Fused Imidazotriazole-Based Therapeutics: A Multidisciplinary Study Against Diabetes-Linked Enzymes Alpha-Amylase and Alpha-Glucosidase Using In Vitro and In Silico Methods
Abstract
1. Introduction
2. Results and Discussion
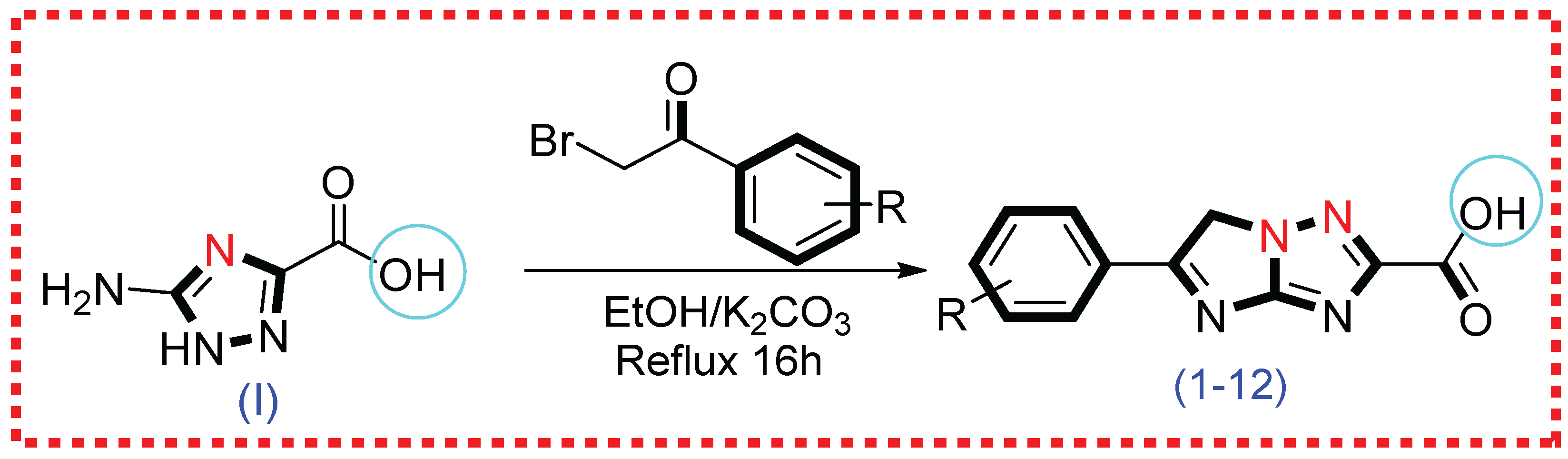
2.1. Synthetic Route
2.2. Biological Efficacy
Structure–Activity Relationship (SAR)
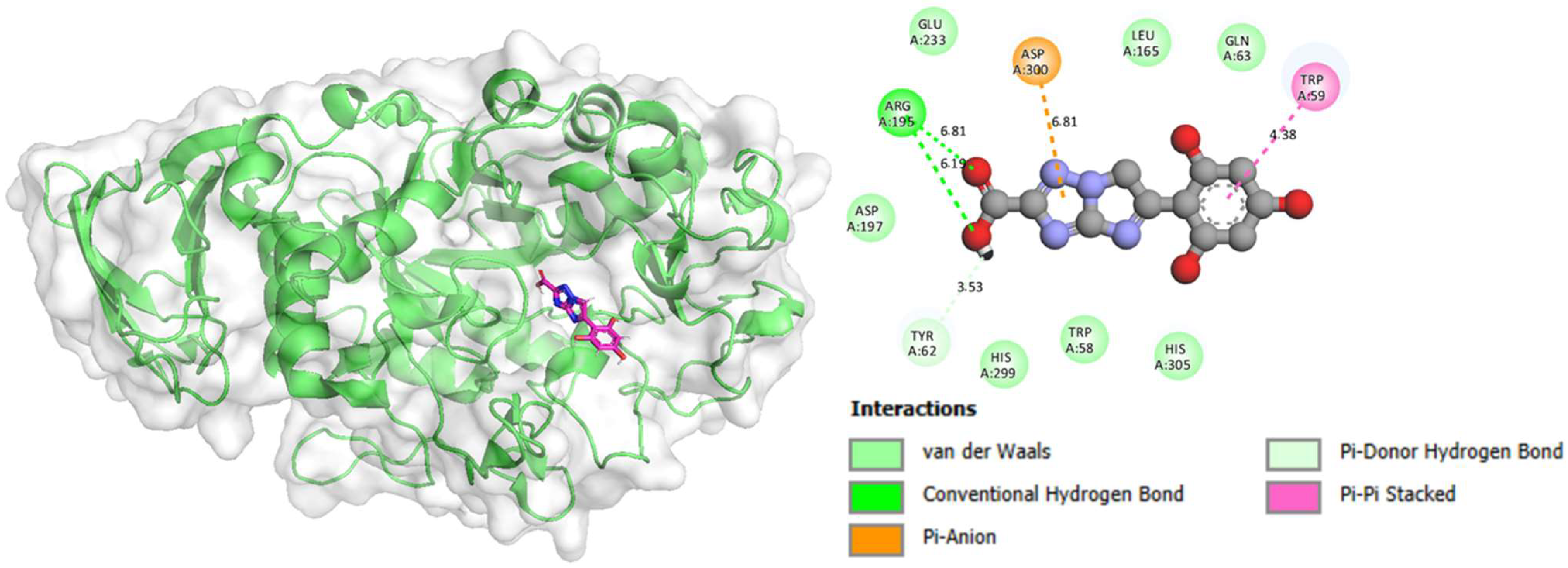
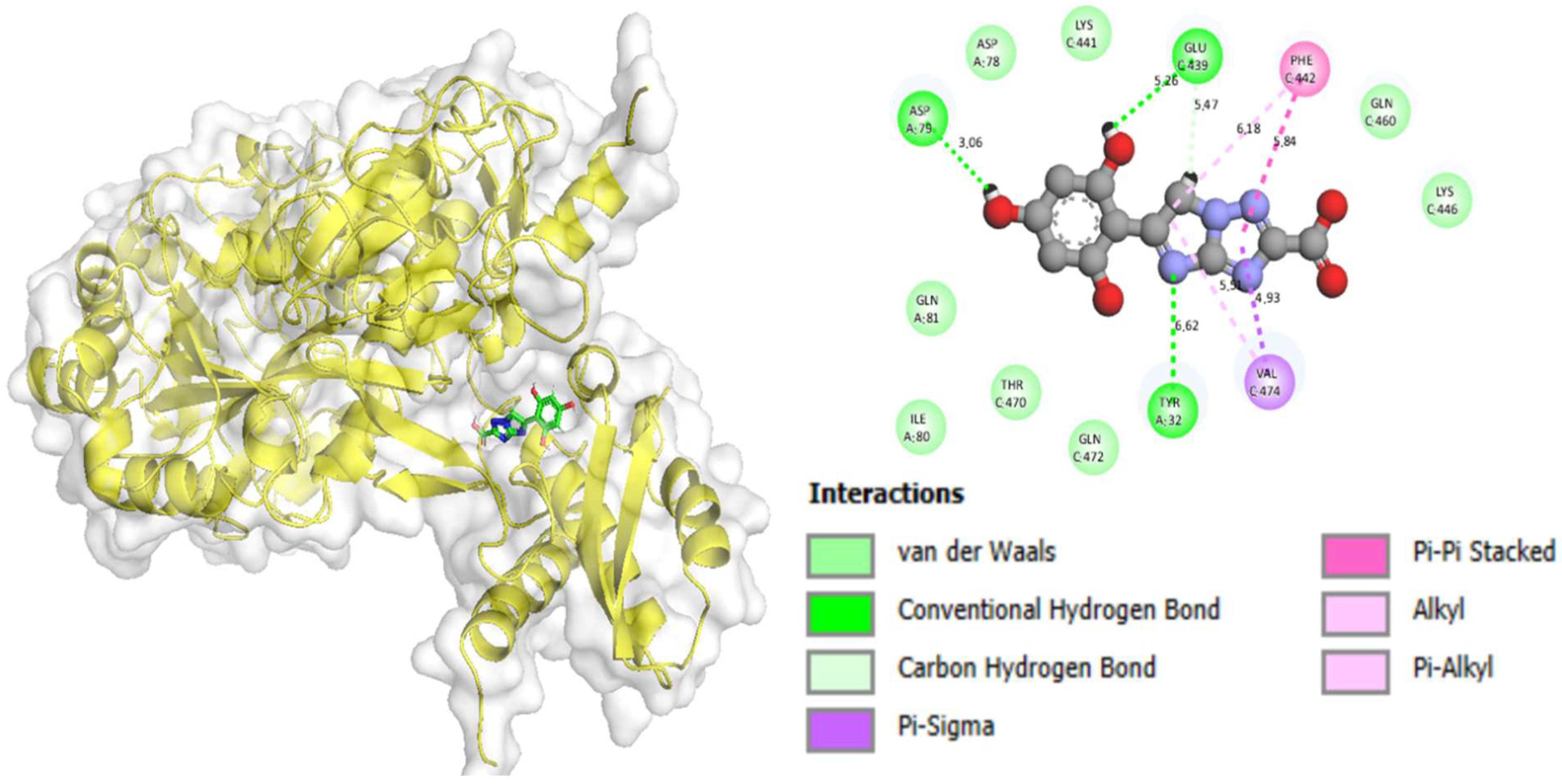
2.3. Molecular Docking
- Center coordinates: (x = 3.625794, y = 64.910854, z = 68.89273);
- Grid box size: (x = 20, y = 20, z = 20).
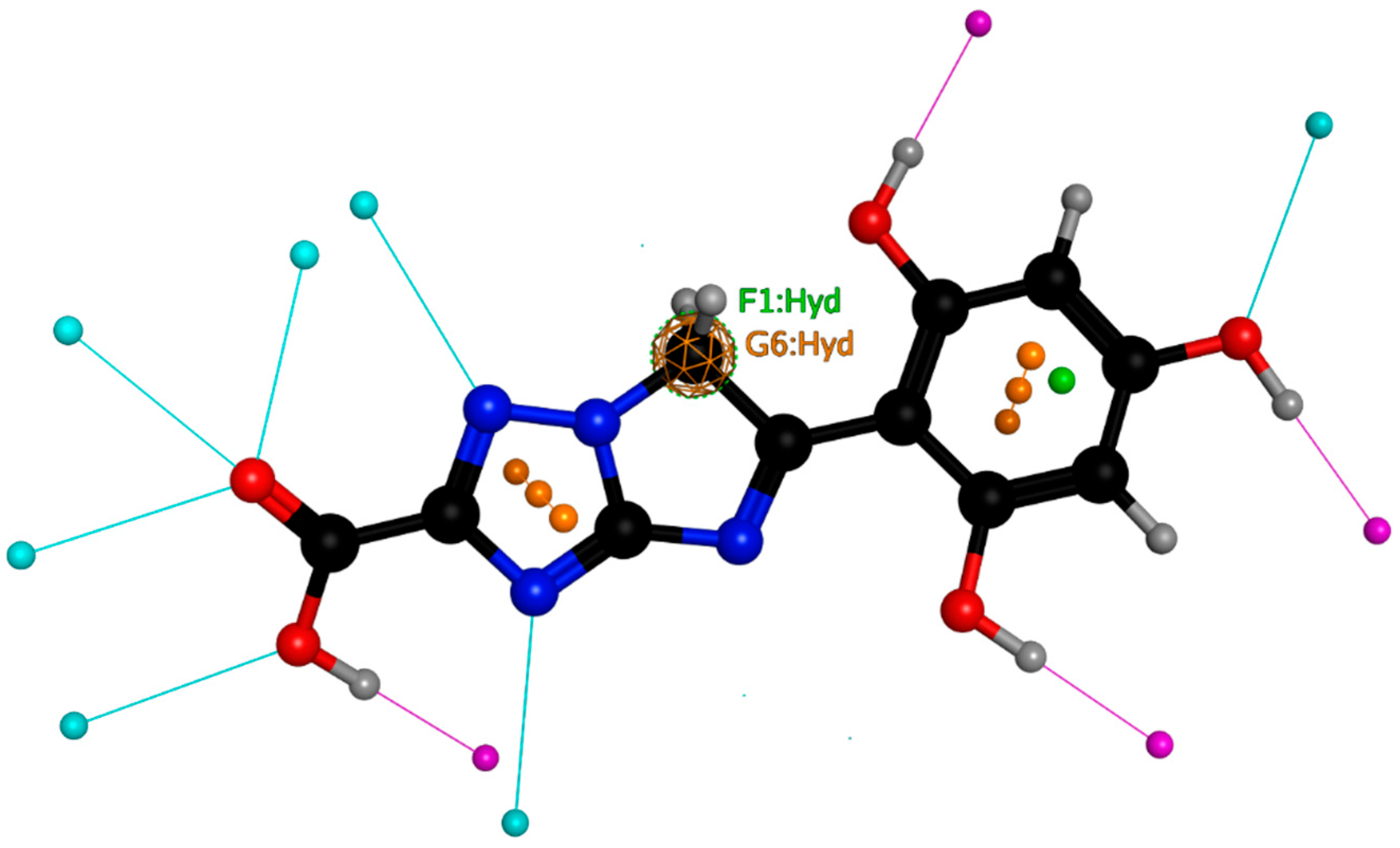
2.4. Pharmacophore Mapping
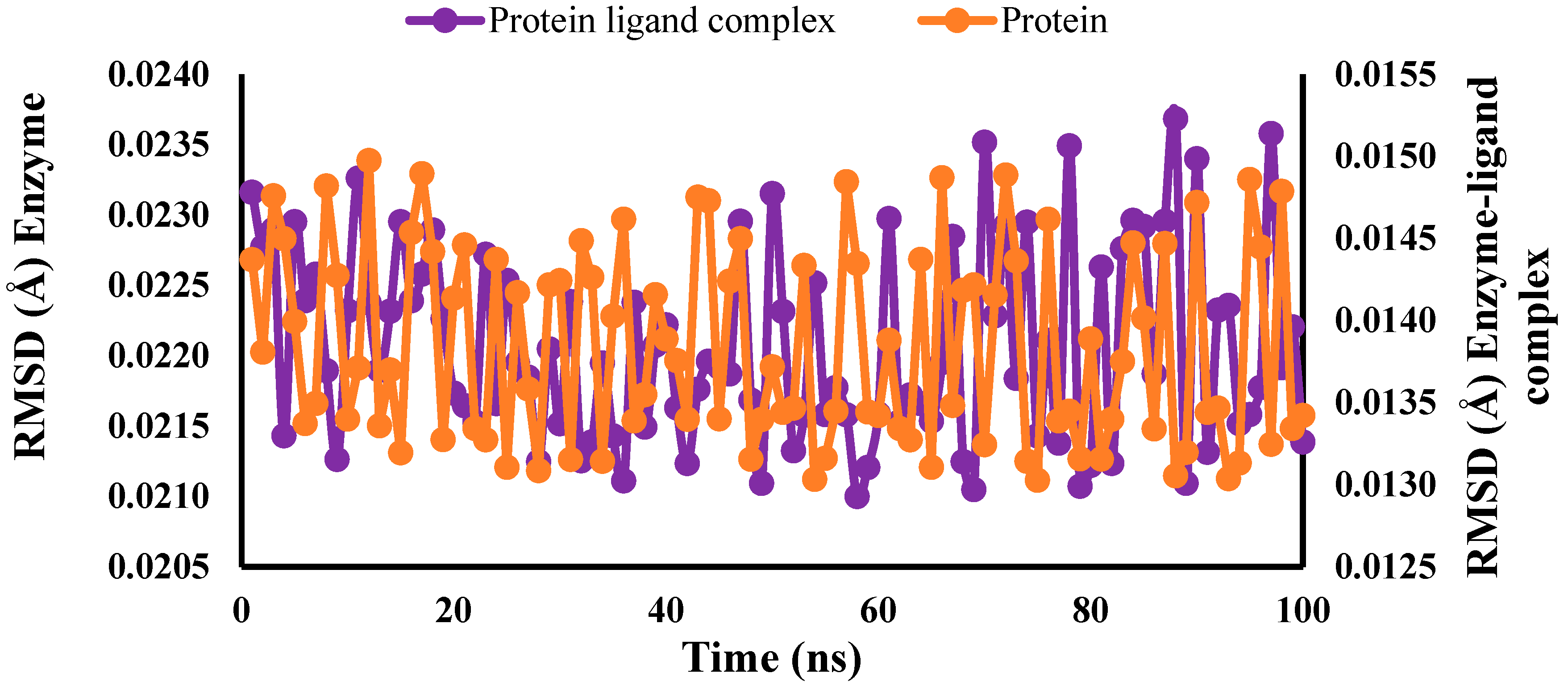
2.5. Molecular Dynamic (MD) Simulation Studies
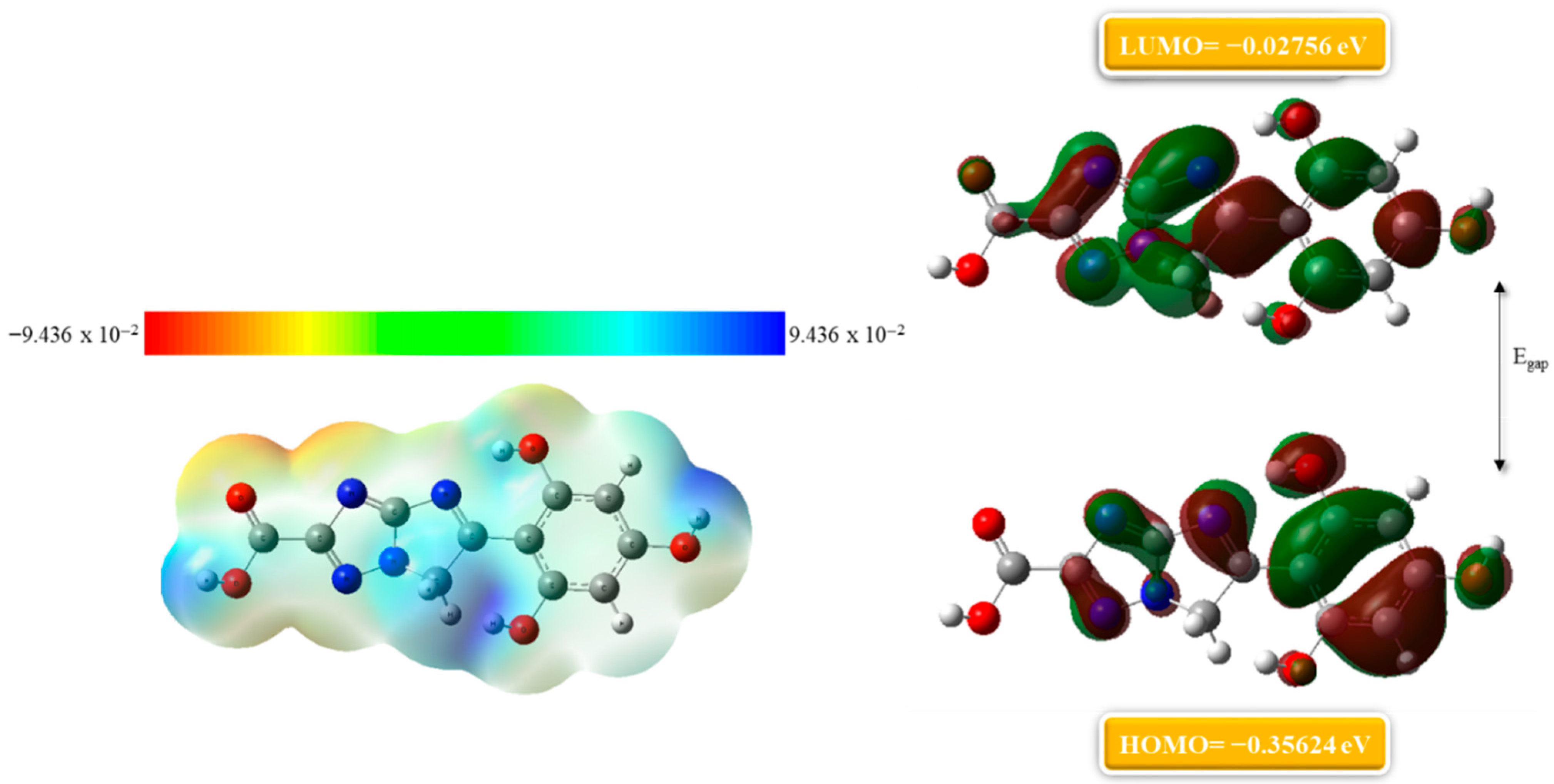
2.6. Density Functional Theory (DFT)
2.6.1. Molecular Electrostatic Potential Map
2.6.2. Frontier Molecular Orbitals (FMO) Analysis
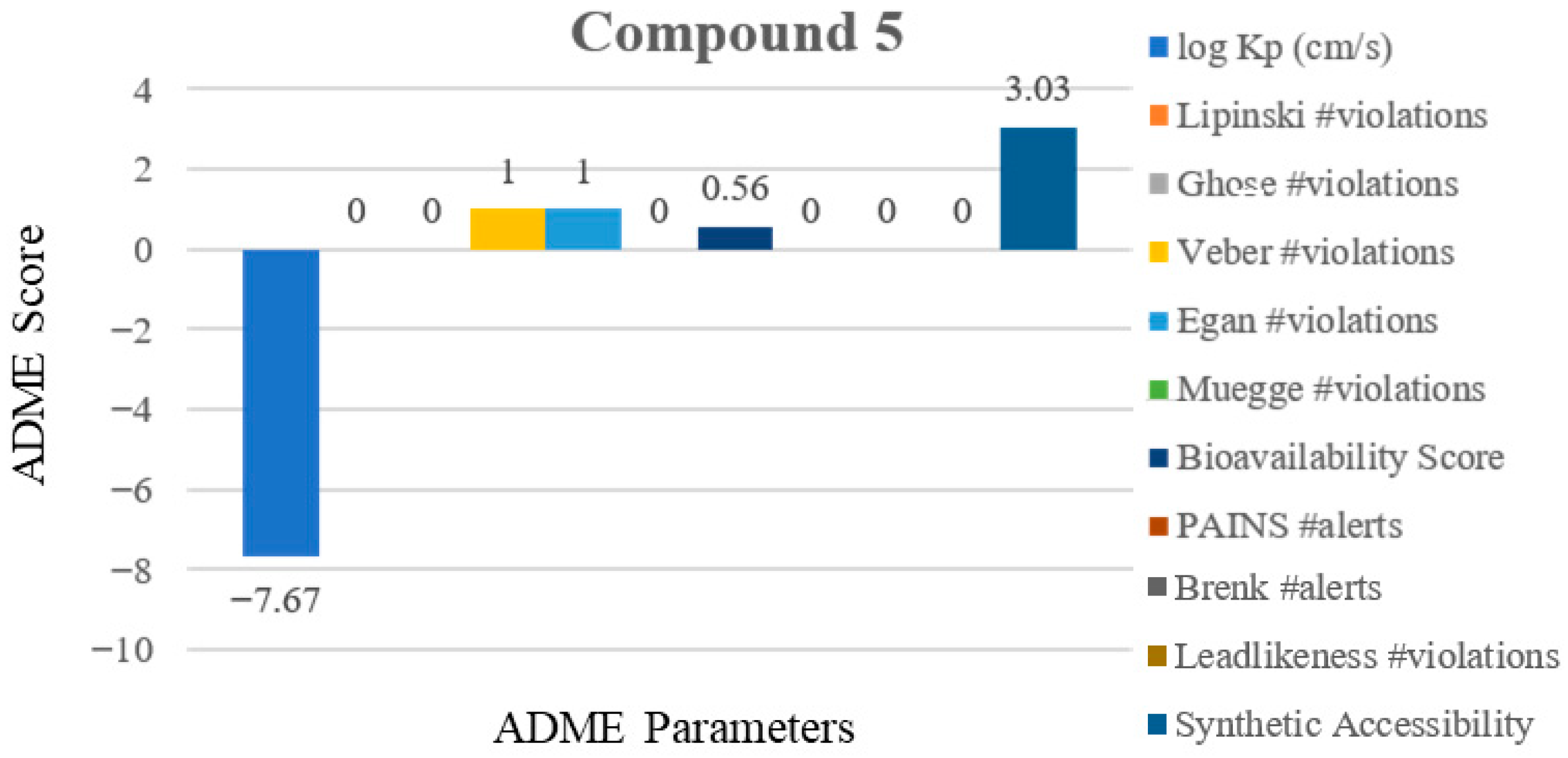
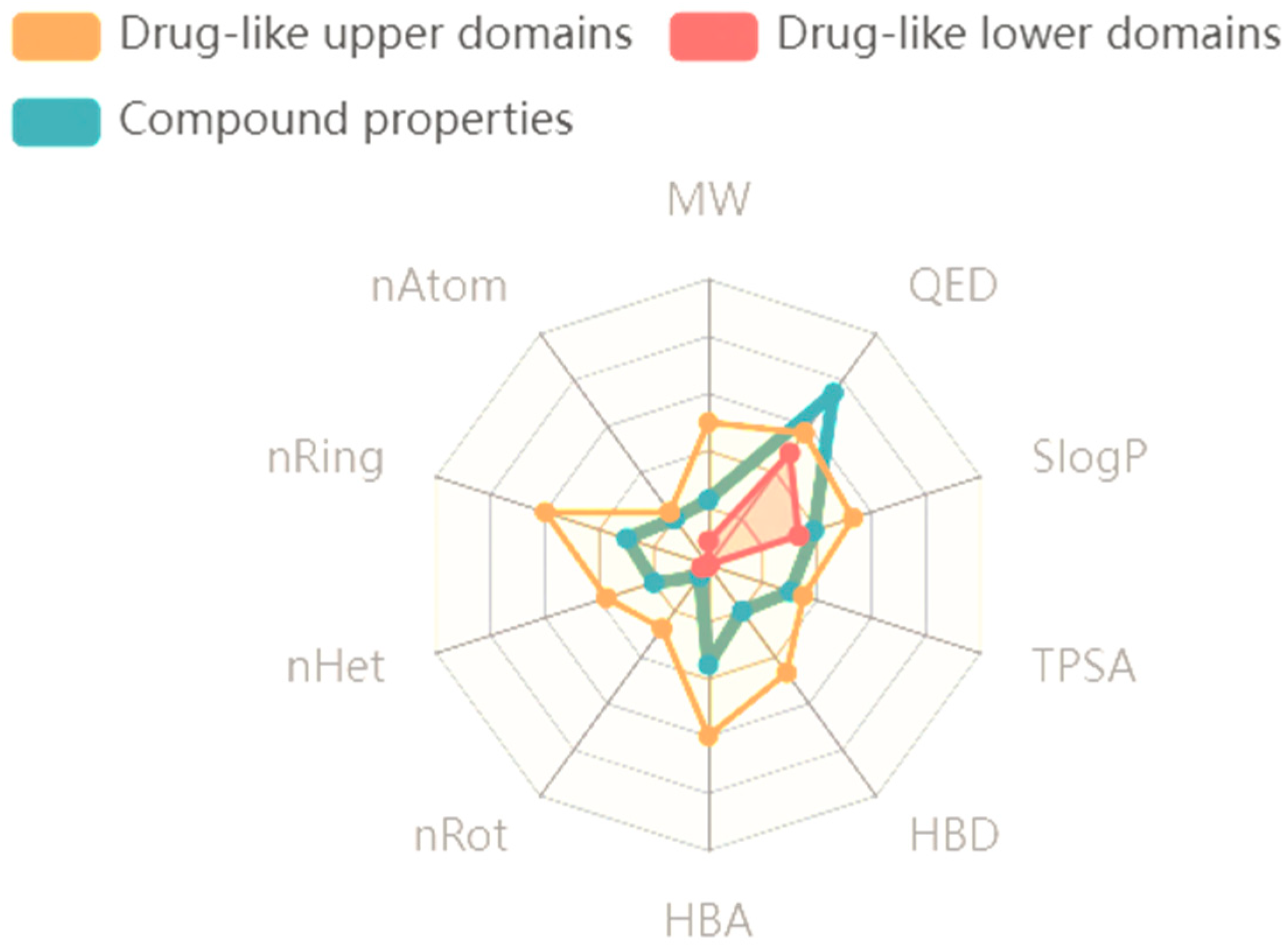
2.7. Absorption Distribution Metabolism Excretion and Toxicity (ADMET) Analysis
3. Materials and Methods
3.1. Materials
3.2. Methodology for the Synthesis of Imidazo–Triazole Conjugates (1–12)
3.3. Spectroscopic Analysis
- 5-(4-Fluoro-2,5-dimethylphenyl)-6H-imidazo{1,2-b]{1,2,4]triazole-2-carboxylic acid (1)
- 5-(m-Tolyl)-6H-imidazo{1,2-b]{1,2,4]triazole-2-carboxylic acid (2)
- 5-(3,5-Dimethyl-4-nitrophenyl)-6H-imidazo{1,2-b]{1,2,4]triazole-2-carboxylic acid (3)
- 5-(2-Bromophenyl)-6H-imidazo{1,2-b]{1,2,4]triazole-2-carboxylic acid (4)
- 5-(2,4,6-Trihydroxyphenyl)-6H-imidazo{1,2-b]{1,2,4]triazole-2-carboxylic acid (5)
- 5-(4-Bromo-3-methylphenyl)-6H-imidazo{1,2-b]{1,2,4]triazole-2-carboxylic acid (6)
- 5-(2,4-Dihydroxy-5-methylphenyl)-6H-imidazo{1,2-b]{1,2,4]triazole-2-carboxylic acid (7)
- 5-(4-Chloro-3,5-dimethylphenyl)-6H-imidazo{1,2-b]{1,2,4]triazole-2-carboxylic acid (8)
- 5-(4-Hydroxy-3,5-dimethylphenyl)-6H-imidazo{1,2-b]{1,2,4]triazole-2-carboxylic acid (9)
- 5-(4-Hydroxy-3-methylphenyl)-6H-imidazo{1,2-b]{1,2,4]triazole-2-carboxylic acid (10)
- 5-(4-Chlorophenyl)-6H-imidazo{1,2-b]{1,2,4]triazole-2-carboxylic acid (11)
- 5-(4-Nitrophenyl)-6H-imidazo{1,2-b]{1,2,4]triazole-2-carboxylic acid (12)
3.4. Assay Protocols of Inhibitory Activity
3.4.1. Alpha-Amylase Inhibition Assay
3.4.2. Alpha-Glucosidase Inhibition Assay
3.5. Molecular Docking Assay Protocols
3.6. Dft Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- American Diabetes Association. Diagnosis and Classification of Diabetes Mellitus. Diabetes Care. 2014, 37, S81–S90. [Google Scholar] [CrossRef]
- Kaul, K.; Tarr, J.M.; Ahmad, S.I.; Kohner, E.M.; Chibber, R. Introduction to diabetes mellitus. In Diabetes; Part of the Book Series: Advances in Experimental Medicine and Biology (AEMB, Volume 771); Springer: New York, NY, USA, 2013; pp. 1–11. [Google Scholar] [CrossRef]
- Alam, S.; Hasan, M.K.; Neaz, S.; Hussain, N.; Hossain, M.F.; Rahman, T. Diabetes Mellitus, insights from epidemiology, biochemistry, risk factors, diagnosis, complications and comprehensive management. Diabetology 2021, 2, 36–50. [Google Scholar] [CrossRef]
- Pawar, S.D.; Thakur, P.; Radhe, B.K.; Jadhav, H.; Behere, V.; Pagar, V. The accuracy of polyuria, polydipsia, polyphagia, and Indian Diabetes Risk Score in adults screened for diabetes mellitus type-II. Med. J. Dr. D.Y. Patil Univ. 2017, 10, 263–267. [Google Scholar] [CrossRef]
- Zheng, Y.; Ley, S.H.; Hu, F.B. Global aetiology and epidemiology of type 2 diabetes mellitus and its complications, Nature. Rev. Endocrinol. 2018, 14, 88–98. [Google Scholar] [CrossRef] [PubMed]
- Edelman, S.V. Type II diabetes mellitus. Adv. Intern. Med. 1998, 43, 449–500. [Google Scholar] [PubMed]
- Katsarou, A.; Gudbjörnsdottir, S.; Rawshani, A.; Dabelea, D.; Bonifacio, E.; Anderson, B.J.; Jacobsen, L.M.; Schatz, D.A.; Lernmark, A. Type 1 diabetes mellitus. Nat. Rev. Dis. Primers 2017, 3, 1–7. [Google Scholar] [CrossRef]
- Olokoba, A.B.; Obateru, O.A.; Olokoba, L.B. Type 2 diabetes mellitus: A review of current trends. Oman Med. J. 2012, 27, 269. [Google Scholar] [CrossRef]
- Saeedi, P.; Petersohn, I.; Salpea, P.; Malanda, B.; Karuranga, S.; Unwin, N.; Colagiuri, S.; Guariguata, L.; Motala, A.A.; Ogurtsova, K.; et al. Global and regional diabetes prevalence estimates for 2019 and projections for 2030 and 2045, Results from the International Diabetes Federation Diabetes Atlas. Diabetes Res. Clin. Pract. 2019, 157, 107843. [Google Scholar] [CrossRef] [PubMed]
- Chentli, F.; Azzoug, S.; Mahgoun, S. Diabetes mellitus in elderly. Indian J. Endocrinol. Metab. 2015, 19, 744–752. [Google Scholar] [CrossRef]
- DeFronzo, R.A. From the Triumvirate to the Ominous Octet: A New Paradigm for the Treatment of Type 2 Diabetes Mellitus. Diabetes 2009, 58, 773–795. [Google Scholar] [CrossRef]
- Ueno, H.; Tsuchimochi, W.; Wang, H.W.; Yamashita, E.; Tsubouchi, C.; Nagamine, K.; Sakoda, H.; Nakazato, M. Effects of miglitol, acarbose, and sitagliptin on plasma insulin and gut peptides in type 2 diabetes mellitus: A crossover study. Diabetes Ther. 2015, 6, 187–196. [Google Scholar] [CrossRef]
- Ali, I.; Lone, M.N.; Aboul-Enein, H.Y. Imidazoles as potential anticancer agents. Med. Chem. Comm. 2017, 8, 1742–1773. [Google Scholar] [CrossRef]
- Alam, M.M. 1, 2, 3-Triazole hybrids as anticancer agents, A review. Arch. Pharm. 2022, 355, 2100158. [Google Scholar] [CrossRef]
- Gao, F.; Wang, T.; Xiao, J.; Huang, G. Antibacterial activity study of 1,2,4-triazole derivatives. Eur. J. Med. Chem. 2019, 173, 274–281. [Google Scholar] [CrossRef]
- Khabnadideh, S.; Rezaei, Z.; Khalafi, A.N.; Bahrinajafi, R.; Mohamadi, R.; Farrokhroz, A.A. Synthesis of N-alkylated derivatives of imidazole as antibacterial agents. Bioorg. Med. Chem. Lett. 2003, 13, 2863–2865. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Fan, L.; Shi, L.; Wang, C.; Pan, Z.; Xu, C.; Yang, G. Synthesis, characterization and antifungal activity of imidazole chitosan derivatives. Carbohydr. Res. 2024, 544, 109238. [Google Scholar] [CrossRef]
- Lass, C.F. Triazole antifungal agents in invasive fungal infections: A comparative review. Drugs 2011, 71, 2405–2419. [Google Scholar] [CrossRef] [PubMed]
- Yousefnejad, F.; Mohammadi-Moghadam-Goozali, M.; Sayahi, M.H.; Halimi, M.; Moazzam, A.; Mohammadi-Khanaposhtani, M.; Mojtabavi, S.; Asadi, M.; Faramarzi, M.A.; Larijani, B.; et al. Design, synthesis, in vitro, and in silico evaluations of benzo [d] imidazole-amide-1,2,3-triazole-N-arylacetamide hybrids as new antidiabetic agents targeting α-glucosidase. Sci. Rep. 2023, 13, 12397. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, T.; Khan, S.; Ali, I.; Abass, K.S.; Alzahrani, E.; Zaki, M.E.; Gomha, S.M.; Alshehri, F.F.; Al Shehri, Z.S.; Kashtoh, H. Workflow-Driven Synthesis, Kinetic Evaluation and In Silico Profiling of Novel Fused Imidazo–Triazoles as Antidiabetic Scaffolds. J. Mol. Struct. 2025, 1349, 143680. [Google Scholar] [CrossRef]
- Sharma, D.; Narasimhan, B.; Kumar, P.; Judge, V.; Narang, R.; De, E.C.; Balzarini, J. Synthesis, antimicrobial and antiviral evaluation of substituted imidazole derivatives. Eur. J. Med Chem. 2009, 44, 2347–2353. [Google Scholar] [CrossRef]
- Kharb, R.; Yar, M.S.; Sharma, P.C. Recent advances and future perspectives of triazole analogs as promising antiviral agents. Mini-Rev. Med. Chem. 2011, 11, 84–96. [Google Scholar] [CrossRef] [PubMed]
- Çapan, İ.; Hawash, M.; Qaoud, M.T.; Jaradat, N. Next-generation carbazole-linked 1,2,4-triazole-thione derivatives: Strategic design, synthesis, molecular docking, and evaluation of antidiabetic potential. ACS Omega 2024, 10, 848–861. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Syal, B.; Seboletswe, P.; Cele, N.; Olofinsan, K.; Singh, P.; Shahidul Islam, M.; Singh, D.; Gupta, P. Synthesis, Biological Evaluation, Molecular Docking and Kinetic Investigation of New 2,4,5-Trisubstituted Imidazole Derivatives as Antidiabetic Agents. ChemistrySelect 2023, 8, e202300924. [Google Scholar] [CrossRef]
- Limbasiya, K.K.; Babariya, J.S.; Modhavadiya, V.A. Imidazol-1,2,3-triazole Derivatives as Novel and Potent Scaffolds of α-Glucosidase Inhibitors: Synthesis and Biological Evaluations. Russ. J. Org. Chem. 2024, 60 (Suppl. S1), S205–S212. [Google Scholar] [CrossRef]
- Azmi, A.; Noori, M.; Ghomi, M.K.; Montazer, M.N.; Iraji, A.; Dastyafteh, N.; Oliyaei, N.; Khoramjouy, M.; Rezaei, Z.; Javanshir, S.; et al. Alpha-glucosidase inhibitory and hypoglycemic effects of imidazole-bearing thioquinoline derivatives with different substituents: In silico, in vitro, and in vivo evaluations. Bioorg. Chem. 2024, 144, 107106. [Google Scholar] [CrossRef]
- Khan, S.; Iqbal, T.; Hussain, R.; Khan, Y.; Fiaz, Z.; Rahim, F.; Darwish, H.W. Synthesis, Characterizations, Anti-Diabetic and Molecular Modeling Approaches of Hybrid Indole-Oxadiazole Linked Thiazolidinone Derivatives. Pharmaceuticals 2024, 17, 1428. [Google Scholar] [CrossRef]
- Khan, S.; Iqbal, S.; Rahim, F.; Shah, M.; Hussain, R.; Alrbyawi, H.; Rehman, W.; Dera, A.A.; Rasheed, L.; Somaily, H.H.; et al. New biologically hybrid pharmacophore thiazolidinone-based indole derivatives: Synthesis, in vitro alpha-amylase and alpha-glucosidase along with molecular docking investigations. Molecules 2022, 27, 6564. [Google Scholar] [CrossRef]
- Khan, S.; Iqbal, T.; Hussain, R.; Alshehri, F.F.; Al Shehri, Z.S.; Gomha, S.M.; Zaki, M.E.; Kashtoh, H. Computationally Guided Design and Synthesis of Pyrimidine–Oxazole Hybrids as Novel Antidiabetic Agents: Kinetic and Molecular Interaction Studies. Comp. Bio. Chem. 2025, 119, 108602. [Google Scholar] [CrossRef]
- Khan, S.; Iqbal, T. A new synthetic approach for determination of thiadiazole clubbed sulfonamide as diabetic therapeutic with in silico modeling and kinetic study. J. Mol. Struct. 2025, 1338, 142261. [Google Scholar] [CrossRef]
- Khan, S.; Hussain, R.; Khan, Y.; Iqbal, T.; Khan, M.B.; Al-Ahmary, K.M.; Mhyawi, S.R.A. Insight into role of triazole derived Schiff base bearing sulfonamide derivatives in targeting Alzheimer’s disease: Synthesis, characterization, in vitro and in silico assessment. J. Mol. Struct. 2024, 1315, 138845. [Google Scholar] [CrossRef]
- Hussain, R.; Ashraf, M.; Khan, S.; Rahim, F.; Rehman, W.; Taha, M.; Sardar, A.; Khan, Y.; Khan, I.; Shah, S.A.A. Molecular modeling, synthesis, and in vitro acetylcholinesterase and butyrylcholinesterase inhibitory activities of novel benzimidazole-bearing thiadiazole derivatives. J. Mol. Struct. 2024, 1295, 136582. [Google Scholar] [CrossRef]
- Khan, S.; Iqbal, T.; Khan, M.B.; Hussain, R.; Islam, M.S.; Dahlous, K.A. Bioactive Pyrazolone Derived Thiazolidinone Scaffolds As a Safe Perspective to Human Health: Anti-Alzheimer’s, DFT, ADMET, and Molecular Docking Insight. ChemistrySelect 2025, 10, e202406105. [Google Scholar] [CrossRef]
- Khan, S.; Iqbal, T.; Rehman, M.U.; Khan, M.B.; Islam, M.S.; Dahlous, K.A. Integrated insight and in silico investigation of hybrid bis-thiazolidinone derivatives along with anti-Alzheimer activity. 3 Biotech 2025, 15, 176. [Google Scholar] [CrossRef]
- Iqbal, T.; Khan, S.; Hussain, R.; Hawsawi, M.B.; Mehmood, T.; Tahir, Y.; Alluhaibi, M.S.; Alharbi, M. Investigation on diabetes and mechanism of thiadiazole–Schiff base as therapeutic hybrid: Enzyme activity and molecular docking studies. ChemistrySelect 2025, 10, 202404291. [Google Scholar] [CrossRef]
- Frisch, M.; Clemente, F. Gaussian 09, Revision A.01; Frisch, M.J., Trucks, G.W., Schlegel, H.B., Scuseria, G.E., Robb, M.A., Cheeseman, J.R., Scalmani, G., Barone, V., Mennucci, B.G.A., Eds.; Petersson, Gaussian, Inc.: Wallingford, CT, USA, 2009. [Google Scholar]
- Khan, A.U.; Muhammad, S.; Khera, R.A.; Shehzad, R.A.; Ayub, K.; Iqbal, J. DFT study of superhalogen (AlF4) doped boron nitride for tuning their nonlinear optical properties. Optik 2021, 231, 166464. [Google Scholar] [CrossRef]
- Esrafili, M.D.; Saeidi, N. DFT calculations on the catalytic oxidation of CO over Si-doped (6,0) boron nitride nanotubes. Struct. Chem. 2016, 27, 595–604. [Google Scholar] [CrossRef]
- Khan, S.; Yar, M.; Kosar, N.; Ayub, K.; Arshad, M.; Zahid, M.N.; Mahmood, T. First-principles study for exploring the adsorption behavior of G-series nerve agents on graphdyine surface. Comput. Theor. Chem. 2020, 1191, 113043. [Google Scholar] [CrossRef]
- Prasad, O.; Sinha, L.; Kumar, N. Theoretical Raman and IR spectra of tegafur and comparison of molecular electrostatic potential surfaces, polarizability and hyperpolarizability of tegafur with 5-fluoro-uracil by density functional theory. J. At. Mol. Sci 2010, 1, 201–214. [Google Scholar] [CrossRef]
- Hagar, M.; Ahmed, H.A.; Aljohani, G.; Alhaddad, O.A. Investigation of some antiviral N-heterocycles as COVID 19 drug: Molecular docking and DFT calculations. Int. J. Mol. Sci. 2020, 21, 3922. [Google Scholar] [CrossRef]
- Liu, H.-B.; Gao, W.-W.; Tangadanchu, V.K.R.; Zhou, C.-H.; Geng, R.-X. Novel aminopyrimidinyl benzimidazoles as potentially antimicrobial agents: Design, synthesis and biological evaluation. Eur. J. Med. Chem. 2018, 143, 66–84. [Google Scholar] [CrossRef] [PubMed]
- Marinho, E.S.; Marinho, M.M. A DFT study of synthetic drug topiroxostat: MEP, HOMO, LUMO. Int. J. Sci. Eng. Res 2016, 7, 1264. [Google Scholar]
- Govindappa, M.; Hemashekhar, B.; Arthikala, M.; Rai, V.R.; Ramachandra, Y.L. Characterization, antibacterial, antioxidant, antidiabetic, anti-inflammatory and antityrosinase activity of green synthesized silver nanoparticles using Calophyllum tomentosum leaves extract. Results Phys. 2018, 9, 400–408. [Google Scholar] [CrossRef]
- Ali, H.; Houghton, P.J.; Soumyanath, A. Amylase inhibitory activity of some Malaysian plants used to treat diabetes; with particular reference to Phyllanthus amarus. J. Ethnopharmacol. 2006, 107, 449–455. [Google Scholar] [CrossRef]
- Korb, O.; Stützle, T.; Exner, T.E. Empirical scoring functions for advanced protein-ligand docking with PLANTS. J. Chem. Inf. Model. 2009, 49, 84–96. [Google Scholar] [CrossRef]
- Frisch, M.J. Gaussian 98, Revision A.9; Gaussian Inc.: Pittsburgh, PA, USA, 1998. [Google Scholar]
- El-Baz, A.F.; Sorour, N.M.; Shetaia, Y.M. Trichosporon jirovecii-mediated synthesis of cadmium sulfide nanoparticles. J. Basic Microbiol. 2016, 56, 520–530. [Google Scholar] [CrossRef] [PubMed]
- Becke, A.D. Density-functional exchange-energy approximation with correct asymptotic behavior. Phys. Rev. A 1988, 38, 3098–3100. [Google Scholar] [CrossRef]
- Lee, C.; Yang, W.; Parr, R.G. Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys. Rev. B 1988, 37, 785–789. [Google Scholar] [CrossRef] [PubMed]
- Flurry, R.L., Jr. Molecular Orbital Theory of Bonding in Organic Molecules; Marcel Dekker: New York, NY, USA, 1968. [Google Scholar]
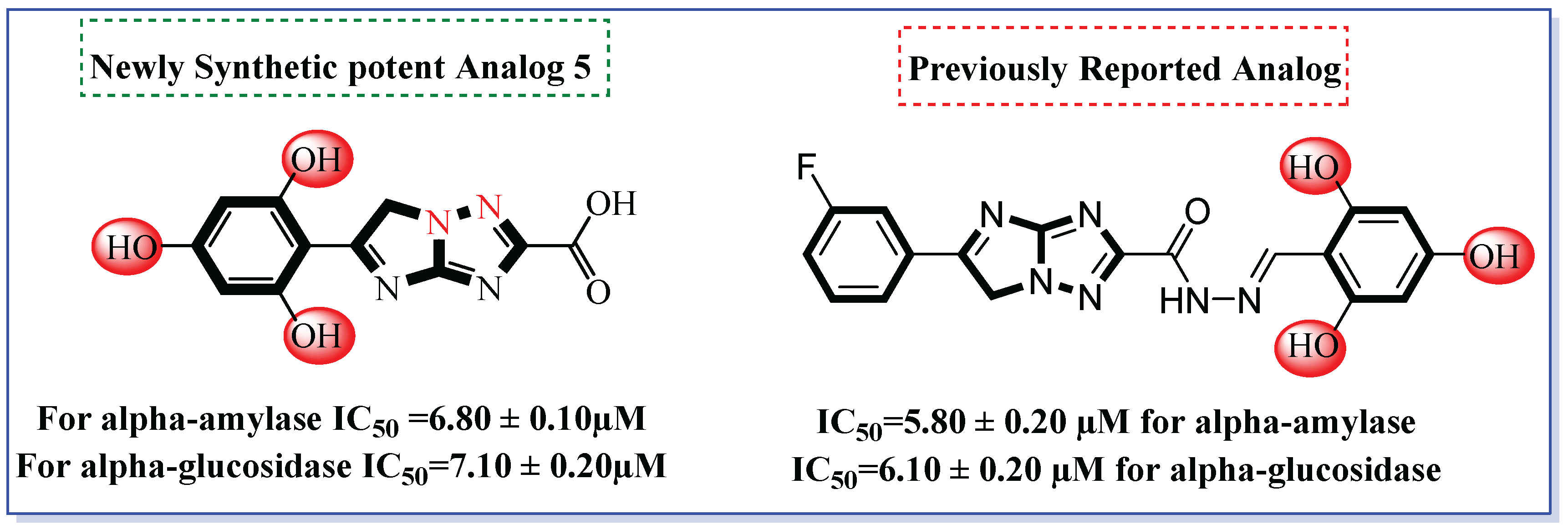
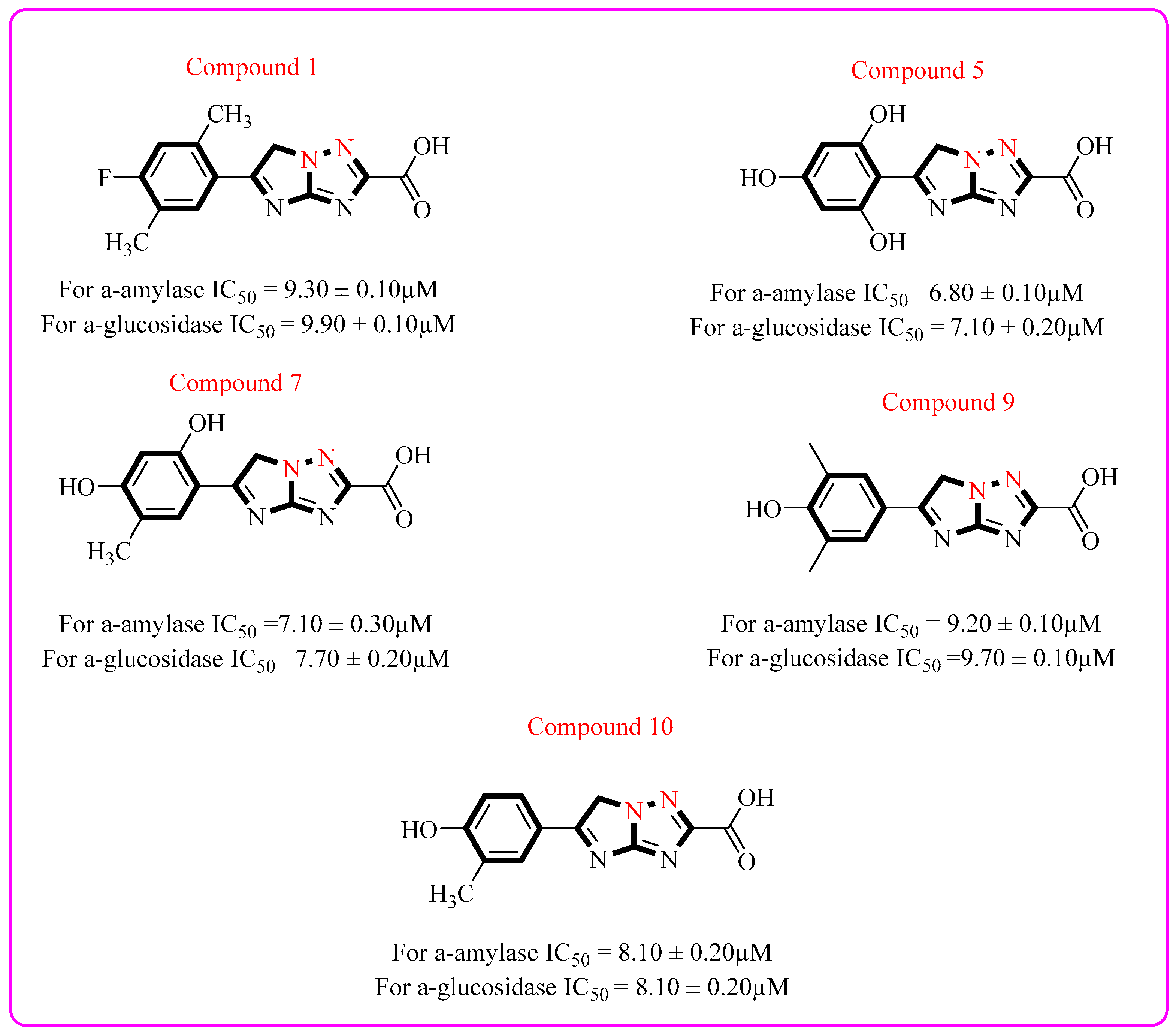

| S/No. | Compounds | IC50 = α-Amylase µM ± SEM | IC50 = α-Glucosidase µM ± SEM |
|---|---|---|---|
| 1 | 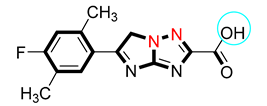 | 9.30 ± 0.10 | 9.90 ± 0.10 |
| 2 | 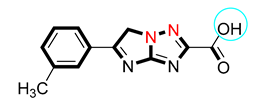 | 13.20 ± 0.20 | 14.10 ± 0.60 |
| 3 | 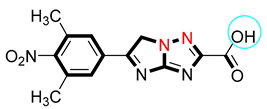 | 19.10 ± 0.20 | 19.70 ± 0.20 |
| 4 |  | 21.40 ± 0.10 | 22.10 ± 0.10 |
| 5 | 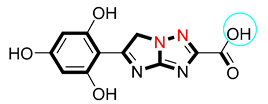 | 6.80 ± 0.10 | 7.10 ± 0.20 |
| 6 | 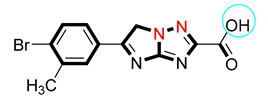 | 20.30 ± 0.40 | 21.10 ± 0.40 |
| 7 | 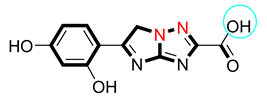 | 7.10 ± 0.30 | 7.70 ± 0.20 |
| 8 | 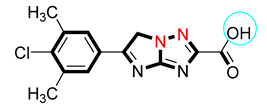 | 11.40 ± 0.20 | 12.10 ± 0.50 |
| 9 | 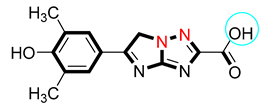 | 9.20 ± 0.10 | 9.70 ± 0.10 |
| 10 | 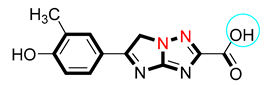 | 8.10 ± 0.20 | 8.10 ± 0.20 |
| 11 | 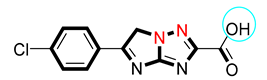 | 13.20 ± 0.10 | 13.60 ± 0.20 |
| 12 | 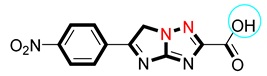 | 17.50 ± 0.60 | 18.10 ± 0.20 |
| Standard Drug Acarbose | 9.40 ± 0.20 | 9.80 ± 0.30 | |
| Compound | Receptor | Interactions | Binding Distance (Å) | Docking Score (kcal/mol) |
|---|---|---|---|---|
| Compound 5 in alpha-amylase complex | ARG-A-195 | H-Bond | 6.19 | −12.44 |
| ARG-A-195 | H-Bond | 6.81 | ||
| TYR-A-62 | C–H Bond | 3.53 | ||
| TRP-A-59 | Pi–Pi Stacked | 4.38 | ||
| ASP-A-300 | Pi-Anion | 6.81 | ||
| Compound 5 in alpha-glucosidase complex | PHE-C-439 | H-Bond | 5.26 | −11.82 |
| PHE-C-439 | C–H Bond | 5.47 | ||
| PHE-C-442 | Pi-Alkyl | 6.18 | ||
| PHE-C-442 | Pi–Pi Stacked | 5.84 | ||
| VAL-C-474 | Pi-Sigma | 4.93 | ||
| VAL-C-474 | Pi-Alkyl | 5.51 | ||
| TYR-A-32 | H-Bond | 6.62 | ||
| ASP-A-79 | H-Bond | 3.06 | ||
| Compound 7 in alpha-amylase complex | ASP-A-197 | H-Bond | 4.27 | −10.30 |
| TYR-A-62 | Pi–Pi Stacked | 4.37 | ||
| LEU-A-165 | Pi-Alkyl | 5.48 | ||
| LEU-A-165 | Pi-Alkyl | 5.29 | ||
| GLN-A-63 | H-Bond | 4.84 | ||
| Compound 7 in alpha-glucosidase complex | ASP-A-79 | H-Bond | 3.76 | −9.55 |
| ASP-A-79 | H-Bond | 4.83 | ||
| GLN-C-472 | H-Bond | 5.85 | ||
| TYR-A-32 | C–H Bond | 5.73 | ||
| TYR-A-32 | C–H Bond | 6.18 | ||
| VAL-C-474 | Pi-Alkyl | 4.00 | ||
| VAL-C-474 | Pi-Sigma | 4.06 | ||
| PHE-C-442 | Pi-Sulfur | 7.09 | ||
| Compound 10 in alpha-amylase complex | TRP-A-59 | Pi–Pi Stacked | 5.80 | −8.76 |
| TRP-A-59 | Pi-Alkyl | 4.07 | ||
| TRP-A-58 | Pi-Alkyl | 6.00 | ||
| TYR-A-62 | Pi–Pi Stacked | 5.02 | ||
| ARG-A-195 | H-Bond | 6.62 | ||
| ASP-A-300 | Pi-Anion | 5.87 | ||
| HIS-A-305 | Pi-Alkyl | 5.97 | ||
| HIS-A-305 | Pi–Pi Stacked | 5.23 | ||
| Compound 10 in alpha-glucosidase complex | GLN-C-472 | H-Bond | 5.32 | −8.00 |
| TYR-A-32 | C–H Bond | 5.69 | ||
| LYS-C-446 | Unfavorable Acceptor-Acceptor | 3.95 | ||
| PHE-C-442 | Pi-Alkyl | 4.71 | ||
| PHE-C-442 | Pi–Pi Stacked | 5.75 | ||
| VAL-C-474 | Pi-Alkyl | 5.06 | ||
| VAL-C-474 | Pi-Alkyl | 4.07 | ||
| VAL-C-474 | Pi-Sigma | 4.47 | ||
| THR-C-470 | C–H Bond | 4.11 | ||
| GLN-A-81 | H-Bond | 3.37 | ||
| GLN-A-81 | H-Bond | 3.63 |
| Property | Model Name | Predicted Value | Unit |
|---|---|---|---|
| Absorption | Water solubility | −2.762 | Numeric (log mol/L) |
| TPSA | 141.06 | Numeric (Å2) | |
| Caco2 permeability | −0.723 | Numeric (log Papp in 10−6 cm/s) | |
| Intestinal absorption (human) | 52.614 | Numeric (% Absorbed) | |
| Skin Permeability | −2.735 | Numeric (log Kp) | |
| P-glycoprotein substrate | Yes | Categorical (Yes/No) | |
| P-glycoprotein I inhibitor | No | Categorical (Yes/No) | |
| P-glycoprotein II inhibitor | No | Categorical (Yes/No) | |
| Distribution | VDss (human) | −0.149 | Numeric (log L/kg) |
| Fraction unbound (human) | 0.378 | Numeric (Fu) | |
| BBB permeability | −1.576 | Numeric (log BB) | |
| CNS permeability | −4.303 | Numeric (log PS) | |
| Metabolism | CYP2D6 substrate | No | Categorical (Yes/No) |
| CYP3A4 substrate | No | Categorical (Yes/No) | |
| CYP1A2 inhibitor | No | Categorical (Yes/No) | |
| CYP2C19 inhibitor | No | Categorical (Yes/No) | |
| CYP2C9 inhibitor | No | Categorical (Yes/No) | |
| CYP2D6 inhibitor | No | Categorical (Yes/No) | |
| CYP3A4 inhibitor | No | Categorical (Yes/No) | |
| Excretion | Total Clearance | 0.499 | Numeric (log ml/min/kg) |
| Renal OCT2 substrate | No | Categorical (Yes/No) | |
| Toxicity | AMES toxicity | No | Categorical (Yes/No) |
| Max. tolerated dose (human) | 0.541 | Numeric (log mg/kg/day) | |
| hERG I inhibitor | No | Categorical (Yes/No) | |
| hERG II inhibitor | No | Categorical (Yes/No) | |
| Oral Rat Acute Toxicity (LD50) | 2.457 | Numeric (mol/kg) | |
| Oral Rat Chronic Toxicity (LOAEL) | 2.828 | Numeric (log mg/kg_bw/day) | |
| Hepatotoxicity | No | Categorical (Yes/No) | |
| Skin Sensitization | No | Categorical (Yes/No) | |
| T. Pyriformis toxicity | 0.285 | Numeric (log ug/L) | |
| Minnow toxicity | 2.363 | Numeric (log mM) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khowdiary, M.M.; Felemban, S. Fused Imidazotriazole-Based Therapeutics: A Multidisciplinary Study Against Diabetes-Linked Enzymes Alpha-Amylase and Alpha-Glucosidase Using In Vitro and In Silico Methods. Pharmaceuticals 2025, 18, 1333. https://doi.org/10.3390/ph18091333
Khowdiary MM, Felemban S. Fused Imidazotriazole-Based Therapeutics: A Multidisciplinary Study Against Diabetes-Linked Enzymes Alpha-Amylase and Alpha-Glucosidase Using In Vitro and In Silico Methods. Pharmaceuticals. 2025; 18(9):1333. https://doi.org/10.3390/ph18091333
Chicago/Turabian StyleKhowdiary, Manal M., and Shifa Felemban. 2025. "Fused Imidazotriazole-Based Therapeutics: A Multidisciplinary Study Against Diabetes-Linked Enzymes Alpha-Amylase and Alpha-Glucosidase Using In Vitro and In Silico Methods" Pharmaceuticals 18, no. 9: 1333. https://doi.org/10.3390/ph18091333
APA StyleKhowdiary, M. M., & Felemban, S. (2025). Fused Imidazotriazole-Based Therapeutics: A Multidisciplinary Study Against Diabetes-Linked Enzymes Alpha-Amylase and Alpha-Glucosidase Using In Vitro and In Silico Methods. Pharmaceuticals, 18(9), 1333. https://doi.org/10.3390/ph18091333






