Computational Development of Multi-Epitope Reovirus Vaccine with Potent Predicted Binding to TLR2 and TLR4
Abstract
1. Introduction
2. Results
2.1. Selection of CTL, HTL, and B Cell Epitopes
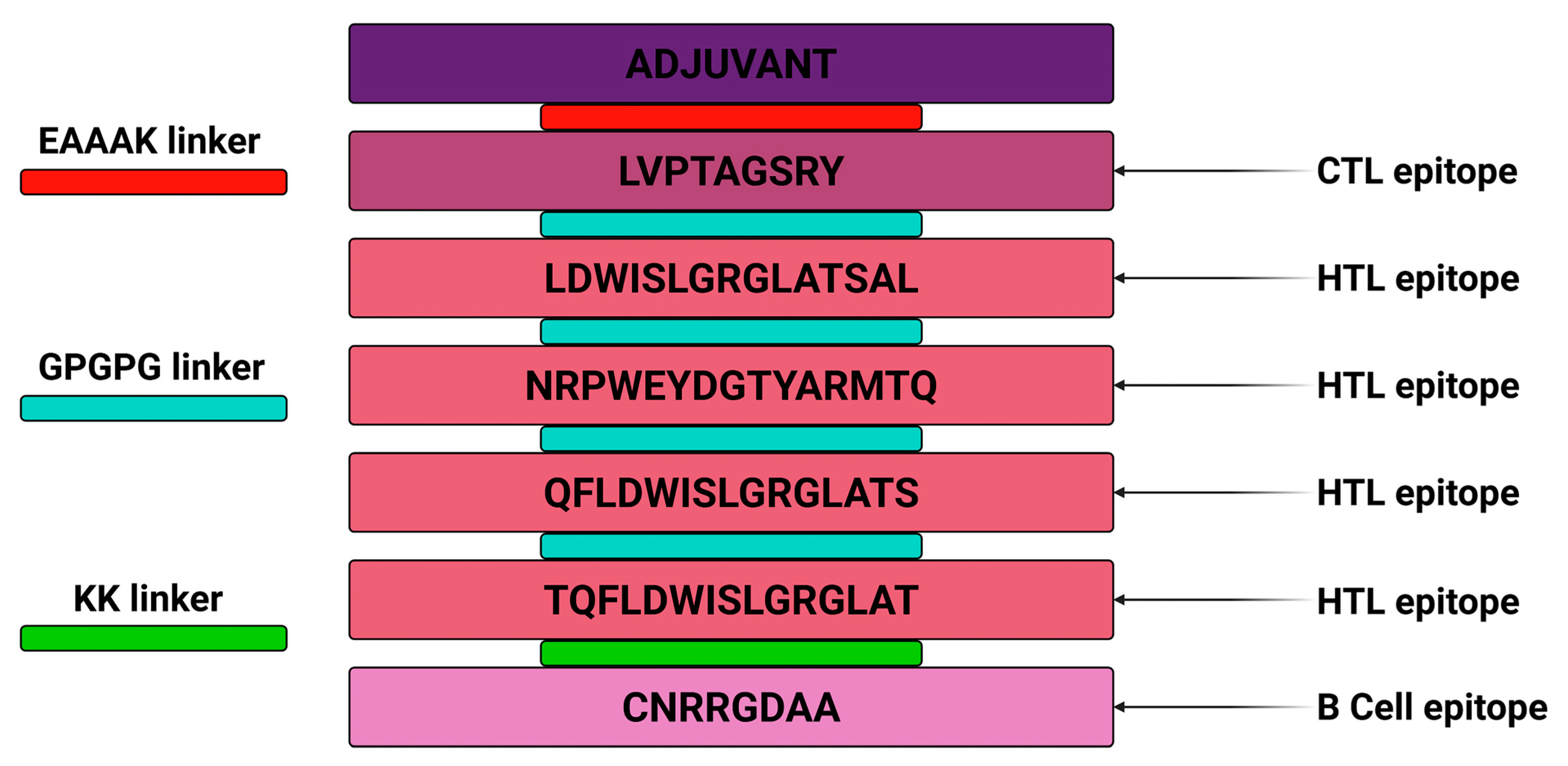
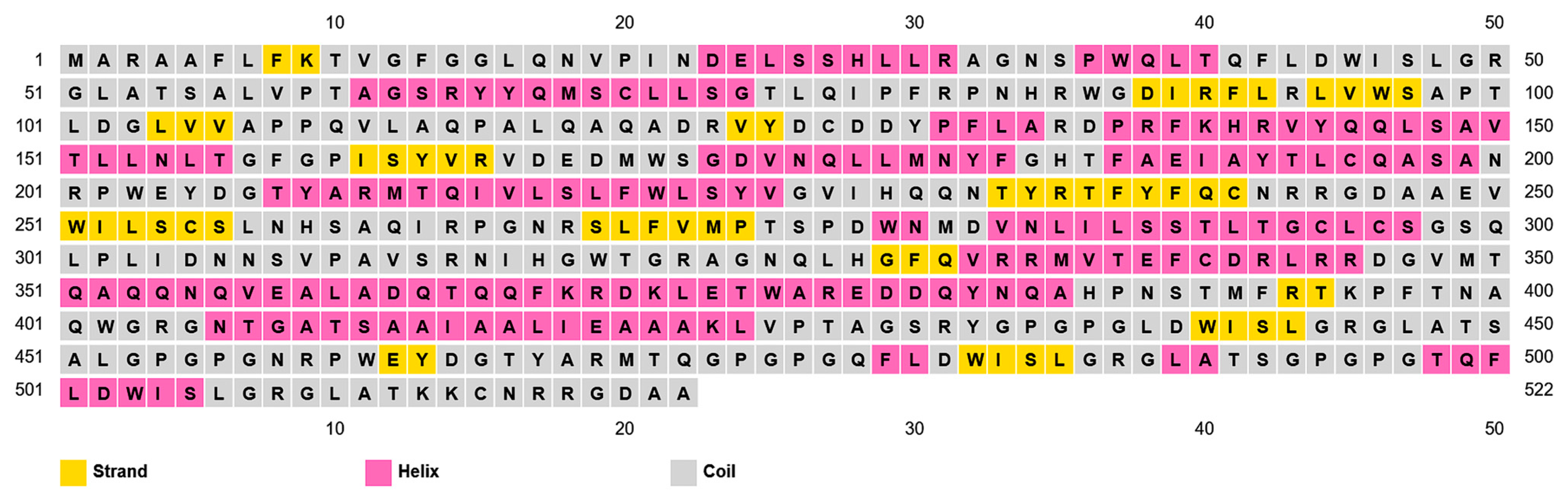
2.2. Vaccine Construction
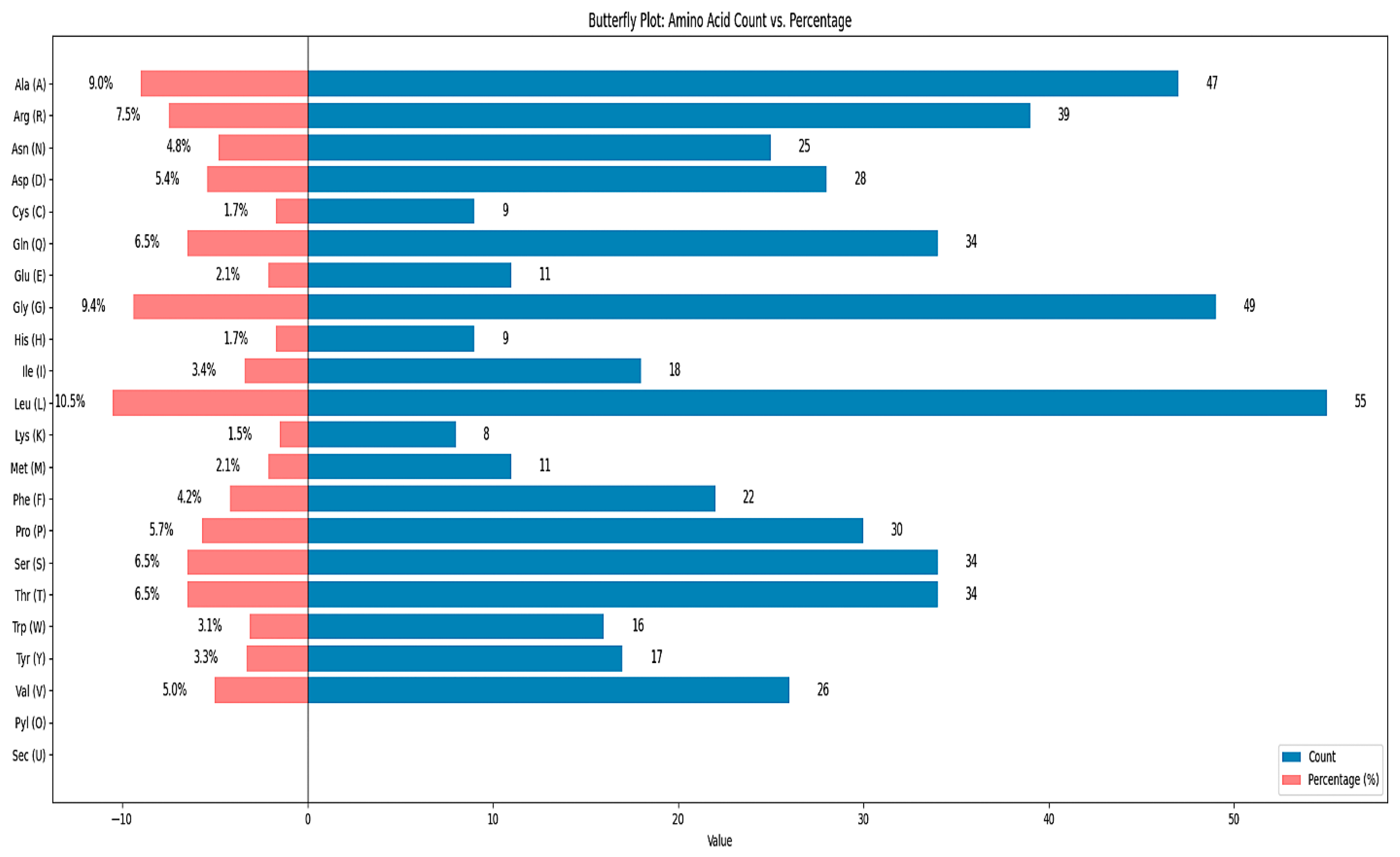
2.3. Vaccine Physiochemical Properties
2.4. Antigenicity, Allergenicity, and Toxicity of Vaccine
2.5. Secondary and Tertiary Structure of Vaccine
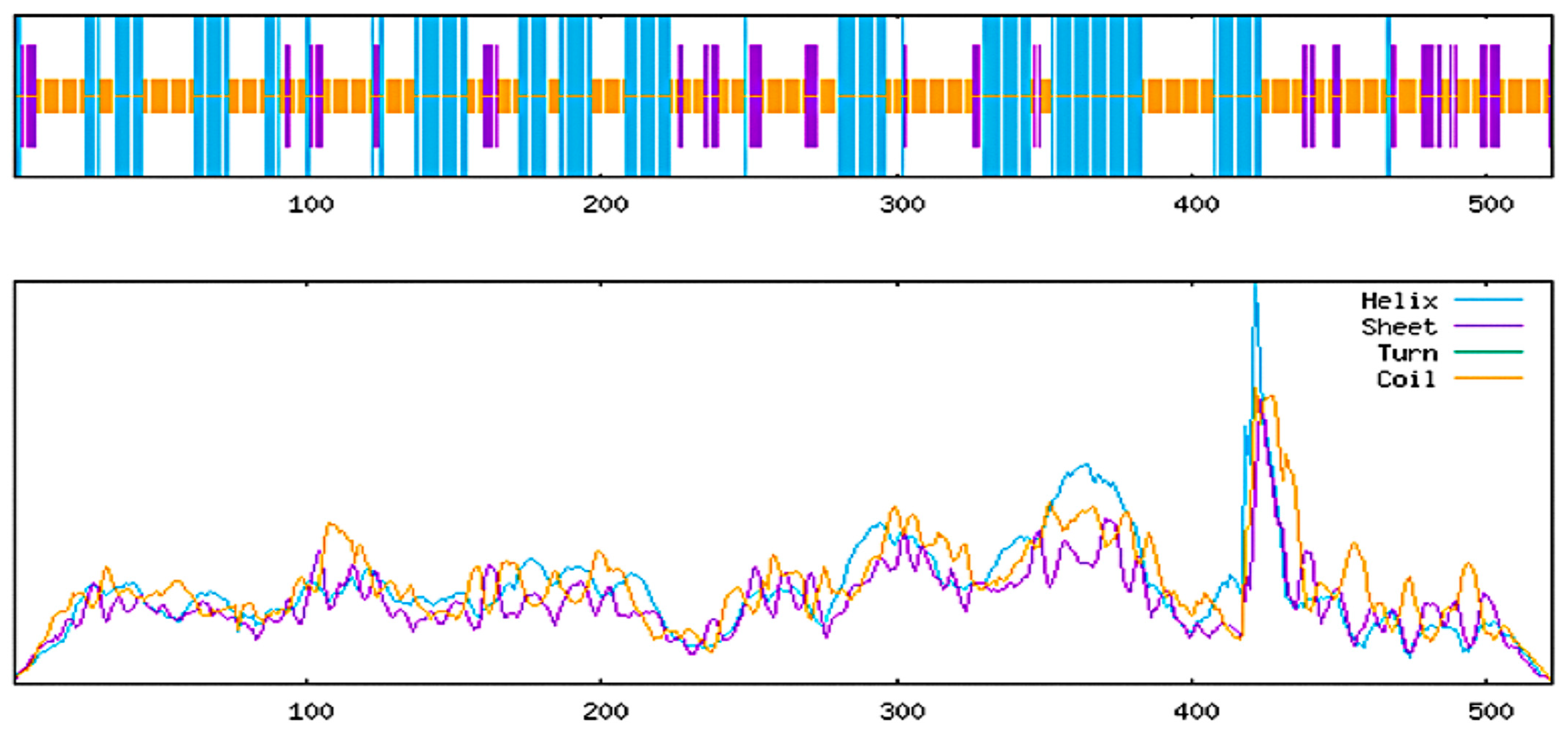
2.5.1. Predicted Secondary Structure
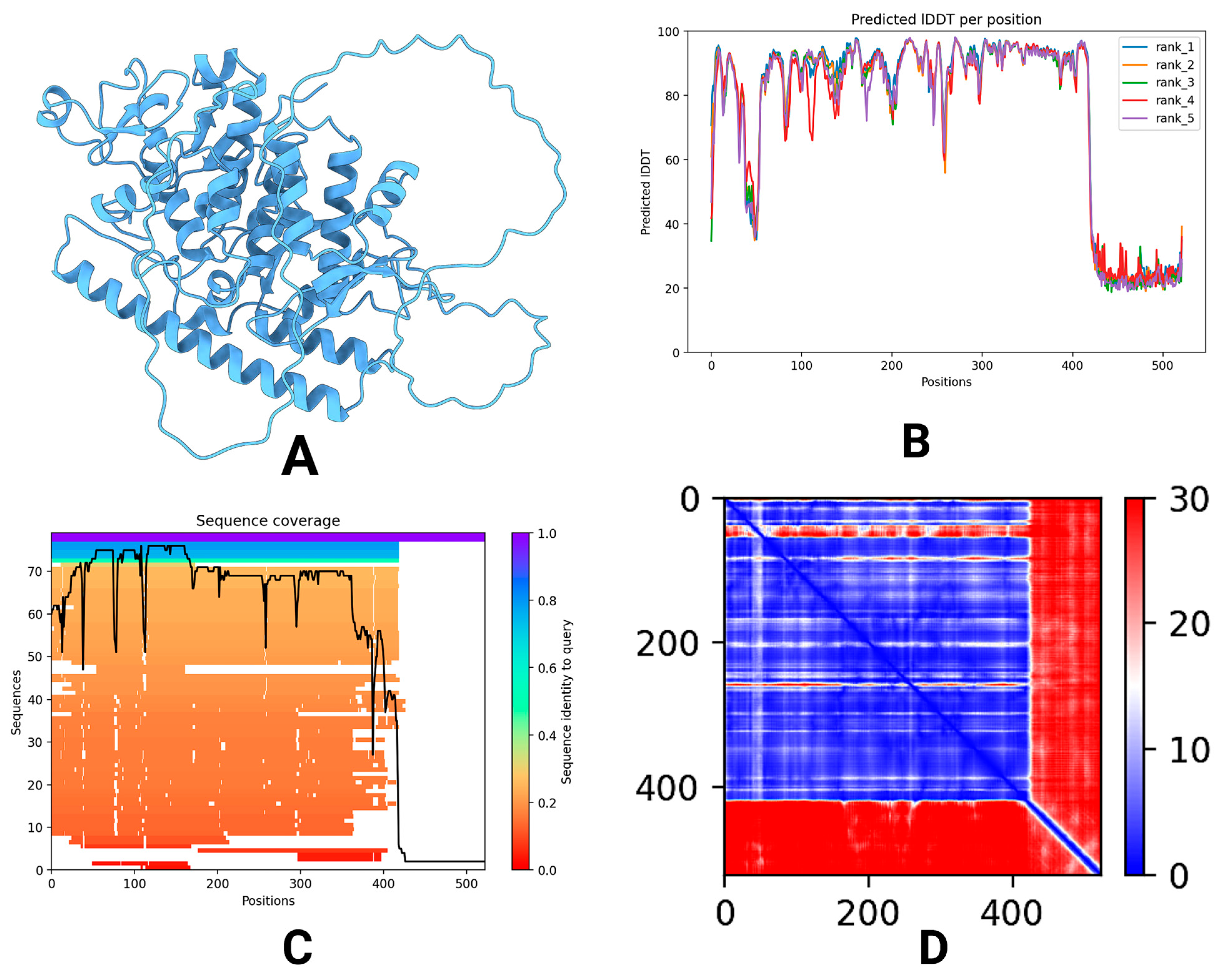
2.5.2. Predicted Tertiary Structure
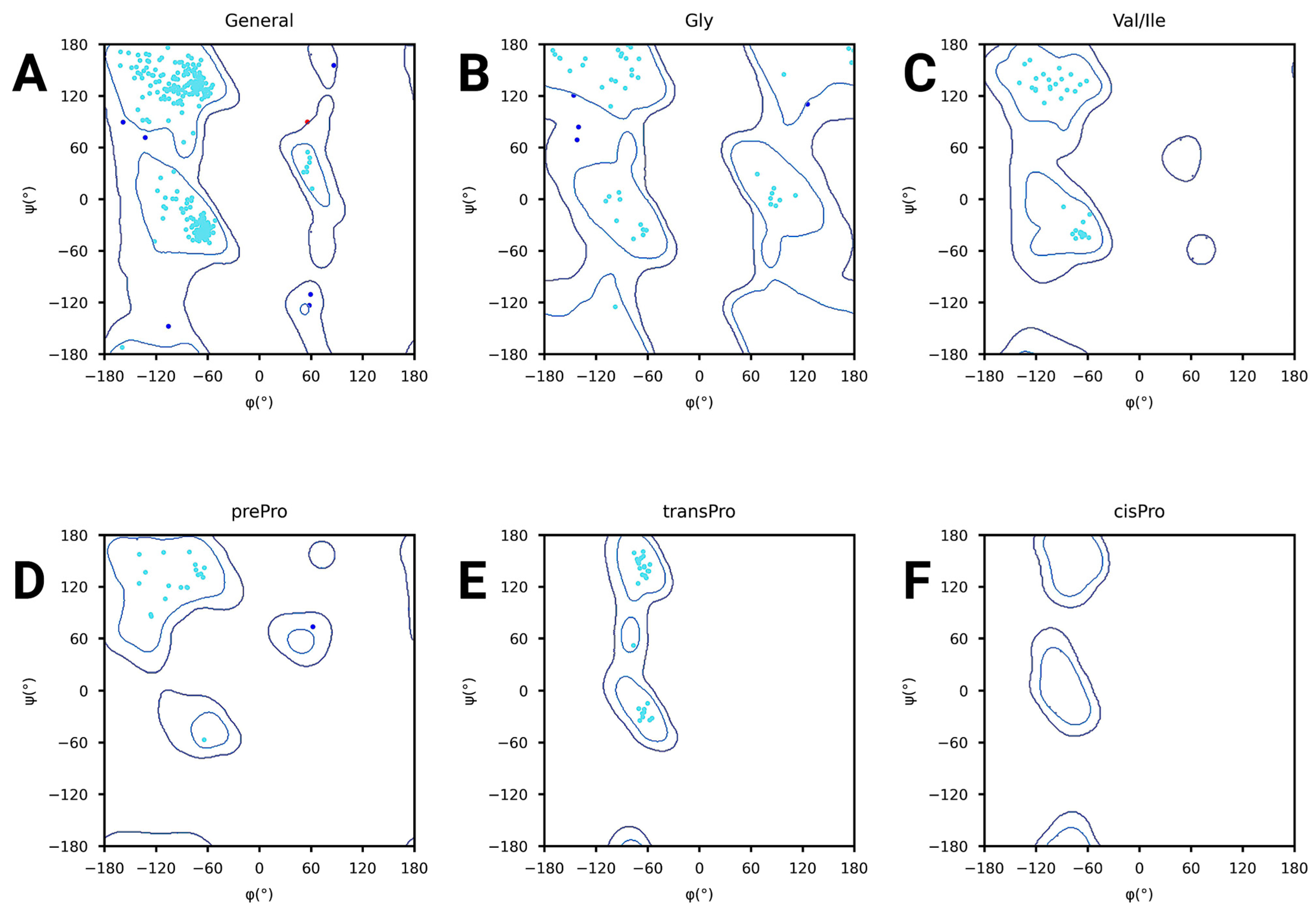
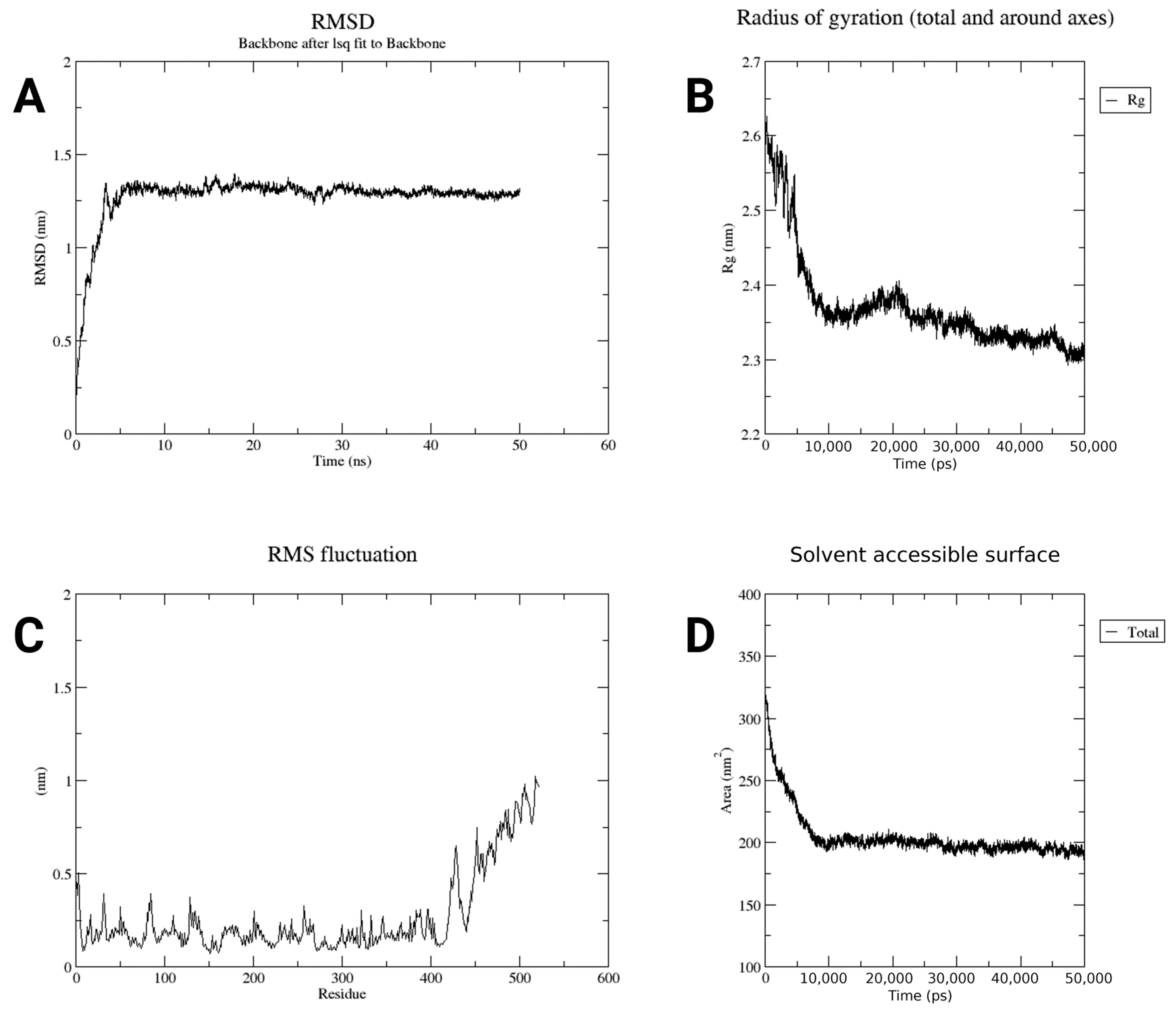
2.6. Structure Quality of Constructed Vaccine
2.7. Simulation of Immune Response
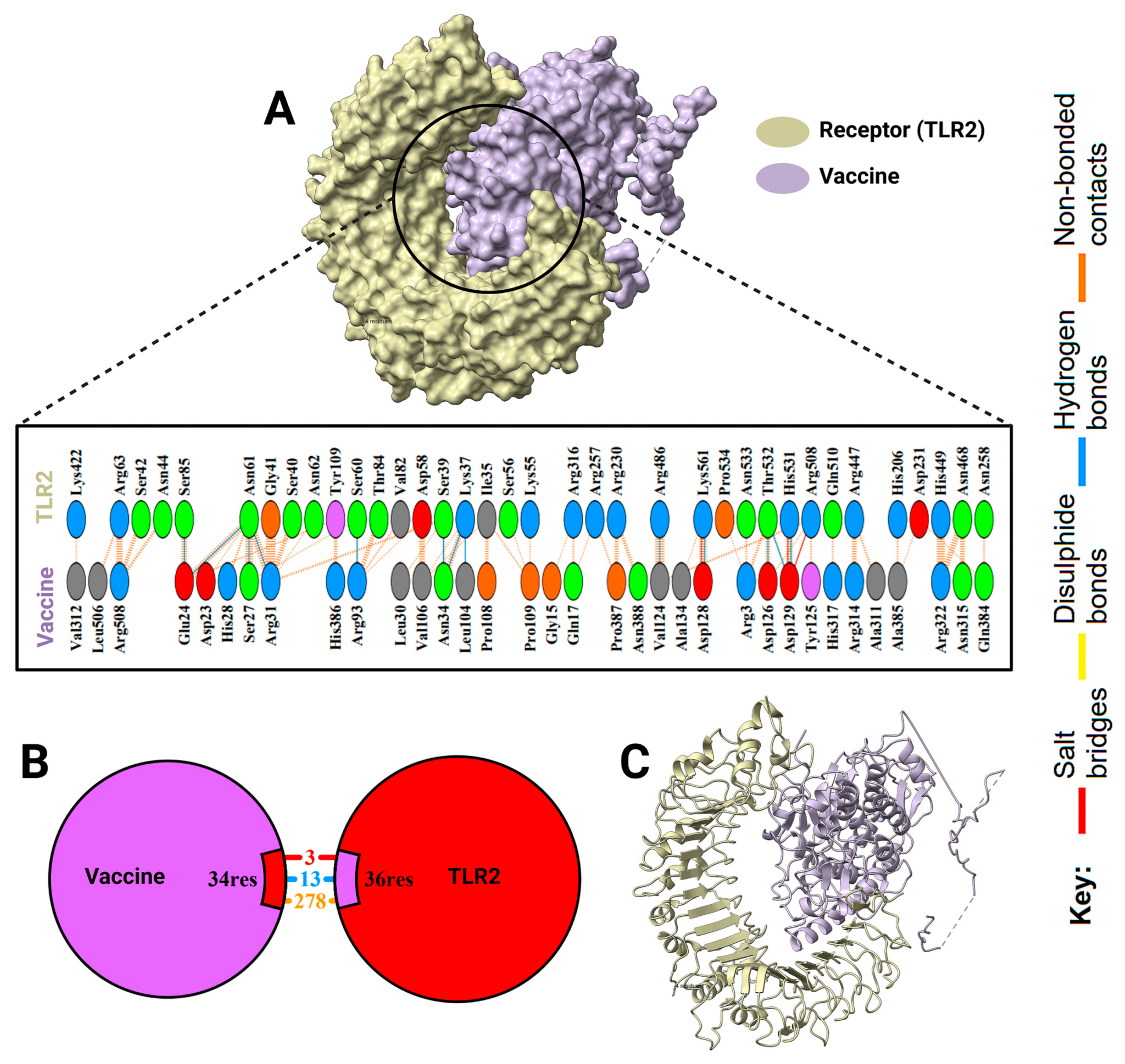
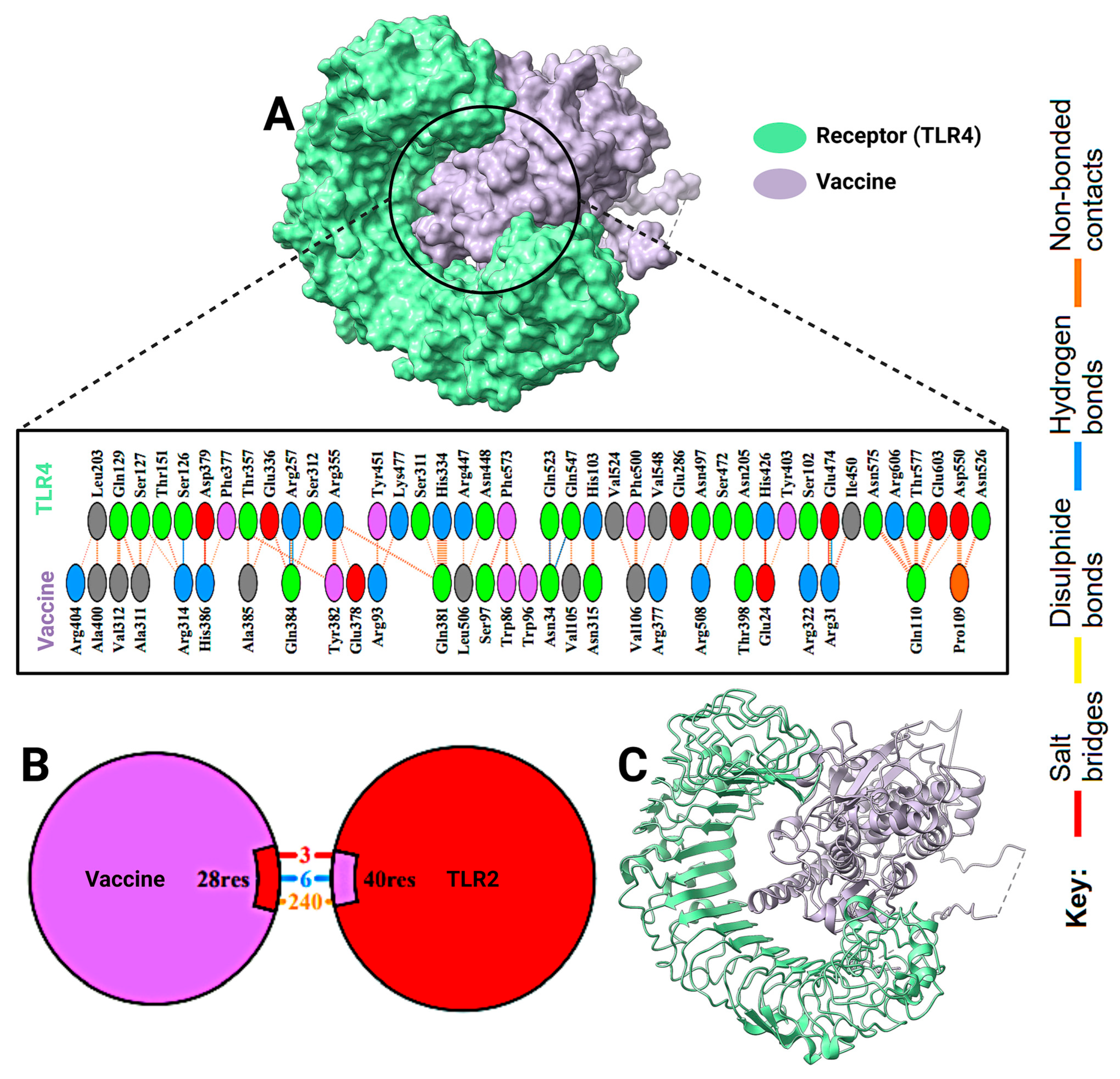
2.8. Docking Results
2.9. Computational Cloning and Virtual Gel Analysis
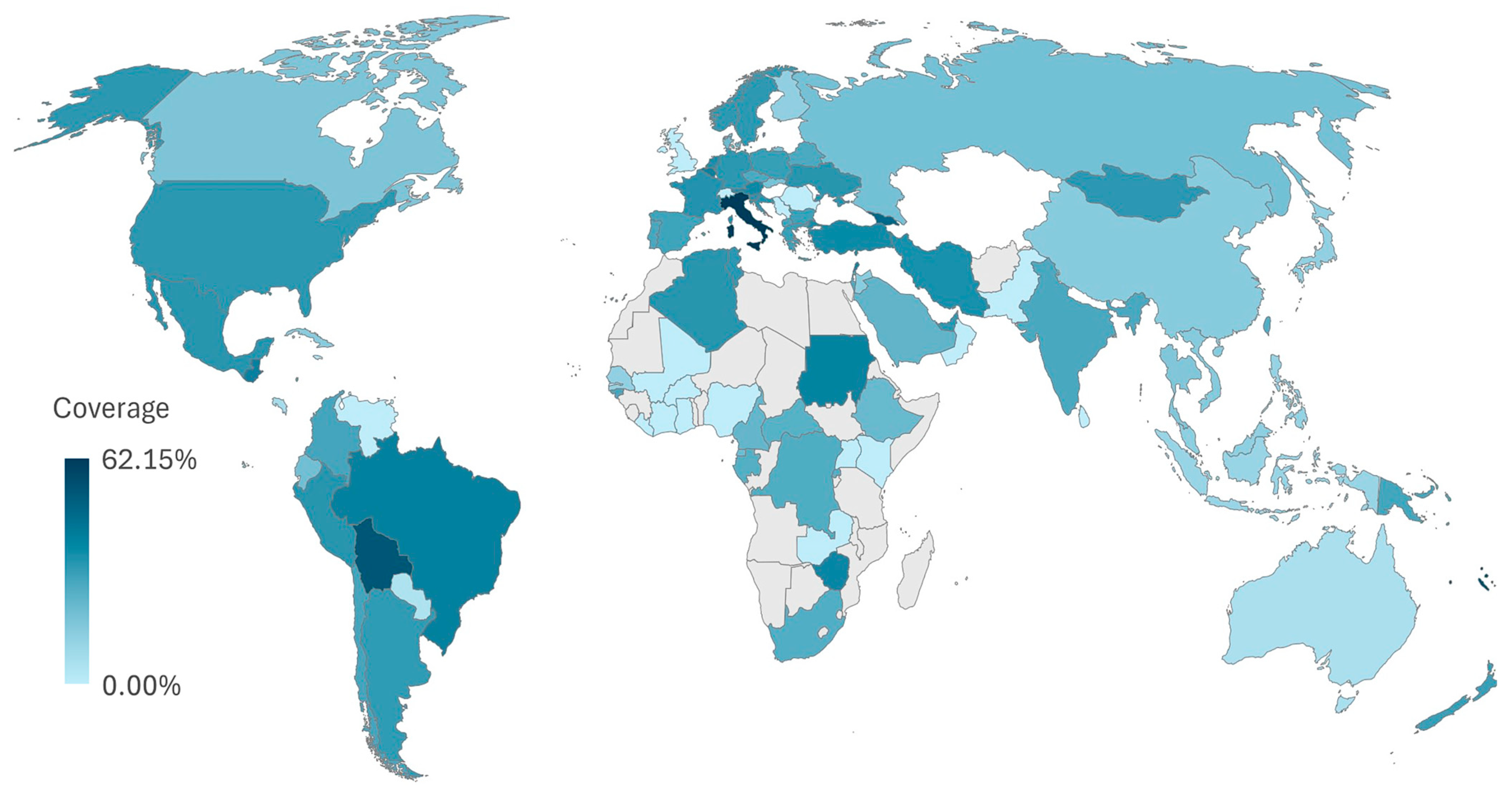
2.10. Vaccine Coverage Worldwide
3. Discussion
4. Materials and Methods
4.1. Protein Retrieval
4.2. Prediction and Evaluation of CTL Epitopes
4.3. Prediction and Evaluation of HTL Epitopes
4.4. Prediction and Evaluation of B Cell Epitopes
4.5. Constructed Vaccined
4.6. Structural and Chemical Attributes
4.7. Antigenicity and Allergenicity of Vaccine
4.8. Secondary and Tertiary Structure Prediction of Vaccine
4.8.1. Secondary Structure
4.8.2. Tertiary Structure Prediction
4.9. Structure Refiner
4.10. Structure Quality of Vaccine
4.10.1. Errat
4.10.2. ProSa-Web
4.10.3. Ramachandran Plotting
4.10.4. Vaccine in Water Simulation
4.11. Immune Response Simulation
4.12. Molecular Docking
4.12.1. Protein Preparation
4.12.2. Docking Setup
4.12.3. Model Analysis and Visualization
4.13. In Silico Cloning and Gel Electrophoresis
4.14. Population Coverage
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Betancourt, W.Q.; Gerba, C.P. Rethinking the Significance of Reovirus in Water and Wastewater. Food Environ. Virol. 2016, 8, 161–173. [Google Scholar] [CrossRef]
- Eledge, M.R.; Zita, M.D.; Boehme, K.W. Reovirus: Friend and Foe. Curr. Clin. Microbiol. Rep. 2019, 6, 132. [Google Scholar] [CrossRef] [PubMed]
- Asri, N.; Mohammadi, S.; Jahdkaran, M.; Rostami-Nejad, M.; Rezaei-Tavirani, M.; Mohebbi, S.R. Viral infections in celiac disease: What should be considered for better management. Clin. Exp. Med. 2025, 25, 25. [Google Scholar] [CrossRef]
- Bouziat, R.; Hinterleitner, R.; Brown, J.J.; Stencel-Baerenwald, J.E.; Ikizler, M.; Mayassi, T.; Meisel, M.; Kim, S.M.; Discepolo, V.; Pruijssers, A.J.; et al. Reovirus infection triggers inflammatory responses to dietary antigens and development of celiac disease. Science (1979) 2017, 356, 44–50. [Google Scholar] [CrossRef]
- Rosen, L.; Evans, H.E.; Spickard, A. REOVIRUS INFECTIONS IN HUMAN VOLUNTEERS. Am. J. Epidemiol. 1963, 77, 29–37. [Google Scholar] [CrossRef] [PubMed]
- DeBiasi, R.L.; Tyler, K.L. Orthoreoviruses and Orbiviruses. Mandell, Douglas, and Bennett’s Principles and Practice of Infectious Diseases 2015, 2, 1848–1850.e1. [Google Scholar] [CrossRef]
- Pan, M.; Alvarez-Cabrera, A.L.; Kang, J.S.; Wang, L.; Fan, C.; Zhou, Z.H. Asymmetric reconstruction of mammalian reovirus reveals interactions among RNA, transcriptional factor µ2 and capsid proteins. Nat. Commun. 2021, 12, 4176. [Google Scholar] [CrossRef] [PubMed]
- Gong, W.; Pan, C.; Cheng, P.; Wang, J.; Zhao, G.; Wu, X. Peptide-Based Vaccines for Tuberculosis. Front. Immunol. 2022, 13, 830497. [Google Scholar] [CrossRef]
- Kuri, P.R.; Goswami, P. Current Update on Rotavirus in-Silico Multiepitope Vaccine Design. ACS Omega 2022, 8, 190–207. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Yadav, P.D.; Cherian, S. Comprehensive immunoinformatics and bioinformatics strategies for designing a multi-epitope based vaccine targeting structural proteins of Nipah virus. Front. Immunol. 2025, 16, 1535322. [Google Scholar] [CrossRef]
- Ma, J.; Qiu, J.; Wang, S.; Ji, Q.; Xu, D.; Wang, H.; Wu, Z.; Liu, Q. A Novel Design of Multi-epitope Vaccine Against Helicobacter pylori by Immunoinformatics Approach. Int. J. Pept. Res. Ther. 2021, 27, 1027–1042. [Google Scholar] [CrossRef] [PubMed]
- Guruprasad, K.; Reddy, B.V.B.; Pandit, M.W. Correlation between stability of a protein and its dipeptide composition: A novel approach for predicting in vivo stability of a protein from its primary sequence. Protein Eng. 1990, 4, 155–161. [Google Scholar] [CrossRef]
- Wang, H.; Zhong, H.; Gao, C.; Zang, J.; Yang, D. The Distinct Properties of the Consecutive Disordered Regions Inside or Outside Protein Domains and Their Functional Significance. Int. J. Mol. Sci. 2021, 22, 10677. [Google Scholar] [CrossRef] [PubMed]
- Carugo, O. pLDDT Values in AlphaFold2 Protein Models Are Unrelated to Globular Protein Local Flexibility. Crystals 2023, 13, 1560. [Google Scholar] [CrossRef]
- Wuyun, Q.; Chen, Y.; Shen, Y.; Cao, Y.; Hu, G.; Cui, W.; Gao, J.; Zheng, W. Recent Progress of Protein Tertiary Structure Prediction. Molecules 2024, 29, 832. [Google Scholar] [CrossRef]
- Asif Rasheed, M.; Awais, M.; Aldhahrani, A.; Althobaiti, F.; Alhazmi, A.; Sattar, S.; Afzal, U.; Ali Baeshen, H.; Ali El Enshasy, H.; Joe Dailin, D.; et al. Designing a highly immunogenic multi epitope based subunit vaccine against Bacillus cereus. Saudi J. Biol. Sci. 2021, 28, 4859–4866. [Google Scholar] [CrossRef]
- Kothiyal, P.; Pant, K.; Singh, P. Immunoinformatics-Based Vaccine Designing. In Advancing Biotechnology: From Science to Therapeutics and Informatics; Springer: Berlin/Heidelberg, Germany, 2025; pp. 203–211. [Google Scholar] [CrossRef]
- Andersen, M.H.; Schrama, D.; Thor Straten, P.; Becker, J.C. Cytotoxic T Cells. J. Investig. Dermatol. 2006, 126, 32–41. [Google Scholar] [CrossRef]
- Huber, S.R.; van Beek, J.; de Jonge, J.; Luytjes, W.; van Baarle, D. T cell responses to viral infections-opportunities for peptide vaccination. Front. Immunol. 2014, 5, 83322. [Google Scholar] [CrossRef]
- Alexander, J.; Fikes, J.; Hoffman, S.; Franke, E.; Sacci, J.; Appella, E.; Chisari, F.V.; Guidotti, L.G.; Chesnut, R.W.; Livingston, B.; et al. The optimization of helper T lymphocyte (HTL) function in vaccine development. Immunol. Res. 1998, 18, 79–92. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Trincado, J.L.; Gomez-Perosanz, M.; Reche, P.A. Fundamentals and Methods for T- and B-Cell Epitope Prediction. J. Immunol. Res. 2017, 2017, 2680160. [Google Scholar] [CrossRef]
- Noorimotlagh, Z.; Karami, C.; Mirzaee, S.A.; Kaffashian, M.; Mami, S.; Azizi, M. Immune and bioinformatics identification of T cell and B cell epitopes in the protein structure of SARS-CoV-2: A systematic review. Int. Immunopharmacol. 2020, 86, 106738. [Google Scholar] [CrossRef] [PubMed]
- Invenção, M.d.C.V.; Macêdo, L.S.d.; Moura, I.A.d.; Santos, L.A.B.d.O.; Espinoza, B.C.F.; Pinho, S.S.d.; Leal, L.R.S.; Santos, D.L.d.; São Marcos, B.d.F.; Elsztein, C.; et al. Design and Immune Profile of Multi-Epitope Synthetic Antigen Vaccine Against SARS-CoV-2: An In Silico and In Vivo Approach. Vaccines 2025, 13, 149. [Google Scholar] [CrossRef]
- Gul, S.; Ahmad, S.; Ullah, A.; Ismail, S.; Khurram, M.; Qamar, M.T.U.; Hakami, A.R.; Alkhathami, A.G.; Alrumaihi, F.; Allemailem, K.S. Designing a Recombinant Vaccine against Providencia rettgeri Using Immunoinformatics Approach. Vaccines 2022, 10, 189. [Google Scholar] [CrossRef]
- Samantaray, M.; Pushan, S.S.; Rajagopalan, M.; Abrol, K.; Basumatari, J.; Murthy, T.P.K.; Ramaswamy, A. Designing a multi-epitope vaccine candidate against pandemic influenza a virus: An immunoinformatics and structural vaccinology approach. Mol. Divers. 2025, 1–20. [Google Scholar] [CrossRef]
- Lester, S.N.; Li, K. Toll-Like Receptors in Antiviral Innate Immunity. J. Mol. Biol. 2014, 426, 1246–1264. [Google Scholar] [CrossRef]
- Oliveira-Nascimento, L.; Massari, P.; Wetzler, L.M. The role of TLR2 ininfection and immunity. Front. Immunol. 2012, 3, 20479. [Google Scholar] [CrossRef]
- Kim, H.J.; Kim, H.; Lee, J.H.; Hwangbo, C. Toll-like receptor 4 (TLR4): New insight immune and aging. Immun. Ageing 2023, 20, 67. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Zhang, Y.; Li, H.; Wang, F.; Yao, S. Toll-like receptor 4: A potential therapeutic target for multiple human diseases. Biomed. Pharmacother. 2023, 166, 115338. [Google Scholar] [CrossRef]
- Mohammadipour, S.; Tavakkoli, H.; Fatemi, S.N.; Sharifi, A.; Mahmoudi, P. Designing a multi-epitope universal vaccine for concurrent infections of SARS-CoV-2 and influenza viruses using an immunoinformatics approach. BMC Infect. Dis. 2025, 25, 688. [Google Scholar] [CrossRef]
- Tahir ul Qamar, M.; Rehman, A.; Tusleem, K.; Ashfaq, U.A.; Qasim, M.; Zhu, X.; Fatima, I.; Shahid, F.; Chen, L.-L. Designing of a next generation multiepitope based vaccine (MEV) against SARS-COV-2: Immunoinformatics and in silico approaches. PLoS ONE 2020, 15, e0244176. [Google Scholar] [CrossRef]
- Jain, R.; Jain, A.; Mauro, E.; LeShane, K.; Densmore, D. ICOR: Improving codon optimization with recurrent neural networks. BMC Bioinform. 2023, 24, 132. [Google Scholar] [CrossRef]
- Larsen, M.V.; Lundegaard, C.; Lamberth, K.; Buus, S.; Lund, O.; Nielsen, M. Large-scale validation of methods for cytotoxic T-lymphocyte epitope prediction. BMC Bioinform. 2007, 8, 424. [Google Scholar] [CrossRef]
- Reynisson, B.; Alvarez, B.; Paul, S.; Peters, B.; Nielsen, M. NetMHCpan-4.1 and NetMHCIIpan-4.0: Improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucleic Acids Res. 2020, 48, 449–454. [Google Scholar] [CrossRef]
- Kupani, M.; Pandey, R.K.; Vashisht, S.; Singh, S.; Prajapati, V.K.; Mehrotra, S. Prediction of an immunogenic peptide ensemble and multi-subunit vaccine for Visceral leishmaniasis using bioinformatics approaches. Heliyon 2023, 9, e22121. [Google Scholar] [CrossRef] [PubMed]
- Dhanda, S.K.; Gupta, S.; Vir, P.; Raghava, G.P. Prediction of IL4 inducing peptides. Clin. Dev. Immunol. 2013, 2013, 263952. [Google Scholar] [CrossRef] [PubMed]
- Singh, O.; Hsu, W.L.; Su, E.C.Y. ILeukin10Pred: A Computational Approach for Predicting IL-10-Inducing Immunosuppressive Peptides Using Combinations of Amino Acid Global Features. Biology 2021, 11, 5. [Google Scholar] [CrossRef]
- Sun, P.; Ju, H.; Liu, Z.; Ning, Q.; Zhang, J.; Zhao, X.; Huang, Y.; Ma, Z.; Li, Y. Bioinformatics resources and tools for conformational B-cell epitope prediction. Comput. Math. Methods Med. 2013, 2013, 943636. [Google Scholar] [CrossRef]
- Martinelli, D.D. In silico vaccine design: A tutorial in immunoinformatics. Healthc. Anal. 2022, 2, 100044. [Google Scholar] [CrossRef]
- Garg, V.K.; Avashthi, H.; Tiwari, A.; Jain, P.A.; Ramkete, P.W.R.; Kayastha, A.M.; Singh, V.K. MFPPI–Multi FASTA ProtParam Interface. Bioinformation 2016, 12, 74. [Google Scholar] [CrossRef]
- Magrane, M.; Consortium, U.P. UniProt Knowledgebase: A hub of integrated protein data. Database 2011, 2011, bar009. [Google Scholar] [CrossRef]
- McGuffin, L.J.; Bryson, K.; Jones, D.T. The PSIPRED protein structure prediction server. Bioinformatics 2000, 16, 404–405. [Google Scholar] [CrossRef]
- Geourjon, C.; Deléage, G. SOPMA: Significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Bioinformatics 1995, 11, 681–684. [Google Scholar] [CrossRef]
- Yang, Z.; Zeng, X.; Zhao, Y.; Chen, R. AlphaFold2 and its applications in the fields of biology and medicine. Signal Transduct. Target. Ther. 2023, 8, 115. [Google Scholar] [CrossRef]
- Adiyaman, R.; Edmunds, N.S.; Genc, A.G.; Alharbi, S.M.A.; McGuffin, L.J. Improvement of protein tertiary and quaternary structure predictions using the ReFOLD refinement method and the AlphaFold2 recycling process. Bioinform. Adv. 2023, 3, vbad078. [Google Scholar] [CrossRef]
- Jumper, J.; Evansm, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Heo, L.; Park, H.; Seok, C. GalaxyRefine: Protein structure refinement driven by side-chain repacking. Nucleic Acids Res. 2013, 41, 384–388. [Google Scholar] [CrossRef]
- Lee, G.R.; Won, J.; Heo, L.; Seok, C. GalaxyRefine2: Simultaneous refinement of inaccurate local regions and overall protein structure. Nucleic Acids Res. 2019, 47, 451–455. [Google Scholar] [CrossRef]
- Laskowski, R.A.; MacArthur, M.W.; Thornton, J.M. Validation of protein models derived from experiment. Curr. Opin. Struct. Biol. 1998, 8, 631–639. [Google Scholar] [CrossRef]
- Colovos, C.; Yeates, T.O. Verification of protein structures: Patterns of nonbonded atomic interactions. Protein Sci. 1993, 2, 1511–1519. [Google Scholar] [CrossRef]
- Wiederstein, M.; Sippl, M.J. ProSA-web: Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007, 35, 407–410. [Google Scholar] [CrossRef]
- Yakubu, A.; De Donato, M.; Imumorin, I.G. Modelling functional and structural impact of non-synonymous single nucleotide polymorphisms of the DQA1 gene of three Nigerian goat breeds. S. Afr. J. Anim. Sci. 2017, 47, 146–156. [Google Scholar] [CrossRef]
- Kwofie, S.K.; Dankwa, B.; Odame, E.A.; Agamah, F.E.; Doe, L.P.A.; Teye, J.; Agyapong, O.; Miller, W.A.; Mosi, L.; Wilson, M.D. In Silico Screening of Isocitrate Lyase for Novel Anti-Buruli Ulcer Natural Products Originating from Africa. Molecules 2018, 23, 1550. [Google Scholar] [CrossRef]
- Kumar, M.; Rathore, R.S. RamPlot: A webserver to draw 2D, 3D and assorted Ramachandran (φ, ψ) maps. J. Appl. Crystallogr. 2025, 58, 630–636. [Google Scholar] [CrossRef]
- Oostenbrink, C.; Villa, A.; Mark, A.E.; Van Gunsteren, W.F. A biomolecular force field based on the free enthalpy of hydration and solvation: The GROMOS force-field parameter sets 53A5 and 53A6. J. Comput. Chem. 2004, 25, 1656–1676. [Google Scholar] [CrossRef]
- Rapin, N.; Lund, O.; Castiglione, F. Immune system simulation online. Bioinformatics 2011, 27, 2013–2014. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Meng, E.C.; Couch, G.S.; Croll, T.I.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 2021, 30, 70–82. [Google Scholar] [CrossRef]
- Yan, Y.; Zhang, D.; Zhou, P.; Li, B.; Huang, S.Y. HDOCK: A web server for protein–protein and protein–DNA/RNA docking based on a hybrid strategy. Nucleic Acids Res. 2017, 45, 365–373. [Google Scholar] [CrossRef]
- Yan, Y.; Tao, H.; He, J.; Huang, S.Y. The HDOCK server for integrated protein–protein docking. Nat. Protoc. 2020, 15, 1829–1852. [Google Scholar] [CrossRef]
- De Beer, T.A.P.; Berka, K.; Thornton, J.M.; Laskowski, R.A. PDBsum additions. Nucleic Acids Res. 2014, 42, 292–296. [Google Scholar] [CrossRef]
- Akhtar, H.; Akhtar, S.; Jan, S.U.; Khan, A.; Us Sahar, N.; Zaidi, S.S.; Qadri, I. Over expression of a synthetic gene encoding interferon lambda using relative synonymous Codon usage bias in Escherichia coli. Pak. J. Pharm. Sci. 2013, 26, 1181–1188. [Google Scholar]
- Grote, A.; Hiller, K.; Scheer, M.; Münch, R.; Nörtemann, B.; Hempel, D.C.; Jahn, D. JCat: A novel tool to adapt codon usage of a target gene to its potential expression host. Nucleic Acids Res. 2005, 33, 526–531. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Wang, P.; Kim, Y.; Haste-Andersen, P.; Beaver, J.; Bourne, P.E.; Bui, H.H.; Buus, S.; Frankild, S.; Greenbaum, J.; et al. Immune epitope database analysis resource (IEDB-AR). Nucleic Acids Res. 2008, 36, 513–518. [Google Scholar] [CrossRef]




| HTL Epitope | IFN | IL4 | IL10 |
|---|---|---|---|
| ANRPWEYDGTYARMT | Positive | Inducer | Inducer |
| AVSRNIHGWTGRAGN | Positive | Inducer | Inducer |
| DDYPFLARDPRFKHR | Positive | Inducer | Inducer |
| DYPFLARDPRFKHRV | Positive | Inducer | Inducer |
| FKHRVYQQLSAVTLL | Positive | Inducer | Inducer |
| FLDWISLGRGLATSA | Positive | Inducer | Inducer |
| LDWISLGRGLATSAL | Positive | Inducer | Inducer |
| NRPWEYDGTYARMTQ | Positive | Inducer | Inducer |
| PFLARDPRFKHRVYQ | Positive | Inducer | Inducer |
| QFLDWISLGRGLATS | Positive | Inducer | Inducer |
| RPWEYDGTYARMTQI | Positive | Inducer | Inducer |
| SANRPWEYDGTYARM | Positive | Inducer | Inducer |
| SRNIHGWTGRAGNQL | Positive | Inducer | Inducer |
| TQFLDWISLGRGLAT | Positive | Inducer | Inducer |
| VSRNIHGWTGRAGNQ | Positive | Inducer | Inducer |
| YPFLARDPRFKHRVY | Positive | Inducer | Inducer |
| Epitope Type | Epitope Sequence |
|---|---|
| CTL epitope | LVPTAGSRY |
| HTL epitope | ANRPWEYDGTYARMT FLDWISLGRGLATSA LDWISLGRGLATSAL NRPWEYDGTYARMTQ QFLDWISLGRGLATS TQFLDWISLGRGLAT |
| B cell epitope | CNRRGDAA |
| Properties | Value |
|---|---|
| Total carbon | 2577 |
| Total hydrogen | 3960 |
| Total nitrogen | 740 |
| Total oxygen | 745 |
| Total sulfur | 20 |
| Total number of atoms | 8042 |
| Formula | C2577H3960N740O745S20 |
| Estimated half-life | 30 h (mammalian reticulocytes, in vitro) >20 h (yeast, in vivo) >10 h (Escherichia coli, in vivo) |
| Instability index | 32.28 (protein is stable) |
| Aliphatic index | 77.99 |
| Grand average of hydropathicity (GRAVY) | −0.277 |
| Number of amino acids | 522 |
| Theoretical pI | 9.02 |
| Molecular weight | 57,869.50 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Noman, A.A.; Alhudhaibi, A.M.; Sharma, P.D.; Jannati, S.Z.; Akhter, T.; Siddika, S.; Khan, K.F.; Taha, T.H.; Alsalamah, S.A.; Abdallah, E.M. Computational Development of Multi-Epitope Reovirus Vaccine with Potent Predicted Binding to TLR2 and TLR4. Pharmaceuticals 2025, 18, 1632. https://doi.org/10.3390/ph18111632
Noman AA, Alhudhaibi AM, Sharma PD, Jannati SZ, Akhter T, Siddika S, Khan KF, Taha TH, Alsalamah SA, Abdallah EM. Computational Development of Multi-Epitope Reovirus Vaccine with Potent Predicted Binding to TLR2 and TLR4. Pharmaceuticals. 2025; 18(11):1632. https://doi.org/10.3390/ph18111632
Chicago/Turabian StyleNoman, Abdullah Al, Abdulrahman Mohammed Alhudhaibi, Pranab Dev Sharma, Sadia Zafur Jannati, Tahamina Akhter, Samira Siddika, Kaniz Fatama Khan, Tarek H. Taha, Sulaiman A. Alsalamah, and Emad M. Abdallah. 2025. "Computational Development of Multi-Epitope Reovirus Vaccine with Potent Predicted Binding to TLR2 and TLR4" Pharmaceuticals 18, no. 11: 1632. https://doi.org/10.3390/ph18111632
APA StyleNoman, A. A., Alhudhaibi, A. M., Sharma, P. D., Jannati, S. Z., Akhter, T., Siddika, S., Khan, K. F., Taha, T. H., Alsalamah, S. A., & Abdallah, E. M. (2025). Computational Development of Multi-Epitope Reovirus Vaccine with Potent Predicted Binding to TLR2 and TLR4. Pharmaceuticals, 18(11), 1632. https://doi.org/10.3390/ph18111632









