Computational Chemistry Advances in the Development of PARP1 Inhibitors for Breast Cancer Therapy
Abstract
1. Introduction
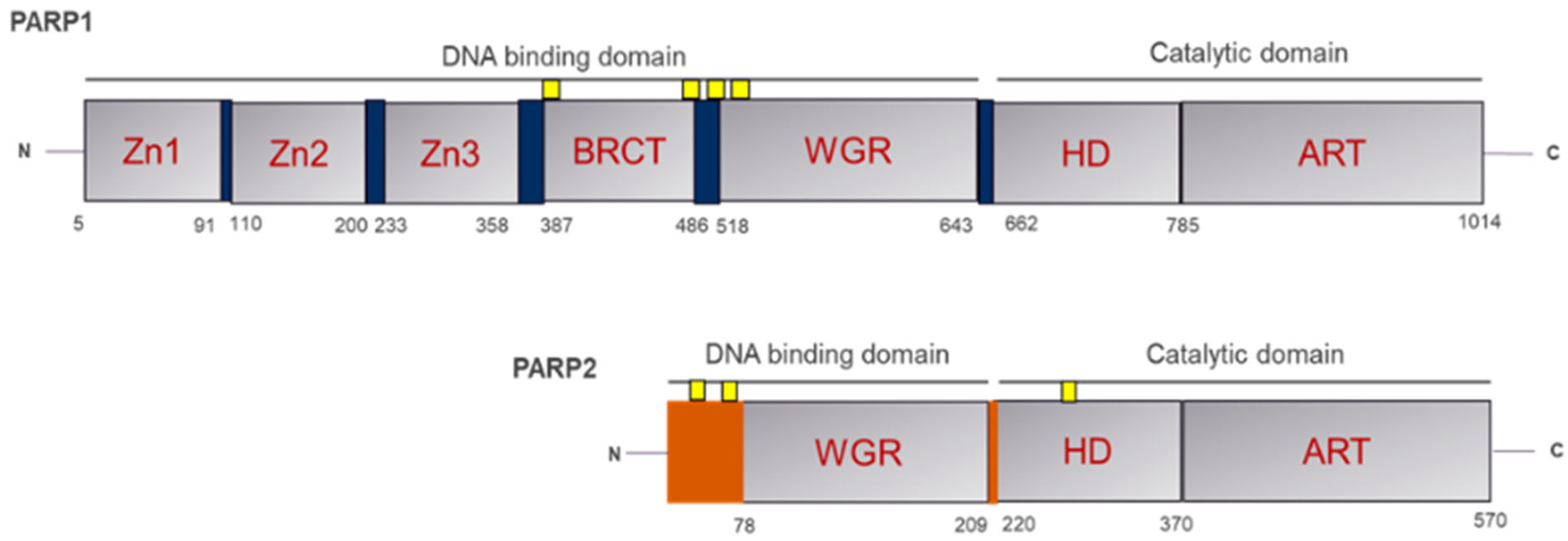
1.1. Overview of PARP1 Structure and Its Biological Significance
1.2. PARP1 in Breast Cancer
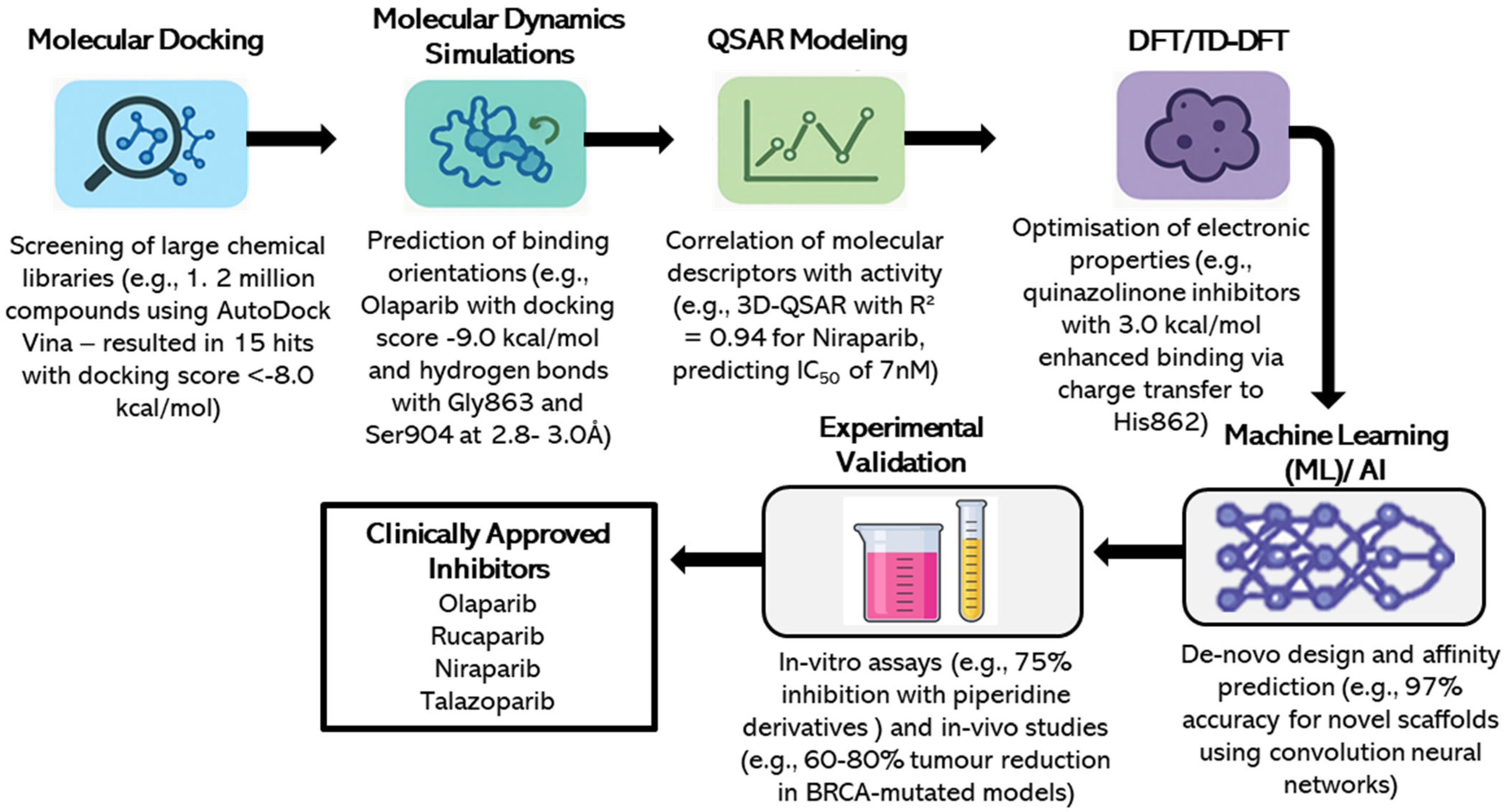
2. Computational Chemistry Methods in Drug Design
2.1. Molecular Docking
2.2. Molecular Dynamics Simulations
2.3. Density Functional Theory (DFT) and TD-DFT Calculations
2.4. Quantitative Structure–Activity Relationship (QSAR) Models
2.5. Virtual Screening and De Novo Design
2.6. Emerging Machine Learning and AI-Driven Approaches
3. Case Studies of PARP1 Inhibitors Discovered Through Computational Methods
3.1. Olaparib
3.2. Rucaparib
3.3. Niraparib
3.4. Talazoparib
3.5. Novel Inhibitors and Emerging Scaffolds
4. Structural Chemistry of PARP1 Inhibitors and Hybrid Scaffolds
5. Challenges in Computational Chemistry for PARP1 Inhibitors
6. Future Directions and Perspectives
6.1. Innovations and Emerging Techniques
6.2. Integrating Computational and Experimental Approaches
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| PARP1 | Poly (ADP-ribose) polymerase 1 |
| DNA | Deoxyribonucleic Acid |
| RNA | Ribonucleic Acid |
| NAD+ | Nicotinamide adenine dinucleotide |
| BRCA | Breast Cancer Gene |
| HRR | Homologous Recombination Repair |
| QSAR | Quantitative Structure Activity Relationship |
| BER | Base Excision Repair |
| MD | Molecular Dynamics |
| DFT | Density Functional Theory |
| TD-DFT | Time Dependent–Density Functional Theory |
| SSB | Single-strand Break |
| DSB | Double-strand Break |
| GPU | Graphical Processing Unit |
| HD | Helical domain |
References
- Hirlekar, B.U.; Nuthi, A.; Singh, K.D.; Murty, U.S.; Dixit, V.A. An overview of compound properties, multiparameter optimization, and computational drug design methods for PARP-1 inhibitor drugs. Eur. J. Med. Chem. 2023, 252, 115300. [Google Scholar] [CrossRef] [PubMed]
- Engbrecht, M.; Mangerich, A. The nucleolus and PARP1 in cancer biology. Cancers 2020, 12, 1813. [Google Scholar] [CrossRef] [PubMed]
- Rudolph, J.; Jung, K.; Luger, K. Inhibitors of PARP: Number crunching and structure gazing. Proc. Natl. Acad. Sci. USA 2022, 119, e2121979119. [Google Scholar] [CrossRef]
- Khizer, H.; Maryam, A.; Ansari, A.; Ahmad, M.S.; Khalid, R.R. Leveraging shape screening and molecular dynamics simulations to optimize PARP1-specific chemo/radio-potentiators for antitumor drug design. Arch. Biochem. Biophys. 2024, 756, 110010. [Google Scholar] [CrossRef]
- Gomatam, A.; Hirlekar, B.U.; Singh, K.D.; Murty, U.S.; Dixit, V.A. Improved QSAR models for PARP-1 inhibition using data balancing, interpretable machine learning, and matched molecular pair analysis. Mol. Divers. 2024, 28, 2135–2152. [Google Scholar] [CrossRef]
- Shahab, M.; Waqas, M.; Fahira, A.; Sharma, B.P.; Zhang, H.; Zheng, G.; Huang, Z. Machine learning-based screening and molecular simulations for discovering novel PARP-1 inhibitors targeting DNA repair mechanisms for breast cancer therapy. Mol. Divers. 2025, 29, 3323–3343. [Google Scholar] [CrossRef]
- Antolin, A.A.; Ameratunga, M.; Banerji, U.; Clarke, P.A.; Workman, P.; Al-Lazikani, B. The kinase polypharmacology landscape of clinical PARP inhibitors. Sci. Rep. 2020, 10, 2585. [Google Scholar] [CrossRef]
- Bhatnagar, A.; Nath, V.; Kumar, N.; Kumar, V. Discovery of novel PARP-1 inhibitors using tandem in silico studies: Integrated Docking, e-pharmacophore, deep learning based de Novo and molecular dynamics simulation approach. J. Biomol. Struct. Dyn. 2024, 42, 3396–3409. [Google Scholar] [CrossRef]
- Ghorab, W.M.; El-Sebaey, S.A.; Ghorab, M.M. Design, synthesis and molecular modeling study of certain quinazolinone derivatives targeting poly (ADP-ribose) polymerase 1 (PARP-1) enzyme as anti-breast cancer and radio-sensitizers. J. Mol. Struct. 2023, 1273, 134358. [Google Scholar] [CrossRef]
- Xue, H.; Bhardwaj, A.; Yin, Y.; Fijen, C.; Ephstein, A.; Zhang, L.; Ding, X.; Pascal, J.M.; VanArsdale, T.L.; Rothenberg, E. A two-step mechanism governing PARP1-DNA retention by PARP inhibitors. Sci. Adv. 2022, 8, eabq0414. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Shaw, D.E.; Adams, P.J.; Azaria, A.; Bank, J.A.; Batson, B.; Bell, A.; Bergdorf, M.; Bhatt, J.; Butts, J.A.; Correia, T.; et al. Anton 3: Twenty microseconds of molecular dynamics simulation in a dedicated supercomputer. Nature 2021, 600, 267–272. [Google Scholar]
- Venugopal, P.P.; M, S.; Chakraborty, D. Theoretical insights into molecular mechanism and energy criteria of PARP-2 enzyme inhibition by benzimidazole analogues. Proteins Struct. Funct. Bioinform. 2021, 89, 988–1004. [Google Scholar] [CrossRef]
- Lin, C.; Liu, C.; Hu, P.; Zou, Z.; Sun, G. Design, synthesis, biological evaluation of novel piperidine-based derivatives as potent poly(ADP-ribose) polymerase-1 (PARP-1) inhibitors. Bioorganic Chem. 2024, 148, 107455. [Google Scholar] [CrossRef]
- Sadybekov, A.V.; Katritch, V. Computational approaches streamlining drug discovery. Nature 2023, 616, 673–685. [Google Scholar] [CrossRef] [PubMed]
- Jin, W.; Barzilay, R.; Jaakkola, T. Junction tree variational autoencoder for molecular graph generation. Int. Conf. Mach. Learn. 2018, 80, 2323–2332. [Google Scholar]
- Qin, H.; Zhang, J.; Zhao, Y.; Zhang, L.; Feng, J.; Zhang, L. Discovery of a potent olaparib–chlorambucil hybrid inhibitor of PARP1 for the treatment of cancer. Front. Pharmacol. 2023, 13, 1054616. [Google Scholar] [CrossRef]
- Akay, M.; Funingana, I.G.; Patel, G.; Mustapha, R.; Gjafa, E.; Ng, T.; Ng, K.; Flynn, M.J. An in-depth review of niraparib in ovarian cancer: Mechanism of action, clinical efficacy and future directions. Oncol. Ther. 2021, 9, 347–364. [Google Scholar] [CrossRef]
- Yi, M.; Dong, B.; Qin, S.; Chu, Q.; Wu, K.; Luo, S. Advances and perspectives of PARP inhibitors. Exp. Hematol. Oncol. 2019, 8, 29. [Google Scholar] [CrossRef]
- Jones, P.; Wilcoxen, K.; Rowley, M.; Toniatti, C. Niraparib: A poly (ADP-ribose) polymerase (PARP) inhibitor for the treatment of tumors with defective homologous recombination. J. Med. Chem. 2015, 58, 3302–3314. [Google Scholar] [CrossRef]
- Cao, Y.; Romero, J.; Olson, J.P.; Degroote, M.; Johnson, P.D.; Kieferová, M.; Kivlichan, I.D.; Menke, T.; Peropadre, B.; Sawaya, N.P.; et al. Quantum chemistry in the age of quantum computing. Chem. Rev. 2019, 119, 10856–10915. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, Z. Integrating multi-omics data with computational modeling for precision oncology. Nat. Rev. Clin. Oncol. 2024, 21, 179–192. [Google Scholar]
- He, C.; Kawaguchi, K.; Toi, M. DNA damage repair functions and targeted treatment in breast cancer. Breast Cancer 2020, 27, 355–362. [Google Scholar] [CrossRef]
- Lin, Y.; Gao, C.; Wang, Z.; Zhang, R.; Chen, Y.; Min, Z. Synthesis and preliminary biological evaluation of new phthalazinone derivatives with PARP-1 and cholinesterase inhibitory activities. Lett. Drug Des. Discov. 2023, 20, 56–70. [Google Scholar]
- Zhu, Q.; Wang, X.; Hu, Y.; He, X.; Gong, G.; Xu, Y. Discovery and SAR study of 2-(1-propylpiperidin-4-yl)-3H-imidazo [4,5-c] pyridine-7-carboxamide: A potent inhibitor of poly(ADP-ribose) polymerase-1 (PARP-1) for the treatment of cancer. Bioorganic Med. Chem. 2025, 23, 6551–6559. [Google Scholar] [CrossRef]
- Wang, Y.Q.; Wang, P.Y.; Wang, Y.T.; Yang, G.F.; Zhang, A.; Miao, Z.H. An update on poly (ADP-ribose) polymerase-1 (PARP-1) inhibitors: Opportunities and challenges in cancer therapy. J. Med. Chem. 2016, 59, 9575–9598. [Google Scholar] [CrossRef]
- Gai, S.; Cao, P.; Zhong, X.; Lin, Y.; Lin, B.; Jiang, M. Designing an anticancer Pd(II) complex as poly (ADP-ribose) polymerase 1 inhibitor. Int. J. Biol. Macromol. 2025, 297, 139885. [Google Scholar] [CrossRef] [PubMed]
- Schaduangrat, N.; Lampa, S.; Simeon, S.; Gleeson, M.P.; Spjuth, O.; Nantasenamat, C. Towards reproducible computational drug discovery. J. Cheminform. 2020, 12, 9. [Google Scholar] [CrossRef] [PubMed]



| PARP1 Inhibitor | Description | Computational Methods | References |
|---|---|---|---|
| Olaparib | First FDA-approved PARP1 inhibitor for BRCA-mutated breast and ovarian cancers; IC50 = 5 nM. | Molecular docking, MD simulations, QSAR, virtual screening | [4,15] |
| Rucaparib | Approved for ovarian cancer; high selectivity for PARP1 (15:1 vs. PARP2); IC50 = 7 nM. | Virtual screening, MD simulations, docking | [1,8] |
| Niraparib | Approved for maintenance therapy in ovarian cancer; IC50 = 7 nM; 65% reduction in cell viability. | Molecular docking, 3D-QSAR, MD simulations | [8] |
| Talazoparib | Highly potent inhibitor for BRCA-mutated breast cancer; IC50 = 1 nM; 62% objective response rate. | Deep learning, docking, MD simulations, TD-DFT | [6] |
| Quinazolinone Analogs | Novel inhibitors with IC50 = 10 nM; promising for BRCA-mutated breast cancer. | TD-DFT, molecular docking, MD simulations | [9] |
| Piperidine Derivatives | Novel scaffold with IC50 = 12 nM; 75% inhibition in vitro. | De Novo design, MD simulations, docking | [14] |
| Olaparib–Chlorambucil | Hybrid inhibitor with synergistic effects; 90% reduction in cell viability in BRCA-deficient cells. | DFT, molecular docking | [17] |
| Drug/Scaffold | Core Scaffold | Key Functional Groups | Lipophilicity (LogP) | Binding Strength | Unique Feature |
|---|---|---|---|---|---|
| Olaparib | Phthalazinone | Carbonyl, piperazine, fluorophenyl | ~2.5 | High | Balanced π-stacking + H-bonds |
| Rucaparib | tricyclic indole-fused lactam | Amide, chlorophenyl, piperazine | ~2.7 | High | Stronger aromatic contacts |
| Niraparib | 2H-indazole-7-carboxamide scaffold | Trifluoromethyl, piperidine, amide | ~3.1 | High | Hydrophobic boosting potency |
| Talazoparib | Tricyclic pyridazinone scaffold | Triazole, lactam, fluorophenyl | ~2.4 | Very High | Exceptional PARP trapping |
| Quinazolinones | Benzopyrimidinone scaffold | Carbonyl, amide, aromatic ring | ~2.3–2.8 | Medium–High | NAD+ mimetics for alternative scaffold |
| Piperidine derivatives | Piperidine hybrids | Flexible amine, aromatic linkers | Variable | Adjustable | Improves solubility & PK |
| Olaparib–Chlorambucil | Hybrid scaffold | Nitrogen mustard + phthalazinone | ~3.0 | Very High | Dual DNA damage + PARP inhibition |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Twala, C.; Govender, P.; Govender, K. Computational Chemistry Advances in the Development of PARP1 Inhibitors for Breast Cancer Therapy. Pharmaceuticals 2025, 18, 1679. https://doi.org/10.3390/ph18111679
Twala C, Govender P, Govender K. Computational Chemistry Advances in the Development of PARP1 Inhibitors for Breast Cancer Therapy. Pharmaceuticals. 2025; 18(11):1679. https://doi.org/10.3390/ph18111679
Chicago/Turabian StyleTwala, Charmy, Penny Govender, and Krishna Govender. 2025. "Computational Chemistry Advances in the Development of PARP1 Inhibitors for Breast Cancer Therapy" Pharmaceuticals 18, no. 11: 1679. https://doi.org/10.3390/ph18111679
APA StyleTwala, C., Govender, P., & Govender, K. (2025). Computational Chemistry Advances in the Development of PARP1 Inhibitors for Breast Cancer Therapy. Pharmaceuticals, 18(11), 1679. https://doi.org/10.3390/ph18111679








