Unraveling the Antihyperglycemic Effects of Dipeptyl Peptidase-4 Inhibitors in Rodents: A Multi-Faceted Approach Combining Effects on Glucose Homeostasis, Molecular Docking, and ADMET Profiling
Abstract
1. Introduction
2. Results
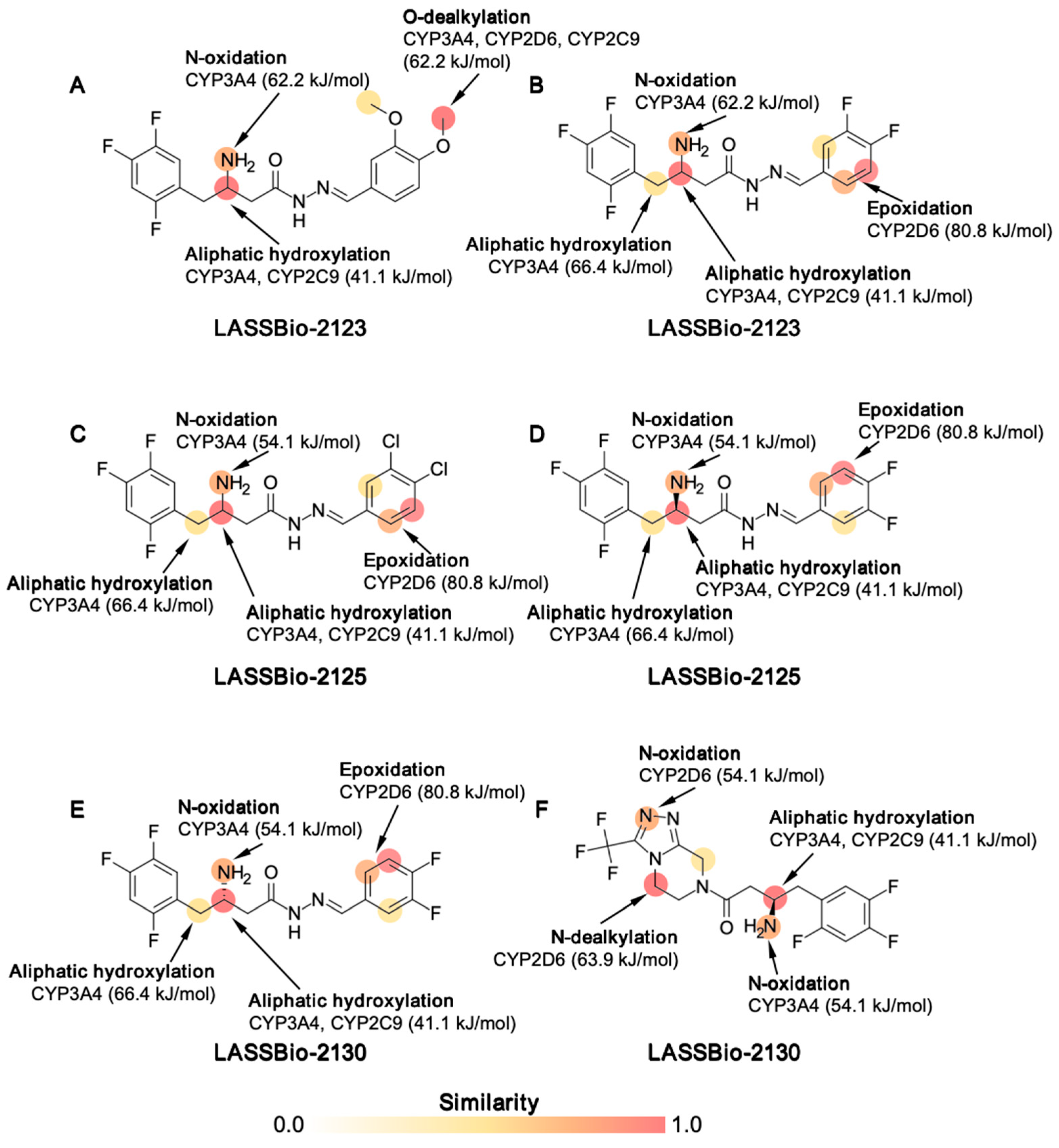
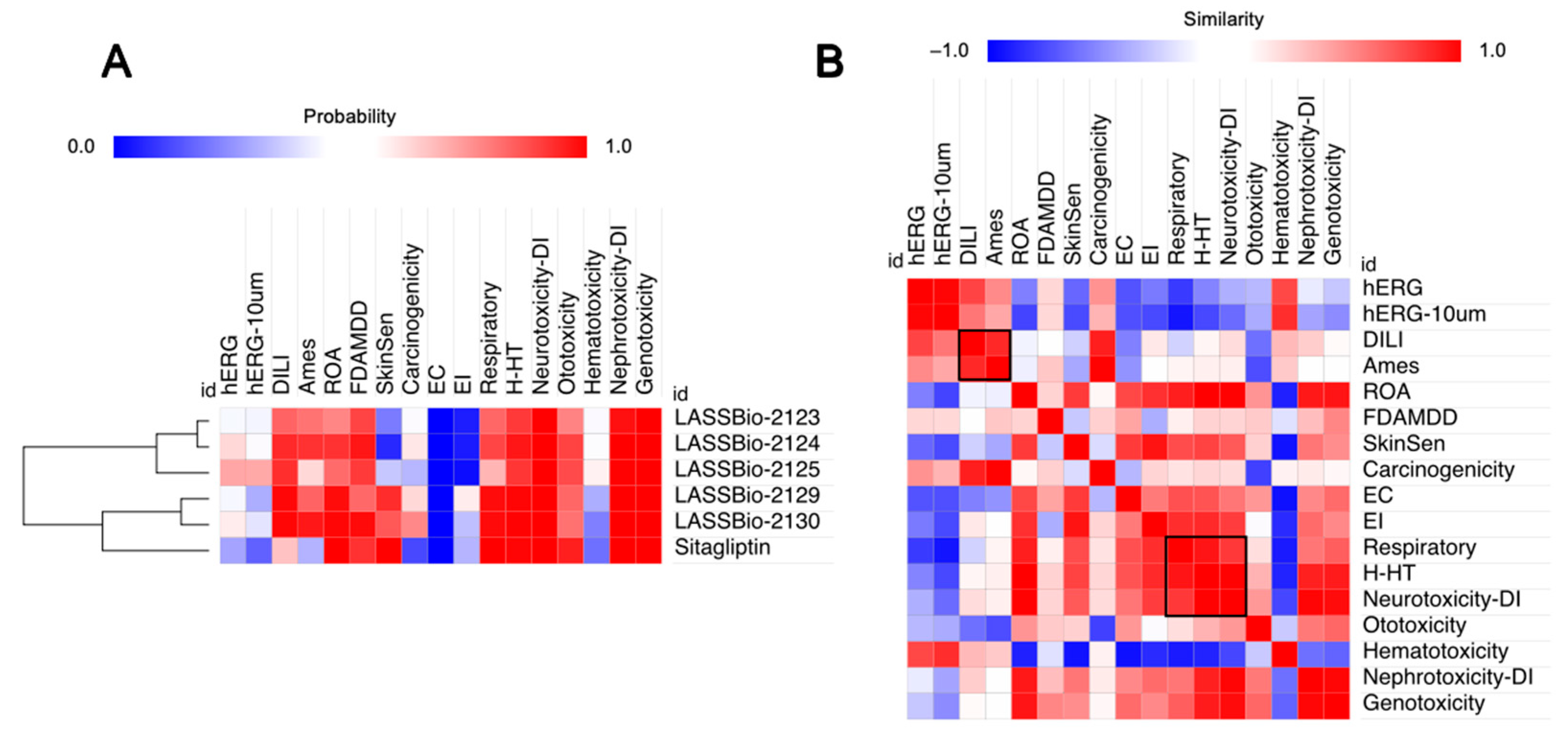
2.1. Effect of β-Aminohydrazines and β-Amino-Acylhydrazones on MPO and ADMET Studies
2.2. Molecular Docking Study on β-Aminohydrazines and β-Amino-Acylhydrazones in Aldose Reductase and Glucokinase Enzymes
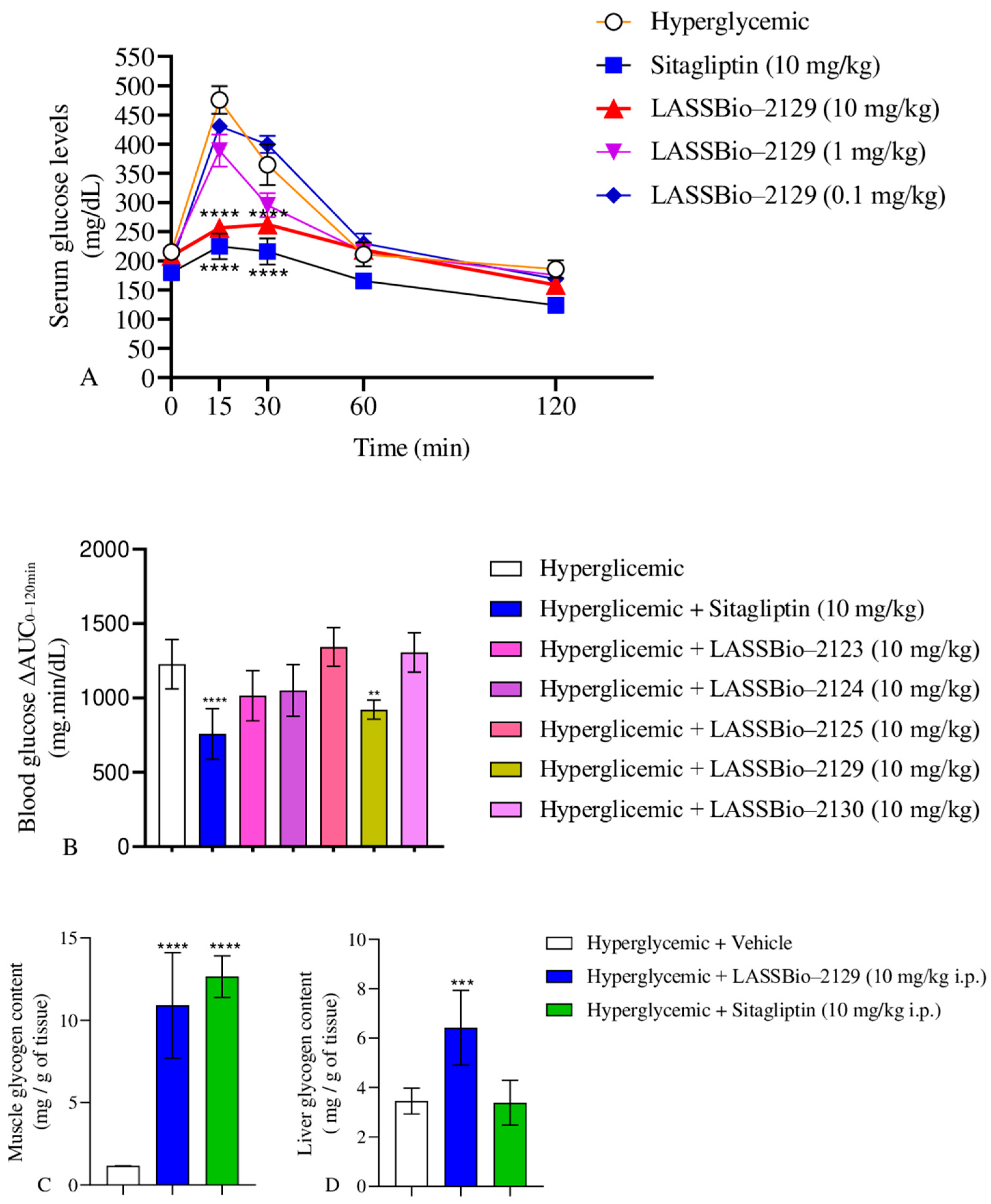
2.3. Effect of β-Aminohydrazines and β-Amino-Acylhydrazones on GTT and Glycogen Content
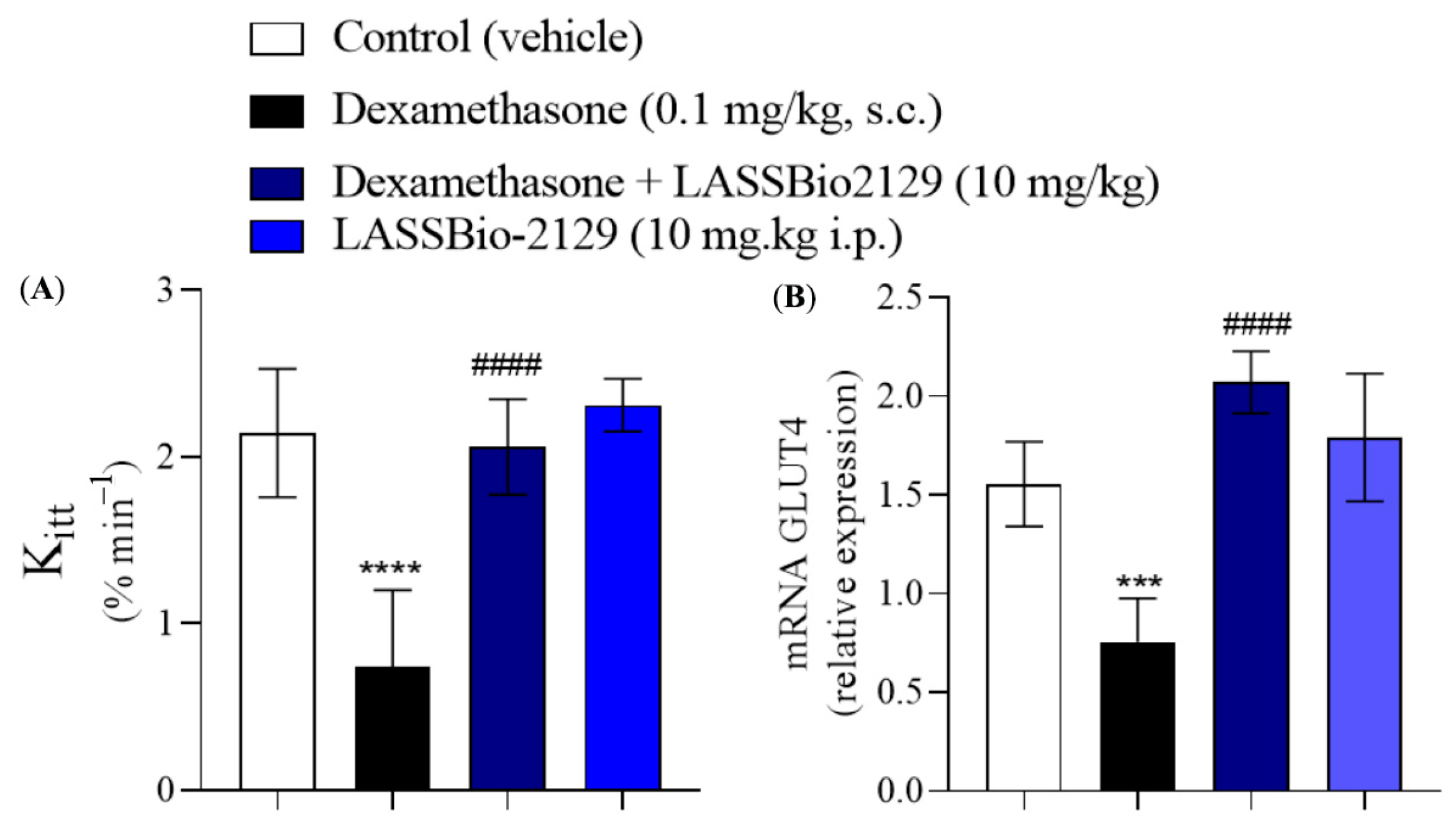
2.4. Effect of the LASSBio-2129 Compound in Dexamethasone-Induced Insulin-Resistant Mice
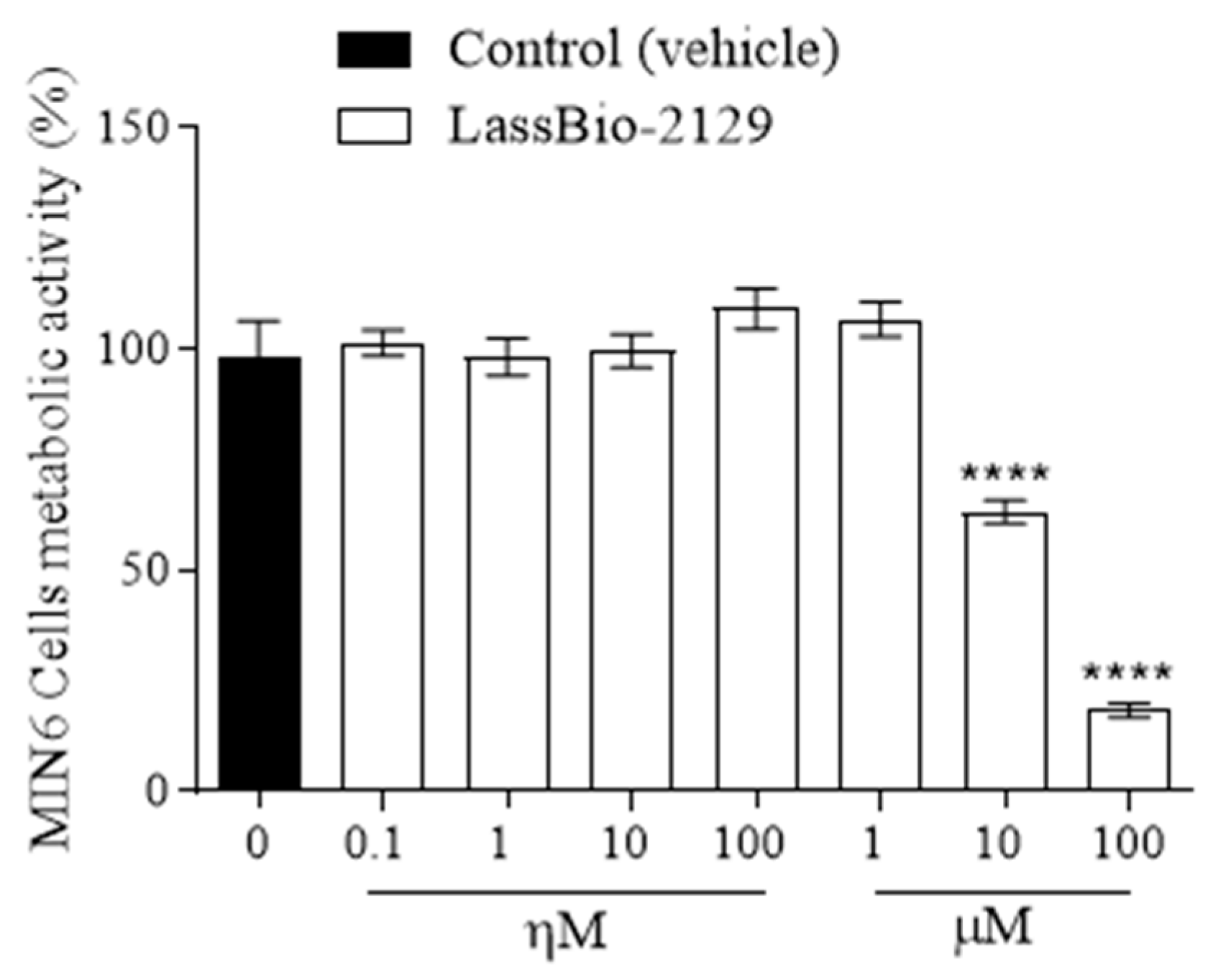
2.5. Effect of the LASSBio-2129 Compound on Cell Viability in MIN6 Cells
3. Discussion
4. Materials and Methods
4.1. Chemical Synthesis of the Dipeptyl Peptidase-4 Inhibitors
4.2. Multiparameter Optimization-Based Absorption, Distribution, Metabolism, Excretion, and Toxicity (ADMET) Studies
4.2.1. Lipophilicity Potential and Hydration Free Energy
4.2.2. PAMPA Predicted Descriptors
4.2.3. Metabolic Stability and Toxicity Prediction
4.3. Molecular Docking Study
4.4. Animals
4.5. Effects of LASSBio-2123, 2124, 2125, 2129 and 2130 on Glucose Tolerance Test
4.6. Glycogen Content Measurements
4.7. Insulin Resistance and Insulin Tolerance Test
4.8. Real-Time PCR
4.9. Cell Culture and Viability Assays
4.10. Data and Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- International Diabetes Federation. IDF Diabetes Atlas, 10th ed.; International Diabetes Federation: Brussels, Belgium, 2021; Available online: https://diabetesatlas.org (accessed on 24 June 2025).
- Mulvihill, E.E.; Drucker, D.J. Pharmacology, Physiology, and Mechanisms of Action of Dipeptidyl Peptidase-4 Inhibitors. Endocr. Rev. 2014, 35, 992–1019. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Jia, Y.; Sun, S.; Meng, L. Adverse Event Profiles of Dipeptidyl Peptidase-4 Inhibitors: Data Mining of the Public Version of the FDA Adverse Event Reporting System. BMC Pharmacol. Toxicol. 2020, 21, 68. [Google Scholar] [CrossRef]
- Lankas, G.R.; Leiting, B.; Roy, R.S.; Eiermann, G.J.; Beconi, M.G.; Biftu, T.; Chan, C.-C.; Edmondson, S.; Feeney, W.P.; He, H.; et al. Dipeptidyl Peptidase IV Inhibition for the Treatment of Type 2 Diabetes: Potential Importance of Selectivity over Dipeptidyl Peptidases 8 and 9. Diabetes 2005, 54, 2988–2994. [Google Scholar] [CrossRef]
- Reina, E.; Franco, L.S.; Carneiro, T.R.; Barreiro, E.J.; Lima, L.M. Stereochemical Insights into β-Amino-N-Acylhydrazones and Their Impact on DPP-4 Inhibition. RSC Adv. 2024, 14, 6617–6626. [Google Scholar] [CrossRef]
- Li, Y.; Yi, H.; Jiang, X.; Li, Z. Synthesis and biological activity of substituted xanthines as DPP-IV inhibitors. Yao Xue Xue Bao 2016, 51, 947–953. [Google Scholar]
- Feng, J.; Zhang, Z.; Wallace, M.B.; Stafford, J.A.; Kaldor, S.W.; Kassel, D.B.; Navre, M.; Shi, L.; Skene, R.J.; Asakawa, T.; et al. Discovery of Alogliptin: A Potent, Selective, Bioavailable, and Efficacious Inhibitor of Dipeptidyl Peptidase IV. J. Med. Chem. 2007, 50, 2297–2300. [Google Scholar] [CrossRef]
- Nordhoff, S.; Cerezo-Gálvez, S.; Deppe, H.; Hill, O.; López-Canet, M.; Rummey, C.; Thiemann, M.; Matassa, V.G.; Edwards, P.J.; Feurer, A. Discovery of Beta-Homophenylalanine Based Pyrrolidin-2-Ylmethyl Amides and Sulfonamides as Highly Potent and Selective Inhibitors of Dipeptidyl Peptidase IV. Bioorg Med. Chem. Lett. 2009, 19, 4201–4203. [Google Scholar] [CrossRef]
- Coumar, M.S.; Chang, C.-N.; Chen, C.-T.; Chen, X.; Chien, C.-H.; Tsai, T.-Y.; Cheng, J.-H.; Wu, H.-Y.; Han, C.-H.; Wu, S.-H.; et al. 3-[2-((2S)-2-Cyano-Pyrrolidin-1-Yl)-2-Oxo-Ethylamino]-3-Methyl-Butyramide Analogues as Selective DPP-IV Inhibitors for the Treatment of Type-II Diabetes. Bioorg Med. Chem. Lett. 2007, 17, 1274–1279. [Google Scholar] [CrossRef]
- Artasensi, A.; Pedretti, A.; Vistoli, G.; Fumagalli, L. Type 2 Diabetes Mellitus: A Review of Multi-Target Drugs. Molecules 2020, 25, 1987. [Google Scholar] [CrossRef]
- Roman, J.; Yuan, Y.; Xu, Y.; Zhu, Q.; Wu, S.; Zhao, F.; Zhou, X.; Meng, S.; Han, D.; Sharp, K.; et al. Functional and Mechanistic Explanation for the Unique Clinical Success of the Glucokinase Activator Dorzagliatin in the Treatment of Type 2 Diabetes. Diabetes 2025, 74, 1374–1384. [Google Scholar] [CrossRef]
- Bhrigu, B.; Sharma, S.; Banik, B.K. Development and Exploration of Organic Compounds as Aldose Reductase Inhibitors: An Overview. Curr. Top. Med. Chem. 2025. ePub ahead of print. [Google Scholar] [CrossRef]
- Bayanati, M.; Ismail Mahboubi Rabbani, M.; Sirous Kabiri, S.; Mir, B.; Rezaee, E.; Tabatabai, S.A. Dipeptidyl Peptidase-4 Inhibitors: A Systematic Review of Structure-Activity Relationship Studies. Iran. J. Pharm. Res. 2024, 23, e151581. [Google Scholar] [CrossRef]
- Pires, D.E.V.; Kaminskas, L.M.; Ascher, D.B. Prediction and Optimization of Pharmacokinetic and Toxicity Properties of the Ligand. In Computational Drug Discovery and Design; Humana Press: New York, NY, USA, 2018; pp. 271–284. ISBN 978-1-4939-7756-7. [Google Scholar]
- Wu, K.; Kwon, S.H.; Zhou, X.; Fuller, C.; Wang, X.; Vadgama, J.; Wu, Y. Overcoming Challenges in Small-Molecule Drug Bioavailability: A Review of Key Factors and Approaches. Int. J. Mol. Sci. 2024, 25, 13121. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.; Bolinger, A.A.; Zhou, J. Highlights on Fluorine-Containing Drugs Approved by U.S. FDA in 2023. Curr. Top. Med. Chem. 2024, 24, 843–849. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Bian, Y.; Baecker, D.; Dhawan, G.; Semghouli, A.; Kiss, L.; Zhang, W.; Sorochinsky, A.E.; Soloshonok, V.A.; Han, J. Fluorine in the Pharmaceutical Industry: FDA-Approved Fluorine-Containing Drugs in 2024. Chemistry 2025, 31, e202500662. [Google Scholar] [CrossRef] [PubMed]
- Gupta, M.; Feng, J.; Bhisetti, G. Experimental and Computational Methods to Assess Central Nervous System Penetration of Small Molecules. Molecules 2024, 29, 1264. [Google Scholar] [CrossRef]
- Ma, X.; Chen, C.; Yang, J. Predictive Model of Blood-Brain Barrier Penetration of Organic Compounds. Acta Pharmacol. Sin. 2005, 26, 500–512. [Google Scholar] [CrossRef]
- Wang, N.-N.; Dong, J.; Deng, Y.-H.; Zhu, M.-F.; Wen, M.; Yao, Z.-J.; Lu, A.-P.; Wang, J.-B.; Cao, D.-S. ADME Properties Evaluation in Drug Discovery: Prediction of Caco-2 Cell Permeability Using a Combination of NSGA-II and Boosting. J. Chem. Inf. Model. 2016, 56, 763–773. [Google Scholar] [CrossRef]
- Fu, L.; Shi, S.; Yi, J.; Wang, N.; He, Y.; Wu, Z.; Peng, J.; Deng, Y.; Wang, W.; Wu, C.; et al. ADMETlab 3.0: An Updated Comprehensive Online ADMET Prediction Platform Enhanced with Broader Coverage, Improved Performance, API Functionality and Decision Support. Nucleic Acids Res. 2024, 52, W422–W431. [Google Scholar] [CrossRef]
- Imberty, A.; Hardman, K.D.; Carver, J.P.; Pérez, S. Molecular Modelling of Protein-Carbohydrate Interactions. Docking of Monosaccharides in the Binding Site of Concanavalin A. Glycobiology 1991, 1, 631–642. [Google Scholar] [CrossRef]
- Das, A.; Swamy Purawarga Matada, G.; Sanjay Dhiwar, P.; Manjunathaiah Raghavendra, N.; Abbas, N.; Singh, E.; Ghara, A.; Prasad Shenoy, G. Molecular Recognition of Some Novel mTOR Kinase Inhibitors to Develop Anticancer Leads by Drug-Likeness, Molecular Docking and Molecular Dynamics Based Virtual Screening Strategy. Comput. Toxicol. 2023, 25, 100257. [Google Scholar] [CrossRef]
- Waring, M.J.; Bennett, S.N.L.; Boyd, S.; Campbell, L.; Davies, R.D.M.; Gerhardt, S.; Hargreaves, D.; Martin, N.G.; Robb, G.R.; Wilkinson, G. Matched Triplicate Design Sets in the Optimisation of Glucokinase Activators—Maximising Medicinal Chemistry Information Content. Med. Chem. Commun. 2013, 4, 657–662. [Google Scholar] [CrossRef]
- Qi, D.; Pulinilkunnil, T.; An, D.; Ghosh, S.; Abrahani, A.; Pospisilik, J.A.; Brownsey, R.; Wambolt, R.; Allard, M.; Rodrigues, B. Single-Dose Dexamethasone Induces Whole-Body Insulin Resistance and Alters Both Cardiac Fatty Acid and Carbohydrate Metabolism. Diabetes 2004, 53, 1790–1797. [Google Scholar] [CrossRef]
- Severino, C.; Brizzi, P.; Solinas, A.; Secchi, G.; Maioli, M.; Tonolo, G. Low-Dose Dexamethasone in the Rat: A Model to Study Insulin Resistance. Am. J. Physiol. Endocrinol. Metab. 2002, 283, E367–E373. [Google Scholar] [CrossRef] [PubMed]
- Su, K.-H.; Chandramouli, V.; Ismail-Beigi, F.; Raymond F Muzic, J. Dexamethasone-Induced Insulin Resistance: Kinetic Modeling Using Novel PET Radiopharmaceutical 6-Deoxy-6-[18F]fluoro-D-glucose. Mol. Imaging Biol. MIB Off. Publ. Acad. Mol. Imaging 2014, 16, 710. [Google Scholar] [CrossRef] [PubMed]
- Urbina, F.; Zorn, K.M.; Brunner, D.; Ekins, S. Comparing the Pfizer Central Nervous System Multiparameter Optimization Calculator and a BBB Machine Learning Model. ACS Chem. Neurosci. 2021, 12, 2247–2253. [Google Scholar] [CrossRef] [PubMed]
- Pettersson, M.; Hou, X.; Kuhn, M.; Wager, T.T.; Kauffman, G.W.; Verhoest, P.R. Quantitative Assessment of the Impact of Fluorine Substitution on P-Glycoprotein (P-Gp) Mediated Efflux, Permeability, Lipophilicity, and Metabolic Stability. J. Med. Chem. 2016, 59, 5284–5296. [Google Scholar] [CrossRef]
- Rizzo, C.; Amata, S.; Pibiri, I.; Pace, A.; Buscemi, S.; Palumbo Piccionello, A. FDA-Approved Fluorinated Heterocyclic Drugs from 2016 to 2022. Int. J. Mol. Sci. 2023, 24, 7728. [Google Scholar] [CrossRef]
- Grygorenko, O.O.; Melnykov, K.P. Fluorinated Building Blocks in Drug Design: New Pathways and Targets. Future Med. Chem. 2024, 16, 1375–1378. [Google Scholar] [CrossRef]
- Dang, N.L.; Hughes, T.B.; Miller, G.P.; Swamidass, S.J. Computationally Assessing the Bioactivation of Drugs by N-Dealkylation. Available online: https://pubs.acs.org/doi/full/10.1021/acs.chemrestox.7b00191 (accessed on 12 June 2025).
- Swanson, K.; Walther, P.; Leitz, J.; Mukherjee, S.; Wu, J.C.; Shivnaraine, R.V.; Zou, J. ADMET-AI: A Machine Learning ADMET Platform for Evaluation of Large-Scale Chemical Libraries. bioRxiv 2023. [Google Scholar] [CrossRef]
- Keshari, A.C.; Thitame, S.N.; Aher, A.A.; Keshari, U.C. Drug-Induced Liver Injury: Mechanisms, Diagnosis, and Management: A Review. J. Pharm. Bioallied Sci. 2025, 17, S55–S58. [Google Scholar] [CrossRef] [PubMed]
- Witkowska, A.B.; Stolarczyk, K.; Fusaro, M.; Leś, A.; Giebułtowicz, J.; Stolarczyk, E.U. Oxidation and Reduction of Hydrazones—Risk Factors Related to the Manufacture and Stability of the Drugs. Int. J. Mol. Sci. 2025, 26, 4295. [Google Scholar] [CrossRef] [PubMed]
- Jaeschke, H.; Ramachandran, A. Central Mechanisms of Acetaminophen Hepatotoxicity: Mitochondrial Dysfunction by Protein Adducts and Oxidant Stress. Drug Metab. Dispos. 2024, 52, 712–721. [Google Scholar] [CrossRef]
- Balestri, F.; Moschini, R.; Mura, U.; Cappiello, M.; Del Corso, A. In Search of Differential Inhibitors of Aldose Reductase. Biomolecules 2022, 12, 485. [Google Scholar] [CrossRef]
- Li, P.; Zhu, D. Clinical Investigation of Glucokinase Activators for the Restoration of Glucose Homeostasis in Diabetes. J. Diabetes 2024, 16, e13544. [Google Scholar] [CrossRef]
- Dougherty, D.A. The Cation−π Interaction in Chemistry and Biology. Chem. Rev. 2025, 125, 2793–2808. [Google Scholar] [CrossRef]
- Wieske, L.H.E.; Erdelyi, M. Halogen Bonds of Halogen(I) Ions—Where Are We and Where to Go? J. Am. Chem. Soc. 2023, 146, 3–18. [Google Scholar] [CrossRef]
- Jayabal, D.; Jayanthi, S.; Thirumalaisamy, R.; Shimu, M.S.S. Molecular Insights of Anti-Diabetic Compounds and Its Hyaluronic Acid Conjugates against Aldose Reductase Enzyme through Molecular Modeling and Simulations Study-a Novel Treatment Option for Inflammatory Diabetes. J. Mol. Model. 2023, 29, 238. [Google Scholar] [CrossRef]
- da Rocha, M.N.; da Fonseca, A.M.; Dantas, A.N.M.; dos Santos, H.S.; Marinho, E.S.; Marinho, G.S. In Silico Study in MPO and Molecular Docking of the Synthetic Drynaran Analogues against Chronic Tinnitus: Modulation of the M1 Muscarinic Acetylcholine Receptor. Mol. Biotechnol. 2024, 66, 254–269. [Google Scholar] [CrossRef]
- Ramírez, E.; Picatoste, B.; González-Bris, A.; Oteo, M.; Cruz, F.; Caro-Vadillo, A.; Egido, J.; Tuñón, J.; Morcillo, M.A.; Lorenzo, Ó. Sitagliptin Improved Glucose Assimilation in Detriment of Fatty-Acid Utilization in Experimental Type-II Diabetes: Role of GLP-1 Isoforms in Glut4 Receptor Trafficking. Cardiovasc. Diabetol. 2018, 17, 12. [Google Scholar] [CrossRef]
- Filippopoulou, F.; Habeos, G.I.; Rinotas, V.; Sophocleous, A.; Sykiotis, G.P.; Douni, E.; Chartoumpekis, D.V. Dexamethasone Administration in Mice Leads to Less Body Weight Gain over Time, Lower Serum Glucose, and Higher Insulin Levels Independently of NRF2. Antioxidants 2021, 11, 4. [Google Scholar] [CrossRef]
- Rojas, J.J.; Pestana-Nobles, R.; Pacheco-Londono, L.C.; Utria-Munive, J.; Galan-Freyle, N.J. Computational Screening Identifies Selective Aldose Reductase Inhibitors with Strong Efficacy and Limited off Target Interactions. Sci. Rep. 2025, 15, 28111. [Google Scholar] [CrossRef] [PubMed]
- Sulis, P.M.; Bittencourt Mendes, A.K.; Fernandes, T.A.; Frederico, M.J.S.; Rey, D.P.; Aragón, M.; Ruparelia, K.C.; Silva, F.R.M.B. Signal Transduction of the Insulin Secretion Induced by the Chalcone Analogue, (E)-3-(Phenyl)-1-(3,4,5-Trimethoxyphenyl)Prop-2-En-1-One, and Its Role in Glucose and Lipid Metabolism. Biochimie 2023, 212, 85–94. [Google Scholar] [CrossRef] [PubMed]
- Frederico, M.J.S.; Cipriani, A.; Heim, J.B.A.; Mendes, A.K.B.; Aragón, M.; Gaspar, J.M.; De Alencar, N.M.N.; Silva, F.R.M.B. Electrophilic Agonists Modulate the Transient Receptor Potential Ankyrin-1 Channels Mediated by Insulin and Glucagon-like Peptide-1 Secretion for Glucose Homeostasis. Pharmaceuticals 2023, 16, 1167. [Google Scholar] [CrossRef] [PubMed]
- Baumeier, C.; Schlüter, L.; Saussenthaler, S.; Laeger, T.; Rödiger, M.; Alaze, S.A.; Fritsche, L.; Häring, H.-U.; Stefan, N.; Fritsche, A.; et al. Elevated Hepatic DPP4 Activity Promotes Insulin Resistance and Non-Alcoholic Fatty Liver Disease. Mol. Metab. 2017, 6, 1254–1263. [Google Scholar] [CrossRef]
- Deacon, C.F. Physiology and Pharmacology of DPP-4 in Glucose Homeostasis and the Treatment of Type 2 Diabetes. Front. Endocrinol. 2019, 10, 80. [Google Scholar] [CrossRef]
- Pires Mendes, C.; Postal, B.G.; Silva Frederico, M.J.; Gonçalves Marques Elias, R.; Aiceles de Medeiros Pinto, V.; da Fonte Ramos, C.; Devantier Neuenfeldt, P.; Nunes, R.J.; Mena Barreto Silva, F.R. Synthesis of a Novel Glibenclamide-Pioglitazone Hybrid Compound and Its Effects on Glucose Homeostasis in Normal and Insulin-Resistant Rats. Bioorg Chem. 2021, 114, 105157. [Google Scholar] [CrossRef]
- Kutoh, E.; Kuto, A.N.; Akiyama, M.; Ozawa, E.; Kurihara, R. Alogliptin: A DPP-4 Inhibitor Modulating Adipose Tissue Insulin Resistance and Atherogenic Lipid. Eur. J. Clin. Pharmacol. 2023, 79, 947–959. [Google Scholar] [CrossRef]
- Oche, J.; Olorundare, O.; Afolabi, S.; Ologe, M.; Njan, A.; Akanbi, O. Comparative Therapeutic Effect of Single/Combined Administration of Saxagliptin, Metformin and Intranasal Insulin on Dexamethasone Induced Insulin Resistance in Albino Wistar Rat Model. Niger. J. Physiol. Sci. 2023, 38, 37–46. [Google Scholar] [CrossRef]
- Wang, T.; Wang, J.; Hu, X.; Huang, X.-J.; Chen, G.-X. Current Understanding of Glucose Transporter 4 Expression and Functional Mechanisms. World J. Biol. Chem. 2020, 11, 76–98. [Google Scholar] [CrossRef]
- Oberhauser, N.; Nurisso, A.; Carrupt, P.-A. MLP Tools: A PyMOL Plugin for Using the Molecular Lipophilicity Potential in Computer-Aided Drug Design. J. Comput. Aided Mol. Des. 2014, 28, 587–596. [Google Scholar] [CrossRef] [PubMed]
- Zafar, A.; Reynisson, J. Hydration Free Energy as a Molecular Descriptor in Drug Design: A Feasibility Study. Mol. Inform. 2016, 35, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Wager, T.T.; Hou, X.; Verhoest, P.R.; Villalobos, A. Moving beyond Rules: The Development of a Central Nervous System Multiparameter Optimization (CNS MPO) Approach to Enable Alignment of Druglike Properties. ACS Chem. Neurosci. 2010, 1, 435–449. [Google Scholar] [CrossRef] [PubMed]
- Marinho, M.M.; da Rocha, M.N.; Magalhães, E.P.; Ribeiro, L.R.; Roberto, C.H.A.; de Queiroz Almeida-Neto, F.W.; Monteiro, M.L.; Nunes, J.V.S.; de Menezes, R.R.P.P.B.; Marinho, E.S.; et al. Insights of Potential Trypanocidal Effect of the Synthetic Derivative (2E)-1-(4-Aminophenyl)-3-(2,4-Dichlorophenyl)Prop-2-En-1-One: In Vitro Assay, MEV Analysis, Quantum Study, Molecular Docking, Molecular Dynamics, MPO Analysis, and Predictive ADMET. Naunyn Schmiedebergs Arch. Pharmacol. 2024, 397, 7797–7818. [Google Scholar] [CrossRef]
- Marinho, E.M.; Batista de Andrade Neto, J.; Silva, J.; Rocha da Silva, C.; Cavalcanti, B.C.; Marinho, E.S.; Nobre Júnior, H.V. Virtual Screening Based on Molecular Docking of Possible Inhibitors of Covid-19 Main Protease. Microb. Pathog. 2020, 148, 104365. [Google Scholar] [CrossRef]
- Filho, R.R.B.X.; Ribeiro, P.R.V.; e Silva, L.M.A.; Pereira, L.L.; Freire, G.A.; Martins, C.B.R.; Wong, D.V.T.; Dionísio, A.P.; de Alencar, N.M.N.; Frederico, M.J.S.; et al. Chemical Composition and Antidiabetic Potential of a Phenolic-Rich Extract from Cashew Fiber. ACS Food Sci. Technol. 2025, 5, 1687–1698. [Google Scholar] [CrossRef]
- Krisman, C.R. A Method for the Colorimetric Estimation of Glycogen with Iodine. Anal. Biochem. 1962, 4, 17–23. [Google Scholar] [CrossRef]
- Bonora, E.; Moghetti, P.; Zancanaro, C.; Cigolini, M.; Querena, M.; Cacciatori, V.; Corgnati, A.; Muggeo, M. Estimates of In Vivo Insulin Action in Man: Comparison of Insulin Tolerance Tests with Euglycemic and Hyperglycemic Glucose Clamp Studies. J. Clin. Endocrinol. Metab. 1989, 68, 374–378. [Google Scholar] [CrossRef]
- Wong, D.V.T.; Holanda, R.B.F.; Cajado, A.G.; Bandeira, A.M.; Pereira, J.F.B.; Amorim, J.O.; Torres, C.S.; Ferreira, L.M.M.; Lopes, M.H.S.; Oliveira, R.T.G.; et al. TLR4 Deficiency Upregulates TLR9 Expression and Enhances Irinotecan-Related Intestinal Mucositis and Late-Onset Diarrhoea. Br. J. Pharmacol. 2021, 178, 4193–4209. [Google Scholar] [CrossRef]
- Mosmann, T. Rapid Colorimetric Assay for Cellular Growth and Survival: Application to Proliferation and Cyto-toxicity Assays. J Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
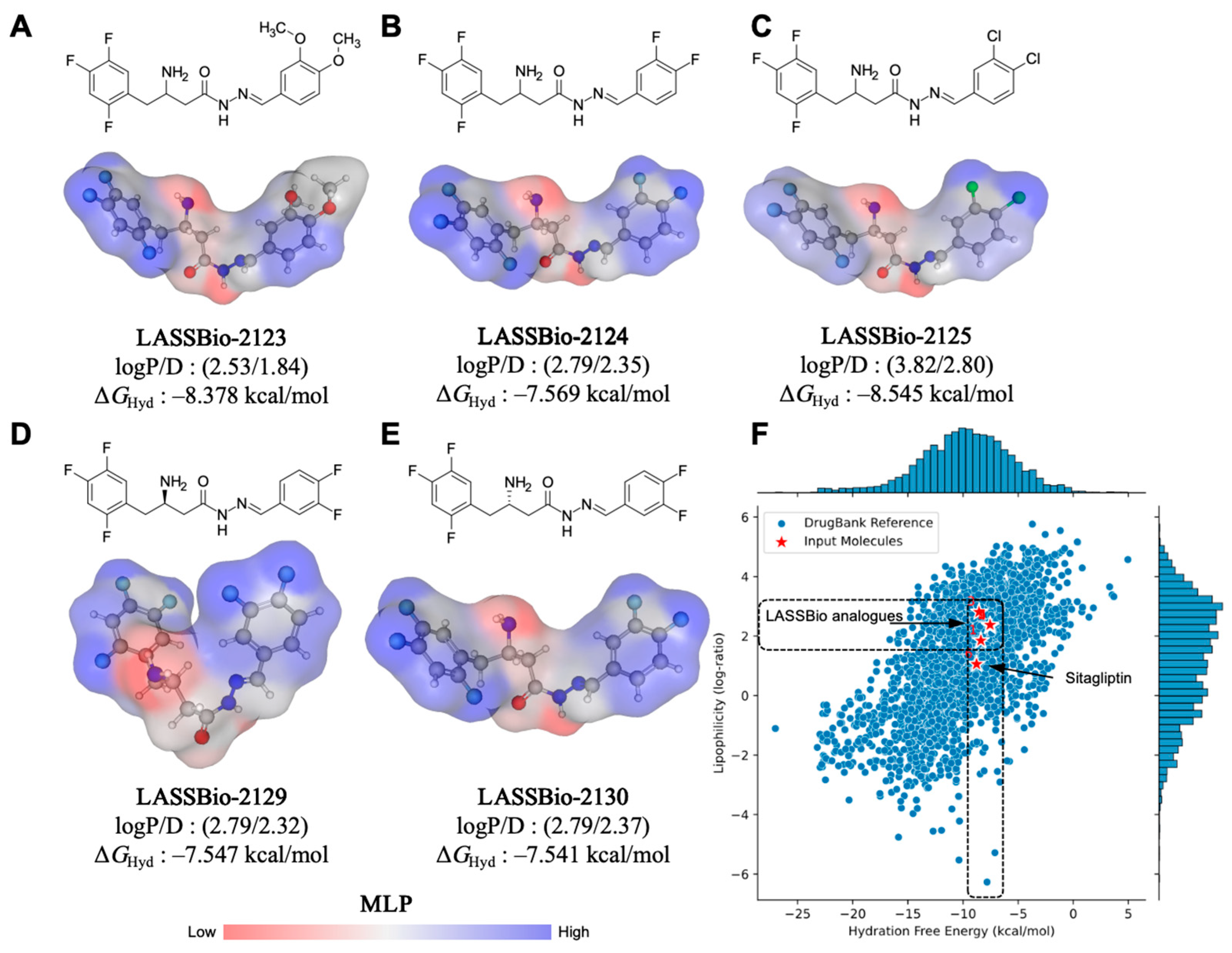
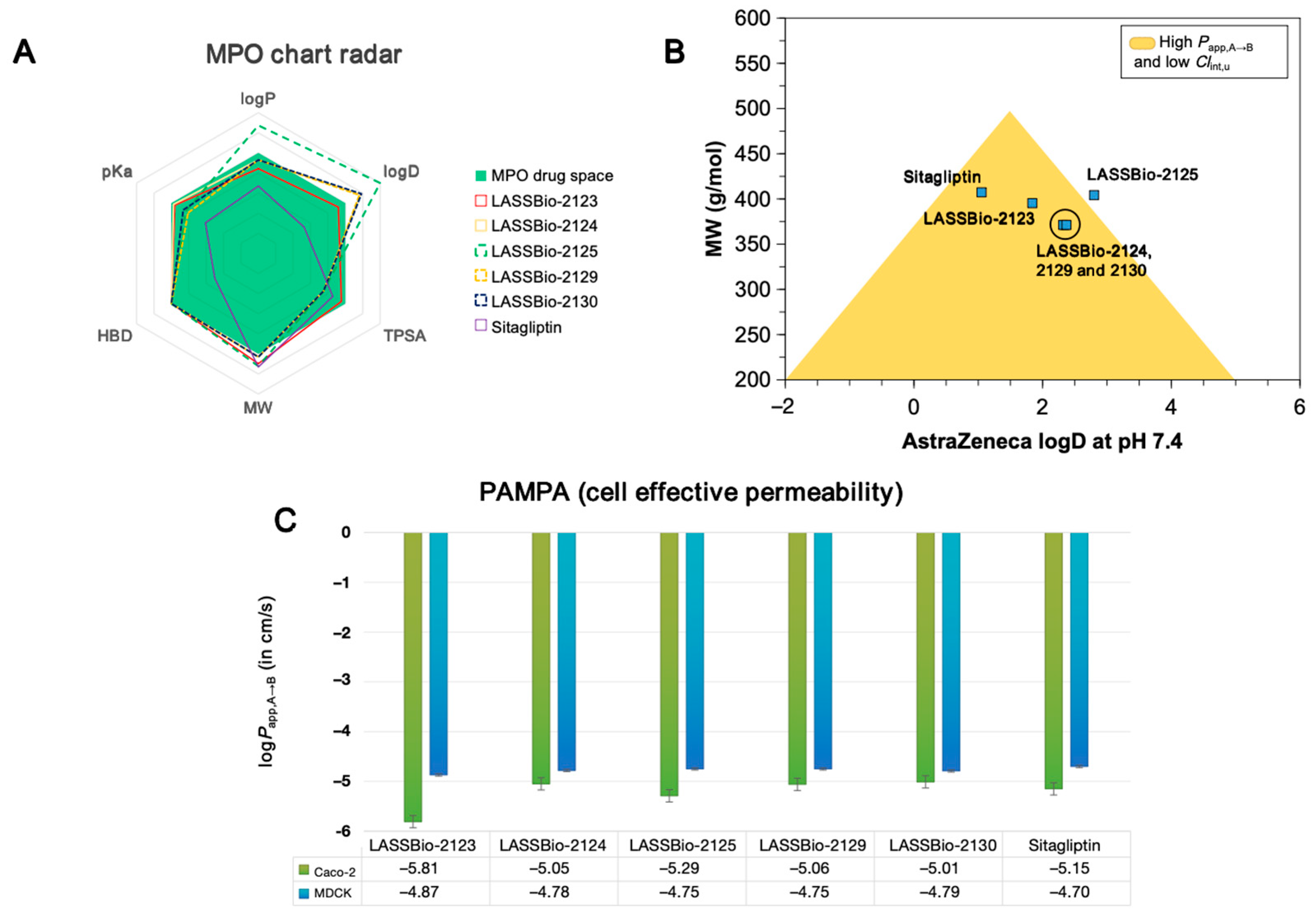
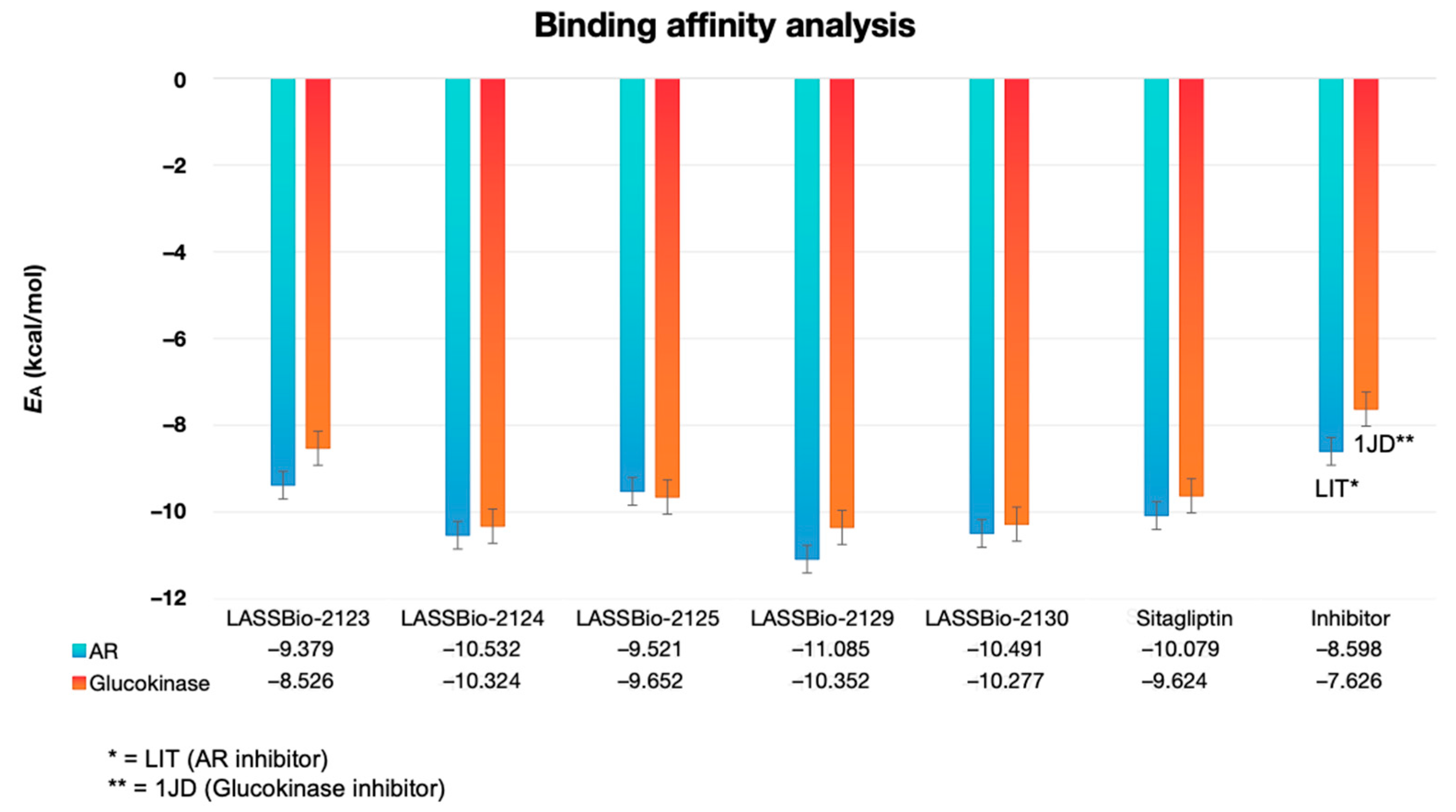
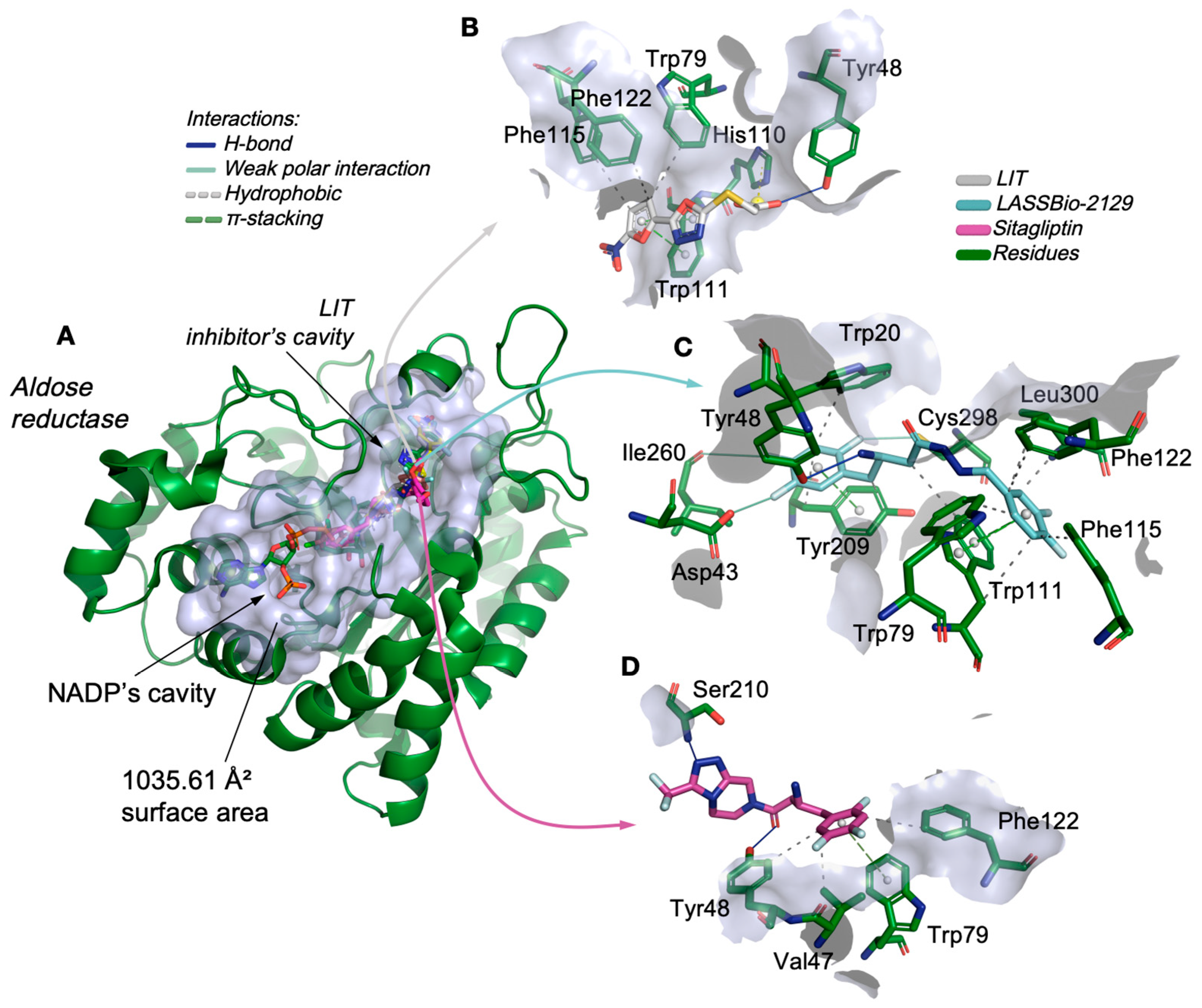
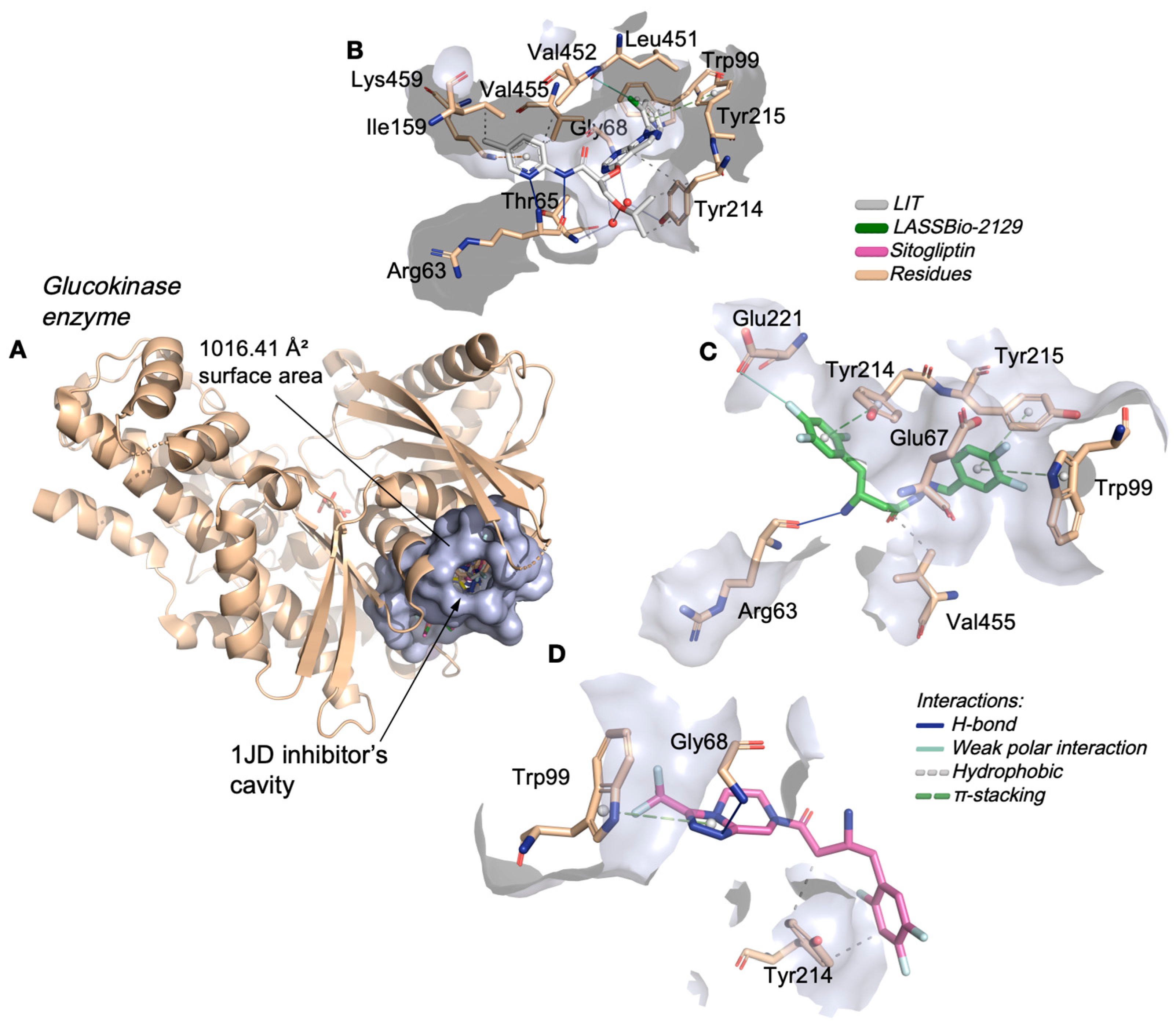
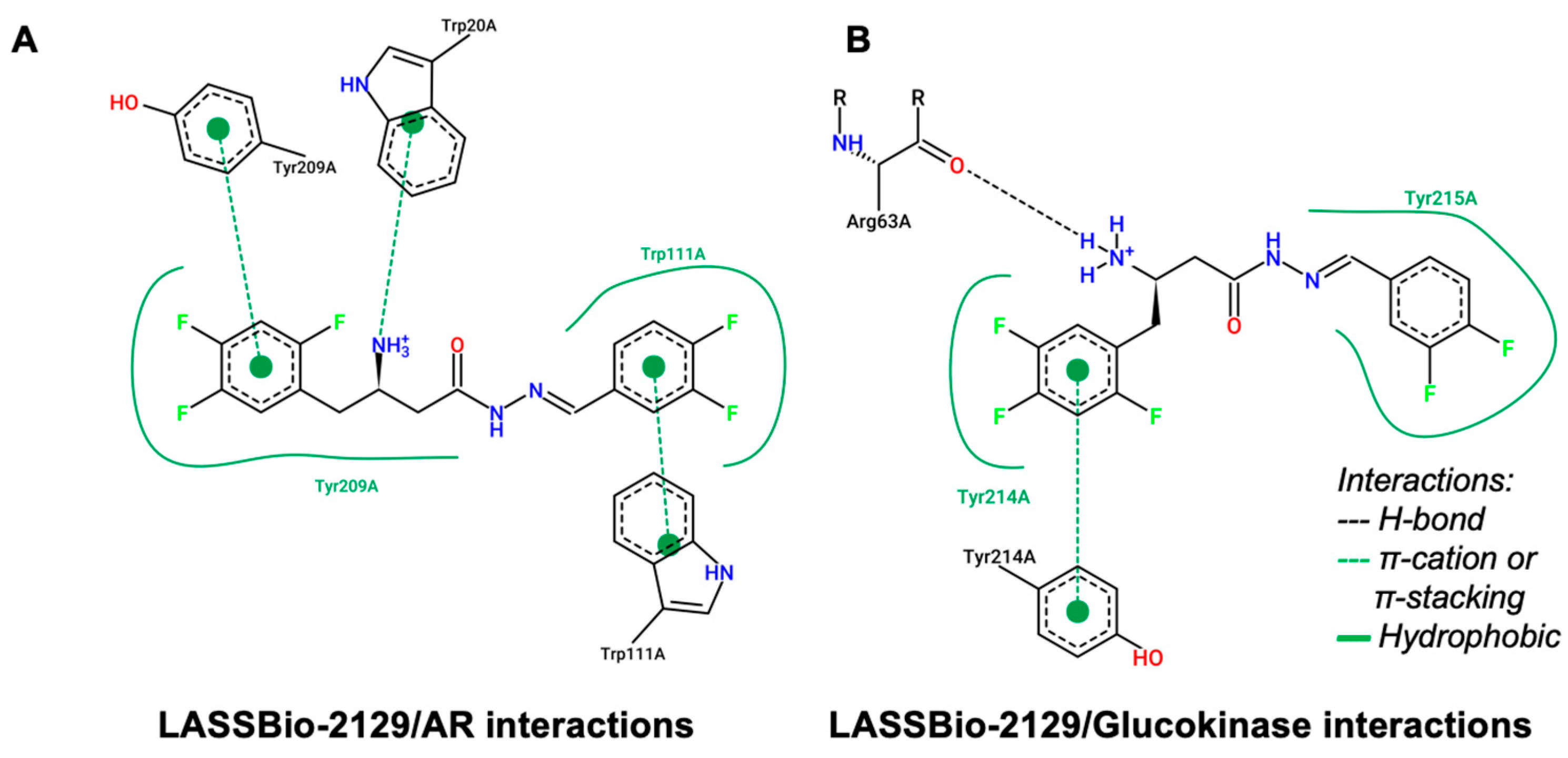
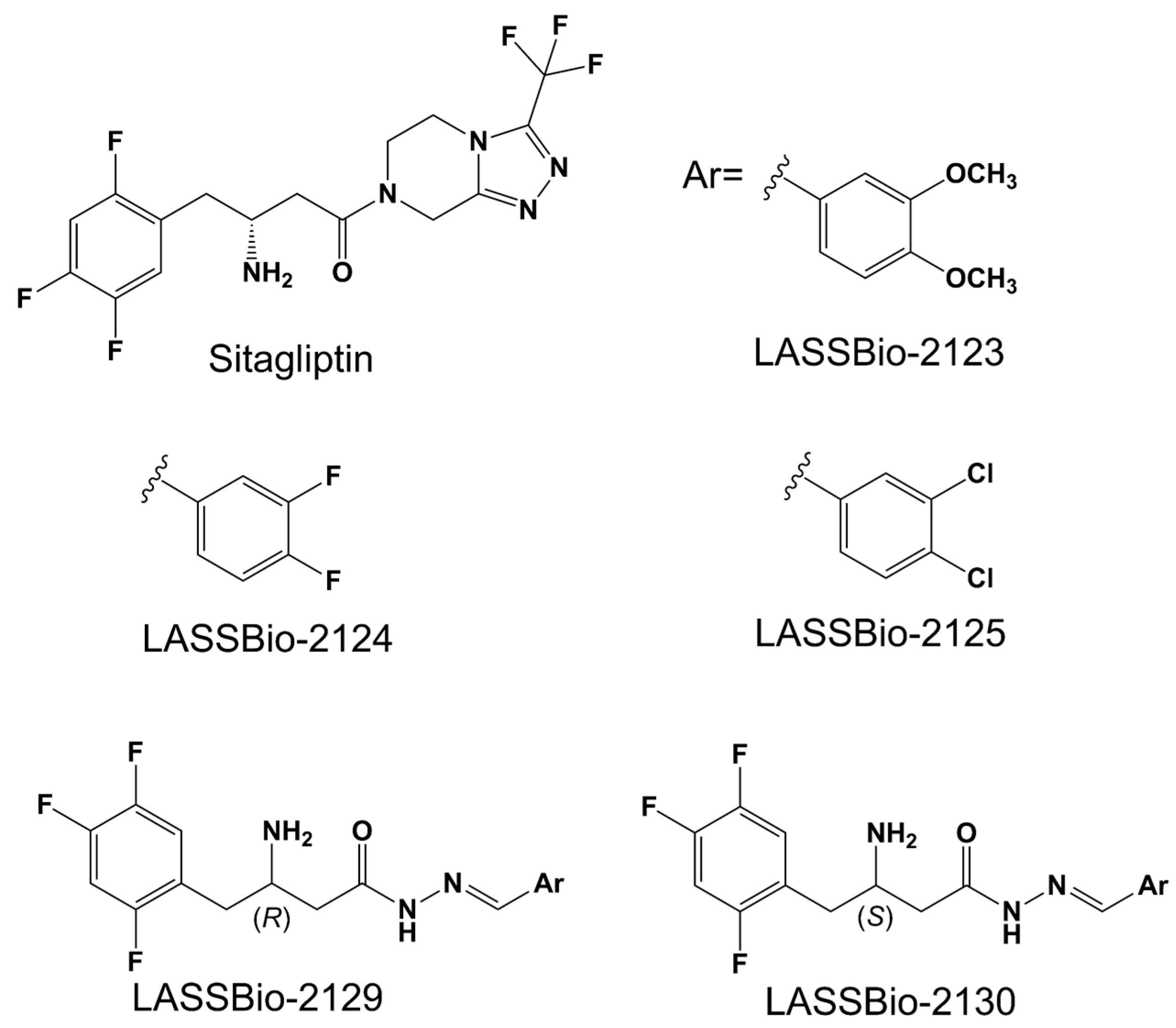
| Compound | logP | AZ logD | TPSA (Å2) | MW (g/mol) | HBD | pKa | MPO score | ∆GHyd |
|---|---|---|---|---|---|---|---|---|
| LASSBio-2123 | 2.53 | 1.84 | 85.94 | 395.38 | 2 | 7.68 | 5.2 | −8.37 |
| LASSBio-2124 | 2.79 | 2.35 | 67.48 | 371.30 | 2 | 7.80 | 5.2 | −7.56 |
| LASSBio-2125 | 3.82 | 2.80 | 67.48 | 404.21 | 2 | 6.68 | 4.4 | −8.54 |
| LASSBio-2129 | 2.79 | 2.32 | 67.48 | 371.30 | 2 | 6.46 | 5.3 | −7.54 |
| LASSBio-2130 | 2.79 | 2.37 | 67.48 | 371.30 | 2 | 6.91 | 5.2 | −7.54 |
| Sitagliptin | 2.01 | 1.05 | 77.04 | 407.31 | 1 | 4.87 | 5.5 | −8.76 |
| Compound | Papp,A→B (cm/s) | Vdss (L/kg) | Clint,u (mL/min/kg) | ClHepa (µL/min/106 cells) | ClMicro (µL/min/mg) | %F | |
|---|---|---|---|---|---|---|---|
| Caco-2 | MDCK | ||||||
| LASSBio-2123 | 1.54 × 10−6 | 1.34 × 10−5 | 4.55 | 5.59 | 26.47 | 39.82 | 0.96 |
| LASSBio-2124 | 8.91 × 10−6 | 1.66 × 10−5 | 3.60 | 6.08 | 17.53 | 7.24 | 0.95 |
| LASSBio-2125 | 5.12 × 10−6 | 1.77 × 10−5 | 4.42 | 5.74 | 33.14 | 32.14 | 0.93 |
| LASSBio-2129 | 8.71 × 10−6 | 1.77 × 10−5 | 7.04 | 6.66 | 17.67 | 6.75 | 0.96 |
| LASSBio-2130 | 9.77 × 10−6 | 1.62 × 10−5 | 2.79 | 5.33 | 18.64 | 7.35 | 0.96 |
| Sitagliptin | 7.07 × 10−6 | 1.99 × 10−5 | 3.84 | 6.51 | −2.59 | 17.37 | 0.93 |
| Target | Compound | RMSD | Ligand–Protein Interactions | |
|---|---|---|---|---|
| Type | Residue (Distance in Å) | |||
| Aldose Reductase | LASSBio-2123 | 1.362 | Hydrophobic | Trp20(3.82), Trp20 (3.09), Val47 (3.31), Tyr48 (3.79), Tyr209 (3.83), Tyr209 (3.83), Ile260 (3.71) |
| H-bond | Tyr48 (2.57), His110 (3.05), Trp111 (2.34), Asn160 (2.17) | |||
| Weak polar interaction | Asp43 (3.15), Gln183 (3.26), Ser214 (3.94), | |||
| LASSBio-2124 | 1.532 | Hydrophobic | Trp111 (3.94), Trp111 (3.81), Phe115 (3.76), Phe122 (2.95), Tyr209 (3.76), Leu300 (3.04), Leu300 (3.27) | |
| H-bond | Trp20 (3.18) | |||
| π-stacking | Trp20 (5.06), Trp111 (3.90), Trp111 (3.65), | |||
| LASSBio-2125 | 1.868 | Hydrophobic | Trp20 (3.58), Trp20 (3.66), Trp79 (3.70), Trp111 (3.60), Tyr209 (3.65), Tyr209 (3.71), Ile260 (3.69) | |
| H-bond | Tyr48 (2.17), His110 (3.11), Gln183 (2.59), | |||
| Weak polar interaction | Ser214 (2.83) | |||
| LASSBio-2129 | 0.858 | Hydrophobic | Trp20 (3.68), Trp79 (3.61), Trp111 (3.97), Trp111 (3.77), Phe115 (3.54), Phe122 (3.72), Tyr209 (3.93), Tyr209 (3.80), Leu300 (3.68), Leu300 (3.41) | |
| H-bond | Tyr48 (2.35), Tyr48 (2.23) | |||
| π-stacking | Trp111 (3.69), Trp111 (3.63), Tyr209 (3.93) | |||
| Weak polar interaction | Asp43 (2.73), Ile260 (3.99), Cys298 (2.97) | |||
| LASSBio-2130 | 1.585 | Hydrophobic | Trp20 (3.94), Trp111 (3.94), Trp111 (3.90), Phe115 (3.72), Phe122 (3.04), Leu300 (3.04), Leu300 (3.29) | |
| H-bond | Trp20 (3.08) | |||
| π-stacking | Trp20 (4.81), Trp111 (3.88), Trp111 (3.64) | |||
| Weak polar interaction | Ser159 (3.73), Asn160 (2.95) | |||
| Sitagliptin * | 1.154 | Hydrophobic | Val47 (3.66), Tyr48 (3.67), Phe122 (3.65) | |
| H-bond | Tyr48 (2.81), Ser210 (2.88) | |||
| π-stacking | Trp79 (4.89) | |||
| LIT ** | 1.866 | Hydrophobic | Trp79 (3.68), Phe115 (3.79), Phe122 (3.86) | |
| H-bond | Tyr48 (2.01) | |||
| π-stacking | Trp111 (3.56), Trp111 (3.50) | |||
| Glucokinase | LASSBio-2123 | 1.238 | Hydrophobic | Thr65 (3.93), Tyr215 (4.00), Glu221 (3.78), Val455 (3.43) |
| H-bond | Thr65 (2.58), Pro66 (2.89), Gly68 (2.38) | |||
| π-stacking | Tyr214 (4.02) | |||
| Weak polar interaction | Glu67 (3.50), Leu451 (3.10) | |||
| LASSBio-2124 | 1.657 | Hydrophobic | Ile211 (3.82), Leu451 (3.34) | |
| H-bond | Thr65 (3.30), Pro66 (3.19), Gly68 (2.29) | |||
| π-stacking | Tyr214 (3.94), Tyr215 (5.10) | |||
| Weak polar interaction | Glu221 (3.49) | |||
| LASSBio-2125 | 1.473 | Hydrophobic | Ile211 (3.81), Tyr214 (3.55), Tyr215 (3.67) | |
| H-bond | Ile211 (2.48) | |||
| π-stacking | Tyr214 (4.09) | |||
| Weak polar interaction | Glu67 (3.91) | |||
| LASSBio-2129 | 1.219 | Hydrophobic | Glu67 (3.94), Val455 (3.71) | |
| H-bond | Arg63 (2.40) | |||
| π-stacking | Trp99 (5.48), Tyr214 (3.90), Tyr215 (5.08) | |||
| Weak polar interaction | Glu221 (3.29) | |||
| LASSBio-2130 | 1.587 | Hydrophobic | Ile211 (3.92), Tyr215 (3.61), Leu451 (3.26) | |
| H-bond | Thr65 (3.20), Pro66 (3.42), Gly68 (2.40) | |||
| π-stacking | Tyr214 (3.98) | |||
| Weak polar interaction | Glu221 (3.41) | |||
| Sitagliptin * | 1.355 | Hydrophobic | Tyr214 (3.57), Tyr214 (3.58) | |
| H-bond | Gly68 (2.27) | |||
| π-stacking | Trp99 (5.48) | |||
| 1JD ** | 1.626 | Hydrophobic | Trp99 (3.98), Ile159 (3.70), Tyr214 (3.88), Tyr214 (3.63), Tyr214 (3.78), Val452 (3.73), Val455 (3.19) | |
| H-bond | Arg63 (1.96), Arg63 (1.81), Gly68 (2.12) | |||
| Water bridges | Thr65 (2.87), Thr65 (3.69), Tyr214 (3.56) | |||
| π-stacking | Trp99 (4.67), Trp99 (4.95), Tyr215 (4.95) | |||
| π-cation | Lys459 (4.60) | |||
| Weak polar interaction | Leu451 (3.83) | |||
| Group | Serum Glucose Levels (mg/dL) Time (min.) | ||||
|---|---|---|---|---|---|
| 0 | 15 | 30 | 60 | 120 | |
| Hyperglycemic | 188.8 ± 9.90 | 475.4 ± 24.15 | 364.7 ± 34.72 | 210.9 ± 20.86 | 185.9 ± 15.25 |
| Sitagliptin | 180.0 ± 12.74 | 224.6 ± 22.33 *** | 216.1 ± 22.48 *** | 165.9 ± 13.60 | 124.1 ± 7.583 |
| 2123 | 172.3 ± 6.55 | 389.3 ± 21.26 * | 210.8 ± 28.72 *** | 190.0 ± 8.60 | 171.0 ± 5.50 |
| 2124 | 180.1 ± 9.51 | 345.3 ± 28.10 | 275.4 ± 28.12 | 210.9 ± 16.75 | 161.4 ± 9.18 |
| 2125 | 208.8 ± 19.24 | 485.4 ± 27.22 | 415.0 ± 28.41 | 198.3 ± 10.44 | 166.6 ± 8.829 |
| 2129 | 208.1 ± 11.78 | 256.3 ± 10.98 *** | 262.4 ± 12.13 *** | 218.7 ± 12.36 | 158.7 ± 9.451 |
| 2130 | 200.2 ± 16.00 | 363.2 ± 16.33 ** | 389.8 ± 25.02 | 349.8 ± 28.60 | 168.2 ± 11.64 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Roriz, R.N.S.; Cardozo, C.J.P.; Freire, G.A.; Martins, C.B.R.; Filho, R.R.B.X.; Pereira, L.L.; Rangel, G.F.P.; Sampaio, T.L.; Ribeiro, L.R.; Silva, G.S.; et al. Unraveling the Antihyperglycemic Effects of Dipeptyl Peptidase-4 Inhibitors in Rodents: A Multi-Faceted Approach Combining Effects on Glucose Homeostasis, Molecular Docking, and ADMET Profiling. Pharmaceuticals 2025, 18, 1589. https://doi.org/10.3390/ph18101589
Roriz RNS, Cardozo CJP, Freire GA, Martins CBR, Filho RRBX, Pereira LL, Rangel GFP, Sampaio TL, Ribeiro LR, Silva GS, et al. Unraveling the Antihyperglycemic Effects of Dipeptyl Peptidase-4 Inhibitors in Rodents: A Multi-Faceted Approach Combining Effects on Glucose Homeostasis, Molecular Docking, and ADMET Profiling. Pharmaceuticals. 2025; 18(10):1589. https://doi.org/10.3390/ph18101589
Chicago/Turabian StyleRoriz, Raquel N. S., Claudia J. P. Cardozo, Gabriela A. Freire, Caio B. R. Martins, Raimundo Rigoberto B. X. Filho, Landerson Lopes Pereira, Gisele F. P. Rangel, Tiago L. Sampaio, Lyanna R. Ribeiro, Gisele Silvestre Silva, and et al. 2025. "Unraveling the Antihyperglycemic Effects of Dipeptyl Peptidase-4 Inhibitors in Rodents: A Multi-Faceted Approach Combining Effects on Glucose Homeostasis, Molecular Docking, and ADMET Profiling" Pharmaceuticals 18, no. 10: 1589. https://doi.org/10.3390/ph18101589
APA StyleRoriz, R. N. S., Cardozo, C. J. P., Freire, G. A., Martins, C. B. R., Filho, R. R. B. X., Pereira, L. L., Rangel, G. F. P., Sampaio, T. L., Ribeiro, L. R., Silva, G. S., Maia, I., Wong, D. V. T., Sousa, D. O. B., de Oliveira, A. C., Reina, E., Lima, L. M., Peláez, W., da Rocha, M. N., Marinho, M. M., ... Frederico, M. J. S. (2025). Unraveling the Antihyperglycemic Effects of Dipeptyl Peptidase-4 Inhibitors in Rodents: A Multi-Faceted Approach Combining Effects on Glucose Homeostasis, Molecular Docking, and ADMET Profiling. Pharmaceuticals, 18(10), 1589. https://doi.org/10.3390/ph18101589









