Protein Levels of 16 Cytochrome P450s and 2 Carboxyl Esterases Using Absolute Quantitative Proteomics: CYP2C9 and CYP3A4 Are the Most Abundant Isoforms in Human Liver and Intestine, Respectively
Abstract
1. Introduction
2. Results
2.1. Patient Demographics
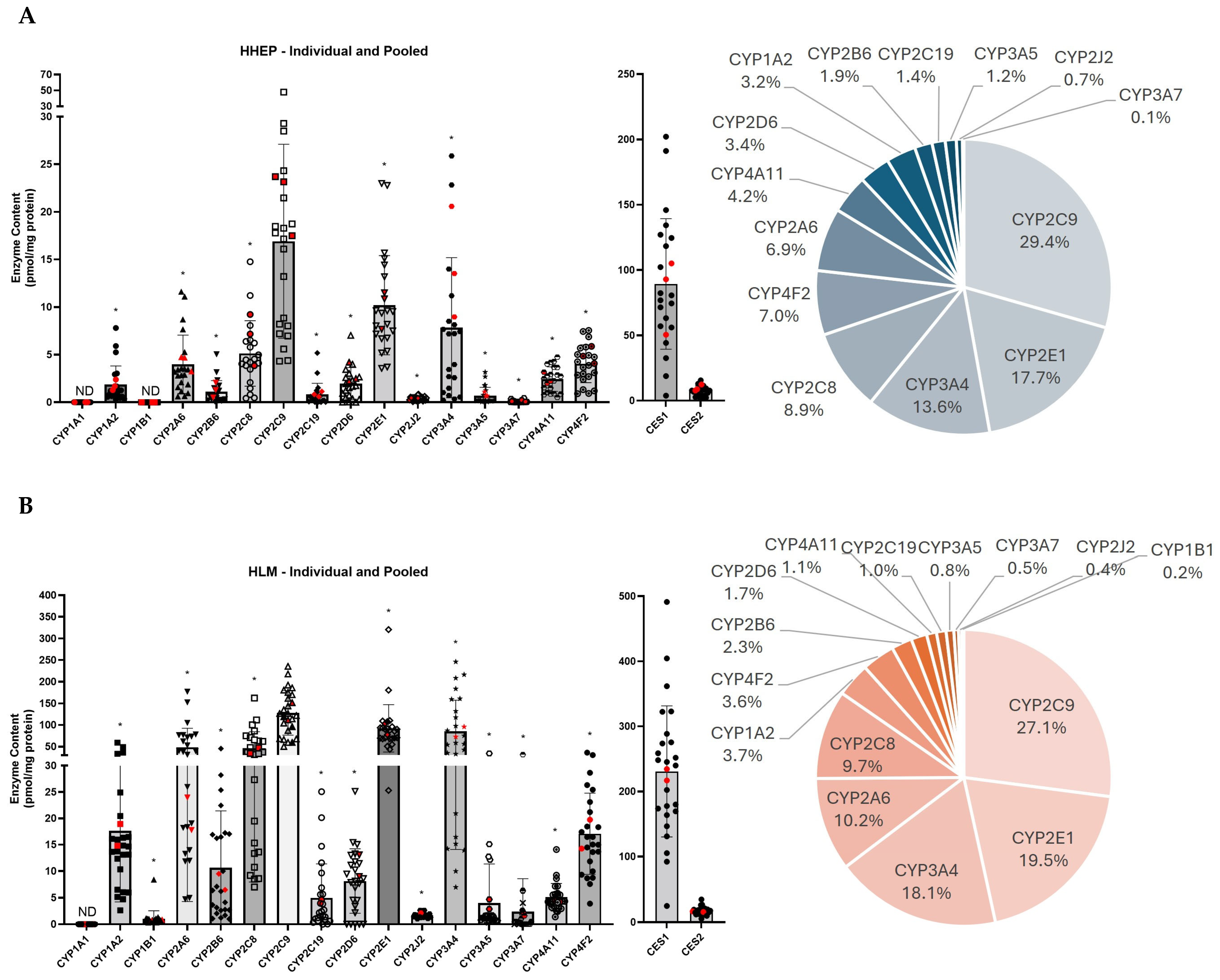
2.2. CYP2C9 Is the Most Expressed CYP450 in Two Different Hepatic Systems
2.3. Protein Expression and Enzyme Activity Are Correlated in Most Liver Samples
2.4. The CYP450 Profiles in Two Different Hepatic Systems Demonstrate High Correlation
3. Discussion
4. Materials and Methods
4.1. Chemicals and Reagents
4.2. Preparation of Human Hepatocyte and Intestinal Mucosa Lysate
4.3. Subjects
4.4. LC-MS/MS-Based Protein Quantification
4.5. Data and Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| A | Asian |
| AA | African-American |
| ACN | Acetonitrile |
| AN | Anoxia |
| AS | American Samoan |
| BSA | Bovine serum albumin |
| C | Caucasian |
| CES | Carboxyl esterase |
| CHIM | Cryopreserved human intestinal mucosa |
| CVA | Cerebrovascular accident |
| CYP450 | Cytochrome P450 |
| DDT | Dithiothreitol |
| F | Female |
| FA | Formic acid |
| GSW | Gunshot wound |
| H | Hispanic |
| H2O | Water |
| HHEP | Human hepatocytes |
| HLM | Human liver microsomes |
| HPLC/MS/MS | High-performance liquid chromatography/tandem mass spectrometry |
| HT | Head trauma |
| IAA | Iodoacetamide |
| LLOQ | Lower limit of quantification |
| M | Male |
| MPER | Mammalian protein extraction reagent |
| ND | Not detected |
| PBS | Phosphate-buffered saline |
| QC | Quality control |
| SD | Standard deviation |
| TFA | Trifluoroacetic acid |
| TPER | Tissue protein extraction reagent |
Appendix A. Supplementary Methods: CES1 and CES2 Quantification Method Validation
References
- Vaja, R.; Rana, M. Drugs and the liver. Anaesth. Intensive Care Med. 2020, 21, 517–523. [Google Scholar] [CrossRef]
- Susa, S.T.; Hussain, A.; Preuss, C.V. Drug Metabolism. In StatPearls [Internet]; StatPearls Publishing: Treasure Island, FL, USA, 2023. Available online: https://www.ncbi.nlm.nih.gov/books/NBK442023/ (accessed on 11 November 2025).
- Wang, D.; Zou, L.; Jin, Q.; Hou, J.; Ge, G.; Yang, L. Human Carboxylesterases: A Comprehensive Review. Acta Pharm. Sin. B 2018, 8, 699–712. [Google Scholar] [CrossRef]
- Achour, B.; Russell, M.R.; Barber, J.; Rostami-Hodjegan, A. Simultaneous Quantification of the Abundance of Several Cytochrome P450 and Uridine 5′-Diphospho-Glucuronosyltransferase Enzymes in Human Liver Microsomes Using Multiplexed Targeted Proteomics. Drug Metab. Dispos. 2014, 42, 500–510. [Google Scholar] [CrossRef]
- Li, A.P.; Alam, N.; Amaral, K.; Ho, M.D.; Loretz, C.; Mitchell, W.; Yang, Q. Cryopreserved Human Intestinal Mucosal Epithelium: A Novel in Vitro Experimental System for the Evaluation of Enteric Drug Metabolism, Cytochrome P450 Induction, and Enterotoxicity. Drug Metab. Dispos. 2018, 46, 1562–1571. [Google Scholar] [CrossRef]
- Li, A.P.; Ho, M.D.; Alam, N.; Mitchell, W.; Wong, S.; Yan, Z.; Kenny, J.R.; Hop, C.E.C.A. Inter-Individual and Inter-Regional Variations in Enteric Drug Metabolizing Enzyme Activities: Results with Cryopreserved Human Intestinal Mucosal Epithelia (Chim) from the Small Intestines of 14 Donors. Pharmacol. Res. Perspect. 2020, 8, e00645. [Google Scholar] [CrossRef]
- Basit, A.; Neradugomma, N.K.; Wolford, C.; Fan, P.W.; Murray, B.; Takahashi, R.H.; Khojasteh, S.C.; Smith, B.J.; Heyward, S.; Totah, R.A.; et al. Characterization of Differential Tissue Abundance of Major Non-Cyp Enzymes in Human. Mol. Pharm. 2020, 17, 4114–4124. [Google Scholar] [CrossRef]
- Ahire, D.S.; Basit, A.; Karasu, M.; Prasad, B. Ultrasensitive Quantification of Drug-Metabolizing Enzymes and Transporters in Small Sample Volume by Microflow Lc-Ms/Ms. J. Pharm. Sci. 2021, 110, 2833–2840. [Google Scholar] [CrossRef]
- Keefer, C.; Chang, G.; Carlo, A.; Novak, J.J.; Banker, M.; Carey, J.; Cianfrogna, J.; Eng, H.; Jagla, C.; Johnson, N.; et al. Mechanistic Insights on Clearance and Inhibition Discordance between Liver Microsomes and Hepatocytes When Clearance in Liver Microsomes Is Higher Than in Hepatocytes. Eur. J. Pharm. Sci. 2020, 155, 105541. [Google Scholar] [CrossRef]
- Di, L.; Keefer, C.; Scott, D.O.; Strelevitz, T.J.; Chang, G.; Bi, Y.A.; Lai, Y.; Duckworth, J.; Fenner, K.; Troutman, M.D.; et al. Mechanistic Insights from Comparing Intrinsic Clearance Values between Human Liver Microsomes and Hepatocytes to Guide Drug Design. Eur. J. Med. Chem. 2012, 57, 441–448. [Google Scholar] [CrossRef]
- Riley, R.J.; McGinnity, D.F.; Austin, R.P. A Unified Model for Predicting Human Hepatic, Metabolic Clearance from in Vitro Intrinsic Clearance Data in Hepatocytes and Microsomes. Drug Metab. Dispos. 2005, 33, 1304–1311. [Google Scholar] [CrossRef]
- Jia, L.; Liu, X. The Conduct of Drug Metabolism Studies Considered Good Practice (Ii): In Vitro Experiments. Curr. Drug Metab. 2007, 8, 822–829. [Google Scholar] [CrossRef]
- Palade, G.E.; Siekevitz, P. Liver Microsomes: An Integrated Morphological and Biochemical Study. J. Biophys. Biochem. Cytol. 1956, 2, 171–200. [Google Scholar] [CrossRef]
- Wisniewski, J.R.; Vildhede, A.; Noren, A.; Artursson, P. In-Depth Quantitative Analysis and Comparison of the Human Hepatocyte and Hepatoma Cell Line Hepg2 Proteomes. J. Proteom. 2016, 136, 234–247. [Google Scholar] [CrossRef]
- Maiers, J.L.; Malhi, H. Endoplasmic Reticulum Stress in Metabolic Liver Diseases and Hepatic Fibrosis. Semin. Liver Dis. 2019, 39, 235–248. [Google Scholar] [CrossRef]
- Xu, M.; Saxena, N.; Vrana, M.; Zhang, H.; Kumar, V.; Billington, S.; Khojasteh, C.; Heyward, S.; Unadkat, J.D.; Prasad, B. Targeted Lc-Ms/Ms Proteomics-Based Strategy to Characterize in Vitro Models Used in Drug Metabolism and Transport Studies. Anal. Chem. 2018, 90, 11873–11882. [Google Scholar] [CrossRef]
- Wisniewski, J.R.; Wegler, C.; Artursson, P. Subcellular Fractionation of Human Liver Reveals Limits in Global Proteomic Quantification from Isolated Fractions. Anal. Biochem. 2016, 509, 82–88. [Google Scholar] [CrossRef]
- Wegler, C.; Matsson, P.; Krogstad, V.; Urdzik, J.; Christensen, H.; Andersson, T.B.; Artursson, P. Influence of Proteome Profiles and Intracellular Drug Exposure on Differences in Cyp Activity in Donor-Matched Human Liver Microsomes and Hepatocytes. Mol. Pharm. 2021, 18, 1792–1805. [Google Scholar] [CrossRef]
- Ohtsuki, S.; Schaefer, O.; Kawakami, H.; Inoue, T.; Liehner, S.; Saito, A.; Ishiguro, N.; Kishimoto, W.; Ludwig-Schwellinger, E.; Ebner, T.; et al. Simultaneous Absolute Protein Quantification of Transporters, Cytochromes P450, and Udp-Glucuronosyltransferases as a Novel Approach for the Characterization of Individual Human Liver: Comparison with Mrna Levels and Activities. Drug Metab. Dispos. 2012, 40, 83–92. [Google Scholar] [CrossRef]
- Michaels, S.; Wang, M.Z. The Revised Human Liver Cytochrome P450 “Pie”: Absolute Protein Quantification of Cyp4f and Cyp3a Enzymes Using Targeted Quantitative Proteomics. Drug Metab. Dispos. 2014, 42, 1241–1251. [Google Scholar] [CrossRef]
- Shimada, T.; Yamazaki, H.; Mimura, M.; Inui, Y.; Guengerich, F.P. Interindividual Variations in Human Liver Cytochrome P-450 Enzymes Involved in the Oxidation of Drugs, Carcinogens and Toxic Chemicals: Studies with Liver Microsomes of 30 Japanese and 30 Caucasians. J. Pharmacol. Exp. Ther. 1994, 270, 414–423. [Google Scholar] [CrossRef]
- Taketani, M.; Shii, M.; Ohura, K.; Ninomiya, S.; Imai, T. Carboxylesterase in the Liver and Small Intestine of Experimental Animals and Human. Life Sci. 2007, 81, 924–932. [Google Scholar] [CrossRef]
- Grangeon, A.; Clermont, V.; Barama, A.; Gaudette, F.; Turgeon, J.; Michaud, V. Development and Validation of an Absolute Protein Assay for the Simultaneous Quantification of Fourteen Cyp450s in Human Microsomes by Hplc-Ms/Ms-Based Targeted Proteomics. J. Pharm. Biomed. Anal. 2019, 173, 96–107. [Google Scholar] [CrossRef]
- Drozdzik, M.; Busch, D.; Lapczuk, J.; Muller, J.; Ostrowski, M.; Kurzawski, M.; Oswald, S. Protein Abundance of Clinically Relevant Drug-Metabolizing Enzymes in the Human Liver and Intestine: A Comparative Analysis in Paired Tissue Specimens. Clin. Pharmacol. Ther. 2018, 104, 515–524. [Google Scholar] [CrossRef]
- Drozdzik, M.; Lapczuk-Romanska, J.; Wenzel, C.; Szelag-Pieniek, S.; Post, M.; Skalski, L.; Kurzawski, M.; Oswald, S. Gene Expression and Protein Abundance of Hepatic Drug Metabolizing Enzymes in Liver Pathology. Pharmaceutics 2021, 13, 1334. [Google Scholar] [CrossRef]
- Couto, N.; Al-Majdoub, Z.M.; Achour, B.; Wright, P.C.; Rostami-Hodjegan, A.; Barber, J. Quantification of Proteins Involved in Drug Metabolism and Disposition in the Human Liver Using Label-Free Global Proteomics. Mol. Pharm. 2019, 16, 632–647. [Google Scholar] [CrossRef]
- Langenfeld, E.; Zanger, U.M.; Jung, K.; Meyer, H.E.; Marcus, K. Mass Spectrometry-Based Absolute Quantification of Microsomal Cytochrome P450 2d6 in Human Liver. Proteomics 2009, 9, 2313–2323. [Google Scholar] [CrossRef]
- Seibert, C.; Davidson, B.R.; Fuller, B.J.; Patterson, L.H.; Griffiths, W.J.; Wang, Y. Multiple-Approaches to the Identification and Quantification of Cytochromes P450 in Human Liver Tissue by Mass Spectrometry. J. Proteome Res. 2009, 8, 1672–1681. [Google Scholar] [CrossRef]
- Wang, M.Z.; Wu, J.Q.; Dennison, J.B.; Bridges, A.S.; Hall, S.D.; Kornbluth, S.; Tidwell, R.R.; Smith, P.C.; Voyksner, R.D.; Paine, M.F.; et al. A Gel-Free Ms-Based Quantitative Proteomic Approach Accurately Measures Cytochrome P450 Protein Concentrations in Human Liver Microsomes. Proteomics 2008, 8, 4186–4196. [Google Scholar] [CrossRef]
- Vildhede, A.; Wisniewski, J.R.; Noren, A.; Karlgren, M.; Artursson, P. Comparative Proteomic Analysis of Human Liver Tissue and Isolated Hepatocytes with a Focus on Proteins Determining Drug Exposure. J. Proteome Res. 2015, 14, 3305–3314. [Google Scholar] [CrossRef]
- Sato, Y.; Miyashita, A.; Iwatsubo, T.; Usui, T. Simultaneous Absolute Protein Quantification of Carboxylesterases 1 and 2 in Human Liver Tissue Fractions Using Liquid Chromatography-Tandem Mass Spectrometry. Drug Metab. Dispos. 2012, 40, 1389–1396. [Google Scholar] [CrossRef]
- Grangeon, A.; Clermont, V.; Barama, A.; Gaudette, F.; Turgeon, J.; Michaud, V. Determination of Cyp450 Expression Levels in the Human Small Intestine by Mass Spectrometry-Based Targeted Proteomics. Int. J. Mol. Sci. 2021, 22, 12791. [Google Scholar] [CrossRef]
- Achour, B.; Barber, J.; Rostami-Hodjegan, A. Expression of Hepatic Drug-Metabolizing Cytochrome P450 Enzymes and Their Intercorrelations: A Meta-Analysis. Drug Metab. Dispos. 2014, 42, 1349–1356. [Google Scholar] [CrossRef] [PubMed]
- Krogstad, V.; Peric, A.; Robertsen, I.; Kringen, M.K.; Wegler, C.; Angeles, P.C.; Hjelmesæth, J.; Karlsson, C.; Andersson, S.; Artursson, P.; et al. A Comparative Analysis of Cytochrome P450 Activities in Paired Liver and Small Intestinal Samples from Patients with Obesity. Drug Metab. Dispos. 2020, 48, 8–17. [Google Scholar] [CrossRef] [PubMed]
- U.S. Department of Health and Human Services; Food and Drug Administration, Center for Drug Evaluation and Research (CDER), Center for Veterinary Medicine (CVM). Bioanalytical Method Validation—Guidance for Industry. 2018. Available online: https://www.fda.gov/media/70858/download (accessed on 11 November 2025).

| Subject | Type | Sex | Age (Years) | Ethnicity | Cause of Death |
|---|---|---|---|---|---|
| HHEP01 | Individual | M | 52 | C | AN |
| HHEP02 | Individual | M | 56 | C | HT |
| HHEP03 | Individual | F | 59 | C | CVA |
| HHEP04 | Individual | F | 48 | C | CVA |
| HHEP05 | Individual | F | 28 | AA | HT |
| HHEP06 | Individual | F | 71 | C | CVA |
| HHEP07 | Individual | F | 22 | C | CVA |
| HHEP08 | Individual | F | 70 | C | CVA |
| HHEP09 | Individual | F | 56 | C | AN |
| HHEP10 | Individual | M | 30 | C | CVA |
| HHEP11 | Individual | M | 55 | C | AN |
| HHEP12 | Individual | M | 45 | C | AN |
| HHEP13 | Individual | M | 61 | C | HT |
| HHEP14 | Individual | M | 31 | H | AN |
| HHEP15 | Individual | F | 56 | C | CVA |
| HHEP16 | Individual | F | 24 | C | HT |
| HHEP17 | Individual | F | 56 | C | CVA |
| HHEP18 | Individual | M | 43 | A | AN |
| HHEP19 | Individual | M | 44 | C | AN |
| HHEP20 | Individual | M | 41 | C | HT |
| HHEP21 | Pool | M (5), F (5) | 7–67 | AA (1), C (9) | AN (3), CVA (6), HT (1) |
| HHEP22 | Pool | M (5), F (5) | 27–67 | AA (2), C (8) | AN (2), CVA (5), HT (3) |
| HHEP23 | Pool | M (10), F (10) | 16–69 | AA (1), C (17), A (2) | AN (10), CVA (3), HT (7) |
| HLM01 | Individual | M | 44 | H | AN |
| HLM02 | Individual | F | 78 | C | CVA |
| HLM03 | Individual | F | 57 | C | HT |
| HLM04 | Individual | M | 71 | C | CVA |
| HLM05 | Individual | M | 21 | C | HT |
| HLM06 | Individual | F | 62 | C | AN |
| HLM07 | Individual | M | 49 | C | HT |
| HLM08 | Individual | M | 45 | C | CVA |
| HLM09 | Individual | F | 55 | C | CVA |
| HLM10 | Individual | F | 49 | C | AN |
| HLM11 | Individual | M | 41 | C | HT |
| HLM12 | Individual | F | 48 | C | CVA |
| HLM13 | Individual | M | 39 | C | HT |
| HLM14 | Individual | M | 39 | C | AN |
| HLM15 | Individual | M | 56 | AA | CVA |
| HLM16 | Individual | M | 35 | AA | CVA |
| HLM17 | Individual | M | 34 | C | GSW |
| HLM18 | Individual | F | 44 | C | CVA |
| HLM19 | Individual | F | 79 | C | CVA |
| HLM20 | Individual | F | 17 | C | AN |
| HLM21 | Individual | M | 48 | C | CVA |
| HLM22 | Individual | F | 34 | H | AN |
| HLM23 | Individual | M | 83 | C | CVA |
| HLM24 | Individual | M | 51 | C | HT |
| HLM25 | Pool | UltraPool HLM 150 | |||
| HLM26 | Pool | UltraPool HLM 150 | |||
| CHIM01 | Individual | F | 49 | C | CVA |
| CHIM02 | Individual | F | 59 | AS | CVA |
| CHIM03 | Individual | M | 38 | C | HT |
| CHIM04 | Individual | Unknown | Unknown | Unknown | Unknown |
| (A) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Individual HHEP | Pooled HHEP | |||||||||||||||||||
| Protein | Mean ± SD | Min–Max | N (20) | Protein | Mean ± SD | Min–Max | N (3) | |||||||||||||
| CYP1A1 | ND | 20 | CYP1A1 | ND | 3 | |||||||||||||||
| CYP1A2 | 1.88 ± 2.10 | 0.30–7.81 | 20 | CYP1A2 | 1.79 ± 0.56 | 1.28–2.39 | 3 | |||||||||||||
| CYP1B1 | ND | 20 | CYP1B1 | ND | 3 | |||||||||||||||
| CYP2A6 | 3.95 ± 3.29 | 0.61–11.6 | 20 | CYP2A6 | 4.23 ± 0.84 | 3.26–4.74 | 3 | |||||||||||||
| CYP2B6 | 1.07 ± 1.24 | 0.08–5.05 | 20 | CYP2B6 | 1.28 ± 0.77 | 0.51–2.06 | 3 | |||||||||||||
| CYP2C19 | 0.82 ± 1.26 | 0.02–51.8 | 20 | CYP2C19 | 0.85 ± 0.27 | 0.60–1.13 | 3 | |||||||||||||
| CYP2C8 | 4.89 ± 3.53 | 0.42–14.8 | 20 | CYP2C8 | 6.75 ± 2.71 | 3.84–9.22 | 3 | |||||||||||||
| CYP2C9 | 16.2 ± 10.7 | 4.32–47.8 | 20 | CYP2C9 | 21.5 ± 3.44 | 17.5–23.7 | 3 | |||||||||||||
| CYP2D6 | 1.80 ± 1.71 | 0.00–7.03 | 20 | CYP2D6 | 2.98 ± 1.07 | 2.24–4.21 | 3 | |||||||||||||
| CYP2E1 | 10.2 ± 5.57 | 3.56–23.0 | 20 | CYP2E1 | 10.0 ± 2.05 | 7.67–11.5 | 3 | |||||||||||||
| CYP2J2 | 0.39 ± 0.22 | 0.067–0.85 | 20 | CYP2J2 | 0.38 ± 0.11 | 0.25–0,46 | 3 | |||||||||||||
| CYP3A4 | 6.85 ± 7.16 | 0.23–25.9 | 20 | CYP3A4 | 14.4 ± 5.85 | 8.97–20.6 | 3 | |||||||||||||
| CYP3A5 | 0.67 ± 0.95 | 0.034–3.16 | 20 | CYP3A5 | 0.81 ± 0.25 | 0.64–1,10 | 3 | |||||||||||||
| CYP3A7 | 0.07 ± 0.12 | 0.00–0.44 | 20 | CYP3A7 | 0.13 ± 0.08 | 0.065–0,23 | 3 | |||||||||||||
| CYP4A11 | 2.43 ± 1.36 | 0.52–4.74 | 20 | CYP4A11 | 2.46 ± 0.62 | 2.0–3,16 | 3 | |||||||||||||
| CYP4F2 | 3.91 ± 2.09 | 0.90–7.55 | 20 | CYP4F2 | 4.93 ± 0.90 | 4.08–5.87 | 3 | |||||||||||||
| Total CYP450 | 55.8 ± 4.40 | Total CYP450 | 72.4 ± 6.04 | |||||||||||||||||
| CES1 | 90.4 ± 52.9 | 3.84–202 | 20 | CES1 | 82.8 ± 28.7 | 50.5–105.2 | 3 | |||||||||||||
| CES2 | 7.27 ± 3.70 | 1.84–15.6 | 20 | CES2 | 9.67 ± 2.45 | 7.78–12.4 | 3 | |||||||||||||
| Total CES | 97.7 ± 58.8 | Total CES | 92.5 ± 51.7 | |||||||||||||||||
| (B) | ||||||||||||||||||||
| Individual HLM | Pooled HLM | |||||||||||||||||||
| Protein | Mean ± SD | Min–Max | N(24) | Protein | Mean ± SD | Min–Max | N(2) | |||||||||||||
| CYP1A1 | ND | 24 | CYP1A1 | ND | 2 | |||||||||||||||
| CYP1A2 | 17.7 ± 14.0 | 2.62–59,2 | 24 | CYP1A2 | 16.9 ± 2.93 | 14.8–18.9 | 2 | |||||||||||||
| CYP1B1 | 1.03 ± 1.59 | 0.34–8.4 | 24 | CYP1B1 | 0.80 ± 0.12 | 0.72–0.89 | 2 | |||||||||||||
| CYP2A6 | 50.6 ± 45.1 | 4.69–177 | 24 | CYP2A6 | 20.9 ± 4.36 | 17.8–24.0 | 2 | |||||||||||||
| CYP2B6 | 10.9 ± 11.2 | 1.01–45.3 | 24 | CYP2B6 | 7.99 ± 2.12 | 6.5–9.5 | 2 | |||||||||||||
| CYP2C19 | 5.02 ± 6.68 | 0.02–25.0 | 24 | CYP2C19 | 4.27 ± 0.46 | 3.9–4.6 | 2 | |||||||||||||
| CYP2C8 | 46.6 ± 39.7 | 7.00–162 | 24 | CYP2C8 | 40.6 ± 9.15 | 34.1–47.0 | 2 | |||||||||||||
| CYP2C9 | 127 ± 53.3 | 50.3–236 | 24 | CYP2C9 | 132 ± 28.2 | 112–152 | 2 | |||||||||||||
| CYP2D6 | 7.90 ± 6.24 | 0.00–25.1 | 24 | CYP2D6 | 11.0 ± 2.88 | 9.0–13.1 | 2 | |||||||||||||
| CYP2E1 | 92.3 ± 56.9 | 25.3–321 | 24 | CYP2E1 | 90.4 ± 18.3 | 77.5–103 | 2 | |||||||||||||
| CYP2J2 | 1.70 ± 0.48 | 1.02–2,6 | 24 | CYP2J2 | 1.82 ± 0.44 | 1.52–2.1 | 2 | |||||||||||||
| CYP3A4 | 85.8 ± 74.6 | 6.96–246.2 | 24 | CYP3A4 | 84.1 ± 16.4 | 72.5–95.7 | 2 | |||||||||||||
| CYP3A5 | 4.00 ± 7.66 | 0.31–34.2 | 24 | CYP3A5 | 3.77 ± 1.28 | 2.86–4.7 | 2 | |||||||||||||
| CYP3A7 | 2.41 ± 6.45 | 0.00–31.4 | 24 | CYP3A7 | 1.95 ± 0.46 | 1.63–2.3 | 2 | |||||||||||||
| CYP4A11 | 5.16 ± 2.69 | 1.41–1.41 | 24 | CYP4A11 | 4.3 ± 0.12 | 4.2–4.4 | 2 | |||||||||||||
| CYP4F2 | 17.1 ± 7.98 | 3.89–36.0 | 24 | CYP4F2 | 17.0 ± 3.88 | 14.2–19.7 | 2 | |||||||||||||
| Total CYP450 | 475 ± 39.7 | Total CYP450 | 437 ± 39.6 | |||||||||||||||||
| CES1 | 231 ± 105 | 24.8–491 | 24 | CES1 | 226 ± 12.3 | 217–234 | 2 | |||||||||||||
| CES2 | 16.7 ± 6.18 | 5.37–34.6 | 24 | CES2 | 16.0 ± 0.67 | 15.5–16.5 | 2 | |||||||||||||
| Total CES | 248 ± 152 | Total CES | 242 ± 148 | |||||||||||||||||
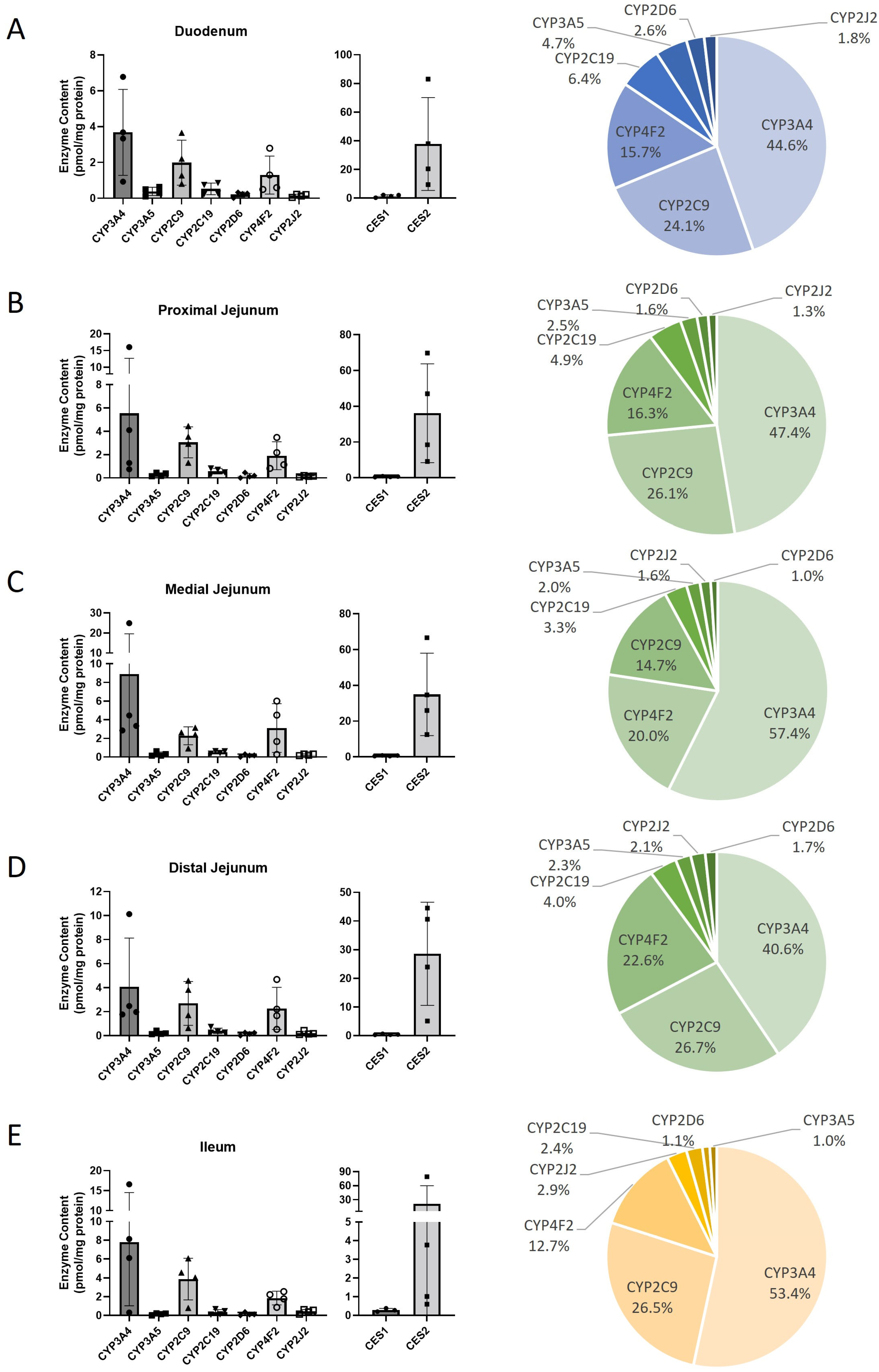
| (C) | ||||||||||||||||||||
| Protein | Duodenum | Proximal jejunum | Medial jejunum | Distal jejunum | Ileum | |||||||||||||||
| Mean ± SD | Min–Max | N (4) | Mean ± SD | Min–Max | N (4) | Mean ± SD | Min–Max | n (4) | Mean ± SD | Min–Max | N (4) | Mean ± SD | Min–Max | N (4) | ||||||
| CYP1A1 | ND | 4 | ND | 4 | ND | 4 | ND | 4 | ND | 4 | ||||||||||
| CYP1A2 | ND | 4 | ND | 4 | ND | 4 | ND | 4 | ND | 4 | ||||||||||
| CYP1B1 | ND | 4 | ND | 4 | ND | 4 | ND | 4 | ND | 4 | ||||||||||
| CYP2A6 | ND | 4 | ND | 4 | ND | 4 | ND | 4 | ND | 4 | ||||||||||
| CYP2B6 | ND | 4 | ND | 4 | ND | 4 | ND | 4 | ND | 4 | ||||||||||
| CYP2C19 | 0.53 ± 0.32 | 0.23–0.86 | 4 | 0.58 ± 0.24 | 0.30–0.82 | 4 | 0.51 ± 0.14 | 0.36–0.63 | 4 | 0.40 ± 0.23 | 0.26–0.74 | 4 | ||||||||
| CYP2C8 | ND | 4 | ND | 4 | ND | 4 | ND | 4 | ND | 4 | ||||||||||
| CYP2C9 | 1.99 ± 1.26 | 0.81–3.66 | 4 | 3.06 ± 1.33 | 1.30–4.45 | 4 | 2.28 ± 0.95 | 0.92–3.14 | 4 | 2.69 ± 1.83 | 0.64–4.58 | 4 | ||||||||
| CYP2D6 | 0.22 ± 0.14 | 0.02–0.33 | 4 | 0.19 ± 0.19 | 0.00–0.46 | 3 | 0.16 ± 0.12 | 0.01–0.31 | 4 | 0.17 ± 0.12 | 0.01–0.27 | 4 | ||||||||
| CYP2E1 | ND | 4 | ND | 4 | ND | 4 | ND | 4 | ND | 4 | ||||||||||
| CYP2J2 | 0.15 ± 0.08 | 0.05–0.22 | 4 | 0.15 ± 0.05 | 0.08–0.20 | 4 | 0.24 ± 0.09 | 0.14–0.32 | 4 | 0.22 ± 0.15 | 0.11–0.43 | 4 | ||||||||
| CYP3A4 | 3.68 ± 2.4 | 0.93–6.77 | 4 | 5.55 ± 7.15 | 0.75–16.1 | 4 | 8.89 ± 10.7 | 2.85–24.9 | 4 | 4.09 ± 4.04 | 1.78–10.1 | 4 | ||||||||
| CYP3A5 | 0.39 ± 0.23 | 0.11–0.64 | 4 | 0.29 ± 0.15 | 0.15–0.45 | 4 | 0.31 ± 0.23 | 0.14–0.31 | 4 | 0.23 ± 0.13 | 0.13–0.42 | 4 | ||||||||
| CYP3A7 | ND | 4 | ND | 4 | ND | 4 | ND | 4 | ND | 4 | ||||||||||
| CYP4A11 | ND | 4 | ND | 4 | ND | 4 | ND | 4 | ND | 4 | ||||||||||
| CYP4F2 | 1.3 ± 1.06 | 0.50–2.79 | 4 | 1.9 ± 1.2 | 0.82–3.47 | 4 | 3.11 ± 2.61 | 0.27–6.0 | 4 | 2.28 ± 1.75 | 0.54–4.68 | 4 | ||||||||
| Total CYP450 | 8.25 ± 1.16 | 11.7 ± 1.78 | 15.5 ± 2.69 | 10.07 ± 1.42 | 14.58 ± 2.45 | |||||||||||||||
| CES1 | 1.51 ± 0.87 | 0.33–2.21 | 4 | 0.60 ± 0.24 | 0.36–0.91 | 4 | 0.61 ± 0.21 | 0.43–0.82 | 4 | 0.40 ± 0.16 | 0.29–0.64 | 4 | ||||||||
| CES2 | 37.73 ± 32.4 | 9.49–83.0 | 4 | 36.1 ± 27.6 | 9.18–69.8 | 4 | 34.9 ± 23.0 | 12.5–66.7 | 4 | 28.6 ± 18.0 | 5.11–44.5 | 4 | ||||||||
| Total CES | 39.2 ± 25.6 | 36.7 ± 25.1 | 35.5 ± 24.3 | 28.9 ± 19.9 | 21.4 ± 14.7 | |||||||||||||||
| (A) | ||||
| CYP450 | Probe | r | p | N |
| CYP1A2 | Phenacetin | 0.788 | <0.0001 | 20 |
| CYP2A6 | Coumarin | 0.7985 | <0.0001 | 20 |
| CYP2B6 | Bupropion | 0.7424 | 0.0002 | 20 |
| CYP2C8 | Amodiaquine | 0.7574 | 0.0001 | 20 |
| CYP2C9 | Diclofenac | 0.3805 | 0.098 | 20 |
| CYP2C19 | Mephenytoin | 0.7003 | 0.0006 | 20 |
| CYP2D6 | Dextromethorphan | 0.8015 | <0.0001 | 20 |
| CYP2E1 | Chlorzoxazone | 0.2915 | 0.2124 | 20 |
| CYP3A4 | Midazolam | 0.5353 | 0.015 | 20 |
| CYP3A4 | Testosterone | 0.7684 | <0.0001 | 20 |
| (B) | ||||
| CYP450 | Probe | r | p | N |
| CYP1A2 | Phenacetin | 0.6814 | 0.0046 | 16 |
| CYP2A6 | Coumarin | 0.9471 | <0.0001 | 16 |
| CYP2B6 | Bupropion | 0.9794 | <0.0001 | 16 |
| CYP2C8 | Amodiaquine | 0.9176 | <0.0001 | 16 |
| CYP2C9 | Diclofenac | 0.65 | 0.0078 | 16 |
| CYP2C19 | Mephenytoin | 0.95 | <0.0001 | 16 |
| CYP2D6 | Dextromethorphan | 0.8291 | 0.0002 | 16 |
| CYP2E1 | Chlorzoxazone | 0.9441 | <0.0001 | 16 |
| CYP3A4 | Midazolam | 0.8168 | 0.0002 | 16 |
| CYP3A4 | Testosterone | 0.9559 | <0.0001 | 16 |
| CYP4A11 | Lauric Acid | 0.7176 | 0.0024 | 16 |
| (C) | ||||
| CYP450/Enzyme | Probe | r | p | N |
| CYP2C9 | Diclofenac | −0.3071 | 0.265 | 15 |
| CYP2C19 | Mephenytoin | 0.7036 | 0.0045 | 15 |
| CYP2D6 | Dextromethorphan | 0.5771 | 0.0266 | 15 |
| CYP3A4 | Midazolam | 0.1321 | 0.6389 | 15 |
| CYP3A4 | Testosterone | 0.2321 | 0.4039 | 15 |
| CYP2J2 | Astemizole | 0.1679 | 0.5492 | 15 |
| CES2 | Irinotecan | −0.0643 | 0.8225 | 15 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Grangeon, A.; Arwood, M.L.; Thacker, D.; Gaudette, F.; Turgeon, J.; Michaud, V. Protein Levels of 16 Cytochrome P450s and 2 Carboxyl Esterases Using Absolute Quantitative Proteomics: CYP2C9 and CYP3A4 Are the Most Abundant Isoforms in Human Liver and Intestine, Respectively. Pharmaceuticals 2025, 18, 1789. https://doi.org/10.3390/ph18121789
Grangeon A, Arwood ML, Thacker D, Gaudette F, Turgeon J, Michaud V. Protein Levels of 16 Cytochrome P450s and 2 Carboxyl Esterases Using Absolute Quantitative Proteomics: CYP2C9 and CYP3A4 Are the Most Abundant Isoforms in Human Liver and Intestine, Respectively. Pharmaceuticals. 2025; 18(12):1789. https://doi.org/10.3390/ph18121789
Chicago/Turabian StyleGrangeon, Alexia, Matthew L. Arwood, David Thacker, Fleur Gaudette, Jacques Turgeon, and Veronique Michaud. 2025. "Protein Levels of 16 Cytochrome P450s and 2 Carboxyl Esterases Using Absolute Quantitative Proteomics: CYP2C9 and CYP3A4 Are the Most Abundant Isoforms in Human Liver and Intestine, Respectively" Pharmaceuticals 18, no. 12: 1789. https://doi.org/10.3390/ph18121789
APA StyleGrangeon, A., Arwood, M. L., Thacker, D., Gaudette, F., Turgeon, J., & Michaud, V. (2025). Protein Levels of 16 Cytochrome P450s and 2 Carboxyl Esterases Using Absolute Quantitative Proteomics: CYP2C9 and CYP3A4 Are the Most Abundant Isoforms in Human Liver and Intestine, Respectively. Pharmaceuticals, 18(12), 1789. https://doi.org/10.3390/ph18121789









