Repurposing Cofilin-Targeting Compounds for Ischemic Stroke Through Cheminformatics and Network Pharmacology
Abstract
1. Introduction
2. Results
2.1. QSAR Model Performance and Statistical Validation
2.2. Cross-Validation and Selection of Hit Models
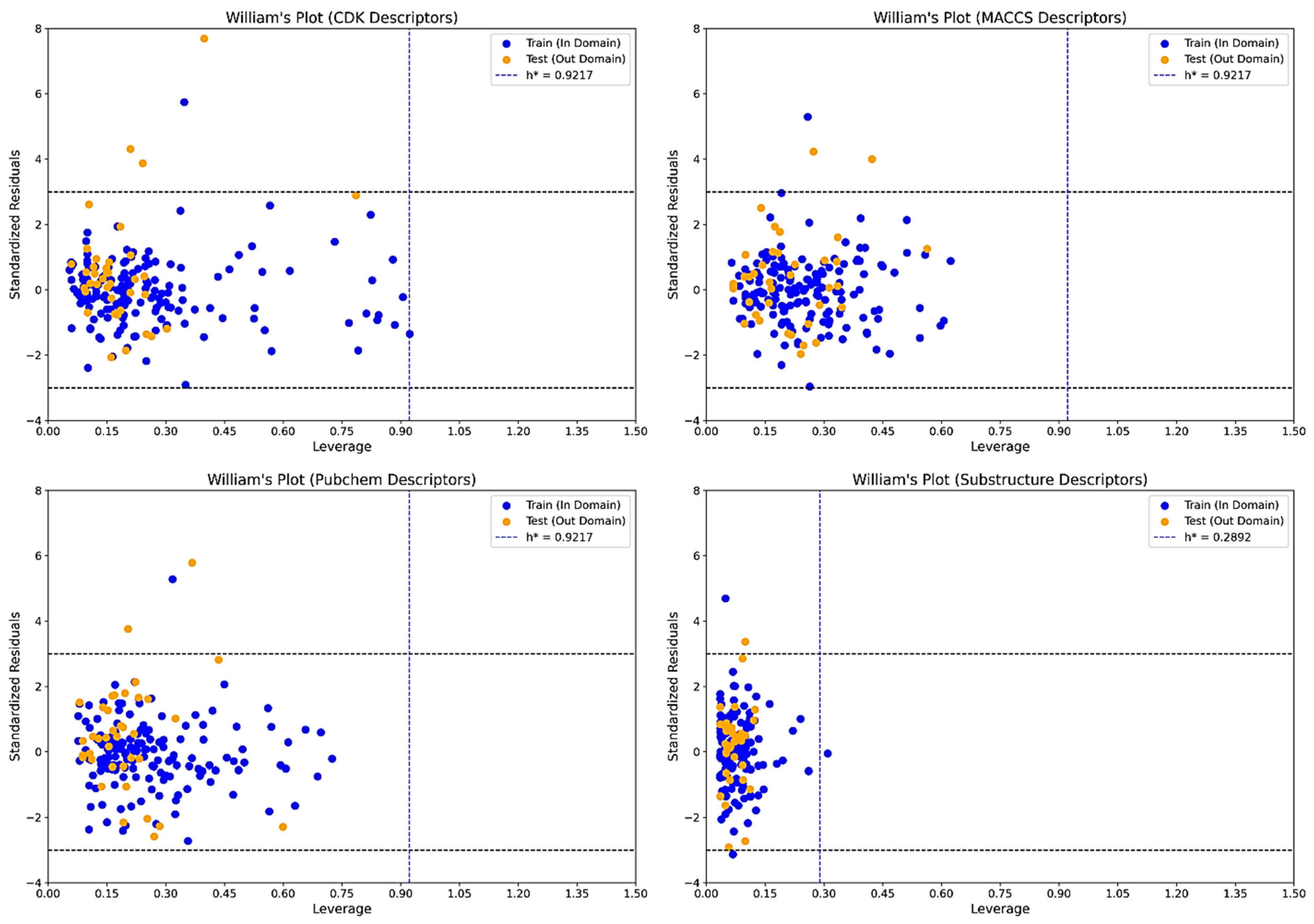
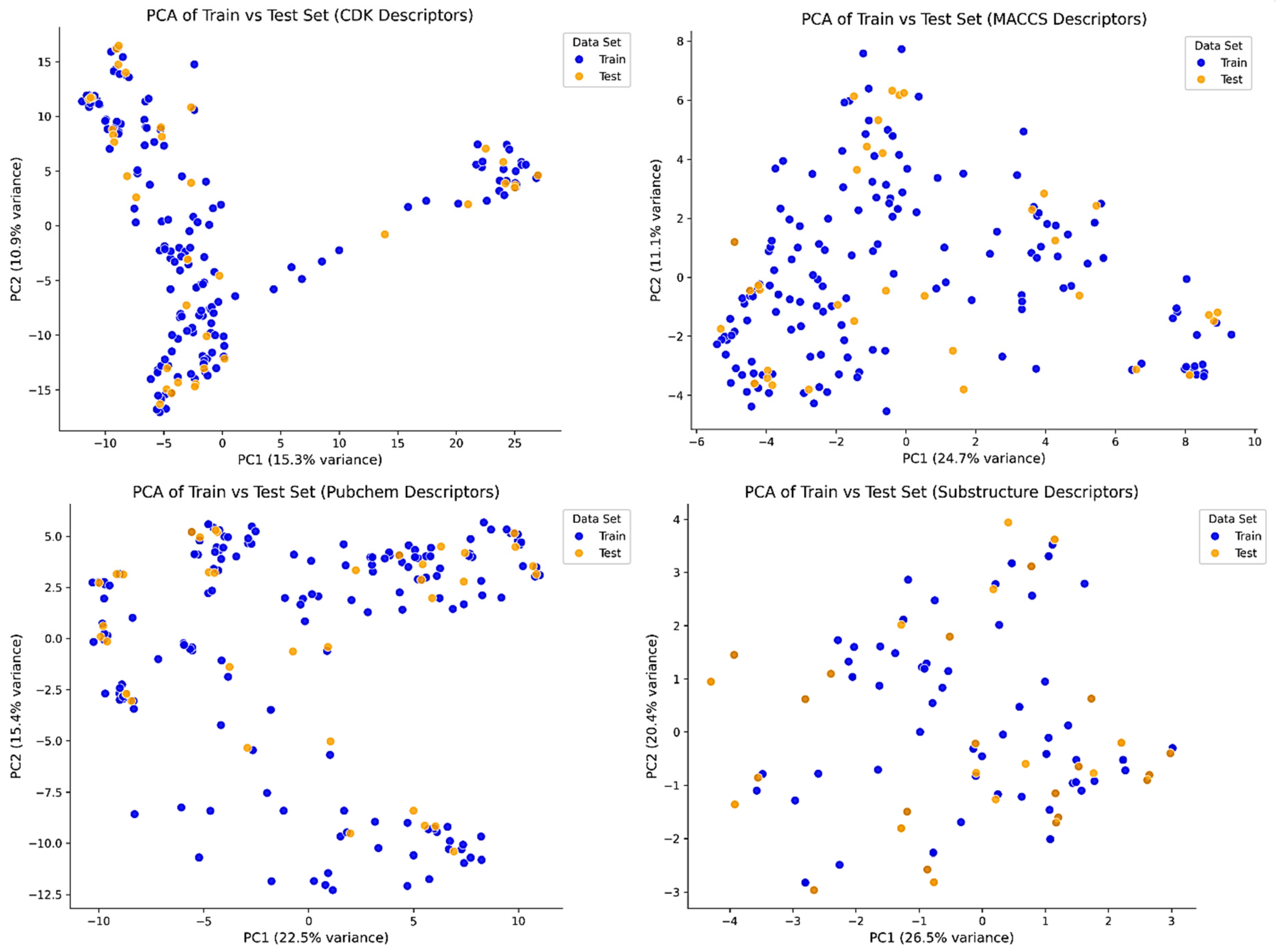
2.3. Applicability Domain Assessment Using William’s Plot
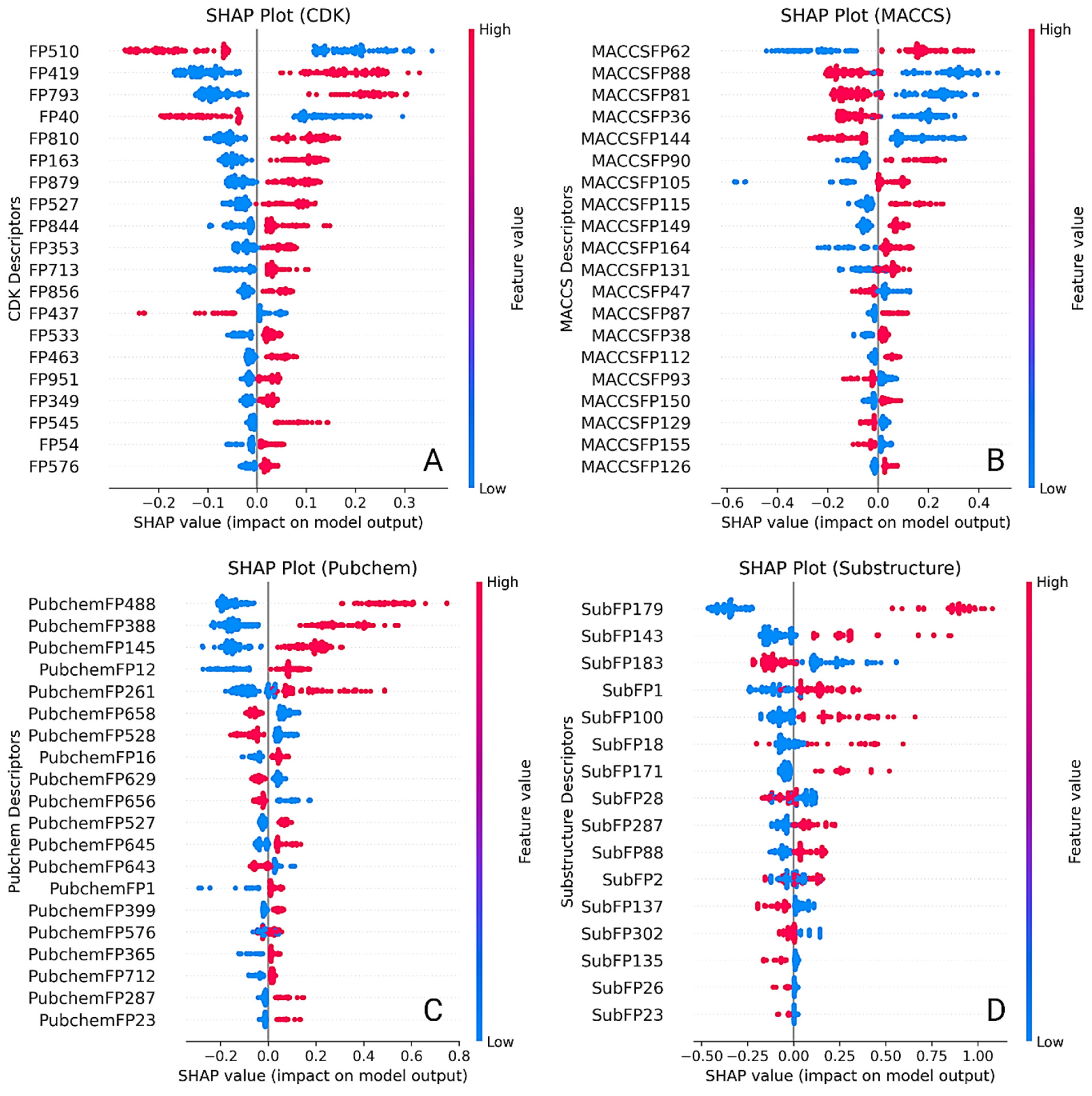
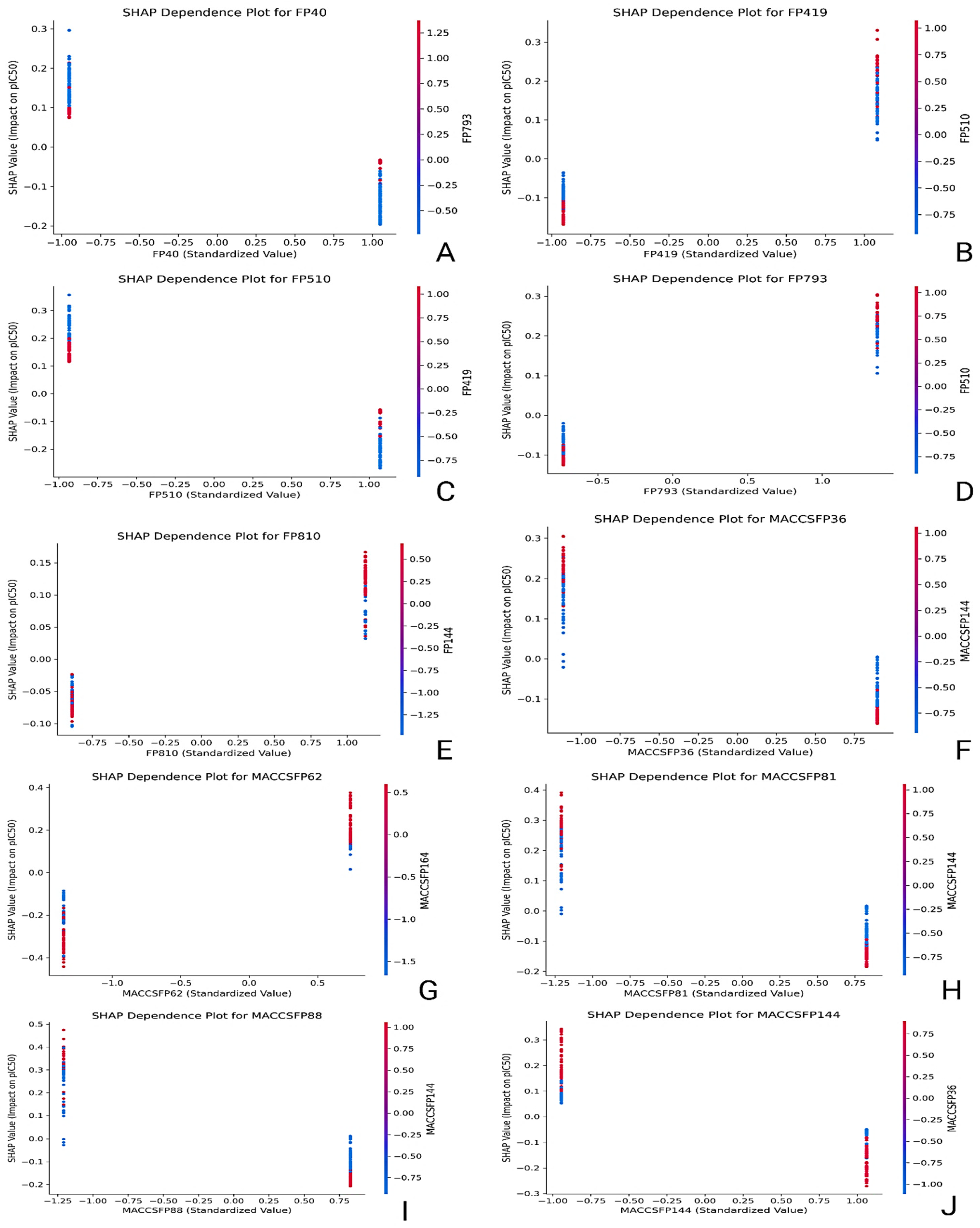
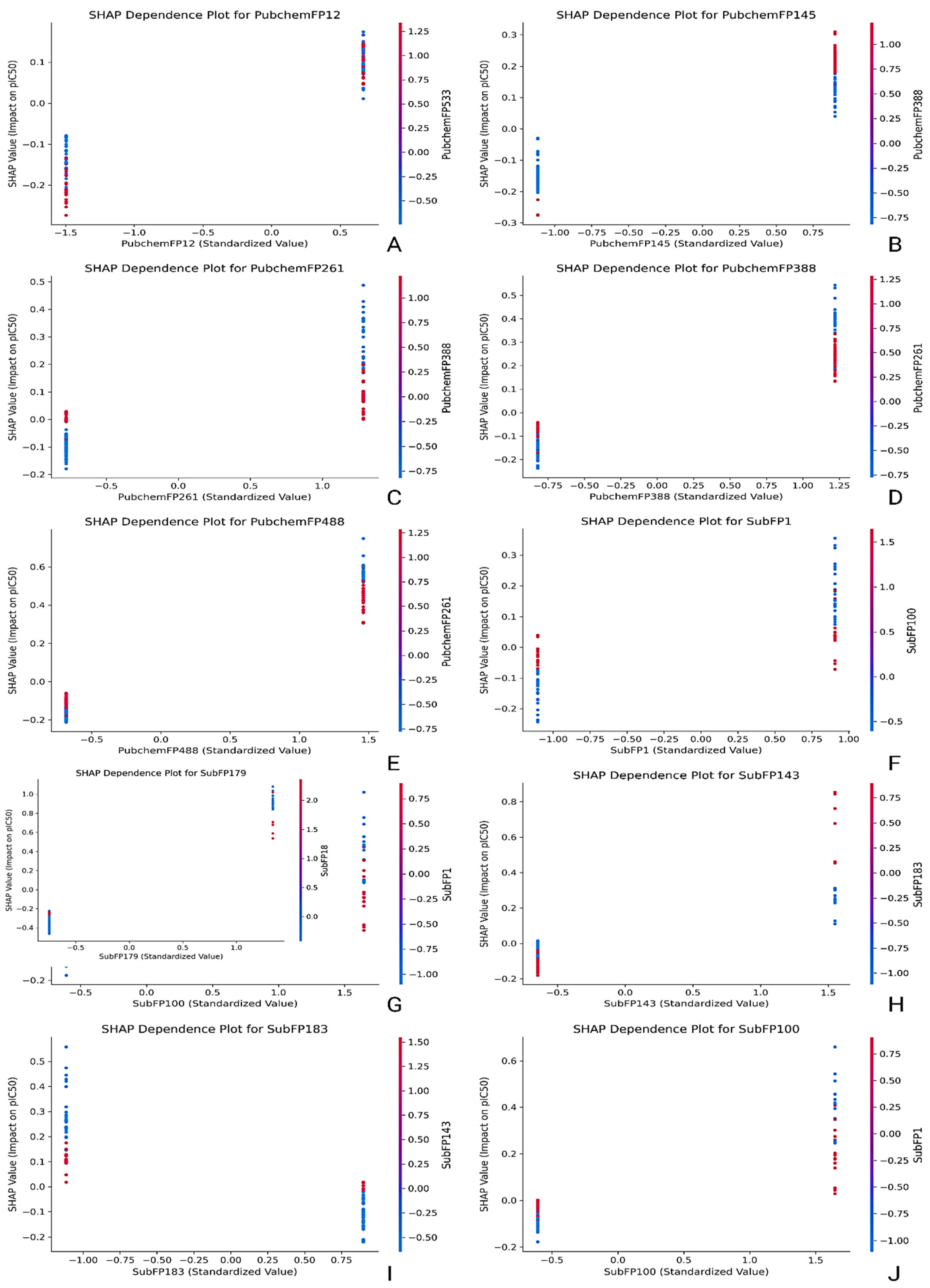
2.4. Feature Importance and Interpretability Using SHAP Analysis
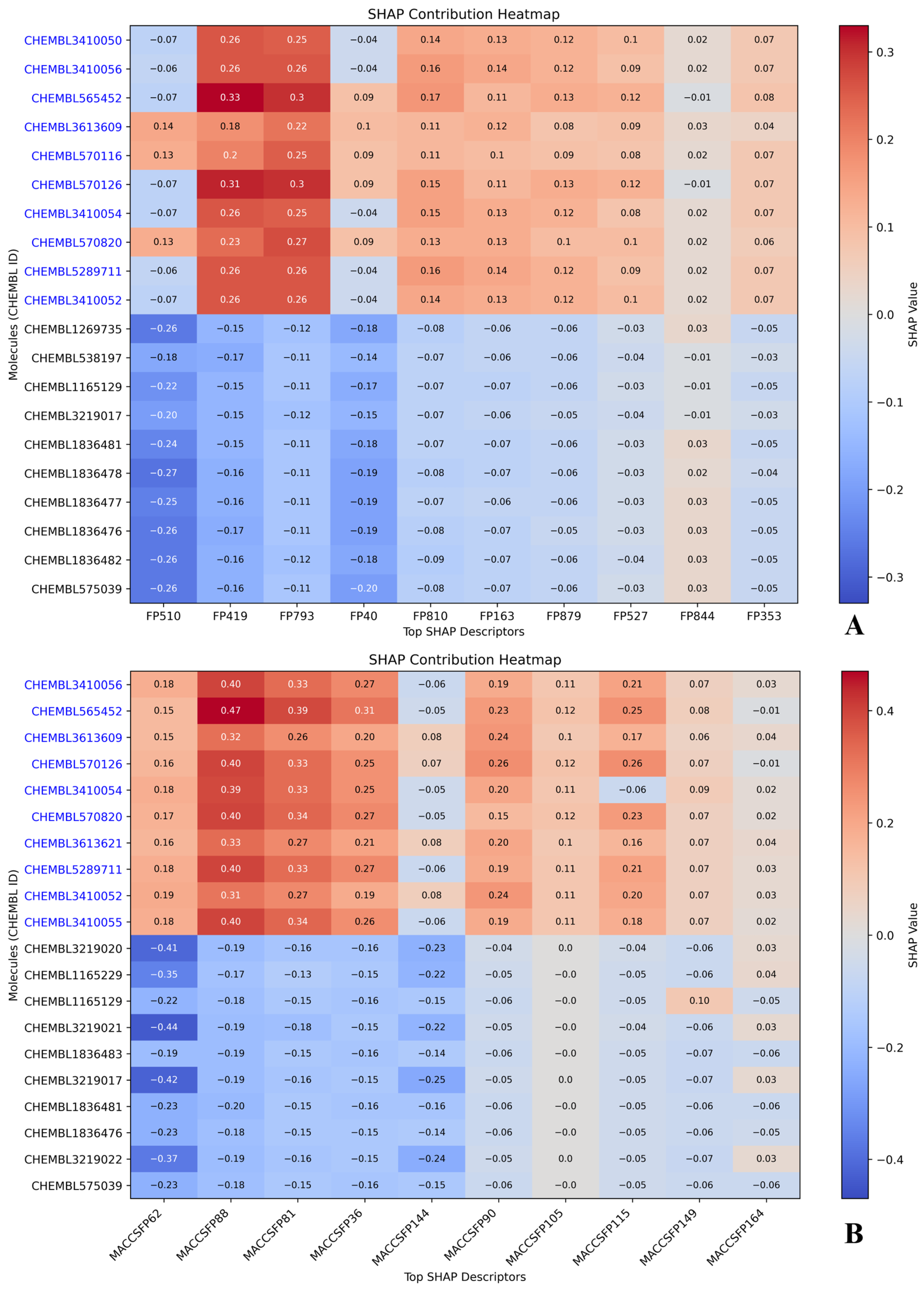
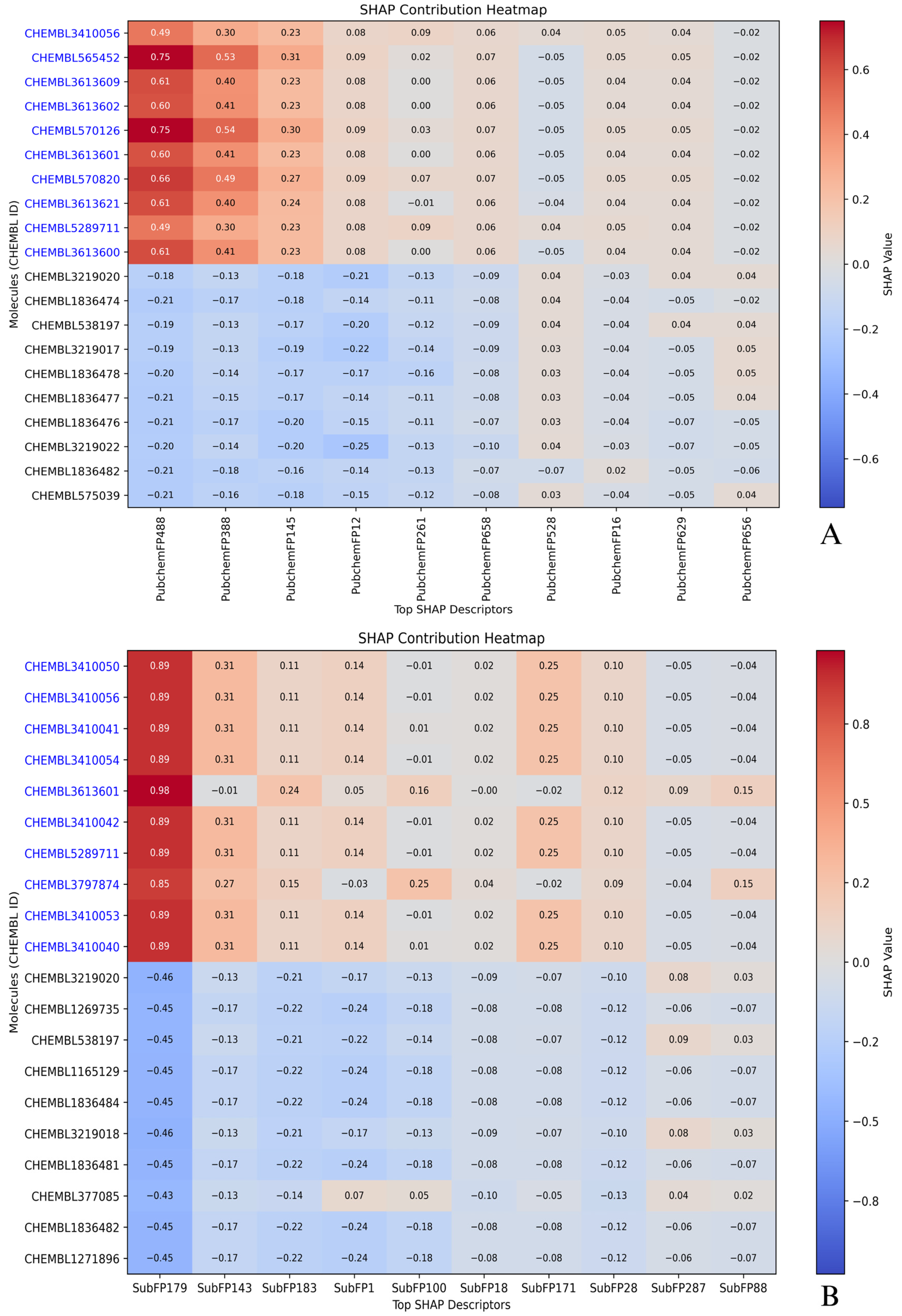
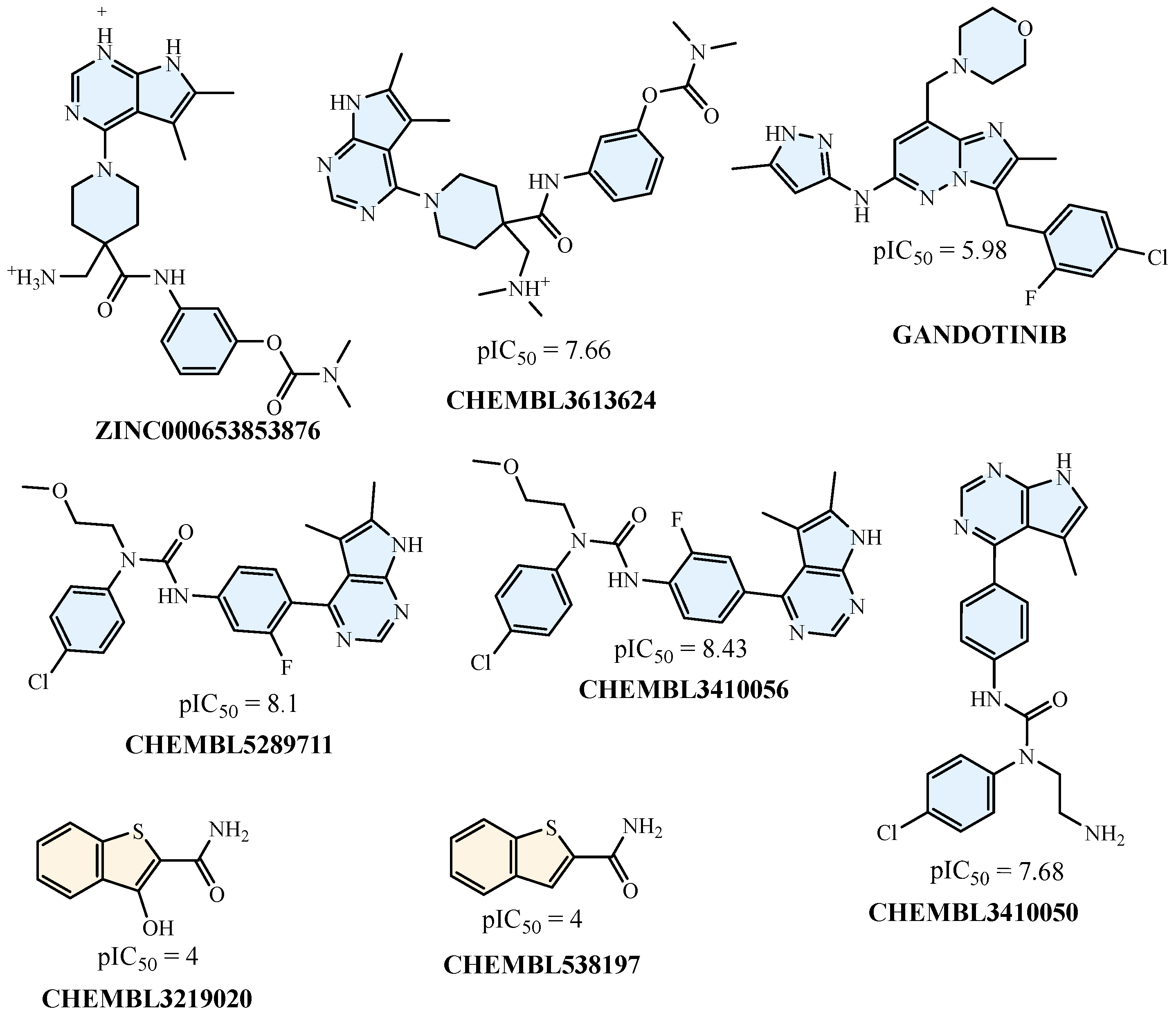
2.5. Compound-Level SHAP Attribution: Active vs. Inactive Profiles
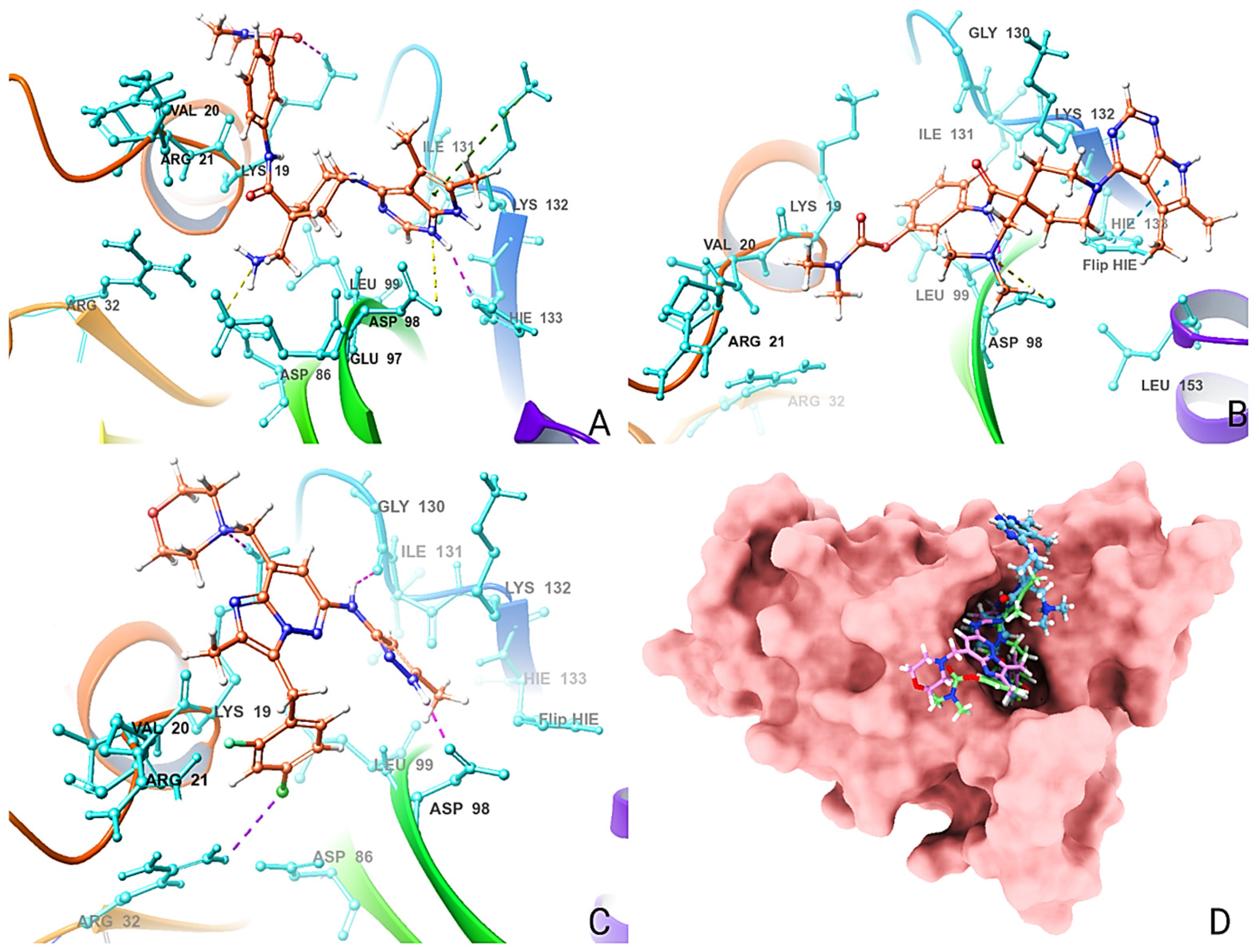
2.6. Molecular Docking Analysis
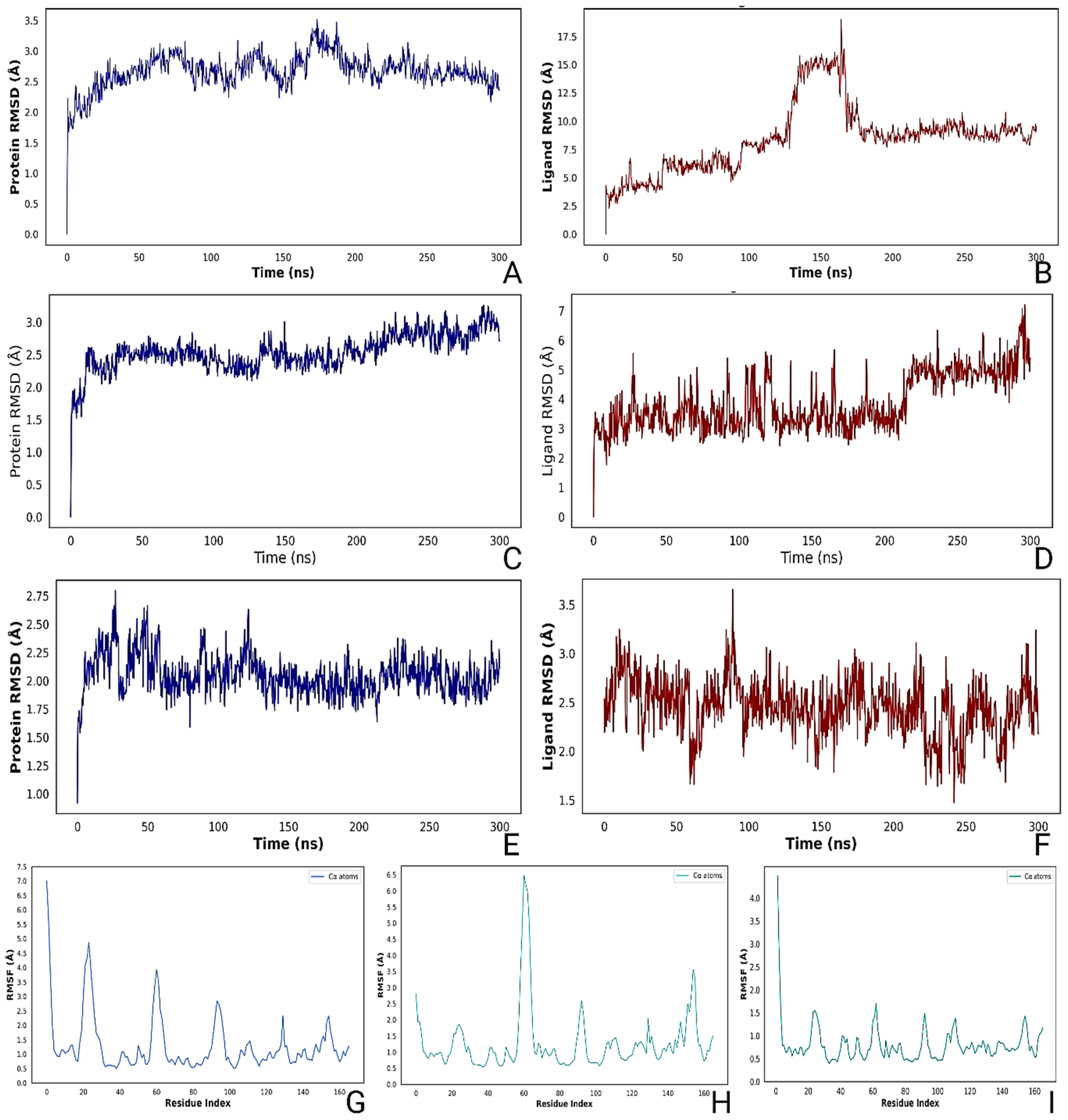
2.7. Molecular Dynamics Simulation and MMGBSA
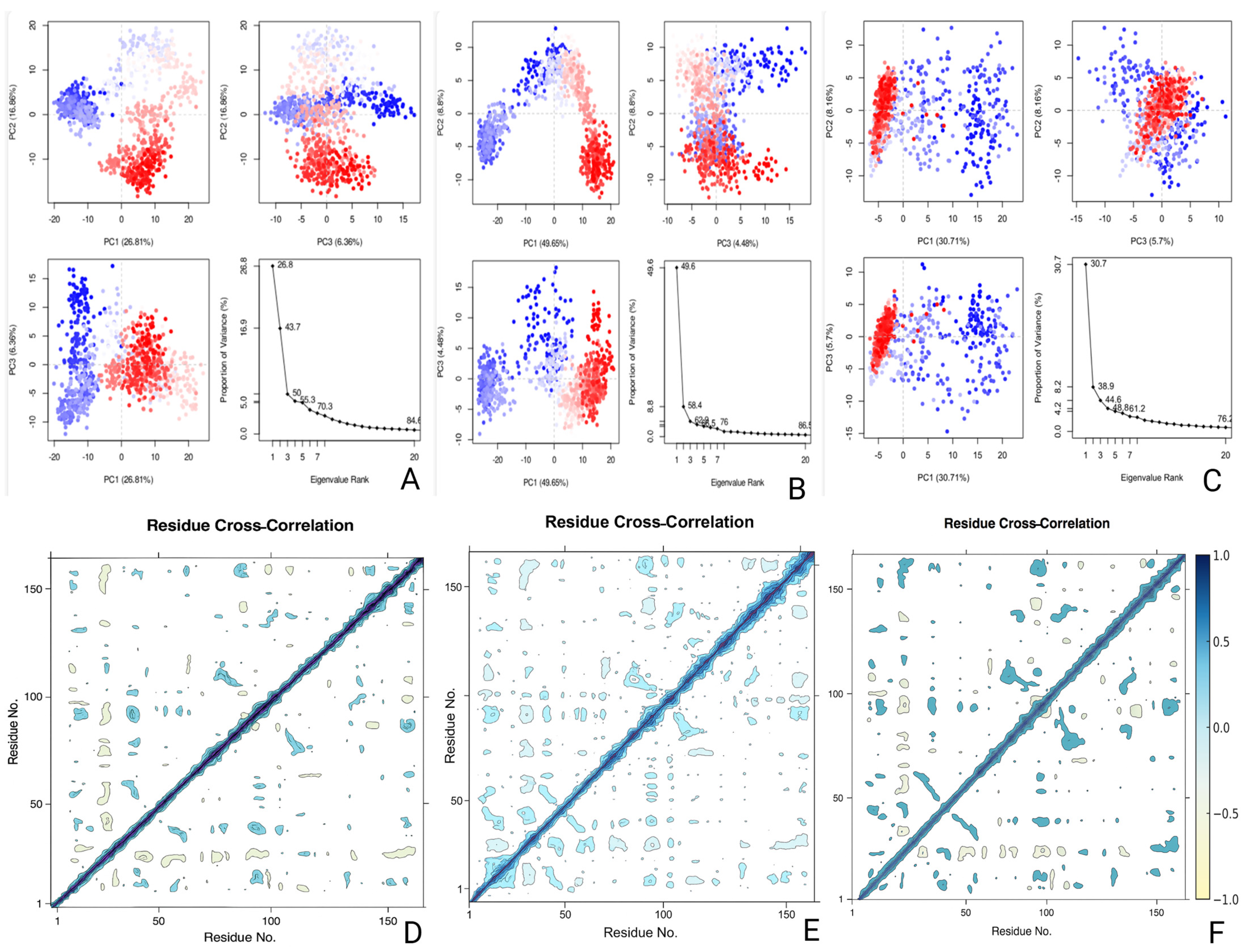
2.8. Energetic and Conformational Insights from MM-GBSA, DCCM, and PCA Analyses
2.9. Network Pharmacology-Based Functional Mapping of Hit Compounds in the Stroke Context
3. Discussion
4. Materials and Methods
4.1. Ligand Dataset Preparation
4.2. Molecular Descriptor Calculation
4.3. Feature Selection and Data Splitting
4.4. QSAR Model Development and Evaluation
4.5. Model Interpretation and Compound Prioritization
4.6. Molecular Docking Studies
4.7. Molecular Dynamics Simulations
4.8. Systematic Network Pharmacology
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pu, L.; Wang, L.; Zhang, R.; Zhao, T.; Jiang, Y.; Han, L. Projected Global Trends in Ischemic Stroke Incidence, Deaths and Disability-Adjusted Life Years From 2020 to 2030. Stroke 2023, 54, 1330–1339. [Google Scholar] [CrossRef] [PubMed]
- Alaqel, S.I.; Khan, A.; Alanazi, M.N.; Nayeem, N.; Ben Khaled, H.; Imran, M. Integrative Transcriptomic and Structural Analysis Identifies PTGS2 as a Key Target in Ischemic Stroke Associated with Neuroinflammation. Mol. Divers. 2025. [Google Scholar] [CrossRef]
- Mathias, K.; Machado, R.S.; Stork, S.; dos Santos, D.; Joaquim, L.; Generoso, J.; Danielski, L.G.; Barichello, T.; Prophiro, J.S.; Petronilho, F. Blood-Brain Barrier Permeability in the Ischemic Stroke: An Update. Microvasc. Res. 2024, 151, 104621. [Google Scholar] [CrossRef] [PubMed]
- Xing, J.; Wang, Y.; Peng, A.; Li, J.; Niu, X.; Zhang, K. The Role of Actin Cytoskeleton CFL1 and ADF/Cofilin Superfamily in Inflammatory Response. Front. Mol. Biosci. 2024, 11, 1408287. [Google Scholar] [CrossRef]
- Hoffmann, L.; Waclawczyk, M.S.; Tang, S.; Hanschmann, E.M.; Gellert, M.; Rust, M.B.; Culmsee, C. Cofilin1 Oxidation Links Oxidative Distress to Mitochondrial Demise and Neuronal Cell Death. Cell Death Dis. 2021, 12, 953. [Google Scholar] [CrossRef] [PubMed]
- Klemmensen, M.M.; Borrowman, S.H.; Pearce, C.; Pyles, B.; Chandra, B. Mitochondrial Dysfunction in Neurodegenerative Disorders. Neurotherapeutics 2024, 21, e00292. [Google Scholar] [CrossRef]
- Sexton, J.A.; Potchernikov, T.; Bibeau, J.P.; Casanova-Sepúlveda, G.; Cao, W.; Lou, H.J.; Boggon, T.J.; De La Cruz, E.M.; Turk, B.E. Distinct Functional Constraints Driving Conservation of the Cofilin N-Terminal Regulatory Tail. Nat. Commun. 2024, 15, 1426. [Google Scholar] [CrossRef]
- Sousa-Squiavinato, A.C.M.; Vasconcelos, R.I.; Gehren, A.S.; Fernandes, P.V.; de Oliveira, I.M.; Boroni, M.; Morgado-Díaz, J.A. Cofilin-1, LIMK1 and SSH1 Are Differentially Expressed in Locally Advanced Colorectal Cancer and According to Consensus Molecular Subtypes. Cancer Cell Int. 2021, 21, 69. [Google Scholar] [CrossRef]
- Paciello, F.; Battistoni, M.; Martini, S.; Simone, C.; Pastore, F.; Sollazzo, R.; Grassi, C.; Ripoli, C. Role of LIMK1-Cofilin-Actin Axis in Dendritic Spine Dynamics in Alzheimer’s Disease. Cell Death Dis. 2025, 16, 431. [Google Scholar] [CrossRef]
- Hemavathy, N.; Ranganathan, S.; Umashankar, V.; Jeyakanthan, J. Computational Development of Allosteric Peptide Inhibitors Targeting LIM Kinases as a Novel Therapeutic Intervention. In Cell Biochemistry and Biophysics; Springer: Berlin/Heidelberg, Germany, 2025. [Google Scholar] [CrossRef]
- Manetti, F. Recent Advances in the Rational Design and Development of LIM Kinase Inhibitors Are Not Enough to Enter Clinical Trials. Eur. J. Med. Chem. 2018, 155, 445–458. [Google Scholar] [CrossRef]
- Prunier, C.; Prudent, R.; Kapur, R.; Sadoul, K.; Lafanechère, L. LIM Kinases: Cofilin and Beyond. Oncotarget 2017, 8, 41749. [Google Scholar] [CrossRef] [PubMed]
- Hamill, S.; Lou, H.J.; Turk, B.E.; Boggon, T.J. Structural Basis for Noncanonical Substrate Recognition of Cofilin/ADF Proteins by LIM Kinases. Mol. Cell 2016, 62, 397–408. [Google Scholar] [CrossRef] [PubMed]
- Klejnot, M.; Gabrielsen, M.; Cameron, J.; Mleczak, A.; Talapatra, S.K.; Kozielski, F.; Pannifer, A.; Olson, M.F. Analysis of the Human Cofilin 1 Structure Reveals Conformational Changes Required for Actin Binding. Acta Crystallogr. Sect. D Biol. Crystallogr. 2013, 69, 1780–1788. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.K.; Su, Y.T.; Ho, Y.C.; Lee, Y.K.; Chu, T.H.; Chen, K.T.; Wu, C.C. HDAC1 Is Involved in Neuroinflammation and Blood-Brain Barrier Damage in Stroke Pathogenesis. J. Inflamm. Res. 2023, 16, 4103–4116. [Google Scholar] [CrossRef]
- Chen, J.S.; Wang, H.K.; Hsu, C.Y.; Su, Y.T.; Chen, J.S.; Liang, C.L.; Hsieh, P.C.H.; Wu, C.C.; Kwan, A.L. HDAC1 Deregulation Promotes Neuronal Loss and Deficit of Motor Function in Stroke Pathogenesis. Sci. Rep. 2021, 11, 16354. [Google Scholar] [CrossRef]
- Qi, D.; Wei, P.; Cui, Y.; Lenahan, C.; Tao, X.; Jin, P. Inhibition of C3a/C3aR by SB290157 Attenuates Neuroinflammation via PKC/P38/NLRP3 Signaling Pathway After Intracerebral Hemorrhage. Neurocrit. Care 2025, 43, 44–58. [Google Scholar] [CrossRef]
- Teertam, S.K.; Prakash Babu, P. Differential Role of SIRT1/MAPK Pathway during Cerebral Ischemia in Rats and Humans. Sci. Rep. 2021, 11, 6339. [Google Scholar] [CrossRef]
- Appunni, S.; Gupta, D.; Rubens, M.; Ramamoorthy, V.; Singh, H.N.; Swarup, V. Deregulated Protein Kinases: Friend and Foe in Ischemic Stroke. Mol. Neurobiol. 2021, 58, 6471–6489. [Google Scholar] [CrossRef]
- Jubaidi, F.F.; Zainalabidin, S.; Taib, I.S.; Abdul Hamid, Z.; Mohamad Anuar, N.N.; Jalil, J.; Mohd Nor, N.A.; Budin, S.B. The Role of PKC-MAPK Signalling Pathways in the Development of Hyperglycemia-Induced Cardiovascular Complications. Int. J. Mol. Sci. 2022, 23, 8582. [Google Scholar] [CrossRef]
- Wei, Y.B.; McCarthy, M.; Ren, H.; Carrillo-Roa, T.; Shekhtman, T.; DeModena, A.; Liu, J.J.; Leckband, S.G.; Mors, O.; Rietschel, M.; et al. A Functional Variant in the Serotonin Receptor 7 Gene (HTR7), Rs7905446, Is Associated with Good Response to SSRIs in Bipolar and Unipolar Depression. Mol. Psychiatry 2019, 25, 1312–1322. [Google Scholar] [CrossRef]
- Zhang, Y.; Yang, L.; Zhang, Q.; Li, C.; Mao, F.; Zhuo, C. The Molecular Mechanisms through Which Psilocybin Prevents Suicide: Evidence from Network Pharmacology and Molecular Docking Analyses. Transl. Psychiatry 2025, 15, 202. [Google Scholar] [CrossRef] [PubMed]
- Gu, M. BPS2025—Cofilin’s Role in the Growth of Cytoskeleton. Biophys. J. 2025, 124, 470a. [Google Scholar] [CrossRef]
- Kovaleva, T.F.; Maksimova, N.S.; Zhukov, I.Y.; Pershin, V.I.; Mukhina, I.V.; Gainullin, M.R. Cofilin: Molecular and Cellular Functions and Its Role in the Functioning of the Nervous System. Neurochem. J. 2019, 13, 11–19. [Google Scholar] [CrossRef]
- Shehjar, F.; Almarghalani, D.A.; Mahajan, R.; Hasan, S.A.M.; Shah, Z.A. The Multifaceted Role of Cofilin in Neurodegeneration and Stroke: Insights into Pathogenesis and Targeting as a Therapy. Cells 2024, 13, 188. [Google Scholar] [CrossRef]
- Cichon, J.; Sun, C.; Chen, B.; Jiang, M.; Chen, X.A.; Sun, Y.; Wang, Y.; Chen, G. Cofilin aggregation blocks intracellular trafficking and induces synaptic loss in hippocampal neurons. J. Biol. Chem. 2012, 287, 3919–3929. [Google Scholar] [CrossRef]
- Ben Zablah, Y.; Merovitch, N.; Jia, Z. The Role of ADF/Cofilin in Synaptic Physiology and Alzheimer’s Disease. Front. Cell Dev. Biol. 2020, 8, 594998. [Google Scholar] [CrossRef]
- Mendez, D.; Gaulton, A.; Bento, A.P.; Chambers, J.; De Veij, M.; Félix, E.; Magariños, M.P.; Mosquera, J.F.; Mutowo, P.; Nowotka, M.; et al. ChEMBL: Towards Direct Deposition of Bioassay Data. Nucleic Acids Res. 2019, 47, D930–D940. [Google Scholar] [CrossRef]
- Yap, C.W. PaDEL-Descriptor: An Open-Source Software to Calculate Molecular Descriptors and Fingerprints. J. Comput. Chem. 2011, 32, 1466–1474. [Google Scholar] [CrossRef]
- Huang, X.; Kroening, D.; Ruan, W.; Sharp, J.; Sun, Y.; Thamo, E.; Wu, M.; Yi, X. A Survey of Safety and Trustworthiness of Deep Neural Networks: Verification, Testing, Adversarial Attack and Defence, and Interpretability. Comput. Sci. Rev. 2020, 37, 100270. [Google Scholar] [CrossRef]
- Jiang, S.; Liang, Y.; Shi, S.; Wu, C.; Shi, Z. Improving Predictions and Understanding of Primary and Ultimate Biodegradation Rates with Machine Learning Models. Sci. Total Environ. 2023, 904, 166623. [Google Scholar] [CrossRef] [PubMed]
- Dutschmann, T.M.; Schlenker, V.; Baumann, K. Chemoinformatic Regression Methods and Their Applicability Domain. Mol. Inform. 2024, 43, e202400018. [Google Scholar] [CrossRef]
- Yang, Y.; Yao, K.; Repasky, M.P.; Leswing, K.; Abel, R.; Shoichet, B.K.; Jerome, S.V. Efficient Exploration of Chemical Space with Docking and Deep Learning. J. Chem. Theory Comput. 2021, 17, 7106–7119. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Wu, C.; Ghoreishi, D.; Chen, W.; Wang, L.; Damm, W.; Ross, G.A.; Dahlgren, M.K.; Russell, E.; Von Bargen, C.D.; et al. OPLS4: Improving Force Field Accuracy on Challenging Regimes of Chemical Space. J. Chem. Theory Comput. 2021, 17, 4291–4300. [Google Scholar] [CrossRef]
- Irwin, J.J.; Shoichet, B.K. ZINC—A Free Database of Commercially Available Compounds for Virtual Screening. J. Chem. Inf. Model. 2005, 45, 177–182. [Google Scholar] [CrossRef]
- Bowers, K.J.; Chow, D.E.; Xu, H.; Dror, R.O.; Eastwood, M.P.; Gregersen, B.A.; Klepeis, J.L.; Kolossvary, I.; Moraes, M.A.; Sacerdoti, F.D.; et al. Scalable Algorithms for Molecular Dynamics Simulations on Commodity Clusters. In Proceedings of the 2006 ACM/IEEE Conference on Supercomputing (SC ‘06), Tampa, FL, USA, 11–17 November 2006. [Google Scholar] [CrossRef]
- Anila, S.; Samsonov, S.A. Benchmarking Water Models in Molecular Dynamics of Protein-Glycosaminoglycan Complexes. J. Chem. Inf. Model. 2024, 64, 1691–1703. [Google Scholar] [CrossRef]
- Kim, M.; Kim, E.; Lee, S.; Kim, J.S.; Lee, S. New Method for Constant—NPT Molecular Dynamics. J. Phys. Chem. A 2019, 123, 1689–1699. [Google Scholar] [CrossRef]
- Yu, H.; Dalby, P.A. A Beginner’s Guide to Molecular Dynamics Simulations and the Identification of Cross-Correlation Networks for Enzyme Engineering. Methods Enzymol. 2020, 643, 15–49. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissTargetPrediction: Updated Data and New Features for Efficient Prediction of Protein Targets of Small Molecules. Nucleic Acids Res. 2019, 47, W357–W364. [Google Scholar] [CrossRef] [PubMed]
- Nickel, J.; Gohlke, B.O.; Erehman, J.; Banerjee, P.; Rong, W.W.; Goede, A.; Dunkel, M.; Preissner, R. SuperPred: Update on Drug Classification and Target Prediction. Nucleic Acids Res. 2014, 42, W26–W31. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Guo, X.; Yun, Y.; Lu, L.; Huang, X.; Jia, S. DisGeNet: A Disease-Centric Interaction Database among Diseases and Various Associated Genes. Database 2025, 2025, baae122. [Google Scholar] [CrossRef]
- Stelzer, G.; Rosen, N.; Plaschkes, I.; Zimmerman, S.; Twik, M.; Fishilevich, S.; Iny Stein, T.; Nudel, R.; Lieder, I.; Mazor, Y.; et al. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Curr. Protoc. Bioinform. 2016, 54, 1–30. [Google Scholar] [CrossRef]
- Davis, A.P.; Wiegers, T.C.; Sciaky, D.; Barkalow, F.; Strong, M.; Wyatt, B.; Wiegers, J.; McMorran, R.; Abrar, S.; Mattingly, C.J. Comparative Toxicogenomics Database’s 20th Anniversary: Update 2025. Nucleic Acids Res. 2025, 53, D1328–D1334. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Nastou, K.; Koutrouli, M.; Kirsch, R.; Mehryary, F.; Hachilif, R.; Hu, D.; Peluso, M.E.; Huang, Q.; Fang, T.; et al. The STRING Database in 2025: Protein Networks with Directionality of Regulation. Nucleic Acids Res. 2025, 53, D730–D737. [Google Scholar] [CrossRef] [PubMed]
- Otasek, D.; Morris, J.H.; Bouças, J.; Pico, A.R.; Demchak, B. Cytoscape Automation: Empowering Workflow-Based Network Analysis. Genome Biol. 2019, 20, 185. [Google Scholar] [CrossRef] [PubMed]
- Shin, M.G.; Pico, A.R. Using Published Pathway Figures in Enrichment Analysis and Machine Learning. BMC Genom. 2023, 24, 713. [Google Scholar] [CrossRef] [PubMed]
- Gable, A.L.; Szklarczyk, D.; Lyon, D.; Matias Rodrigues, J.F.; Von Mering, C. Systematic Assessment of Pathway Databases, Based on a Diverse Collection of User-Submitted Experiments. Brief. Bioinform. 2022, 23, bbac355. [Google Scholar] [CrossRef]


| Descriptor Set | MACCS | CDK | PubChem | Substructure |
|---|---|---|---|---|
| Algorithm | Bagging | Ridge (Pace-like) | Gradient Boosting | Gradient Boosting |
| R (Train) | 0.975 | 0.999 | 0.975 | 0.886 |
| R2 (Train) | 0.943 | 0.999 | 0.947 | 0.784 |
| RMSE (Train) | 0.333 | 0.032 | 0.32 | 0.647 |
| MAE (Train) | 0.236 | 0.015 | 0.23 | 0.453 |
| R (Test) | 0.883 | 0.835 | 0.834 | 0.826 |
| R2 (Test) | 0.764 | 0.674 | 0.661 | 0.662 |
| RMSE (Test) | 0.673 | 0.792 | 0.807 | 0.806 |
| MAE (Test) | 0.501 | 0.571 | 0.54 | 0.559 |
| Precision | 1 | 1 | 1 | 1 |
| Recall | 0.684 | 0.789 | 0.632 | 0.789 |
| F1 Score | 0.812 | 0.882 | 0.774 | 0.882 |
| Before Hyperparameter Tuning | ||||
|---|---|---|---|---|
| Descriptors | Model | RÂ2 Mean  ± SD | RMSE Mean  ± SD | MAE Mean  ± SD |
| CDK | Random Forest | 0.5674 Â ± 0.2124 | 0.8373 Â ± 0.2109 | 0.6437 Â ± 0.1338 |
| CDK | SVR | 0.5805 Â ± 0.2233 | 0.8239 Â ± 0.1954 | 0.6377 Â ± 0.1288 |
| CDK | Gradient Boosting | 0.4894 Â ± 0.3605 | 0.8795 Â ± 0.2641 | 0.6442 Â ± 0.1466 |
| CDK | KNN | 0.4489 Â ± 0.3967 | 0.9237 Â ± 0.2292 | 0.6935 Â ± 0.1483 |
| CDK | PLS Regression | 0.5272 Â ± 0.3817 | 0.8446 Â ± 0.2748 | 0.6219 Â ± 0.1766 |
| MACCS | Random Forest | 0.5414 Â ± 0.2264 | 0.8570 Â ± 0.2082 | 0.6491 Â ± 0.1374 |
| MACCS | SVR | 0.4871 Â ± 0.2934 | 0.8979 Â ± 0.2244 | 0.6902 Â ± 0.1638 |
| MACCS | Gradient Boosting | 0.3942 Â ± 0.2867 | 0.9875 Â ± 0.1955 | 0.7467 Â ± 0.1289 |
| MACCS | KNN | 0.3999 Â ± 0.4142 | 0.9489 Â ± 0.2511 | 0.7174 Â ± 0.1848 |
| PubChem | Random Forest | 0.5768 Â ± 0.2296 | 0.8206 Â ± 0.2157 | 0.6109 Â ± 0.1319 |
| PubChem | SVR | 0.5556 Â ± 0.2236 | 0.8486 Â ± 0.1984 | 0.6498 Â ± 0.1362 |
| PubChem | Gradient Boosting | 0.5281 Â ± 0.2857 | 0.8562 Â ± 0.2406 | 0.6218 Â ± 0.1451 |
| Substructure | Random Forest | 0.4756 Â ± 0.2937 | 0.9098 Â ± 0.2312 | 0.6941 Â ± 0.1582 |
| Substructure | SVR | 0.5276 Â ± 0.2665 | 0.8667 Â ± 0.2321 | 0.6845 Â ± 0.1626 |
| Substructure | KNN | 0.4212 Â ± 0.3797 | 0.9486 Â ± 0.2470 | 0.7390 Â ± 0.1915 |
| Substructure | Gradient Boosting | 0.4772 Â ± 0.3235 | 0.9068 Â ± 0.2419 | 0.6854 Â ± 0.1726 |
| After Hyperparameter Tuning | ||||
| Descriptors | Model | RÂ2 Mean  ± SD | RMSE Mean  ± SD | MAE Mean  ± SD |
| CDK | Random Forest | 0.5642 Â ± 0.2298 | 0.8341 Â ± 0.2174 | 0.6447 Â ± 0.1427 |
| CDK | SVR | 0.5805 Â ± 0.2233 | 0.8239 Â ± 0.1954 | 0.6377 Â ± 0.1288 |
| CDK | Gradient Boosting | 0.5059 Â ± 0.3210 | 0.8725 Â ± 0.2622 | 0.6438 Â ± 0.1530 |
| CDK | KNN | 0.5150 Â ± 0.2978 | 0.8727 Â ± 0.1976 | 0.6659 Â ± 0.1156 |
| CDK | PLS Regression | 0.4772 Â ± 0.3149 | 0.9068 Â ± 0.2265 | 0.7244 Â ± 0.1625 |
| MACCS | Random Forest | 0.5575 Â ± 0.2120 | 0.8447 Â ± 0.2076 | 0.6596 Â ± 0.1384 |
| MACCS | SVR | 0.5042 Â ± 0.2505 | 0.8932 Â ± 0.1906 | 0.7230 Â ± 0.1437 |
| MACCS | Gradient Boosting | 0.4473 Â ± 0.2564 | 0.9431 Â ± 0.2093 | 0.7375 Â ± 0.1509 |
| PubChem | Random Forest | 0.5826 Â ± 0.2291 | 0.8107 Â ± 0.2113 | 0.6184 Â ± 0.1377 |
| PubChem | SVR | 0.5591 Â ± 0.1905 | 0.8533 Â ± 0.1897 | 0.6950 Â ± 0.1282 |
| PubChem | Gradient Boosting | 0.5587 Â ± 0.2424 | 0.8307 Â ± 0.2395 | 0.6277 Â ± 0.1501 |
| Substructure | Random Forest | 0.5029 Â ± 0.2550 | 0.8944 Â ± 0.2190 | 0.6969 Â ± 0.1513 |
| Substructure | SVR | 0.5285 Â ± 0.2487 | 0.8746 Â ± 0.2138 | 0.7081 Â ± 0.1515 |
| Substructure | Gradient Boosting | 0.4765 Â ± 0.3351 | 0.9045 Â ± 0.2388 | 0.7020 Â ± 0.1669 |
| Gene | Functional Cluster | Stroke-Related Role | Enriched Pathways (FDR < 0.0001) |
|---|---|---|---|
| MAPK1 | Intracellular signaling kinase | Neuroinflammation, apoptosis, BBB integrity | MAPK signaling, inflammatory mediator regulation |
| PRKCB | Intracellular signaling kinase | Inflammatory cascades, neuronal death | Calcium signaling, PKC signaling |
| PRKCG | Intracellular signaling kinase | BBB regulation, oxidative stress | Calcium signaling, PKC signaling |
| HDAC1 | Epigenetic regulator | Chromatin remodeling, neuronal survival | Histone modification, DNA repair |
| HTR1A | Serotonin receptor | Vasodilation, synaptic plasticity | Serotonergic synapse |
| HTR2A | Serotonin receptor | Vasodilation, platelet aggregation | Serotonergic synapse |
| HTR2C | Serotonin receptor | Cognitive recovery, mood modulation | Serotonergic synapse |
| HTR7 | Serotonin receptor | Neurovascular regulation, post-stroke depression | Serotonergic synapse |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alaqel, S.I.; Khan, A.; Alanazi, M.N.; Nayeem, N.; Khaled, H.B.; Imran, M. Repurposing Cofilin-Targeting Compounds for Ischemic Stroke Through Cheminformatics and Network Pharmacology. Pharmaceuticals 2025, 18, 1323. https://doi.org/10.3390/ph18091323
Alaqel SI, Khan A, Alanazi MN, Nayeem N, Khaled HB, Imran M. Repurposing Cofilin-Targeting Compounds for Ischemic Stroke Through Cheminformatics and Network Pharmacology. Pharmaceuticals. 2025; 18(9):1323. https://doi.org/10.3390/ph18091323
Chicago/Turabian StyleAlaqel, Saleh I., Abida Khan, Mashael N. Alanazi, Naira Nayeem, Hayet Ben Khaled, and Mohd Imran. 2025. "Repurposing Cofilin-Targeting Compounds for Ischemic Stroke Through Cheminformatics and Network Pharmacology" Pharmaceuticals 18, no. 9: 1323. https://doi.org/10.3390/ph18091323
APA StyleAlaqel, S. I., Khan, A., Alanazi, M. N., Nayeem, N., Khaled, H. B., & Imran, M. (2025). Repurposing Cofilin-Targeting Compounds for Ischemic Stroke Through Cheminformatics and Network Pharmacology. Pharmaceuticals, 18(9), 1323. https://doi.org/10.3390/ph18091323








