Abstract
Peanut allergy, a prevalent and potentially severe condition affecting millions worldwide, has been linked to specific human leukocyte antigens (HLAs), suggesting increased susceptibility. Employing an immunoinformatic strategy, we developed a “logo model” based on amino acid frequencies in the peptide binding core and used it to predict peptides originating from 28 known peanut allergens binding to HLA-DRB1*03:01, one of the susceptibility alleles. These peptides hold promise for immunotherapy in HLA-DRB1*03:01 carriers, offering reduced allergenicity compared to whole proteins. By targeting essential epitopes, immunotherapy can modulate immune responses with minimal risk of severe reactions. This precise approach could induce immune tolerance with fewer adverse effects, presenting a safer and more effective treatment for peanut allergy and other allergic conditions.
1. Introduction
A food allergy is a condition in which the immune system reacts abnormally to certain foods, triggering a range of mild symptoms like hives, itching, and swelling to more severe reactions like anaphylaxis, which can be life-threatening. Common food allergens originate from peanuts, tree nuts, shellfish, milk, eggs, wheat, and soy [1]. Diagnosis often involves skin prick tests or blood tests, and management typically requires the patient to strictly avoid the allergen and carry emergency medication like epinephrine [2].
Peanut allergy is one of the most common and potentially severe food allergies, affecting millions worldwide [3]. It occurs when the immune system mistakenly identifies the proteins in peanuts as harmful, triggering an allergic reaction. At present, 17 peanut allergens have been identified (http://www.allergen.org) [4]. Sensitization to Ara h 1, Ara h 2, and Ara h 3 has been detected in 57% to 90% of American peanut-allergic patients, in 37% to 74% of Swedish patients, and in 16% to 42% of Spanish patients [5]. Ara h 2 stands out as the predominant peanut allergen, with its specific IgE serving as the most effective serological marker for diagnosing peanut allergy [6]. On the other hand, Ara h 3 emerges as one of the principal allergenic proteins from peanuts, being recognized via serum IgE testing in approximately 45% of peanut-allergic patients [7]. Ara h 3 constitutes 19% of the total protein content in peanut extracts and is classified as a major peanut allergen. Despite ongoing research, there is currently no cure for peanut allergy, highlighting the importance of awareness, education, and preparedness in dealing with this condition.
The association between peanut allergy and the human leukocyte antigen (HLA) system, also known as human major histocompatibility complex (MHC) antigens, has been a subject of interest in allergy research [8,9,10,11,12,13,14]. HLAs bind to peptide fragments derived from pathogens or self-proteins and present them to T-cells for inspection. This process is pivotal for the activation of immune responses, including adaptive immunity. Variations in HLA genes influence individual susceptibility to diseases, transplantation compatibility, and vaccine efficacy. Studies have identified a potential link between peanut allergy and certain HLA alleles, such as HLA-DRB1*08 [8], DQB1*04 [8,13], DQB1*06:03 [13], and HLA-DRB1*03:01 [8,9,13]. Research suggests that individuals carrying these alleles may have an increased susceptibility to peanut allergy due to their immune response to specific peanut proteins.
Allergy immunotherapy, also known as allergy shots or allergy desensitization, is a treatment aimed at reducing allergic reactions by gradually exposing the immune system to allergens. This therapy involves injecting increasing doses of allergens over time, allowing the body to build up a tolerance and reduce its allergic response. It is commonly used to treat allergies to pollen [15], dust mites [16], pet dander [17], and certain insect venoms [18]. Immunotherapy can alleviate symptoms and, in some cases, provide long-term relief. While it requires commitment and patience due to its gradual nature, allergy immunotherapy has been shown to be effective in reducing the severity of allergic reactions and improving the quality of life of many allergy sufferers.
Immunotherapy for peanut allergy aims to reduce allergic reactions upon accidental exposure to peanuts. Studies have shown promising results, with many participants experiencing an increase in their tolerance to peanuts [19,20,21,22,23,24,25,26]. However, immunotherapy carries risks, including potential allergic reactions during treatment. Its long-term efficacy and safety remain under investigation, but immunotherapy offers hope for individuals with peanut allergy, potentially providing a life-changing treatment option.
Here, we applied an immunoinformatic approach to derive a model for the prediction of peptides binding to HLA-DRB1*03:01, one of the HLA alleles susceptible to peanut allergy. As the model is based on amino acid frequencies in the peptide binding core, like the visual representation of amino acid sequence conservation in proteins (logo graphs) [27], we named it the “logo model” [28]. In the sequence logo, each position is depicted by a stack of letters, in which the size of the letters indicates their frequency within the sequences. Next, we used the derived logo model to identify the high-affinity binding peptides among 28 known peanut allergens [4]. We hypothesize that these high-affinity binders are suitable for the immunotherapy of patients carrying HLA-DRB1*03:01. Peptides used for immunotherapy offer a promising alternative to whole proteins due to their reduced allergenicity. The immunotherapy shots can be designed to contain only the essential epitopes necessary for immune modulation, minimizing the risk of triggering severe allergic reactions. Additionally, peptides are less likely to cross-link IgE antibodies, which are responsible for allergic responses. By targeting specific immune pathways, peptides can induce immune tolerance more selectively and with fewer adverse effects than whole proteins. This targeted approach holds potential to enable safer and more effective immunotherapy for allergic conditions, including peanut allergy.
2. Results
2.1. Logo Model Generation
The logo model consists of two quantitative matrices (QMs)—one for the binders and one for the non-binders. The QMs are focused on the peptide binding core, which is why nonamers were chosen as the training peptides. The QM for the binders was derived from a training set of 105 nonamer peptides binding to HLA-DRB1*03:01 (Table S1). The nonamers were derived from a set of peptide binders to HLA-DRB1*03:01 freely available in NNAlign 2.0 [29]. The peptide data in NNAlign were retrieved from the Immune Epitope Database [30].
A set of 154 non-binders to HLA-DRB1*03:01 was retrieved from the Immune Epitope Database [30] as well. The non-binding peptides were of different lengths, and each peptide was presented as a set of overlapping nonamers, under the presumption that if a peptide is a non-binder, any nonamer derived from it is also a non-binder. Thus, the initial pool of non-binding nonamers consisted of 1187 peptides, a number that decreased to 1123 after eliminating duplicates within the set and those present in the positive set. To mitigate bias in the selection of training and test sets, this final set of non-binders was randomly divided 10 times into a training set of 105 nonamers and a test set of 1018 nonamers. The training sets were used to derive ten QMs for non-binders, and the test sets were used to validate them. The sets of non-binders are given in Table S2.
The amino acid frequencies at each position were mean normalized within the nonamer binding core, according to the following formula:
where Xi is the frequency of amino acid i at a given position, Xmean is the mean frequency for all positions, and Xmax and Xmin are the maximum and the minimum frequencies, respectively, for all positions. The normalized values are constrained within the range of [–1, 1]. They are arranged into a QM, which measures nine positions of 20 amino acids (Table 1). Amino acids with positive coefficients favor binding to HLA-DRB1*03:01, while those with negative coefficients do not. Amino acids with coefficients around zero are neutral.

Table 1.
Quantitative matrix (QM) for binding nonamers to HLA-DRB1*03:01.
Figure 1 (left) displays the sequence logo for HLA-DRB1*03:01, generated from the training set of 105 binding nonamers, while Figure 1 (right) illustrates the sequence logo according to NNAlign [29]. There is a strong consensus between the two logos regarding the preferred anchor positions of p1, p6, and p9. The only difference arises in the preferences for the anchor position p4. In the NNAlign logo, the favoured amino acids at p4 are Asp, Ser, Ala, Asn, and Glu. Only Ala and Ser align with the preferences derived from the logo model. This difference could be explained by the composition of the training set used to derive the positive QM of the logo model. Among the 105 binding nonamers, Asp appears in only five peptides, Asn in one, and Glu in two, while Ala, Arg, and Leu each appear in 14 peptides.
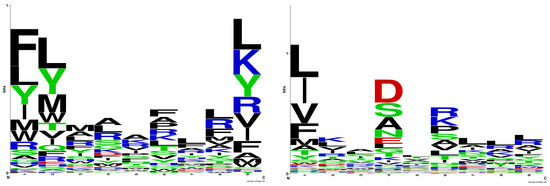
Figure 1.
Sequence logo for HLA-DRB1*03:01 derived in the present study (left); sequence logo for HLA-DRB1*03:01 according to NNAlign (right). The sequence logos were generated by WebLogo (https://weblogo.berkeley.edu, accessed on 23 March 2024).
Ten QMs were developed based on each training set of 105 non-binding nonamers (Table S2). The QM for the binders was employed to compute the binding score (BS) for a tested peptide by summarizing the values of the corresponding amino acids at each position. Similarly, the QMs for the non-binders were used to calculate the non-binding score (NBS) for a tested peptide. In this case, our interpretation is as follows: amino acids with positive coefficients favor non-binding to HLA-DRB1*03:01, while those with negative coefficients favor binding. Amino acids with coefficients around zero are neutral. If the BS is higher than the NBS, the nonamer is classified as a binder; otherwise, it is identified as a non-binder.
2.2. Logo Model Validation
The derived logo model was validated on a test set of binding peptides to HLA-DRB1*03:01 distinct from the peptides included in the training set. The binding peptides were sourced from Immune Epitope Database [30] on 19 February 2024 and contained 10,321 records. After removing internal duplicates and the duplicates present in the training set of the binders, the final test set comprised 7814 binding peptides of different lengths (Table S1). The ten test sets of 1018 non-binders derived as described above were used to validate the corresponding ten QMs for the non-binders derived from the ten training sets (Table S2).
Since the derived QMs cover only the binding core composed of nine residues, binding peptides of varying lengths in the test set were represented as sets of overlapping nonamers. The BS and NBS for each nonamer were predicted using the QMs for the binders and non-binders. If the BS > NBS, the nonamer was classified as a binder; otherwise, it was classified as a non-binder. To classify the parent peptide as a binder, the presence of at least one binding nonamer in the peptide sequence was required.
The validation of the QM for the binders and the 10 QMs for the non-binders is given in Table S2. The sensitivity varied from 0.862 to 0.980, the specificity ranged from 0.628 to 0.707, the accuracy ranged from 0.844 to 0.941, the precision ranged from 0.952 to 0.958, Matthew’s correlation coefficient (MCC) values ranged from 0.451 to 0.689, and F1 scores ranged from 0.907 to 0.967. Among the best-performing QMs for the non-binders, QM1 gave the highest sensitivity, accuracy, MCC, and F1, while QM6 showed the highest specificity and precision but the worst sensitivity and MCC. Based on the highest MCC, we selected QM1 as the best-performing QM for the non-binders. MCC is an efficient predictor estimator given the proportions of positive and negative elements in the datasets [31]. QM1 for the non-binders is given in Table 2. Using all the non-binders as a training set for derivation of a QM for the non-binders did not improve the predictions for the binders or non-binders (Table S2). The results from the validation of the logo model for HLA-DRB1*03:01 are summarized in Table 3.

Table 2.
Quantitative matrix (QM1) for non-binding nonamers to HLA-DRB1*03:01.

Table 3.
Validation of the logo model for prediction of peptide binding to HLA-DRB1*03:01, applying the QM for binders and QM1 for non-binders.
2.3. Prediction of Peptide Binders to HLA-DRB1*03:01 among Peanut Allergens
The information about the peanut allergens was collected from www.allergen.org [4] with the allergen source being Arachis hypogaea. Seventeen allergens were found, some with multiple variants. In total, 28 protein sequences were obtained from the UniProt [32] or GenBank [33] databases (Table 4).

Table 4.
Peanut allergens used in this study [4].
Each protein was presented as a set of overlapping nonamers, and the BS and NBS were calculated for each nonamer using the derived QMs for the binders and non-binders to HLA-DRB1*03:01. If the BS > NBS, the nonamer was classified as a binder; otherwise, it was classified as a non-binder. Strong binders were defined as those with a BS greater than two (Table 5).

Table 5.
Predicted strong binding nonamers to HLA-DRB1*03:01 originating from peanut allergens. Strong binders are defined as those with a BS greater than 2.
The predictions of the logo model identified a set of 17 robust binders derived from peanut allergens. A notable relationship exists between strong binders to MHC and T-cell epitopes [34]. The enhanced binding affinity between peptides and MHC augments the probability of efficient T-cell activation, thereby facilitating antigen presentation and subsequent immune responses. With this understanding, it is feasible that these high binders to HLA-DRB1*03:01 could serve as T-cell epitopes and hold promise as potential candidates for allergy immunotherapy.
3. Discussion
MHC proteins play a pivotal role in antigen presentation, a fundamental process in adaptive immunity. These intracellular molecules bind peptide fragments derived from pathogens or self-proteins and display them on the surface of antigen-presenting cells for recognition by T-cells. MHC class I molecules present endogenous antigens to CD8+ cytotoxic T-cells, aiding in the detection and elimination of infected or aberrant cells. Conversely, MHC class II molecules present exogenous antigens to CD4+ helper T-cells, orchestrating immune responses by activating various effector cells. MHC-mediated antigen presentation is crucial for immune surveillance and response. Evolutionary pressure shapes the polymorphism of the peptide binding site of MHCs. Diverse MHC alleles increase the range of antigens presented to T-cells, enhancing the immune surveillance of the populations. The highest quantity of the identified MHCs is observed in humans, representing one of the most diverse examples within vertebrates. Presently, the IMGT/HLA database contains structural data on 26,610 HLA class I and 11,398 HLA class II proteins [35], with these figures steadily increasing over time.
HLA polymorphism underlies one’s susceptibility to and protection against infections, autoimmune diseases, and allergenicity. Certain HLA alleles confer a heightened susceptibility to specific infections, while others offer protective effects [36,37]. Moreover, HLA variants are implicated in autoimmune diseases, in which aberrant immune responses target self-tissues [38,39]. Specific HLA alleles are associated with an increased susceptibility or resistance to specific infections. For instance, HLA-B27 is linked to a higher risk of developing severe symptoms from infections like Klebsiella pneumoniae [40]. Conversely, HLA-B57 has been shown to provide some level of protection against HIV progression, as individuals with this allele tend to control the virus better and have a slower disease progression [41]. HLA-DRB1*01:04 is strongly associated with rheumatoid arthritis [42], while HLA-DQB1*06:02 is linked to narcolepsy [43].
The development of allergies is also influenced by HLA polymorphism. Specific HLA alleles can predispose individuals to sensitization to certain allergens. For instance, HLA-DRB1*11 is associated with an increased risk of developing allergies to certain pollens [44], while HLA-DQ2 and HLA-DQ8 are linked to celiac disease [45], a condition triggered by an immune response to gluten. HLA diversity contributes to variations in allergenicity, affecting allergic sensitization and responses [46,47,48,49,50]. The association of peanut allergy to several HLA class II alleles is one of the many proofs supporting the interplay between genetics and immune responses.
Here, we describe a universal method for the quantitative assessment of peptide–protein interactions based on the amino acid frequencies in the peptide binding cores. The proposed method is not entirely novel. This is why it was named after the sequence logo graphs initially proposed by Schneider and Stephens in 1990 [27]. The novelty of our method lies in the following:
- Mode of quantification: Schneider and Stephens use Shannon entropy to quantify the nucleic acid/amino acid frequency at a given position. In contrast, we apply mean normalization;
- Functionality: While the sequence logo method was developed as a graphical tool for visualizing patterns in aligned sequences, our method generates quantitative matrices used to calculate binding and non-binding scores. Based on these scores, peptides are classified as binders or non-binders to a given protein.
We have already applied this method to identify peptides that cause celiac disease [28]. The method is universal and can be applied to quantify various peptide–protein interactions. It holds significant potential and is expected to be utilized by us and other researchers in future studies.
Here, the method was applied to derive a logo model for peptide binding predictions to one specific HLA allele, namely HLA-DRB1*03:01, which is associated with peanut allergy. The QM for the binders to HLA-DRB1*03:01 (Table 1) revealed amino acid preferences at the peptide anchor positions of p1, p4p, p6, and p9.
The most preferred peptide residue in pocket 1 is Phe, followed by Leu, Tyr, and Ile. Pocket 1 stands out as the most crucial pocket in HLA-DR alleles [51,52,53]. It is deep, hydrophobic, and favours residues such as Phe, Trp, Tyr, Leu, Ile, and Met. In HLA-DRB1*03:01, pocket 1 consists of ten hydrophobic residues: Phe24α, Ile31α, Phe32α, Trp43α, Ala52α, Phe54α, Tyr78β, Val85β, Val86β, and Phe89β, and only one polar one: Asn82β (Figure 2a). The composition of the pocket includes the residues within 6 Å around the side chain of p1 in the binding ligand GGI1G2S3D4N5K6V7T8R9RGG from the X-ray structure of HLA-DRA*01:01/HLA-DRB1*03:01 (PDB ID: 7N19; [54]). The Ile at p1 from the original ligand was modified to Phe and flexibly docked to the binding site. Phe (Phe3) comfortably fits into this hydrophobic pocket 1, whereas charged residues like Lys, Glu, and Asp are unfavourable (Table 2).
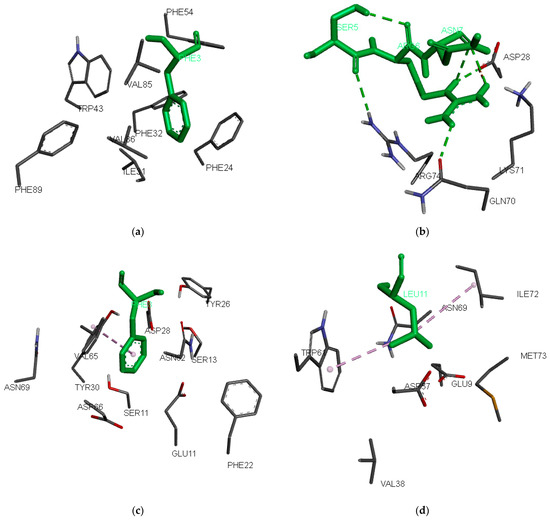
Figure 2.
Preferred anchor peptide residues inside the binding pockets: (a) 1, (b) 4, (c) 6, and (d) 9. The peptide residues are indicated by green-colored sticks, while the protein residues are indicated by element-colored lines. The docking poses were generated using AutoDock Vina v. 1.2.0 [55]. Hydrogen bonds are indicated with green dashed lines, and hydrophobic interactions are indicated with purple dashed lines.
Binding pocket 4 consists of the residues Thr12β, Ser13β, Glu14β, Tyr26β, Leu27β, Asp28β, Tyr30β, Gln70β, Lys71β, Arg74β, Tyr78β, and Asn82β. It is deep and polar because eleven of the twelve residues are polar or charged. While preferences for negatively charged peptide anchors at p4 are expected, our QM for the binders surprisingly favours Ala, Arg, and Leu. Among the 105 binding nonamers from the training set, Ala, Arg, and Leu each appear in 14 peptides, whereas only five peptides feature Asp and two peptides feature Glu at p4. Notably, the positive electrostatic potential of Arg74β does not deter the binding of Arg in pocket 4. A closer examination reveals that Arg74β forms a hydrogen bond with the backbone oxygen atom of Ser at p3 (Ser5), while Arg at p4 (Arg6) forms three hydrogen bonds: two with the backbone oxygen of Asn at p5 (Asn7) and one with Gln70β (Figure 2b).
At peptide anchor position 6, the most preferred amino acids are Phe, Ala, Arg, and Pro. Pocket 6 is composed of Gln9α, Glu11α, Phe22α, Asn62α, Val65α, Asp66α, Asn69α, Ser11β, Thr12β, Ser13β, Tyr26β, Asp28β, Arg29β, Tyr30β, and Lys71β. Phe (Phe8) fits well here, forming hydrophobic interactions with Val65 (Figure 2c). Arg and Lys also are well accepted in this pocket because of the negatively charged Glu11α and Asp66α.
Pocket 9 is the second most important pocket in HLA-DR alleles. In HLA-DRB1*03:01, it consists of Val65α, Asn69α, Leu70α, Ile72α, Met73α, Arg76α, Glu9β, Tyr10β, Tyr30β, Phe31β, His32β, Glu35β, Asn37β, Val38β, Leu53β, Gly54β, Asp57β, Tyr60β, and Trp61β. The preferences here are for Leu, Lys, Tyr, and Arg. Leu at p9 (Leu11) forms hydrophobic interactions with Trp61 and Ile72 (Figure 2d). Lys and Arg are also considered favourable in this pocket due to the presence of negatively charged Glu9β and Asp57β.
Further, we employed the logo model to analyse the protein sequences of 28 known peanut allergens. This analysis aimed to pinpoint the most probable strong binders to HLA-DRB1*03:01. Our hypothesis posits that these strong binding nonamers may serve as T-cell epitopes. The pathophysiology of peanut allergy involves the participation of peanut-specific CD4+ T-cells. Three T-cell epitopes associated with HLA-DRB1*03:01 originating from Ara h 1, Ara h 2, and Ara h 3 are available in the Immune Epitope Database [30] (Table 6). They contain between four and seven predicted binding nonamers to HLA-DRB1*03:01, with the strongest of them having a BS between 0.363 and 1.437. The measured binding affinities to HLA-DRB1*03:01 are known for only two of them. The peptide ARQQWELQGDRRCQS originating from Ara h 2.0101 is a weak binder (IC50 > 500 nM). The best predicted nonamer from this peptide has a very low BS of 0.363. In contrast, the peptide EFLEQAFQVDDRQIV from Ara h 3.0101 is a moderate binder with an IC50 of 147 nM, and the nonamer predicted to be the best has a BS of 1.437.

Table 6.
Known T-cell epitopes and predicted strong binding nonamers to HLA-DRB1*03:01 originating from peanut allergens.
The identification of T-cell epitopes from peanut allergens is crucial for the development of peptide immunotherapeutic agents. There is no definitive evidence indicating whether peptides with strong or moderate HLA binding affinities are more suitable for allergy immunotherapy. It was found that an affinity threshold of approximately 500 nM (preferably 50 nM or less) apparently determines the capacity of a peptide epitope to elicit a CTL response [58]. Strong binders (IC50 < 50 nM) are more effective at stimulating T-helper (Th) cells because they bind more tightly to HLA class II molecules and are presented more effectively to these cells. This can potentially lead to a stronger immune response and more effective immunotherapy [59]. However, strong binders can sometimes trigger an overly vigorous immune response, increasing the risk of adverse effects or unintended immune reactions. They might also be less specific, potentially leading to broader or more unpredictable responses [60]. On the other hand, moderate binders (500 nM < IC50 < 50 nM) might produce a more controlled and potentially safer immune response. They can still be effective at stimulating Th cells but have a lower risk of overstimulation. This can result in fewer side effects and more manageable therapy. However, they may be less effective at inducing a strong immune response compared to strong binders, which could potentially result in less efficacy in some cases [61].
The choice between strong and moderate binders might depend on how specific the allergen is and how it interacts with the immune system. Strong binders might be favoured for complete desensitization, while moderate binders might be chosen for symptom management. The decision will typically be based on balancing efficacy with safety, as well as considering individual patient factors and therapeutic goals [62].
By administering peptide immunotherapeutic agents in gradually escalating doses, they could potentially desensitize the immune system of patients carrying HLA-DRB1*03:01, thereby increasing their tolerance to peanuts. However, to validate this hypothesis, rigorous clinical trials are imperative. These trials would ascertain whether the identified nonamers indeed induce tolerance and mitigate allergic reactions in patients with peanut allergy. The outcomes of such trials could have profound implications for peanut allergy management, potentially offering a novel immunotherapeutic approach for individuals susceptible to peanut-induced allergic responses. Additionally, understanding the efficacy and safety of this approach would contribute significantly to personalized medicine strategies tailored to specific HLA genotypes.
4. Materials and Methods
4.1. Datasets
The positive training set, consisting of binding peptides, was sourced from the freely available peptide dataset in the NNAlign tool [29]. The original dataset comprises 2042 peptides of various lengths binding to HLA-DRB1*03:01. Given the logo model’s focus on the peptide binding core, we specifically chose nonamer binders, resulting in a collection of 105 nonamers.
The test set of binding peptides was collected from Immune Epitope Database [30] with the following settings: Epitope: Linear peptide; Host: Human; Assay: MHC Ligand; Outcome: Positive; and MHC Restriction: HLA-DRA*01:01/HLA-DRB1*03:01. The initial dataset exported on 19 February 2024 contained 10,321 records. Peptide binding affinities were measured either qualitatively or quantitatively by direct or competitive fluorescence assays on cellular or purified MHC, by competitive radioactivity assays on purified MHC, by mass spectrometry on cellular MHC, or by high-throughput multiplexed assays.
The non-binding peptides were collected from Immune Epitope Database [30] with the following settings: Epitope: Linear peptide; Host: Human; Assay: MHC Ligand; Outcome: Negative; and MHC Restriction: HLA-DRA*01:01/HLA-DRB1*03:01. The initial dataset was exported on 1 March 2024, and contained 154 peptides of different lengths. Each peptide was presented as a set of overlapping nonamers, and a pool of 1187 nonamers was generated subsequently and reduced to 1123 after removing the duplicates.
The peptide datasets used in the study are given in Tables S1 and S2.
4.2. Statistical Analysis
The performance of the two QMs of the logo model was assessed on the external test sets. True positives (TPs) correspond to peptides correctly identified as binders, while true negatives (TNs) are peptides correctly recognized as non-binders. False positives (FPs) occur when non-binders are incorrectly predicted as binders, and false negatives (FNs) are non-binders incorrectly identified as binders. On this basis, the following measures were calculated:
4.3. Docking Protocol
The X-ray structure of HLA-DRA*01:01/HLA-DRB1*03:01 in complex with the Aspergillus nidulans epitope GGI1G2S3D4N5K6V7T8R9RG (PDB ID: 7N19) [48] was utilized in the molecular docking calculations. The original peptide underwent mutation with preferred anchor amino acids, specifically Phe, Arg, Phe, and Leu at positions 1, 4, 6, and 9, respectively, via a single amino acid substitution method. AutoDock Vina v.1.2.0 of AutoDock Suite (Center for Computational Structural Biology, CCSB, La Jolla, CA, USA) [49] was employed for docking, with the following settings: grid box coordinates: x-center: −1.306 Å, y-center: 12.876 Å, and z-center: 15.26 Å; number of points: at x-dimension: 28, y-dimension: 42, and z-dimension 32; standard exhaustiveness (8); and energy range (3). A flexible docking procedure was implemented with fifty-one residues within 6 Å radius from the original peptide in the X-ray structure set as flexible during the calculations. They were Gln9α, Glu11α, Phe22α, Phe24α, Ile31α, Phe32α, Trp43α, Phe51α, Ser53α, Phe54α, Glu55α, Asn62α, Val65α, Asp66α, Asn69α, Leu70α, Ile72α, Met73α, Arg76α, Glu9β, Tyr10β, Ser11β, Thr12β, Ser13β, Glu14β, Tyr26β, Leu27β, Asp28β, Arg29β, Tyr30β, Phe31β, His32β, Glu35β, Asn37β, Val38β, Phe47β, Leu53β, Pro56β, Asp57β, Tyr60β, Trp61β, Leu67β, Gln70β, Lys71β, Arg74β, Tyr78β, His81β, Asn82β, Val85β, Val86β, and Phe89β. During docking calculations, the peptide side chains were flexible while their backbones were kept rigid.
5. Conclusions
The peptides identified in the present study offer potential for epitope-based immunotherapy targeting HLA-DRB1*03:01-restricted peanut allergy patients, potentially reducing the risk of irrelevant protein targeting and minimizing undesirable immune responses. Targeted immunotherapy, delivered via subcutaneous injections or orally, allows for gradual immune tolerance induction with personalized dosing. However, clinical validation is essential. Peptide immunotherapy, like any other immunotherapy, has risks including allergic reactions, autoimmune responses, or cytokine release syndrome. Ensuring peptide purity and specificity is crucial to prevent unintended immune activation or off-target effects, while proper handling is essential to maintain therapeutic efficacy and safety. In our opinion, the most suitable peptide candidates for the immunotherapy of peanut allergy have yet to be discovered. Immunoinformatic approaches could serve as valuable tools in this discovery process, helping to identify and predict peptide-HLA binding affinities, T-cell receptor interactions, and potential immunogenicity.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ph17081097/s1, Table S1: Training and test sets of binding peptides to HLA-DRB1*03:01. Table S2: Training sets, QMs and test sets of non-binding peptides to HLA-DRB1*03:01.
Author Contributions
Conceptualization, I.D. (Irini Doytchinova); methodology, I.D. (Irini Doytchinova); software, I.D. (Irini Doytchinova); validation, I.D. (Irini Doytchinova) and I.D. (Ivan Dimitrov); investigation, I.D. (Irini Doytchinova), M.A., S.S. and I.D. (Ivan Dimitrov); writing—original draft preparation, I.D. (Irini Doytchinova); writing—review and editing, I.D. (Irini Doytchinova), M.A., S.S. and I.D. (Ivan Dimitrov); visualization, I.D. (Irini Doytchinova); supervision, I.D. (Ivan Dimitrov); project administration, I.D. (Ivan Dimitrov); funding acquisition, I.D. (Ivan Dimitrov). All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the project GIANT LEAPS—Gap resolutIon in sAfety, NuTritional, alLergenicity and Environmental assessments to promote Alternative Protein utilization and the dietary Shift (HORIZON-CL6-2021-FARM2FORK-01, Grant 101059632/2022)—and conducted at the Centre of Excellence for Informatics and ICT, supported by the Science and Education for Smart Growth Operational Program and co-financed by the European Union through the European Structural and Investment funds (Grant No. BG05M2OP001-1.001-0003).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Materials, further inquiries can be directed to the corresponding author. All datasets used in the study and the derived models are published in this paper.
Acknowledgments
During the preparation of this work, the authors used ChatGPT (free version) for English editing. After using this tool, the authors reviewed and edited the content as needed and take full responsibility for the content of the publication.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Abbreviations
BS: binding score; FN: false negative; FP: false positive; HLA: human leukocyte antigen; MCC: Matthew’s correlation coefficient; MHC: major histocompatibility complex; NBS: non-binding score; QM: quantitative matrix; TN: true negative; TP: true positive.
References
- Peters, R.L.; Krawiec, M.; Koplin, J.J.; Santos, A.F. Update on Food Allergy. Pediatr. Allergy Immunol. 2021, 32, 647–657. [Google Scholar]
- Muraro, A.; de Silva, D.; Halken, S.; Worm, M.; Khaleva, E.; Arasi, S.; Dunn-Galvin, A.; Nwaru, B.I.; De Jong, N.W.; Rodríguez Del Río, P.; et al. Managing Food Allergy: GA2LEN Guideline 2022. World Allergy Organ. J. 2022, 15, 100687. [Google Scholar]
- Mahr, T.A.; Lieberman, J.A.; Haselkorn, T.; Damle, V.; Ali, Y.; Chidambaram, A.; Griffin, N.M.; Sublett, J.W. Characteristics of Peanut Allergy Diagnosis in a US Health Care Claims Database (2011–2017). J. Allergy Clin. Immunol. Pract. 2021, 9, 1683–1694.e5. [Google Scholar]
- Supported by WHO/IUIS Allergen Nomenclature Sub-Committee. Available online: www.allergen.org (accessed on 20 March 2024).
- Vereda, A.; van Hage, M.; Ahlstedt, S.; Ibañez, M.D.; Cuesta-Herranz, J.; van Odijk, J.; Wickman, M.; Sampson, H.A. Peanut allergy: Clinical and immunologic differences among patients from 3 different geographic regions. J. Allergy Clin. Immunol. 2011, 127, 603–607. [Google Scholar] [PubMed]
- Hemmings, O.; Du Toit, G.; Radulovic, S.; Lack, G.; Santos, A.F. Ara h 2 is the dominant peanut allergen despite similarities with Ara h 6. J. Allergy Clin. Immunol. 2020, 146, 621–630. [Google Scholar] [PubMed]
- Rabjohn, P.; Helm, E.M.; Stanley, J.S.; West, C.M.; Sampson, H.A.; Burks, A.W.; Bannon, G.A. Molecular cloning and epitope analysis of the peanut allergen Ara h 3. J. Clin. Investig. 1999, 103, 535–542. [Google Scholar] [PubMed]
- Howell, W.M.; Turner, S.J.; Hourihane, J.O.; Dean, T.P.; Warner, J.O. HLA Class II DRB1, DQB1 and DPB1 Genotypic Associations with Peanut Allergy: Evidence from a Family-Based and Case-Control Study. Clin. Exp. Allergy 1998, 28, 156–162. [Google Scholar]
- Pascal, M.; Konstantinou, G.N.; Masilamani, M.; Lieberman, J.; Sampson, H.A. In Silico Prediction of Ara h 2 T Cell Epitopes in Peanut-Allergic Children. Clin. Exp. Allergy 2013, 43, 116–127. [Google Scholar]
- Hemler, J.A.; Phillips, E.J.; Mallal, S.A.; Kendall, P.L. The Evolving Story of Human Leukocyte Antigen and the Immunogenetics of Peanut Allergy. Ann. Allergy Asthma Immunol. 2015, 115, 471–476. [Google Scholar]
- Martino, D.J.; Ashley, S.; Koplin, J.; Ellis, J.; Saffery, R.; Dharmage, S.C.; Gurrin, L.; Matheson, M.C.; Kalb, B.; Marenholz, I.; et al. Genome-wide Association Study of Peanut Allergy Reproduces Association with Amino Acid Polymorphisms in HLA-DRB1. Clin. Exp. Allergy 2017, 47, 217–223. [Google Scholar]
- Asai, Y.; Eslami, A.; van Ginkel, C.D.; Akhabir, L.; Wan, M.; Yin, D.; Ellis, G.; Ben-Shoshan, M.; Marenholz, I.; Martino, D.; et al. A Canadian Genome-Wide Association Study and Meta-Analysis Confirm HLA as a Risk Factor for Peanut Allergy Independent of Asthma. J. Allergy Clin. Immunol. 2018, 141, 1513–1516. [Google Scholar]
- Kostara, M.; Chondrou, V.; Sgourou, A.; Douros, K.; Tsabouri, S. HLA Polymorphisms and Food Allergy Predisposition. J. Pediatr. Genet. 2020, 9, 77–86. [Google Scholar]
- Kanchan, K.; Grinek, S.; Bahnson, H.T.; Ruczinski, I.; Shankar, G.; Larson, D.; Du Toit, G.; Barnes, K.C.; Sampson, H.A.; Suarez-Farinas, M.; et al. HLA Alleles and Sustained Peanut Consumption Promote IgG4 Responses in Subjects Protected from Peanut Allergy. J. Clin. Investig. 2022, 132, e152070. [Google Scholar]
- Jutel, M.; Jaeger, L.; Suck, R.; Meyer, H.; Fiebig, H.; Cromwell, O. Allergen-specific Immunotherapy with Recombinant Grass Pollen Allergens. J. Allergy Clin. Immunol. 2005, 116, 608–613. [Google Scholar] [PubMed]
- Yonekura, S.; Okamoto, Y.; Sakurai, D.; Horiguchi, S.; Hanazawa, T.; Nakano, A.; Kudou, F.; Nakamaru, Y.; Honda, K.; Hoshioka, A.; et al. Sublingual Immunotherapy with House Dust Extract for House Dust-Mite Allergic Rhinitis in Children. Allergol. Int. 2010, 59, 381–388. [Google Scholar]
- Clark, J.; White, N.D. Immunotherapy for Cat Allergies: A Potential Strategy to Scratch Back. Am. J. Lifestyle Med. 2017, 11, 310–313. [Google Scholar] [PubMed]
- Ruëff, F.; Bauer, A.; Becker, S.; Bircher, A.J.; Ebner, C.; Jung, K.; Klimek, L.; Koberne, F.; Koletzko, S.; Lepp, U.; et al. Diagnosis and Treatment of Hymenoptera Venom Allergy: S2k Guideline of the German Society of Allergology and Clinical Immunology (DGAKI) in Collaboration with the Arbeitsgemeinschaft für Berufs- und Umwelt-dermatologie e.V. (ABD), the Medical Association of German Allergologists (AeDA), the German Society of Dermatology (DDG), the German Society of Oto-Rhino-Laryngology, Head and Neck Surgery (DGHNOKC), the German Society of Pediatrics and Adolescent Medicine (DGKJ), the Society for Pediatric Allergy and Environmental Medicine (GPA), German Respiratory Society (DGP), and the Austrian Society for Allergy and Immunology (ÖGAI). Allergol. Select. 2023, 7, 154–190. [Google Scholar] [PubMed]
- Ponda, P.; Cerise, J.E.; Navetta-Modrov, B.; Zhang, C.; Goldfarb, J.; Irizar, H.; Goldberg, J.D.; Wang, M.; Zhang, S.; Sampson, H.A. The Age-Specific Microbiome of Children with Milk, Egg, and Peanut Allergy. Ann. Allergy Asthma Immunol. 2024, 133, 203–210.e6. [Google Scholar] [PubMed]
- Jones, S.M.; Kim, E.H.; Nadeau, K.C.; Nowak-Wegrzyn, A.; Wood, R.A.; Sampson, H.A.; Scurlock, A.M.; Chinthrajah, S.; Wang, J.; Pesek, R.D.; et al. Efficacy and Safety of Oral Immunotherapy in Children Aged 1–3 Years with Peanut Allergy (the Immune Tolerance Network IMPACT Trial): A Randomised Placebo-Controlled Study. Lancet 2022, 399, 359–371. [Google Scholar]
- Kim, E.H.; Keet, C.A.; Virkud, Y.V.; Bird, J.A.; Beyer, K.; Leickly, F.; Liu, A.H.; Petroni, D.; Sampson, H.A.; Wood, R.A.; et al. Open-Label Study of the Efficacy, Safety, and Durability of Peanut Sublingual Immunotherapy in Peanut-Allergic Children. J. Allergy Clin. Immunol. 2023, 151, 1558–1565.e6. [Google Scholar]
- Du Toit, G.; Brown, K.R.; Vereda, A.; Irani, A.M.; Tilles, S.; Ratnayake, A.; Jones, S.M.; Vickery, B.P. Oral Immunotherapy for Peanut Allergy in Children 1 to Less Than 4 Years of Age. NEJM Evid. 2023, 2, EVIDoa2300145. [Google Scholar] [PubMed]
- Greenhawt, M.; Shaker, M.; Abrams, E.M. Peanut Oral Immunotherapy in Very Young Children. Lancet 2022, 399, 336–337. [Google Scholar]
- Kim, E.H.; Bird, J.A.; Keet, C.A.; Virkud, Y.V.; Herlihy, L.; Ye, P.; Smeekens, J.M.; Guo, R.; Yue, X.; Penumarti, A.; et al. Desensitization and Remission after Peanut Sublingual Immunotherapy in 1- to 4-Year-Old Peanut-Allergic Children: A Randomized, Placebo-Controlled Trial. J. Allergy Clin. Immunol. 2024, 153, 173–181.e10. [Google Scholar]
- Tedner, S.G.; Asarnoj, A.; Thulin, H.; Westman, M.; Konradsen, J.R.; Nilsson, C. Food Allergy and Hypersensitivity Reactions in Children and Adults—A Review. J. Intern. Med. 2022, 291, 283–302. [Google Scholar] [PubMed]
- Fleischer, D.M.; Shreffler, W.G.; Campbell, D.E.; Green, T.D.; Anvari, S.; Assa’ad, A.; Bégin, P.; Beyer, K.; Bird, J.A.; Brown-Whitehorn, T.; et al. Long-Term, Open-Label Extension Study of the Efficacy and Safety of Epicutaneous Immunotherapy for Peanut Allergy in Children: PEOPLE 3-Year Results. J. Allergy Clin. Immunol. 2020, 146, 863–874. [Google Scholar] [PubMed]
- Schneider, T.D.; Stephens, R.M. Sequence Logos: A New Way to Display Consensus Sequences. Nucleic Acids Res. 1990, 18, 6097–6100. [Google Scholar] [PubMed]
- Doytchinova, I.; Atanasova, M.; Fernandez, A.; Moreno, F.J.; Koning, F.; Dimitrov, I. Modeling Peptide-Protein Interactions by a Logo-Based Method: Application in Peptide-HLA Binding Predictions. Molecules 2024, 29, 284. [Google Scholar] [CrossRef]
- Nielsen, M.; Andreatta, M. NNAlign: A Platform to Construct and Evaluate Artificial Neural Network Models of Receptor-Ligand Interactions. Nucleic Acids Res. 2017, 45, W344–W349. [Google Scholar]
- Vita, R.; Mahajan, S.; Overton, J.A.; Dhanda, S.K.; Martini, S.; Cantrell, J.R.; Wheeler, D.K.; Sette, A.; Peters, B. The Immune Epitope Database (IEDB): 2018 Update. Nucleic Acids Res. 2019, 47, D339–D343. [Google Scholar]
- Chicco, D.; Jurman, G. The Matthews Correlation Coefficient (MCC) Should Replace the ROC AUC as the Standard Metric for Assessing Binary Classification. BioData Min. 2023, 16, 4. [Google Scholar]
- UniProt Consortium. UniProt: The Universal Protein Knowledgebase in 2023. Nucleic Acids Res. 2023, 51, D523–D531. [Google Scholar]
- Benson, D.A.; Cavanaugh, M.; Clark, K.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Sayers, E.W. GenBank. Nucleic Acids Res. 2013, 41, D36–D42. [Google Scholar]
- Celis, E.; Tsai, V.; Crimi, C.; DeMars, R.; Wentworth, P.A.; Chesnut, R.W.; Grey, H.M.; Sette, A.; Serra, H.M. Induction of anti-tumor cytotoxic T lymphocytes in normal humans using primary cultures and synthetic peptide epitopes. Proc. Natl. Acad. Sci. USA 1994, 91, 2105–2109. [Google Scholar] [PubMed]
- Barker, D.J.; Maccari, G.; Georgiou, X.; Cooper, M.A.; Flicek, P.; Robinson, J.; Marsh, S.G.E. The IPD-IMGT/HLA Database. Nucleic Acids Res. 2023, 51, D1053–D1060. [Google Scholar] [PubMed]
- MacDonald, K.S.; Embree, J.E.; Nagelkerke, N.J.; Castillo, J.; Ramhadin, S.; Njenga, S.; Oyug, J.; Ndinya-Achola, J.; Barber, B.H.; Bwayo, J.J.; et al. The HLA A2/6802 supertype is associated with reduced risk of perinatal human immunodeficiency virus type 1 transmission. J. Infect. Dis. 2001, 183, 503–506. [Google Scholar]
- Rallón, N.; Restrepo, C.; Vicario, J.L.; Del Romero, J.; Rodríguez, C.; García-Samaniego, J.; García, M.; Cabello, A.; Górgolas, M.; Benito, J.M. Human leucocyte antigen (HLA)-DQB103:02 and HLA-A02:01 have opposite patterns in their effects on susceptibility to HIV infection. HIV Med. 2017, 18, 587–594. [Google Scholar] [PubMed]
- MacDonald, K.S.; Fowke, K.R.; Kimani, J.; Dunand, V.A.; Nagelkerke, N.J.; Ball, T.B.; Oyugi, J.; Njagi, E.; Gaur, L.K.; Brunham, R.C.; et al. Influence of HLA supertypes on susceptibility and resistance to human immunodeficiency virus type 1 infection. J. Infect. Dis. 2000, 181, 1581–1589. [Google Scholar]
- Noble, J.A.; Valdes, A.M.; Thomson, G.; Erlich, H.A. The HLA class II locus DPB1 can influence susceptibility to type 1 diabetes. Diabetes 2000, 49, 121–125. [Google Scholar]
- Zhang, L.; Zhang, Y.J.; Chen, J.; Yang, M.; Wei, W.; Wang, Y.; Lu, J. The Association of HLA-B27 and Klebsiella pneumoniae in Ankylosing Spondylitis: A Systematic Review. Microb. Pathog. 2018, 117, 49–54. [Google Scholar]
- Lobos, C.A.; Downing, J.; D’Orsogna, L.J.; Chatzileontiadou, D.S.M.; Gras, S. Protective HLA-B57: T Cell and Natural Killer Cell Recognition in HIV Infection. Biochem. Soc. Trans. 2022, 50, 1329–1339. [Google Scholar]
- Ge, C.; Weisse, S.; Xu, B.; Thiel, M.; Chalouni, C.; Travis, A.M.; Kowal, C.; Elwood, J.; Brawn-Cinani, B.; Turner, D.; et al. Key Interactions in the Trimolecular Complex Consisting of the Rheumatoid Arthritis-Associated DRB1*04:01 Molecule, the Major Glycosylated Collagen II Peptide and the T-Cell Receptor. Ann. Rheum. Dis. 2022, 81, 480–489. [Google Scholar]
- Capittini, C.; De Silvestri, A.; Terzaghi, M.; Pasi, A.; Zucconi, M.; Luisi, C.; De Amici, M.; Beri, R.; Tinelli, C.; Milani, G.P.; et al. Correlation between HLA-DQB1*06:02 and Narcolepsy with and without Cataplexy: Approving a Safe and Sensitive Genetic Test in Four Major Ethnic Groups. A Systematic Meta-Analysis. Sleep Med. 2018, 52, 150–157. [Google Scholar] [PubMed]
- D’Amato, M.; Scotto d’Abusco, A.; Maggi, E.; Parmiani, S.; Ferrara, A.; Pene, J.; Richter, A.; Kontou-Fili, K.; Dente, F.L.; Capron, F.; et al. Association of Responsiveness to the Major Pollen Allergen of Parietaria officinalis with HLA-DRB1* Alleles: A Multicenter Study. Hum. Immunol. 1996, 46, 100–106. [Google Scholar]
- Aboulaghras, S.; Piancatelli, D.; Taghzouti, K.; Benkirane, H.; Cantalupo, G.; Bellanti, F.; Lancellotti, S.; Strohmenger, L.; Caserta, V.; Sorrentino, M.C.; et al. Meta-Analysis and Systematic Review of HLA DQ2/DQ8 in Adults with Celiac Disease. Int. J. Mol. Sci. 2023, 24, 1188. [Google Scholar] [CrossRef]
- Howell, W.M.; Holgate, S.T. HLA genetics and allergic disease. Thorax 1995, 50, 815–818. [Google Scholar] [PubMed]
- Kontakioti, E.; Domvri, K.; Papakosta, D.; Daniilidis, M. HLA and asthma phenotypes/endotypes: A review. Hum. Immunol. 2014, 75, 930–939. [Google Scholar]
- Dimitrov, I.; Doytchinova, I. Associations between main food allergens and HLA-DR/DQ polymorphism. Int. Arch. Allergy Immunol. 2016, 169, 33–39. [Google Scholar]
- Gheerbrant, H.; Guillien, A.; Vernet, R.; Lupinek, C.; Pison, C.; Pin, I.; Demenais, F.; Nadif, R.; Bousquet, J.; Pickl, W.F.; et al. Associations between specific IgE sensitization to 26 respiratory allergen molecules and HLA class II alleles in the EGEA cohort. Allergy 2021, 76, 2575–2586. [Google Scholar]
- Germundson, D.L.; Nookala, S.; Smith, N.A.; Warda, Y.; Nagamoto-Combs, K. HLA-II Alleles Influence Physical and Behavioral Responses to a Whey Allergen in a Transgenic Mouse Model of Cow’s Milk Allergy. Front. Allergy 2022, 3, 870513. [Google Scholar]
- Southwood, S.; Sidney, J.; Kondo, A.; del Guercio, M.F.; Appella, E.; Hoffman, S.; Kubo, R.T.; Chesnut, R.W.; Grey, H.M.; Sette, A. Several common HLA-DR types share largely overlapping peptide binding repertoires. J. Immunol. 1998, 160, 3363–3373. [Google Scholar]
- Doytchinova, I.A.; Flower, D.R. In silico identification of supertypes for Class II Major Histocompatibility Complexes. J. Immunol. 2005, 174, 7085–7095. [Google Scholar]
- Stern, L.J.; Calvo-Calle, J.M. HLA-DR: Molecular insights and vaccine design. Curr. Pharm. Des. 2009, 15, 3249–3261. [Google Scholar]
- Greaves, S.A.; Ravindran, A.; Santos, R.G.; Chen, L.; Falta, M.T.; Wang, Y.; Mitchell, A.M.; Atif, S.M.; Mack, D.G.; Tinega, A.N.; et al. CD4+ T cells in the lungs of acute sarcoidosis patients recognize an Aspergillus nidulans epitope. J. Exp. Med. 2021, 218, e20210785. [Google Scholar]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [PubMed]
- DeLong, J.H.; Simpson, K.H.; Wambre, E.; James, E.A.; Robinson, D.; Kwok, W.W. Ara h 1-reactive T cells in individuals with peanut allergy. J. Allergy Clin. Immunol. 2011, 127, 1211–1218.e3. [Google Scholar] [PubMed]
- Birrueta, G.; Tripple, V.; Pham, J.; Manohar, M.; James, E.A.; Kwok, W.W.; Nadeau, K.C.; Sette, A.; Peters, B.; Schulten, V. Peanut-specific T cell responses in patients with different clinical reactivity. PLoS ONE 2018, 13, e0204620. [Google Scholar]
- Sette, A.; Vitiello, A.; Reherman, B.; Fowler, P.; Nayersina, R.; Kast, W.M.; Melief, C.J.; Oseroff, C.; Yuan, L.; Ruppert, J.; et al. The relationship between class I binding affinity and immunogenicity of potential cytotoxic T cell epitopes. J. Immunol. 1994, 153, 5586–5592. [Google Scholar]
- Baumgaertner, P.; Schmidt, J.; Costa-Nunes, C.; Bordry, N.; Guillaume, P.; Luescher, I.; Speiser, D.; Rufer, N.; Hebeisen, M. CD8 T Cell Function and Cross-Reactivity Explored by Stepwise Increased Peptide-HLA versus TCR Affinity. Front. Immunol. 2022, 13, 973986. [Google Scholar]
- Arunachalam, A.B. Vaccines Induce Homeostatic Immunity, Generating Several Secondary Benefits. Vaccines 2024, 12, 396. [Google Scholar] [CrossRef] [PubMed]
- Hamley, I.W. Peptides for Vaccine Development. ACS Appl. Bio Mater. 2022, 5, 905–944. [Google Scholar]
- Gupta, K.; Kumar, S.; Das, M.; Dwivedi, P.D. Peptide Based Immunotherapy: A Pivotal Tool for Allergy Treatment. Int. Immunopharmacol. 2014, 19, 391–398. [Google Scholar] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).