Humulus lupulus (Hop)-Derived Chemical Compounds Present Antiproliferative Activity on Various Cancer Cell Types: A Meta-Regression Based Panoramic Meta-Analysis
Abstract
1. Introduction
2. Results
2.1. Selection and Characteristics of Studies
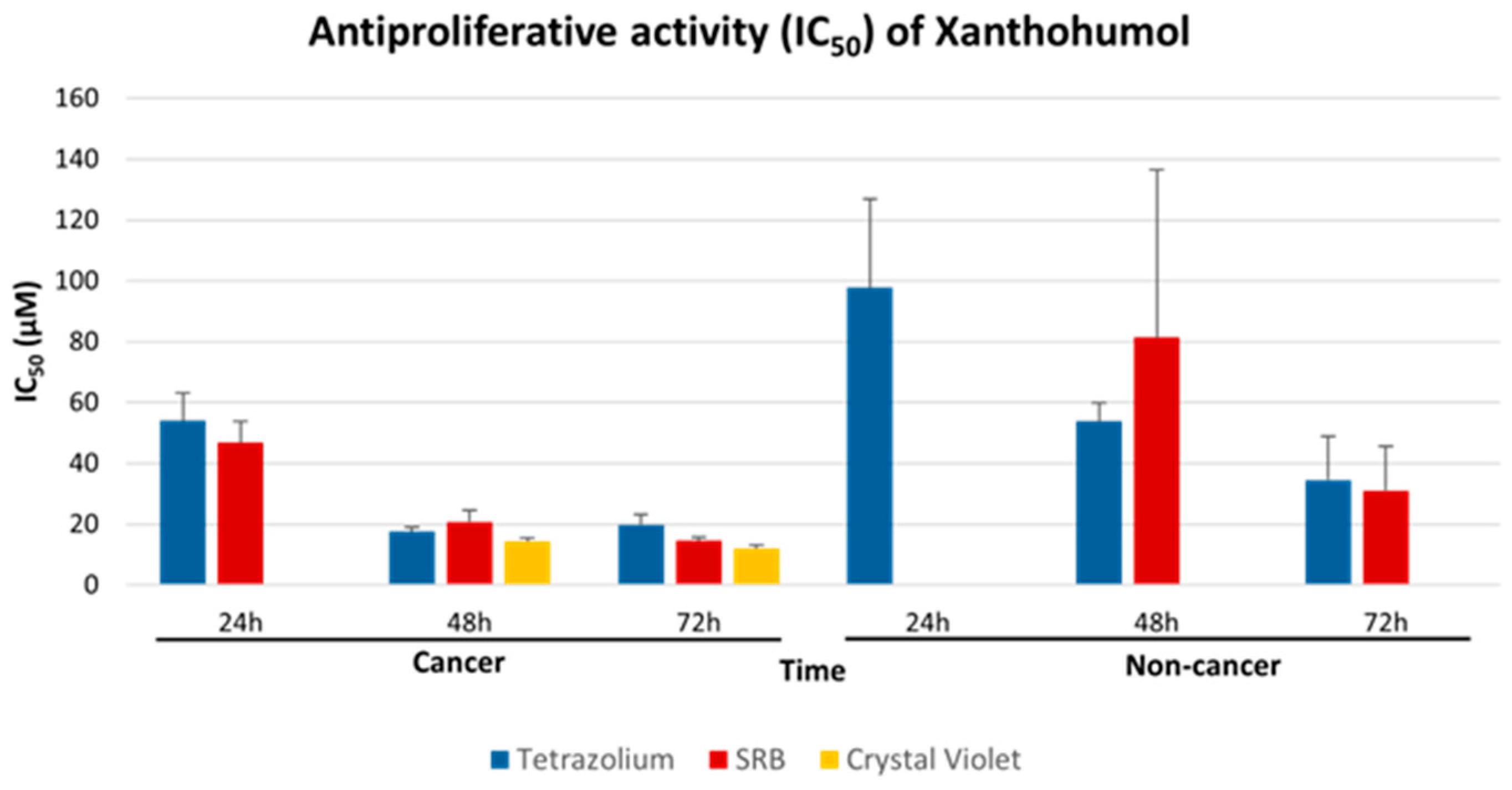
2.2. Interchangeability of Tetrazolium-Based, SRB and CV Assays
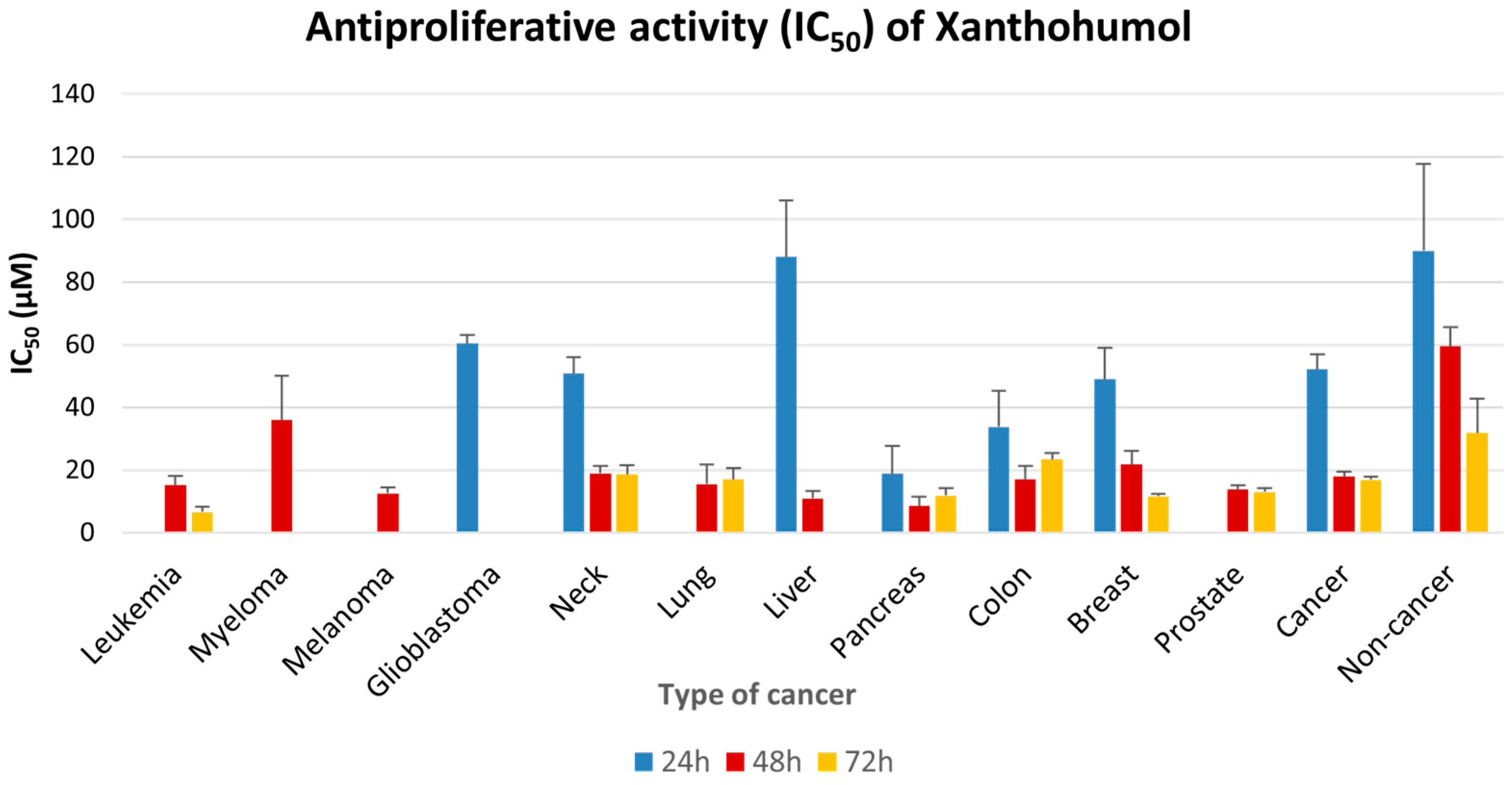
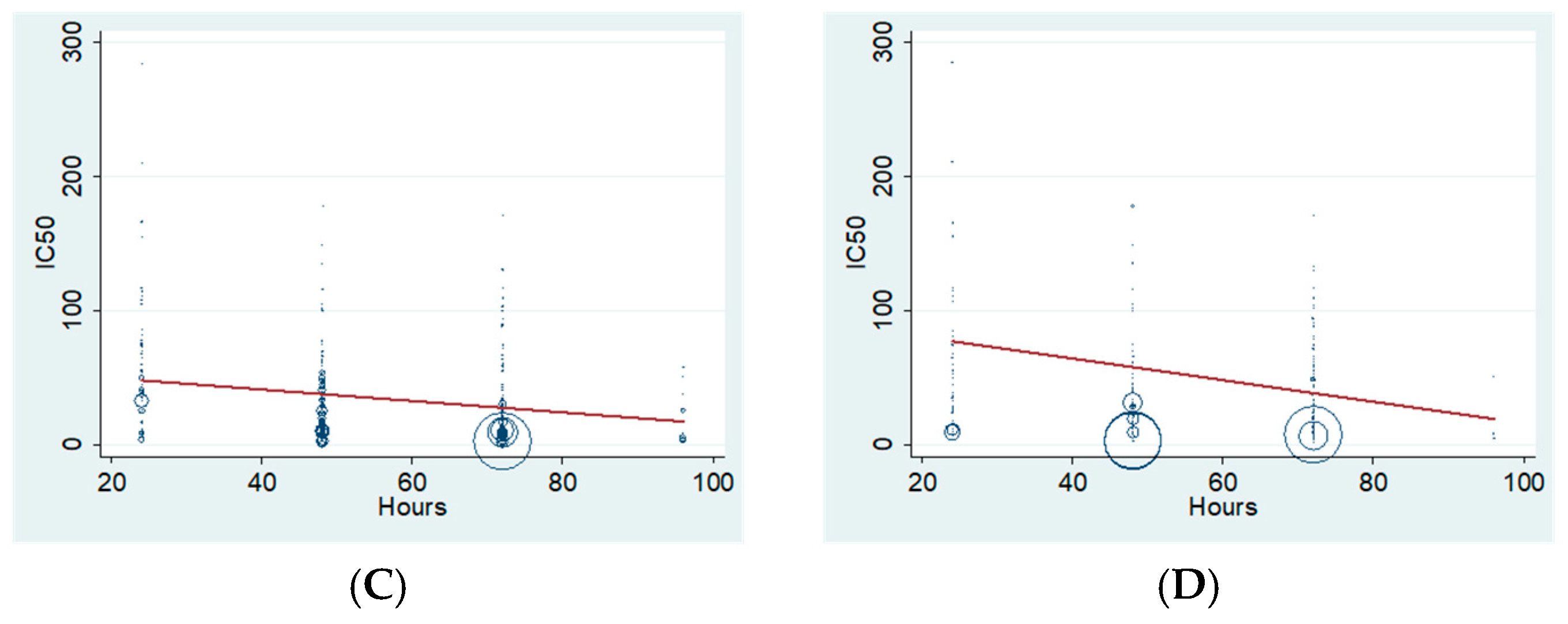
2.3. Antiproliferative Effect of XN Increases with Time of Incubation
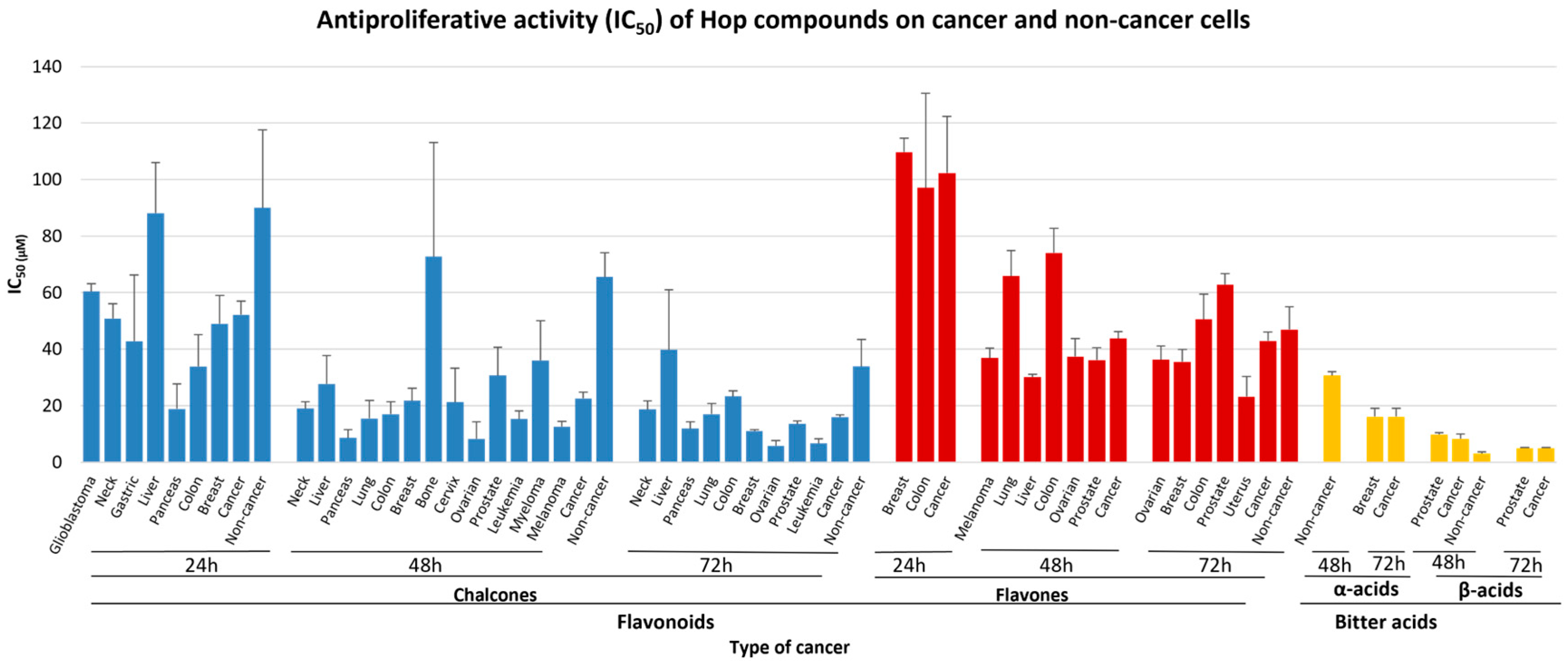
2.4. Antiproliferative Potential of Hops Flavonoids, Bitter Acids and Crude Extracts
3. Discussion
4. Materials and Methods
4.1. Literature Search Strategy
4.2. Study Selection Criteria
4.3. Studies’ Outcomes and Data Extraction
4.4. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| MTT | 3-[4,5-dimethylthiazol-2-yl]-2,5 diphenyl tetrazolium bromide |
| SRB | Sulfohodamine B |
| XTT | 2,3-bis-(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide |
| MTS | 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium |
| WST | Water-Soluble Tetrazolium Salt |
| CV | Crystal Violet |
| PRISMA | Preferred Reporting Items for Systematic Reviews and Meta-Analyses |
| SMD | Standardized Mean Difference |
| IC50 | Half Maximal Inhibitory Concentration |
| XN | Xanthohumol |
| IXN | Isoxanthohumol |
| DMX | Desmethylxanthohumol |
| 8-PN | 8-prenylnaringenin |
| 6-PN | 6-prenylnaringenin |
References
- Carbone, K.; Gervasi, F. An Updated Review of the Genus Humulus: A Valuable Source of Bioactive Compounds for Health and Disease Prevention. Plants 2022, 11, 3434. [Google Scholar] [CrossRef]
- Fukuda, T.; Ohya, R.; Kobayashi, K.; Ano, Y. Matured Hop Bitter Acids in Beer Improve Lipopolysaccharide-Induced Depression-Like Behavior. Front. Neurosci. 2019, 13, 41. [Google Scholar] [CrossRef] [PubMed]
- Gerhauser, C.; Alt, A.; Heiss, E.; Gamal-Eldeen, A.; Klimo, K.; Knauft, J.; Neumann, I.; Scherf, H.-R.; Frank, N.; Bartsch, H.; et al. Cancer Chemopreventive Activity of Xanthohumol, a Natural Product Derived from Hop. Mol. Cancer Ther. 2002, 1, 959–969. [Google Scholar] [PubMed]
- Hitzman, R.T.; Dunlap, T.L.; Howell, C.E.; Chen, S.-N.; Vollmer, G.; Pauli, G.F.; Bolton, J.L.; Dietz, B.M. 6-Prenylnaringenin from Hops Disrupts ERα-Mediated Downregulation of CYP1A1 to Facilitate Estrogen Detoxification. Chem. Res. Toxicol. 2020, 33, 2793–2803. [Google Scholar] [CrossRef]
- Moureu, S.; Jacquin, J.; Samaillie, J.; Deweer, C.; Rivière, C.; Muchembled, J. Antifungal Activity of Hop Leaf Extracts and Xanthohumol on Two Strains of Venturia Inaequalis with Different Sensitivities to Triazoles. Microorganisms 2023, 11, 1605. [Google Scholar] [CrossRef]
- Rozalski, M.; Micota, B.; Sadowska, B.; Stochmal, A.; Jedrejek, D.; Wieckowska-Szakiel, M.; Rozalska, B. Antiadherent and Antibiofilm Activity of Humulus lupulus L. Derived Products: New Pharmacological Properties. BioMed Res. Int. 2013, 2013, 101089. [Google Scholar] [CrossRef]
- Stompor, M.; Dancewicz, K.; Gabryś, B.; Anioł, M. Insect Antifeedant Potential of Xanthohumol, Isoxanthohumol, and Their Derivatives. J. Agric. Food Chem. 2015, 63, 6749–6756. [Google Scholar] [CrossRef]
- Yamaguchi, N.; Satoh-Yamaguchi, K.; Ono, M. In Vitro Evaluation of Antibacterial, Anticollagenase, and Antioxidant Activities of Hop Components (Humulus lupulus) Addressing Acne Vulgaris. Phytomedicine 2009, 16, 369–376. [Google Scholar] [CrossRef]
- Power, F.B.; Tutin, F.; Rogerson, H. CXXXV.—The Constituents of Hops. J. Chem. Soc. Trans. 1913, 103, 1267–1292. [Google Scholar] [CrossRef]
- Stevens, J.F.; Page, J.E. Xanthohumol and Related Prenylflavonoids from Hops and Beer: To Your Good Health! Phytochemistry 2004, 65, 1317–1330. [Google Scholar] [CrossRef] [PubMed]
- Logan, I.E.; Miranda, C.L.; Lowry, M.B.; Maier, C.S.; Stevens, J.F.; Gombart, A.F. Antiproliferative and Cytotoxic Activity of Xanthohumol and Its Non-Estrogenic Derivatives in Colon and Hepatocellular Carcinoma Cell Lines. Int. J. Mol. Sci. 2019, 20, 1203. [Google Scholar] [CrossRef]
- Klimek, K.; Tyśkiewicz, K.; Miazga-Karska, M.; Dębczak, A.; Rój, E.; Ginalska, G. Bioactive Compounds Obtained from Polish “Marynka” Hop Variety Using Efficient Two-Step Supercritical Fluid Extraction and Comparison of Their Antibacterial, Cytotoxic, and Anti-Proliferative Activities In Vitro. Molecules 2021, 26, 2366. [Google Scholar] [CrossRef] [PubMed]
- Viegas, O.; Žegura, B.; Pezdric, M.; Novak, M.; Ferreira, I.M.P.L.V.O.; Pinho, O.; Filipič, M. Protective Effects of Xanthohumol against the Genotoxicity of Heterocyclic Aromatic Amines MeIQx and PhIP in Bacteria and in Human Hepatoma (HepG2) Cells. Food Chem. Toxicol. 2012, 50, 949–955. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, R.J.; Miranda, C.L.; Stevens, J.F.; Deinzer, M.L.; Buhler, D.R. Influence of prenylated and non-prenylated flavonoids on liver microsomal lipid peroxidation and oxidative injury in rat hepatocytes. Food Chem. Toxicol. 2001, 39, 437–445. [Google Scholar] [CrossRef]
- Lu, X.; Geng, J.; Zhang, J.; Miao, J.; Liu, M. Xanthohumol, a Prenylated Flavonoid from Hops, Induces Caspase-Dependent Degradation of Oncoprotein BCR-ABL in K562 Cells. Antioxidants 2019, 8, 402. [Google Scholar] [CrossRef]
- Hsieh, M.-Y.; Hsieh, M.-J.; Lo, Y.-S.; Lin, C.-C.; Chuang, Y.-C.; Chen, M.-K.; Chou, M.-C. Xanthohumol Targets the JNK1/2 Signaling Pathway in Apoptosis of Human Nasopharyngeal Carcinoma Cells. Environ. Toxicol. 2022, 37, 1509–1520. [Google Scholar] [CrossRef]
- Bocquet, L.; Sahpaz, S.; Bonneau, N.; Beaufay, C.; Mahieux, S.; Samaillie, J.; Roumy, V.; Jacquin, J.; Bordage, S.; Hennebelle, T.; et al. Phenolic Compounds from Humulus lupulus as Natural Antimicrobial Products: New Weapons in the Fight against Methicillin Resistant Staphylococcus Aureus, Leishmania Mexicana and Trypanosoma Brucei Strains. Molecules 2019, 24, 1024. [Google Scholar] [CrossRef]
- Rój, E.; Tadić, V.M.; Mišić, D.; Žižović, I.; Arsić, I.; Dobrzyńska-Inger, A.; Kostrzewa, D. Supercritical Carbon Dioxide Hops Extracts with Antimicrobial Properties. Open Chem. 2015, 13, 000010151520150131. [Google Scholar] [CrossRef]
- Sahin, S.; Eulenburg, V.; Kreis, W.; Villmann, C.; Pischetsrieder, M. Three-Step Test System for the Identification of Novel GABAA Receptor Modulating Food Plants. Plant Foods Hum. Nutr. 2016, 71, 355–360. [Google Scholar] [CrossRef]
- Zanoli, P.; Zavatti, M.; Rivasi, M.; Brusiani, F.; Losi, G.; Puia, G.; Avallone, R.; Baraldi, M. Evidence That the β-Acids Fraction of Hops Reduces Central GABAergic Neurotransmission. J. Ethnopharmacol. 2007, 109, 87–92. [Google Scholar] [CrossRef]
- Skehan, P.; Storeng, R.; Scudiero, D.; Monks, A.; McMahon, J.; Vistica, D.; Warren, J.T.; Bokesch, H.; Kenney, S.; Boyd, M.R. New Colorimetric Cytotoxicity Assay for Anticancer-Drug Screening. JNCI J. Natl. Cancer Inst. 1990, 82, 1107–1112. [Google Scholar] [CrossRef] [PubMed]
- Mosmann, T. Rapid Colorimetric Assay for Cellular Growth and Survival: Application to Proliferation and Cytotoxicity Assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
- Feoktistova, M.; Geserick, P.; Leverkus, M. Crystal Violet Assay for Determining Viability of Cultured Cells. Cold Spring Harb. Protoc. 2016, 2016, pdb.prot087379. [Google Scholar] [CrossRef]
- Ioannidis, J.P.A.; Fanelli, D.; Dunne, D.D.; Goodman, S.N. Meta-Research: Evaluation and Improvement of Research Methods and Practices. PLoS Biol. 2015, 13, e1002264. [Google Scholar] [CrossRef]
- Papaefthimiou, M.; Kontou, P.I.; Bagos, P.G.; Braliou, G.G. Integration of Antioxidant Activity Assays Data of Stevia Leaf Extracts: A Systematic Review and Meta-Analysis. Antioxidants 2024, 13, 692. [Google Scholar] [CrossRef] [PubMed]
- Pashler, H.; Harris, C.R. Is the Replicability Crisis Overblown? Three Arguments Examined. Perspect. Psychol. Sci. 2012, 7, 531–536. [Google Scholar] [CrossRef]
- Yin, S.; Song, M.; Zhao, R.; Liu, X.; Kang, W.K.; Lee, J.M.; Kim, Y.E.; Zhang, C.; Shim, J.-H.; Liu, K.; et al. Xanthohumol Inhibits the Growth of Keratin 18-Overexpressed Esophageal Squamous Cell Carcinoma in Vitro and in Vivo. Front. Cell Dev. Biol. 2020, 8, 366. [Google Scholar] [CrossRef] [PubMed]
- Ho, K.-H.; Kuo, T.-C.; Lee, Y.-T.; Chen, P.-H.; Shih, C.-M.; Cheng, C.-H.; Liu, A.-J.; Lee, C.-C.; Chen, K.-C. Xanthohumol Regulates miR-4749-5p-Inhibited RFC2 Signaling in Enhancing Temozolomide Cytotoxicity to Glioblastoma. Life Sci. 2020, 254, 117807. [Google Scholar] [CrossRef]
- Scagliarini, A.; Mathey, A.; Aires, V.; Delmas, D. Xanthohumol, a Prenylated Flavonoid from Hops, Induces DNA Damages in Colorectal Cancer Cells and Sensitizes SW480 Cells to the SN38 Chemotherapeutic Agent. Cells 2020, 9, 932. [Google Scholar] [CrossRef]
- Stompor, M.; Świtalska, M.; Wietrzyk, J. The Influence of a Single and Double Biotinylation of Xanthohumol on Its Anticancer Activity. Acta Biochim. Pol. 2019, 66, 559–565. [Google Scholar] [CrossRef]
- Sławińska-Brych, A.; Zdzisińska, B.; Czerwonka, A.; Mizerska-Kowalska, M.; Dmoszyńska-Graniczka, M.; Stepulak, A.; Gagoś, M. Xanthohumol Exhibits Anti-Myeloma Activity in Vitro through Inhibition of Cell Proliferation, Induction of Apoptosis via the ERK and JNK-Dependent Mechanism, and Suppression of sIL-6R and VEGF Production. Biochim. Biophys. Acta BBA-Gen. Subj. 2019, 1863, 129408. [Google Scholar] [CrossRef] [PubMed]
- Koosha, S.; Mohamed, Z.; Sinniah, A.; Ibrahim, Z.A.; Seyedan, A.; Alshawsh, M.A. Antiproliferative and Apoptotic Activities of 8-Prenylnaringenin against Human Colon Cancer Cells. Life Sci. 2019, 232, 116633. [Google Scholar] [CrossRef]
- Krajnović, T.; Drača, D.; Kaluđerović, G.N.; Dunđerović, D.; Mirkov, I.; Wessjohann, L.A.; Maksimović-Ivanić, D.; Mijatović, S. The Hop-Derived Prenylflavonoid Isoxanthohumol Inhibits the Formation of Lung Metastasis in B16-F10 Murine Melanoma Model. Food Chem. Toxicol. Int. J. Publ. Br. Ind. Biol. Res. Assoc. 2019, 129, 257–268. [Google Scholar] [CrossRef]
- Roehrer, S.; Stork, V.; Ludwig, C.; Minceva, M.; Behr, J. Analyzing Bioactive Effects of the Minor Hop Compound Xanthohumol C on Human Breast Cancer Cells Using Quantitative Proteomics. PLoS ONE 2019, 14, e0213469. [Google Scholar] [CrossRef]
- Bartmańska, A.; Tronina, T.; Popłoński, J.; Milczarek, M.; Filip-Psurska, B.; Wietrzyk, J. Highly Cancer Selective Antiproliferative Activity of Natural Prenylated Flavonoids. Molecules 2018, 23, 2922. [Google Scholar] [CrossRef]
- Wei, S.; Sun, T.; Du, J.; Zhang, B.; Xiang, D.; Li, W. Xanthohumol, a Prenylated Flavonoid from Hops, Exerts Anticancer Effects against Gastric Cancer in Vitro. Oncol. Rep. 2018, 40, 3213–3222. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, D.O.; Freitas, J.; Nogueira, P.; Henriques, S.N.; Carmo, A.M.; Castro, M.A.; Guido, L.F. Xanthohumol Inhibits Cell Proliferation and Induces Apoptosis in Human Thyroid Cells. Food Chem. Toxicol. Int. J. Publ. Br. Ind. Biol. Res. Assoc. 2018, 121, 450–457. [Google Scholar] [CrossRef] [PubMed]
- Ho, K.-H.; Chang, C.-K.; Chen, P.-H.; Wang, Y.-J.; Chang, W.-C.; Chen, K.-C. miR-4725-3p Targeting Stromal Interacting Molecule 1 Signaling Is Involved in Xanthohumol Inhibition of Glioma Cell Invasion. J. Neurochem. 2018, 146, 269–288. [Google Scholar] [CrossRef]
- Popłoński, J.; Turlej, E.; Sordon, S.; Tronina, T.; Bartmańska, A.; Wietrzyk, J.; Huszcza, E. Synthesis and Antiproliferative Activity of Minor Hops Prenylflavonoids and New Insights on Prenyl Group Cyclization. Molecules 2018, 23, 776. [Google Scholar] [CrossRef]
- Sun, Z.; Zhou, C.; Liu, F.; Zhang, W.; Chen, J.; Pan, Y.; Ma, L.; Liu, Q.; Du, Y.; Yang, J.; et al. Inhibition of Breast Cancer Cell Survival by Xanthohumol via Modulation of the Notch Signaling Pathway in Vivo and in Vitro. Oncol. Lett. 2018, 15, 908–916. [Google Scholar] [CrossRef]
- Saito, K.; Matsuo, Y.; Imafuji, H.; Okubo, T.; Maeda, Y.; Sato, T.; Shamoto, T.; Tsuboi, K.; Morimoto, M.; Takahashi, H.; et al. Xanthohumol Inhibits Angiogenesis by Suppressing Nuclear Factor-κB Activation in Pancreatic Cancer. Cancer Sci. 2018, 109, 132–140. [Google Scholar] [CrossRef]
- Stompor, M.; Świtalska, M.; Podgórski, R.; Uram, Ł.; Aebisher, D.; Wietrzyk, J. Synthesis and Biological Evaluation of 4′-O-Acetyl-Isoxanthohumol and Its Analogues as Antioxidant and Antiproliferative Agents. Acta Biochim. Pol. 2017, 64, 577–583. [Google Scholar] [CrossRef]
- Liu, M.; Yin, H.; Qian, X.; Dong, J.; Qian, Z.; Miao, J. Xanthohumol, a Prenylated Chalcone from Hops, Inhibits the Viability and Stemness of Doxorubicin-Resistant MCF-7/ADR Cells. Molecules 2016, 22, 36. [Google Scholar] [CrossRef]
- Gallo, C.; Dallaglio, K.; Bassani, B.; Rossi, T.; Rossello, A.; Noonan, D.M.; D’Uva, G.; Bruno, A.; Albini, A. Hop Derived Flavonoid Xanthohumol Inhibits Endothelial Cell Functions via AMPK Activation. Oncotarget 2016, 7, 59917–59931. [Google Scholar] [CrossRef]
- Chen, P.-H.; Chang, C.-K.; Shih, C.-M.; Cheng, C.-H.; Lin, C.-W.; Lee, C.-C.; Liu, A.-J.; Ho, K.-H.; Chen, K.-C. The miR-204-3p-Targeted IGFBP2 Pathway Is Involved in Xanthohumol-Induced Glioma Cell Apoptotic Death. Neuropharmacology 2016, 110 Pt A, 362–375. [Google Scholar] [CrossRef] [PubMed]
- Lempereur, M.; Majewska, C.; Brunquers, A.; Wongpramud, S.; Valet, B.; Janssens, P.; Dillemans, M.; Van Nedervelde, L.; Gallo, D. Tetrahydro-Iso-Alpha Acids Antagonize Estrogen Receptor Alpha Activity in MCF-7 Breast Cancer Cells. Int. J. Endocrinol. 2016, 2016, 9747863. [Google Scholar] [CrossRef] [PubMed]
- Yoo, Y.B.; Park, K.S.; Kim, J.B.; Kang, H.J.; Yang, J.H.; Lee, E.K.; Kim, H.Y. Xanthohumol Inhibits Cellular Proliferation in a Breast Cancer Cell Line (MDA-MB231) through an Intrinsic Mitochondrial-Dependent Pathway. Indian J. Cancer 2014, 51, 518–523. [Google Scholar] [CrossRef]
- Krajnović, T.; Kaluđerović, G.N.; Wessjohann, L.A.; Mijatović, S.; Maksimović-Ivanić, D. Versatile Antitumor Potential of Isoxanthohumol: Enhancement of Paclitaxel Activity in Vivo. Pharmacol. Res. 2016, 105, 62–73. [Google Scholar] [CrossRef] [PubMed]
- Yong, W.K.; Ho, Y.F.; Malek, S.N.A. Xanthohumol Induces Apoptosis and S Phase Cell Cycle Arrest in A549 Non-Small Cell Lung Cancer Cells. Pharmacogn. Mag. 2015, 11 (Suppl. 2), S275–S283. [Google Scholar] [CrossRef]
- Zhang, B.; Chu, W.; Wei, P.; Liu, Y.; Wei, T. Xanthohumol Induces Generation of Reactive Oxygen Species and Triggers Apoptosis through Inhibition of Mitochondrial Electron Transfer Chain Complex I. Free Radic. Biol. Med. 2015, 89, 486–497. [Google Scholar] [CrossRef]
- Sławińska-Brych, A.; Król, S.K.; Dmoszyńska-Graniczka, M.; Zdzisińska, B.; Stepulak, A.; Gagoś, M. Xanthohumol Inhibits Cell Cycle Progression and Proliferation of Larynx Cancer Cells in Vitro. Chem. Biol. Interact. 2015, 240, 110–118. [Google Scholar] [CrossRef]
- Jiang, W.; Zhao, S.; Xu, L.; Lu, Y.; Lu, Z.; Chen, C.; Ni, J.; Wan, R.; Yang, L. The Inhibitory Effects of Xanthohumol, a Prenylated Chalcone Derived from Hops, on Cell Growth and Tumorigenesis in Human Pancreatic Cancer. Biomed. Pharmacother. Biomedecine Pharmacother. 2015, 73, 40–47. [Google Scholar] [CrossRef]
- Zenger, K.; Dutta, S.; Wolff, H.; Genton, M.G.; Kraus, B. In Vitro Structure-Toxicity Relationship of Chalcones in Human Hepatic Stellate Cells. Toxicology 2015, 336, 26–33. [Google Scholar] [CrossRef] [PubMed]
- Yong, W.K.; Abd Malek, S.N. Xanthohumol Induces Growth Inhibition and Apoptosis in ca Ski Human Cervical Cancer Cells. Evid.-Based Complement. Altern. Med. ECAM 2015, 2015, 921306. [Google Scholar] [CrossRef] [PubMed]
- Mouratidis, P.X.E.; Colston, K.W.; Tucknott, M.L.; Tyrrell, E.; Pirianov, G. An Investigation into the Anticancer Effects and Mechanism of Action of Hop β-Acid Lupulone and Its Natural and Synthetic Derivatives in Prostate Cancer Cells. Nutr. Cancer 2013, 65, 1086–1092. [Google Scholar] [CrossRef]
- Boncler, M.; Różalski, M.; Krajewska, U.; Podsędek, A.; Watala, C. Comparison of PrestoBlue and MTT Assays of Cellular Viability in the Assessment of Anti-Proliferative Effects of Plant Extracts on Human Endothelial Cells. J. Pharmacol. Toxicol. Methods 2014, 69, 9–16. [Google Scholar] [CrossRef]
- Tronina, T.; Bartmańska, A.; Milczarek, M.; Wietrzyk, J.; Popłoński, J.; Rój, E.; Huszcza, E. Antioxidant and Antiproliferative Activity of Glycosides Obtained by Biotransformation of Xanthohumol. Bioorg. Med. Chem. Lett. 2013, 23, 1957–1960. [Google Scholar] [CrossRef]
- Tronina, T.; Bartmańska, A.; Filip-Psurska, B.; Wietrzyk, J.; Popłoński, J.; Huszcza, E. Fungal Metabolites of Xanthohumol with Potent Antiproliferative Activity on Human Cancer Cell Lines in Vitro. Bioorg. Med. Chem. 2013, 21, 2001–2006. [Google Scholar] [CrossRef] [PubMed]
- Allsopp, P.; Possemiers, S.; Campbell, D.; Gill, C.; Rowland, I. A Comparison of the Anticancer Properties of Isoxanthohumol and 8-Prenylnaringenin Using in Vitro Models of Colon Cancer. BioFactors 2013, 39, 441–447. [Google Scholar] [CrossRef]
- Kang, Y.; Park, M.A.; Heo, S.-W.; Park, S.-Y.; Kang, K.W.; Park, P.-H.; Kim, J.-A. The Radio-Sensitizing Effect of Xanthohumol Is Mediated by STAT3 and EGFR Suppression in Doxorubicin-Resistant MCF-7 Human Breast Cancer Cells. Biochim. Biophys. Acta 2013, 1830, 2638–2648. [Google Scholar] [CrossRef]
- Hemachandra, L.P.; Madhubhani, P.; Chandrasena, R.; Esala, P.; Chen, S.-N.; Main, M.; Lankin, D.C.; Scism, R.A.; Dietz, B.M.; Pauli, G.F.; et al. Hops (Humulus lupulus) Inhibits Oxidative Estrogen Metabolism and Estrogen-Induced Malignant Transformation in Human Mammary Epithelial Cells (MCF-10A). Cancer Prev. Res. 2012, 5, 73–81. [Google Scholar] [CrossRef]
- Deeb, D.; Gao, X.; Jiang, H.; Arbab, A.S.; Dulchavsky, S.A.; Gautam, S.C. Growth Inhibitory and Apoptosis-Inducing Effects of Xanthohumol, a Prenylated Chalone Present in Hops, in Human Prostate Cancer Cells. Anticancer Res. 2010, 30, 3333–3339. [Google Scholar] [PubMed]
- Negrão, R.; Costa, R.; Duarte, D.; Taveira Gomes, T.; Mendanha, M.; Moura, L.; Vasques, L.; Azevedo, I.; Soares, R. Angiogenesis and Inflammation Signaling Are Targets of Beer Polyphenols on Vascular Cells. J. Cell. Biochem. 2010, 111, 1270–1279. [Google Scholar] [CrossRef]
- Wesołowska, O.; Wiśniewski, J.; Sroda, K.; Krawczenko, A.; Bielawska-Pohl, A.; Paprocka, M.; Duś, D.; Michalak, K. 8-Prenylnaringenin Is an Inhibitor of Multidrug Resistance-Associated Transporters, P-Glycoprotein and MRP1. Eur. J. Pharmacol. 2010, 644, 32–40. [Google Scholar] [CrossRef] [PubMed]
- Strathmann, J.; Klimo, K.; Sauer, S.W.; Okun, J.G.; Prehn, J.H.M.; Gerhäuser, C. Xanthohumol-Induced Transient Superoxide Anion Radical Formation Triggers Cancer Cells into Apoptosis via a Mitochondria-Mediated Mechanism. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2010, 24, 2938–2950. [Google Scholar] [CrossRef]
- Dorn, C.; Weiss, T.S.; Heilmann, J.; Hellerbrand, C. Xanthohumol, a Prenylated Chalcone Derived from Hops, Inhibits Proliferation, Migration and Interleukin-8 Expression of Hepatocellular Carcinoma Cells. Int. J. Oncol. 2010, 36, 435–441. [Google Scholar] [CrossRef]
- Mendes, V.; Monteiro, R.; Pestana, D.; Teixeira, D.; Calhau, C.; Azevedo, I. Xanthohumol Influences Preadipocyte Differentiation: Implication of Antiproliferative and Apoptotic Effects. J. Agric. Food Chem. 2008, 56, 11631–11637. [Google Scholar] [CrossRef] [PubMed]
- Ho, Y.-C.; Liu, C.-H.; Chen, C.-N.; Duan, K.-J.; Lin, M.-T. Inhibitory Effects of Xanthohumol from Hops (Humulus lupulus L.) on Human Hepatocellular Carcinoma Cell Lines. Phytother. Res. 2008, 22, 1465–1468. [Google Scholar] [CrossRef]
- Monteghirfo, S.; Tosetti, F.; Ambrosini, C.; Stigliani, S.; Pozzi, S.; Frassoni, F.; Fassina, G.; Soverini, S.; Albini, A.; Ferrari, N. Antileukemia Effects of Xanthohumol in Bcr/Abl-Transformed Cells Involve Nuclear Factor-kappaB and P53 Modulation. Mol. Cancer Ther. 2008, 7, 2692–2702. [Google Scholar] [CrossRef]
- Koo, J.-H.; Kim, H.T.; Yoon, H.-Y.; Kwon, K.-B.; Choi, I.-W.; Jung, S.H.; Kim, H.-U.; Park, B.-H.; Park, J.-W. Effect of Xanthohumol on Melanogenesis in B16 Melanoma Cells. Exp. Mol. Med. 2008, 40, 313–319. [Google Scholar] [CrossRef]
- Lee, S.H.; Kim, H.J.; Lee, J.S.; Lee, I.-S.; Kang, B.Y. Inhibition of Topoisomerase I Activity and Efflux Drug Transporters’ Expression by Xanthohumol. from Hops. Arch. Pharm. Res. 2007, 30, 1435–1439. [Google Scholar] [CrossRef]
- Yang, J.-Y.; Della-Fera, M.A.; Rayalam, S.; Baile, C.A. Effect of Xanthohumol and Isoxanthohumol on 3T3-L1 Cell Apoptosis and Adipogenesis. Apoptosis Int. J. Program. Cell Death 2007, 12, 1953–1963. [Google Scholar] [CrossRef]
- Monteiro, R.; Faria, A.; Azevedo, I.; Calhau, C. Modulation of Breast Cancer Cell Survival by Aromatase Inhibiting Hop (Humulus lupulus L.) Flavonoids. J. Steroid Biochem. Mol. Biol. 2007, 105, 124–130. [Google Scholar] [CrossRef] [PubMed]
- Plazar, J.; Zegura, B.; Lah, T.T.; Filipic, M. Protective Effects of Xanthohumol against the Genotoxicity of Benzo(a)Pyrene (BaP), 2-Amino-3-Methylimidazo[4,5-f]Quinoline (IQ) and Tert-Butyl Hydroperoxide (t-BOOH) in HepG2 Human Hepatoma Cells. Mutat. Res. 2007, 632, 1–8. [Google Scholar] [CrossRef]
- Delmulle, L.; Bellahcène, A.; Dhooge, W.; Comhaire, F.; Roelens, F.; Huvaere, K.; Heyerick, A.; Castronovo, V.; De Keukeleire, D. Anti-Proliferative Properties of Prenylated Flavonoids from Hops (Humulus lupulus L.) in Human Prostate Cancer Cell Lines. Phytomedicine Int. J. Phytother. Phytopharm. 2006, 13, 732–734. [Google Scholar] [CrossRef] [PubMed]
- Colgate, E.C.; Miranda, C.L.; Stevens, J.F.; Bray, T.M.; Ho, E. Xanthohumol, a Prenylflavonoid Derived from Hops Induces Apoptosis and Inhibits NF-kappaB Activation in Prostate Epithelial Cells. Cancer Lett. 2007, 246, 201–209. [Google Scholar] [CrossRef] [PubMed]
- Dietz, B.M.; Kang, Y.-H.; Liu, G.; Eggler, A.L.; Yao, P.; Chadwick, L.R.; Pauli, G.F.; Farnsworth, N.R.; Mesecar, A.D.; van Breemen, R.B.; et al. Xanthohumol Isolated from Humulus lupulus Inhibits Menadione-Induced DNA Damage through Induction of Quinone Reductase. Chem. Res. Toxicol. 2005, 18, 1296–1305. [Google Scholar] [CrossRef]
- Pan, L.; Becker, H.; Gerhäuser, C. Xanthohumol Induces Apoptosis in Cultured 40-16 Human Colon Cancer Cells by Activation of the Death Receptor- and Mitochondrial Pathway. Mol. Nutr. Food Res. 2005, 49, 837–843. [Google Scholar] [CrossRef]
- Miranda, C.L.; Stevens, J.F.; Helmrich, A.; Henderson, M.C.; Rodriguez, R.J.; Yang, Y.-H.; Deinzer, M.L.; Barnes, D.W.; Buhler, D.R. Antiproliferative and Cytotoxic Effects of Prenylated Flavonoids from Hops (Humulus lupulus) in Human Cancer Cell Lines. Food Chem. Toxicol. 1999, 37, 271–285. [Google Scholar] [CrossRef]
- Cragg, G.M.; Newman, D.J.; Snader, K.M. Natural Products in Drug Discovery and Development. J. Nat. Prod. 1997, 60, 52–60. [Google Scholar] [CrossRef]
- Talib, W.H.; Mahasneh, A.M. Antiproliferative Activity of Plant Extracts Used Against Cancer in Traditional Medicine. Sci. Pharm. 2010, 78, 33–45. [Google Scholar] [CrossRef]
- Nuutinen, T. Medicinal Properties of Terpenes Found in Cannabis Sativa and Humulus lupulus. Eur. J. Med. Chem. 2018, 157, 198–228. [Google Scholar] [CrossRef] [PubMed]
- Fricker, S.P.; Buckley, R.G. Comparison of Two Colorimetric Assays as Cytotoxicity Endpoints for an in Vitro Screen for Antitumour Agents. Anticancer Res. 1996, 16, 3755–3760. [Google Scholar]
- Keepers, Y.P.; Pizao, P.E.; Peters, G.J.; van Ark-Otte, J.; Winograd, B.; Pinedo, H.M. Comparison of the Sulforhodamine B Protein and Tetrazolium (MTT) Assays for in Vitro Chemosensitivity Testing. Eur. J. Cancer Clin. Oncol. 1991, 27, 897–900. [Google Scholar] [CrossRef] [PubMed]
- Rubinstein, L.V.; Shoemaker, R.H.; Paull, K.D.; Simon, R.M.; Tosini, S.; Skehan, P.; Scudiero, D.A.; Monks, A.; Boyd, M.R. Comparison of In Vitro Anticancer-Drug-Screening Data Generated With a Tetrazolium Assay Versus a Protein Assay Against a Diverse Panel of Human Tumor Cell Lines. JNCI J. Natl. Cancer Inst. 1990, 82, 1113–1117. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, R.; Bhuia, M.S.; Al Hasan, M.S.; Hossain Snigdha, S.; Afrin, S.; Büsselberg, D.; Habtemariam, S.; Sönmez Gürer, E.; Sharifi-Rad, J.; Ahmed Aldahish, A.; et al. Anticancer Potential of Phytochemicals Derived from Mangrove Plants: Comprehensive Mechanistic Insights. Food Sci. Nutr. 2024, 12, 6174–6205. [Google Scholar] [CrossRef]
- Fankam, A.G.; Kuete, V. Screening Methods of Anticancer Agents from Natural Source. In Advances in Botanical Research; Elsevier: Amsterdam, The Netherlands, 2024; Volume 111, pp. 53–82. [Google Scholar] [CrossRef]
- Pappa, S.A.; Kontou, P.I.; Bagos, P.G.; Braliou, G.G. Urine-Based Molecular Diagnostic Tests for Leishmaniasis Infection in Human and Canine Populations: A Meta-Analysis. Pathogens 2021, 10, 269. [Google Scholar] [CrossRef]
- Papaefthimiou, M.; Kontou, P.I.; Bagos, P.G.; Braliou, G.G. Antioxidant Activity of Leaf Extracts from Stevia Rebaudiana Bertoni Exerts Attenuating Effect on Diseased Experimental Rats: A Systematic Review and Meta-Analysis. Nutrients 2023, 15, 3325. [Google Scholar] [CrossRef]
- Tapari, A.; Braliou, G.G.; Papaefthimiou, M.; Mavriki, H.; Kontou, P.I.; Nikolopoulos, G.K.; Bagos, P.G. Performance of Antigen Detection Tests for SARS-CoV-2: A Systematic Review and Meta-Analysis. Diagnostics 2022, 12, 1388. [Google Scholar] [CrossRef]
- Papakostidis, C.; Giannoudis, P.V. Meta-Analysis. What Have We Learned? Injury 2023, 54, S30–S34. [Google Scholar] [CrossRef]
- Kontou, P.I.; Braliou, G.G.; Dimou, N.L.; Nikolopoulos, G.; Bagos, P.G. Antibody Tests in Detecting SARS-CoV-2 Infection: A Meta-Analysis. Diagnostics 2020, 10, 319. [Google Scholar] [CrossRef]
- Gurevitch, J.; Koricheva, J.; Nakagawa, S.; Stewart, G. Meta-Analysis and the Science of Research Synthesis. Nature 2018, 555, 175–182. [Google Scholar] [CrossRef] [PubMed]
- Nozawa, H.; Tazumi, K.; Sato, K.; Yoshida, A.; Takata, J.; Arimoto-Kobayashi, S.; Kondo, K. Inhibitory Effects of Beer on Heterocyclic Amine-Induced Mutagenesis and PhIP-Induced Aberrant Crypt Foci in Rat Colon. Mutat. Res. Toxicol. Environ. Mutagen. 2004, 559, 177–187. [Google Scholar] [CrossRef] [PubMed]
- Guo, D.; Zhang, B.; Liu, S.; Jin, M. Xanthohumol Induces Apoptosis via Caspase Activation, Regulation of Bcl-2, and Inhibition of PI3K/Akt/mTOR-Kinase in Human Gastric Cancer Cells. Biomed. Pharmacother. 2018, 106, 1300–1306. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.-H.; Sun, T.-L.; Xiang, D.-X.; Wei, S.-S.; Li, W.-Q. Anticancer Activity and Mechanism of Xanthohumol: A Prenylated Flavonoid From Hops (Humulus lupulus L.). Front. Pharmacol. 2018, 9, 530. [Google Scholar] [CrossRef]
- Thongchot, S.; Thanee, M.; Loilome, W.; Techasen, A.; Boonmars, T.; Sa-Ngiamwibool, P.; Titapun, A.; Yongvanit, P.; Isidoro, C.; Namwat, N. Curative Effect of Xanthohumol Supplementation during Liver Fluke-Associated Cholangiocarcinogenesis: Potential Involvement of Autophagy. J. Tradit. Complement. Med. 2019, 10, 230–235. [Google Scholar] [CrossRef]
- ZHAO, X.; JIANG, K.; LIANG, B.; HUANG, X. Anticancer Effect of Xanthohumol Induces Growth Inhibition and Apoptosis of Human Liver Cancer through NF-κB/P53-Apoptosis Signaling Pathway. Oncol. Rep. 2016, 35, 669–675. [Google Scholar] [CrossRef]
- Mi, X.; Wang, C.; Sun, C.; Chen, X.; Huo, X.; Zhang, Y.; Li, G.; Xu, B.; Zhang, J.; Xie, J.; et al. Xanthohumol Induces Paraptosis of Leukemia Cells through P38 Mitogen Activated Protein Kinase Signaling Pathway. Oncotarget 2017, 8, 31297–31304. [Google Scholar] [CrossRef]
- Anzures-Cabrera, J.; Higgins, J.P.T. Graphical Displays for Meta-Analysis: An Overview with Suggestions for Practice. Res. Synth. Methods 2010, 1, 66–80. [Google Scholar] [CrossRef]
- Paez, A. Gray Literature: An Important Resource in Systematic Reviews. J. Evid.-Based Med. 2017, 10, 233–240. [Google Scholar] [CrossRef]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G. Preferred Reporting Items for Systematic Reviews and Meta-Analyses: The PRISMA Statement. PLoS Med. 2009, 6, e1000097. [Google Scholar] [CrossRef]
- Forero, D.A.; Lopez-Leon, S.; González-Giraldo, Y.; Bagos, P.G. Ten Simple Rules for Carrying out and Writing Meta-Analyses. PLoS Comput. Biol. 2019, 15, e1006922. [Google Scholar] [CrossRef] [PubMed]
- Donders, A.R.T.; Van Der Heijden, G.J.M.G.; Stijnen, T.; Moons, K.G.M. Review: A Gentle Introduction to Imputation of Missing Values. J. Clin. Epidemiol. 2006, 59, 1087–1091. [Google Scholar] [CrossRef]
- Hopewell, S.; McDonald, S.; Clarke, M.J.; Egger, M. Grey Literature in Meta-analyses of Randomized Trials of Health Care Interventions. Cochrane Database Syst. Rev. 2007, 2007, MR000010. [Google Scholar] [CrossRef]
- Aleshin, V.A.; Artiukhov, A.V.; Oppermann, H.; Kazantsev, A.V.; Lukashev, N.V.; Bunik, V.I. Mitochondrial Impairment May Increase Cellular NAD(P)H: Resazurin Oxidoreductase Activity, Perturbing the NAD(P)H-Based Viability Assays. Cells 2015, 4, 427–451. [Google Scholar] [CrossRef] [PubMed]
- Kabakov, A.E.; Gabai, V.L. Cell Death and Survival Assays. In Chaperones: Methods and Protocols; Calderwood, S.K., Prince, T.L., Eds.; Springer: New York, NY, USA, 2018. [Google Scholar] [CrossRef]
- Nowak, E.; Kammerer, S.; Küpper, J.-H. ATP-Based Cell Viability Assay Is Superior to Trypan Blue Exclusion and XTT Assay in Measuring Cytotoxicity of Anticancer Drugs Taxol and Imatinib, and Proteasome Inhibitor MG-132 on Human Hepatoma Cell Line HepG2. Clin. Hemorheol. Microcirc. 2018, 69, 327–336. [Google Scholar] [CrossRef]
- Präbst, K.; Engelhardt, H.; Ringgeler, S.; Hübner, H. Basic Colorimetric Proliferation Assays: MTT, WST, and Resazurin. In Cell Viability Assays: Methods and Protocols; Gilbert, D.F., Friedrich, O., Eds.; Springer: New York, NY, USA, 2017. [Google Scholar] [CrossRef]
- Vichai, V.; Kirtikara, K. Sulforhodamine B Colorimetric Assay for Cytotoxicity Screening. Nat. Protoc. 2006, 1, 1112–1116. [Google Scholar] [CrossRef]
- Weir, C.J.; Butcher, I.; Assi, V.; Lewis, S.C.; Murray, G.D.; Langhorne, P.; Brady, M.C. Dealing with Missing Standard Deviation and Mean Values in Meta-Analysis of Continuous Outcomes: A Systematic Review. BMC Med. Res. Methodol. 2018, 18, 25. [Google Scholar] [CrossRef] [PubMed]
- Furukawa, T.A.; Barbui, C.; Cipriani, A.; Brambilla, P.; Watanabe, N. Imputing Missing Standard Deviations in Meta-Analyses Can Provide Accurate Results. J. Clin. Epidemiol. 2006, 59, 7–10. [Google Scholar] [CrossRef] [PubMed]
- DerSimonian, R.; Laird, N. Meta-Analysis in Clinical Trials. Control. Clin. Trials 1986, 7, 177–188. [Google Scholar] [CrossRef]
- Thompson, S.G.; Higgins, J.P.T. How Should Meta-Regression Analyses Be Undertaken and Interpreted? Stat. Med. 2002, 21, 1559–1573. [Google Scholar] [CrossRef]
- Stata User’s Guide Release 13; StataCorp LP: College Station, TX, USA, 2013.
- Deeks, J.J.; Altman, D.G.; Bradburn, M.J. Statistical Methods for Examining Heterogeneity and Combining Results from Several Studies in Meta-Analysis. In Systematic Reviews in Health Care; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2001; pp. 285–312. [Google Scholar] [CrossRef]
- Harbord, R.; Higgins, J. Meta-Regression in Stata. Stata J. 2008, 8, 493–519. [Google Scholar] [CrossRef]
- Kiofentzoglou, D.; Andronidou, E.M.; Kontou, P.I.; Bagos, P.G.; Braliou, G.G. Antimicrobial Activity of Chemical Hop (Humulus lupulus) Compounds: A Systematic Review and Meta-Analysis. Appl. Sci. 2025, 15, 7806. [Google Scholar] [CrossRef]



| Author Reference | Assay | IC50 (nM or mg/mL for Extracts) | SD | Hours | Number of Replicates | Cell Line | Cell Type | Cancer/Normal | Compound or Type of Extract |
|---|---|---|---|---|---|---|---|---|---|
| Hsieh et al. [16] | MTT | 56.00 | 7.30 | 24 | 3 | NPC-039 | Human neck cancer | Neck | Xanthohumol |
| Hsieh et al. [16] | MTT | 56.00 | 7.30 | 24 | 3 | NPC-039 | Human neck cancer | Neck | Xanthohumol |
| Hsieh et al. [16] | MTT | 45.60 | 7.30 | 24 | 3 | NPC-BM | Human neck cancer | Neck | Xanthohumol |
| Hsieh et al. [16] | MTT | 34.90 | 6.00 | 24 | 3 | Human normal nasopharyngeal | Non-cancer | Xanthohumol | |
| Hsieh et al. [16] | MTT | 22.80 | 7.30 | 48 | 3 | NPC-039 | Human neck cancer | Neck | Xanthohumol |
| Hsieh et al. [16] | MTT | 18.10 | 7.30 | 48 | 3 | NPC-BM | Human neck cancer | Neck | Xanthohumol |
| Hsieh et al. [16] | MTT | 20.00 | 6.00 | 48 | 3 | Human normal nasopharyngeal | Non-cancer | Xanthohumol | |
| Hsieh et al. [16] | MTT | 19.50 | 7.30 | 72 | 3 | NPC-039 | Human neck cancer | Neck | Xanthohumol |
| Hsieh et al. [16] | MTT | 17.80 | 7.30 | 72 | 3 | NPC-BM | Human neck cancer | Neck | Xanthohumol |
| Hsieh et al. [16] | MTT | 20.00 | 6.00 | 72 | 3 | Human normal nasopharyngeal | Non-cancer | Xanthohumol | |
| Klimek et al. [12] | MTT | 74.94 | 2.62 | 24 | 3 | BJ | Human normal skin fibroblast | Non-cancer | Xanthohumol |
| Klimek et al. [12] | MTT | 34.36 | 2.89 | 72 | 3 | MCF-7 | Human breast cancer | Breast | Xanthohumol |
| Klimek et al. [12] | MTT | 20.85 | 2.99 | 72 | 3 | A549 | Human lung cancer | Lung | Xanthohumol |
| Klimek et al. [12] | MTT | 102.59 | 3.48 | 72 | 3 | HepG2 | Human liver cancer | Liver | Xanthohumol |
| Klimek et al. [12] | MTT | 48.67 | 1.35 | 72 | 3 | BJ | Human normal skin fibroblast | Non-cancer | Xanthohumol |
| Klimek et al. [12] | MTT | 155.70 | 4.23 | 24 | 3 | BJ | Human normal skin fibroblast | Non-cancer | Hops dynamic supercritical fluid extract (SFE) |
| Klimek et al. [12] | MTT | 45.17 | 3.58 | 72 | 3 | A549 | Human lung cancer | Lung | Hops dynamic supercritical fluid extract (SFE) |
| Klimek et al. [12] | MTT | 66.48 | 2.97 | 72 | 3 | MCF-7 | Human breast cancer | Breast | Hops dynamic supercritical fluid extract (SFE) |
| Klimek et al. [12] | MTT | 26.27 | 1.56 | 72 | 3 | HepG2 | Human liver cancer | Liver | Hops dynamic supercritical fluid extract (SFE) |
| Klimek et al. [12] | MTT | 104.30 | 4.16 | 72 | 3 | BJ | Human normal skin fibroblast | Non-cancer | Hops dynamic supercritical fluid extract (SFE) |
| Hitzman et al. [4] | MTT | 76.00 | 6.91 | 24 | 3 | MCF-7 | Human breast cancer | Breast | Xanthohumol |
| Hitzman et al. [4] | MTT | 32.80 | 0.40 | 24 | 3 | MCF-7 | Human breast cancer | Breast | Spent hops ethanolic extract, LC-MS characterized (33.20% XH, 1.22% 6-PN, 1.11% isoxanthohumol and 0.28% 8-PN) |
| Hitzman et al. [4] | MTT | 105.00 | 6.91 | 24 | 3 | MCF-7 | Human breast cancer | Breast | 6-prenylnaringenin |
| Hitzman et al. [4] | MTT | 115.00 | 8.20 | 24 | 3 | MCF-7 | Human breast cancer | Breast | 8-prenylnaringenin |
| Yin et al. [27] | MTT | 7.90 | 7.30 | 24 | 3 | KYSE30 | Human esophageal cancer | Esophagus | Xanthohumol |
| Yin et al. [27] | MTT | 3.10 | 7.30 | 48 | 3 | KYSE30 | Human esophageal cancer | Esophagus | Xanthohumol |
| Yin et al. [27] | MTT | 2.60 | 7.30 | 72 | 3 | KYSE30 | Human esophageal cancer | Esophagus | Xanthohumol |
| Ho et al. [28] | MTT | 60.00 | 7.30 | 24 | 3 | U87-MG | Human glioblastoma | Glioblastoma | Xanthohumol |
| Ho et al. [28] | MTT | 68.10 | 7.30 | 24 | 3 | A172 | Human glioblastoma | Glioblastoma | Xanthohumol |
| Scagliarini et al. [29] | Crystal Violet | 39.00 | 6.48 | 24 | 3 | HT-29 | Human colon cancer | Colon | Xanthohumol |
| Scagliarini et al. [29] | Crystal Violet | 12.00 | 2.8 | 48 | 3 | HT-29 | Human colon cancer | Colon | Xanthohumol |
| Scagliarini et al. [29] | Crystal Violet | 22.00 | 6.49 | 48 | 3 | SW480 | Human colon cancer | Colon | Xanthohumol |
| Scagliarini et al. [29] | Crystal Violet | 12.00 | 3.57 | 48 | 3 | SW620 | Human colon cancer | Colon | Xanthohumol |
| Scagliarini et al. [29] | Crystal Violet | 10.00 | 1.75 | 72 | 3 | HT-29 | Human colon cancer | Colon | Xanthohumol |
| Scagliarini et al. [29] | Crystal Violet | 20.00 | 3.3 | 72 | 3 | SW480 | Human colon cancer | Colon | Xanthohumol |
| Scagliarini et al. [29] | Crystal Violet | 7.00 | 1.38 | 72 | 3 | SW620 | Human colon cancer | Colon | Xanthohumol |
| Stompor et al. [30] | SRB | 8.8 | 1.16 | 72 | 4 | MCF-7 | Human breast cancer | Breast | Xanthohumol |
| Stompor et al. [30] | SRB | 18.9 | 4.6 | 72 | 4 | MDA-MB-231 | Human breast cancer | Breast | Xanthohumol |
| Stompor et al. [30] | SRB | 8.7 | 1.3 | 72 | 4 | 4T1 | Murine breast cancer | Breast | Xanthohumol |
| Stompor et al. [30] | SRB | 21.5 | 2.7 | 72 | 4 | HepG2 | Human liver cancer | Liver | Xanthohumol |
| Stompor et al. [30] | SRB | 12.6 | 8.8 | 72 | 4 | BALB/3T3 | Murine normal fibroblasts | Non-cancer | Xanthohumol |
| Stompor et al. [30] | SRB | 21.1 | 4.3 | 72 | 4 | MCF-10A | Human normal breast | Non-cancer | Xanthohumol |
| Lu et al. [15] | MTT | 39.82 | 1.50 | 24 | 3 | K562 | Human leukemia | Leukemia | Xanthohumol |
| Lu et al. [15] | MTT | 19.56 | 0.77 | 48 | 3 | K562 | Human leukemia | Leukemia | Xanthohumol |
| Lu et al. [15] | MTT | 4.43 | 1.20 | 72 | 3 | K562 | Human leukemia | Leukemia | Xanthohumol |
| Lu et al. [15] | MTT | 10.00 | 1.50 | 72 | 3 | K562/ADR | Human leukemia, adriamycin-resistant | Leukemia | Xanthohumol |
| Sławińska-Brych et al. [31] | MTT | 50.12 | 7.30 | 48 | 3 | U266 | Human myeloma | Myeloma | Xanthohumol |
| Sławińska-Brych et al. [31] | MTT | 21.85 | 7.30 | 48 | 3 | RPMI8226 | Human myeloma | Myeloma | Xanthohumol |
| Sławińska-Brych et al. [31] | MTT | 38.40 | 7.30 | 96 | 3 | U266 | Human myeloma | Myeloma | Xanthohumol |
| Sławińska-Brych et al. [31] | MTT | 8.24 | 7.30 | 96 | 3 | RPMI8226 | Human myeloma | Myeloma | Xanthohumol |
| Koosha et al. [32] | MTT | 166.68 | 8.20 | 24 | 3 | HCT116 | Human colon cancer | Colon | 8-prenylnaringenin |
| Koosha et al. [32] | MTT | 70.00 | 2.90 | 48 | 3 | HCT116 | Human colon cancer | Colon | 8-prenylnaringenin |
| Koosha et al. [32] | MTT | 58.49 | 4.10 | 72 | 3 | HCT116 | Human colon cancer | Colon | 8-prenylnaringenin |
| Krajnović et al. [33] | Crystal violet | 42.00 | 5.19 | 48 | 3 | B16-F10 | Murine melanoma | Melanoma | Isoxanthohumol |
| Krajnović et al. [33] | MTT | 30.00 | 4.05 | 48 | 3 | B16-F10 | Murine melanoma | Melanoma | Isoxanthohumol |
| Bocquet et al. [17] | MTT | 7.10 | 0.80 | 48 | 3 | HepG2 | Human liver cancer | Liver | Xanthohumol |
| Bocquet et al. [17] | MTT | 29.40 | 2.60 | 48 | 3 | MG-63 | Human osteosarcoma | Bone | Xanthohumol |
| Bocquet et al. [17] | MTT | 19.50 | 0.50 | 48 | 3 | WI-38 | Human normal lung fibroblast | Non-cancer | Xanthohumol |
| Bocquet et al. [17] | MTT | 9.60 | 0.50 | 48 | 3 | J774 | Mouse monocyte macrophage | Non-cancer | Xanthohumol |
| Bocquet et al. [17] | MTT | 31.40 | 8.10 | 72 | 3 | MG-63 | Human osteosarcoma | Bone | Hydro-alcoholic extract: ethanol/water (9:1; v/v) 3 successive macerations of 4 h and 1 o/n, stirring in the dark |
| Bocquet et al. [17] | MTT | 6.80 | 2.50 | 72 | 3 | HepG2 | Human liver cancer | Liver | Hydro-alcoholic extract: ethanol/water (9:1; v/v) 3 successive macerations of 4 h and 1 o/n, stirring in the dark |
| Bocquet et al. [17] | MTT | 7.60 | 0.10 | 72 | 3 | WI-38 | Human normal lung fibroblast | Non-cancer | Hydro-alcoholic extract: ethanol/water (9:1; v/v) 3 successive macerations of 4 h and 1 o/n, stirring in the dark |
| Bocquet et al. [17] | MTT | 19.70 | 2.80 | 72 | 3 | J774 | Mouse monocyte macrophage | Non-cancer | Hydro-alcoholic extract: ethanol/water (9:1; v/v) 3 successive macerations of 4 h and 1 o/n, stirring in the dark |
| Bocquet et al. [17] | MTT | 2.60 | 0.10 | 48 | 3 | WI-38 | Human normal lung fibroblast | Non-cancer | Lupulone |
| Bocquet et al. [17] | MTT | 3.60 | 0.10 | 48 | 3 | J774 | Mouse monocyte macrophage | Non-cancer | Lupulone |
| Bocquet et al. [17] | MTT | 2.90 | 0.50 | 48 | 3 | HepG2 | Human liver cancer | Liver | Lupulone |
| Bocquet et al. [17] | MTT | 10.40 | 0.40 | 48 | 3 | MG-63 | Human osteosarcoma | Bone | Lupulone |
| Bocquet et al. [17] | MTT | 29.00 | 2.30 | 48 | 3 | WI-38 | Human normal lung fibroblast | Non-cancer | Humulone |
| Bocquet et al. [17] | MTT | 31.70 | 0.30 | 48 | 3 | J774 | Mouse monocyte macrophage | Non-cancer | Humulone |
| Bocquet et al. [17] | MTT | 178.50 | 2.50 | 48 | 3 | WI-38 | Human normal lung fibroblast | Non-cancer | Desmethylxanthohumol |
| Bocquet et al. [17] | MTT | 28.50 | 1.00 | 48 | 3 | J774 | Mouse monocyte macrophage | Non-cancer | Desmethylxanthohumol |
| Bocquet et al. [17] | MTT | 65.90 | 2.90 | 48 | 3 | HepG2 | Human liver cancer | Liver | Desmethylxanthohumol |
| Bocquet et al. [17] | MTT | 116.20 | 3.30 | 48 | 3 | MG-63 | Human osteosarcoma | Bone | Desmethylxanthohumol |
| Roehrer et al. [34] | MTS | 12.25 | 6.91 | 48 | 3 | MCF-7 | Human breast cancer | Breast | Xanthohumol |
| Roehrer et al. [34] | MTS | 8.80 | 7.30 | 96 | 3 | MCF-7 | Human breast cancer | Breast | Xanthohumol |
| Logan et al. [11] | SRB | 40.8 | 1.4 | 24 | 5 | HCT116 | Human colon cancer | Colon | Xanthohumol |
| Logan et al. [11] | SRB | 50.2 | 1.4 | 24 | 5 | HT-29 | Human colon cancer | Colon | Xanthohumol |
| Logan et al. [11] | SRB | 25.4 | 1.1 | 24 | 5 | HepG2 | Human liver cancer | Liver | Xanthohumol |
| Logan et al. [11] | SRB | 37.2 | 1.5 | 24 | 5 | Huh7 | Human liver cancer | Liver | Xanthohumol |
| Bartmańska et al. [35] | SRB | 10.84 | 0.32 | 72 | 5 | MCF-7 | Human breast cancer | Breast | Xanthohumol |
| Bartmańska et al. [35] | SRB | 8.46 | 3.19 | 72 | 5 | MDA-MB-231 | Human breast cancer | Breast | Xanthohumol |
| Bartmańska et al. [35] | SRB | 7.99 | 2.77 | 72 | 5 | T-47D | Human breast cancer | Breast | Xanthohumol |
| Bartmańska et al. [35] | SRB | 9.42 | 0.25 | 72 | 5 | HT-29 | Human colon cancer | Colon | Xanthohumol |
| Bartmańska et al. [35] | SRB | 2.06 | 1.03 | 72 | 5 | A2780 | Human ovarian cancer | Ovarian | Xanthohumol |
| Bartmańska et al. [35] | SRB | 8.21 | 0.83 | 72 | 5 | A2780 | Human ovarian cancer | Ovarian | Xanthohumol |
| Bartmańska et al. [35] | SRB | 6.49 | 2.14 | 72 | 5 | DU145 | Human prostate cancer | Prostate | Xanthohumol |
| Bartmańska et al. [35] | SRB | 8.61 | 1.11 | 72 | 5 | PC-3 | Human prostate cancer | Prostate | Xanthohumol |
| Bartmańska et al. [35] | SRB | 9.57 | 4.23 | 72 | 5 | HLMEC | Human lung microvascular endothelial | Non-cancer | Xanthohumol |
| Bartmańska et al. [35] | SRB | 55.95 | 27.31 | 72 | 5 | MCF-10A | Human normal breast | Non-cancer | Xanthohumol |
| Bartmańska et al. [35] | SRB | 43.25 | 4.37 | 72 | 5 | MCF-7 | Human breast cancer | Breast | 6-prenylnaringenin |
| Bartmańska et al. [35] | SRB | 62.64 | 19.54 | 72 | 5 | MDA-MB-231 | Human breast cancer | Breast | 6-prenylnaringenin |
| Bartmańska et al. [35] | SRB | 16.01 | 3.74 | 72 | 5 | T-47D | Human breast cancer | Breast | 6-prenylnaringenin |
| Bartmańska et al. [35] | SRB | 64.61 | 17.07 | 72 | 5 | HT-29 | Human colon cancer | Colon | 6-prenylnaringenin |
| Bartmańska et al. [35] | SRB | 44.16 | 14.71 | 72 | 5 | A2780 | Human ovarian cancer | Ovarian | 6-prenylnaringenin |
| Bartmańska et al. [35] | SRB | 81.73 | 17.68 | 72 | 5 | A2780 | Human ovarian cancer | Ovarian | 6-prenylnaringenin |
| Bartmańska et al. [35] | SRB | 79.56 | 8.89 | 72 | 5 | DU145 | Human prostate cancer | Prostate | 6-prenylnaringenin |
| Bartmańska et al. [35] | SRB | 75.53 | 29.79 | 72 | 5 | PC-3 | Human prostate cancer | Prostate | 6-prenylnaringenin |
| Bartmańska et al. [35] | SRB | 13.69 | 5.16 | 72 | 5 | HLMEC | Human lung microvascular endothelial | Non-cancer | 6-prenylnaringenin |
| Bartmańska et al. [35] | SRB | 110.06 | 32.95 | 72 | 5 | MCF-10A | Human normal breast | Non-cancer | 6-prenylnaringenin |
| Bartmańska et al. [35] | SRB | 49.53 | 7.36 | 72 | 5 | MCF-7 | Human breast cancer | Breast | 8-prenylnaringenin |
| Bartmańska et al. [35] | SRB | 63.81 | 7.27 | 72 | 5 | MDA-MB-231 | Human breast cancer | Breast | 8-prenylnaringenin |
| Bartmańska et al. [35] | SRB | 26.71 | 9.7 | 72 | 5 | T-47D | Human breast cancer | Breast | 8-prenylnaringenin |
| Bartmańska et al. [35] | SRB | 89.84 | 3.42 | 72 | 5 | HT-29 | Human colon cancer | Colon | 8-prenylnaringenin |
| Bartmańska et al. [35] | SRB | 25.91 | 8.32 | 72 | 5 | A2780 | Human ovarian cancer | Ovarian | 8-prenylnaringenin |
| Bartmańska et al. [35] | SRB | 66.37 | 10.14 | 72 | 5 | A2780 | Human ovarian cancer | Ovarian | 8-prenylnaringenin |
| Bartmańska et al. [35] | SRB | 60.58 | 6.66 | 72 | 5 | DU145 | Human prostate cancer | Prostate | 8-prenylnaringenin |
| Bartmańska et al. [35] | SRB | 51.36 | 11.31 | 72 | 5 | PC-3 | Human prostate cancer | Prostate | 8-prenylnaringenin |
| Bartmańska et al. [35] | SRB | 23.91 | 10.86 | 72 | 5 | HLMEC | Human lung microvascular endothelial | Non-cancer | 8-prenylnaringenin |
| Bartmańska et al. [35] | SRB | 90.72 | 19.8 | 72 | 5 | MCF-10A | Human normal breast | Non-cancer | 8-prenylnaringenin |
| Bartmańska et al. [35] | SRB | 16.73 | 0.88 | 72 | 5 | MCF-7 | Human breast cancer | Breast | Isoxanthohumol |
| Bartmańska et al. [35] | SRB | 43.34 | 10.32 | 72 | 5 | MDA-MB-231 | Human breast cancer | Breast | Isoxanthohumol |
| Bartmańska et al. [35] | SRB | 26.75 | 6.44 | 72 | 5 | T-47D | Human breast cancer | Breast | Isoxanthohumol |
| Bartmańska et al. [35] | SRB | 30.59 | 1 | 72 | 5 | HT-29 | Human colon cancer | Colon | Isoxanthohumol |
| Bartmańska et al. [35] | SRB | 7.93 | 1.65 | 72 | 5 | A2780 | Human ovarian cancer | Ovarian | Isoxanthohumol |
| Bartmańska et al. [35] | SRB | 11.65 | 1.44 | 72 | 5 | A2780 | Human ovarian cancer | Ovarian | Isoxanthohumol |
| Bartmańska et al. [35] | SRB | 59.17 | 5.73 | 72 | 5 | DU145 | Human prostate cancer | Prostate | Isoxanthohumol |
| Bartmańska et al. [35] | SRB | 53.24 | 10.59 | 72 | 5 | PC-3 | Human prostate cancer | Prostate | Isoxanthohumol |
| Bartmańska et al. [35] | SRB | 12.5 | 5.65 | 72 | 5 | HLMEC | Human lung microvascular endothelial | Non-cancer | Isoxanthohumol |
| Bartmańska et al. [35] | SRB | 72.12 | 21.66 | 72 | 5 | MCF-10A | Human normal breast | Non-cancer | Isoxanthohumol |
| Bartmańska et al. [35] | SRB | 130.79 | 6.11 | 72 | 5 | MCF-7 | Human breast cancer | Breast | Naringenin |
| Bartmańska et al. [35] | SRB | 166.09 | 82.44 | 72 | 5 | MDA-MB-231 | Human breast cancer | Breast | Naringenin |
| Bartmańska et al. [35] | SRB | 104.53 | 48.31 | 72 | 5 | T-47D | Human breast cancer | Breast | Naringenin |
| Bartmańska et al. [35] | SRB | 130.8 | 28.19 | 72 | 5 | HT-29 | Human colon cancer | Colon | Naringenin |
| Bartmańska et al. [35] | SRB | 100.05 | 4.77 | 72 | 5 | A2780 | Human ovarian cancer | Ovarian | Naringenin |
| Bartmańska et al. [35] | SRB | 109.23 | 16.98 | 72 | 5 | A2780 | Human ovarian cancer | Ovarian | Naringenin |
| Bartmańska et al. [35] | SRB | 133.66 | 12.92 | 72 | 5 | DU145 | Human prostate cancer | Prostate | Naringenin |
| Bartmańska et al. [35] | SRB | 171.23 | 28.78 | 72 | 5 | PC-3 | Human prostate cancer | Prostate | Naringenin |
| Bartmańska et al. [35] | SRB | 117.24 | 32.27 | 72 | 5 | HLMEC | Human lung microvascular endothelial | Non-cancer | Naringenin |
| Bartmańska et al. [35] | SRB | 187.1 | 72.41 | 72 | 5 | MCF-10A | Human normal breast | Non-cancer | Naringenin |
| Bartmańska et al. [35] | SRB | 10.07 | 2.31 | 72 | 5 | MCF-7 | Human breast cancer | Breast | α,β-dihydroxanthohumol |
| Bartmańska et al. [35] | SRB | 10.02 | 3.26 | 72 | 5 | MDA-MB-231 | Human breast cancer | Breast | α,β-dihydroxanthohumol |
| Bartmańska et al. [35] | SRB | 7.27 | 3.05 | 72 | 5 | T-47D | Human breast cancer | Breast | α,β-dihydroxanthohumol |
| Bartmańska et al. [35] | SRB | 12.23 | 2.99 | 72 | 5 | HT-29 | Human colon cancer | Colon | α,β-dihydroxanthohumol |
| Bartmańska et al. [35] | SRB | 1.8 | 0.64 | 72 | 5 | A2780 | Human ovarian cancer | Ovarian | α,β-dihydroxanthohumol |
| Bartmańska et al. [35] | SRB | 11.59 | 3.36 | 72 | 5 | A2780 | Human ovarian cancer | Ovarian | α,β-dihydroxanthohumol |
| Bartmańska et al. [35] | SRB | 12.96 | 4.2 | 72 | 5 | DU145 | Human prostate cancer | Prostate | α,β-dihydroxanthohumol |
| Bartmańska et al. [35] | SRB | 16.27 | 5.22 | 72 | 5 | PC-3 | Human prostate cancer | Prostate | α,β-dihydroxanthohumol |
| Bartmańska et al. [35] | SRB | 14.17 | 4.24 | 72 | 5 | HLMEC | Human lung microvascular endothelial | Non-cancer | α,β-dihydroxanthohumol |
| Bartmańska et al. [35] | SRB | 72.05 | 8.55 | 72 | 5 | MCF-10A | Human normal breast | Non-cancer | α,β-dihydroxanthohumol |
| Wei et al. [36] | MTS | 16.04 | 7.30 | 24 | 3 | AGS | Human gastric cancer | Gastric | Xanthohumol |
| Wei et al. [36] | MTS | 111.16 | 7.30 | 24 | 3 | SGC-7901 | Human gastric cancer | Gastric | Xanthohumol |
| Wei et al. [36] | MTS | 35.81 | 7.30 | 24 | 3 | MGC-803 | Human gastric cancer | Gastric | Xanthohumol |
| Wei et al. [36] | EdU | 8.00 | 7.30 | 24 | 3 | AGS | Human gastric cancer | Gastric | Xanthohumol |
| Wei et al. [36] | MTS | 285.26 | 6.00 | 24 | 3 | GES-1 | Human normal gastric | Non-cancer | Xanthohumol |
| Carvalho et al. [37] | SRB | 85.5 | 8.92 | 24 | 3 | TPC-1 | Human thyroid cancer | Thyroid | Xanthohumol |
| Carvalho et al. [37] | SRB | 59 | 8.92 | 48 | 3 | TPC-1 | Human thyroid cancer | Thyroid | Xanthohumol |
| Carvalho et al. [37] | SRB | 48.5 | 8.92 | 72 | 3 | TPC-1 | Human thyroid cancer | Thyroid | Xanthohumol |
| Ho et al. [38] | MTT | 53.70 | 7.30 | 24 | 3 | M059K | Human glioblastoma | Glioblastoma | Xanthohumol |
| Ho et al. [38] | MTT | 55.60 | 7.30 | 24 | 3 | U87-MG | Human glioblastoma | Glioblastoma | Xanthohumol |
| Popłoński et al. [39] | SRB | 8.1 | 0.8 | 72 | 4 | MCF-7 | Human breast cancer | Breast | Xanthohumol |
| Popłoński et al. [39] | SRB | 10.1 | 1.1 | 72 | 4 | HT-29 | Human colon cancer | Colon | Xanthohumol |
| Popłoński et al. [39] | SRB | 7 | 1.5 | 72 | 4 | PC-3 | Human prostate cancer | Prostate | Xanthohumol |
| Sun et al. [40] | MTT | 39.40 | 6.91 | 24 | 6 | MCF-7 | Human breast cancer | Breast | Xanthohumol |
| Sun et al. [40] | MTT | 33.30 | 7.30 | 24 | 6 | MDA-MB-231 | Human breast cancer | Breast | Xanthohumol |
| Sun et al. [40] | MTT | 19.60 | 6.91 | 48 | 6 | MCF-7 | Human breast cancer | Breast | Xanthohumol |
| Sun et al. [40] | MTT | 21.50 | 7.30 | 48 | 6 | MDA-MB-231 | Human breast cancer | Breast | Xanthohumol |
| Sun et al. [40] | MTT | 61.10 | 6.00 | 48 | 6 | h-TERT-BJ | Human normal skin fibroblast | Non-cancer | Xanthohumol |
| Sun et al. [40] | MTT | 135.30 | 6.00 | 48 | 6 | MCF-10A | Human normal breast | Non-cancer | Xanthohumol |
| Saito et al. [41] | WST-1 | 17.00 | 7.30 | 72 | 6 | BxPC-3 | Human pancreatic cancer | Pancreas | Xanthohumol |
| Saito et al. [41] | WST-1 | 15.90 | 7.30 | 72 | 6 | MIA PaCa-2 | Human pancreatic cancer | Pancreas | Xanthohumol |
| Saito et al. [41] | WST-1 | 12.90 | 7.30 | 72 | 6 | AsPC-1 | Human pancreatic cancer | Pancreas | Xanthohumol |
| Stompor et al. [42] | SRB | 30 | 3.8 | 72 | 3 | MCF-7 | Human breast cancer | Breast | Isoxanthohumol |
| Stompor et al. [42] | SRB | 29.7 | 4.2 | 72 | 3 | A549 | Human lung cancer | Lung | Isoxanthohumol |
| Stompor et al. [42] | SRB | 8.96 | 1.5 | 72 | 3 | LoVo | Human colon cancer | Colon | Isoxanthohumol |
| Stompor et al. [42] | SRB | 26.8 | 4 | 72 | 3 | LoVo | Human colon cancer | Colon | Isoxanthohumol |
| Stompor et al. [42] | SRB | 16 | 3.6 | 72 | 3 | MES-SA | Human uterine cancer | Uterus | Isoxanthohumol |
| Stompor et al. [42] | SRB | 30.4 | 4.1 | 72 | 3 | MES-SA | Human uterine cancer | Uterus | Isoxanthohumol |
| Stompor et al. [42] | SRB | 37.1 | 3.8 | 72 | 3 | MCF-10A | Human normal breast | Non-cancer | Isoxanthohumol |
| Stompor et al. [42] | XTT | 15.60 | 4.05 | 72 | 3 | U-118 MG | Human glioblastoma | Glioblastoma | Isoxanthohumol |
| Liu et al. [43] | MTT | 81.45 | 6.91 | 24 | 3 | MCF-7 | Human breast cancer | Breast | Xanthohumol |
| Liu et al. [43] | MTT | 78.33 | 7.30 | 24 | 3 | MCF-7/ADR | Human breast cancer, doxorubicin-resistant | Breast | Xanthohumol |
| Liu et al. [43] | MTT | 34.02 | 3.45 | 48 | 3 | MCF-7 | Human breast cancer | Breast | Xanthohumol |
| Liu et al. [43] | MTT | 33.71 | 3.12 | 48 | 3 | MCF-7/ADR | Human breast cancer, doxorubicin-resistant | Breast | Xanthohumol |
| Liu et al. [43] | MTT | 11.22 | 0.95 | 72 | 3 | MCF-7 | Human breast cancer | Breast | Xanthohumol |
| Liu et al. [43] | MTT | 11.37 | 1.15 | 72 | 3 | MCF-7/ADR | Human breast cancer, doxorubicin-resistant | Breast | Xanthohumol |
| Gallo et al. [44] | MTT | 18.30 | 6.00 | 96 | 3 | HUVEC | Human umbilical vein endothelial | Non-cancer | Xanthohumol |
| Chen et al. [45] | MTT | 64.80 | 7.30 | 24 | 3 | U87-MG | Human glioblastoma | Glioblastoma | Xanthohumol |
| Chen et al. [45] | MTT | 19.70 | 7.30 | 48 | 3 | U87-MG | Human glioblastoma | Glioblastoma | Xanthohumol |
| Chen et al. [45] | MTT | 13.10 | 7.30 | 72 | 3 | U87-MG | Human glioblastoma | Glioblastoma | Xanthohumol |
| Lempereur et al. [46] | Crystal Violet | 4.10 | 11.60 | 72 | 6 | MCF-7 | Human breast cancer | Breast | Tetrahydro Iso-Alpha Acids |
| Lempereur et al. [46] | Crystal Violet | 20.60 | 11.60 | 72 | 6 | MDA-MB-231 | Human breast cancer | Breast | Tetrahydro Iso-Alpha Acids |
| Lempereur et al. [46] | Crystal Violet | 15.30 | 11.60 | 72 | 6 | MCF-7 | Human breast cancer | Breast | α-acids |
| Lempereur et al. [46] | Crystal Violet | 15.70 | 11.60 | 72 | 6 | MDA-MB-231 | Human breast cancer | Breast | α-acids |
| Lempereur et al. [46] | Crystal Violet | 13.10 | 11.60 | 72 | 6 | MCF-7 | Human breast cancer | Breast | Iso-α-acids |
| Lempereur et al. [46] | Crystal Violet | 13.70 | 11.60 | 72 | 6 | MDA-MB-231 | Human breast cancer | Breast | Iso-α-acids |
| Lempereur et al. [46] | Crystal Violet | 29.90 | 11.60 | 72 | 6 | MDA-MB-231 | Human breast cancer | Breast | Dihydro-iso-alpha acids |
| Yoo et al. [47] | MTT | 16.80 | 7.30 | 48 | 4 | MDA-MB-231 | Human breast cancer | Breast | Xanthohumol |
| Krajnović et al. [48] | Crystal Violet | 15.77 | 1.74 | 48 | 3 | A375 | Human melanoma | Melanoma | Xanthohumol |
| Krajnović et al. [48] | Crystal Violet | 9.97 | 2.32 | 48 | 3 | B16 | Murine melanoma | Melanoma | Xanthohumol |
| Krajnović et al. [48] | Crystal Violet | 48.30 | 11.6 | 48 | 3 | A375 | Human melanoma | Melanoma | 8-prenylnaringenin |
| Krajnović et al. [48] | Crystal Violet | 38.55 | 8.84 | 48 | 3 | B16 | Murine melanoma | Melanoma | 8-prenylnaringenin |
| Krajnović et al. [48] | Crystal Violet | 24.18 | 1.43 | 48 | 3 | A375 | Human melanoma | Melanoma | Isoxanthohumol |
| Krajnović et al. [48] | Crystal Violet | 21.88 | 5.19 | 48 | 3 | B16 | Murine melanoma | Melanoma | Isoxanthohumol |
| Krajnović et al. [48] | MTT | 15.00 | 1.15 | 48 | 3 | A375 | Human melanoma | Melanoma | Xanthohumol |
| Krajnović et al. [48] | MTT | 8.70 | 0.99 | 48 | 3 | B16 | Murine melanoma | Melanoma | Xanthohumol |
| Krajnović et al. [48] | MTT | 27.80 | 3.82 | 48 | 3 | A375 | Human melanoma | Melanoma | 8-prenylnaringenin |
| Krajnović et al. [48] | MTT | 40.85 | 0.78 | 48 | 3 | B16 | Murine melanoma | Melanoma | 8-prenylnaringenin |
| Krajnović et al. [48] | MTT | 22.90 | 0.78 | 48 | 3 | A375 | Human melanoma | Melanoma | Isoxanthohumol |
| Krajnović et al. [48] | MTT | 22.15 | 4.05 | 48 | 3 | B16 | Murine melanoma | Melanoma | Isoxanthohumol |
| Yong et al. [49] | SRB | 74.06 | 3.43 | 24 | 3 | A549 | Human lung cancer | Lung | Xanthohumol |
| Yong et al. [49] | SRB | 25.48 | 0.52 | 48 | 3 | A549 | Human lung cancer | Lung | Xanthohumol |
| Yong et al. [49] | SRB | 149.2 | 8.59 | 48 | 3 | MRC-5 | Human normal lung fibroblast | Non-cancer | Xanthohumol |
| Yong et al. [49] | SRB | 13.5 | 1.42 | 72 | 3 | A549 | Human lung cancer | Lung | Xanthohumol |
| Yong et al. [49] | SRB | 94.38 | 3.07 | 72 | 3 | MRC-5 | Human normal lung fibroblast | Non-cancer | Xanthohumol |
| Zhang et al. [50] | CCK-8 | 7.90 | 7.30 | 48 | 3 | HeLa | Human cervical cancer | Cervix | Xanthohumol |
| Zhang et al. [50] | CCK-8 | 8.60 | 2.99 | 48 | 3 | A549 | Human lung cancer | Lung | Xanthohumol |
| Zhang et al. [50] | CCK-8 | 32.00 | 6.00 | 48 | 3 | MCF-10A | Human normal breast | Non-cancer | Xanthohumol |
| Zhang et al. [50] | CCK-8 | 53.60 | 4.05 | 48 | 3 | A549 | Human lung cancer | Lung | Isoxanthohumol |
| Zhang et al. [50] | CCK-8 | 46.30 | 4.05 | 48 | 3 | HeLa | Human cervical cancer | Cervix | Isoxanthohumol |
| Sławińska-Brych et al. [51] | MTT | 12.30 | 7.30 | 48 | 3 | RK33 | Human neck cancer | Neck | Xanthohumol |
| Sławińska-Brych et al. [51] | MTT | 22.50 | 7.30 | 48 | 3 | RK45 | Human neck cancer | Neck | Xanthohumol |
| Sławińska-Brych et al. [51] | MTT | 100.00 | 6.00 | 48 | 3 | HSF | Human normal skin fibroblast | Non-cancer | Xanthohumol |
| Sławińska-Brych et al. [51] | MTT | 105.00 | 6.00 | 48 | 3 | OLN93 | Rat oligodendroglia | Non-cancer | Xanthohumol |
| Jiang et al. [52] | MTS | 10.00 | 7.30 | 24 | 3 | PANC-1 | Human pancreatic cancer | Pancreas | Xanthohumol |
| Jiang et al. [52] | MTS | 27.70 | 7.30 | 24 | 3 | BxPC-3 | Human pancreatic cancer | Pancreas | Xanthohumol |
| Jiang et al. [52] | MTS | 8.10 | 7.30 | 48 | 3 | PANC-1 | Human pancreatic cancer | Pancreas | Xanthohumol |
| Jiang et al. [52] | MTS | 9.10 | 7.30 | 48 | 3 | BxPC-3 | Human pancreatic cancer | Pancreas | Xanthohumol |
| Jiang et al. [52] | MTS | 5.60 | 7.30 | 72 | 3 | BxPC-3 | Human pancreatic cancer | Pancreas | Xanthohumol |
| Jiang et al. [52] | MTS | 4.40 | 7.30 | 72 | 3 | PANC-1 | Human pancreatic cancer | Pancreas | Xanthohumol |
| Zenger et al. [53] | MTT | 65.00 | 4.00 | 24 | 3 | HSC | Human hepatic stellate | Non-cancer | Xanthohumol |
| Yong and Abd Malek [54] | SRB | 59.96 | 2.76 | 24 | 2 | Ca Ski | Human cervical cancer | Cervix | Xanthohumol |
| Yong and Abd Malek [54] | SRB | 34.01 | 1.6 | 48 | 2 | Ca Ski | Human cervical cancer | Cervix | Xanthohumol |
| Yong and Abd Malek [54] | SRB | 20.08 | 1.58 | 72 | 2 | Ca Ski | Human cervical cancer | Cervix | Xanthohumol |
| Mouratidis et al. [55] | MTT | 10.50 | 0.50 | 48 | 3 | PC-3 | Human prostate cancer | Prostate | Lupulone |
| Mouratidis et al. [55] | MTT | 9.00 | 0.50 | 48 | 3 | DU145 | Human prostate cancer | Prostate | Lupulone |
| Mouratidis et al. [55] | MTT | 5.00 | 0.50 | 72 | 3 | PC-3 | Human prostate cancer | Prostate | Lupulone |
| Mouratidis et al. [55] | MTT | 5.00 | 0.50 | 72 | 3 | DU145 | Human prostate cancer | Prostate | Lupulone |
| Boncler et al. [56] | MTT | 9.60 | 0.40 | 24 | 4 | HUVEC | Human umbilical vein endothelial | Non-cancer | Spent hops, after SFE extraction of hops, were dried and then extracted with acetone:water (70:30; v/v) |
| Tronina et al. [57] | SRB | 10.95 | 1.03 | 72 | 4 | MCF-7 | Human breast cancer | Breast | Xanthohumol |
| Tronina et al. [57] | SRB | 91.31 | 8.92 | 72 | 4 | HT-29 | Human colon cancer | Colon | Xanthohumol |
| Tronina et al. [57] | SRB | 10.67 | 1.06 | 72 | 4 | PC-3 | Human prostate cancer | Prostate | Xanthohumol |
| Tronina et al. [58] | SRB | 10.95 | 1.03 | 72 | 4 | MCF-7 | Human breast cancer | Breast | Xanthohumol |
| Tronina et al. [58] | SRB | 91.31 | 8.92 | 72 | 4 | HT-29 | Human colon cancer | Colon | Xanthohumol |
| Tronina et al. [58] | SRB | 10.67 | 1.06 | 72 | 4 | PC-3 | Human prostate cancer | Prostate | Xanthohumol |
| Tronina et al. [58] | SRB | 26.54 | 12.68 | 72 | 4 | MCF-7 | Human breast cancer | Breast | Isoxanthohumol |
| Tronina et al. [58] | SRB | 88.82 | 4.15 | 72 | 4 | HT-29 | Human colon cancer | Colon | Isoxanthohumol |
| Tronina et al. [58] | SRB | 71.32 | 19.59 | 72 | 4 | PC-3 | Human prostate cancer | Prostate | Isoxanthohumol |
| Tronina et al. [58] | SRB | 9.15 | 0.62 | 72 | 4 | MCF-7 | Human breast cancer | Breast | α,β-dihydroxanthohumol |
| Tronina et al. [58] | SRB | 74.41 | 23.44 | 72 | 4 | HT-29 | Human colon cancer | Colon | α,β-dihydroxanthohumol |
| Tronina et al. [58] | SRB | 14.73 | 3.88 | 72 | 4 | PC-3 | Human prostate cancer | Prostate | α,β-dihydroxanthohumol |
| Allsopp et al. [59] | MTT | 70.00 | 8.20 | 24 | 3 | CaCo-2 | Human colon cancer | Colon | 8-prenylnaringenin |
| Allsopp et al. [59] | MTT | 55.00 | 4.05 | 24 | 3 | CaCo-2 | Human colon cancer | Colon | Isoxanthohumol |
| Kang et al. [60] | MTT | 16.60 | 6.91 | 24 | 3 | MCF-7 | Human breast cancer | Breast | Xanthohumol |
| Kang et al. [60] | MTT | 18.00 | 7.30 | 24 | 3 | MCF-7/ADR | Human breast cancer, doxorubicin-resistant | Breast | Xanthohumol |
| Kang et al. [60] | MTT | 35.20 | 7.30 | 24 | 3 | HT-29 | Human colon cancer | Colon | Xanthohumol |
| Viegas et al. [13] | MTT | 117.10 | 3.48 | 24 | 3 | HepG2 | Human liver cancer | Liver | Xanthohumol |
| Hemachandra et al. [61] | MTT | 11.00 | 0.50 | 24 | 3 | MCF-10A | Human normal breast | Non-cancer | Ethanolic extract: Spent hops, after SFE extraction of hops, were dried and then extracted with ethanol |
| Deeb et al. [62] | MTS | 26.50 | 7.30 | 72 | 3 | LNCaP | Human prostate cancer | Prostate | Xanthohumol |
| Deeb et al. [62] | MTS | 29.00 | 7.30 | 72 | 3 | DU145 | Human prostate cancer | Prostate | Xanthohumol |
| Deeb et al. [62] | MTS | 31.00 | 7.30 | 72 | 3 | C4-2 | Human prostate cancer | Prostate | Xanthohumol |
| Deeb et al. [62] | MTS | 25.10 | 7.30 | 72 | 3 | PC-3 | Human prostate cancer | Prostate | Xanthohumol |
| Negrão et al. [63] | MTT | 24.00 | 6.00 | 24 | 3 | HUVEC | Human umbilical vein endothelial | Non-cancer | Xanthohumol |
| Negrão et al. [63] | MTT | 12.00 | 6.00 | 24 | 3 | HASMC | Human aortic smooth muscle | Non-cancer | Xanthohumol |
| Negrão et al. [63] | MTT | 28.00 | 4.05 | 24 | 3 | HUVEC | Human umbilical vein endothelial | Non-cancer | Isoxanthohumol |
| Wesołowska et al. [64] | SRB | 33 | 3.42 | 72 | 3 | LoVo | Human colon cancer | Colon | 8-prenylnaringenin |
| Wesołowska et al. [64] | SRB | 55 | 3.42 | 72 | 3 | LoVo | Human colon cancer | Colon | 8-prenylnaringenin |
| Strathmann et al. [65] | SRB | 6.7 | 0.2 | 72 | 3 | BPH-1 | Human benign prostatic hyperplasia | Non-cancer | Xanthohumol |
| Dorn et al. [66] | XTT | 20.00 | 5.00 | 72 | 3 | HepG2 | Human liver cancer | Liver | Xanthohumol |
| Dorn et al. [66] | XTT | 15.00 | 5.00 | 72 | 3 | Huh7 | Human liver cancer | Liver | Xanthohumol |
| Mendes et al. [67] | SRB | 26 | 27.31 | 24 | 3 | 3T3-L1 | Murine preadipocyte | Non-cancer | Xanthohumol |
| Mendes et al. [67] | SRB | 12 | 27.31 | 48 | 3 | 3T3-L1 | Murine preadipocyte | Non-cancer | Xanthohumol |
| Mendes et al. [67] | SRB | 17 | 27.31 | 72 | 3 | 3T3-L1 | Murine preadipocyte | Non-cancer | Xanthohumol |
| Ho et al. [68] | MTT | 166.00 | 3.00 | 24 | 3 | HA22T/VGH | Human liver cancer | Liver | Xanthohumol |
| Ho et al. [68] | MTT | 108.00 | 5.00 | 24 | 3 | Hep3B | Human liver cancer | Liver | Xanthohumol |
| Ho et al. [68] | MTT | 211.00 | 6.00 | 24 | 3 | AML12 | Murine normal liver | Non-cancer | Xanthohumol |
| Monteghirfo et al. [69] | MTT | 10.00 | 1.50 | 48 | 3 | K562 | Human leukemia | Leukemia | Xanthohumol |
| Monteghirfo et al. [69] | MTT | 16.00 | 1.50 | 48 | 3 | Mononuclear cells from CML patients | Leukemia | Xanthohumol | |
| Monteghirfo et al. [69] | MTT | 5.40 | 1.50 | 72 | 3 | K562 | Human leukemia | Leukemia | Xanthohumol |
| Koo et al. [70] | MTT | 8.60 | 1.15 | 24 | 3 | B16-F10 | Murine melanoma | Melanoma | Xanthohumol |
| Lee et al. [71] | SRB | 12.13 | 3.43 | 48 | 3 | A549 | Human lung cancer | Lung | Xanthohumol |
| Lee et al. [71] | SRB | 10.15 | 8.92 | 48 | 3 | HCT15 | Human colon cancer | Colon | Xanthohumol |
| Lee et al. [71] | SRB | 14.39 | 8.92 | 48 | 3 | SK-MEL-2 | Human melanoma | Melanoma | Xanthohumol |
| Lee et al. [71] | SRB | 16 | 1.03 | 48 | 3 | SK-OV-3 | Human ovarian cancer | Ovarian | Xanthohumol |
| Lee et al. [71] | SRB | 100.17 | 3.42 | 48 | 3 | HCT15 | Human colon cancer | Colon | 8-prenylnaringenin |
| Lee et al. [71] | SRB | 66.39 | 11.31 | 48 | 3 | A549 | Human lung cancer | Lung | 8-prenylnaringenin |
| Lee et al. [71] | SRB | 102.23 | 11.31 | 48 | 3 | SK-MEL-2 | Human melanoma | Melanoma | 8-prenylnaringenin |
| Lee et al. [71] | SRB | 75.2 | 10.14 | 48 | 3 | SK-OV-3 | Human ovarian cancer | Ovarian | 8-prenylnaringenin |
| Lee et al. [71] | SRB | 63.48 | 4 | 48 | 3 | HCT15 | Human colon cancer | Colon | Isoxanthohumol |
| Lee et al. [71] | SRB | 77.59 | 4.2 | 48 | 3 | A549 | Human lung cancer | Lung | Isoxanthohumol |
| Lee et al. [71] | SRB | 40.34 | 19.59 | 48 | 3 | SK-MEL-2 | Human melanoma | Melanoma | Isoxanthohumol |
| Lee et al. [71] | SRB | 27.93 | 1.65 | 48 | 3 | SK-OV-3 | Human ovarian cancer | Ovarian | Isoxanthohumol |
| Yang et al. [72] | MTS | 75.00 | 14.14 | 24 | 8 | 3T3-L1 | Murine preadipocyte | Non-cancer | Xanthohumol |
| Yang et al. [72] | MTS | 53.00 | 9.90 | 48 | 8 | 3T3-L1 | Murine preadipocyte | Non-cancer | Xanthohumol |
| Monteiro et al. [73] | SRB | 7.1 | 4.6 | 72 | 9 | Sk-Br-3 | Human breast cancer | Breast | Xanthohumol |
| Monteiro et al. [73] | SRB | 22.6 | 9.7 | 72 | 9 | Sk-Br-3 | Human breast cancer | Breast | 8-prenylnaringenin |
| Monteiro et al. [73] | SRB | 41 | 12.68 | 72 | 9 | Sk-Br-3 | Human breast cancer | Breast | Isoxanthohumol |
| Plazar et al. [74] | MTT | 75.00 | 3.48 | 24 | 5 | HepG2 | Human liver cancer | Liver | Xanthohumol |
| Delmulle et al. [75] | WST-1 | 13.20 | 1.10 | 48 | 3 | PC-3 | Human prostate cancer | Prostate | Xanthohumol |
| Delmulle et al. [75] | WST-1 | 12.30 | 1.10 | 48 | 3 | DU145 | Human prostate cancer | Prostate | Xanthohumol |
| Delmulle et al. [75] | WST-1 | 18.40 | 1.20 | 48 | 3 | PC-3 | Human prostate cancer | Prostate | 6-prenylnaringenin |
| Delmulle et al. [75] | WST-1 | 29.10 | 1.10 | 48 | 3 | DU145 | Human prostate cancer | Prostate | 6-prenylnaringenin |
| Delmulle et al. [75] | WST-1 | 33.50 | 1.00 | 48 | 3 | PC-3 | Human prostate cancer | Prostate | 8-prenylnaringenin |
| Delmulle et al. [75] | WST-1 | 43.10 | 1.20 | 48 | 3 | DU145 | Human prostate cancer | Prostate | 8-prenylnaringenin |
| Delmulle et al. [75] | WST-1 | 45.20 | 1.10 | 48 | 3 | PC-3 | Human prostate cancer | Prostate | Isoxanthohumol |
| Delmulle et al. [75] | WST-1 | 47.40 | 1.10 | 48 | 3 | DU145 | Human prostate cancer | Prostate | Isoxanthohumol |
| Delmulle et al. [75] | WST-1 | 49.90 | 1.00 | 48 | 3 | PC-3 | Human prostate cancer | Prostate | Desmethylxanthohumol |
| Delmulle et al. [75] | WST-1 | 53.80 | 1.10 | 48 | 3 | DU145 | Human prostate cancer | Prostate | Desmethylxanthohumol |
| Colgate et al. [76] | MTT | 24.00 | 7.30 | 48 | 4 | PC-3 | Human prostate cancer | Prostate | Xanthohumol |
| Colgate et al. [76] | MTT | 5.00 | 6.00 | 48 | 4 | BPH-1 | Human benign prostatic hyperplasia | Non-cancer | Xanthohumol |
| Dietz et al. [77] | Crystal Violet | 30.70 | 7.6 | 48 | 3 | Hepa 1c1c7 | Murine liver cancer | Liver | Xanthohumol |
| Dietz et al. [77] | Crystal Violet | 30.70 | 2.9 | 48 | 3 | Hepa 1c1c7 | Murine liver cancer | Liver | Isoxanthohumol |
| Pan et al. [78] | SRB | 4.1 | 0.9 | 24 | 3 | HCT116 | Human colon cancer | Colon | Xanthohumol |
| Pan et al. [78] | SRB | 3.6 | 0.6 | 48 | 3 | HCT116 | Human colon cancer | Colon | Xanthohumol |
| Pan et al. [78] | SRB | 2.6 | 0.1 | 72 | 3 | HCT116 | Human colon cancer | Colon | Xanthohumol |
| Gerhauser et al. [3] | Crystal Violet | 7.40 | 1.4 | 48 | 3 | Hepa 1c1c7 | Murine liver cancer | Liver | Xanthohumol |
| Gerhauser et al. [3] | Crystal Violet | 29.90 | 1.9 | 48 | 3 | Hepa 1c1c7 | Murine liver cancer | Liver | Isoxanthohumol |
| Miranda et al. [79] | SRB | 13.3 | 1.16 | 48 | 4 | MCF-7 | Human breast cancer | Breast | Xanthohumol |
| Miranda et al. [79] | SRB | 46 | 8.92 | 48 | 4 | HT-29 | Human colon cancer | Colon | Xanthohumol |
| Miranda et al. [79] | SRB | 0.52 | 1.03 | 48 | 4 | A2780 | Human ovarian cancer | Ovarian | Xanthohumol |
| Miranda et al. [79] | SRB | 3.47 | 1.16 | 96 | 4 | MCF-7 | Human breast cancer | Breast | Xanthohumol |
| Miranda et al. [79] | SRB | 51.1 | 8.92 | 96 | 4 | HT-29 | Human colon cancer | Colon | Xanthohumol |
| Miranda et al. [79] | SRB | 5.22 | 1.03 | 96 | 4 | A2780 | Human ovarian cancer | Ovarian | Xanthohumol |
| Miranda et al. [79] | SRB | 15.3 | 12.68 | 48 | 4 | MCF-7 | Human breast cancer | Breast | Isoxanthohumol |
| Miranda et al. [79] | SRB | 62.5 | 4.15 | 48 | 4 | HT-29 | Human colon cancer | Colon | Isoxanthohumol |
| Miranda et al. [79] | SRB | 18 | 1.65 | 48 | 4 | A2780 | Human ovarian cancer | Ovarian | Isoxanthohumol |
| Miranda et al. [79] | SRB | 4.69 | 12.68 | 96 | 4 | MCF-7 | Human breast cancer | Breast | Isoxanthohumol |
| Miranda et al. [79] | SRB | 25.7 | 1.65 | 96 | 4 | A2780 | Human ovarian cancer | Ovarian | Isoxanthohumol |
| Miranda et al. [79] | SRB | 57.8 | 4.15 | 96 | 4 | HT-29 | Human colon cancer | Colon | Isoxanthohumol |
| Time (h) | Cancer | Non-Cancer | ||
|---|---|---|---|---|
| p-Value | Number of Studies | p-Value | Number of Studies | |
| 24 | 0.777 | 37 | 0.517 | 9 |
| 48 | 0.350 | 46 | 0.356 | 12 |
| 72 | 0.702 | 52 | 0.913 | 9 |
| Compound | Time (h) | Number of Experiments | Number of Studies | SMD | 95% CI | p-Value | Type of Cancer |
|---|---|---|---|---|---|---|---|
| Xanthohumol | 48 | 12 | 2 | 0.62 | 0.00, 1.79 | 0.30 | Melanoma |
| Isoxanthohumol | 48 | 18 | 3 | 1.00 | 0.18, 4.98 | 0.16 | Melanoma |
| 8-prenylnaringenin | 48 | 12 | 2 | 0.88 | 0.00, 3.55 | 0.52 | Melanoma |
| Group of Compounds | Time | Number of Studies | Type of Cancer | IC50 (μΜ/ μg/mL) * | 95% CI | p-Value | I2 (%) |
|---|---|---|---|---|---|---|---|
| Chalcones | 24 | 5 | Glioblastoma | 60.44 | 55.13, 65.75 | 0.000 | 51.6 |
| 24 | 2 | Neck | 50.80 | 40.61, 60.99 | 0.000 | 67.2 | |
| 24 | 4 | Gastric | 42.75 | 0.00, 88.89 | 0.069 | 99.2 | |
| 24 | 6 | Liver | 88.08 | 52.80, 123.35 | 0.000 | 99.7 | |
| 24 | 2 | Pancreas | 18.85 | 1.50, 36.20 | 0.033 | 88.7 | |
| 24 | 5 | Colon | 33.83 | 11.56, 56.11 | 0.003 | 99.9 | |
| 24 | 7 | Breast | 48.97 | 29.38, 68.57 | 0.000 | 98.1 | |
| 24 | 37 | Cancer | 52.16 | 42.66, 61.66 | 0.000 | 99.8 | |
| 24 | 9 | Non-cancer | 90.03 | 35.94, 144.11 | 0.001 | 99.9 | |
| Chalcones | 48 | 3 | Leukemia | 15.22 | 9.55, 20.89 | 0.000 | 98.0 |
| 48 | 2 | Myeloma | 35.99 | 8.28, 63.69 | 0.011 | 95.6 | |
| 48 | 5 | Melanoma | 12.54 | 8.89, 16.20 | 0.000 | 94.3 | |
| 48 | 4 | Neck | 18.93 | 14.11, 23.74 | 0.000 | 26.4 | |
| 48 | 3 | Lung | 15.50 | 3.18, 27.81 | 0.014 | 98.5 | |
| 48 | 4 | Liver | 27.69 | 7.89, 47.48 | 0.006 | 99.7 | |
| 48 | 2 | Pancreas | 8.60 | 2.76, 14.44 | 0.004 | 0.0 | |
| 48 | 6 | Colon | 17.02 | 8.62, 25.41 | 0.000 | 96.7 | |
| 48 | 2 | Bone | 72.79 | 0.00, 157.86 | 0.093 | 99.9 | |
| 48 | 7 | Breast | 21.75 | 13.26, 30.24 | 0.000 | 97.1 | |
| 48 | 2 | Cervix | 21.27 | 0.00, 46.85 | 0.103 | 97.2 | |
| 48 | 2 | Ovarian | 8.26 | 0.00, 23.43 | 0.286 | 99.7 | |
| 48 | 5 | Prostate | 30.67 | 10.98, 50.37 | 0.002 | 99.9 | |
| 48 | 50 | Cancer | 22.54 | 18.06, 27.01 | 0.000 | 99.7 | |
| 48 | 14 | Non-cancer | 65.53 | 48.75, 82.32 | 0.000 | 99.9 | |
| Chalcones | 72 | 3 | Leukemia | 6.59 | 3.26, 9.91 | 0.000 | 92.5 |
| 72 | 2 | Neck | 18.65 | 12.81, 24.49 | 0.000 | 0.0 | |
| 72 | 2 | Lung | 17.02 | 9.82, 24.21 | 0.000 | 93.2 | |
| 72 | 4 | Liver | 39.79 | 0.00, 83.02 | 0.071 | 99.7 | |
| 72 | 5 | Pancreas | 11.86 | 7.16, 16.56 | 0.000 | 59.8 | |
| 72 | 10 | Colon | 23.29 | 19.58, 27.00 | 0.000 | 99.8 | |
| 72 | 17 | Breast | 10.97 | 9.79, 12.14 | 0.000 | 94.9 | |
| 72 | 4 | Ovarian | 5.73 | 1.90, 9.57 | 0.003 | 98.7 | |
| 72 | 12 | Prostate | 13.51 | 11.37, 15.64 | 0.000 | 99.8 | |
| 72 | 63 | Cancer | 15.93 | 14.42, 17.43 | 0.000 | 99.8 | |
| 72 | 11 | Non-cancer | 33.89 | 15.15, 52.62 | 0.000 | 99.9 | |
| Flavones | 24 | 3 | Colon | 97.16 | 31.77, 162.55 | 0.004 | 99.6 |
| 24 | 2 | Breast | 109.68 | 99.90, 119.45 | 0.000 | 61.7 | |
| 24 | 5 | Cancer | 102.26 | 62.78, 141.74 | 0.000 | 99.3 | |
| Flavones | 48 | 12 | Melanoma | 36.81 | 29.99, 43.62 | 0.000 | 98.9 |
| 48 | 3 | Lung | 65.83 | 47.94, 83.71 | 0.000 | 96.1 | |
| 48 | 2 | Liver | 30.14 | 28.34, 31.94 | 0.000 | 0.0 | |
| 48 | 4 | Colon | 74.05 | 56.91, 91.19 | 0.000 | 98.7 | |
| 48 | 3 | Ovarian | 37.41 | 25.11, 49.72 | 0.000 | 98.6 | |
| 48 | 6 | Prostate | 36.12 | 27.47, 44.77 | 0.000 | 99.6 | |
| 48 | 32 | Cancer | 43.76 | 38.89, 48.64 | 0.000 | 99.4 | |
| Flavones | 72 | 9 | Colon | 50.55 | 33.01, 68.09 | 0.000 | 99.7 |
| 72 | 2 | Uterus | 23.16 | 9.04, 37.27 | 0.001 | 95.2 | |
| 72 | 13 | Breast | 35.55 | 27.15, 43.96 | 0.000 | 97.9 | |
| 72 | 6 | Ovarian | 36.19 | 26.65, 45.74 | 0.000 | 98.3 | |
| 72 | 7 | Prostate | 62.84 | 55.13, 70.54 | 0.000 | 80.5 | |
| 72 | 39 | Cancer | 42.95 | 36.91, 48.99 | 0.000 | 99.4 | |
| 72 | 7 | Non-cancer | 46.96 | 31.08, 62.84 | 0.000 | 96.9 | |
| α-acids | 48 | 2 | Non-cancer | 30.67 | 28.10, 33.24 | 0.000 | 75.4 |
| α-acids | 72 | 7 | Breast | 16.06 | 10.24, 21.88 | 0.000 | 63.7 |
| 72 | 7 | Cancer | 16.06 | 10.24, 21.88 | 0.000 | 63.7 | |
| β-acids | 48 | 2 | Prostate | 9.75 | 8.28, 11.22 | 0.000 | 92.6 |
| 48 | 4 | Cancer | 8.20 | 4.77, 11.63 | 0.000 | 99.4 | |
| 48 | 2 | Non-cancer | 3.10 | 2.12, 4.08 | 0.000 | 99.3 | |
| β-acids | 72 | 2 | Prostate | 5.00 | 4.60, 5.40 | 0.000 | 0.0 |
| 72 | 2 | Cancer | 5.00 | 4.60, 5.40 | 0.000 | 0.0 | |
| Hops crude extract ** | 24 | 3 | Non-cancer | 57.90 | 41.20, 74.60 | 0.000 | 99.9 |
| Hops crude extract ** | 72 | 2 | Liver | 16.57 | 0.00, 35.65 | 0.089 | 99.2 |
| 72 | 5 | Cancer | 35.23 | 15.20, 55.26 | 0.001 | 99.5 | |
| 72 | 3 | Non-cancer | 43.80 | 0.00, 87.78 | 0.051 | 99.9 |
| 24 h | 48 h | 72 h | ||||
|---|---|---|---|---|---|---|
| Compounds | p-Value | Number of Studies (Cancer/Non-Cancer) | p-Value | Number of Studies (Cancer/Non-Cancer) | p-Value | Number of Studies (Cancer/Non-Cancer) |
| Chalcones | 0.049 | (37/9) | 0.000 | (50/14) | 0.019 | (63/11) |
| Xanthohumol | 0.049 | (37/9) | 0.000 | (46/12) | 0.088 | (52/9) |
| α,β-dihydroxanthohumol | 0.046 | (11/2) | ||||
| Desmethylxanthohumol | 0.566 | (4/2) | ||||
| Flavones | 0.712 | (39/7) | ||||
| 8-prenylnaringenin | 0.777 | (12/2) | ||||
| 6-prenylnaringenin | 0.947 | (8/2) | ||||
| Isoxanthohumol | 0.760 | (19/3) | ||||
| β-acids | 0.129 | (4/2) | ||||
| Lupulone | 0.129 | (4/2) | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsionkis, G.; Andronidou, E.M.; Kontou, P.I.; Tamposis, I.A.; Tegopoulos, K.; Pergantas, P.; Grigoriou, M.E.; Skavdis, G.; Bagos, P.G.; Braliou, G.G. Humulus lupulus (Hop)-Derived Chemical Compounds Present Antiproliferative Activity on Various Cancer Cell Types: A Meta-Regression Based Panoramic Meta-Analysis. Pharmaceuticals 2025, 18, 1139. https://doi.org/10.3390/ph18081139
Tsionkis G, Andronidou EM, Kontou PI, Tamposis IA, Tegopoulos K, Pergantas P, Grigoriou ME, Skavdis G, Bagos PG, Braliou GG. Humulus lupulus (Hop)-Derived Chemical Compounds Present Antiproliferative Activity on Various Cancer Cell Types: A Meta-Regression Based Panoramic Meta-Analysis. Pharmaceuticals. 2025; 18(8):1139. https://doi.org/10.3390/ph18081139
Chicago/Turabian StyleTsionkis, Georgios, Elisavet M. Andronidou, Panagiota I. Kontou, Ioannis A. Tamposis, Konstantinos Tegopoulos, Panagiotis Pergantas, Maria E. Grigoriou, George Skavdis, Pantelis G. Bagos, and Georgia G. Braliou. 2025. "Humulus lupulus (Hop)-Derived Chemical Compounds Present Antiproliferative Activity on Various Cancer Cell Types: A Meta-Regression Based Panoramic Meta-Analysis" Pharmaceuticals 18, no. 8: 1139. https://doi.org/10.3390/ph18081139
APA StyleTsionkis, G., Andronidou, E. M., Kontou, P. I., Tamposis, I. A., Tegopoulos, K., Pergantas, P., Grigoriou, M. E., Skavdis, G., Bagos, P. G., & Braliou, G. G. (2025). Humulus lupulus (Hop)-Derived Chemical Compounds Present Antiproliferative Activity on Various Cancer Cell Types: A Meta-Regression Based Panoramic Meta-Analysis. Pharmaceuticals, 18(8), 1139. https://doi.org/10.3390/ph18081139









