Synthesis, Structure, and Stability of Copper(II) Complexes Containing Imidazoline-Phthalazine Ligands with Potential Anticancer Activity
Abstract
1. Introduction
2. Results and Discussion
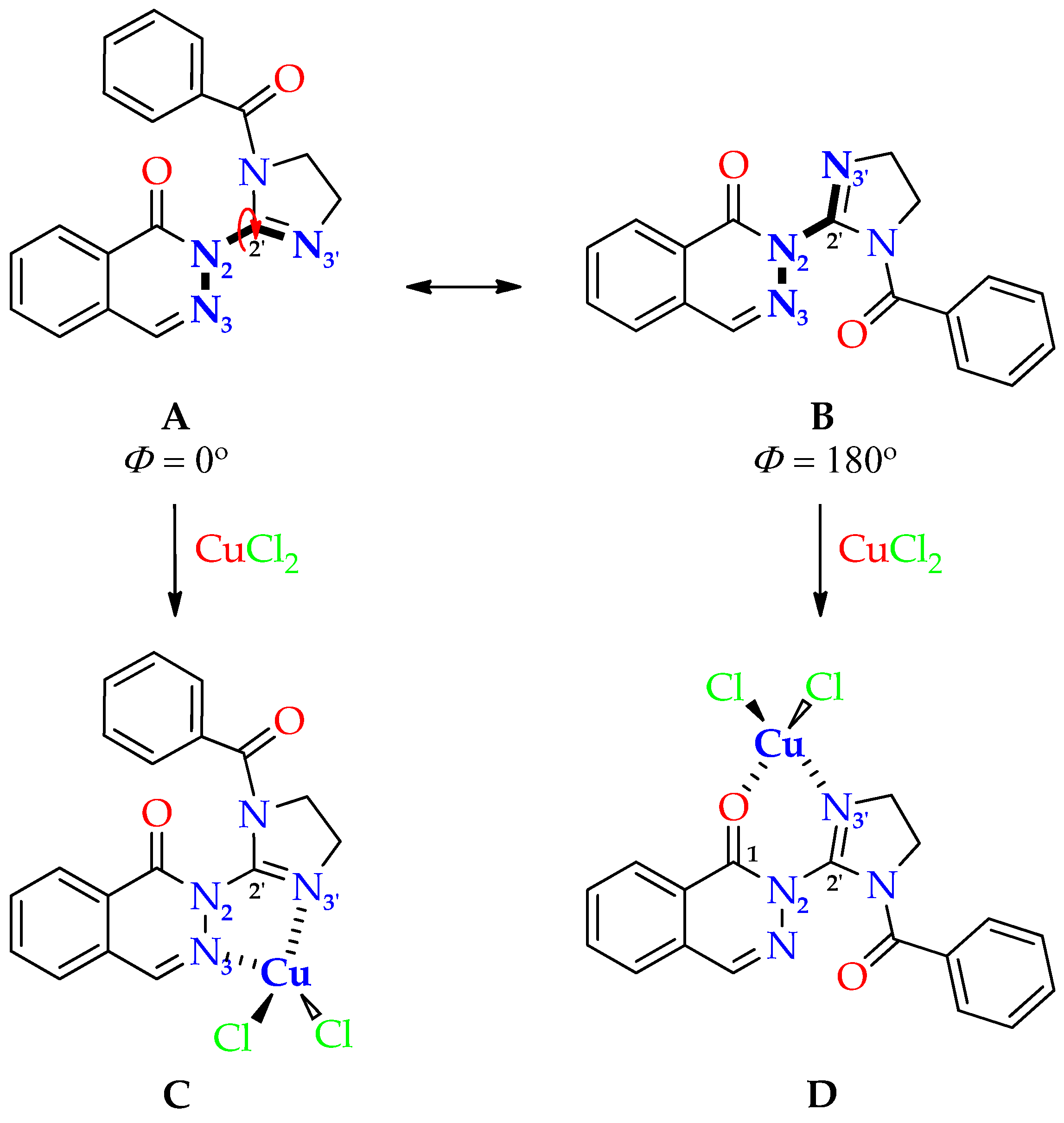
2.1. Chemistry
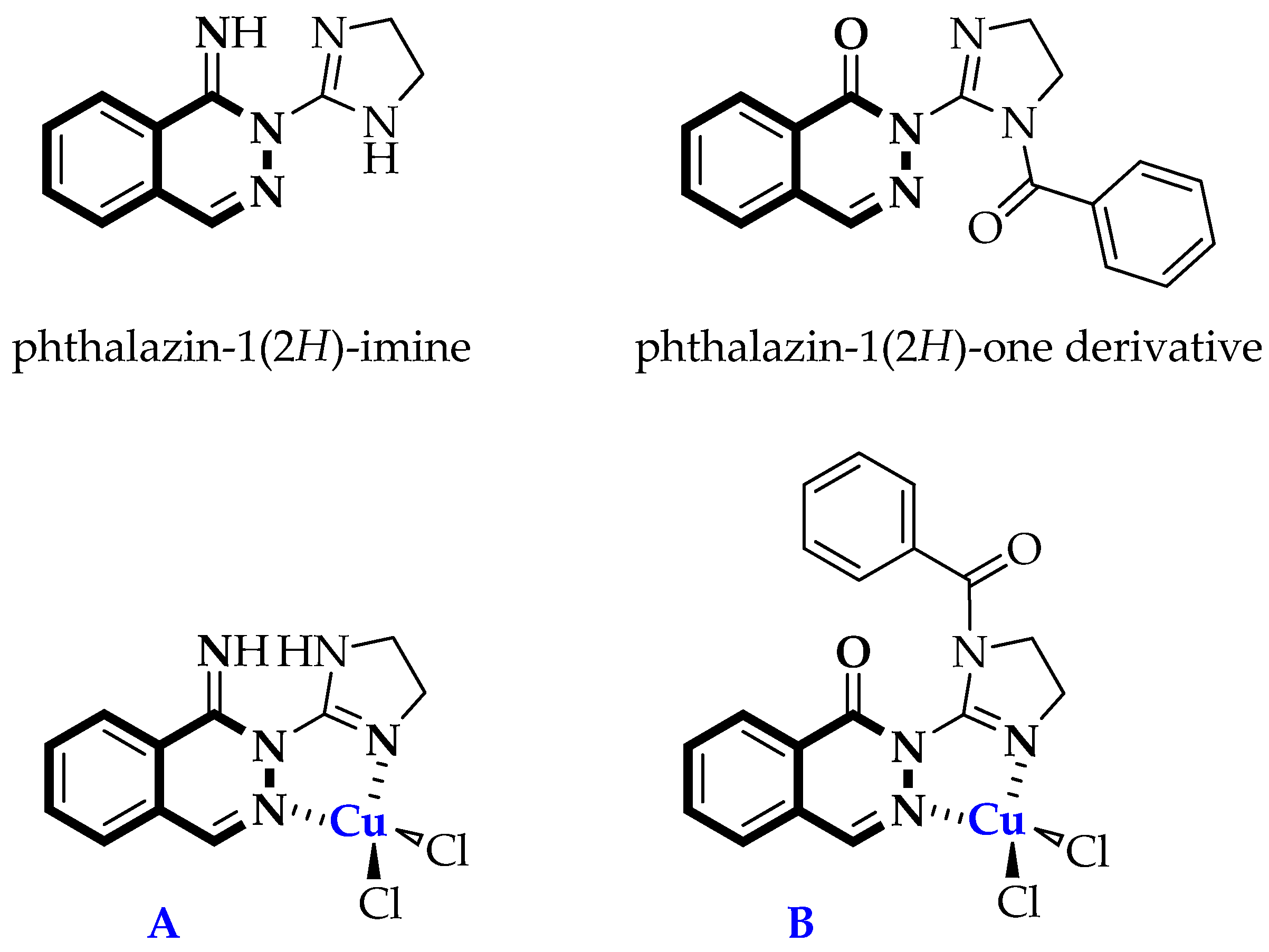
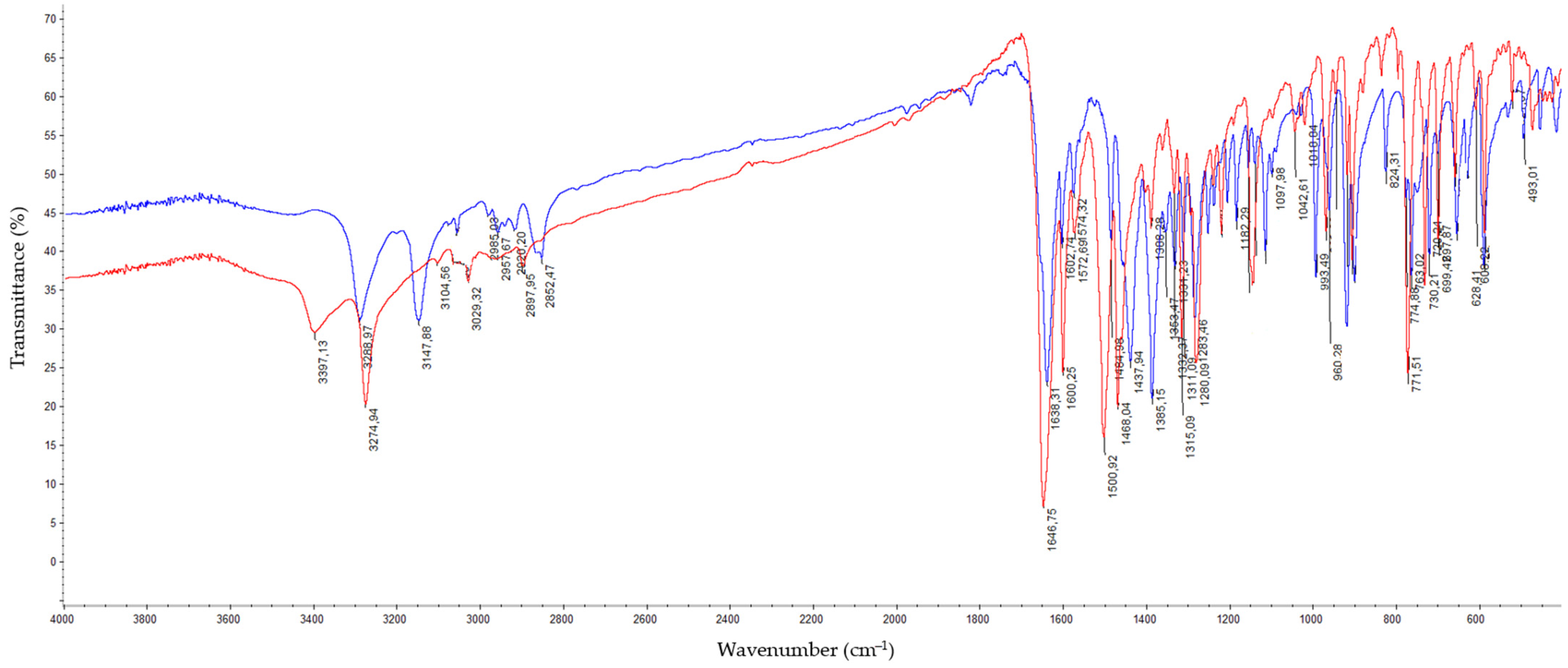
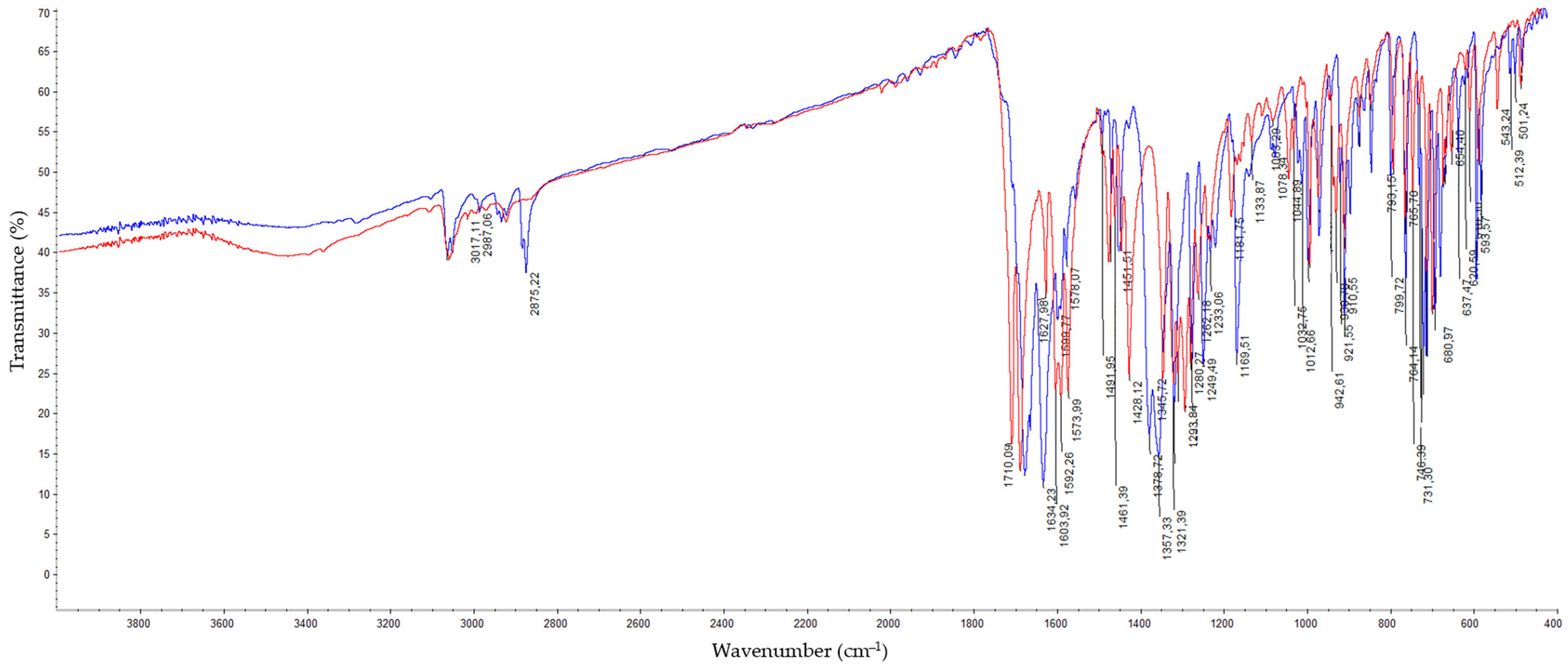
Synthesis of Dichloro[2-(4,5-dihydro-1H-imidazol-2-yl)phthalazin-1(2H)-imine]copper(II) (C1) and Dichloro[2-(1-benzoyl-4,5-dihydro-1H-imidazol-2-yl)phthalazin-1(2H)-one]copper(II) (C2)
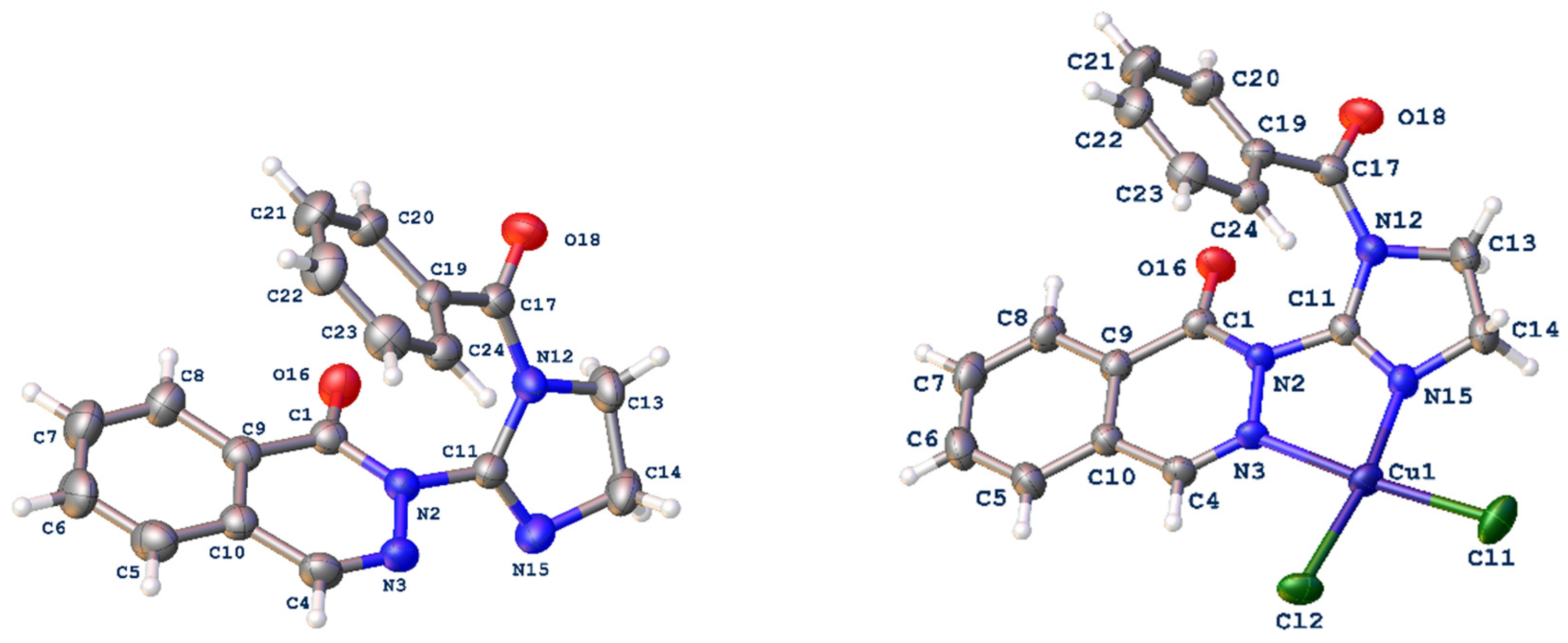
2.2. Structure of Copper(II) Complexes C1 and C2
Crystallographic Studies of Ligand L3 and Copper(II) Complex C2
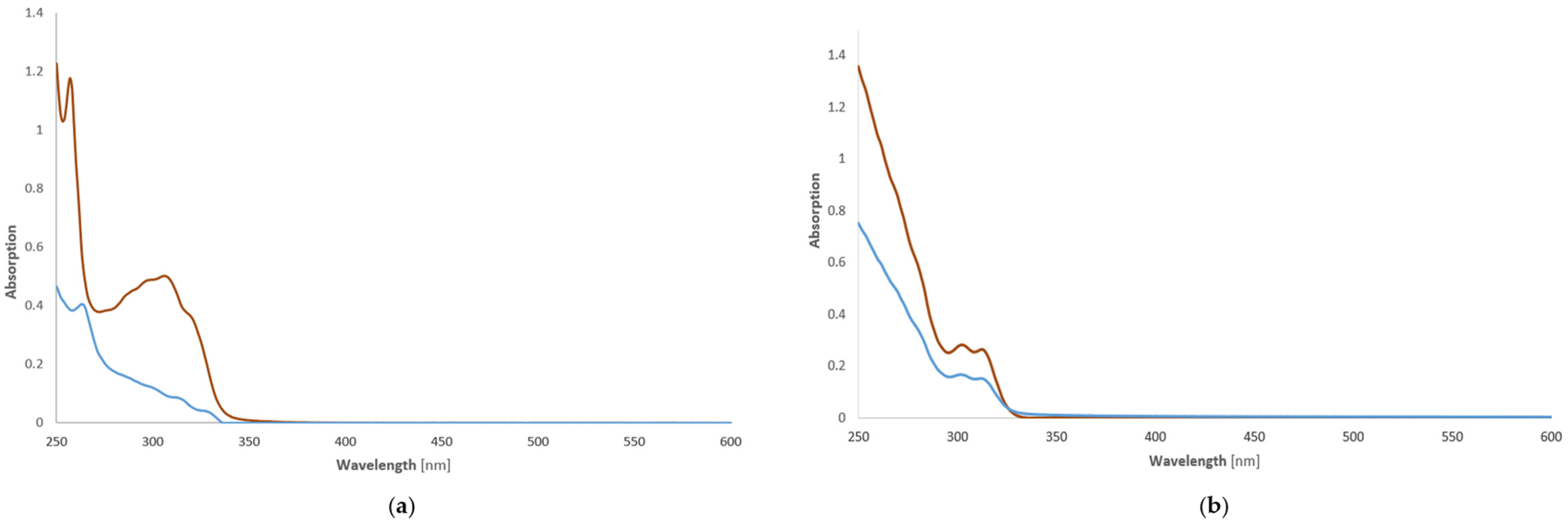
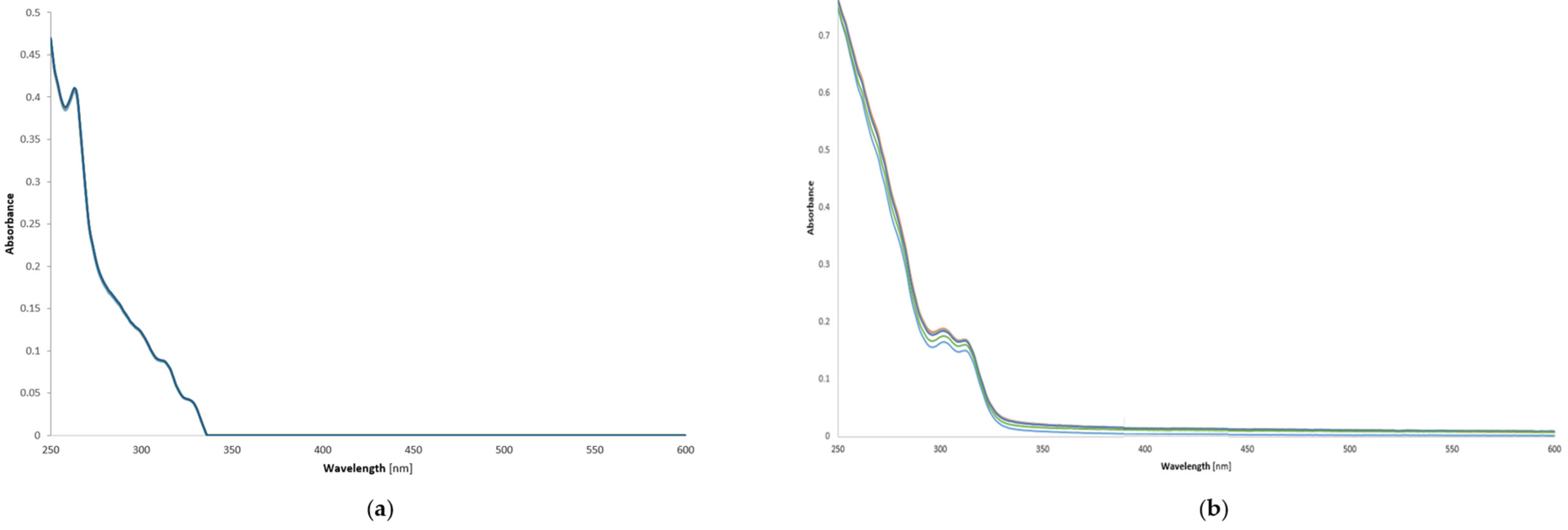
2.3. Stability of Copper(II) Complexes—UV-Vis Investigations of Compounds C1 and C2 in Phosphate-Buffered Saline Solution (PBS)
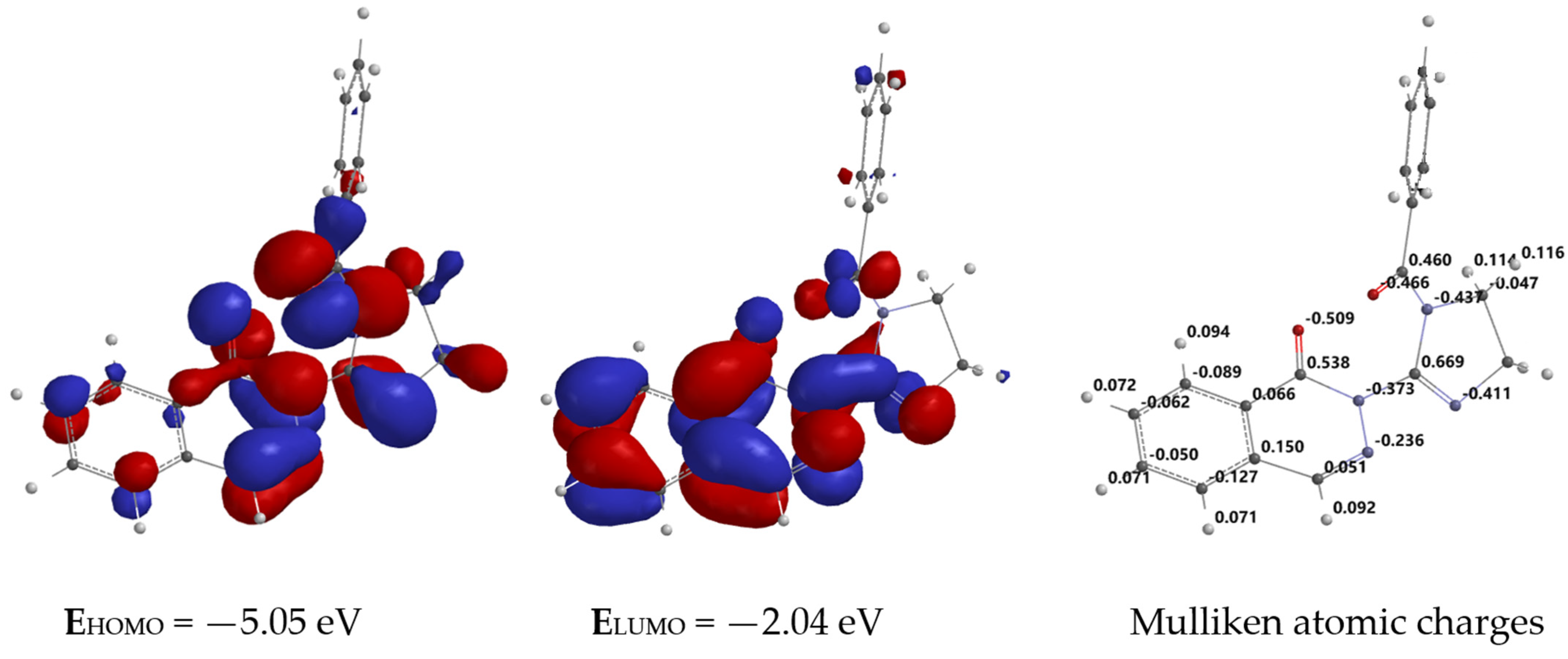
2.4. In Silico Molecular Modeling of Ligand L3
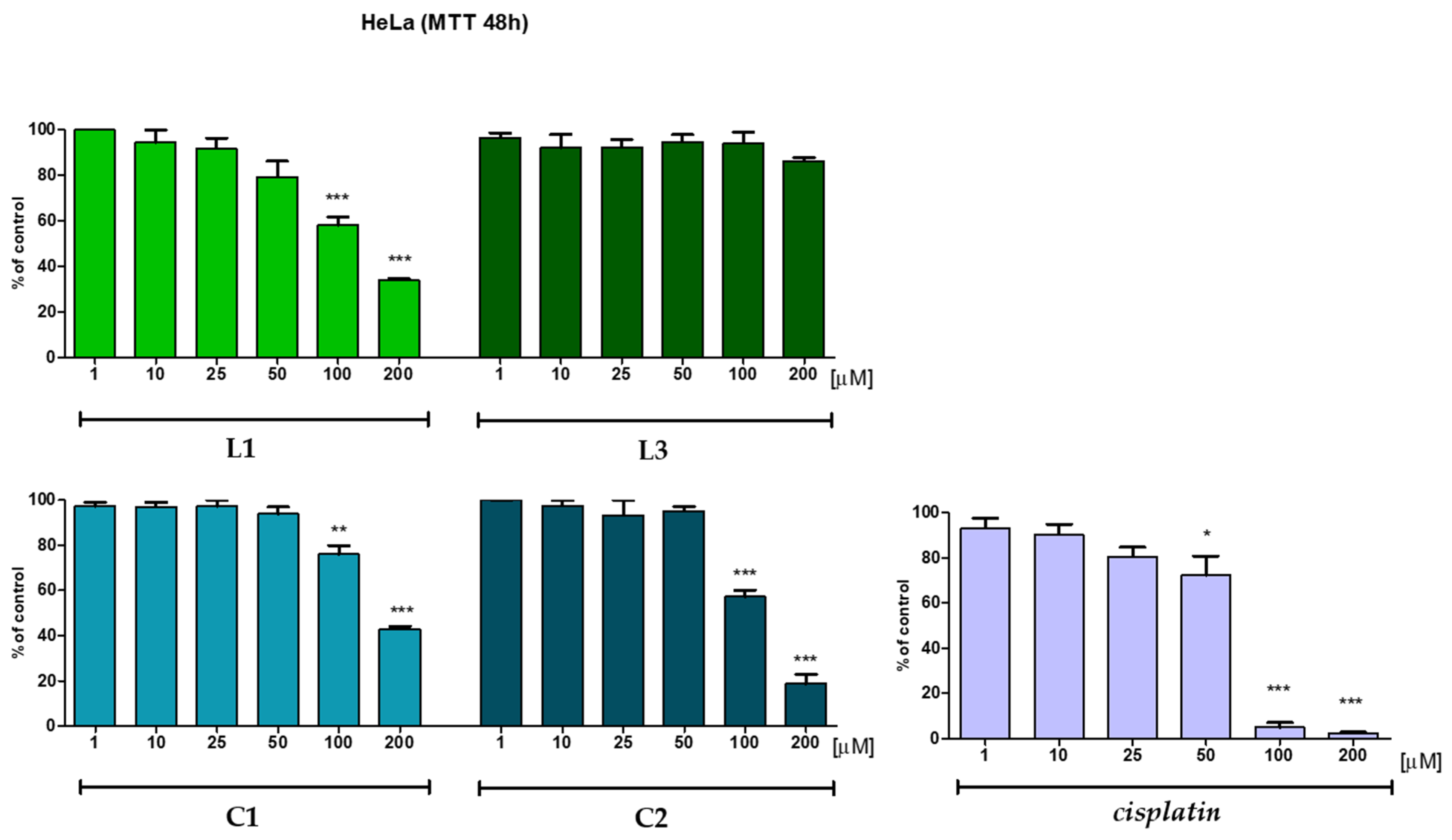
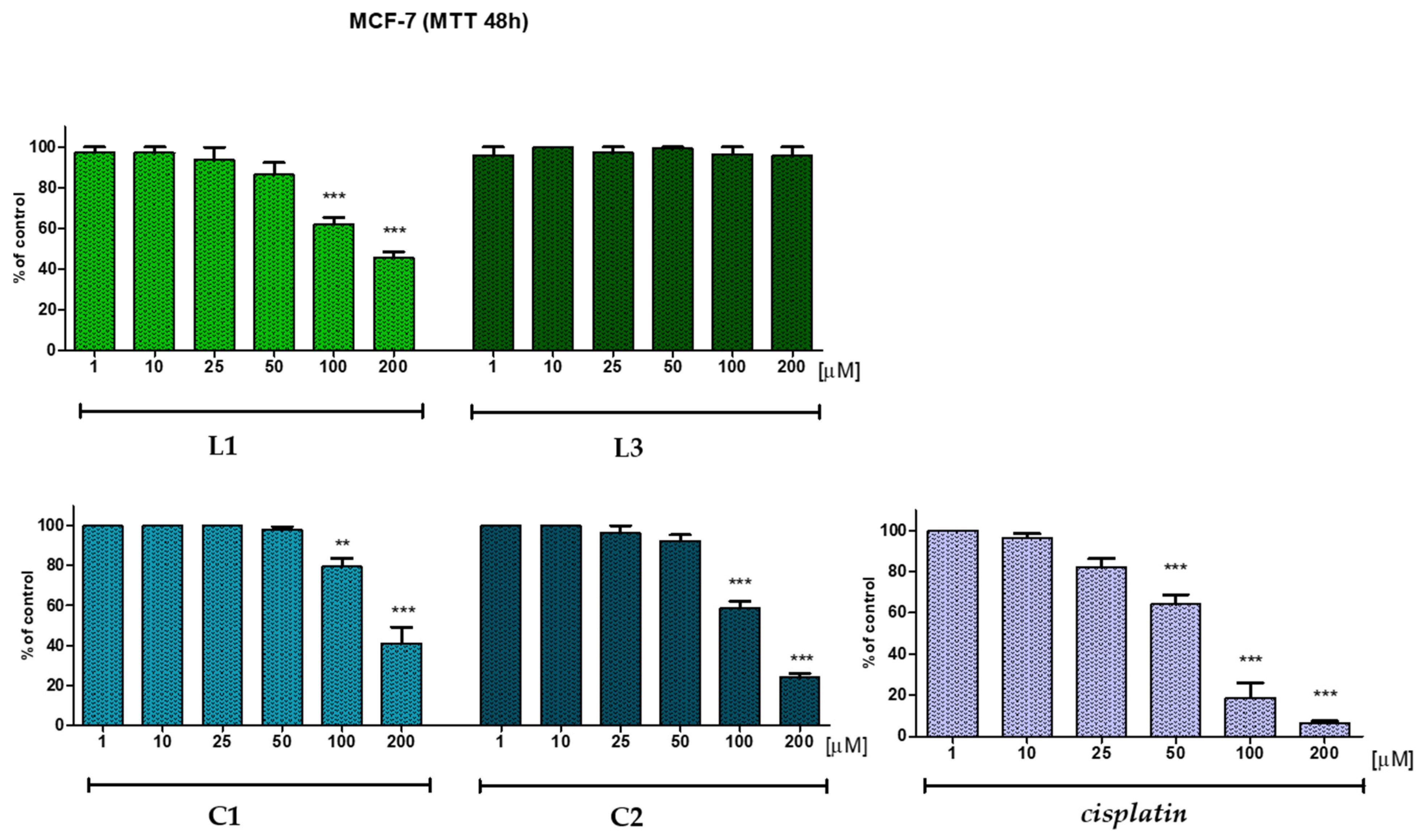
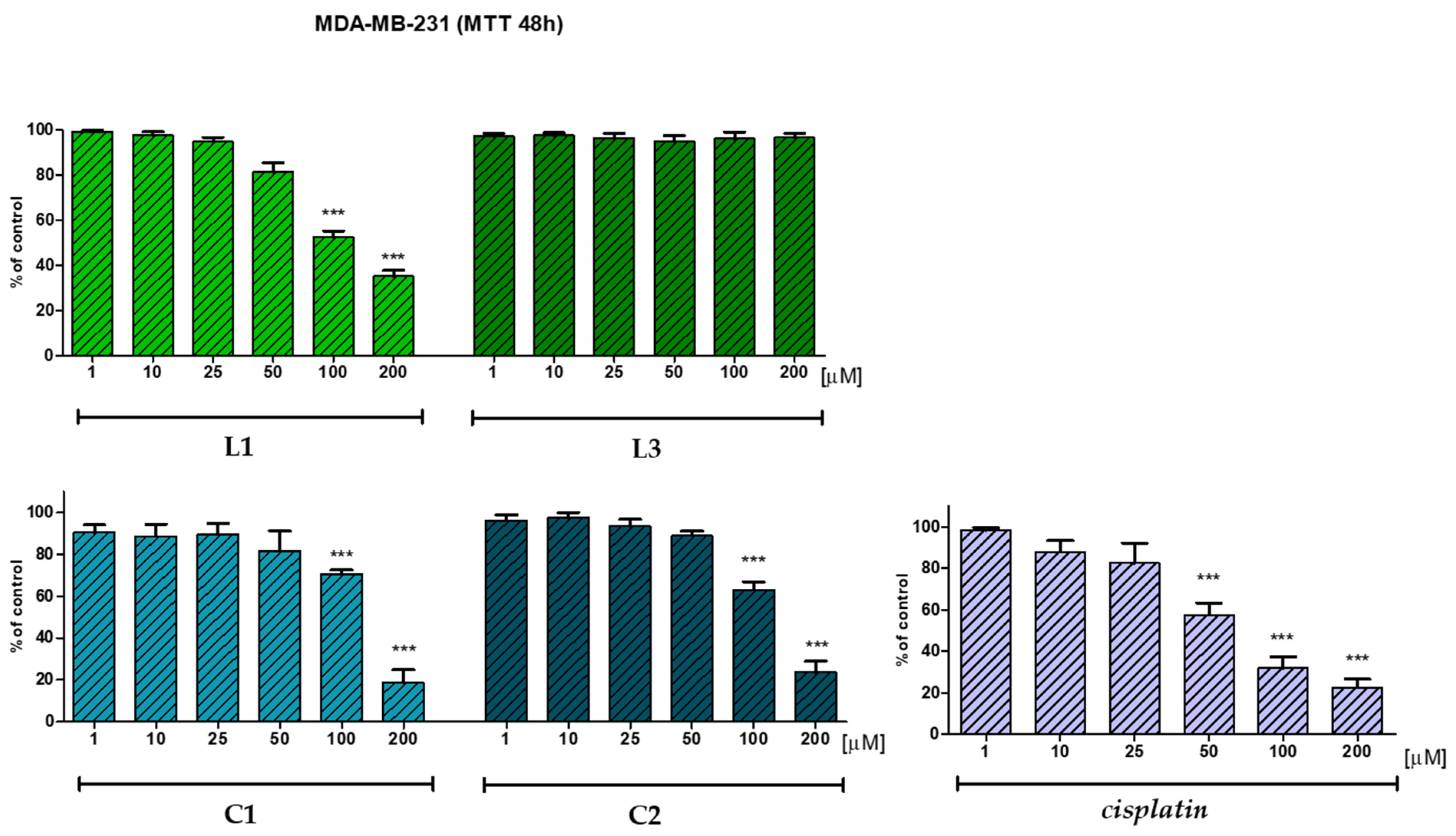
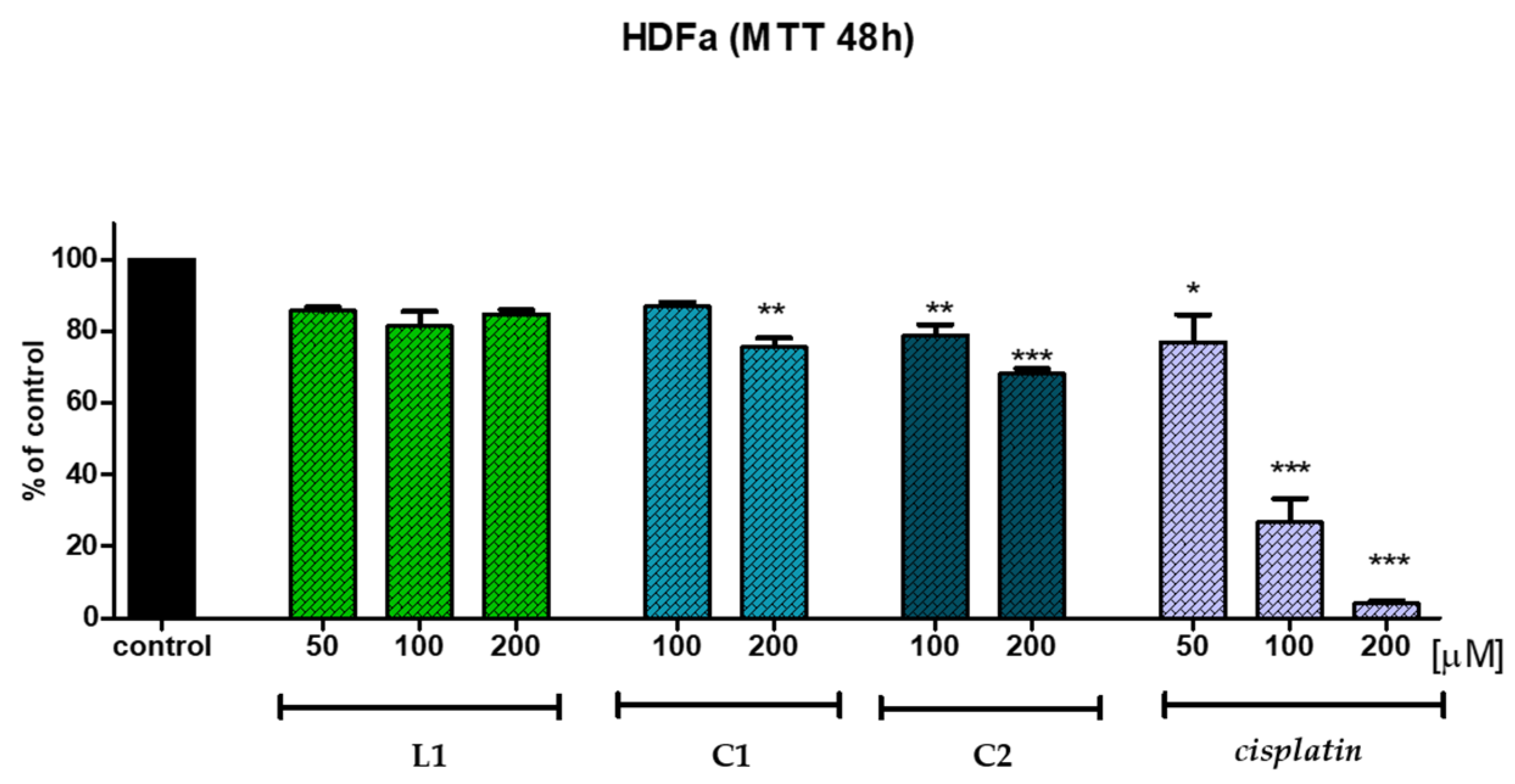
2.5. In Vitro Anticancer Potential
2.6. Antimicrobial Activity
2.7. Determination of Antioxidant Activity
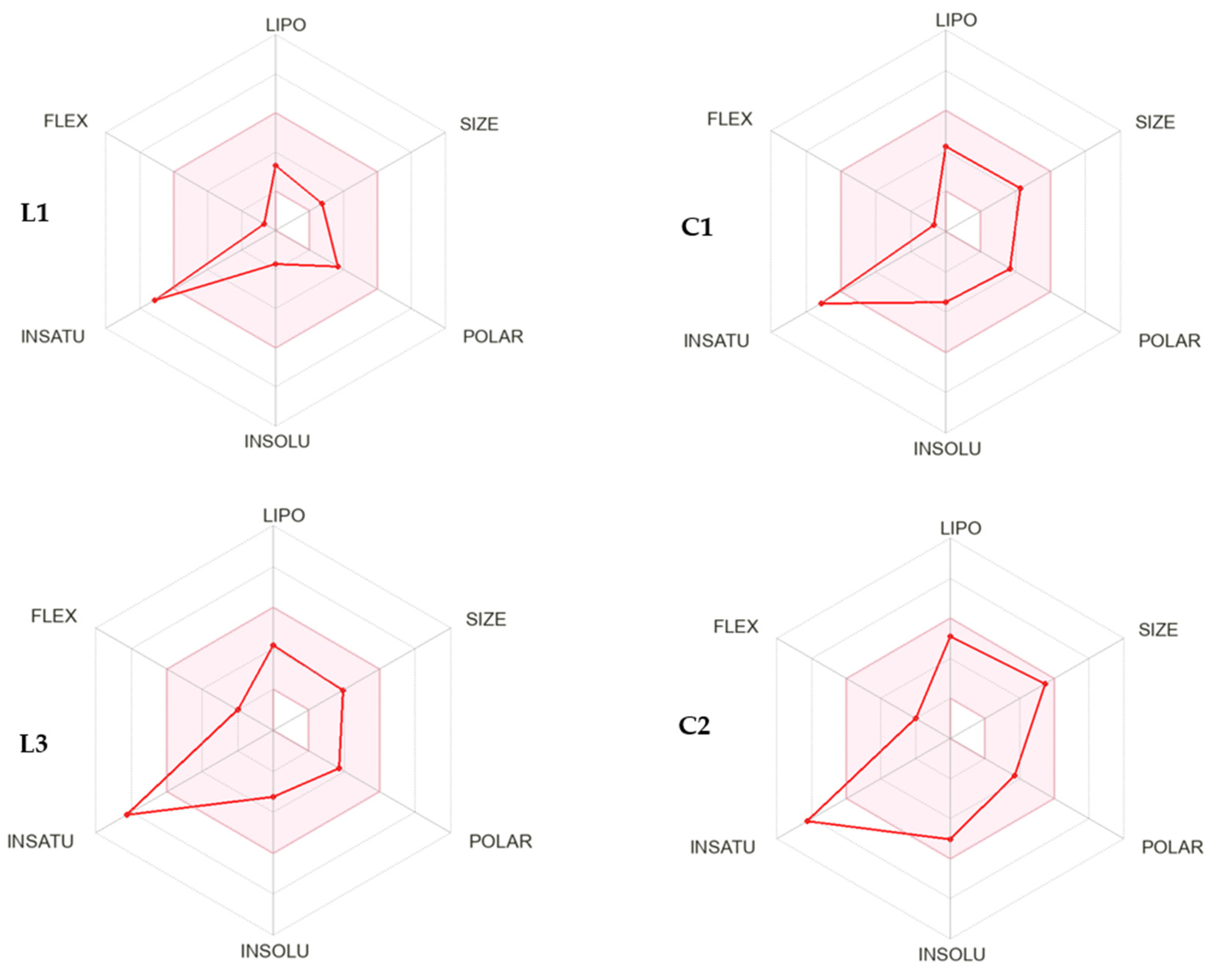
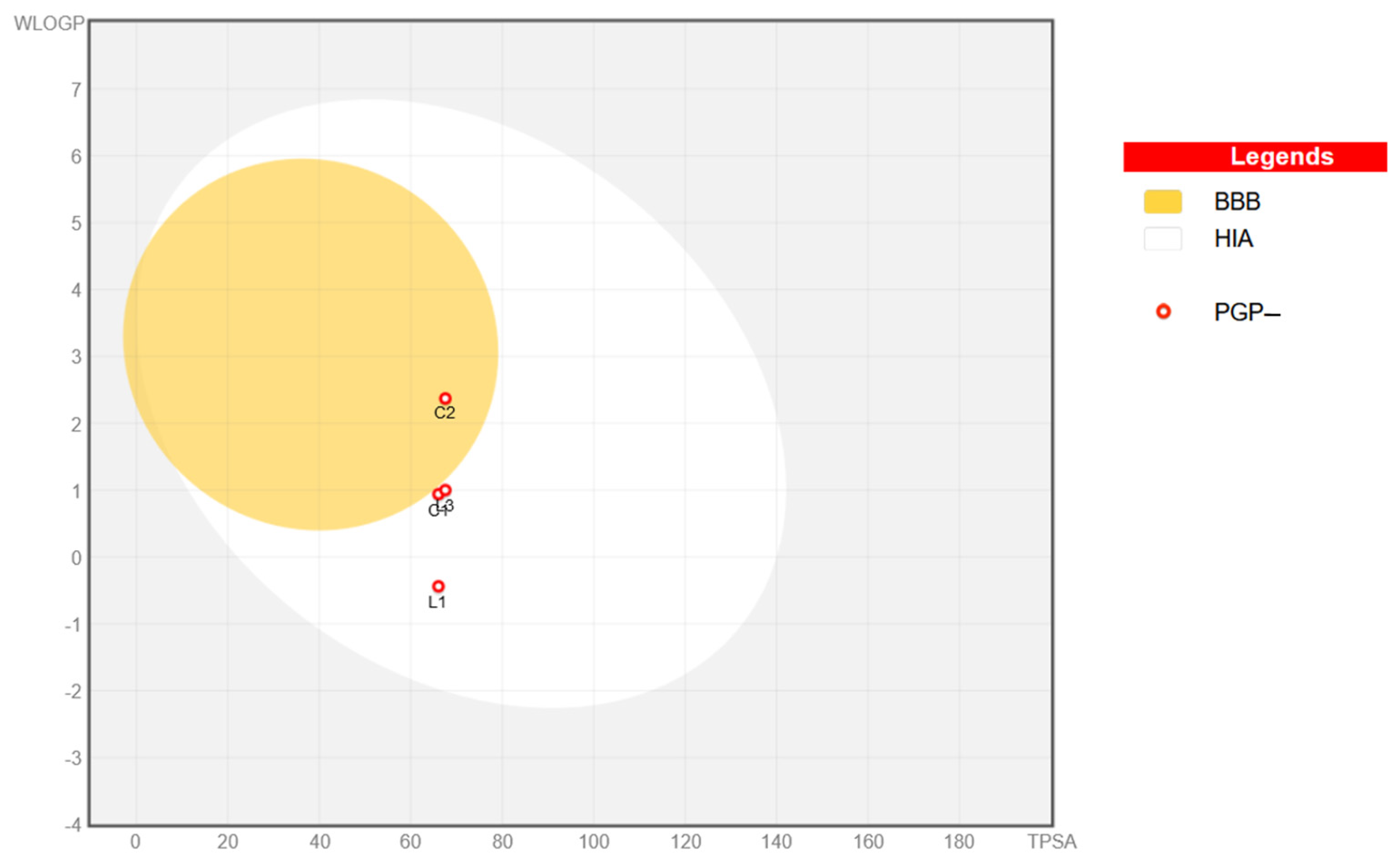
2.8. In Silico Physicochemical, Pharmacokinetic, and Drug-Likeness Predictions for Ligands L1 and L3 and Cu(II) Complexes C1 and C2
3. Materials and Methods
3.1. Experimental Part
3.1.1. General Information
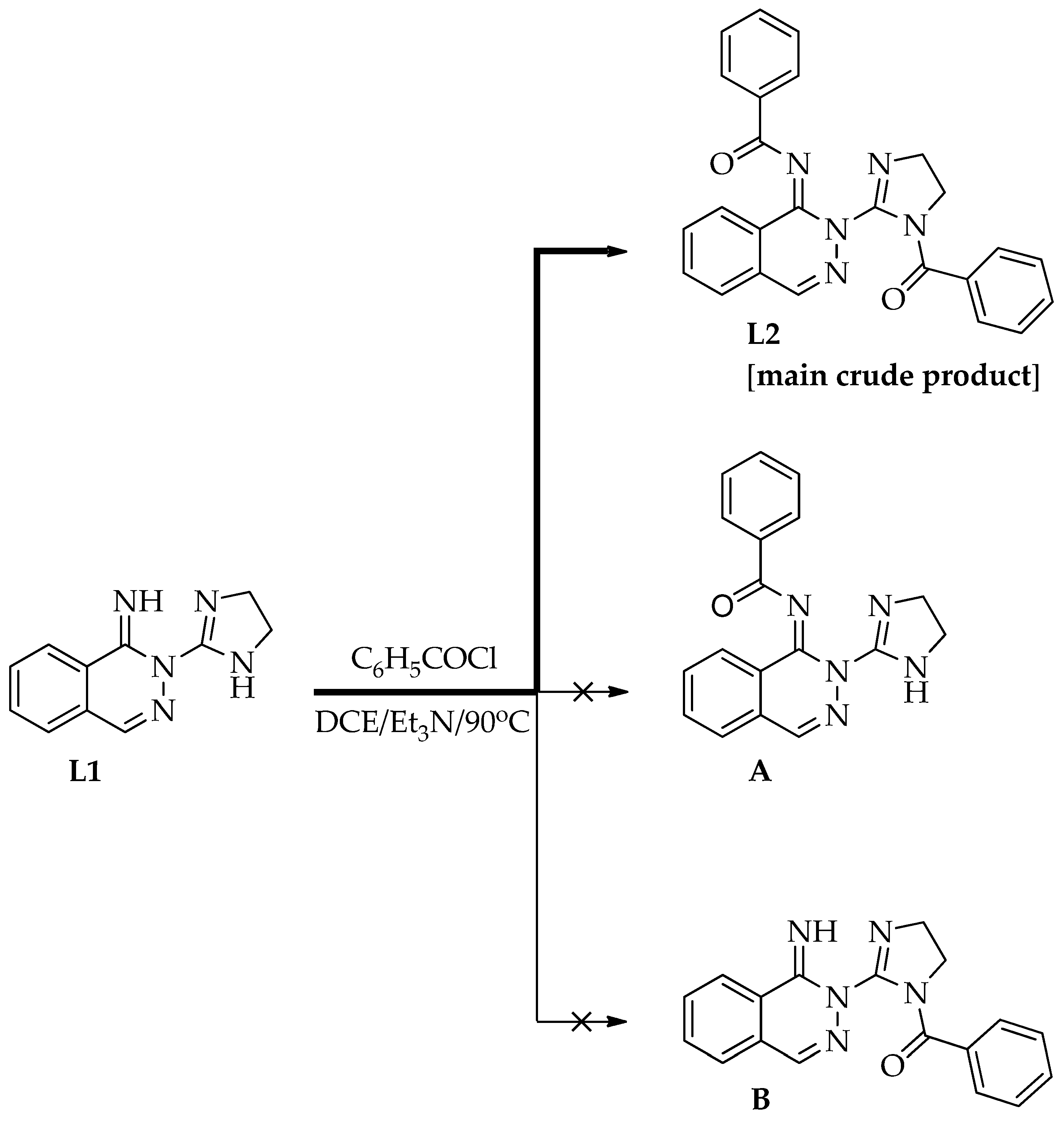
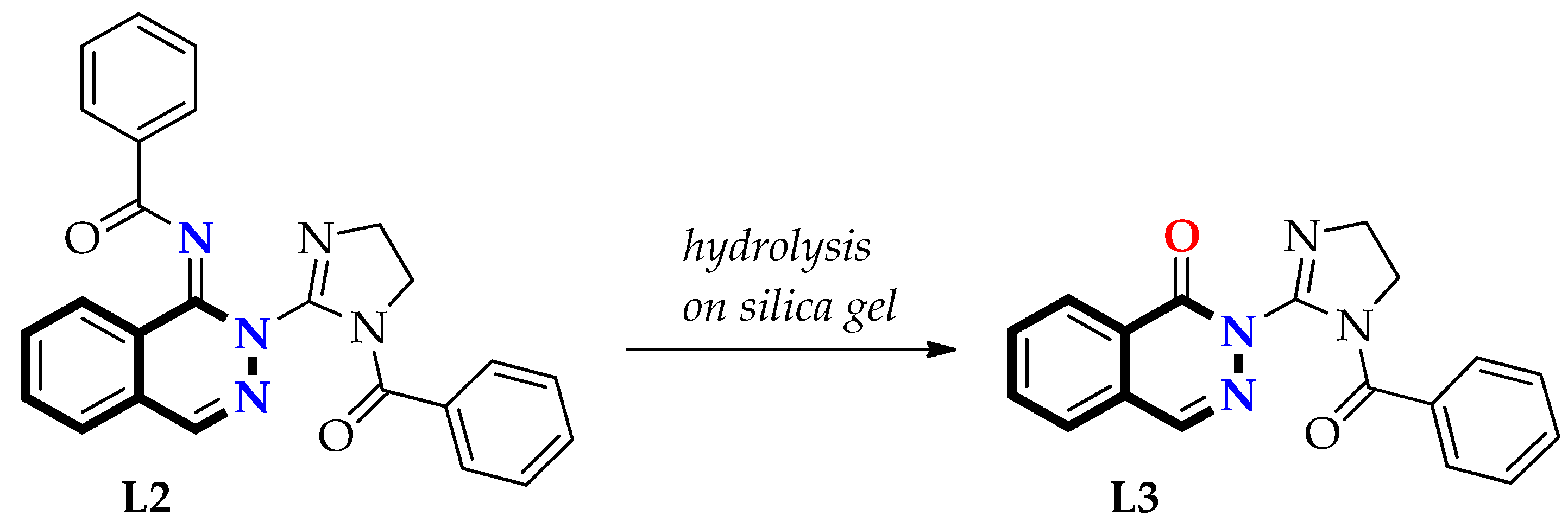
3.1.2. Preparation of Free Ligands: N-(2-(1-Benzoyl-4,5-dihydro-1H-imidazol-2-yl)phthalazin-1(2H)-ylidene)benzamide (L2) and 2-(1-Benzoyl-4,5-dihydro-1H-imidazol-2-yl)phthalazin-1(2H)-one (L3)
First Step—Synthesis of Ligand L2 and Its Purification
Second Step—Synthesis of Ligand L3
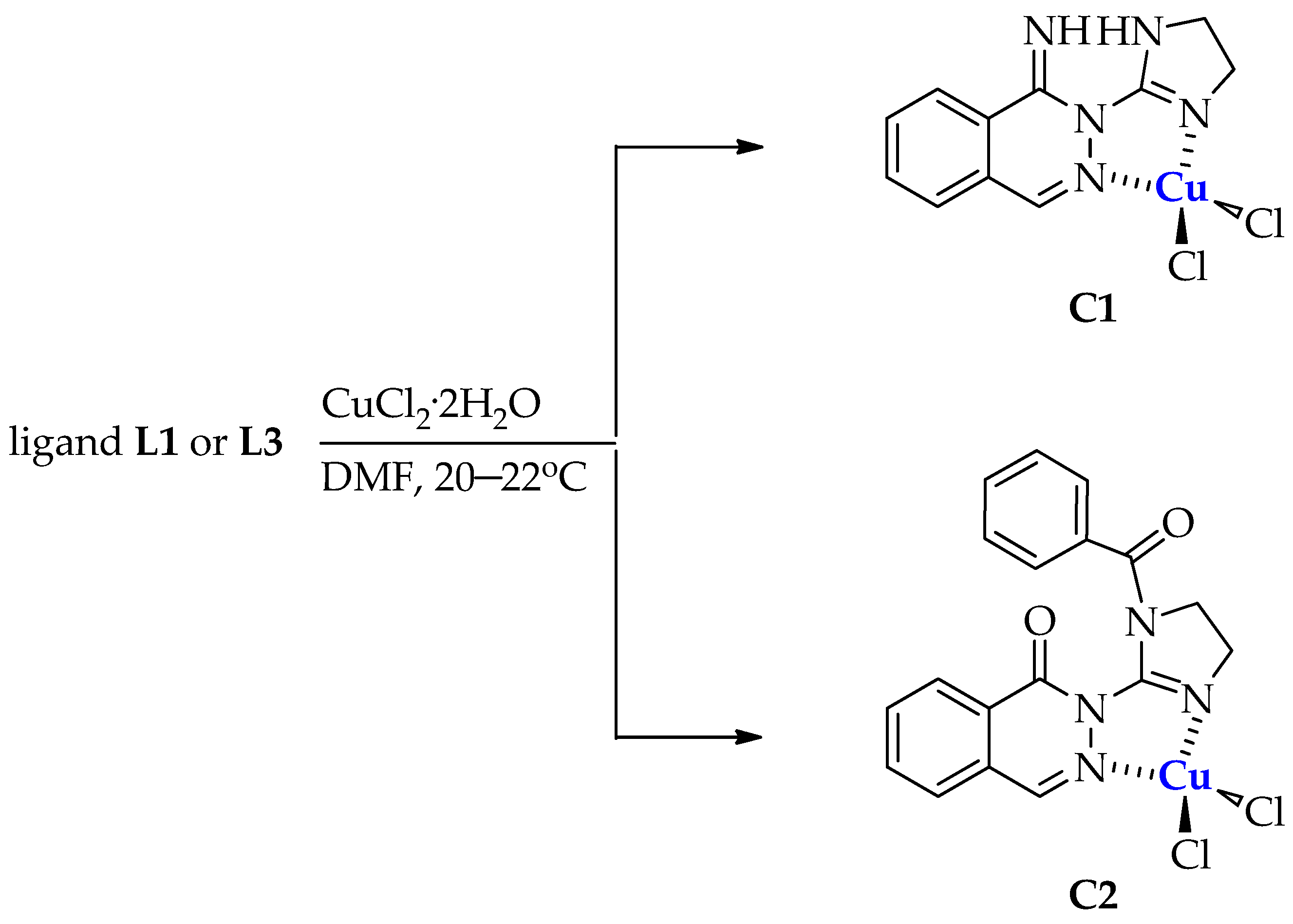
3.1.3. Synthesis of Copper(II) Complexes C1 and C2
Synthesis of Dichloro[2-(4,5-dihydro-1H-imidazol-2-yl)phthalazin-1(2H)-imine]copper(II) (C1)
Synthesis of Dichloro[2-(1-benzoyl-4,5-dihydro-1H-imidazol-2-yl)phthalazin-1(2H)-one]copper(II) (C2)
3.2. Stability Studies of Dichloro[2-(4,5-dihydro-1H-imidazol-2-yl)phthalazin-1(2H)-imine]copper(II) (C1) and Dichloro[2-(1-benzoyl-4,5-dihydro-1H-imidazol-2-yl)phthalazin-1(2H)-one]copper(II) (C2)
3.3. Biology
3.3.1. Evaluation of In Vitro Cytotoxicity
Cell Culture
The MTT Assay
Statistical Analysis
3.3.2. Determination of Minimum Inhibitory Concentration (MIC) and Minimum Bactericidal/Fungicidal Concentration (MBC/MFC)
3.3.3. Antioxidant Studies
Materials
DPPH Radical Scavenging Assay
ABTS Radical Scavenging Assay
FRAP Test
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hill, D.P.; Harper, A.; Malcolm, J.; McAndrews, M.S.; Mockus, S.M.; Patterson, S.E.; Reynolds, T.; Baker, E.J.; Bult, C.J.; Chesler, E.J.; et al. Cisplatin-Resistant triple-negative breast cancer subtypes: Multiple mechanisms of resistance. BMC Cancer 2019, 19, 1039. [Google Scholar] [CrossRef] [PubMed]
- Jhan, J.R.; Andrechek, E.R. Triple-Negative breast cancer and the potential for targeted therapy. Pharmacogenomics 2017, 18, 1595–1609. [Google Scholar] [CrossRef]
- Aysola, K.; Desai, A.; Welch, C.; Xu, J.; Qin, Y.; Reddy, V.; Matthews, R.; Owens, C.; Okoli, J.; Beech, D.J.; et al. Triple negative breast cancer—An overview. Hered. Genet. 2013, 2013, 1. [Google Scholar] [CrossRef]
- Penault, F.; Viale, G. Pathological and molecular diagnosis of triple-negative breast cancer: A clinical perspective. Ann. Oncol. 2012, 23, 19–22. [Google Scholar] [CrossRef]
- Lehmann, B.D.; Bauer, J.A.; Chen, X.; Sanders, M.E.; Chakravarthy, A.B.; Shyr, Y.; Pietenpol, J.A. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J. Clin. Investig. 2011, 121, 2750–2767. [Google Scholar] [CrossRef]
- Catalano, A.; Iacopetta, D.; Ceramella, J.; Mariconda, A.; Rosano, C.; Scumaci, D.; Saturnino, C.; Longo, P.; Sinicropi, M.S. New achievements for the treatment of triple-negative breast cancer. Appl. Sci. 2022, 12, 5554. [Google Scholar] [CrossRef]
- Muhammad, N.; Hanif, M.; Yang, P. Beyond cisplatin: New frontiers in metallodrugs for hard to treat triple negative breast cancer. Coord. Chem. Rev. 2024, 499, 215507. [Google Scholar] [CrossRef]
- Nayeem, N.; Contel, M. Exploring the potential of metallodrugs as chemotherapeutics for triple negative breast cancer. Chem. Eur. J. 2021, 27, 8891–8917. [Google Scholar] [CrossRef]
- Ferraro, M.G.; Bocchetti, M.; Riccardi, C.; Trifuoggi, M.; Paduano, L.; Montesarchio, D.; Misso, G.; Santamaria, R.; Piccolo, M.; Irace, C. Triple negative breast cancer preclinical therapeutic management by a cationic ruthenium-based nucleolipid nanosystem. Int. J. Mol. Sci. 2023, 24, 6473. [Google Scholar] [CrossRef]
- Abdel-Mohsen, M.A.; Badawy, A.M.; Abu-Youssef, M.A.; Yehia, M.A.; Lobna Shamaa, L.D.; Abdullah Mohamed, S. Influence of copper(I) nicotinate complex on the Notch1 signaling pathway in triple negative breast cancer cell lines. Sci. Rep. 2024, 14, 2522. [Google Scholar] [CrossRef]
- Arojojoye, A.S.; Olelewe, C.; Gukathasan, S.; Kim, J.H.; Vekaria, H.; Parkin, S.; Sullivan, P.G.; Awuah, S.G. Serum-stable gold(III) bisphosphine complex induces mild mitochondrial uncoupling and in vivo antitumor potency in triple negative breast cancer. J. Med. Chem. 2023, 66, 7868–7879. [Google Scholar] [CrossRef] [PubMed]
- Katkova, S.A.; Bunev, A.S.; Gasanov, R.E.; Khochenkov, D.A.; Kulsha, A.V.; Ivashkevich, O.A.; Serebryanskaya, T.V.; Kinzhalov, M.A. Metal-(acyclic diaminocarbene) complexes demonstrate nanomolar antiproliferative activity against triple-negative breast cancer. Chem. A Eur. J. 2024, 30, e202400101. [Google Scholar] [CrossRef]
- Muggia, F.M.; Bonetti, A.; Hoeschele, J.D.; Rozencweig, M.B.; Howell, S.B. Platinum antitumor complexes: 50 Years since discovery Barnett Rosenberg’s. J. Clin. Oncol. 2015, 33, 4219–4226. [Google Scholar] [CrossRef] [PubMed]
- Baek, D.W.; Park, J.Y.; Lee, S.J.; Chae, Y.S. Impressive effect of cisplatin monotherapy on a patient with heavily pretreated triple-negative breast cancer with poor performance. Yeungnam Univ. J. Med. 2020, 37, 230–235. [Google Scholar] [CrossRef] [PubMed]
- Date, T.; Kuche, K.; Chaudhari, D.; Ghadi, R.; Sahel, K.D.; Chitkara, D.; Jain, S. Hitting multiple cellular targets in triple-negative breast cancer using dual-action cisplatin(IV) prodrugs for safer synergistic chemotherapy. ACS Biomater. Sci. Eng. 2022, 8, 2349–2362. [Google Scholar] [CrossRef]
- Sirohi, B.; Arnedos, M.; Popat, S.; Walsh, G.; Johnston, S.; Smith, I.E. Platinum-Based chemotherapy in triple-negative breast cancer. Ann. Oncol. 2008, 19, 1847–1852. [Google Scholar] [CrossRef]
- Byrski, T.; Dent, R.; Blecharz, P.; Foszczynska-Kloda, M.; Gronwald, J.; Huzarski, T.; Cybulski, C.; Marczyk, E.; Chrzan, R.; Eisen, A.; et al. Results of a phase II open-label, non-randomized trial of cisplatin chemotherapy in patients with BRCA1-positive metastatic breast cancer. Breast Cancer Res. 2012, 14, R110. [Google Scholar] [CrossRef]
- Wahba, H.A.; El-Hadaad, H.A. Current approaches in treatment of triple-negative breast cancer. Cancer Biol. Med. 2015, 12, 106–116. [Google Scholar] [CrossRef]
- Agnello, L.; Tortorella, S.; d’Argenio, A.; Carbone, C.; Camorani, S.; Locatelli, E.; Auletta, L.; Sorrentino, D.; Fedele, M.; Zannetti, A.; et al. Optimizing cisplatin delivery to triple-negative breast cancer through novel EGFR aptamer-conjugated polymeric nanovectors. J. Exp. Clin. Cancer Res. 2021, 40, 239. [Google Scholar] [CrossRef]
- Ghosh, S. Cisplatin: The first metal based anticancer drug. Bioorg. Chem. 2019, 88, 102925. [Google Scholar] [CrossRef]
- Ranasinghe, R.; Mathai, M.L.; Zulli, A. Cisplatin for cancer therapy and overcoming chemoresistance. Heliyon 2022, 8, e10608. [Google Scholar] [CrossRef]
- Abdolmaleki, S.; Aliabadi, A.; Khaksar, S. Unveiling the promising anticancer effect of copper-based compounds: A comprehensive review. J. Cancer Res. Clin. Oncol. 2024, 150, 213. [Google Scholar] [CrossRef] [PubMed]
- Vančo, J.; Trávníček, Z.; Hošek, J.; Malina, T.; Dvořák, Z. Copper(II) complexes containing natural flavonoid pomiferin show considerable in vitro cytotoxicity and anti-inflammatory effects. Int. J. Mol. Sci. 2021, 22, 7626. [Google Scholar] [CrossRef]
- Santini, C.; Pellei, M.; Gandin, V.; Porchia, M.; Tisato, F.; Marzano, C. Advances in copper complexes as anticancer agents. Chem. Rev. 2014, 114, 815–862. [Google Scholar] [CrossRef] [PubMed]
- Denoyer, D.; Clatworthy, S.A.S.; Cater, M.A. Copper complexes in cancer therapy. Met. Ions Life Sci. 2018, 18, 469–506. [Google Scholar] [CrossRef]
- Kellett, A.; Molphy, Z.; McKee, V.; Slator, C. Recent advances in anticancer copper compounds. Chem. Pharm. 2019, 4, 91–119. [Google Scholar] [CrossRef]
- Zehra, S.; Tabassum, S.; Arjmand, F. Biochemical pathways of copper complexes: Progress over the past 5 years. Drug Discov. Today 2021, 26, 1086–1096. [Google Scholar] [CrossRef]
- Galindo-Murillo, R.; García-Ramos, J.K.; Ruiz-Azuara, L.; Cheatham, T.E.; Cortés-Guzmán, F. Intercalation processes of copper complexes in DNA. Nucleic Acids Res. Spec. Publ. 2015, 43, 5364–5376. [Google Scholar] [CrossRef]
- Devi, C.S.; Thulasiram, B.; Aerva, R.R.; Nagababu, P. Recent advances in copper intercalators as anticancer agents. J. Fluoresc. 2018, 28, 1195–1205. [Google Scholar] [CrossRef]
- Mahendiran, D.; Amuthakala, S.; Bhuvanesh, N.S.P.; Kumar, R.S.; Rahiman, A.K. Copper complexes as prospective anticancer agents: In vitro and in vivo evaluation, selective targeting of cancer cells by DNA damage and S phase arrest. RSC Adv. 2018, 8, 16973–16990. [Google Scholar] [CrossRef]
- Erxleben, A. Interactions of copper complexes with nucleic acids. Coord. Chem. Rev. 2018, 360, 92–121. [Google Scholar] [CrossRef]
- Zehra, S.; Roisnel, T.; Arjmand, F. Enantiomeric amino acid Schiff base copper(II) complexes as a new class of RNA-targeted metallo-intercalators: Single X-ray crystal structural details, comparative in vitro DNA/RNA binding profile, cleavage, and cytotoxicity. ACS Omega 2019, 4, 7691–7705. [Google Scholar] [CrossRef]
- Bollu, V.S.; Bathini, T.; Barui, A.K.; Roy, A.; Ragi, N.C.; Maloth, S.; Sripadi, P.; Sreedhar, B.; Nagababu, P.; Patra, C.R. Design of DNA-intercalators based copper(II) complexes, investigation of their potential anti-cancer activity and sub-chronic toxicity. Mater. Sci. Eng. 2019, 105, 110079. [Google Scholar] [CrossRef]
- Khan, R.A.; Usman, M.; Dhivya, R.; Balaji, P.; Alsalme, A.; AlLohedan, H.; Arjmand, F.; AlFarhan, K.; Akbarsha, M.A.; Marchetti, F.; et al. Heteroleptic copper(I) complexes of “scorpionate” bis-pyrazolyl carboxylate ligand with auxiliary phosphine as potential anticancer agents: An insight into cytotoxic mode. Sci. Rep. 2017, 7, 45229–45246. [Google Scholar] [CrossRef] [PubMed]
- Molinaro, C.; Martoriati, A.; Pelinski, L.; Cailliau, K. Cooper complexes as anticancer agents targeting topoisomerases I and II. Cancers 2020, 12, 2863–2889. [Google Scholar] [CrossRef] [PubMed]
- Patel, R.N.; Shukla, K.K.; Singh, A.; Choudhary, M.; Chauhan, U.K.; Dwivedi, S. Copper(II) complexes as superoxide dismutase mimics: Synthesis, characterization, crystal structure and bioactivity of copper(II) complexes. Inorganica Chim. Acta 2009, 362, 4891–4898. [Google Scholar] [CrossRef]
- Sîrbu, A.; Palamarciuc, O.; Babak, M.V.; Lim, J.M.; Ohui, K.; Enyedy, E.A.; Shova, S.; Darvasiová, D.; Rapta, P.; Ang, W.H.; et al. Copper(II) thiosemicarbazone complexes induce marked ROS accumulation and promote nrf2-mediated antioxidant response in highly resistant breast cancer cells. Dalton Trans. 2017, 46, 3833–3847. [Google Scholar] [CrossRef]
- Shi, X.; Chen, Z.; Wang, Y.; Guo, Z.; Wang, X. Hypotoxic copper complexes with potent anti-metastatic and anti-angiogenic activities against cancer cells. Dalton Trans. 2018, 47, 5049–5054. [Google Scholar] [CrossRef]
- Vincent, A.; Fores, J.R.; Tauziet, E.; Quévrain, E.; Dancs, Á.; Conte-Daban, A.; Bernard, A.S.; Pelupessy, P.; Coulibaly, K.; Seksik, P.; et al. An easy-to-implement combinatorial. Chem. Commun. 2020, 56, 399–402. [Google Scholar] [CrossRef]
- Chen, D.; Cui, Q.C.; Yang, H.; Dou, Q.P. Disulfiram, a clinically used anti-alcoholism drug and copper-binding agent, induces apoptotic cell death in breast cancer cultures and xenografts via inhibition of the proteasome activity. Cancer Res. 2006, 66, 10425–10433. [Google Scholar] [CrossRef]
- Rada, J.P.; Forte, J.; Gontard, G.; Corce, V.; Slamain, M.; Rey, N.A. Isoxazole-Derived Aroylhydrazones and their dinuclear copper(II) complexes show antiproliferative activity on breast cancer cells with a potentially alternative mechanism of action. ChemBioChem 2020, 21, 2474–2486. [Google Scholar] [CrossRef] [PubMed]
- Cvek, B.; Milacic, V.; Taraba, J.; Dou, Q.P. Ni(II), Cu(II), and Zn(II) diethyldithiocarbamate complexes show various activities against the proteasome in breast cancer cells. J. Med. Chem. 2008, 51, 6256–6258. [Google Scholar] [CrossRef] [PubMed]
- Li, D.D.; Yagüe, E.; Wang, L.Y.; Dai, L.L.; Yang, Z.B.; Zhi, S.; Zhang, N.; Zhao, X.M.; Hu, Y.H. Novel copper complexes that inhibit the proteasome and trigger apoptosis in triple-negative breast cancer cells. ACS Med. Chem. Lett. 2019, 10, 1328–1335. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Hu, C.; Quan, Y.; Ye, X.; Shi, X.; Guo, Z.; Wang, X. A copper complex that combats triple negative breast cancer by restraining angiogenesis. Dalton Trans. 2023, 52, 7626–7634. [Google Scholar] [CrossRef]
- Alajroush, D.R.; Smith, C.B.; Anderson, B.F.; Oyeyemi, I.T.; Beebe, S.J.; Holder, A.A. A comparison of in vitro studies between cobalt(III) and copper(II) complexes with thiosemicarbazone ligands to treat triple negative breast cancer. Inorganica Chim. Acta 2024, 562, 121898. [Google Scholar] [CrossRef]
- El-Adl, K.; Ibrahim, M.K.; Khedr, F.; Abulkhair, H.S.; Eissa, I.H. N-substituted-4-phenylphthalazin-1-amine-derived VEGFR-2 inhibitors: Design, synthesis, molecular docking, and anticancer evaluation studies. Arch. Pharm. 2021, 354, e2000219. [Google Scholar] [CrossRef]
- El-Gazzar, M.G.; Aly, H.M. Anticancer evaluation and docking study of new bifunctional phthalazine derivatives. Curr. Org. Synth. 2018, 15, 414–422. [Google Scholar] [CrossRef]
- El-Helby, A.G.A.; Ayyad, R.R.A.; Sakr, H.; El-Adl, K.; Ali, M.M.; Khedr, F. Design, synthesis, molecular docking, and an-ticancer activity of phthalazine derivatives as VEGFR-2 inhibitors. Arch. Pharm. Chem. Life Sci. 2017, 350, e1700240. [Google Scholar] [CrossRef]
- Behalo, M.S.; El-Karim, I.A.G.; Rafaat, R. Synthesis of novel phthalazine derivatives as potential anticancer and antioxidant agents based on 1-chloro-4-(4-phenoxyphenyl)phthalazine. J. Heterocycl. Chem. 2017, 54, 3591–3599. [Google Scholar] [CrossRef]
- Li, J.; Zhao, Y.F.; Yuan, X.Y.; Xu, J.-X.; Gong, P. Synthesis and anticancer activities of novel 1,4-disubstituted phthalazines. Molecules 2006, 11, 574–582. [Google Scholar] [CrossRef]
- Fong, P.C.; Boss, D.S.; Yap, T.A.; Tutt, A.; Wu, P.; Mergui-Roelvink, M.; Mortimer, P.; Swaisland, H.; Lau, A.; O’Connor, M.J.; et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N. Engl. J. Med. 2009, 361, 123–134. [Google Scholar] [CrossRef] [PubMed]
- Hobbs, E.A.; Litton, J.K.; Yap, T.A. Development of the PARP inhibitor talazoparib for the treatment of advanced BRCA1 and BRCA2 mutated breast cancer. Expert Opin. Pharmacother. 2021, 22, 1825–1837. [Google Scholar] [CrossRef]
- Sączewski, F.; Dziemidowicz-Borys, E.; Bednarski, P.J.; Grünert, R.; Gdaniec, M.; Tabin, P. Synthesis, crystal structure and biological activities of copper(II) complexes with chelating bidentate 2-substituted benzimidazole ligands. J. Inorg. Biochem. 2006, 100, 1389–1398. [Google Scholar] [CrossRef] [PubMed]
- Balewski, Ł.; Sączewski, F.; Bednarski, P.J.; Gdaniec, M.; Borys, E.; Makowska, A. Structural diversity of copper(II) com-plexes with N-(2-pyridyl)imidazolidin-2-ones(thiones) and their in vitro antitumor activity. Molecules 2014, 19, 17026–17051. [Google Scholar] [CrossRef] [PubMed]
- Makowska, A.; Sączewski, F.; Bednarski, P.J.; Gdaniec, M.; Balewski, Ł.; Warmbier, M.; Kornicka, A. Synthesis, structure and cytotoxic properties of copper(II) complexes of 2-iminocoumarins bearing a 1,3,5-triazine or benzoxazole/benzothiazole moiety. Molecules 2022, 27, 7155. [Google Scholar] [CrossRef]
- Balewski, Ł.; Plech, T.; Korona-Głowniak, I.; Hering, A.; Szczesio, M.; Olczak, A.; Bednarski, P.J.; Kokoszka, J.; Kornicka, A. Copper(II) complexes with 1-(isoquinolin-3-yl)heteroalkyl-2-ones: Synthesis, structure and evaluation of anticancer, antimicrobial and antioxidant potential. Int. J. Mol. Sci. 2024, 25, 8. [Google Scholar] [CrossRef]
- Balewski, Ł.; Gdaniec, M.; Hering, A.; Furman, C.; Ghinet, A.; Kokoszka, J.; Ordyszewska, A.; Kornicka, A. Synthesis and structure of novel hybrid compounds containing phthalazin-1(2H)-imine and 4,5-dihydro-1H-imidazole cores and their sulfonyl derivatives with potential biological activities. Int. J. Mol. Sci. 2024, 25, 11495. [Google Scholar] [CrossRef]
- Sączewski, F.; Gdaniec, M. Syntheses of novel fused heterocyclic systems by reactions of 1,2-dihydro-2-(4,5-dihydroimidazol-2-yl)phthalazin-l-o1 with active methylene compounds. Liebigs Ann. 1996, 10, 1487–1695. [Google Scholar]
- Layer, R.W. The chemistry of imines. Chem. Rev. 1963, 63, 489–510. [Google Scholar] [CrossRef]
- Cordes, E.H.; Jencks, W.P. On the mechanism of Schiff base formation and hydrolysis. J. Am. Chem. Soc. 1962, 84, 832–837. [Google Scholar] [CrossRef]
- Qi, J.; Zheng, Y.; Li, B.; Ai, Y.; Chen, M.; Zheng, X. Pyridoxal hydrochloride thiosemicarbazones with copper ions inhibit cell division via Topo-I and Topo-IIα. J. Inorg. Biochem. 2022, 232, 111816. [Google Scholar] [CrossRef]
- Gurgul, I.; Hricovíniová, J.; Mazuryk, O.; Hricovíniová, Z.; Brindell, M. Enhancement of the cytotoxicity of quinazolinone Schiff base derivatives with copper coordination. Inorganics 2023, 11, 391. [Google Scholar] [CrossRef]
- Gul, N.S.; Khan, T.M.; Chen, M.; Huang, K.B.; Hou, C.; Choudhary, M.I.; Liang, H.; Chen, Z.F. New copper complexes inducing bimodal death through apoptosis and autophagy in A549 cancer cells. J. Inorg. Biochem. 2020, 213, 111260. [Google Scholar] [CrossRef] [PubMed]
- Heffeter, P.; Jungwirth, U.; Jakupec, M.; Hartinger, C.; Galanski, M.; Elbling, L.; Micksche, M.; Keppler, B. Resistance against novel anticancer metal compounds: Differences and similarities. Drug Resist. 2008, 11, 1–16. [Google Scholar] [CrossRef]
- Zhao, L.; Chai, C.; Petz, W.; Frenking, G. Carbones and carbon atom as ligands in transition metal complexes. Molecules 2020, 25, 4943. [Google Scholar] [CrossRef]
- Cheng, P.; Liao, D.; Yan, S.; Jiang, Z.; Wang, G. Paramagnetic and diamagnetic binuclear copper(II) complexes with different bidentate exogenous bridges: Synthesis, spectroscopy and magnetic properties. Transit. Met. Chem. 1996, 21, 515–518. [Google Scholar] [CrossRef]
- Allen, H.; Baalham, C.A.; Lommerse, J.P.M.; Raithby, P.R. Carbonyl-carbonyl interactions can be competitive with hydrogen bonds. Acta Crystallogr. 1998, B54, 320–329. [Google Scholar] [CrossRef]
- Singh, S.K.; Das, A. The n→π* interaction: A rapidly emerging non-covalent interaction. Phys. Chem. Chem. Phys. 2015, 17, 9596–9612. [Google Scholar] [CrossRef]
- Newberry, R.W.; Raines, R.T. The n→π* interaction. Acc. Chem. Res. 2017, 50, 1838–1846. [Google Scholar] [CrossRef]
- Sahariah, B.; Sarma, B.K. Relative orientation of the carbonyl groups determines the nature of orbital interactions in carbonyl-carbonyl short contacts. Chem. Sci. 2019, 10, 909–917. [Google Scholar] [CrossRef]
- Spartan ‘08 for Windows, Wavefunction, 18401 Von Karman Ave., Suite 370, Irvine, CA 92612, USA. Available online: www.wavefunction.com (accessed on 1 December 2024).
- Storr, T.; Thompson, K.H.; Orvig, C. Design of targeting ligands in medicinal inorganic chemistry. Chem. Soc. Rev. 2006, 35, 534–544. [Google Scholar] [CrossRef] [PubMed]
- Ndagi, U.; Mhlongo, N.; Soliman, M.E. Metal complexes in cancer therapy—An update from drug de-sign perspective. Drug Des. Dev. Ther. 2017, 3, 599–616. [Google Scholar] [CrossRef]
- The European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 14.0. 2024. Available online: www.eucast.org (accessed on 1 December 2024).
- The European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs for Antifungal Agents, Version 10.0. 2020. Available online: www.eucast.org/astoffungi/clinicalbreakpointsforantifungals/ (accessed on 1 December 2024).
- Parker, A.; Lawson, M.A.E.; Vaux, L.; Pin, C. Host-Microbe interaction in the gastrointestinal tract. Environ. Microbiol. 2018, 20, 2337–2353. [Google Scholar] [CrossRef] [PubMed]
- Gomaa, E.Z. Human gut microbiota/microbiome in health and diseases: A review. Antonie Van Leeuwenhoek 2020, 113, 2019–2040. [Google Scholar] [CrossRef]
- Aburto, M.R.; Cryan, J.F. Gastrointestinal and brain barriers: Unlocking gates of communication across the microbiota-gut-brain axis. Nat. Rev. Gastroenterol. Hepatol. 2024, 21, 222–247. [Google Scholar] [CrossRef]
- Carloni, S.; Rescigno, M. The gut-brain vascular axis in neuroinflammation. Semin. Immunol. 2023, 69, 101802. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.X.; Yang, L.L.; Yao, X.Q. Gut microbiota-host lipid crosstalk in Alzheimer’s disease: Implications for disease progression and therapeutics. Mol. Neurodegener. 2024, 19, 35. [Google Scholar] [CrossRef]
- Wang, X.; Yuan, W.; Yang, C.; Wang, Z.; Zhang, J.; Xu, D.; Sun, X.; Sun, W. Emerging role of gut microbiota in autoimmune diseases. Front. Immunol. 2024, 15, 1365554. [Google Scholar] [CrossRef]
- Singh, K.; Bhori, M.; Kasu, Y.A.; Bhat, G.; Marar, T. Antioxidants as precision weapons in war against cancer chemotherapy induced toxicity—Exploring the armoury of obscurity. Saudi Pharm. J. 2018, 26, 177–190. [Google Scholar] [CrossRef]
- Marino, P.; Pepe, G.; Basilicata, M.G.; Vestuto, V.; Marzocco, S.; Autore, G.; Procino, A.; Gomez-Monterrey, I.M.; Manfra, M.; Campiglia, P. Potential role of natural antioxidant products in oncological diseases. Antioxidants 2023, 12, 704. [Google Scholar] [CrossRef]
- Luo, M.; Zhou, L.; Huang, Z.; Li, B.; Nice, E.C.; Xu, J.; Huang, C. Antioxidant therapy in cancer: Rationale and progress. Antioxidants 2022, 11, 1128. [Google Scholar] [CrossRef]
- Alsamri, H.; Al Dhaheri, Y.; Iratni, R. Targeting triple-negative breast cancer by the phytopolyphenol carnosol: ROS-dependent mechanisms. Antioxidants 2023, 12, 1349. [Google Scholar] [CrossRef] [PubMed]
- Alaouna, M.; Penny, C.; Hull, R.; Molefi, T.; Chauke-Malinga, N.; Khanyile, R.; Makgoka, M.; Bida, M.; Dlamini, Z. Overcoming the challenges of phytochemicals in triple negative breast cancer therapy: The path forward. Plants 2023, 12, 2350. [Google Scholar] [CrossRef] [PubMed]
- Seager, A.L.; Shah, U.K.; Mikhail, J.M.; Nelson, B.C.; Marquis, B.J.; Doak, S.H.; Johnson, G.E.; Griffiths, S.M.; Carmichael, P.L.; Scott, S.J.; et al. Pro-Oxidant induced DNA damage in human lymphoblastoid cells: Homeostatic mechanisms of genotoxic tolerance. Toxicol. Sci. 2012, 128, 387–397. [Google Scholar] [CrossRef]
- Martin-Cordero, C.; Leon-Gonzalez, A.J.; Calderon-Montano, J.M.; Burgos-Moron, E.; Lopez-Lazaro, M. Pro-Oxidant natural products as anticancer agents. Curr. Drug Targets 2012, 13, 1006–1028. [Google Scholar] [CrossRef] [PubMed]
- Prasanna, S.; Doerksen, R.J. Topological polar surface area: A useful descriptor in 2D-QSAR. Curr. Med. Chem. 2009, 16, 21–41. [Google Scholar] [CrossRef] [PubMed]
- Bytheway, I.; Darley, M.G.; Popelier, P.L. The calculation of polar surface area from first principles: An application of quantum chemical topology to drug design. ChemMedChem 2008, 3, 445–453. [Google Scholar] [CrossRef]
- Elmaidomy, A.H.; Mohammed, R.; Owis, A.I.; Hetta, M.H.; AboulMagd, A.M.; Siddique, A.B.; Abdelmohsen, U.R.; Rateb, M.E.; El Sayed, K.A.; Hassan, H.M. Triple-Negative breast cancer suppressive activities, antioxidants and pharmacophore model of new acylated rhamnopyranoses from Premna odorata. RSC Adv. 2020, 10, 10584–10598. [Google Scholar] [CrossRef]
- Rigaku Oxford Diffraction. CrysAlisPro Software System, version 1.171.38.43c; Rigaku Corporation: Oxford, UK, 2015.
- Sheldrick, G.M. SHELXT-Integrated Space-Group and Crystal-Structure Determination. Acta Crystallogr. 2015, A71, 3–8. [Google Scholar] [CrossRef]
- Sheldrick, G.M. Crystal structure refinement with SHELXL. Acta Crystallogr. 2015, C71, 3–8. [Google Scholar]
- Dolomanov, O.V.; Bourhis, L.J.; Gildea, R.J.; Howard, J.A.K.; Puschmann, H. OLEX2: A complete structure solution, refinement and analysis program. J. Appl. Crystallogr. 2009, 42, 339–341. [Google Scholar] [CrossRef]
- Hałasa, R.; Turecka, K.; Orlewska, C.; Werel, W. Comparison of fluorescence optical respirometry and microbroth dilution methods for testing antimicrobial compounds. J. Microbiol. Methods 2014, 107, 98–105. [Google Scholar] [CrossRef] [PubMed]
- Turecka, K.; Chylewska, A.; Kawiak, A.; Waleron, K. Antifungal activity and mechanism of action of the Co(III) coordination complexes with diamine chelate ligands against reference and clinical strains of Candida spp. Front. Microbiol. 2018, 9, 1594. [Google Scholar] [CrossRef] [PubMed]
- Bułakowska, A.; Sławiński, J.; Hałasa, R.; Hering, A.; Gucwa, M.; Ochocka, J.R.; Stefanowicz-Hajduk, J. An In Vitro Antimicrobial, Anticancer and Antioxidant Activity of N-[(2-Arylmethylthio)phenylsulfonyl]cinnamamide Derivatives. Molecules 2023, 28, 3087. [Google Scholar] [CrossRef] [PubMed]
- Thring, T.S.A.; Hili, P.; Naughton, D.P. Anti-collagenase, anti-elastase and anti-oxidant activities of extracts from 21 plants. BMC Complement. Altern. Med. 2009, 9, 27. [Google Scholar] [CrossRef]
- Ray, P.D.; Huang, B.W.; Tsuji, Y. Reactive oxygen species (ROS) homeostasis and redox regulation in cellular signaling. Cell. Signal. 2012, 24, 981–990. [Google Scholar] [CrossRef]
- Sandhaus, S.; Taylor, R.; Edwards, T.; Huddleston, A.; Wooten, Y.; Venkatraman, R.; Weber, R.T.; González-Sarrías, A.; Martin, P.M.; Cagle, P.; et al. A novel copper(II) complex identified as a potent drug against colorectal and breast cancer cells and as a poison inhibitor for human topoisomerase IIα. Inorg. Chem. Commun. 2016, 64, 45–49. [Google Scholar] [CrossRef]
- Singh, V.; Verma, S.; Fatima, F.; Samanta, S.K.; Varadwaj, P.K.; Sahoo, A.K. In silico study of a small bioactive molecule targeting topoisomerase II and p53-MDM2 complex in triple-negative breast cancer. ACS Omega 2023, 8, 38025–38037. [Google Scholar] [CrossRef]
- Myeza, N.; Slabber, C.; Munro, O.Q.; Sookai, S.; Zacharias, S.C.; Martins-Furness, C.; Harmse, L. An 8-aminoquinoline-naphthyl copper complex causes apoptotic cell death by modulating the expression of apoptotic regulatory proteins in breast cancer cells. Eur. J. Pharmacol. 2024, 978, 176764. [Google Scholar] [CrossRef]



| Compound Reference | L3 | C2 |
|---|---|---|
| Chemical formula | C18H14N4O2 | C18H14N4O2Cl2Cu |
| Formula mass | 318.33 | 452.77 |
| Crystal system | triclinic | triclinic |
| a/Å | 8.3830(3) | 7.38342(18) |
| b/Å | 8.7581(2) | 10.0507(3) |
| c/Å | 10.8035(3) | 12.8296(3) |
| α/° | 102.174(3) | 91.220(2) |
| β° | 102.877(3) | 91.9414(18) |
| ɣ° | 91.121(2) | 110.026(2) |
| Unit cell volume/Å3 | 754.00(4) | 893.44(4) |
| Space group | P-1 | P-1 |
| Z | 2 | 2 |
| Radiation type | Mo Kα | Cu Kα |
| Absorption coefficient, μ/mm−1 | 0.095 | 4.673 |
| No. of refl. measured, 2Θmax/° | 20,402, 52.7 | 22,327, 153.1 |
| No. of independent reflections | 3074 | 3714 |
| Rint | 0.0182 | 0.0276 |
| Final R1 values (I > 2σ(I)) | 0.0402 | 0.0266 |
| Final wR(F2) values (I > 2σ(I)) | 0.0906 | 0.0734 |
| Final R1 values (all data) | 0.0453 | 0.0285 |
| Final wR(F2) values (all data) | 0.1008 | 0.0751 |
| Goodness of fit on F2 | 1.037 | 1.059 |
| Compound Reference | L3 | C2 |
|---|---|---|
| dO···C (Å) | 2.9547(17) | 2.655(2) |
| ΘO···C=O (°) | 114.31(10) | 113.42(13) |
| TC=O···C=O (°) | 169.83(12) | 169.82(16) |
| Compound Reference | L3 | C2 |
|---|---|---|
| Bond lengths (Å) | ||
| C1-O16 | 1.2057(16) | 1.209(2) |
| C1-N2 | 1.3846(17) | 1.417(2) |
| N2-N3 | 1.3966(16) | 1.4006(18) |
| N2-C11 | 1.4142(17) | 1.397(2) |
| C11-N15 | 1.2612(18) | 1.283(2) |
| C11-N12 | 1.3963(17) | 1.367(2) |
| N12-C17 | 1.3788(18) | 1.408(2) |
| C17-O18 | 1.2146(16) | 1.207(2) |
| Cu1-Cl1 | 2.2262(5) | |
| Cu1-Cl2 | 2.2420(5) | |
| Cu1-N3 | 2.0844(13) | |
| Cu1-N15 | 1.9634(15) | |
| Bond angles (°) | ||
| C4 N3 N2 | 115.31(11) | 118.03(13) |
| C11 N15 C14 | 105.60(12) | 107.55(14) |
| C11 N2 C1 | 119.15(11) | 123.44(13) |
| N12 C11 N2 | 120.63(11) | 125.00(14) |
| C11 N12 C17 | 131.66(11) | 133.28(14) |
| Torsion angles (°) | ||
| N3 N2 C11 N15 | 55.60(18) | 14.3(2) |
| C11 N12 C17 C19 | 36.1(2) | 30.4(3) |
| N12 C17 C19 C24 | 37.58(19) | 24.7(2) |
| HeLa Cell Line | ||||||
|---|---|---|---|---|---|---|
| Conc. [μg/mL] | 1 | 10 | 25 | 50 | 100 | 200 |
| Compound | Mean % of Control | |||||
| L1 | 100 | 94.5 | 91.8 | 79.3 | 58.1 | 34.1 |
| C1 | 97.3 | 97.1 | 97.4 | 93.9 | 91.0 | 56.4 |
| L3 | 96.5 | 92.1 | 92.4 | 94.8 | 94.1 | 86.4 |
| C2 | 99.9 | 97.5 | 93.3 | 95.2 | 77.7 | 18.8 |
| Cisplatin* | 93.1 | 90.3 | 80.6 | 72.3 | 5.0 | 2.5 |
| MCF-7 Cell Line | ||||||
|---|---|---|---|---|---|---|
| Conc. [μg/mL] | 1 | 10 | 25 | 50 | 100 | 200 |
| Compound | Mean % of Control | |||||
| L1 | 97.3 | 97.2 | 93.7 | 86.4 | 62.1 | 45.7 |
| C1 | 100 | 100 | 100 | 97.9 | 86.2 | 49.2 |
| L3 | 95.9 | 100 | 97.2 | 99.4 | 96.5 | 95.6 |
| C2 | 100 | 100 | 96.4 | 92.5 | 73.7 | 42.5 |
| Cisplatin* | 100 | 96.4 | 81.9 | 64.1 | 18.8 | 6.5 |
| MDA-MB-231 Cell Line | ||||||
|---|---|---|---|---|---|---|
| Conc. [μg/mL] | 1 | 10 | 25 | 50 | 100 | 200 |
| Compound | Mean % of Control | |||||
| L1 | 99.3 | 97.8 | 95.0 | 81.4 | 52.7 | 35.2 |
| C1 | 90.5 | 88.8 | 89.6 | 81.7 | 80.5 | 18.7 |
| L3 | 97.2 | 97.6 | 96.6 | 94.9 | 96.2 | 96.7 |
| C2 | 96.2 | 97.5 | 93.5 | 88.9 | 70.5 | 23.6 |
| Cisplatin* | 98.2 | 87.7 | 82.4 | 57.6 | 31.9 | 22.6 |
| HDFa Cell Line | |||
|---|---|---|---|
| Conc. [μg/mL] | 50 | 100 | 200 |
| Compound | Mean % of Control | ||
| L1 | 85.6 | 81.3 | 84.8 |
| C1 | - | 86.9 | 75.6 |
| C2 | - | 78.8 | 68.1 |
| Cisplatin* | 76.8 | 26.8 | 4.1 |
| Compound | ABTS | DPPH | FRAP |
|---|---|---|---|
| L1 | 23.63 ± 0.28 | 197.89 ± 1.79 | NR * |
| L3 | 198.66 ± 1.33 | 427.34 ± 2.25 | 214.28 ± 0.2 |
| C1 | 253.92 ± 2.15 | 63.35 ± 0.25 | 156.01 ± 0.35 |
| C2 | 400.37 ± 0.87 | 78.33 ± 1.13 | 158.32 ± 0.21 |
| Ascorbic acid | 15.68 ± 0.8 | 12.75 ± 0.55 | 10.17 ± 0.51 |
| Physicochemical Properties | Lipophilicity | Water Sol. | Pharmacokinetics | Drug-Likeness | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mol. Wt. | ROTB (n) | HBA (n) | HBD (n) | TPSA | CLogP | Solubility Class | GI abs. | BBB perm. | CYP1A2 * | CYP2C19 * | Lipinski Filter | |
| Rule | <500 | <10 | <10 | <5 | - | <5 | - | - | - | - | - | - |
| L1 | 213.24 | 1 | 3 | 2 | 66.06 | 1.07 | soluble (v) | high | No | Yes | No | Yes(0) |
| L3 | 318.33 | 3 | 4 | 0 | 59.60 | 2.17 | soluble | high | No | Yes | Yes | Yes(0) |
| C1 | 347.69 | 1 | 3 | 2 | 66.05 | 1.41 | soluble | high | No | Yes | No | Yes(0) |
| C2 | 452.78 | 3 | 4 | 0 | 67.56 | 2.39 | soluble (m) | high | Yes | Yes | No | Yes(0) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Balewski, Ł.; Inkielewicz-Stępniak, I.; Gdaniec, M.; Turecka, K.; Hering, A.; Ordyszewska, A.; Kornicka, A. Synthesis, Structure, and Stability of Copper(II) Complexes Containing Imidazoline-Phthalazine Ligands with Potential Anticancer Activity. Pharmaceuticals 2025, 18, 375. https://doi.org/10.3390/ph18030375
Balewski Ł, Inkielewicz-Stępniak I, Gdaniec M, Turecka K, Hering A, Ordyszewska A, Kornicka A. Synthesis, Structure, and Stability of Copper(II) Complexes Containing Imidazoline-Phthalazine Ligands with Potential Anticancer Activity. Pharmaceuticals. 2025; 18(3):375. https://doi.org/10.3390/ph18030375
Chicago/Turabian StyleBalewski, Łukasz, Iwona Inkielewicz-Stępniak, Maria Gdaniec, Katarzyna Turecka, Anna Hering, Anna Ordyszewska, and Anita Kornicka. 2025. "Synthesis, Structure, and Stability of Copper(II) Complexes Containing Imidazoline-Phthalazine Ligands with Potential Anticancer Activity" Pharmaceuticals 18, no. 3: 375. https://doi.org/10.3390/ph18030375
APA StyleBalewski, Ł., Inkielewicz-Stępniak, I., Gdaniec, M., Turecka, K., Hering, A., Ordyszewska, A., & Kornicka, A. (2025). Synthesis, Structure, and Stability of Copper(II) Complexes Containing Imidazoline-Phthalazine Ligands with Potential Anticancer Activity. Pharmaceuticals, 18(3), 375. https://doi.org/10.3390/ph18030375








