Impact of Genetic Variation in Adrenergic Receptors on β-Blocker Effectiveness and Safety in Cardiovascular Disease Management: A Systematic Review
Abstract
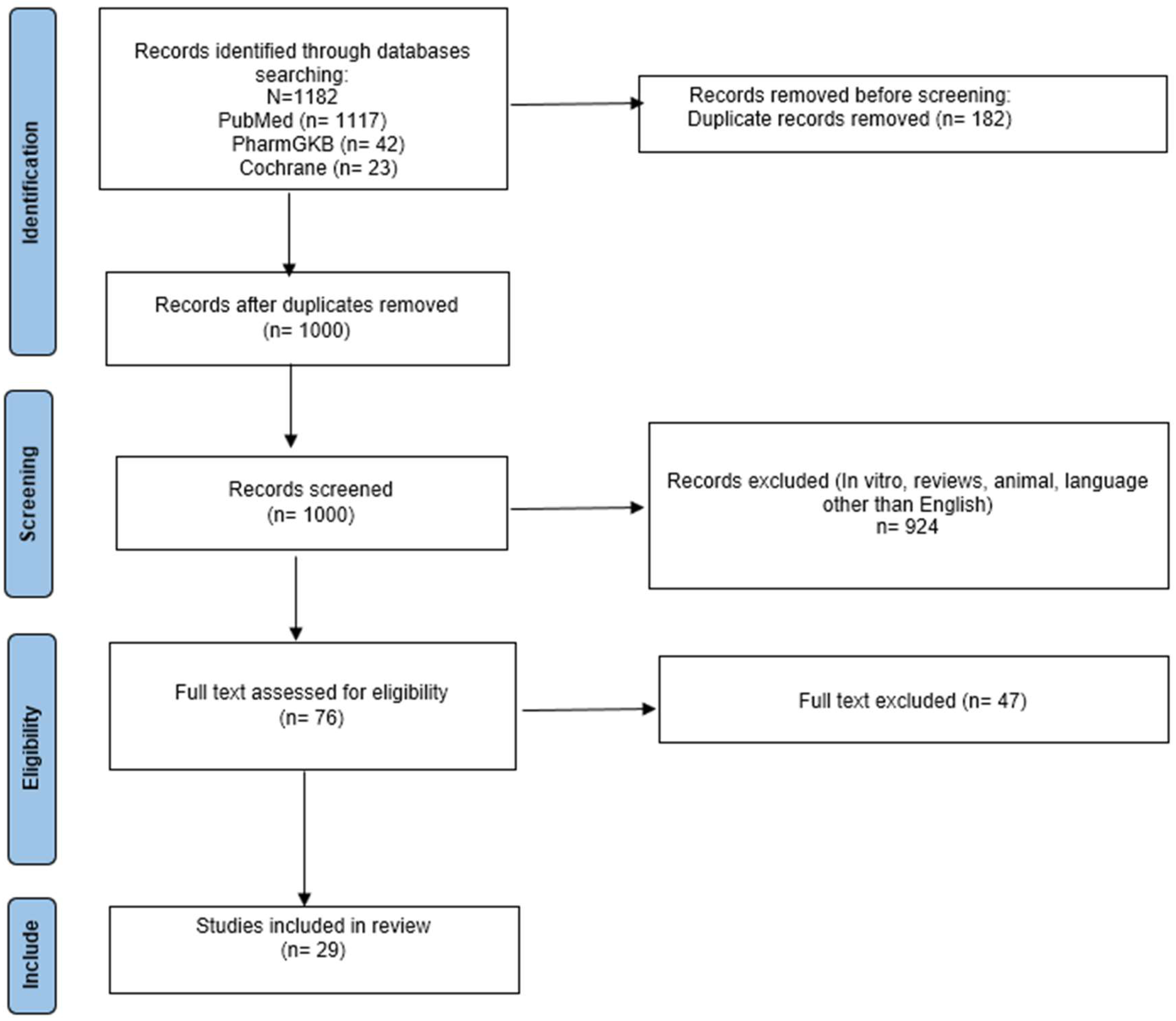
1. Introduction
2. Materials and Methods
2.1. Search Strategy
2.2. Study Selection and Inclusion
2.3. Level of Evidence
3. Results
3.1. ADRB1 rs1801253 (NG_012187.1: g.6251 G>C, NP_000675.1: p. Gly389Arg)
3.2. ADRB1 rs1801252 (NG_012187.1: g.5231 A>G; NP_000675.1: p.Ser49Gly)
3.3. ADRB2 rs1042714 (NG_016421.2: g.5318 G>C; NP_000015.2: p.Glu27Gln)
3.4. ADRB2 rs1042713 (NG_016421.2: g.5285 G>A; NP_000015.2: p.Gly16Arg)
4. Discussion
4.1. ADRB1 Gene
4.1.1. ADRB1 rs1801253 (NG_012187.1: g.6251 G>C, NP_000675.1: p.Gly389Arg)
4.1.2. ADRB1 rs1801252 (NG_012187.1: g.5231 A>G; NP_000675.1: p.Ser49Gly)
4.2. β-2 Adrenergic Receptor (ADRB2)
4.2.1. ADRB2 rs1042714 (NG_016421.2: g.5318 G>C; NP_000015.2: p.Glu27Gln)
4.2.2. ADRB2 rs1042713 (NG_016421.2: g.5285 G>A; NP_000015.2: p.Gly16Arg)
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Wang, C.; Xing, J.; Zhao, B.; Wang, Y.; Zhang, L.; Wang, Y.; Zheng, M.; Liu, G. The Effects of High-Intensity Interval Training on Exercise Capacity and Prognosis in Heart Failure and Coronary Artery Disease: A Systematic Review and Meta-Analysis. Cardiovasc. Ther. 2022, 2022, 4273809. [Google Scholar] [CrossRef]
- Reiter, M.J. Cardiovascular drug class specificity: Beta-blockers. Prog. Cardiovasc. Dis. 2004, 47, 11–33. [Google Scholar] [CrossRef]
- Shahin, M.H.; Conrado, D.J.; Gonzalez, D.; Gong, Y.; Lobmeyer, M.T.; Beitelshees, A.L.; Boerwinkle, E.; Gums, J.G.; Chapman, A.; Turner, S.T.; et al. Genome-Wide Association Approach Identified Novel Genetic Predictors of Heart Rate Response to β-Blockers. J. Am. Heart Assoc. 2018, 7, e006463. [Google Scholar] [CrossRef]
- Bonten, T.N.; Plaizier, C.E.I.; Snoep, J.-J.D.; Stijnen, T.; Dekkers, O.M.; van der Bom, J.G. Effect of β-blockers on platelet aggregation: A systematic review and meta-analysis. Br. J. Clin. Pharmacol. 2014, 78, 940–949. [Google Scholar] [CrossRef]
- Shin, J.; Johnson, J.A. Pharmacogenetics of β-Blockers. Pharmacotherapy 2007, 27, 874–887. [Google Scholar] [CrossRef] [PubMed]
- Alhayek, S.; Preuss, C.V. Beta 1 Receptors. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2024. Available online: http://www.ncbi.nlm.nih.gov/books/NBK532904/ (accessed on 4 September 2024).
- Snyder, E.M.; Johnson, B.D.; Joyner, M.J. Genetics of β2-Adrenergic Receptors and the Cardiopulmonary Response to Exercise. Exerc. Sport. Sci. Rev. 2008, 36, 98–105. [Google Scholar] [CrossRef] [PubMed]
- Farzam, K.; Jan, A. Beta Blockers. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2024. Available online: http://www.ncbi.nlm.nih.gov/books/NBK532906/ (accessed on 20 August 2024).
- Martinez, A.; Lakkimsetti, M.; Maharjan, S.; Aslam, M.A.; Basnyat, A.; Kafley, S.; Reddy, S.S.; Ahmed, S.S.; Razzaq, W.; Adusumilli, S.; et al. Beta-Blockers and Their Current Role in Maternal and Neonatal Health: A Narrative Review of the Literature. Cureus 2023, 15, e44043. [Google Scholar] [CrossRef] [PubMed]
- Weir, M.R. Beta-blockers in the treatment of hypertension: Are there clinically relevant differences? Postgrad. Med. 2009, 121, 90–98. [Google Scholar] [CrossRef]
- CPIC®. Guideline for Beta-Blockers and CYP2D6, ADRB1, ADRB2, ADRA2C, GRK4, and GRK5—CPIC. Available online: https://cpicpgx.org/guidelines/cpic-guideline-for-beta-blockers/ (accessed on 15 September 2024).
- Lynch, T.; Price, A. The Effect of Cytochrome P450 Metabolism on Drug Response, Interactions, and Adverse Effects. Available online: https://www.aafp.org/pubs/afp/issues/2007/0801/p391.html (accessed on 8 December 2024).
- Thomas, C.D.; Johnson, J.A. Pharmacogenetic factors affecting β-blocker metabolism and response. Expert. Opin. Drug Metab. Toxicol. 2020, 16, 953–964. [Google Scholar] [CrossRef]
- Castaño-Amores, C.; Díaz-Villamarín, X.; Pérez-Gutiérrez, A.M.; Antúnez-Rodríguez, A.; Pozo-Agundo, A.; Moreno-Escobar, E.; Sánchez-Ramos, J.G.; Martínez-González, L.J.; Dávila-Fajardo, C.L. Pharmacogenetic polymorphisms affecting bisoprolol response. Biomed. Pharmacother. 2021, 142, 112069. [Google Scholar] [CrossRef]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G. PRISMA Group Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. PLoS Med. 2009, 6, e1000097. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Xiao, T.; Chen, L.; Xie, S.; Deng, M.; Wu, D. The Association of ADRB1 and CYP2D6 Polymorphisms With Antihypertensive Effects and Analysis of Their Contribution to Hypertension Risk. Am. J. Med. Sci. 2018, 355, 235–239. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Li, G.; Deng, M.; Song, W.; Huang, X.; Guo, X.; Wu, Z.; Wu, S.; Xu, J. Associations between ADRB1 and CYP2D6 gene polymorphisms and the response to β-blocker therapy in hypertension. J. Int. Med. Res. 2015, 43, 424–434. [Google Scholar] [CrossRef] [PubMed]
- Si, D.; Wang, J.; Xu, Y.; Chen, X.; Zhang, M.; Zhou, H. Association of common polymorphisms in β1-adrenergic receptor with antihypertensive response to carvedilol. J. Cardiovasc. Pharmacol. 2014, 64, 306–309. [Google Scholar] [CrossRef]
- Liu, J.; Liu, Z.-Q.; Yu, B.-N.; Xu, F.-H.; Mo, W.; Zhou, G.; Liu, Y.-Z.; Li, Q.; Zhou, H.-H. beta1-Adrenergic receptor polymorphisms influence the response to metoprolol monotherapy in patients with essential hypertension. Clin. Pharmacol. Ther. 2006, 80, 23–32. [Google Scholar] [CrossRef]
- Johnson, J.A.; Zineh, I.; Puckett, B.J.; McGorray, S.P.; Yarandi, H.N.; Pauly, D.F. Beta 1-adrenergic receptor polymorphisms and antihypertensive response to metoprolol. Clin. Pharmacol. Ther. 2003, 74, 44–52. [Google Scholar] [CrossRef]
- Sofowora, G.G.; Dishy, V.; Muszkat, M.; Xie, H.G.; Kim, R.B.; Harris, P.A.; Prasad, H.C.; Byrne, D.W.; Nair, U.B.; Wood, A.J.J.; et al. A common beta1-adrenergic receptor polymorphism (Arg389Gly) affects blood pressure response to beta-blockade. Clin. Pharmacol. Ther. 2003, 73, 366–371. [Google Scholar] [CrossRef]
- Suonsyrjä, T.; Donner, K.; Hannila-Handelberg, T.; Fodstad, H.; Kontula, K.; Hiltunen, T.P. Common genetic variation of beta1- and beta2-adrenergic receptor and response to four classes of antihypertensive treatment. Pharmacogenet. Genom. 2010, 20, 342–345. [Google Scholar] [CrossRef]
- Liu, J.; Liu, Z.-Q.; Tan, Z.-R.; Chen, X.-P.; Wang, L.-S.; Zhou, G.; Zhou, H.-H. Gly389Arg polymorphism of beta1-adrenergic receptor is associated with the cardiovascular response to metoprolol. Clin. Pharmacol. Ther. 2003, 74, 372–379. [Google Scholar] [CrossRef]
- Cotarlan, V.; Brofferio, A.; Gerhard, G.S.; Chu, X.; Shirani, J. Impact of β(1)- and β(2)-adrenergic receptor gene single nucleotide polymorphisms on heart rate response to metoprolol prior to coronary computed tomographic angiography. Am. J. Cardiol. 2013, 111, 661–666. [Google Scholar] [CrossRef]
- Rau, T.; Düngen, H.-D.; Edelmann, F.; Waagstein, F.; Lainščak, M.; Dimković, S.; Apostolović, S.; Nešković, A.N.; Haverkamp, W.; Gelbrich, G.; et al. Impact of the β1-adrenoceptor Arg389Gly polymorphism on heart-rate responses to bisoprolol and carvedilol in heart-failure patients. Clin. Pharmacol. Ther. 2012, 92, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Kurnik, D.; Cunningham, A.J.; Sofowora, G.G.; Kohli, U.; Li, C.; Friedman, E.A.; Muszkat, M.; Menon, U.B.; Wood, A.J.; Stein, C.M. GRK5 Gln41Leu polymorphism is not associated with sensitivity to β1-adrenergic blockade in humans. Pharmacogenomics 2009, 10, 1581. [Google Scholar] [CrossRef]
- Metra, M.; Covolo, L.; Pezzali, N.; Zacà, V.; Bugatti, S.; Lombardi, C.; Bettari, L.; Romeo, A.; Gelatti, U.; Giubbini, R.; et al. Role of beta-adrenergic receptor gene polymorphisms in the long-term effects of beta-blockade with carvedilol in patients with chronic heart failure. Cardiovasc. Drugs Ther. 2010, 24, 49–60. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Bi, Y.; Xu, Y. Effects of metoprolol on beta1 adrenergic receptor polymorphism and receptor density in urban Chinese patients with heart failure. Chin. Med. J. (Engl.) 2007, 120, 1720–1723. [Google Scholar] [PubMed]
- Baudhuin, L.M.; Miller, W.L.; Train, L.; Bryant, S.; Hartman, K.A.; Phelps, M.; Larock, M.; Jaffe, A.S. Relation of ADRB1, CYP2D6, and UGT1A1 polymorphisms with dose of, and response to, carvedilol or metoprolol therapy in patients with chronic heart failure. Am. J. Cardiol. 2010, 106, 402–408. [Google Scholar] [CrossRef]
- Lee, H.-Y.; Chung, W.-J.; Jeon, H.-K.; Seo, H.-S.; Choi, D.-J.; Jeon, E.-S.; Kim, J.-J.; Shin, J.H.; Kang, S.-M.; Lim, S.C.; et al. Impact of the β-1 adrenergic receptor polymorphism on tolerability and efficacy of bisoprolol therapy in Korean heart failure patients: Association between β adrenergic receptor polymorphism and bisoprolol therapy in heart failure (ABBA) study. Korean J. Intern. Med. 2016, 31, 277–287. [Google Scholar] [CrossRef]
- Fiuzat, M.; Neely, M.L.; Starr, A.Z.; Kraus, W.E.; Felker, G.M.; Donahue, M.; Adams, K.; Piña, I.L.; Whellan, D.; O’Connor, C.M. Association between adrenergic receptor genotypes and beta-blocker dose in heart failure patients: Analysis from the HF-ACTION DNA substudy. Eur. J. Heart Fail. 2013, 15, 258–266. [Google Scholar] [CrossRef]
- Parikh, K.S.; Fiuzat, M.; Davis, G.; Neely, M.; Blain-Nelson, P.; Whellan, D.J.; Abraham, W.T.; Adams, K.F.; Felker, G.M.; Liggett, S.B.; et al. Dose Response of β-Blockers in Adrenergic Receptor Polymorphism Genotypes. Circ. Genom. Precis. Med. 2018, 11, e002210. [Google Scholar] [CrossRef]
- Biolo, A.; Clausell, N.; Santos, K.G.; Salvaro, R.; Ashton-Prolla, P.; Borges, A.; Rohde, L.E. Impact of beta1-adrenergic receptor polymorphisms on susceptibility to heart failure, arrhythmogenesis, prognosis, and response to beta-blocker therapy. Am. J. Cardiol. 2008, 102, 726–732. [Google Scholar] [CrossRef]
- Guerra, L.A.; Lteif, C.; Arwood, M.J.; McDonough, C.W.; Dumeny, L.; Desai, A.A.; Cavallari, L.H.; Duarte, J.D. Genetic polymorphisms in ADRB2 and ADRB1 are associated with differential survival in heart failure patients taking β-blockers. Pharmacogenom. J. 2022, 22, 62–68. [Google Scholar] [CrossRef]
- Terra, S.G.; Hamilton, K.K.; Pauly, D.F.; Lee, C.R.; Patterson, J.H.; Adams, K.F.; Schofield, R.S.; Belgado, B.S.; Hill, J.A.; Aranda, J.M.; et al. Beta1-adrenergic receptor polymorphisms and left ventricular remodeling changes in response to beta-blocker therapy. Pharmacogenet. Genom. 2005, 15, 227–234. [Google Scholar] [CrossRef]
- Magvanjav, O.; McDonough, C.W.; Gong, Y.; McClure, L.A.; Talbert, R.L.; Horenstein, R.B.; Shuldiner, A.R.; Benavente, O.R.; Mitchell, B.D.; Johnson, J.A.; et al. Pharmacogenetic Associations of β1-Adrenergic Receptor Polymorphisms With Cardiovascular Outcomes in the SPS3 Trial (Secondary Prevention of Small Subcortical Strokes). Stroke 2017, 48, 1337–1343. [Google Scholar] [CrossRef] [PubMed]
- Iaccarino, G.; Trimarco, V.; Lanni, F.; Cipolletta, E.; Izzo, R.; Arcucci, O.; De Luca, N.; Di Renzo, G. beta-Blockade and increased dyslipidemia in patients bearing Glu27 variant of beta2 adrenergic receptor gene. Pharmacogenom. J. 2005, 5, 292–297. [Google Scholar] [CrossRef]
- Isaza, C.A.; Henao, J.; Sánchez, J.C.; Porras, G.L.; Cardona, J.; Bedoya, G. Beta-2-adrenergic receptor polymorphisms and changes in lipids induced by metoprolol. Pharmacology 2007, 80, 279–285. [Google Scholar] [CrossRef]
- Kaye, D.M.; Smirk, B.; Williams, C.; Jennings, G.; Esler, M.; Holst, D. Beta-adrenoceptor genotype influences the response to carvedilol in patients with congestive heart failure. Pharmacogenetics 2003, 13, 379–382. [Google Scholar] [CrossRef]
- Fayed, M.S.; Saleh, M.A.; Sabri, N.A.; Elkholy, A.A. β1-Adrenergic Receptor Polymorphisms: A Possible Genetic Predictor of Bisoprolol Response in Acute Coronary Syndrome. Future Sci. OA 2023, 9, FSO895. [Google Scholar] [CrossRef]
- Aleong, R.G.; Sauer, W.H.; Davis, G.; Murphy, G.A.; Port, J.D.; Anand, I.S.; Fiuzat, M.; O’Connor, C.M.; Abraham, W.T.; Liggett, S.B.; et al. Prevention of atrial fibrillation by bucindolol is dependent on the beta1389 Arg/Gly adrenergic receptor polymorphism. JACC Heart Fail. 2013, 1, 338–344. [Google Scholar] [CrossRef] [PubMed]
- Zaugg, M.; Bestmann, L.; Wacker, J.; Lucchinetti, E.; Boltres, A.; Schulz, C.; Hersberger, M.; Kälin, G.; Furrer, L.; Hofer, C.; et al. Adrenergic receptor genotype but not perioperative bisoprolol therapy may determine cardiovascular outcome in at-risk patients undergoing surgery with spinal block: The Swiss Beta Blocker in Spinal Anesthesia (BBSA) study: A double-blinded, placebo-controlled, multicenter trial with 1-year follow-up. Anesthesiology 2007, 107, 33–44. [Google Scholar] [CrossRef]
- Isaza, C.; Henao, J.; Ramirez, E.; Cuesta, F.; Cacabelos, R. Polymorphic variants of the beta2-adrenergic receptor (ADRB2) gene and ADRB2-related propanolol-induced dyslipidemia in the Colombian population. Methods Find. Exp. Clin. Pharmacol. 2005, 27, 237–244. [Google Scholar] [CrossRef]
- Shahin, M.H.; El Rouby, N.; Conrado, D.J.; Gonzalez, D.; Gong, Y.; Lobmeyer, M.T.; Beitelshees, A.L.; Boerwinkle, E.; Gums, J.G.; Chapman, A.; et al. β2-Adrenergic Receptor Gene Affects the Heart Rate Response of β-Blockers: Evidence From 3 Clinical Studies. J. Clin. Pharmacol. 2019, 59, 1462–1470. [Google Scholar] [CrossRef]
- Lanfear, D.E.; Jones, P.G.; Marsh, S.; Cresci, S.; McLeod, H.L.; Spertus, J.A. Beta2-adrenergic receptor genotype and survival among patients receiving beta-blocker therapy after an acute coronary syndrome. JAMA 2005, 294, 1526–1533. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Cardiovascular Diseases (CVDs). Available online: https://www.who.int/news-room/fact-sheets/detail/cardiovascular-diseases-(cvds) (accessed on 5 August 2024).
- Dézsi, C.A.; Szentes, V. The Real Role of β-Blockers in Daily Cardiovascular Therapy. Am. J. Cardiovasc. Drugs 2017, 17, 361–373. [Google Scholar] [CrossRef] [PubMed]
- ADRB1 Adrenoceptor Beta 1 [Homo Sapiens (Human)]—Gene—NCBI. Available online: https://www.ncbi.nlm.nih.gov/gene/153 (accessed on 14 September 2023).
- rs1801253 RefSNP Report—dbSNP—NCBI. Available online: https://www.ncbi.nlm.nih.gov/snp/rs1801253 (accessed on 20 August 2024).
- PharmGKB. Very Important Pharmacogene: ADRB1. Available online: https://www.pharmgkb.org/vip/PA166170369 (accessed on 19 August 2024).
- rs1801253 (SNP)—Population Genetics—Homo_Sapiens—Ensembl Genome Browser 112. Available online: https://www.ensembl.org/Homo_sapiens/Variation/Population?db=core;r=10:114044797-114045797;v=rs1801253;vdb=variation;vf=654397587 (accessed on 15 September 2024).
- rs1801252 RefSNP Report—dbSNP—NCBI. Available online: https://www.ncbi.nlm.nih.gov/snp/rs1801252 (accessed on 20 August 2024).
- rs1801252 (SNP)—Population Genetics—Homo_Sapiens—Ensembl Genome Browser 112. Available online: https://www.ensembl.org/Homo_sapiens/Variation/Population?db=core;r=10:114043777-114044777;v=rs1801252;vdb=variation;vf=654397586 (accessed on 15 September 2024).
- ADRB2 Adrenoceptor Beta 2 [Homo Sapiens (Human)]—Gene—NCBI. Available online: https://www.ncbi.nlm.nih.gov/gene/154 (accessed on 14 September 2023).
- PharmGKB. Very Important Pharmacogene: ADRB2. Available online: https://www.pharmgkb.org/vip/PA166165410 (accessed on 19 August 2024).
- rs1042714 RefSNP Report—dbSNP—NCBI. Available online: https://www.ncbi.nlm.nih.gov/snp/rs1042714 (accessed on 20 August 2024).
- Kushnir, V.M.; Cassell, B.; Gyawali, C.P.; Newberry, R.D.; Kibe, P.; Nix, B.D.; Sabzpoushan, A.; Kanuri, N.D.; Sayuk, G.S. Genetic variation in the beta-2 adrenergic receptor (ADRB2) predicts functional gastrointestinal diagnoses and poorer health-related quality of life. Aliment. Pharmacol. Ther. 2013, 38, 313–323. [Google Scholar] [CrossRef] [PubMed][Green Version]
- rs1042714 (SNP)—Population Genetics—Homo_Sapiens—Ensembl Genome Browser 112. Available online: https://www.ensembl.org/Homo_sapiens/Variation/Population?db=core;r=5:148826410-148827410;v=rs1042714;vdb=variation;vf=336458548 (accessed on 15 September 2024).
- rs1042713 RefSNP Report—dbSNP—NCBI. Available online: https://www.ncbi.nlm.nih.gov/snp/rs1042713 (accessed on 20 August 2024).
- rs1042713 (SNP)—Population Genetics—Homo_Sapiens—Ensembl Genome Browser 112. 2025. Available online: https://www.ensembl.org/Homo_sapiens/Variation/Population?db=core;r=5:148826377-148827377;v=rs1042713;vdb=variation;vf=336458547 (accessed on 15 September 2024).
| Author (Year) | Study Type | Population | Drug Used (Monotherapy or Combination) | Outcome | Association | Genotyping of CYP2D6 | Significance | ADRB Blockade |
|---|---|---|---|---|---|---|---|---|
| Efficacy | ||||||||
| Chen L et al. (2018) [16] | Retrospective study | 261 hypertensive patients | Metoprolol (monotherapy) | Antihypertensive effect | Patients with G/G genotype (Gly/Gly) showed higher metoprolol antihypertensive effects compared to patients with G/C genotype (Gly/Arg) (numeric values not provided). | Yes | p = 0.027 | G (Gly): higher C (Arg): lower |
| Wu D et al. (2015) [17] | Retrospective study | 93 hypertensive patients | Metoprolol (monotherapy) | Antihypertensive effect | Patients with the C/C genotype (Arg/Arg) showed greater SBP reduction after treatment compared to those with the G/C genotype (Gly/Arg) (127 mmHg vs. 132 mmHg). | Yes | p < 0.05 | G (Gly): lower C (Arg): higher |
| Si D et al. (2014) [18] | Prospective study | 87 hypertensive patients | Carvedilol (monotherapy) | Antihypertensive effect | Patients with the C/C genotype (Arg/Arg) showed a four-fold greater reduction in DBP with carvedilol compared to those with the G/G genotype (Gly/Gly) (10.61 mmHg vs. 2.62 mmHg). | No | p = 0.013 | G (Gly): lower C (Arg): higher |
| Liu J et al. (2006) [19] | Prospective clinical trial | 223 hypertensive patients | Metoprolol (monotherapy) | Antihypertensive effect | Patients with G/G (Gly/Gly) or G/C (Gly/Arg) genotype showed a lower reduction in SBP after metoprolol treatment compared to those with C/C genotype (Arg/Arg) (G/G: 1.10% ± 1.50%, G/C: 2.80% ± 4.70%, C/C: 10.40% ± 4.00%). | No | p < 0.001 (G/G vs. C/C) p = 0.001 (G/C vs. C/C) | G (Gly): lower C (Arg): higher |
| Patients with C/C (Arg/Arg) or G/C (Gly/Arg) genotype showed a greater reduction in DBP after metoprolol treatment compared to those with G/G genotype (Gly/Gly) (C/C: 6.10% ± 4.30%, G/C: 2.20% ± 4.20%, G/G: 0.90% ± 4.00%). | p < 0.001 | G (Gly): lower C (Arg): higher | ||||||
| Patients with G/G genotype (Gly/Gly) showed a lower reduction in mean arterial pressure after metoprolol treatment compared to those with G/C (Gly/Arg) or C/C (Arg/Arg) genotype (G/G: 1.00% ± 2.50%; G/C: 2.50% ± 3.00%, C/C: 8.10% ± 3.50%). | p < 0.001 | G (Gly): lower C (Arg): higher | ||||||
| Johnson JA et al. (2003) [20] | Prospective clinical trial | 40 hypertensive patients | Metoprolol (monotherapy) | Antihypertensive effect | Patients with C/C genotype (Arg/Arg) showed a two-fold greater reduction in 24 h DBP after metoprolol treatment compared to those with G/G (Gly/Gly) or G/C (Gly/Arg) genotype (−12.00% ± 8.60% vs. −5.10% 7.80%). * | No | p = 0.012 | G (Gly): lower C (Arg): higher |
| Patients with the C/C genotype (Arg/Arg) showed a 6.5 mmHg greater absolute reduction in 24 h DBP compared to those with G/G (Gly/Gly) or G/C (Gly/Arg) genotype (95% CI −1.70 to −11.30 mmHg). * | p = 0.009 | G (Gly): lower C (Arg): higher | ||||||
| Patients with C/C genotype (Arg/Arg) showed an 8.6 mmHg greater reduction in daytime DBP compared to those with G/G (Gly/Gly) or G/C (Gly/Arg) genotype (95% CI −3.50 to −13.60 mmHg). * | p = 0.0014 | G (Gly): lower C (Arg): higher | ||||||
| Sofowora GG et al. (2003) [21] | Prospective study | 400 healthy volunteers | Atenolol (monotherapy) | Antihypertensive effect | Patients with the G/G genotype (Gly/Gly) showed a lower reduction in resting SBP in response to atenolol compared to those with the C/C genotype (Arg/Arg) (G/G: 0.20 ± 1.70 mm Hg, C/C: 8.70 ± 1.30 mm Hg). | No | p = 0.001 | G (Gly): lower C (Arg): higher |
| Patients with the G/G (Gly/Gly) genotype showed a lower reduction in mean arterial pressure in response to atenolol compared to those with the C/C genotype (Arg/Arg) (G/G: 2.00 ± 1.70 mmHg, C/C: 7.20 ± 1.00 mmHg). | p = 0.009 | G (Gly): lower C (Arg): higher | ||||||
| Suonsyrjä T et al. (2010) [22] | Randomized Controlled Trial | 233 hypertensive patients | Bisoprolol (monotherapy) | Antihypertensive effect | Patients with the G/G genotype (Gly/Gly) showed a greater SBP reduction in response to bisoprolol compared to those with the C/C genotype (Arg/Arg) (−15.80 mmHg (−16.80, −9.10) vs. −10.30 mmHg (−14.30, −6.40)). | No | p = 0.003 | G (Gly): higher C (Arg): lower |
| Patients with the G/G genotype (Gly/Gly) showed a greater DBP reduction in response to bisoprolol compared to those with the C/C genotype (Arg/Arg) (−11.20 mmHg (−14.40, −9.00) vs. −7.90 mmHg (−10.60, −5.40)). | p = 0.003 | G (Gly): higher C (Arg): lower | ||||||
| Fayed MS et al. (2023) [40] | Prospective study | 77 new onset acute coronary syndrome patients | Bisoprolol (combination with ACEi, nitroglycerin, spironolactone, loop diuretics, clopidogrel, aspirin, and statins) | Antihypertensive effect | Patients with the C/C genotype (Arg/Arg) showed a greater DBP reduction compared to those with the G/G (Gly/Gly) or G/C (Gly/Arg) genotype (−9.5% ± 9.7% vs. −0.80% ± 11.5%). $ | No | p = 0.00015 | G (Gly): lower C (Arg): higher |
| Patients with the C/C genotype (Arg/Arg) showed an 8 mm Hg greater absolute reduction in 24 h DBP compared to those with the G/G (Gly/Gly) or G/C (Gly/Arg) genotype (95% CI: −8.8 to −7.3 mm Hg). $ | p = 0.00012 | G (Gly): lower C (Arg): higher | ||||||
| Patients with G/G (Gly/Gly) and G/C (Gly/Arg) showed a lower reduction in SBP compared to those with the C/C genotype (Arg/Arg) (−0.76% ± 8.7% vs. −8.5% ± 7.8%). @ | p = 0.000218 | G (Gly): lower C (Arg): higher | ||||||
| Patients with the G/G genotype (Gly/Gly) showed a −9.6 mmHg lower absolute reduction in 24 h SBP compared to those with the C/C (Arg/Arg) and G/C (Gly/Arg) genotypes (95% CI: −10.5 to −8.7 mmHg). @ | p = 0.00012 | G(Gly): lower C (Arg): higher | ||||||
| Liu J et al. (2003) [23] | Prospective study | 123 male volunteers | Metoprolol (monotherapy) | Antihypertensive effect | Volunteers with the G/G (Gly/Gly) or G/C (Gly/Arg) genotype showed a lower SBP reduction at the 3 dosage levels of metoprolol (75, 150, 225 mg) compared to those with the C/C genotype (Arg/Arg) (4.60% ± 0.50% vs. 5.90% ±0.70%, 6.00% ± 0.80% vs. 9.20% ±1.00%, 9.90%± 0.90% vs. 11.60% ± 1.20%, respectively) | No | p = 0.011 | G (Gly): lower C (Arg): higher |
| HR | Volunteers with the C/C genotype (Arg/Arg) showed a greater reduction in resting HR at the 3 dosage levels of metoprolol (75, 150, 225 mg) compared to those with the G/G (Gly/Gly) or G/C (Gly/Arg) genotype (6.30% ± 0.80% vs. 4.10% ± 0.70%, 10.10% ± 1.00% vs. 6.20% ± 1.10%, and 14.40% ±1.40% vs. 10.90% ±1.30%, respectively). | p = 0.008 | G (Gly): lower C (Arg): higher | |||||
| HR | Volunteers with the C/C genotype (Arg/Arg) showed a greater reduction in exercise HR at the 3 dosage levels of metoprolol (75, 150, 225 mg) compared to those with the G/G (Gly/Gly) or G/C (Gly/Arg) genotype (8.90% 0.50% vs. 6.60% 0.70%, 14.00% 0.90% vs. 11.70% 1.00%, and 20.10% 1.50% vs. 16.40% 1.30%, respectively). | p = 0.017 | G (Gly): lower C (Arg): higher | |||||
| Cotarlan V et al. (2013) [24] | Prospective study | 201 patients scheduled for non-invasive coronary computed tomographic angiography | Metoprolol (monotherapy) | HR | Patients with the G/G genotype (Gly/Gly) showed a greater percentage of non-response rate (HR > 60 beats/min) compared to those with the C/C (Arg/Arg) or G/C (Gly/Arg) genotype (45% vs. 17%). | No | p = 0.020 | G (Gly): lower C (Arg): higher |
| Rau T et al. (2012) [25] | Prospective study | 876 patients with HF | Bisoprolol and carvedilol (monotherapy) | HR | Patients with the C/C genotype (Arg/Arg) showed greater HR during titration in carvedilol treatment compared to those with the G/G (Gly/Gly) and G/C (Gly/Arg) genotypes (90 beats/min vs. 77 beats/min). | No | p < 0.0001 | G (Gly): higher C (Arg): lower |
| Kurnik D et al. (2009) [26] | Prospective study | 154 healthy volunteers | Atenolol (monotherapy) | HR | Volunteers with the C/C (Arg/Arg) or G/C (Arg/Gly) genotype showed greater HR reduction after atenolol treatment compared to those with the G/G (Gly/Gly) genotype (numeric values not provided). | No | 95% CI:11.7 (3.80–19.5) p = 0.040 | G (Gly): lower C (Arg): higher |
| Metra M et al. (2010) [27] | Prospective study | 183 patients with HF | Carvedilol (combination with ACEi, diuretics, or aldosterone antagonists) | LVEF | Patients with the C/C (Arg/Arg) or G/C (Gly/Arg) genotype showed a higher rise in LVEF after treatment compared to those with the G/G genotype (Gly/Gly) (C/C: 7.80 ± 7.60%, G/C: 9.00 ± 11.40%, C/C: 4.10 ± 7.60% units). | No | p = 0.0847 (G/G vs. C/C) p = 0.1058 (G/C vs. C/C) | G (Gly): lower C (Arg): higher |
| Luo M et al. (2007) [28] | Prospective study | 156 patients with HF | Metoprolol (combination with digitalis, ACEi, and diuretics) | LVEF | Patients with the C/C genotype (Arg/Arg) showed a higher difference in LVEF before and after metoprolol treatment compared to those with the G/C genotype (Gly/Arg) (4.60% ± 2.98% vs. 1.90% ± 2.04%). | No | p = 0.027 | G (Gly): lower C (Arg): higher |
| Baudhuin LM et al. (2010) [29] | Retrospective study | 93 patients with HF | Metoprolol and carvedilol (combination with ACEIs, ARBs, and diuretics) | Dose–response | Patients with the G/G genotype (Gly/Gly) needed an approximately 25 mg higher carvedilol daily dose compared to those with the G/C (Gly/Arg) genotype. | Yes | p = 0.020 | G (Gly): lower C (Arg): higher |
| Lee HY et al. (2016) [30] | Prospective study | 100 patients with HF | Bisoprolol (combination with ACEi, ARB, Spironolactone, loop diuretics, and digoxin aspirin) | Dose–response | Patients with the C/C (Arg/Arg) genotype required higher doses of bisoprolol compared to those with the G/G (Gly/Gly) or G/C (Gly/Arg) genotype (5.26 ± 2.62 mg vs. 3.96 ± 2.05 mg). | No | p = 0.022 | G (Gly): higher C (Arg): lower |
| Parikh KS et al. (2018) [32] | Retrospective analysis of RCTs (BEST, HF-ACTION) | 1040 patients under β-blocker treatment | Bucindolol (monotherapy) | CVM/HF hospitalization | Patients with the C/C genotype (Arg/Arg) showed a higher CVM/HF hospitalization reduction at higher β-blocker doses compared to those with the G/G (Gly/Gly) or G/C (Gly/Arg) genotype (C/C: 24%, G/G + G/C:32%). There were no significant differences for no/low-dose β-blockers (C/C: 35%, G/G + G/C:34%). | No | p = 0.026 | G (Gly): lower C (Arg): higher |
| Biolo A et al. (2008) [33] | Prospective study | 201 patients with HF | Metoprolol and carvedilol (combination with ACEi, diuretics, and spironolactone) | MCE | Patients with the C/C genotype (Arg/Arg) showed a higher prevalence of non-sustained ventricular tachycardia compared to those with the G/G genotype (Gly/Gly) (48% vs. 17%). | No | p = 0.015 | G (Gly): higher C (Arg): lower |
| Aleong RG et al. (2013) [41] | Randomized controlled clinical trial | 1040 patients with HF | Bucindilol (combination with digoxine) | MCE | Patients with the C/C genotype (Arg/Arg) showed a lower incidence of new-onset arterial fibrillation in bucindolol treatment compared to those with the G/G (Gly/Gly) or G/C (Gly/Arg) genotype. | No | Hazard Ratio = 0.26 [95% CI: 0.12–0.57], p = 0.0003 | G (Gly): lower C (Arg): higher |
| Safety | ||||||||
| Fiuzat M et al. (2013) [31] | Randomized, multicenter trial | 902 patients under β-blocker treatment | Carvedilol and metoprolol(combination with loop diuretics) | ACM | Patients with the C/C genotype (Arg/Arg) receiving low-dose β-blockers showed a two-fold higher risk of death compared to those receiving high doses. There were no significant differences in risk between patients receiving low vs. high dose β-blockers among patients with the G/G genotype (Gly/Gly). | No | Hazard Ratio = 2.09; p = 0.015 Hazard Ratio = 0.91; p = 0.73 | NA |
| Parikh KS et al. (2018) [32] | Retrospective analysis of RCTs (BEST, HF-ACTION) | 1040 patients under β-blocker treatment | Bucindolol (monotherapy) | ACM | Patients with the C/C genotype (Arg/Arg) showed a 46% ACM reduction at higher bucindolol doses compared to those with the G/G (Gly/Gly) or G/C (Gly/Arg) genotype (C/C: 10%, G/G + G/C: 19%). There were no significant differences for no/low-dose bucindolol (C/C:21%, G/G + G/C:20%). | No | Hazard Ratio= 0.54; p = 0.018 | NA |
| 957 patients with HF | Various beta-blockers: carvedilol, metoprolol, bisoprolol, and atenolol (combination with ACEIs, ARBs, and Aldosterone Receptor) | ACM | Patients with the C/C genotype (Arg/Arg) showed a greater ACM at lower β-blocker doses compared to those with the G/G (Gly/Gly) or G/C (Gly/Arg) genotype (C/C:21%, G/G + G/C:14%). There were no significant differences for high-dose β-blockers (C/C:10%, G/G + G/C:13%). | Hazard Ratio = 0.83; p = 0.015 | NA | |||
| Zaugg M et al. (2007) [42] | Double-blinded, placebo-controlled, multicenter trial | 224 patients undergoing surgery with a spinal block | Bisoprolol (combination with Ca2 antagonists, diuretics, ACEi, ARBs, nitrates, and statins) | Adverse events | Patients with the G/G genotype (Gly/Gly) showed a higher number of adverse events compared to those with the C/C genotype (Arg/Arg) (24 of 74 [32.40%] vs. 21 of 112 [18.70%]). | No | Hazard Ratio = 1.87 [95% CI: 1.04–3.35] p = 0.040 | G (Gly): higher C (Arg): lower |
| Guerra LA et al. (2022) [34] | Retrospective study | 308 patients with HF | Metoprolol and carvedilol (combination with ACEIs, ARBs, and diuretics) | Survival | Patients with the C/C genotype (Arg/Arg) showed a higher survival rate at higher β-blocker doses compared to those with the G/G (Gly/Gly) genotype. | No | p = 0.023 | NA |
| Outcome | Total Number of Articles | Outcome Variation Associated with Higher Receptor Blockade | Higher Blockade with C Allele (Arg) | Higher Blockade with G Allele (Gly) | Level of Evidence |
|---|---|---|---|---|---|
| Arterial pressure | 3 | Arterial pressure reduction | [19,21] | [16] | Moderate (2:1) |
| SBP | 6 | SBP reduction | [17,19,21,23,40] | [22] | High (5:1) |
| DBP | 5 | DBP reduction | [18,19,20,40] | [22] | High (4:1) |
| HR | 5 * | HR reduction | [23,24,26] | [25] | High (4:1) |
| LVEF | 2 | LVEF increase | [27,28] | Moderate (2:0) | |
| Dose requirements | 2 | Dose reduction | [29] | [30] | Low (1:1) |
| CVM and heart failure hospitalization | 1 | Lower mortality and hospitalization | [32] | Low (1:0) | |
| MCE | 2 | Lower incidence | [41] | [33] | Low (1:1) |
| Adverse events | 1 | Higher incidence | [42] | Low (0:1) | |
| Overall | 27 $ | 20 | 7 | Moderate (20:7) |
| Author (Year) | Study Type | Population | Drug Used (Monotherapy or Combination) | Outcome | Association | Genotyping of CYP2D6 | Significance | Receptor Blockade |
|---|---|---|---|---|---|---|---|---|
| Liu J et al. (2006) [19] | Prospective clinical trial | 223 hypertensive patients | Metoprolol (monotherapy) | Antihypertensive effect | Patients with the A/A genotype (Ser/Ser) showed a greater SBP reduction in response to metoprolol compared to those with the A/G genotype (Ser/Gly) (8.40% ± 3.20% vs. 5.30% ± 5.20%). | No | p = 0.047 | A (Ser): higher G (Gly): lower |
| Suonsyrjä T et al. (2010) [22] | RCT | 233 hypertensive men | Bisoprolol (monotherapy) | Antihypertensive effect | Patients with the A/A genotype (Ser/Ser) showed a greater SBP reduction in response to bisoprolol compared to those with the A/G genotype (Ser/Gly) (−11.50 (−15.50, −7.00) mmHg vs. −9.90 (−13.40, −6.20) mmHg). | No | p = 0.04, p = 0.02 (Mann–Whitney U test and multivariate analysis) | A (Ser): higher G (Gly): lower |
| Cotarlan V et al. (2013) [24] | Prospective study | 201 patients scheduled for non-invasive coronary computed tomographic angiography | Metoprolol (monotherapy) | HR | Patients with the G/G (Gly/Gly) or G/A (Ser/Gly) genotype showed a higher percentage of non-response rate (HR > 60 beats/min) compared to those with the A/A genotype (Ser/Ser) (29% vs. 15%). | No | p = 0.037 | A (Ser): higher G (Gly): lower |
| Terraa SG et al. (2005) [35] | Prospective study | 61 β-blocker naïve patients with systolic HF | Metoprolol (combination with ACEi, ARBs, furosemide, digoxin, spironolactone, or antiplatelet therapy) | LVEDD | Patients with G/G (Gly/Gly) and G/A (Ser/Gly) showed an LVEDD decrease after 6 months of metoprolol treatment (from 65 ± 13 mm to 63 ± 12 mm) compared to those with the A/A genotype (Ser/Ser), who showed an LVEDD increase (from 61 ± 900 mm to 63 ± 90 mm). | No | p = 0.030 | A (Ser): lower G (Gly): higher |
| Magvanjav O et al. (2017) [36] | Retrospective study | 926 hypertensive patients | Not specified | MCE | Patients with the G/G (Gly/Gly) or G/A (Ser/Gly) genotype showed a higher cumulative incidence of MCE compared to those with A/A (Ser/Ser) genotype (15.70% vs. 7.60%). | No | p = 0.018 | A (Ser): higher G (Gly): lower |
| Outcome | Total Number of Articles | Outcome Variation Associated with Higher Receptor Blockade | Enhanced Receptor Blockade with A Allele (Ser) | Enhanced Receptor Blockade with G Allele (Gly) | Level of Evidence |
|---|---|---|---|---|---|
| SBP | 2 | SBP reduction | [19,22] | Moderate (2:0) | |
| HR | 1 | HR reduction | [24] | Low (1:0) | |
| LVEDD | 1 | LVEDD reduction | [35] | Low (0:1) | |
| MCE | 1 | Lower incidence | [36] | Low (1:0) | |
| Overall | 5 | 4 | 1 | High (4:1) |
| Author (Year) | Study Type | Population | Drug | Outcome | Association | Genotyping of CYP2D6 | Significance | Higher/Lower Blockade |
|---|---|---|---|---|---|---|---|---|
| Efficacy | ||||||||
| Laccarino G et al. (2005) [37] | Prospective study | 1050 hypertensive patients | Atenolol and metoprolol (combination with Statin, fibrates, diuretics, ACEi, and ARBs) | Dyslipidemia | Patients with the G/G genotype (Glu/Glu) showed a higher incidence of dyslipidemia compared to those with the C/C genotype (Gln/Gln) (48.40% vs. 37.30%). | No | p < 0.050 | NA |
| TG levels | Patients with the G/G genotype (Glu/Glu) showed higher serum TG levels compared to those with patients with the G/C Glu/Gln or C/C (Gln/Gln) genotypes (Gln/Gln: 12.90%; Gln/Glu: 18.60%; Glu/Glu: 25.00%). | p < 0.020 | NA | |||||
| Isaza C et al. (2005) [43] | Prospective study | 141 healthy volunteers | Propranolol (monotherapy) | HDL-C levels | Volunteers with the C/C genotype (Gln/Gln) showed lower propranolol-induced HDL-C levels (baseline: 37.80 ± 4.40 mg/dL vs. post-propranolol: 31.40 ± 6.20 mg/dL) compared to those with the G/G genotype (Glu/Glu) (baseline: 42.30 ± 18.90 mg/dL vs. post-propranolol: 40.00 ± 19.30 mg/dL). | No | p = 0.005 | NA |
| TG levels | Volunteers with the G/G genotype (Glu/Glu) showed higher propranolol-induced TG levels (baseline: 119.80 ± 85.90 mg/dL vs. post-propranolol: 242.30 ± 179.80 mg/dL) compared to those with the C/C genotype (Gln/Gln) (baseline: 173.00 ± 105.60 mg/dL vs. post-propranolol: 169.10 ± 97.30 mg/dL). | p = 0.012 | NA | |||||
| Isaza CA et al. (2007) [38] | Prospective study | 105 hypertensive patients | Metoprolol (monotherapy) | TC levels | Patients with the C/C genotype (Gln/Gln) showed lower TC levels during metoprolol treatment (pretreatment: 217 ± 45 mg/dL, during treatment: 208 ± 41 mg/dL), with no changes in patients with the G/C genotype (Glu/Gln) (pretreatment: 199 ± 32 mg/dL, during treatment: 206 ± 42 mg/dL). | No | p = 0.030 | NA |
| TG levels | Patients with the G/C genotype (Glu/Gln) showed lower TG levels with metoprolol therapy (pretreatment: 199 ± 55 mg/dL during treatment: 260.00 ± 7.10 mg/dL), with no changes in patients with the C/C genotype (Gln/Gln) (pretreatment: 215 ± 132 mg/dL, during treatment: 212 ± 148 mg/dL). | p = 0.025 | NA | |||||
| Shahin MH et al. (2019) [44] | RCT | 768 hypertensive patients | Atenolol and metoprolol (combination with hydrochlorothiazide and amlodipine) | HR | Patients with the G/G genotype (Glu/Glu) showed a greater HR reduction in response to atenolol and metoprolol compared to those with the G/C (Glu/Gln) or C/C (Gln/Gln) genotype (numeric values not provided). | Yes | β = −0.83 p = 0.010 (atenolol); β = −1.59 p = 0.0007 (metoprolol) | C (Gln): lower G (Glu): higher |
| 368 hypertensive patients | ||||||||
| Kaye DM et al. (2003) [39] | Prospective study | 80 patients with HF | Carvedilol (combination with ACEi, diuretics, and digoxin) | LVEF | Patients with the G/G (Glu/Glu) or G/C (Glu/Gln) genotype showed higher improvement in LVEF compared to those with the C/C (Gln/Gln) genotype (36% vs. 26%). | No | p = 0.003 | C (Gln): lower G (Glu): higher |
| Metra et al. (2010) [27] | Prospective study | 183 patients with HF | Carvedilol (ACEi, diuretics, or aldosterone antagonists) | LVEF | Patients with the G/G genotype (Glu/Glu) showed a larger LVEF increase compared to patients with the C/C (Gln/Gln) or G/C (Glu/Gln) genotype, considered together or separately (G/G: +13.00 ± 12.20%, C/C: +7.10 ± 8.10%, G/C: +8.30 ± 11.40%, G/C + C/C: +7.60 ± 9.60% units). | No | p = 0.022 | C (Gln): lower G (Glu): higher |
| Decline in pulmonary wedge pressure | Patients with the G/G genotype (Glu/Glu) showed a larger decline in pulmonary wedge pressure compared to patients with the G/C (Glu/Gln) or C/C (Gln/Gln) genotype at rest (G/G: −10 ± 10 mm Hg, G/C: −6 ± 10 mm Hg, C/C: −5 ± 7 mm Hg) and at peak exercise (G/G: −12 ± 9 mm Hg, G/C:−7 ± 10, C/C: −5 ± 7 mm Hg). | p = 0.027 (rest); p = 0.015 (exercise) | G (Gln): lower G (Glu): higher | |||||
| Safety | ||||||||
| Guerra LA et al. (2022) [34] | Retrospective study | 308 patients with HF | Metoprolol and Carvedilol (combination with ACEi, ARBs, or diuretics) | Survival | Patients with the G/G genotype (Glu/Glu) showed higher survival rates at higher β-blocker doses compared to those with the C/C genotype (Gln/Gln) (0.85 vs. 0.65). | No | p = 0.030 | NA |
| Lanfear DE et al. (2005) [45] | Prospective study | 735 patients with acute coronary syndrome | Metoprolol (combination with aspirin, ACEi or ARBs, statins, nitrates, and diuretics) | Survival | Patients with the G/C (Glu/Gln) or G/G (Glu/Glu) genotypes showed higher survival rates compared to those with C/C genotype (Gln/Gln) (3-year Kaplan–Meier death rates: 6%, 11%, and 16%, respectively). | No | p = 0.030 | NA |
| Outcome | Total Number of Articles | Outcome Variation Associated with Higher Receptor Blockade | Enhanced Receptor Blockade with G Allele (Glu) as Higher Blockade | Enhanced Receptor Blockade with C Allele (Gln) | Level of Evidence |
|---|---|---|---|---|---|
| HR | 1 | HR reduction | [44] | Low (1:0) | |
| LVEF | 2 | LVEF increase | [27,39] | Moderate (2:0) | |
| Pulmonary wedge pressure | 2 * | Pulmonary wedge pressure reduction | [27] | Moderate (2:0) | |
| Overall | 5 | 5 | 0 | Moderate $ (5:0) |
| Author (Year) | Type of Study | Population | Drug | Outcome | Association | Genotyping of CYP2D6 | Significance | Receptor Blockade |
|---|---|---|---|---|---|---|---|---|
| Efficacy | ||||||||
| Shahin MH et al. (2019) [44] | RCT | 757 hypertensive patients | Atenolol and metoprolol (combination with hydrochlorothiazide and amlodipine) | HR | Patients with the A/A genotype (Arg/Arg) showed higher HR lowering response to atenolol compared to those with the G/A (Gly/Arg) or G/G (Gly/Gly) genotype (numeric values not provided). | Yes | β = −0.70 p = 0.040 | G (Gly): lower A (Arg): higher |
| 368 hypertensive patients | Patients with the A/A genotype (Arg/Arg) showed higher HR lowering response to atenolol compared to those with the G/A (Gly/Arg) or G/G (Gly/Gly) genotype (numeric values not provided). | β = −1.15 p = 0.030 | G (Gly): lower A (Arg): higher | |||||
| Safety | ||||||||
| Lanfear DE et al. (2005) [45] | Prospective study | 735 patients with acute coronary syndrome | Metoprolol (combination with aspirin, ACEi, ARBs, statins, nitrates, and diuretics) | ACM | Patients with the A/A genotype (Arg/Arg) showed higher death rates compared to those with the G/A (Gly/Arg) and G/G (Gly/Gly) genotypes (20% vs. 10%). | No | p = 0.005 | NA |
| Outcome | Total Number of Articles | Outcome Variation Associated with Higher Receptor Blockade | Enhanced Receptor Blockade with A Allele (Arg) | Enhanced Receptor Blockade with G Allele (Gly) | Level of Evidence |
|---|---|---|---|---|---|
| HR | 2 * | HR reduction | [44] | Moderate (2:0) | |
| Overall | 2 | 2 | 0 | Low $ (2:0) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abbes, H.; Soria-Chacartegui, P.; Omezzine, A.; Abad-Santos, F.; Zubiaur, P. Impact of Genetic Variation in Adrenergic Receptors on β-Blocker Effectiveness and Safety in Cardiovascular Disease Management: A Systematic Review. Pharmaceuticals 2025, 18, 493. https://doi.org/10.3390/ph18040493
Abbes H, Soria-Chacartegui P, Omezzine A, Abad-Santos F, Zubiaur P. Impact of Genetic Variation in Adrenergic Receptors on β-Blocker Effectiveness and Safety in Cardiovascular Disease Management: A Systematic Review. Pharmaceuticals. 2025; 18(4):493. https://doi.org/10.3390/ph18040493
Chicago/Turabian StyleAbbes, Houwaida, Paula Soria-Chacartegui, Asma Omezzine, Francisco Abad-Santos, and Pablo Zubiaur. 2025. "Impact of Genetic Variation in Adrenergic Receptors on β-Blocker Effectiveness and Safety in Cardiovascular Disease Management: A Systematic Review" Pharmaceuticals 18, no. 4: 493. https://doi.org/10.3390/ph18040493
APA StyleAbbes, H., Soria-Chacartegui, P., Omezzine, A., Abad-Santos, F., & Zubiaur, P. (2025). Impact of Genetic Variation in Adrenergic Receptors on β-Blocker Effectiveness and Safety in Cardiovascular Disease Management: A Systematic Review. Pharmaceuticals, 18(4), 493. https://doi.org/10.3390/ph18040493








