistance-bypassing tubulin inhibitor of cancer cells. Molecules 2022, 27, 5858. [Google Scholar] [CrossRef]
- Mahmoud, N.; Hegazy, M.F.; Wadie, W.; Elbadawi, M.; Fleischer, E.; Klinger, A.; Bringmann, G.; Khayyal, M.T.; Efferth, T. Naphthoquinone derivatives as P-glycoprotein inducers in inflammatory bowel disease: 2D monolayers, 3D spheroids, and in vivo models. Pharmacol. Res. 2022, 179, 106233. [Google Scholar] [CrossRef]
- Efferth, T.; Davey, M.; Olbrich, A.; Rücker, G.; Gebhart, E.; Davey, R. Activity of drugs from traditional Chinese medicine toward sensitive and MDR1- or MRP1-overexpressing multidrug-resistant human CCRF-CEM leukemia cells. Blood Cells Mol. Dis. 2002, 28, 160–168. [Google Scholar] [CrossRef]
- Pinto-Garcia, L.; Efferth, T.; Torres, A.; Hoheisel, J.D.; Youns, M. Berberine inhibits cell growth and mediates caspase-independent cell death in human pancreatic cancer cells. Planta Med. 2010, 76, 1155–1161. [Google Scholar] [CrossRef] [PubMed]
- Maeng, H.J.; Yoo, H.J.; Kim, I.W.; Song, I.S.; Chung, S.J.; Shim, C.K. P-glycoprotein-mediated transport of berberine across Caco-2 cell monolayers. J. Pharm. Sci. 2002, 91, 2614–2621. [Google Scholar] [CrossRef]
- Pan, G.Y.; Wang, G.J.; Liu, X.D.; Fawcett, J.P.; Xie, Y.Y. The involvement of P-glycoprotein in berberine absorption. Pharmacol. Toxicol. 2002, 91, 193–197. [Google Scholar] [CrossRef]
- Ma, X.; Zhou, J.; Zhang, C.X.; Li, X.Y.; Li, N.; Ju, R.J.; Shi, J.F.; Sun, M.G.; Zhao, W.Y.; Mu, L.M.; et al. Modulation of drug-resistant membrane and apoptosis proteins of breast cancer stem cells by targeting berberine liposomes. Biomaterials 2013, 34, 4452–4465. [Google Scholar] [CrossRef]
- Qian, K.; Tang, C.Y.; Chen, L.Y.; Zheng, S.; Zhao, Y.; Ma, L.S.; Xu, L.; Fan, L.H.; Yu, J.D.; Tan, H.S.; et al. Berberine reverses breast cancer multidrug resistance based on fluorescence pharmacokinetics in vitro and in vivo. ACS Omega 2021, 6, 10645–10654. [Google Scholar] [CrossRef] [PubMed]
- Seupel, R.; Hemberger, Y.; Feineis, D.; Xu, M.; Seo, E.J.; Efferth, T.; Bringmann, G. Ancistrocyclinones A and B, unprecedented pentacyclic N,C-coupled naphthylisoquinoline alkaloids, from the Chinese liana Ancistrocladus tectorius. Org. Biomol. Chem. 2018, 16, 1581–1590. [Google Scholar] [CrossRef]
- Shitan, N.; Tanaka, M.; Terai, K.; Ueda, K.; Yazaki, K. Human MDR1 and MRP1 recognize berberine as their transport substrate. Biosci. Biotechnol. Biochem. 2007, 71, 242–245. [Google Scholar] [CrossRef]
- Kim, J.B.; Ko, E.; Han, W.; Shin, I.; Park, S.Y.; Noh, D.Y. Berberine diminishes the side population and ABCG2 transporter expression in MCF-7 breast cancer cells. Planta Med. 2008, 74, 1693–1700. [Google Scholar] [CrossRef]
- Tan, K.W.; Li, Y.; Paxton, J.W.; Birch, N.P.; Scheepens, A. Identification of novel dietary phytochemicals inhibiting the efflux transporter breast cancer resistance protein (BCRP/ABCG2). Food Chem. 2013, 138, 2267–2274. [Google Scholar] [CrossRef]
- Beck, W.T.; Qian, X.D. Photoaffinity substrates for P-glycoprotein. Biochem. Pharmacol. 1992, 43, 89–93. [Google Scholar] [CrossRef]
- Efferth, T.; Koch, E. Complex interactions between phytochemicals. The multi-target therapeutic concept of phytotherapy. Curr. Drug Targets 2011, 12, 122–132. [Google Scholar] [CrossRef]
- Huang, B.; Wen, G.; Li, R.; Wu, M.; Zou, Z. Integrated network pharmacology, bioinformatics, and molecular docking to explore the mechanisms of berberine regulating autophagy in breast cancer. Medicine 2023, 102, e35070. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Chen, J.; Feng, W.; Wang, C.; Chen, M.; Li, Y.; Chen, J.; Liu, X.; Liu, Q.; Tian, J. Berberine ameliorates iron levels and ferroptosis in the brain of 3 × Tg-AD mice. Phytomedicine 2023, 118, 154962. [Google Scholar] [CrossRef] [PubMed]
- Maksum, I.P.; Rustaman, R.; Deawati, Y.; Rukayadi, Y.; Utami, A.R.; Nafisa, Z.K. Study of the antidiabetic mechanism of berberine compound on FOXO1 transcription factor through molecular docking and molecular dynamics simulations. J. Mol. Model. 2024, 30, 260. [Google Scholar] [CrossRef] [PubMed]
- Kakoti, B.B.; Zothantluanga, J.H.; Deka, K.; Halder, R.K.; Roy, D. In silico design and computational screening of berberine derivatives for potential antidiabetic activity through allosteric activation of the AMPK pathway. In Silico Pharmacol. 2025, 13, 12. [Google Scholar] [CrossRef]
- Hientz, K.; Mohr, A.; Bhakta-Guha, D.; Efferth, T. The role of p53 in cancer drug resistance and targeted chemotherapy. Oncotarget 2017, 8, 8921–8946. [Google Scholar] [CrossRef]
- Choi, M.S.; Oh, J.H.; Kim, S.M.; Jung, H.Y.; Yoo, H.S.; Lee, Y.M.; Moon, D.C.; Han, S.B.; Hong, J.T. Berberine inhibits p53-dependent cell growth through induction of apoptosis of prostate cancer cells. Int. J. Oncol. 2009, 34, 1221–1230. [Google Scholar] [CrossRef]
- Katiyar, S.K.; Meeran, S.M.; Katiyar, N.; Akhtar, S. p53 cooperates berberine-induced growth inhibition and apoptosis of non-small cell human lung cancer cells in vitro and tumor xenograft growth in vivo. Mol. Carcinog. 2009, 48, 24–37. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhang, X.; Liu, A.; Liu, S.; Zhang, L.; Wu, B.; Hu, Q. Berberine induces apoptosis in p53-null leukemia cells by down-regulating XIAP at the post-transcriptional level. Cell Physiol. Biochem. 2013, 32, 1213–1224. [Google Scholar] [CrossRef]
- Liu, Z.; Chen, Y.; Gao, H.; Xu, W.; Zhang, C.; Lai, J.; Liu, X.; Sun, Y.; Huang, H. Berberine inhibits cell proliferation by interfering with wild-type and mutant p53 in human glioma cells. Onco Targets Ther. 2020, 13, 12151–12162. [Google Scholar] [CrossRef] [PubMed]
- Fefilova, E.; Kirdeeva, Y.; Parfenyev, S.; Daks, A.; Fedorova, O.; Sorokina, M.; Ha, N.X.; Huong, T.T.; Loc, V.T.; Hai, P.T.; et al. MDM2 up-regulates the energy metabolism in NSCLC in a p53-independent manner. Biochem. Biophys. Res. Commun. 2025, 743, 151169. [Google Scholar] [CrossRef]
- Nicholson, R.I.; Gee, J.M.W.; Harper, M.E. EGFR and cancer prognosis. Eur. J. Cancer 2001, 37, 9–15. [Google Scholar] [CrossRef]
- Juchum, M.; Günther, M.; Laufer, S.A. Fighting cancer drug resistance: Opportunities and challenges for mutation-specific EGFR inhibitors. Drug Resist. Updat. 2015, 20, 12–28. [Google Scholar] [CrossRef] [PubMed]
- Sigismund, S.; Avanzato, D.; Lanzetti, L. Emerging functions of the EGFR in cancer. Mol. Oncol. 2018, 12, 3–20. [Google Scholar] [CrossRef]
- Wang, J.; Yang, S.; Cai, X.; Dong, J.; Chen, Z.; Wang, R.; Zhang, S.; Cao, H.; Lu, D.; Jin, T.; et al. Berberine inhibits EGFR signaling and enhances the antitumor effects of EGFR inhibitors in gastric cancer. Oncotarget 2016, 7, 76076–76086. [Google Scholar] [CrossRef]
- Jabbarzadeh Kaboli, P.; Leong, M.P.; Ismail, P.; Ling, K.H. Antitumor effects of berberine against EGFR, ERK1/2, P38 and AKT in MDA-MB231 and MCF-7 breast cancer cells using molecular modelling and in vitro study. Pharmacol. Rep. 2019, 71, 13–23. [Google Scholar] [CrossRef]
- Yan, G.; Efferth, T. Broad-spectrum cross-resistance to anticancer drugs mediated by epidermal growth factor receptor. Anticancer Res. 2019, 39, 3585–3593. [Google Scholar] [CrossRef]
- Yan, G.; Saeed, M.E.M.; Foersch, S.; Schneider, J.; Roth, W.; Efferth, T. Relationship between EGFR expression and subcellular localization with cancer development and clinical outcome. Oncotarget 2019, 10, 1918–1931. [Google Scholar] [CrossRef]
- Scharnhorst, V.; van der Eb, A.J.; Jochemsen, A.G. WT1 proteins: Functions in growth and differentiation. Gene 2001, 273, 141–161. [Google Scholar] [CrossRef]
- Ning, H.; Lu, W.; Jia, Q.; Wang, J.; Yao, T.; Lv, S.; Li, Y.; Wen, H. Discovery of oxyepiberberine as a novel tubulin polymerization inhibitor and an anti-colon cancer agent against LS-1034 cells. Investig. New Drugs 2021, 39, 386–393. [Google Scholar] [CrossRef]
- Hermawan, A.; Putri, H.; Utomo, R.Y. Comprehensive bioinformatics study reveals targets and molecular mechanism of hesperetin in overcoming breast cancer chemoresistance. Mol. Divers. 2020, 24, 933–947. [Google Scholar] [CrossRef]
- Mofers, A.; Selvaraju, K.; Gubat, J.; D’Arcy, P.; Linder, S. Identification of proteasome inhibitors using analysis of gene expression profiles. Eur. J. Pharmacol. 2020, 889, 173709. [Google Scholar] [CrossRef]
- Colombo, G.M.; Corsello, S.M. Modernizing the NCI60 cell line screen for phenotypic drug discovery in the 21st Century. Cancer Res. 2024, 84, 2397–2399. [Google Scholar] [CrossRef] [PubMed]
- Lamprou, I.; Tsolou, A.; Kakouratos, C.; Mitrakas, A.G.; Xanthopoulou, E.T.; Kassela, K.; Karakasiliotis, I.; Zois, C.E.; Giatromanolaki, A.; Koukourakis, M.I. Suppressed PLIN3 frequently occurs in prostate cancer, promoting docetaxel resistance via intensified autophagy, an event reversed by chloroquine. Med. Oncol. 2021, 38, 116. [Google Scholar] [CrossRef] [PubMed]
- Yao, D.; Xia, S.; Jin, C.; Zhao, W.; Lan, W.; Liu, Z.; Xiu, Y. Feedback activation of GATA1/miR-885-5p/PLIN3 pathway decreases sunitinib sensitivity in clear cell renal cell carcinoma. Cell Cycle 2020, 19, 2195–2206. [Google Scholar] [CrossRef] [PubMed]
- Saafan, H.; Foerster, S.; Parra-Guillen, Z.P.; Hammer, E.; Michaelis, M.; Cinatl, J., Jr.; Völker, U.; Fröhlich, H.; Kloft, C.; Ritter, C.A. Utilising the EGFR interactome to identify mechanisms of drug resistance in non-small cell lung cancer—Proof of concept towards a systems pharmacology approach. Eur. J. Pharm. Sci. 2016, 94, 20–32. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Lu, W.; Kulkarni, B.; Pejovic, T.; Yan, X.; Chiang, J.H.; Hood, L.; Odunsi, K.; Lin, B. Analysis of chemotherapy response programs in ovarian cancers by the next-generation sequencing technologies. Gynecol. Oncol. 2010, 117, 159–169. [Google Scholar] [CrossRef]
- Wang, R.; Zhang, L. Identification and functional characterization of essential genes related to gefitinib sensitivity in lung adenocarcinoma. Curr. Med. Chem. 2025, 32, 5024–5042. [Google Scholar] [CrossRef]
- Lu, J.; Böttcher, M.; Walther, T.; Mougiakakos, D.; Zenz, T.; Huber, W. Energy metabolism is co-determined by genetic variants in chronic lymphocytic leukemia and influences drug sensitivity. Haematologica 2019, 104, 1830. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wu, Y.; Hou, C.; Li, F. Circ_0072088 knockdown contributes to cisplatin sensitivity and inhibits tumor progression by miR-944/LASP1 axis in non-small cell lung cancer. J. Gene Med. 2022, 24, e3414. [Google Scholar] [CrossRef]
- Ran, M.; Zhou, Y.; Guo, Y.; Huang, D.; Zhang, S.L.; Tam, K.Y. Cytosolic malic enzyme and glucose-6-phosphate dehydrogenase modulate redox balance in NSCLC with acquired drug resistance. FEBS J. 2023, 290, 4792–4809. [Google Scholar] [CrossRef]
- Davis, T.; van Niekerk, G.; Peres, J.; Prince, S.; Loos, B.; Engelbrecht, A.M. Doxorubicin resistance in breast cancer: A novel role for the human protein AHNAK. Biochem. Pharmacol. 2018, 148, 174–183. [Google Scholar] [CrossRef]
- Hsu, Y.C.; Chen, H.Y.; Yuan, S.; Yu, S.L.; Lin, C.H.; Wu, G.; Yang, P.C.; Li, K.C. Genome-wide analysis of three-way interplay among gene expression, cancer cell invasion and anti-cancer compound sensitivity. BMC Med. 2013, 11, 106. [Google Scholar] [CrossRef] [PubMed]
- Park, K.S.; Chun, J.L.; Kim, E.Y.; Lee, J.H.; Lee, H.S.; Yamauchi, N.; Lee, K.B.; Kim, M.K. Protective effect of trehalose on canine spermatozoa in cryopreservation. J. Fac. Agric. Kyushu Univ. 2018, 63, 53–60. [Google Scholar] [CrossRef]
- Castro-Guijarro, A.C.; Sanchez, A.M.; Flamini, M.I. Potential biomarkers associated with prognosis and trastuzumab response in HER2+ breast cancer. Cancers 2023, 15, 4374. [Google Scholar] [CrossRef]
- Zhang, H.; Zheng, Y. LPCAT1 is transcriptionally regulated by FOXA1 to promote breast cancer progression and paclitaxel resistance. Oncol. Lett. 2023, 25, 134. [Google Scholar] [CrossRef]
- Wen, Y.; Chelariu-Raicu, A.; Umamaheswaran, S.; Nick, A.M.; Stur, E.; Hanjra, P.; Jiang, D.; Jennings, N.B.; Chen, X.; Corvigno, S.; et al. Endothelial p130cas confers resistance to anti-angiogenesis therapy. Cell Rep. 2022, 38, 110301. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.; Wang, L.; Zhang, J.; Dong, W.; Zhang, T.; Ni, Y.; Cao, H.; Wang, K.; Li, Y.; Wang, Y.; et al. ARRB1 enhances the chemosensitivity of lung cancer through the mediation of DNA damage response. Oncol. Rep. 2017, 37, 761–767. [Google Scholar] [CrossRef]
- Kallifatidis, G.; Smith, D.K.; Morera, D.S.; Gao, J.; Hennig, M.J.; Hoy, J.J.; Pearce, R.F.; Dabke, I.R.; Li, J.; Merseburger, A.S.; et al. β-arrestins regulate stem cell-like phenotype and response to chemotherapy in bladder cancer. Mol. Cancer Ther. 2019, 18, 801–811. [Google Scholar] [CrossRef]
- Karabay, A.Z.; Ozkan, T.; Karadag Gurel, A.; Koc, A.; Hekmatshoar, Y.; Sunguroglu, A.; Aktan, F.; Buyukbingöl, Z. Identification of exosomal microRNAs and related hub genes associated with imatinib resistance in chronic myeloid leukemia. Naunyn Schmiedebergs Arch. Pharmacol. 2024, 397, 9701–9721. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, W.; Ni, P.; Li, A.; Zhou, J.; Xu, S. MiR-99a and MiR-491 regulate cisplatin resistance in human gastric cancer cells by targeting CAPNS1. Int. J. Biol. Sci. 2016, 12, 1437–1447. [Google Scholar] [CrossRef] [PubMed]
- Derakhshani, A.; Rezaei, Z.; Safarpour, H.; Sabri, M.; Mir, A.; Sanati, M.A.; Vahidian, F.; Gholamiyan Moghadam, A.; Aghadoukht, A.; Hajiasgharzadeh, K.; et al. Overcoming trastuzumab resistance in HER2-positive breast cancer using combination therapy. J. Cell Physiol. 2020, 235, 3142–3156. [Google Scholar] [CrossRef] [PubMed]
- Chang, N.; Ge, N.; Zhao, Y.; Yang, L.; Qin, W.; Cui, Y. Hsa_circ_0007142 contributes to cisplatin resistance in esophageal squamous cell carcinoma via miR-494-3p/LASP1 axis. J. Clin. Lab. Anal. 2022, 36, e24304. [Google Scholar] [CrossRef]
- Zhong, C.; Chen, Y.; Tao, B.; Peng, L.; Peng, T.; Yang, X.; Xia, X.; Chen, L. LIM and SH3 protein 1 regulates cell growth and chemosensitivity of human glioblastoma via the PI3K/AKT pathway. BMC Cancer 2018, 18, 722. [Google Scholar] [CrossRef]
- Gámez-Chiachio, M.; Molina-Crespo, Á.; Ramos-Nebot, C.; Martinez-Val, J.; Martinez, L.; Gassner, K.; Llobet, F.J.; Soriano, M.; Hernandez, A.; Cordani, M.; et al. Gasdermin B over-expression modulates HER2-targeted therapy resistance by inducing protective autophagy through Rab7 activation. J. Exp. Clin. Cancer Res. 2022, 41, 285. [Google Scholar] [CrossRef]
- Zheng, Y.; Yuan, D.; Zhang, F.; Tang, R. A systematic pan-cancer analysis of the gasdermin (GSDM) family of genes and their correlation with prognosis, the tumor microenvironment, and drug sensitivity. Front. Genet. 2022, 13, 926796. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Xia, M.; Wei, L.; Guo, K.; Sun, R.; Liu, Y.; Qiu, C.; Jiang, J. ABX-1431 inhibits the development of endometrial adenocarcinoma and reverses progesterone resistance by targeting MGLL. Cell Death Dis. 2022, 13, 1067. [Google Scholar] [CrossRef]
- Kawasaki, S.; Ohtsuka, H.; Sato, Y.; Douchi, D.; Sato, M.; Ariake, K.; Masuda, K.; Fukase, K.; Mizuma, M.; Nakagawa, K.; et al. Silencing of LRRFIP1 enhances the sensitivity of gemcitabine in pancreatic cancer cells by activating JNK/c-Jun signaling. Pancreatology 2021, 21, 771–778. [Google Scholar] [CrossRef]
- Li, Y.; Li, W.; Yang, Y.; Lu, Y.; He, C.; Hu, G.; Liu, H.; Chen, J.; He, J.; Yu, H. MicroRNA-21 targets LRRFIP1 and contributes to VM-26 resistance in glioblastoma multiforme. Brain Res. 2009, 1286, 13–18. [Google Scholar] [CrossRef]
- Gao, X.; Qian, J.; Zhang, Y.; Wang, H.; Cui, J.; Yang, Y. Analysis of differential membrane proteins related to matrix stiffness-mediated metformin resistance in hepatocellular carcinoma cells. Proteome Sci. 2023, 21, 14. [Google Scholar] [CrossRef]
- Mieczkowska, I.K.; Pantelaiou-Prokaki, G.; Prokakis, E.; Schmidt, G.E.; Müller-Kirschbaum, L.C.; Werner, M.; Sen, M.; Velychko, T.; Jannasch, K.; Dullin, C.; et al. Decreased PRC2 activity supports the survival of basal-like breast cancer cells to cytotoxic treatments. Cell Death Dis. 2021, 12, 1118. [Google Scholar] [CrossRef]
- Xiao, H.; Chen, H.; Zhang, L.; Duolikun, M.; Zhen, B.; Kuerban, S.; Li, X.; Wang, Y.; Chen, L.; Lin, J. Cytoskeletal gene alterations linked to sorafenib resistance in hepatocellular carcinoma. World J. Surg. Oncol. 2024, 22, 152. [Google Scholar] [CrossRef] [PubMed]
- Frottin, F.; Bienvenut, W.V.; Bignon, J.; Jacquet, E.; Vaca Jacome, A.S.; Van Dorsselaer, A.; Cianferani, S.; Carapito, C.; Meinnel, T.; Giglione, C. MetAP1 and MetAP2 drive cell selectivity for a potent anti-cancer agent in synergy, by controlling glutathione redox state. Oncotarget 2016, 7, 63306–63323. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Cai, X.; Liu, L.; Li, A.; Huang, H.; Fu, Y.; Dai, Z.; Sun, Y. A novel platinum-resistance-related gene signature in ovarian cancer: Identification and patient-derived organoids verification. Curr. Cancer Drug Targets 2025, 25, 64–84. [Google Scholar] [CrossRef]
- Kim, J.S.; Lee, J.H.; Jeon, S.R.; Kim, Y.; Jeon, S.H.; Wu, H.G. Identification of genes involved in EGF-induced apoptosis using CRISPR/Cas9 knockout screening: Implications for novel therapeutic targets in EGFR-overexpressing cancers. Cancer Res. Treat. 2023, 55, 737–745. [Google Scholar] [CrossRef]
- Sun, A.; Yang, H.; Li, T.; Luo, J.; Zhou, L.; Chen, R.; Han, L.; Lin, Y. Molecular mechanisms, targets and clinical potential of berberine in regulating metabolism: A review focussing on databases and molecular docking studies. Front. Pharmacol. 2024, 15, 1368950. [Google Scholar] [CrossRef]
- Lo, Y.W.; Lin, S.T.; Chang, S.J.; Chan, C.H.; Lyu, K.W.; Chang, J.F.; May, E.W.; Lin, D.Y.; Chou, H.C.; Chan, H.L. Mitochondrial proteomics with siRNA knockdown to reveal ACAT1 and MDH2 in the development of doxorubicin-resistant uterine cancer. J. Cell Mol. Med. 2015, 19, 744–759. [Google Scholar] [CrossRef]
- Liu, Q.; Harvey, C.T.; Geng, H.; Xue, C.; Chen, V.; Beer, T.M.; Qian, D.Z. Malate dehydrogenase 2 confers docetaxel resistance via regulations of JNK signaling and oxidative metabolism. Prostate 2013, 73, 1028–1037. [Google Scholar] [CrossRef]
- Ortega Duran, M.; Shaheed, S.U.; Sutton, C.W.; Shnyder, S.D. A proteomic investigation to discover candidate proteins involved in novel mechanisms of 5-fluorouracil resistance in colorectal cancer. Cells 2024, 13, 342. [Google Scholar] [CrossRef]
- Jung, S.; Myagmarjav, D.; Jo, T.; Lee, S.; Han, S.; Quynh, N.T.N.; Anh, N.H.; Vu, S.H.; Choi, Y.; Lee, M.S. Inhibitory role of TRIP-Br1 oncoprotein in anticancer drug-mediated programmed cell death via mitophagy activation. Int. J. Biol. Sci. 2022, 18, 3859–3873. [Google Scholar] [CrossRef]
- Miglietta, S.; Sollazzo, M.; Gherardi, I.; Milioni, S.; Cavina, B.; Marchio, L.; De Luise, M.; Coada, C.A.; Fiorillo, M.; Perrone, A.M.; et al. Mitochondrial chaperonin DNAJC15 promotes vulnerability to ferroptosis of chemoresistant ovarian cancer cells. Open Biol. 2025, 15, 240151. [Google Scholar] [CrossRef] [PubMed]
- Kleih, M.; Böpple, K.; Dong, M.; Gaißler, A.; Heine, S.; Olayioye, M.A.; Aulitzky, W.E.; Essmann, F. Direct impact of cisplatin on mitochondria induces ROS production that dictates cell fate of ovarian cancer cells. Cell Death Dis. 2019, 10, 851. [Google Scholar] [CrossRef]
- Lozinski, M.; Bowden, N.A.; Graves, M.C.; Fay, M.; Day, B.W.; Stringer, B.W.; Tooney, P.A. Transcriptomic profiling of dna damage response in patient-derived glioblastoma cells before and after radiation and temozolomide treatment. Cells 2022, 11, 1215. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Hu, K.; Liu, Y.; Liu, L.; Tang, J.; Qin, X. Knockdown of replication protein A 3 induces protective autophagy and enhances cisplatin sensitivity in lung adenocarcinoma by inhibiting AKT/mTOR signaling via binding to cyclin-dependent kinases regulatory subunit 2. Drug Dev. Res. 2022, 83, 1589–1599. [Google Scholar] [CrossRef]
- Lei, Y.; Wang, D.; Chen, W.; Tian, X.; Wei, J. FOXM1/NCAPH activates glycolysis to promote colon adenocarcinoma stemness and 5-FU resistance. Anticancer Drugs 2023, 34, 929–938. [Google Scholar] [CrossRef] [PubMed]
- Shimomura, H.; Sasahira, T.; Nakashima, C.; Kurihara-Shimomura, M.; Kirita, T. Non-SMC condensin I complex subunit H (NCAPH) is associated with lymphangiogenesis and drug resistance in oral squamous cell carcinoma. J. Clin. Med. 2019, 9, 72. [Google Scholar] [CrossRef]
- Zhan, S.J.; Liu, B.; Linghu, H. Identifying genes as potential prognostic indicators in patients with serous ovarian cancer resistant to carboplatin using integrated bioinformatics analysis. Oncol. Rep. 2018, 39, 2653–2663. [Google Scholar] [CrossRef]
- Xie, Q.; Xiao, Y.S.; Jia, S.C.; Zheng, J.X.; Du, Z.C.; Chen, Y.C.; Chen, M.T.; Liang, Y.K.; Lin, H.Y.; Zeng, D. FABP7 is a potential biomarker to predict response to neoadjuvant chemotherapy for breast cancer. Cancer Cell Int. 2020, 20, 562. [Google Scholar] [CrossRef] [PubMed]
- Pak, B.J.; Li, Q.; Kerbel, R.S.; Ben-David, Y. TYRP2-mediated resistance to cis-diamminedichloroplatinum (II) in human melanoma cells is independent of tyrosinase and TYRP1 expression and melanin content. Melanoma Res. 2000, 10, 499–505. [Google Scholar] [CrossRef] [PubMed]
- Hartman, M.L.; Czyz, M. TYRP1 mRNA level is stable and MITF-M-independent in drug-naïve, vemurafenib- and trametinib-resistant BRAFV600E melanoma cells. Arch. Dermatol. Res. 2020, 312, 385–392. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.; Zhang, G.; Luo, W.; Xu, M.; Peng, R.; Du, Z.; Liu, Y.; Bai, Z.; Xiao, X.; Qin, S. PROTAC technology: From drug development to probe technology for target deconvolution. Eur. J. Med. Chem. 2024, 276, 116725. [Google Scholar] [CrossRef]
- Li, G.; Shi, Q.; Wu, Q.; Sui, X. Target identification of natural products in cancer with chemical proteomics and artificial intelligence approaches. Cancer Biol. Med. 2025, 22, 549–597. [Google Scholar] [CrossRef]
- Guedeney, N.; Cornu, M.; Schwalen, F.; Kieffer, C.; Voisin-Chiret, A.S. PROTAC technology: A new drug design for chemical biology with many challenges in drug discovery. Drug Discov. Today 2023, 28, 103395. [Google Scholar] [CrossRef]
- He, S.J.; Li, J.; Zhou, J.C.; Yang, Z.Y.; Liu, X.; Ge, Y.W. Chemical proteomics accelerates the target discovery of natural products. Biochem. Pharmacol. 2024, 230, 116609. [Google Scholar] [CrossRef]
- Efferth, T.; Konkimalla, V.B.; Wang, Y.-F.; Sauerbrey, A.; Meinhardt, S.; Zintl, F.; Mattern, J.; Volm, M. Prediction of broad spectrum resistance of tumors towards anticancer drugs. Clin. Cancer Res. 2008, 14, 2405–2412. [Google Scholar] [CrossRef]
- Özenver, N.; Dawood, M.; Fleischer, E.; Klinger, A.; Efferth, T. Chemometric and transcriptomic profiling, microtubule disruption and cell death induction by secalonic acid in tumor cells. Molecules 2020, 25, 3224. [Google Scholar] [CrossRef]
- Alley, M.C.; Scudiero, D.A.; Monks, A.; Hursey, M.L.; Czerwinski, M.J.; Fine, D.L.; Abbott, B.J.; Mayo, J.G.; Shoemaker, R.H.; Boyd, M.R. Feasibility of drug screening with panels of human tumor cell lines using a microculture tetrazolium assay. Cancer Res. 1988, 48, 589–601. [Google Scholar] [PubMed]
- Lorenzi, P.L.; Reinhold, W.C.; Varma, S.; Hutchinson, A.A.; Pommier, Y.; Chanock, S.J.; Weinstein, J.N. DNA fingerprinting of the NCI-60 cell line panel. Mol. Cancer Ther. 2009, 8, 713–724. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.; Wilson, I.; Orton, T.; Pognan, F. Investigation of the Alamar Blue (resazurin) fluorescent dye for the assessment of mammalian cell cytotoxicity. Eur. J. Biochem. 2000, 267, 5421–5542. [Google Scholar] [CrossRef] [PubMed]
- Özenver, N.; Abdelfatah, S.; Klinger, A.; Fleischer, E.; Efferth, T. Identification and characterization of deschloro-chlorothricin obtained from a large natural product library targeting aurora A kinase in multiple myeloma. Investig. New Drugs 2021, 39, 348–361. [Google Scholar] [CrossRef]
| Gene | Cell Line | IC50 (µM) | Degree of Resistance |
|---|---|---|---|
| ABCB1 | CCRF-CEM | 5 ± 1 | 28.2 |
| CEM/ADR5000 | 141 ± 9 | ||
| ABCB5 | HEK293 | 2 ± 0 | 18.5 |
| HEK293-ABCB5 | 37 ± 3 | ||
| ABCG2 | MDA-MB-231 | 17 ± 1 | 2.41 |
| MDA-MB-231-BCRP | 41 ± 7 | ||
| TP53 | HCT-116 p53+/+ | 19 ± 1 | 0.94 |
| HCT-116 p53–/– | 18 ± 0 | ||
| EGFR | U87.MG | 7 ± 0 | 3.57 |
| U87.MGΔEGFR | 25 ± 13 |
| Drug Resistance Mechanism | p- or r-Value | Berberine Chloride (log10IC50, M) | Control Drug (log10IC50, M) |
|---|---|---|---|
| ABCB1 Expression | Epirubicin | ||
| 7q21 (Chromosomal | r-value | * 0.389 | * 0.447 |
| Locus of ABCB1 Gene) | p-value | * 0.005 | * 3.55 × 10−4 |
| ABCB1 Expression | r-value | * 0.531 | * 0.588 |
| (Microarray) | p-value | * 5.15 × 10−5 | * 0.588.82 × 10−6 |
| ABCB1 Expression | r-value | *0.476 | * 0.410 |
| (RT-PCR) | p-value | * 4.15 × 10−4 | * 1.54 × 10−3 |
| Rhodamine 123 | r-value | * 0.553 | * 0.526 |
| Accumulation | p-value | * 1.86 × 10−5 | * 1.12 × 10−5 |
| ABCB5 Expression | Maytansine | ||
| ABCB5 Expression | r-value | −0.098 | * 0.454 |
| (Microarray) | p-value | 0.258 | * 6.67 × 10−4 |
| ABCB5 Expression | r-value | −0.184 | * 0.402 |
| (RT-PCR) | p-value | 0.109 | * 0.0026 |
| ABCC1 Expression | Vinblastine | ||
| DNA Gene | r-value | 0.294 | * 0.429 |
| Copy Number | p-value | 0.021 | * 0.001 |
| ABCC1 Expression | r-value | 0.296 | * 0.398 |
| (Microarray) | p-value | 0.022 | * 0.003 |
| ABCC1 Expression | r-value | 0.124 | 0.299 |
| (RT-PCR) | p-value | 0.196 | * 0.036 |
| ABCG2 Expression | Pancratistatin | ||
| ABCG2 Expression | r-value | −0.011 | * 0.323 |
| (Microarray) | p-value | 0.471 | * 0.006 |
| ABCG2 Expression | r-value | −0.045 | * 0.346 |
| (Western Blot) | p-value | 0.382 | * 0.004 |
| EGFR Expression | Erlotinib | ||
| EGFR Expression | r-value | 0.298 | * −0.458 |
| (Microarray) | p-value | 0.021 | * 1.15 × 10−4 |
| EGFR Expression | r-value | * 0.336 | * −0.379 |
| (PCR Slot Blot) | p-value | * 0.011 | * 0.002 |
| EGFR Expression | r-value | 0.292 | * −0.376 |
| (Protein Array) | p-value | 0.023 | * 0.002 |
| WT1 Expression | Melphalan | ||
| WT1 Expression | r-value | * −0.319 | * −0.346 |
| (Microarray) | p-value | * 0.014 | * 0.004 |
| TP53 Mutation | 5-Fluorouracil | ||
| TP53 Mutation | r-value | 0.024 | * −0.502 |
| (cDNA Sequencing) | p-value | 0.450 | * 3.50 × 10−5 |
| TP53 Function | r-value | 0.144 | * −0.436 |
| (Yeast Functional Assay) | p-value | 0.175 | * 5.49 × 10−4 |
| NRAS Mutation | Doxorubicin | ||
| Codon 12 mutation | r-value | −0.16071 | * −0.424 |
| (cDNA Sequencing) | p-value | 0.155 | * 9.61 × 10−4 |
| Code | Name | Function | Category |
|---|---|---|---|
| PLIN3 | Perilipin 3 | Mannose 6-phosphate receptor required for endosome-to-Golgi transport | Metabolism |
| MVP | Major vault protein/lung resistance-related protein | Multi-subunit ribonucleoprotein structures involved in nucleo-cytoplasmic transport | Transport function |
| TLDC1 | MTOR-associated protein, Eak-7 homolog | TOR signaling, positive regulation of protein localization to the lysosome | Others |
| AP2B1 | Adaptor-related protein complex 2 subunit β1 | Links clathrin to receptors in coated vesicles. | Others |
| DYNC1LI2 | Dynein cytoplasmic 1 light intermediate chain 2 | Microtubule-associated motor protein | Cytoskeleton |
| PFKP | Phosphofructokinase, platelet | Glycolysis regulation | Metabolism |
| ME1 | Malic enzyme 1 | Generates NADPH for fatty acid biosynthesis; links the glycolytic and citric acid cycles | Metabolism |
| AHNAK | Desmoyokin | Structural scaffold protein involved in metastasis | Cancer development and metastasis |
| VCL | Vinculin | Cytoskeletal protein associated with cell–cell and cell–matrix junctions; involved in anchoring F-actin to the membrane | Cytoskeleton |
| GAPVD1 | GTPase-activating protein and VPS9 domain-containing protein 1 | Regulation of protein transport | Transport function |
| LPCAT1 | Lysophosphatidylcholine acyltransferase 1 | Phospholipid metabolism; involved in tumor progression | Cancer development and metastasis |
| TNKS1BP1 | Tankyrase-1-binding protein | Double-strand break repair and regulation of protein phosphorylation | DNA repair |
| ARRB1 | Arrestin β1 | Agonist-mediated desensitization of G-protein-coupled receptors; regulation of receptor-mediated immune functions | Others |
| CAPNS1 | Calpain small subunit 1 | Calcium-dependent cysteine proteinase; involved in apoptosis, proliferation, migration, adhesion, and autophagy | Others |
| ERBB2 | V-Erb-B2 avian erythroblastic leukemia viral oncogene homolog 2 | Oncogene; regulation of proliferation | Cancer development and metastasis |
| LASP1 | LIM and SH3 domain protein 1 | cAMP and cGMP-dependent signaling protein that binds to the actin cytoskeleton at extensions of the cell membrane | Cytoskeleton |
| GSDMD | Gasdermin D | Tumor suppressor; regulation of epithelial proliferation | Cancer development and metastasis |
| MGLL | Monoglyceride lipase | Serine hydrolase, role in carcinogenesis and metastasis | Cancer development and metastasis |
| LRRFIP1 | Leucine-rich repeat flightless- interacting protein 1 | DNA-binding transcription repressor activity | Others |
| TAX1BP3 | Tax1 (human T-cell leukemia virus type I) binding protein 3 | Promotes protein–protein interactions that affect cell signaling, adhesion, protein scaffolding, and receptor and ion transporter functions; involved in metastasis | Metastasis |
| RBBP7 | Retinoblastoma binding protein 7, chromatin remodeling factor | Regulation of cell proliferation and differentiation | Chromosomal function |
| CECR5 | Haloacid dehalogenase-like hydrolase domain containing 5 | Involved in glycerophospholipid biosynthesis | Metabolism |
| ACTG1 | Actin γ1 | Cell motility and maintenance of the cytoskeleton | Cytoskeleton |
| SCAMP3 | Secretory carrier membrane protein 3 | Carrier to the cell surface in post-Golgi recycling pathways; protein trafficking in endosomal pathways | Transport function |
| METAP2 | Methionyl aminopeptidase 2 | Methionyl aminopeptidase protects the α subunit of eukaryotic initiation factor 2 from inhibitory phosphorylation; involved in cancer | Cancer development and metastasis |
| SUPT6H | Suppressor of Ty 6 (SPT6) homolog (S. cerevisiae) | Regulation of transcription elongation by RNA polymerase II and transcription elongation-coupled chromatin remodeling | Chromosomal function |
| DDX20 | DEAD/H (Asp-Glu-Ala-Asp/His)Box helicase 20 | Putative RNA helicase involved in translation initiation, nuclear and mitochondrial splicing, ribosome and spliceosome assembly, regulation of cell growth and division | Mitochondrial function |
| MDH2 | Malate dehydrogenase 2, mitochondrial | Oxidation of malate to oxaloacetate; role in the malate-aspartate shuttle that operates in the metabolic coordination between cytosol and mitochondria | Mitochondrial function |
| RAP1GDS1 | Rap1 GTPase-GDP dissociation stimulator 1 | Stimulatory GDP/GTP exchange protein; regulates mitochondrial dynamics | Mitochondrial function |
| TIMM23 | Translocase of the inner mitochondrial membrane 23 | Transport of transit peptide-containing proteins across the inner mitochondrial membrane | Mitochondrial function |
| RPA3 | Replication protein A3 | DNA repair and DNA replication | DNA repair |
| HIST2H2AB | Histone cluster 2 H2A family member B | Replication-dependent histone responsible for the nucleosome structure of the chromosomal fiber | Chromosomal function |
| SRRT | Serrate RNA effector molecule homolog (Arabidopsis) | Involved in primary miRNA processing | Others |
| SMN1 | Survival of motor neuron 1, telomeric | The coding gene is part of a 500 kb inverted duplication on chromosome 5q13. | Others |
| DNAJC11 | DnaJ heat shock protein family (Hsp40) member C11 | Involved in cristae formation | Mitochondrial function |
| WFS1 | Wolframin ER transmembrane glycoprotein | Regulation of cellular Ca2+ homeostasis in the endoplasmic reticulum | Others |
| NCAPH | Non-SMC condensin I complex subunit H | Involved in the conversion of interphase chromatin into condensed chromosomes; associated with mitotic chromosomes | Chromosomal function |
| FLAD1 | Flavin adenine dinucleotide synthetase 1 homolog (S. cerevisiae) | Catalyzes the adenylation of flavin mononucleotide (FMN) to form flavin adenine dinucleotide (FAD) coenzyme | Metabolism |
| FABP7 | Fatty acid-binding protein 7 | Binds long-chain fatty acids | Metabolism |
| TYRP1 | Tyrosinase-related protein 1 | Role in the melanin biosynthetic pathway | Others |
| Protein | Involved Drugs | Tumor Type | Reference |
|---|---|---|---|
| PLIN3 | Docetaxel | Prostate Ca | [90] |
| Sunitinib | Renal clear cell Ca | [91] | |
| MVP | |||
| AP2B1 | Erlotinib | Non-small cell lung cancer | [92] |
| Cisplatin | Ovarian Ca | [93] | |
| PFKP | Gefitinib | Lung adenocarcinoma | [94] |
| Rotenone, navitoclax, and orlistat | Chronic lymphocytic leukemia | [95] | |
| Sunitinib | Renal clear cell Ca | [96] | |
| ME1 | Gefitinib | Non-small cell lung cancer | [97] |
| AHNAK | Doxorubicin | Breast Ca | [98] |
| Paclitaxel, docetaxel, erlotinib, everolimus, and dasatinib | Diverse | [99] | |
| VCL | Doxorubicin | Breast Ca | [100] |
| Trastuzumab | Breast Ca | [101] | |
| LPCAT1 | Paclitaxel | Breast Ca | [102] |
| TNKS1BP1 | Bevacizumab | Ovarian Ca | [103] |
| ARRB1 | Cisplatin, etoposide | Non-small cell lung cancer | [104] |
| Gemcitabine | Bladder Ca | [105] | |
| Imatinib | Chronic myeloid leukemia | [106] | |
| CAPNS1 | Cisplatin | Gastric cancer | [107] |
| ERBB2 | Trastuzumab | Breast Ca | [108] |
| LASP1 | Cisplatin | Esophageal squamous cell Ca | [109] |
| Cisplatin | Non-small cell lung cancer | [96] | |
| Temozolomide | Glioblastoma | [110] | |
| GSDMD | HER2-targeting drugs | Breast and gastroesophageal Ca | [111] |
| Nelarabine, fluphenazine, dexrazoxane, bortezomib, midostaurin, and vincristine. | Renal clear cell Ca | [112] | |
| MGLL | Progesterone | Endometrial adeno Ca | [113] |
| LRRFIP1 | Gemcitabine | Pancreas Ca | [114] |
| Teniposide | Glioblastoma | [115] | |
| TAX1BP3 | Metformin | Hepatocellular Ca | [116] |
| RBBP7 | Cyclophosphamide, doxorubicin, and 5-fluorouracil | Basal-like breast cancer | [117] |
| ACTG1 | Sorafenib | Hepatocellular Ca | [118] |
| METAP2 | Fumagillin | Diverse | [119] |
| SUPT6H | Cisplatin | Ovarian Ca | [120] |
| DDX20 | Epidermal growth factor | Lung Ca | [121] |
| MDH2 | Ripretinib | Gastrointestinal stromal tumor | [122] |
| Doxorubicin | Uterine cancer | [123] | |
| Docetaxel | Prostate Ca | [124] | |
| RAP1GDS1 | 5-Fluorouracil | Colorectal Ca | [125] |
| TIMM23 | Staurosporine | Breast Ca | [126] |
| Cisplatin | Ovarian Ca | [127] | |
| Cisplatin | High-grade serous ovarian Ca | [128] | |
| RPA3 | Temozolomide | Glioblastoma | [129] |
| Cisplatin | Lung adenocarcinoma | [130] | |
| NCAPH | 5-Fluorouracil | Colon adenocarcinoma | [131] |
| Cisplatin | Oral Squamous Cell Ca | [132] | |
| Carboplatin | Serous ovarian cancer | [133] | |
| FABP7 | Anthracyclines and taxanes | Breast Ca | [134] |
| TYRP1 | Cisplatin | Melanoma | [135] |
| Vemurafenib and trametinib | Melanoma | [136] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Özenver, N.; T. Ali, N.; Yücer, R.; Lei, X.; Bringmann, G.; Efferth, T.; Dawood, M. Profiling the Complexity of Resistance Factors in Cancer Cells Towards Berberine and Its Derivatives. Pharmaceuticals 2026, 19, 27. https://doi.org/10.3390/ph19010027
Özenver N, T. Ali N, Yücer R, Lei X, Bringmann G, Efferth T, Dawood M. Profiling the Complexity of Resistance Factors in Cancer Cells Towards Berberine and Its Derivatives. Pharmaceuticals. 2026; 19(1):27. https://doi.org/10.3390/ph19010027
Chicago/Turabian StyleÖzenver, Nadire, Nadeen T. Ali, Rümeysa Yücer, Xiao Lei, Gerhard Bringmann, Thomas Efferth, and Mona Dawood. 2026. "Profiling the Complexity of Resistance Factors in Cancer Cells Towards Berberine and Its Derivatives" Pharmaceuticals 19, no. 1: 27. https://doi.org/10.3390/ph19010027
APA StyleÖzenver, N., T. Ali, N., Yücer, R., Lei, X., Bringmann, G., Efferth, T., & Dawood, M. (2026). Profiling the Complexity of Resistance Factors in Cancer Cells Towards Berberine and Its Derivatives. Pharmaceuticals, 19(1), 27. https://doi.org/10.3390/ph19010027








