Design, Synthesis, and Biological Evaluation of 5′,7-Disubstituted 7-Deaza-adenosine Analogues as Irreversible Pan-FGFR Inhibitors
Abstract
1. Introduction
2. Results and Discussion
2.1. Chemistry
2.2. Biological Evaluation
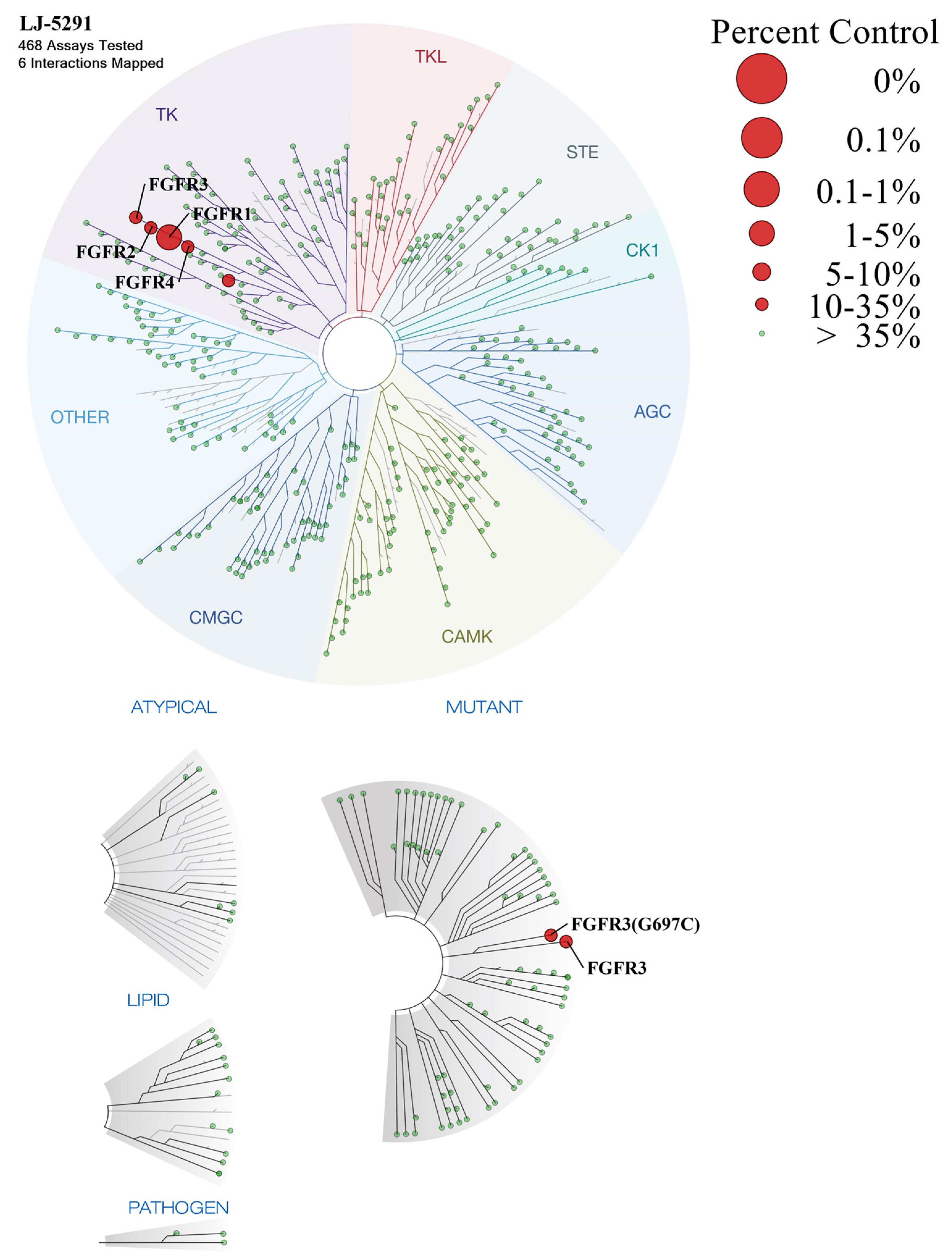
2.2.1. Biochemical Potencies of Compounds 5a–d, 6a–h
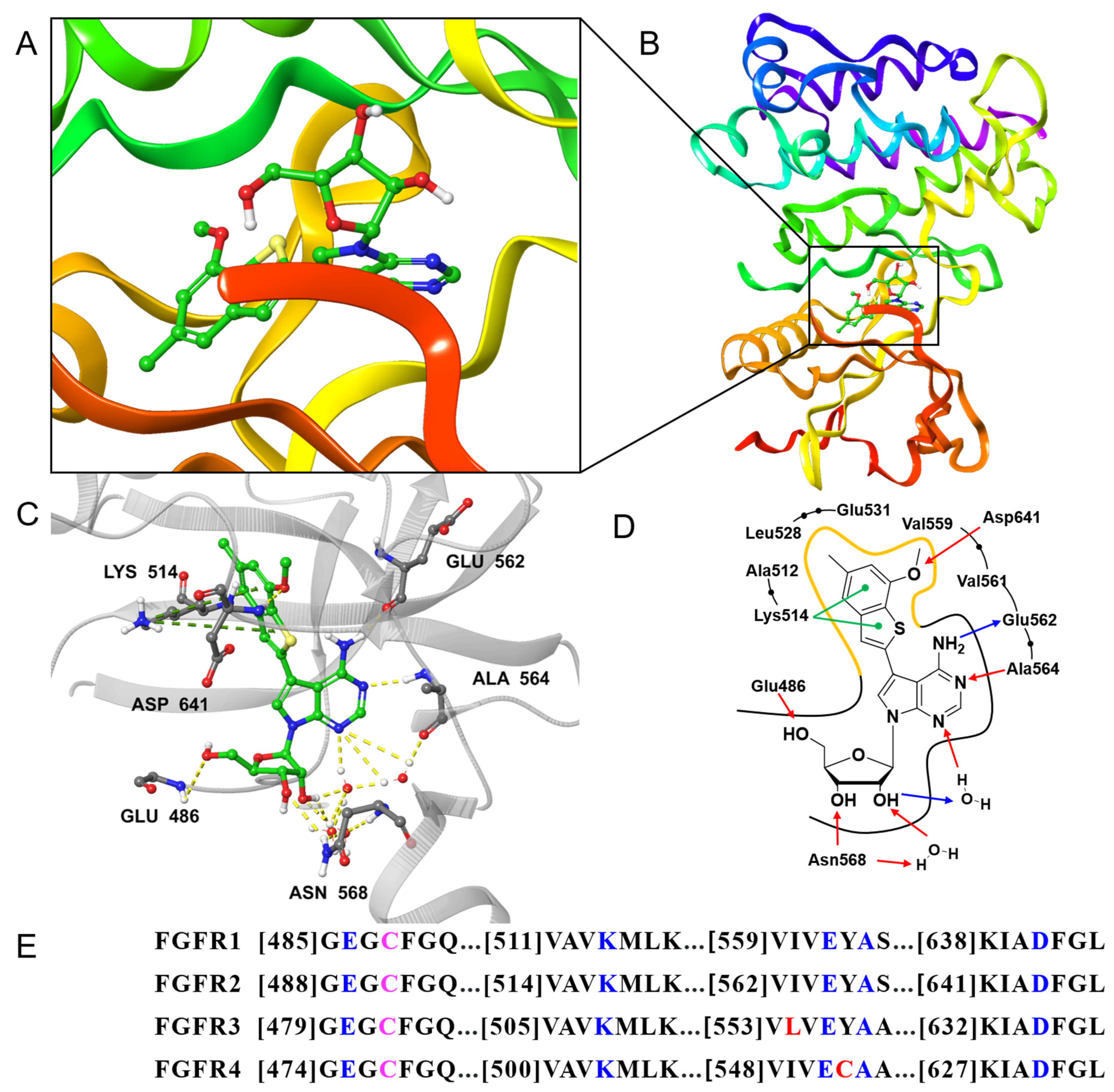
2.2.2. Crystal Structure of FGFR1 in Complex with Compound 6h
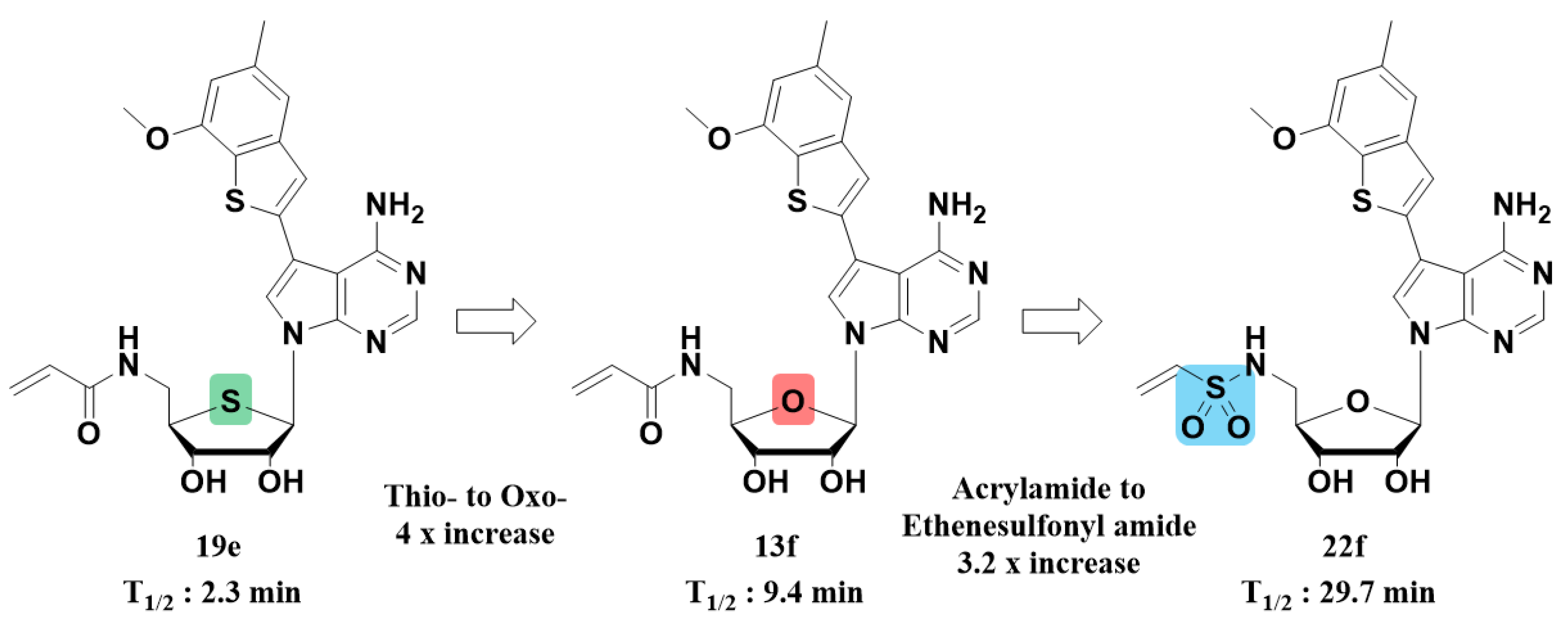
2.2.3. Structures, Biochemical Potencies, and Antiproliferation Efficacy of Compounds 13a–f, ent-13f, 19a–e
2.2.4. Structures, Biochemical Potencies, and Antiproliferation Efficacy of Compounds 21, and 22a–f
2.2.5. Anticancer Activity of Compounds 13a–f, 19b–e, 21, and 22a–f
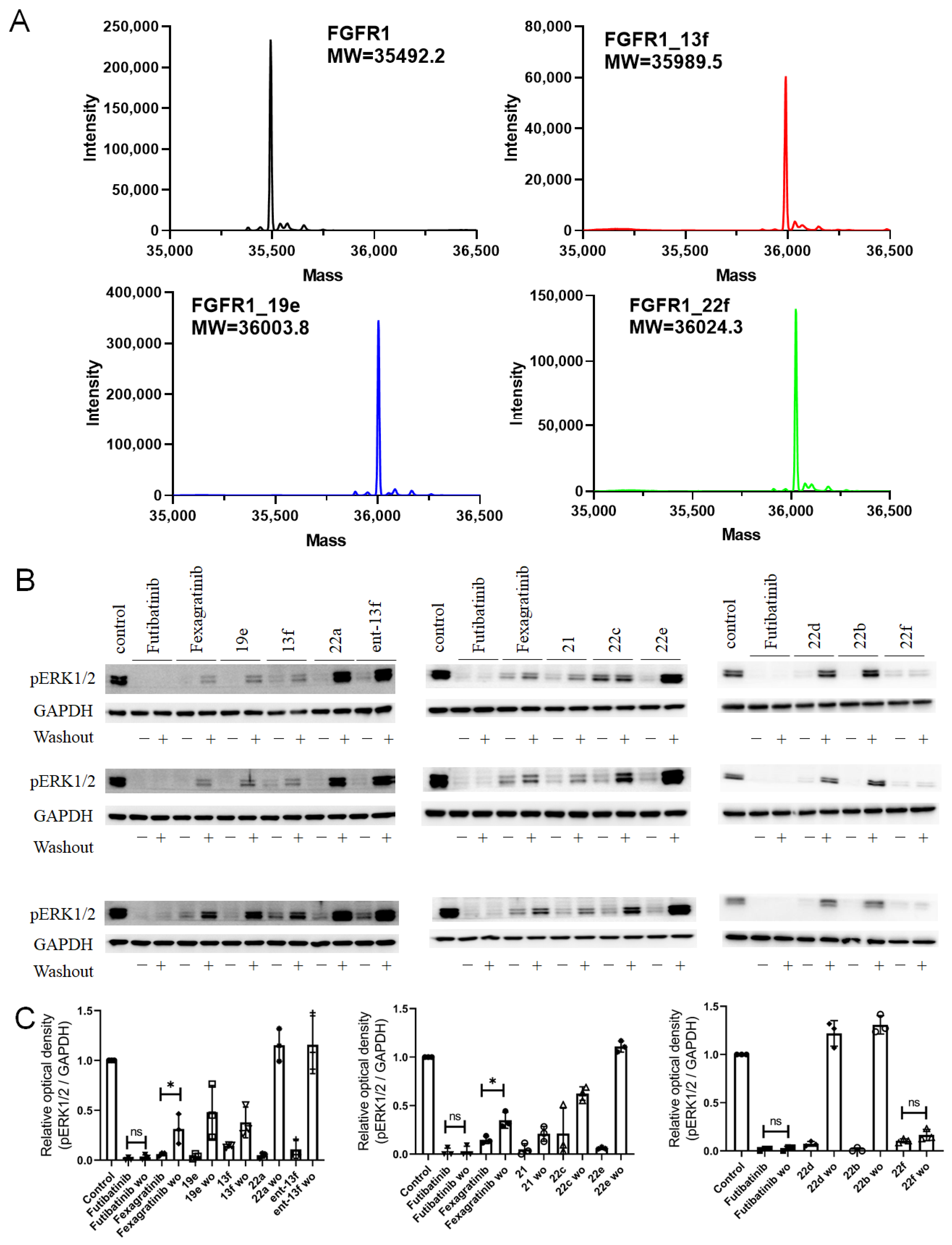
2.2.6. Covalent Binding Confirmation via Intact Mass Spectrometry and Western Blot Analysis
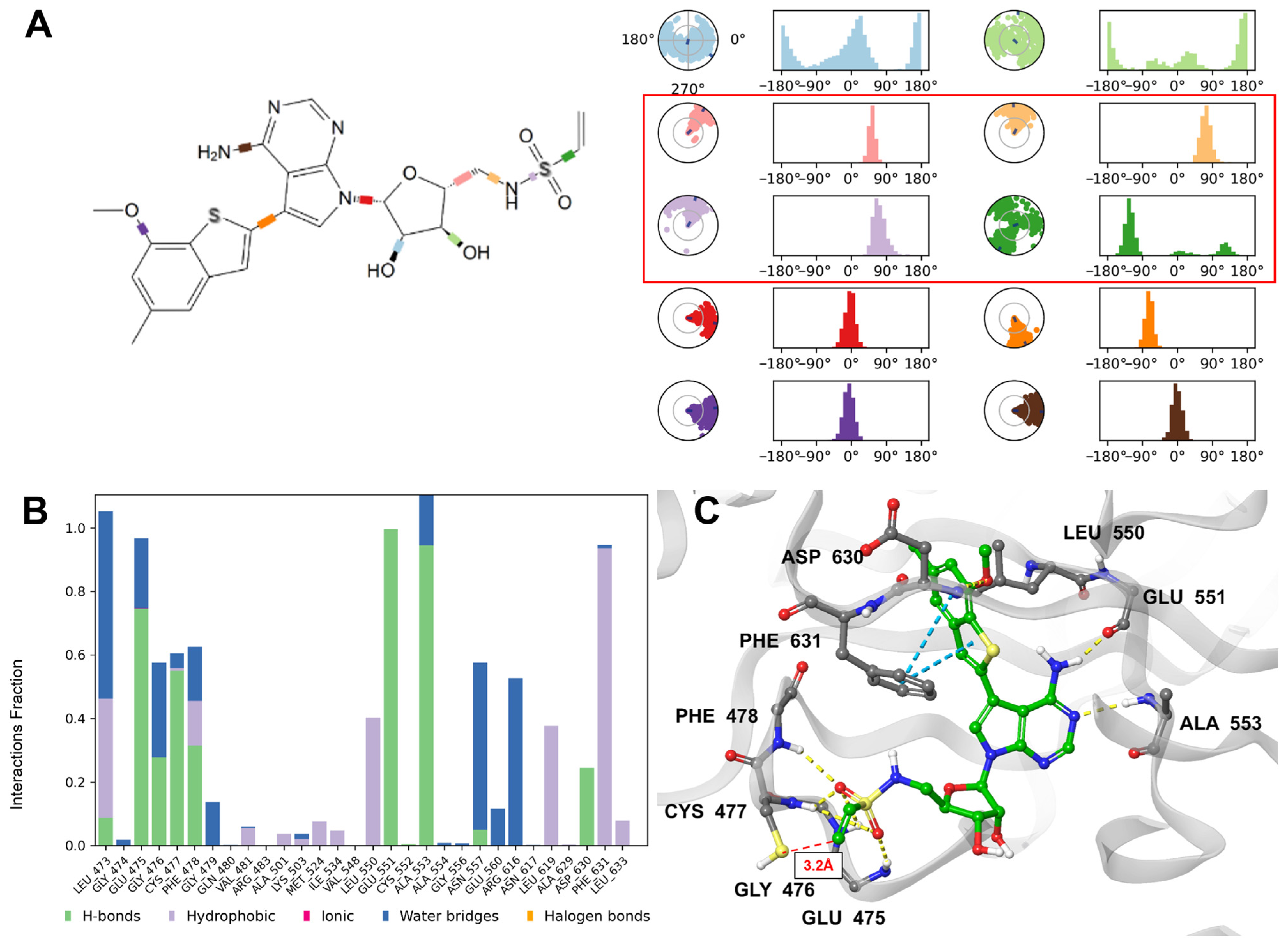
2.2.7. Analysis of Molecular Dynamics
2.2.8. In Vitro ADME /Tox Profiling of Compound 22f
3. Materials and Methods
3.1. Chemistry
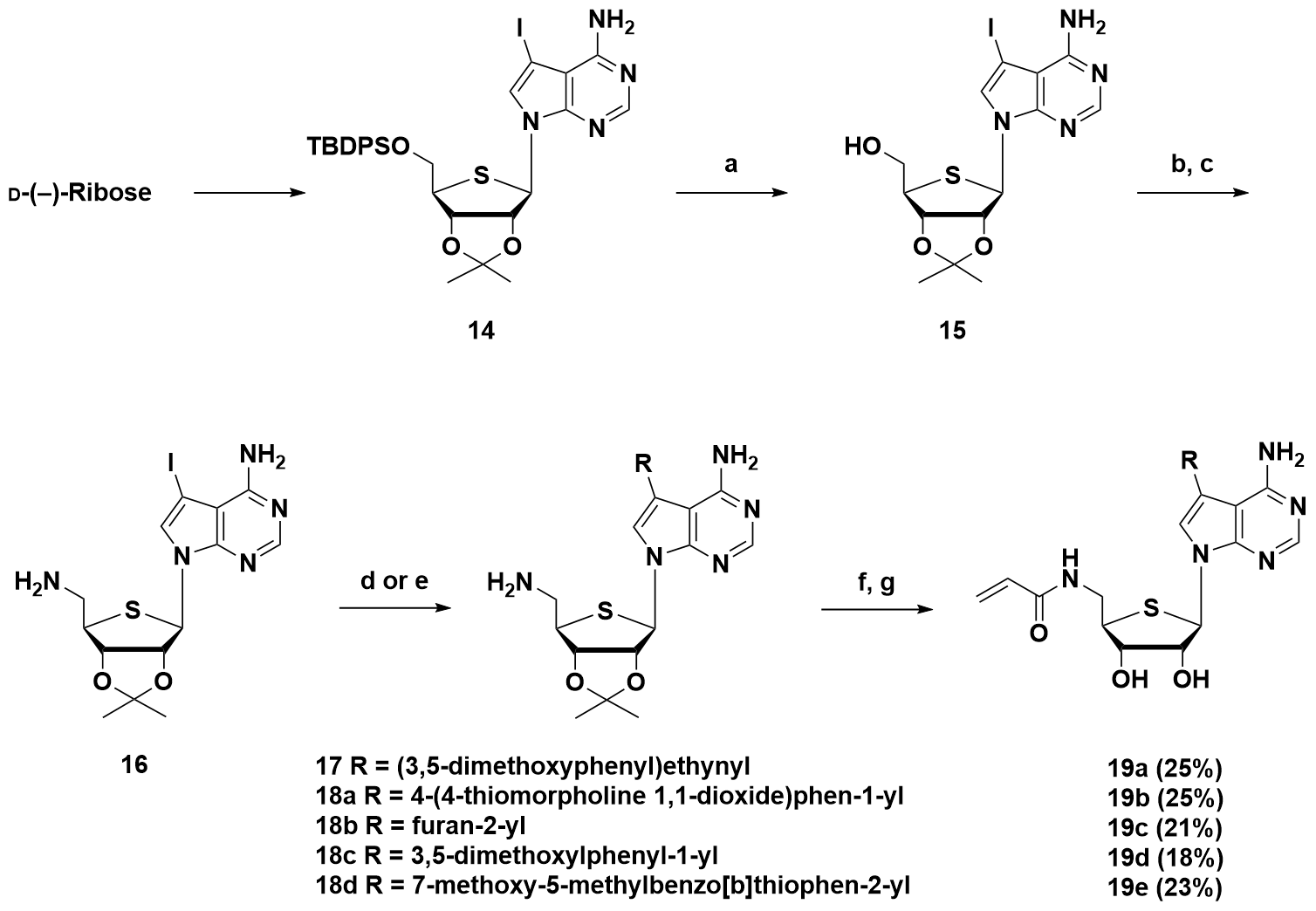
3.1.1. Synthesis of Compound 14
3.1.2. Synthesis of 7-Substituted-7-deaza Adenosine Derivatives (5a–d, 6a–h)
(3R,4S,5R)-5-(((Tert-butyldimethylsilyl)oxy)methyl)tetrahydrofuran-2,3,4-triol (1) and (3R,4S,5S)-5-(((Tert-butyldimethylsilyl)oxy)methyl)tetrahydrofuran-2,3,4-triol (ent-1)
(2R,3S,4R,5R)-2-(((Tert-butyldimethylsilyl)oxy)methyl)-5-(4-chloro-5-iodo-7H-pyrrolo[2,3-d]pyrimidin-7-yl)tetrahydrofuran-3,4-diol (2) and (2S,3S,4R,5S)-2-(((Tert-butyldimethylsilyl)oxy)methyl)-5-(4-chloro-5-iodo-7H-pyrrolo[2,3-d]pyrimidin-7-yl)tetrahydrofuran-3,4-diol (ent-2)
(2R,3R,4S,5R)-2-(4-Amino-5-iodo-7H-pyrrolo[2,3-d]pyrimidin-7-yl)-5-(((tert-butyldimethylsilyl)oxy)methyl)tetrahydrofuran-3,4-diol (3)
(2R,3R,4S,5R)-2-(4-Amino-5-iodo-7H-pyrrolo[2,3-d]pyrimidin-7-yl)-5-(hydroxymethyl)tetrahydrofuran-3,4-diol (4)
3.1.3. General Procedure for Sonogashira Coupling for the Preparation of 5a–d
(2R,3R,4S,5R)-2-(4-Amino-5-(phenylethynyl)-7H-pyrrolo[2,3-d]pyrimidin-7-yl)-5-(hydroxymethyl)tetrahydrofuran-3,4-diol (5a)
(2R,3R,4S,5R)-2-(4-Amino-5-((3-methoxy-5-methylphenyl)ethynyl)-7H-pyrrolo[2,3-d]pyrimidin-7-yl)-5-(hydroxymethyl)tetrahydrofuran-3,4-diol (5b)
(2R,3R,4S,5R)-2-(4-Amino-5-((3,5-dimethoxyphenyl)ethynyl)-7H-pyrrolo[2,3-d]pyrimidin-7-yl)-5-(hydroxymethyl)tetrahydrofuran-3,4-diol (5c)
(2R,3R,4S,5R)-2-(4-Amino-5-((3-methoxy-5-methylphenyl)ethynyl)-7H-pyrrolo[2,3-d]pyrimidin-7-yl)-5-(hydroxymethyl)tetrahydrofuran-3,4-diol (5d)
3.1.4. General Procedure for Suzuki Coupling for the Preparation of 6a–h
(2R,3R,4S,5R)-2-(5-([1,1′-Biphenyl]-4-yl)-4-amino-7H-pyrrolo[2,3-d]pyrimidin-7-yl)-5-(hydroxymethyl)tetrahydrofuran-3,4-diol (6a)
(2R,3R,4S,5R)-2-(4-Amino-5-(3,5-dimethoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-7-yl)-5-(hydroxymethyl)tetrahydrofuran-3,4-diol (6b)
(2R,3R,4S,5R)-2-(4-Amino-5-(thiophen-2-yl)-7H-pyrrolo[2,3-d]pyrimidin-7-yl)-5-(hydroxymethyl)tetrahydrofuran-3,4-diol (6c)
(2R,3R,4S,5R)-2-(4-Amino-5-(benzofuran-2-yl)-7H-pyrrolo[2,3-d]pyrimidin-7-yl)-5-(hydroxymethyl)tetrahydrofuran-3,4-diol (6d)
(2R,3R,4S,5R)-2-(4-Amino-5-(benzo[b]thiophen-2-yl)-7H-pyrrolo[2,3-d]pyrimidin-7-yl)-5-(hydroxymethyl)tetrahydrofuran-3,4-diol (6e)
(2R,3R,4S,5R)-2-(4-Amino-5-(1H-indol-2-yl)-7H-pyrrolo[2,3-d]pyrimidin-7-yl)-5-(hydroxymethyl)tetrahydrofuran-3,4-diol (6f)
5-(4-Amino-7-((2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)tetrahydrofuran-2-yl)-7H-pyrrolo[2,3-d]pyrimidin-5-yl)thiophene-2-carbaldehyde (6g)
(2R,3R,4S,5R)-2-(4-Amino-5-(7-methoxy-5-methylbenzo[b]thiophen-2-yl)-7H-pyrrolo[2,3-d]pyrimidin-7-yl)-5-(hydroxymethyl)tetrahydrofuran-3,4-diol (6h)
3.1.5. Synthesis of Enantiomer of 7-Substituted-7-deaza adenosine Derivatives
7-((3aR,4R,6R,6aR)-6-(((Tert-butyldimethylsilyl)oxy)methyl)-2,2-dimethyltetrahydrofuro[3,4-d][1,3]dioxol-4-yl)-4-chloro-5-iodo-7H-pyrrolo[2,3-d]pyrimidine (7) and 7-((3aR,4S,6S,6aR)-6-(((Tert-butyldimethylsilyl)oxy)methyl)-2,2-dimethyltetrahydrofuro[3,4-d][1,3]dioxol-4-yl)-4-chloro-5-iodo-7H-pyrrolo[2,3-d]pyrimidine (ent-7)
7-((3aR,4R,6R,6aR)-6-(((Tert-butyldimethylsilyl)oxy)methyl)-2,2-dimethyltetrahydrofuro[3,4-d][1,3]dioxol-4-yl)-5-iodo-7H-pyrrolo[2,3-d]pyrimidin-4-amine (8) and 7-((3aR,4S,6S,6aR)-6-(((Tert-butyldimethylsilyl)oxy)methyl)-2,2-dimethyltetrahydrofuro[3,4-d][1,3]dioxol-4-yl)-5-iodo-7H-pyrrolo[2,3-d]pyrimidin-4-amine (ent-8)
((3aR,4R,6R,6aR)-6-(4-Amino-5-iodo-7H-pyrrolo[2,3-d]pyrimidin-7-yl)-2,2-dimethyltetrahydrofuro[3,4-d][1,3]dioxol-4-yl)methanol (9) ((3aS,4S,6S,6aS)-6-(4-Amino-5-iodo-7H-pyrrolo[2,3-d]pyrimidin-7-yl)-2,2-dimethyltetrahydrofuro[3,4-d][1,3]dioxol-4-yl)methanol (ent-9)
7-((3aR,4R,6R,6aR)-6-(Aminomethyl)-2,2-dimethyltetrahydrofuro[3,4-d][1,3]dioxol-4-yl)-5-iodo-7H-pyrrolo[2,3-d]pyrimidin-4-amine (10) and 7-((3aS,4S,6S,6aS)-6-(Aminomethyl)-2,2-dimethyltetrahydrofuro[3,4-d][1,3]dioxol-4-yl)-5-iodo-7H-pyrrolo[2,3-d]pyrimidin-4-amine (ent-10)
3.1.6. General Procedure for Sonogashira Coupling for the Preparation of 11a,b
3.1.7. General Procedure for Suzuki Coupling for the Preparation of 12a–d and ent-12d
5-([1,1′-Biphenyl]-4-yl)-7-((3aR,4R,6R,6aR)-6-(aminomethyl)-2,2-dimethyltetrahydrofuro[3,4-d][1,3]dioxol-4-yl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine (12a)
7-((3aR,4R,6R,6aR)-6-(aminomethyl)-2,2-dimethyltetrahydrofuro[3,4-d][1,3]dioxol-4-yl)-5-(1H-indol-2-yl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine (12b)
7-((3aR,4R,6R,6aR)-6-(Aminomethyl)-2,2-dimethyltetrahydrofuro[3,4-d][1,3]dioxol-4-yl)-5-(3,5-dimethoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine (12c)
7-((3aR,4R,6R,6aR)-6-(Aminomethyl)-2,2-dimethyltetrahydrofuro[3,4-d][1,3]dioxol-4-yl)-5-(7-methoxy-5-methylbenzo[b]thiophen-2-yl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine (12d) and 7-((3aS,4S,6S,6aS)-6-(Aminomethyl)-2,2-dimethyltetrahydrofuro[3,4-d][1,3]dioxol-4-yl)-5-(7-methoxy-5-methylbenzo[b]thiophen-2-yl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine (ent-12d)
3.1.8. General Procedure for EDC Coupling and Acetonide Deprotection for the Preparation of 13a–f and ent-13f
N-(((2R,3S,4R,5R)-5-(4-Amino-5-((3,5-dimethoxyphenyl)ethynyl)-7H-pyrrolo[2,3-d]pyrimidin-7-yl)-3,4-dihydroxytetrahydrofuran-2-yl)methyl)acrylamide (13a)
N-(((2R,3S,4R,5R)-5-(4-Amino-5-((3-methoxy-5-methylphenyl)ethynyl)-7H-pyrrolo[2,3-d]pyrimidin-7-yl)-3,4-dihydroxytetrahydrofuran-2-yl)methyl)acrylamide (13b)
N-(((2R,3S,4R,5R)-5-(5-([1,1′-Biphenyl]-4-yl)-4-amino-7H-pyrrolo[2,3-d]pyrimidin-7-yl)-3,4-dihydroxytetrahydrofuran-2-yl)methyl)acrylamide (13c)
N-(((2R,3S,4R,5R)-5-(4-Amino-5-(1H-indol-2-yl)-7H-pyrrolo[2,3-d]pyrimidin-7-yl)-3,4-dihydroxytetrahydrofuran-2-yl)methyl)acrylamide (13d)
N-(((2R,3S,4R,5R)-5-(4-Amino-5-(3,5-dimethoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-7-yl)-3,4-dihydroxytetrahydrofuran-2-yl)methyl)acrylamide (13e)
N-(((2R,3S,4R,5R)-5-(4-Amino-5-(7-methoxy-5-methylbenzo[b]thiophen-2-yl)-7H-pyrrolo[2,3-d]pyrimidin-7-yl)-3,4-dihydroxytetrahydrofuran-2-yl)methyl)acrylamide (13f)
N-(((2S,3R,4S,5S)-5-(4-Amino-5-(7-methoxy-5-methylbenzo[b]thiophen-2-yl)-7H-pyrrolo[2,3-d]pyrimidin-7-yl)-3,4-dihydroxytetrahydrofuran-2-yl)methyl)acrylamide (ent-13f)
3.1.9. Synthesis of 5′-Acrylamide-7-substituted-7-deaza-4′-thioadenosine Derivatives (19a–e)
((3aS,4R,6R,6aR)-6-(4-Amino-5-iodo-7H-pyrrolo[2,3-d]pyrimidin-7-yl)-2,2-dimethyltetrahydrothieno[3,4-d][1,3]dioxol-4-yl)methanol (15)
7-((3aR,4R,6R,6aS)-6-(Aminomethyl)-2,2-dimethyltetrahydrothieno[3,4-d][1,3]dioxol-4-yl)-5-iodo-7H-pyrrolo[2,3-d]pyrimidin-4-amine (16)
3.1.10. Procedure for Sonogashira Coupling for the Preparation of 17
3.1.11. General Procedure for Suzuki Coupling for the Preparation of 18a–d
3.1.12. General Procedure for EDC Coupling and Acetonide Deprotection for the Preparation of 19a–e
N-(((2R,3S,4R,5R)-5-(4-Amino-5-((3,5-dimethoxyphenyl)ethynyl)-7H-pyrrolo[2,3-d]pyrimidin-7-yl)-3,4-dihydroxytetrahydrothiophen-2-yl)methyl)acrylamide (19a)
N-(((2R,3S,4R,5R)-5-(4-Amino-5-(4-(1,1-dioxidothiomorpholino)phenyl)-7H-pyrrolo[2,3-d]pyrimidin-7-yl)-3,4-dihydroxytetrahydrothiophen-2-yl)methyl)acrylamide (19b)
N-(((2R,3S,4R,5R)-5-(4-Amino-5-(furan-2-yl)-7H-pyrrolo[2,3-d]pyrimidin-7-yl)-3,4-dihydroxytetrahydrothiophen-2-yl)methyl)acrylamide (19c)
N-(((2R,3S,4R,5R)-5-(4-Amino-5-(3,5-dimethoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-7-yl)-3,4-dihydroxytetrahydrothiophen-2-yl)methyl)acrylamide (19d)
N-(((2R,3S,4R,5R)-5-(4-Amino-5-(7-methoxy-5-methylbenzo[b]thiophen-2-yl)-7H-pyrrolo[2,3-d]pyrimidin-7-yl)-3,4-dihydroxytetrahydrothiophen-2-yl)methyl)acrylamide (19e)
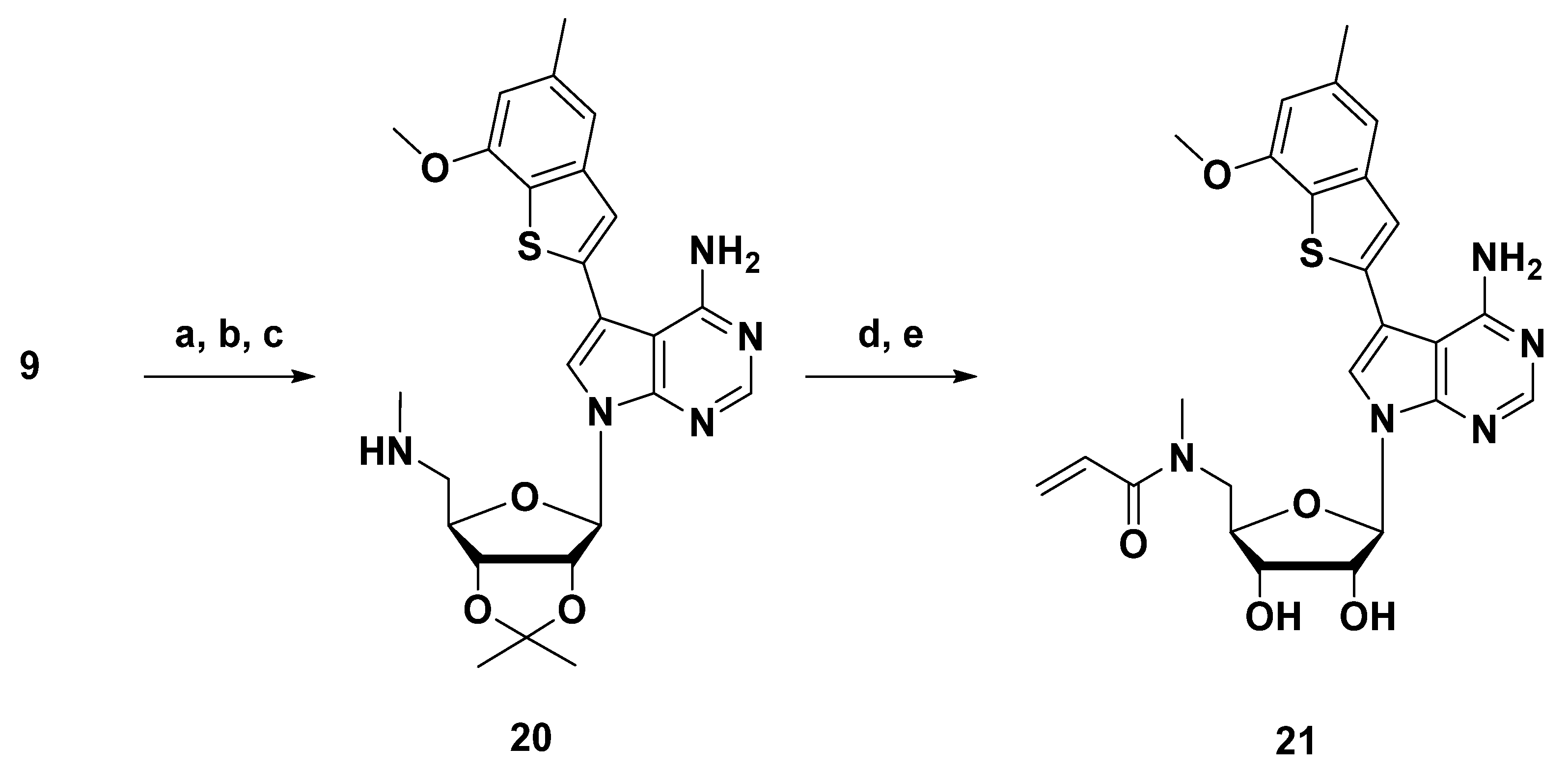
3.1.13. Synthesis of N-Methyl-5′-acrylamide-7-substituted-7-deaza Adenosine 21
7-((3aR,4R,6R,6aR)-2,2-Dimethyl-6-((methylamino)methyl)tetrahydrofuro[3,4-d][1,3]dioxol-4-yl)-5-(7-methoxy-5-methylbenzo[b]thiophen-2-yl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine (20)
(2R,3R,4S,5R)-2-(4-Amino-5-(7-methoxy-5-methylbenzo[b]thiophen-2-yl)-7H-pyrrolo[2,3-d]pyrimidin-7-yl)-5-((methylamino)methyl)tetrahydrofuran-3,4-diol (21)
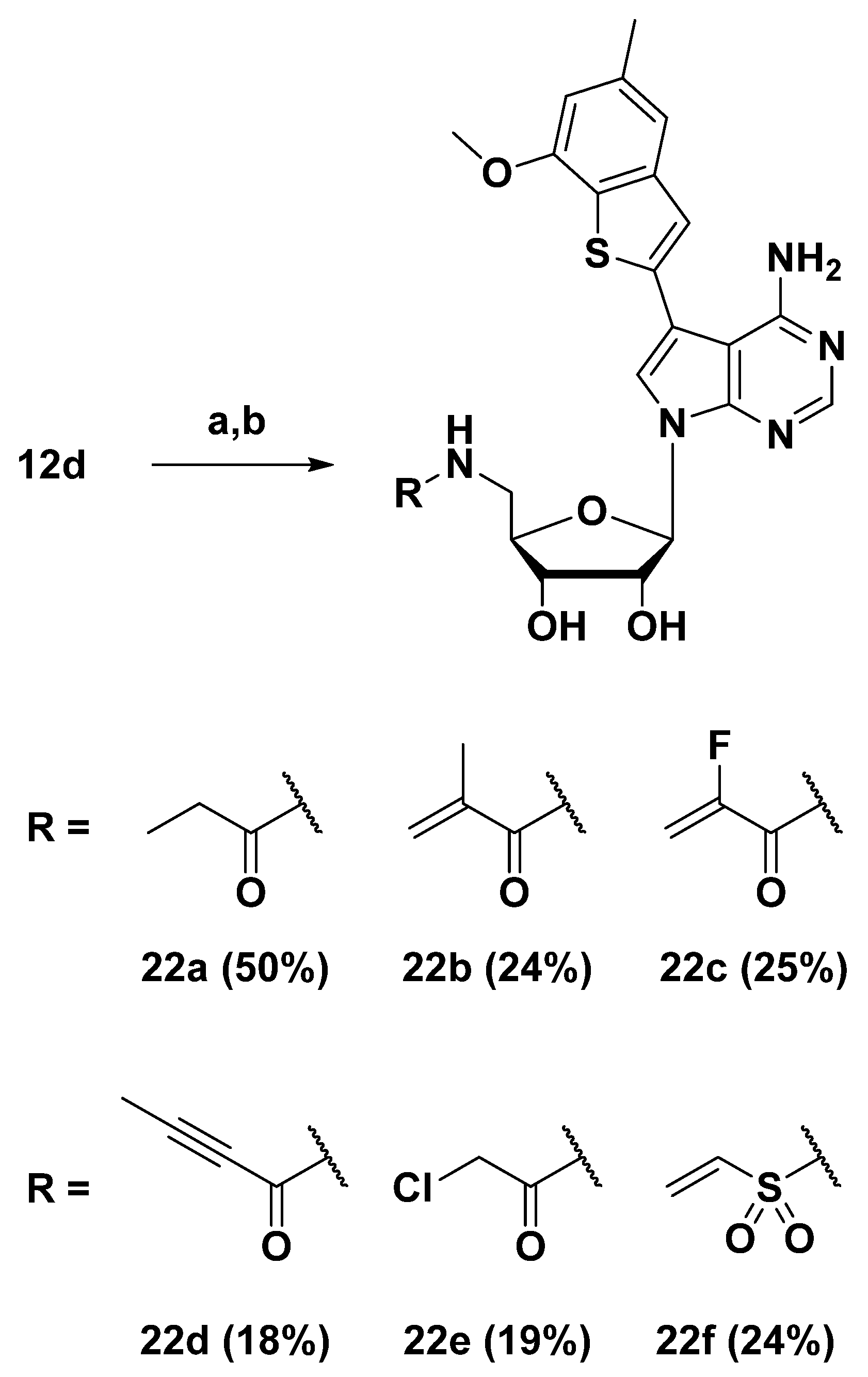
3.1.14. General Procedure for EDC Coupling and Acetonide Deprotection for the Preparation of 22a–e
N-(((2R,3S,4R,5R)-5-(4-Amino-5-(7-methoxy-5-methylbenzo[b]thiophen-2-yl)-7H-pyrrolo[2,3-d]pyrimidin-7-yl)-3,4-dihydroxytetrahydrofuran-2-yl)methyl)propionamide (22a)
N-(((2R,3S,4R,5R)-5-(4-Amino-5-(7-methoxy-5-methylbenzo[b]thiophen-2-yl)-7H-pyrrolo[2,3-d]pyrimidin-7-yl)-3,4-dihydroxytetrahydrofuran-2-yl)methyl)methacrylamide (22b)
N-(((2R,3S,4R,5R)-5-(4-Amino-5-(7-methoxy-5-methylbenzo[b]thiophen-2-yl)-7H-pyrrolo[2,3-d]pyrimidin-7-yl)-3,4-dihydroxytetrahydrofuran-2-yl)methyl)-2-fluoroacrylamide (22c)
N-(((2R,3S,4R,5R)-5-(4-Amino-5-(7-methoxy-5-methylbenzo[b]thiophen-2-yl)-7H-pyrrolo[2,3-d]pyrimidin-7-yl)-3,4-dihydroxytetrahydrofuran-2-yl)methyl)but-2-ynamide (22d)
N-(((2R,3S,4R,5R)-5-(4-Amino-5-(7-methoxy-5-methylbenzo[b]thiophen-2-yl)-7H-pyrrolo[2,3-d]pyrimidin-7-yl)-3,4-dihydroxytetrahydrofuran-2-yl)methyl)-2-chloroacetamide (22e)
3.1.15. N-(((2R,3S,4R,5R)-5-(4-Amino-5-(7-methoxy-5-methylbenzo[b]thiophen-2-yl)-7H-pyrrolo[2,3-d]pyrimidin-7-yl)-3,4-dihydroxytetrahydrofuran-2-yl)methyl)ethenesulfonamide (22f)
3.2. Biological and Computational Methods
3.2.1. Kinase Inhibition Assay for FGFR1, FGFR2, FGFR3, and FGFR4
3.2.2. Kinase Selectivity Profiling Assay
3.2.3. FGFR1 Kinase Cloning and Protein Expression
3.2.4. FGFR1 Protein Purification
3.2.5. FGFR1 Crystallization and Soaking Experiments
3.2.6. Cell Culture
3.2.7. Cell Proliferation Assay Using IncuCyte Live-Cell Imaging
3.2.8. Cell Culture (SRB Assay)
3.2.9. Cell Proliferation Assay (SRB Assay)
3.2.10. Intact Mass Analysis of FGFR1-Ligand Covalent Complexes
3.2.11. Western Blot
3.2.12. Molecular Dynamics Simulation
3.2.13. CYP Inhibition Assay
3.2.14. In Vitro Microsomal Stability Assay
3.2.15. In Vitro Plasma Metabolic Stability Assay
3.2.16. In Vitro hERG Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| FGFR | Fibroblast growth factor receptor |
| FGF | Fibroblast Growth Factor |
| TKI | Tyrosine Kinase Inhibitor |
| SAR | Structure–Activity Relationship |
| IC50 | Half maximal inhibitory concentration |
| Kd | Dissociation constant |
| MD | Molecular Dynamics |
| ERK | Extracellular Signal-Regulated Kinase |
| MEK | Mitogen-Activated Protein Kinase Kinase |
| MAPK | Mitogen-Activated Protein Kinase |
| PI3K | Phosphoinositide 3-kinase |
| AKT | Protein Kinase B |
| TACC3 | Transforming Acidic Coiled-Coil containing protein 3 |
| TNBC | Triple-Negative Breast Cancer |
| NSCLC | Non-Small Cell Lung Cancer |
| HCT116 | Human colorectal carcinoma cell line |
| RT4 | Human bladder cancer cell line |
| SRB | Sulforhodamine B |
| ADME | Absorption, Distribution, Metabolism, and Excretion |
| CYP | Cytochrome P450 |
| hERG | Human Ether-à-go-go-Related Gene (potassium channel) |
| MS | Mass Spectrometry |
| HRMS | High-Performance Liquid Chromatography |
| UV | Ultraviolet |
| ESI | Electrospray Ionization |
| FAB | Fast Atom Bombardment |
| CLint | Intrinsic Clearance |
| SEM | Standard Error of the Mean |
| SD | Standard Deviation |
| EDC | 1-Ethyl-3-(3-dimethylaminopropyl)carbodiimmide |
| DMAP | 4-Dimethylaminopyridine |
| DIAD | Diisopropyl azodicarboxylate |
| DBU | 1,8-Diazabicyclo[5.4.0]undec-7-ene |
| PEt3 | Triethylphosphine |
| TBSCl | tert-Butyldimethylsilyl chloride |
| TBAF | Tetra-n-butylammonium fluoride |
| TBDPS | tert-Butyldiphenylsilyl |
| DPPA | Diphenylphosphoryl azide |
| TCDI | 1,1′-Thiocarbonyldiimidazole |
| DMSO | Dimethyl sulfoxide |
| ACN | Acetonitrile |
| EtOAc | Ethyl acetate |
| THF | Tetrahydrofuran |
| DMF | Dimethylformamide |
| MeOH | Methanol |
References
- Babina, I.S.; Turner, N.C. Advances and challenges in targeting FGFR signalling in cancer. Nat. Rev. Cancer 2017, 17, 318–332. [Google Scholar] [CrossRef] [PubMed]
- Helsten, T.; Elkin, S.; Arthur, E.; Tomson, B.N.; Carter, J.; Kurzrock, R. The FGFR landscape in cancer: Analysis of 4853 tumors by next-generation sequencing. Clin. Cancer Res. 2016, 22, 259–267. [Google Scholar] [CrossRef]
- Imamura, T. Physiological functions and underlying mechanisms of fibroblast growth factor (FGF) family members: Recent findings and implications for their pharmacological application. Biol. Pharm. Bull. 2014, 37, 1081–1089. [Google Scholar] [CrossRef] [PubMed]
- Qin, A.; Johnson, A.; Ross, J.S.; Miller, V.A.; Ali, S.M.; Schrock, A.B.; Gadgeel, S.M. Detection of known and novel FGFR fusions in non-small cell lung cancer by comprehensive genomic profiling. J. Thorac. Oncol. 2019, 14, 54–62. [Google Scholar] [CrossRef]
- Tiong, K.H.; Mah, L.Y.; Leong, C.-O. Functional roles of fibroblast growth factor receptors (FGFRs) signaling in human cancers. Apoptosis 2013, 18, 1447–1468. [Google Scholar] [CrossRef]
- Su, N.; Jin, M.; Chen, L. Role of FGF/FGFR signaling in skeletal development and homeostasis: Learning from mouse models. Bone Res. 2014, 2, 14003. [Google Scholar] [CrossRef]
- Casadei, C.; Dizman, N.; Schepisi, G.; Cursano, M.C.; Basso, U.; Santini, D.; Pal, S.K.; De Giorgi, U. Targeted therapies for advanced bladder cancer: New strategies with FGFR inhibitors. Ther. Adv. Med. Oncol. 2019, 11, 1758835919890285. [Google Scholar] [CrossRef]
- Mazzaferro, V.; El-Rayes, B.F.; Busset, M.D.D.; Cotsoglou, C.; Harris, W.P.; Damjanov, N.; Masi, G.; Rimassa, L.; Personeni, N.; Braiteh, F.; et al. Derazantinib (ARQ 087) in advanced or inoperable FGFR2 gene fusion-positive intrahepatic cholangiocarcinoma. Br. J. Cancer 2019, 120, 165–171. [Google Scholar] [CrossRef]
- Goyal, L.; Shi, L.; Liu, L.Y.; Fece de la Cruz, F.; Lennerz, J.K.; Raghavan, S.; Leschiner, I.; Elagina, L.; Siravegna, G.; Ng, R.W.; et al. TAS-120 overcomes resistance to ATP-competitive FGFR inhibitors in patients with FGFR2 fusion-positive intrahepatic cholangiocarcinoma. Cancer Discov. 2019, 9, 1064–1079. [Google Scholar] [CrossRef]
- Taylor, J.G.; Cheuk, A.T.; Tsang, P.S.; Chung, J.-Y.; Song, Y.K.; Desai, K.; Yu, Y.; Chen, Q.-R.; Shah, K.; Youngblood, V.; et al. Identification of FGFR4-activating mutations in human rhabdomyosarcomas that promote metastasis in xenotransplanted models. J. Clin. Investig. 2009, 119, 3395–3407. [Google Scholar] [CrossRef] [PubMed]
- Gozgit, J.M.; Wong, M.J.; Moran, L.; Wardwell, S.; Mohemmad, Q.K.; Narasimhan, N.I.; Shakespeare, W.C.; Wang, F.; Clackson, T.; Rivera, V.M. Ponatinib (AP24534), a multitargeted pan-FGFR inhibitor with activity in multiple FGFR-amplified or mutated cancer models. Mol. Cancer Ther. 2012, 11, 690–699. [Google Scholar] [CrossRef]
- Renhowe, P.A.; Pecchi, S.; Shafer, C.M.; Machajewski, T.D.; Jazan, E.M.; Taylor, C.; Antonios-McCrea, W.; McBride, C.M.; Frazier, K.; Wiesmann, M.; et al. Design, structure–activity relationships and in vivo characterization of 4-amino-3-benzimidazol-2-ylhydroquinolin-2-ones: A novel class of receptor tyrosine kinase inhibitors. J. Med. Chem. 2009, 52, 278–292. [Google Scholar] [CrossRef]
- Bello, E.; Colella, G.; Scarlato, V.; Oliva, P.; Berndt, A.; Valbusa, G.; Serra, S.C.; D’Incalci, M.; Cavaletti, E.; Giavazzi, R.; et al. E-3810 is a potent dual inhibitor of VEGFR and FGFR that exerts antitumor activity in multiple preclinical models. Cancer Res. 2011, 71, 1396–1405. [Google Scholar] [CrossRef] [PubMed]
- Roth, G.J.; Heckel, A.; Colbatzky, F.; Handschuh, S.; Kley, J.; Lehmann-Lintz, T.; Lotz, R.; Tonsch-Grunt, U.; Walter, R.; Hilberg, F. Design, synthesis, and evaluation of indolinones as triple angiokinase inhibitors and the discovery of a highly specific 6-methoxycarbonyl-substituted indolinone (BIBF 1120). J. Med. Chem. 2009, 52, 4466–4480. [Google Scholar] [CrossRef]
- Dieci, M.V.; Arnedos, M.; Andre, F.; Soria, J.C. Fibroblast growth factor receptor inhibitors as a cancer treatment: From a biologic rationale to medical perspectives. Cancer Discov. 2013, 3, 264–279. [Google Scholar] [CrossRef] [PubMed]
- Yanochko, G.M.; Vitsky, A.; Heyen, J.R.; Hirakawa, B.; Lam, J.L.; May, J.; Nichols, T.; Sace, F.; Trajkovic, D.; Blasi, E. Pan-FGFR inhibition leads to blockade of FGF23 signaling, soft tissue mineralization, and cardiovascular dysfunction. Toxicol. Sci. 2013, 135, 451–464. [Google Scholar] [CrossRef]
- Gavine, P.R.; Mooney, L.; Kilgour, E.; Thomas, A.P.; Al-Kadhimi, K.; Beck, S.; Rooney, C.; Coleman, T.; Baker, D.; Mellor, M.J.; et al. AZD4547: An orally bioavailable, potent, and selective inhibitor of the fibroblast growth factor receptor tyrosine kinase family. Cancer Res. 2012, 72, 2045–2056. [Google Scholar] [CrossRef] [PubMed]
- Quinn, D.I.; Petrylak, D.P.; Bellmunt, J.; Necchi, A.; Gurney, H.; Lee, J.-L.; Van Der Heijden, M.S.; Rosenbaum, E.; Penel, N.; Pang, S.-T.; et al. FORT-1: Phase II/III study of rogaratinib versus chemotherapy in patients with locally advanced or metastatic urothelial carcinoma selected based on FGFR1/3 mRNA expression. J. Clin. Oncol. 2020, 38 (Suppl. S6), 489. [Google Scholar] [CrossRef]
- Piha-Paul, S.A.; Hierro, C.; Boni, V.; Moreno, V.; Hahn, N.M.; Bitting, R.L.; Bauer, T.M.; Aggarwal, R.R.; Gourlay, S.; Smith, P.; et al. A phase; multicenter, dose-escalation study of PRN1371, an irreversible covalent FGFR1–4 kinase inhibitor, in patients with advanced solid tumors, followed by expansion cohorts in patients with FGFR genetic alterations. J. Clin. Oncol. 2017, 35 (Suppl. S15), TPS2616. [Google Scholar] [CrossRef]
- Javle, M.M.; Roychowdhury, S.; Kelley, R.K.; Sadeghi, S.; Macarulla, T.; Waldschmidt, D.T.; Goyal, L.; Borbath, I.; El-Khoueiry, A.B.; Yong, W.-P.; et al. Final results from a phase II study of infigratinib (BGJ398), an FGFR-selective tyrosine kinase inhibitor, in patients with previously treated advanced cholangiocarcinoma harboring an FGFR2 gene fusion or rearrangement. J. Clin. Oncol. 2021, 39 (Suppl. S3), 265. [Google Scholar] [CrossRef]
- Sootome, H.; Fujita, H.; Ito, K.; Ochiiwa, H.; Fujioka, Y.; Ito, K.; Miura, A.; Sagara, T.; Ito, S.; Ohsawa, H.; et al. Futibatinib is a novel irreversible FGFR 1–4 inhibitor that shows selective antitumor activity against FGFR-deregulated tumors. Cancer Res. 2020, 80, 4986–4997. [Google Scholar] [CrossRef]
- Guo, Y.; Yuan, C.; Ding, W.; Gao, Y.; Zhu, X.; Ying, J.; Sun, M.; Zhang, J.; Zhuang, Z.; Huang, Y.; et al. Gunagratinib, a highly selective irreversible FGFR inhibitor, in patients with previously treated locally advanced or metastatic cholangiocarcinoma harboring FGFR pathway alterations: A phase IIa dose-expansion study. J. Clin. Oncol. 2023, 41. [Google Scholar] [CrossRef]
- Zhang, P.; Yue, L.; Leng, Q.Q.; Chang, C.; Gan, C.; Ye, T.; Cao, D. Targeting FGFR for cancer therapy. J. Hematol. Oncol. 2024, 17, 39. [Google Scholar] [CrossRef]
- Mashelkar, K.K.; Byun, W.S.; Ko, H.; Sung, K.; Tripathi, S.K.; An, S.; Yum, Y.A.; Kwon, J.Y.; Kim, M.; Kim, G.; et al. Discovery of a novel template, 7-substituted 7-deaza-4′-thioadenosine derivatives as multi-kinase inhibitors. Pharmaceuticals 2021, 14, 1290. [Google Scholar] [CrossRef]
- Kwon, E.-J.; Mashelkar, K.K.; Seo, J.; Shin, Y.-Z.; Sung, K.; Jang, S.C.; Cheon, S.W.; Lee, H.; Kim, G.; Han, B.W.; et al. In silico discovery of 5′-modified 7-deoxy-7-ethynyl-4′-thioadenosine as a HASPIN inhibitor and its synergistic anticancer effect with the PLK1 inhibitor. ACS Cent. Sci. 2023, 9, 1140–1149. [Google Scholar] [CrossRef]
- Chen, Z.; Wannere, C.S.; Corminboeuf, C.; Puchta, R.; von Ragué Schleyer, P. Nucleus-independent chemical shifts (NICS) as an aromaticity criterion. Chem. Rev. 2005, 105, 3842–3888. [Google Scholar] [CrossRef] [PubMed]
- Motherwell, W.B.; Moreno, R.B.; Pavlakos, I.; Arendorf, J.R.T.; Arif, T.; Tizzard, G.J.; Coles, S.J.; Aliev, A.E. Noncovalent interactions of π systems with sulfur: The atomic chameleon of molecular recognition. Angew. Chem. Int. Ed. 2018, 57, 1193–1198. [Google Scholar] [CrossRef] [PubMed]
- Jo, J.-C.; Choi, E.K.; Shin, J.-S.; Moon, J.-H.; Hong, S.-W.; Lee, H.-R.; Kim, S.-M.; Jung, S.-A.; Lee, D.-H.; Jung, S.H.; et al. Targeting FGFR pathway in human hepatocellular carcinoma: Expressing pFGFR and pMET for antitumor activity. Mol. Cancer Ther. 2015, 14, 2613–2622. [Google Scholar] [CrossRef] [PubMed]
- Huynh, H.; Chow, P.K.H.; Tai, W.M.; Choo, S.P.; Chung, A.Y.F.; Ong, H.S.; Soo, K.C.; Ong, R.; Linnartz, R.; Shi, M.M. Dovitinib demonstrates antitumor and antimetastatic activities in xenograft models of hepatocellular carcinoma. J. Hepatol. 2012, 56, 595–601. [Google Scholar] [CrossRef]
- Luo, Q.; Zhang, L.; Hao, Y.; Xu, C.; Wang, X.; Jia, Z.; Xie, X.; Huang, Z.; Gao, X.; Chen, Y.; et al. Erdafitinib inhibits the tumorigenicity of MDA-MB-231 triple-negative breast cancer cells by inducing TRIM25/ubiquitin-dependent degradation of FGFR4. Breast Cancer Res. 2025, 27, 128. [Google Scholar] [CrossRef]
- Berger, W.; Setinek, U.; Mohr, T.; Kindas-Mügge, I.; Vetterlein, M.; Dekan, G.; Eckersberger, F.; Caldas, C.; Micksche, M. Evidence for a role of FGF-2 and FGF receptors in the proliferation of non-small cell lung cancer cells. Int. J. Cancer 1999, 83, 415–423. [Google Scholar] [CrossRef]
- Fischer, H.; Taylor, N.; Allerstorfer, S.; Grusch, M.; Sonvilla, G.; Holzmann, K.; Setinek, U.; Elbling, L.; Cantonati, H.; Grasl-Kraupp, B.; et al. Fibroblast growth factor receptor-mediated signals contribute to the malignant phenotype of non-small cell lung cancer cells: Therapeutic implications and synergism with epidermal growth factor receptor inhibition. Mol. Cancer Ther. 2008, 7, 3408–3419. [Google Scholar] [CrossRef]
- Göke, F.; Göke, A.; von Mässenhausen, A.; Franzen, A.; Sharma, R.; Kirsten, R.; Böhm, D.; Kristiansen, G.; Stenzinger, A.; Wynes, M.; et al. Fibroblast growth factor receptor 1 as a putative therapy target in colorectal cancer. Digestion 2013, 88, 172–181. [Google Scholar] [CrossRef]
- Lau, D.K.; Collin, J.P.; Mariadason, J.M. Clinical developments and challenges in treating FGFR2-driven gastric cancer. Biomedicines 2004, 12, 1117. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.-L.; Liu, Y.-L.; Chen, J.-Z. Computational simulation studies on the binding selectivity of 1-(1H-benzimidazol-5-yl)-5-aminopyrazoles in complexes with FGFR1 and FGFR4. Molecules 2018, 23, 767. [Google Scholar] [CrossRef]
- Tucker, J.A.; Klein, T.; Breed, J.; Breeze, A.L.; Overman, R.; Phillips, C.; Norman, R.A. Structural insights into FGFR kinase isoform selectivity: Diverse binding modes of AZD4547 and Ponatinib in complex with FGFR1 and FGFR4. Structure 2014, 22, 1764–1774. [Google Scholar] [CrossRef] [PubMed]
- Dolomanov, O.V.; Bourhis, L.J.; Gildea, R.J.; Howard, J.A.K.; Puschmann, H. OLEX2: A Complete Structure Solution, Refinement and Analysis Program. J. Appl. Cryst. 2009, 42, 339–341. [Google Scholar] [CrossRef]
- Sheldrick, G.M. SHELXT—Integrated space-group and crystal-structure determination. Acta Cryst. 2015, A71, 3–8. [Google Scholar] [CrossRef]
- Sheldrick, G.M. A Short History of SHELX. Acta Crystallographica. Acta Cryst. 2008, A64, 112–122. [Google Scholar] [CrossRef]



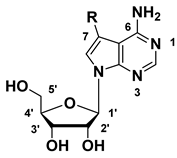
| Comp. | R | Inhibition Rate (%) at 1 μM a/IC50 (nM) | |||
|---|---|---|---|---|---|
| FGFR1 | FGFR2 | FGFR3 | FGFR4 | ||
| 5a | 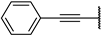 | 82 ± 4% | 61 ± 4% | 50 ± 1% | −4 ± 2% |
| 5b | 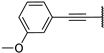 | 98 ± 2% | 97 ± 1% | 95 ± 4% | 25 ± 3% |
| 5c | 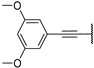 | 99 ± 2% | 96 ± 0% | 98 ± 0% | 25 ± 9% |
| 5d | 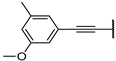 | 99 ± 2% | 99 ± 1% | 98 ± 1% | 22 ± 1% |
| 6a | 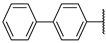 | 37 ± 0% | 17 ± 8% | 20 ± 1% | −11 ± 4% |
| 6b | 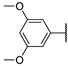 | 102 ± 0% | 100 ± 1% | 101 ± 1% | 88 ± 2% |
| 6c | 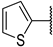 | 99 ± 0% | 90 ± 1% | 92 ± 3% | 28 ± 5% |
| 6d | 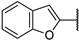 | 80 ± 2% | 29 ± 2% | 26 ± 4% | 5 ± 6% |
| 6e | 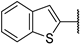 | 98 ± 1% | 100 ± 0% | 98 ± 1% | 83 ± 1% |
| 6f | 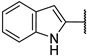 | 82 ± 5% | 82 ± 5% | 57 ± 5% | 15 ± 5% |
| 6g | 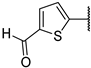 | 75 ± 3% | 75 ± 3% | 63 ± 3% | 4 ± 2% |
| 6h | 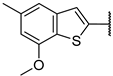 | 101 ± 1% 2 | 102 ± 1% 4 | 100 ± 0% 6 | 73 ± 2% 299 |
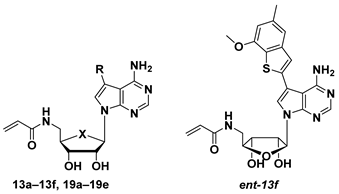

| Comp. | X | R | Inhibition Rate (%) at 0.1 μM b/IC50 (nM) c | IC50 (μM) d | ||||
|---|---|---|---|---|---|---|---|---|
| FGFR1 | FGFR2 | FGFR3 | FGFR4 | HCT116 (FGFR1 Amplification) | RT4 (FGFR3-TACC3 | |||
| 13a | O | 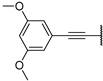 | 49 ± 2% 81 | 53 ± 2% | 64 ± 2% 57 | −5 ± 4% | >1000 | 19.05 |
| 13b | O | 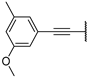 | 84 ± 0% 25 | 67 ± 5% | 66 ± 2% 57 | −7 ± 5% | 7.33 | 21.69 |
| 13c | O | 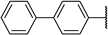 | 2 ± 10% | −2 ± 9% | −2 ± 1% >1000 | −1 ± 1% | 17.39 | 8.37 |
| 13d | O | 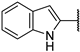 | 30 ± 6% | 9 ± 2% | 10 ± 2% >1000 | −7 ± 3% | 8.81 | 32.87 |
| 13e | O | 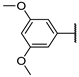 | 20 ± 6% | 27 ± 2% | 17 ± 3% | 0 ± 6% | >1000 | N.D. |
| 13f | O | 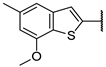 | 100 ± 0% 2 | 97 ± 0% 4 | 97 ± 1% 3 | 45 ± 5% 102 | 1.89 | 10.59 |
| ent-13f | O | 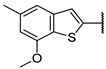 | 88 ± 0% 19 | 71 ± 3% 51 | 84 ± 0% 17 | 8 ± 5% >1000 | 3.51 | N.D. |
| 19a | S | 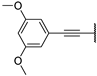 | 88 ± 1% 20 | 75 ± 3% | −5 ± 1% | −5 ± 3% | 12.03 | 16.42 |
| 19b | S | 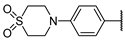 | −14 ± 6% >1000 | 5 ± 5% | 8 ± 8% >1000 | 1 ± 2% | >1000 | >50 |
| 19c | S | 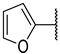 | 16 ± 2% 285 | 14 ± 1% | −7 ± 5% >1000 | 13 ± 4% | >1000 | >50 |
| 19d | S | 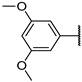 | 59 ± 4% 76 | 50 ± 1% | 24 ± 5% 284 | 7 ± 4% | 19.88 | >50 |
| 19e | S | 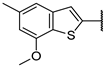 | 100 ± 1% 0.9 | 98 ± 1% 4 | 100 ± 0% 3 | 64 ± 3% 61 | 0.52 | 1.52 |
| Futibatinib | 6 | 61 | 12 | >1000 | 15.05 | 3.90 | ||
| Fexagratinib | 0.7 | 4 | 2 | 82 | 4.10 | 15.05 | ||
| Comp. | Human | Rat | Mouse | |||
|---|---|---|---|---|---|---|
| T1/2 (min) | CLint (mic) (μL/min/mg) | T1/2 (min) | CLint (mic) (μL/min/mg) | T1/2 (min) | CLint (mic) (μL/min/mg) | |
| 6h | 67.7 | 20.5 | 64.9 | 21.4 | 69.2 | 20.0 |
| 13f | 9.4 | 148.1 | 26.3 | 52.6 | 19.1 | 72.6 |
| 19e | 2.3 | 606.1 | 8.3 | 167.6 | 5.1 | 273.3 |
| Testosterone | 17.4 ± 2.3 | 80.4 ± 11.5 | 1.6 ± 0.2 | 827.2 ± 93.2 | 6.3 ± 1.5 | 233.4 ± 31.2 |
| Diclofenac | 5.6 ± 0.6 | 258.3 ± 27.9 | 16.8 ± 3.7 | 98.7 ± 23.5 | 41.3 ± 5.6 | 32.4 ± 4.2 |
| Propafenone | 7.0 ± 1.5 | 202.6 ± 38.9 | 1.5 ± 0.2 | 946.6 ± 97.5 | 2.9 ± 0.7 | 550.3 ± 163.2 |
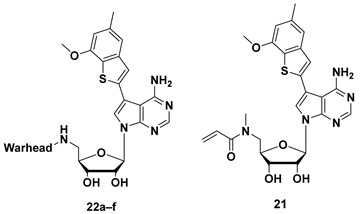

| Comp. | Warhead | Inhibition Rate (%) at 0.1 μM a/IC50 ± SEM (nM) b | IC50 (μM) c | ||||
|---|---|---|---|---|---|---|---|
| FGFR1 | FGFR2 | FGFR3 | FGFR4 | HCT116 (FGFR1 Amplification) | RT4 (FGFR3-TACC3 | ||
| 13f | 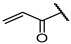 | 100 ± 0% 2 | 97 ± 0% 4 | 97 ± 1% 3 | 45 ± 5% 102 | 1.89 | 10.59 |
| 21 | 98 ± 0% 3 | 89 ± 1% 16 | 95 ± 0% 8 | 40 ± 2% 151 | 5.03 | 6.30 | |
| 22a | 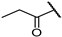 | 98 ± 1% 3 | 91 ± 1% 8 | 94 ± 3% 8 | 43 ± 0% 137 | 2.08 | 10.87 |
| 22b | 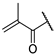 | 99 ± 1% | 91 ± 2% | 101% | 92 ± 4% | 2.73 | 1.22 |
| 22c | 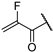 | 99 ± 0% | 94 ± 0% | 97 ± 2% | 77 ± 1% | 1.68 | 1.43 |
| 22d | 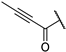 | 99 ± 1% | 95 ± 2% | 96 ± 1% | 35 ± 2% | 1.63 | 0.07 |
| 22e | 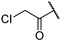 | 100 ± 0% | 97 ± 1% | 97 ± 0% | 78 ± 1% | 0.65 | 10.33 |
| 22f | 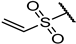 | 98 ± 1% 1 | 95 ± 0% 8 | 97 ± 0% 5 | 64 ± 5% 53 | 0.58 | 0.67 |
| Futibatinib | 6 | 61 | 12 | >1000 | 15.05 | 3.90 | |
| Fexagratinib | 0.7 | 4 | 2 | 82 | 4.10 | 15.05 | |
| Comp. | Human | Rat | Mouse | |||
|---|---|---|---|---|---|---|
| T1/2 (min) | CLint (mic) (μL/min/mg) | T1/2 (min) | CLint (mic) (μL/min/mg) | T1/2 (min) | CLint (mic) (μL/min/mg) | |
| 21 | 12.8 | 108.5 | 11.8 | 117.1 | 12.1 | 114.4 |
| 22a | 12.6 | 109.9 | 23.8 | 58.2 | 24.7 | 56.0 |
| 22f | 29.7 | 46.7 | 18.2 | 76.0 | 27.7 | 50.0 |
| Futibatinib | 57.3 | 24.2 | 40.8 | 34.0 | 60.5 | 22.9 |
| Testosterone | 17.4 ± 2.3 | 80.4 ± 11.5 | 1.6 ± 0.2 | 827.2 ± 93.2 | 6.3 ± 1.5 | 233.4 ± 31.2 |
| Diclofenac | 5.6 ± 0.6 | 258.3 ± 27.9 | 16.8 ± 3.7 | 98.7 ± 23.5 | 41.3 ± 5.6 | 32.4 ± 4.2 |
| Propafenone | 7.0 ± 1.5 | 202.6 ± 38.9 | 1.5 ± 0.2 | 946.6 ± 97.5 | 2.9 ± 0.7 | 550.3 ± 163.2 |
| Comp. | IC50 (μM) a | ||||
|---|---|---|---|---|---|
| SK-HEP-1 | MDA-MB-231 | A549 | HCT116 | SNU-638 | |
| 13a | 9.60 | >50 | 48.83 | >50 | 29.13 |
| 13b | 3.58 | 7.05 | 12.69 | 37.88 | 13.29 |
| 13f | 0.26 | 0.61 | 0.69 | 1.12 | 0.82 |
| 19b | >50 | >50 | >50 | >50 | >50 |
| 19c | 26.10 | >50 | >50 | 33.50 | >50 |
| 19d | >50 | >50 | >50 | >50 | >50 |
| 19e | 0.55 | 0.62 | 0.82 | 0.93 | 0.75 |
| 21 | 0.60 | 0.81 | 1.21 | 1.96 | 1.42 |
| 22a | 1.24 | 2.36 | 3.77 | 4.88 | 5.00 |
| 22b | 0.98 | 3.33 | 2.68 | 4.63 | 2.85 |
| 22c | 0.50 | 1.25 | 1.15 | 1.68 | 1.49 |
| 22d | 0.51 | 0.78 | 1.61 | 1.43 | 0.85 |
| 22e | 0.36 | 0.91 | 0.88 | 0.92 | 0.75 |
| 22f | 0.57 | 0.65 | 0.64 | 0.92 | 0.41 |
| Etoposide | 0.69 | 7.75 | 0.17 | 0.96 | 0.21 |
| Parameters | Results |
|---|---|
| CYP Inhibition a: IC50 (μM) | >50/>50/20.7/10.8/>50 |
| Metabolic Stability in Plasma b: T1/2 (min) | 47.3/32.6/37.3 |
| hERG inhibition: IC50 (μM) | >30 |
| Human oral absorption c (%) | 41.1 |
| Serum albumin binding c (logKHSA) | −0.177 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, J.H.; Tran, P.T.; Ko, H.L.; Mun, S.; Jang, S.C.; Moon, D.H.; Han, J.; Kim, J.; Kim, G.; Choi, H.; et al. Design, Synthesis, and Biological Evaluation of 5′,7-Disubstituted 7-Deaza-adenosine Analogues as Irreversible Pan-FGFR Inhibitors. Pharmaceuticals 2025, 18, 1745. https://doi.org/10.3390/ph18111745
Park JH, Tran PT, Ko HL, Mun S, Jang SC, Moon DH, Han J, Kim J, Kim G, Choi H, et al. Design, Synthesis, and Biological Evaluation of 5′,7-Disubstituted 7-Deaza-adenosine Analogues as Irreversible Pan-FGFR Inhibitors. Pharmaceuticals. 2025; 18(11):1745. https://doi.org/10.3390/ph18111745
Chicago/Turabian StylePark, Jung Hoon, Phuong Thao Tran, Hye Lin Ko, Seonghee Mun, Sung Chul Jang, Dong Hyun Moon, Jaeho Han, Jieun Kim, Gibae Kim, Hongseok Choi, and et al. 2025. "Design, Synthesis, and Biological Evaluation of 5′,7-Disubstituted 7-Deaza-adenosine Analogues as Irreversible Pan-FGFR Inhibitors" Pharmaceuticals 18, no. 11: 1745. https://doi.org/10.3390/ph18111745
APA StylePark, J. H., Tran, P. T., Ko, H. L., Mun, S., Jang, S. C., Moon, D. H., Han, J., Kim, J., Kim, G., Choi, H., Kim, S. W., Kim, M., Lee, S. K., Han, B. W., Kang, K. W., & Jeong, L. S. (2025). Design, Synthesis, and Biological Evaluation of 5′,7-Disubstituted 7-Deaza-adenosine Analogues as Irreversible Pan-FGFR Inhibitors. Pharmaceuticals, 18(11), 1745. https://doi.org/10.3390/ph18111745










