Can O-GIcNAc Transferase (OGT) Complex Be Used as a Target for the Treatment of Hematological Malignancies?
Abstract
1. Introduction
| Cancer Types | Mechanism | Physiological Effects | Expression | Reference(s) |
|---|---|---|---|---|
| CLL | OGT-induced c-myc, p53, and Akt O-GlcNAcylation. | Decline | [26] | |
| OGT expression increased. | Increase in resistance. | [27] | ||
| pre-B-ALL | OGT-mediated increase in O-GlcNAcylation, which in turn activates the PI3K/AKT/c-Myc cascade. | Inhibition of apoptosis. | Rise | [20] |
| DLBCL | Increased OGT mRNA expression. | Reduction in patients survival. | Decline | [28] |
| Silencing OGT results in decreased activity of NF-kB and NFATc. | Growth inhibition. | [28] | ||
| CML | OGT-induced STAT5 O-GlcNAcylation. | Promotion of cell proliferation. | Decline | [29] |
| APL | OGT-mediated O-GlcNAc-dependent upregulation of LGALS12. | Helping to maintain cell stemness. | Rise | [30] |
| AML | Inhibition of OGT and OGA, causing O-GlcNAcylation imbalance. | Causing inhibition of stem/progenitor cell self-renewal in CD34+ HSPC and AML cells (KG-1) and driving myeloid differentiation. | [31] | |
| OSMI-1, BADGP, and siRNA-mediated OGT inhibition. | Inducing cell death. | Decline | [32] | |
| Decreased protein levels of pNF-κB and pAKT and caspase-9/caspase-3 cascade. | Improvement inchemotherapy sensitivity. | [33] | ||
| Increased OGT expression. | Enhancement of resistance to BTZ. | [34] |
2. OGT
3. OGT/HCF-1 Complex
4. NSL Complex
5. OGT/TET Complex
6. PR-DUB Complex
7. Small Molecules Targeting OGT
8. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Haslauer, T.; Greil, R.; Zaborsky, N.; Geisberger, R. CAR T-Cell Therapy in Hematological Malignancies. Int. J. Mol. Sci. 2021, 22, 8996. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA A Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Xing, Y.; Zhang, Q.; Xie, J.; Huang, D.; Gu, H.; He, P.; Zhou, M.; Xu, S.; Pang, X.; et al. Synthesis and Biological Evaluation of B-Cell Lymphoma 6 Inhibitors of N-Phenyl-4-pyrimidinamine Derivatives Bearing Potent Activities against Tumor Growth. J. Med. Chem. 2020, 63, 676–695. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Li, J.; Xu, L.; Găman, M.A.; Zou, Z. The genesis and evolution of acute myeloid leukemia stem cells in the microenvironment: From biology to therapeutic targeting. Cell Death Discov. 2022, 8, 397. [Google Scholar] [CrossRef] [PubMed]
- Barreca, M.; Spanò, V.; Rocca, R.; Bivacqua, R.; Gualtieri, G.; Raimondi, M.V.; Gaudio, E.; Bortolozzi, R.; Manfreda, L.; Bai, R.; et al. Identification of pyrrolo[3′,4′:3,4]cyclohepta[1,2-d][1,2]oxazoles as promising new candidates for the treatment of lymphomas. Eur. J. Med. Chem. 2023, 254, 115372. [Google Scholar] [CrossRef] [PubMed]
- Haselager, M.V.; Kater, A.P.; Eldering, E. Proliferative Signals in Chronic Lymphocytic Leukemia; What Are We Missing? Front. Oncol. 2020, 10, 592205. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Wang, Z.; Zhao, J.; Wu, F. Lymphadenopathy Due to Kimura’s Disease Mimicking Lymphoma on FDG PET/CT. Clin. Nucl. Med. 2019, 44, 299–300. [Google Scholar] [CrossRef] [PubMed]
- Proytcheva, M. Juvenile myelomonocytic leukemia. Semin. Diagn. Pathol. 2011, 28, 298–303. [Google Scholar] [CrossRef]
- Singh, A.; Hungund, B.; Kumar, L.; Pattanshetti, M. Clinico-haematological profile of patients with bicytopenia. Pathology 2018, 50, 540–548. [Google Scholar] [CrossRef]
- Chen, X.; Zhang, L.; He, H.; Sun, Y.; Shen, Q.; Shi, L. Increased O-GlcNAcylation induces myocardial hypertrophy. Vitr. Cell. Dev. Biol. Anim. 2020, 56, 735–743. [Google Scholar] [CrossRef]
- Liu, J.; Hao, Y.; He, Y.; Li, X.; Sun, D.E.; Zhang, Y.; Yang, P.Y.; Chen, X. Quantitative and Site-Specific Chemoproteomic Profiling of Protein O-GlcNAcylation in the Cell Cycle. ACS Chem. Biol. 2021, 16, 1917–1923. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Qian, K. Protein O-GlcNAcylation: Emerging mechanisms and functions. Nat. Rev. Mol. Cell Biol. 2017, 18, 452–465. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhang, J.; Dong, H.; Kong, Y.; Guan, Y. Emerging field: O-GlcNAcylation in ferroptosis. Front. Mol. Biosci. 2023, 10, 1203269. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.J. Advances in Strategies and Tools Available for Interrogation of Protein O-GlcNAcylation. ChemBioChem A Eur. J. Chem. Biol. 2021, 22, 3010–3026. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.H.; Liao, C.C.; Chen, M.Y.; Chou, T.Y. Feedback Regulation of O-GlcNAc Transferase through Translation Control to Maintain Intracellular O-GlcNAc Homeostasis. Int. J. Mol. Sci. 2021, 22, 3463. [Google Scholar] [CrossRef] [PubMed]
- Ye, L.; Ding, W.; Xiao, D.; Jia, Y.; Zhao, Z.; Ao, X.; Wang, J. O-GlcNAcylation: Cellular physiology and therapeutic target for human diseases. MedComm 2023, 4, e456. [Google Scholar] [CrossRef] [PubMed]
- Na, H.J.; Akan, I.; Abramowitz, L.K.; Hanover, J.A. Nutrient-Driven O-GlcNAcylation Controls DNA Damage Repair Signaling and Stem/Progenitor Cell Homeostasis. Cell Rep. 2020, 31, 107632. [Google Scholar] [CrossRef]
- Sun, L.; Lv, S.; Song, T. O-GlcNAcylation links oncogenic signals and cancer epigenetics. Discov. Oncol. 2021, 12, 54. [Google Scholar] [CrossRef]
- Chatham, J.C.; Zhang, J.; Wende, A.R. Role of O-Linked N-Acetylglucosamine Protein Modification in Cellular (Patho)Physiology. Physiol. Rev. 2021, 101, 427–493. [Google Scholar] [CrossRef]
- Zhang, B.; Zhou, P.; Li, X.; Shi, Q.; Li, D.; Ju, X. Bitterness in sugar: O-GlcNAcylation aggravates pre-B acute lymphocytic leukemia through glycolysis via the PI3K/Akt/c-Myc pathway. Am. J. Cancer Res. 2017, 7, 1337–1349. [Google Scholar]
- Hurrish, K.H.; Su, Y.; Patel, S.; Ramage, C.L.; Carter, J.L.; Edwards, H.; Buck, S.A.; Wiley, S.E.; Hüttemann, M.; Polin, L.; et al. Enhancing anti-AML activity of venetoclax by isoflavone ME-344 through suppression of OXPHOS and/or purine biosynthesis. Res. Sq. 2023. [Google Scholar] [CrossRef] [PubMed]
- Jaworska, M.; Szczudło, J.; Pietrzyk, A.; Shah, J.; Trojan, S.E.; Ostrowska, B.; Kocemba-Pilarczyk, K.A. The Warburg effect: A score for many instruments in the concert of cancer and cancer niche cells. Pharmacol. Rep. 2023, 75, 876–890. [Google Scholar] [CrossRef] [PubMed]
- Parker, M.P.; Peterson, K.R.; Slawson, C. O-GlcNAcylation and O-GlcNAc Cycling Regulate Gene Transcription: Emerging Roles in Cancer. Cancers 2021, 13, 1666. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Lai, M.K.P.; Arumugam, T.V.; Jo, D.G. O-GlcNAcylation as a Therapeutic Target for Alzheimer’s Disease. Neuromol. Med. 2020, 22, 171–193. [Google Scholar] [CrossRef]
- Hu, J.; Gao, Q.; Yang, Y.; Xia, J.; Zhang, W.; Chen, Y.; Zhou, Z.; Chang, L.; Hu, Y.; Zhou, H.; et al. Hexosamine biosynthetic pathway promotes the antiviral activity of SAMHD1 by enhancing O-GlcNAc transferase-mediated protein O-GlcNAcylation. Theranostics 2021, 11, 805–823. [Google Scholar] [CrossRef] [PubMed]
- Spaner, D.E. O-GlcNAcylation in Chronic Lymphocytic Leukemia and Other Blood Cancers. Front. Immunol. 2021, 12, 772304. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Böttcher, M.; Walther, T.; Mougiakakos, D.; Zenz, T.; Huber, W. Energy metabolism is co-determined by genetic variants in chronic lymphocytic leukemia and influences drug sensitivity. Haematologica 2019, 104, 1830–1840. [Google Scholar] [CrossRef]
- Wu, J.; Meng, F.; Ran, D.; Song, Y.; Dang, Y.; Lai, F.; Yang, L.; Deng, M.; Song, Y.; Zhu, J. The Metabolism and Immune Environment in Diffuse Large B-Cell Lymphoma. Metabolites 2023, 13, 734. [Google Scholar] [CrossRef]
- Zheng, L.; Yang, Q.; Li, F.; Zhu, M.; Yang, H.; Tan, T.; Wu, B.; Liu, M.; Xu, C.; Yin, J.; et al. The Glycosylation of Immune Checkpoints and Their Applications in Oncology. Pharmaceuticals 2022, 15, 1451. [Google Scholar] [CrossRef]
- Decourcelle, A.; Loison, I.; Baldini, S.; Leprince, D.; Dehennaut, V. Evidence of a compensatory regulation of colonic O-GlcNAc transferase and O-GlcNAcase expression in response to disruption of O-GlcNAc homeostasis. Biochem. Biophys. Res. Commun. 2020, 521, 125–130. [Google Scholar] [CrossRef]
- Luanpitpong, S.; Rodboon, N.; Samart, P.; Janan, M.; Klaihmon, P.; Lorthongpanich, C.; U-Pratya, Y.; Issaragrisil, S. Inhibition of O-GlcNAcase Inhibits Hematopoietic and Leukemic Stem Cell Self-Renewal and Drives Dendritic Cell Differentiation via STAT3/5 Signaling. Stem Cells 2022, 40, 1078–1093. [Google Scholar] [CrossRef] [PubMed]
- Walter, L.A.; Lin, Y.H.; Halbrook, C.J.; Chuh, K.N.; He, L.; Pedowitz, N.J.; Batt, A.R.; Brennan, C.K.; Stiles, B.L.; Lyssiotis, C.A.; et al. Inhibiting the Hexosamine Biosynthetic Pathway Lowers O-GlcNAcylation Levels and Sensitizes Cancer to Environmental Stress. Biochemistry 2020, 59, 3169–3179. [Google Scholar] [CrossRef] [PubMed]
- Lam, C.; Low, J.Y.; Tran, P.T.; Wang, H. The hexosamine biosynthetic pathway and cancer: Current knowledge and future therapeutic strategies. Cancer Lett. 2021, 503, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Tibullo, D.; Giallongo, C.; Romano, A.; Vicario, N.; Barbato, A.; Puglisi, F.; Parenti, R.; Amorini, A.M.; Wissam Saab, M.; Tavazzi, B.; et al. Mitochondrial Functions, Energy Metabolism and Protein Glycosylation are Interconnected Processes Mediating Resistance to Bortezomib in Multiple Myeloma Cells. Biomolecules 2020, 10, 696. [Google Scholar] [CrossRef] [PubMed]
- Kapuria, V.; Rohrig, U.F.; Waridel, P.; Lammers, F.; Borodkin, V.S.; van Aalten, D.M.F.; Zoete, V.; Herr, W. The conserved threonine-rich region of the HCF-1(PRO) repeat activates promiscuous OGT:UDP-GlcNAc glycosylation and proteolysis activities. J. Biol. Chem. 2018, 293, 17754–17768. [Google Scholar] [CrossRef] [PubMed]
- de Queiroz, R.M.; Moon, S.H.; Prives, C. O-GlcNAc transferase regulates p21 protein levels and cell proliferation through the FoxM1-Skp2 axis in a p53-independent manner. J. Biol. Chem. 2022, 298, 102289. [Google Scholar] [CrossRef] [PubMed]
- Itkonen, H.M.; Loda, M.; Mills, I.G. O-GlcNAc Transferase—An Auxiliary Factor or a Full-blown Oncogene? Mol. Cancer Res. 2021, 19, 555–564. [Google Scholar] [CrossRef]
- Lubas, W.A.; Frank, D.W.; Krause, M.; Hanover, J.A. O-Linked GlcNAc transferase is a conserved nucleocytoplasmic protein containing tetratricopeptide repeats. J. Biol. Chem. 1997, 272, 9316–9324. [Google Scholar] [CrossRef]
- Kreppel, L.K.; Blomberg, M.A.; Hart, G.W. Dynamic glycosylation of nuclear and cytosolic proteins. Cloning and characterization of a unique O-GlcNAc transferase with multiple tetratricopeptide repeats. J. Biol. Chem. 1997, 272, 9308–9315. [Google Scholar] [CrossRef]
- Martinez, M.; Renuse, S.; Kreimer, S.; O’Meally, R.; Natov, P.; Madugundu, A.K.; Nirujogi, R.S.; Tahir, R.; Cole, R.; Pandey, A.; et al. Quantitative Proteomics Reveals that the OGT Interactome Is Remodeled in Response to Oxidative Stress. Mol. Cell. Proteom. 2021, 20, 100069. [Google Scholar] [CrossRef]
- Song, T.; Zou, Q.; Yan, Y.; Lv, S.; Li, N.; Zhao, X.; Ma, X.; Liu, H.; Tang, B.; Sun, L. DOT1L O-GlcNAcylation promotes its protein stability and MLL-fusion leukemia cell proliferation. Cell Rep. 2021, 36, 109739. [Google Scholar] [CrossRef] [PubMed]
- Kampa-Schittenhelm, K.M.; Haverkamp, T.; Bonin, M.; Tsintari, V.; Bühring, H.J.; Haeusser, L.; Blumenstock, G.; Dreher, S.T.; Ganief, T.; Akmut, F.; et al. Epigenetic activation of O-linked β-N-acetylglucosamine transferase overrides the differentiation blockage in acute leukemia. EBioMedicine 2020, 54, 102678. [Google Scholar] [CrossRef] [PubMed]
- Kositzke, A.; Fan, D.; Wang, A.; Li, H.; Worth, M.; Jiang, J. Elucidating the protein substrate recognition of O-GlcNAc transferase (OGT) toward O-GlcNAcase (OGA) using a GlcNAc electrophilic probe. Int. J. Biol. Macromol. 2021, 169, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Chong, P.A.; Nosella, M.L.; Vanama, M.; Ruiz-Arduengo, R.; Forman-Kay, J.D. Exploration of O-GlcNAc transferase glycosylation sites reveals a target sequence compositional bias. J. Biol. Chem. 2023, 299, 104629. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wan, R.; Zou, Z.; Lao, L.; Shao, G.; Zheng, Y.; Tang, L.; Yuan, Y.; Ge, Y.; He, C.; et al. O-GlcNAcylation determines the translational regulation and phase separation of YTHDF proteins. Nat. Cell Biol. 2023, 25, 1676–1690. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, O.; Affar, M.; Masclef, L.; Echbicheb, M.; Gushul-Leclaire, M.; Estavoyer, B.; Vocadlo, D.J.; Affar, E.B. Immunoprecipitation and Western blot-based detection of protein O-GlcNAcylation in cells. STAR Protoc. 2022, 3, 101108. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Tomic, J.; Wen, F.; Shaha, S.; Bahlo, A.; Harrison, R.; Dennis, J.W.; Williams, R.; Gross, B.J.; Walker, S.; et al. Aberrant O-GlcNAcylation characterizes chronic lymphocytic leukemia. Leukemia 2010, 24, 1588–1598. [Google Scholar] [CrossRef]
- Giménez, N.; Schulz, R.; Higashi, M.; Aymerich, M.; Villamor, N.; Delgado, J.; Juan, M.; López-Guerra, M.; Campo, E.; Rosich, L.; et al. Targeting IRAK4 disrupts inflammatory pathways and delays tumor development in chronic lymphocytic leukemia. Leukemia 2020, 34, 100–114. [Google Scholar] [CrossRef]
- Kim, U.; Shin, H.Y. Genomic Mutations of the STAT5 Transcription Factor Are Associated with Human Cancer and Immune Diseases. Int. J. Mol. Sci. 2022, 23, 11297. [Google Scholar] [CrossRef]
- Sermikli, B.P.; Aydogdu, G.; Yilmaz, E. Role of the O-GlcNAc modification on insulin resistance and endoplasmic reticulum stress in 3T3-L1 cells. Mol. Biol. Rep. 2020, 47, 5927–5942. [Google Scholar] [CrossRef]
- Asthana, A.; Ramakrishnan, P.; Vicioso, Y.; Zhang, K.; Parameswaran, R. Hexosamine Biosynthetic Pathway Inhibition Leads to AML Cell Differentiation and Cell Death. Mol. Cancer Ther. 2018, 17, 2226–2237. [Google Scholar] [CrossRef] [PubMed]
- Julien, E.; Herr, W. A switch in mitotic histone H4 lysine 20 methylation status is linked to M phase defects upon loss of HCF-1. Mol. Cell 2004, 14, 713–725. [Google Scholar] [CrossRef] [PubMed]
- Popay, T.M.; Wang, J.; Adams, C.M.; Howard, G.C.; Codreanu, S.G.; Sherrod, S.D.; McLean, J.A.; Thomas, L.R.; Lorey, S.L.; Machida, Y.J.; et al. MYC regulates ribosome biogenesis and mitochondrial gene expression programs through its interaction with host cell factor-1. eLife 2021, 10, e60191. [Google Scholar] [CrossRef] [PubMed]
- Itkonen, H.M.; Urbanucci, A.; Martin, S.E.; Khan, A.; Mathelier, A.; Thiede, B.; Walker, S.; Mills, I.G. High OGT activity is essential for MYC-driven proliferation of prostate cancer cells. Theranostics 2019, 9, 2183–2197. [Google Scholar] [CrossRef]
- Lee, K.H.; Kim, B.C.; Jeong, C.W.; Ku, J.H.; Kim, H.H.; Kwak, C. MLL5, a histone modifying enzyme, regulates androgen receptor activity in prostate cancer cells by recruiting co-regulators, HCF1 and SET1. BMB Rep. 2020, 53, 634–639. [Google Scholar] [CrossRef]
- Singh, M.; Bacolla, A.; Chaudhary, S.; Hunt, C.R.; Pandita, S.; Chauhan, R.; Gupta, A.; Tainer, J.A.; Pandita, T.K. Histone Acetyltransferase MOF Orchestrates Outcomes at the Crossroad of Oncogenesis, DNA Damage Response, Proliferation, and Stem Cell Development. Mol. Cell. Biol. 2020, 40, e00232-20. [Google Scholar] [CrossRef] [PubMed]
- Deplus, R.; Delatte, B.; Schwinn, M.K.; Defrance, M.; Méndez, J.; Murphy, N.; Dawson, M.A.; Volkmar, M.; Putmans, P.; Calonne, E.; et al. TET2 and TET3 regulate GlcNAcylation and H3K4 methylation through OGT and SET1/COMPASS. EMBO J. 2013, 32, 645–655. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Yang, Y.; Qiu, R.; Zhang, K.; Teng, X.; Liu, R.; Wang, Y. Proteomic analysis of the OGT interactome: Novel links to epithelial-mesenchymal transition and metastasis of cervical cancer. Carcinogenesis 2018, 39, 1222–1234. [Google Scholar] [CrossRef]
- Scheuermann, J.C.; de Ayala Alonso, A.G.; Oktaba, K.; Ly-Hartig, N.; McGinty, R.K.; Fraterman, S.; Wilm, M.; Muir, T.W.; Müller, J. Histone H2A deubiquitinase activity of the Polycomb repressive complex PR-DUB. Nature 2010, 465, 243–247. [Google Scholar] [CrossRef]
- Campagne, A.; Lee, M.K.; Zielinski, D.; Michaud, A.; Le Corre, S.; Dingli, F.; Chen, H.; Shahidian, L.Z.; Vassilev, I.; Servant, N.; et al. BAP1 complex promotes transcription by opposing PRC1-mediated H2A ubiquitylation. Nat. Commun. 2019, 10, 348. [Google Scholar] [CrossRef]
- Cao, L.; Xia, X.; Kong, Y.; Jia, F.; Yuan, B.; Li, R.; Li, Q.; Wang, Y.; Cui, M.; Dai, Z.; et al. Deregulation of tumor suppressive ASXL1-PTEN/AKT axis in myeloid malignancies. J. Mol. Cell Biol. 2020, 12, 688–699. [Google Scholar] [CrossRef] [PubMed]
- Whisenhunt, T.R.; Yang, X.; Bowe, D.B.; Paterson, A.J.; Van Tine, B.A.; Kudlow, J.E. Disrupting the enzyme complex regulating O-GlcNAcylation blocks signaling and development. Glycobiology 2006, 16, 551–563. [Google Scholar] [CrossRef] [PubMed]
- Hein, M.Y.; Hubner, N.C.; Poser, I.; Cox, J.; Nagaraj, N.; Toyoda, Y.; Gak, I.A.; Weisswange, I.; Mansfeld, J.; Buchholz, F.; et al. A human interactome in three quantitative dimensions organized by stoichiometries and abundances. Cell 2015, 163, 712–723. [Google Scholar] [CrossRef] [PubMed]
- Giles, A.C.; Desbois, M.; Opperman, K.J.; Tavora, R.; Maroni, M.J.; Grill, B. A complex containing the O-GlcNAc transferase OGT-1 and the ubiquitin ligase EEL-1 regulates GABA neuron function. J. Biol. Chem. 2019, 294, 6843–6856. [Google Scholar] [CrossRef]
- Berthier, A.; Vinod, M.; Porez, G.; Steenackers, A.; Alexandre, J.; Yamakawa, N.; Gheeraert, C.; Ploton, M.; Maréchal, X.; Dubois-Chevalier, J.; et al. Combinatorial regulation of hepatic cytoplasmic signaling and nuclear transcriptional events by the OGT/REV-ERBα complex. Proc. Natl. Acad. Sci. USA 2018, 115, E11033–E11042. [Google Scholar] [CrossRef] [PubMed]
- Thomas, J.F.; Valencia-Sánchez, M.I.; Tamburri, S.; Gloor, S.L.; Rustichelli, S.; Godínez-López, V.; De Ioannes, P.; Lee, R.; Abini-Agbomson, S.; Gretarsson, K.; et al. Structural basis of histone H2A lysine 119 deubiquitination by Polycomb Repressive Deubiquitinase BAP1/ASXL1. bioRxiv 2023. [Google Scholar] [CrossRef] [PubMed]
- Salewskij, K.; Gross-Thebing, T.; Ing-Simmons, E.; Duethorn, B.; Rieger, B.; Fan, R.; Chen, R.; Govindasamy, N.; Brinkmann, H.; Kremer, L.; et al. Ronin governs the metabolic capacity of the embryonic lineage for post-implantation development. EMBO Rep. 2021, 22, e53048. [Google Scholar] [CrossRef]
- Essawy, A.; Jo, S.; Beetch, M.; Lockridge, A.; Gustafson, E.; Alejandro, E.U. O-linked N-acetylglucosamine transferase (OGT) regulates pancreatic α-cell function in mice. J. Biol. Chem. 2021, 296, 100297. [Google Scholar] [CrossRef]
- Emerson, F.J.; Chiu, C.; Lin, L.Y.; Riedel, C.G.; Zhu, M.; Lee, S.S. The chromatin factors SET-26 and HCF-1 oppose the histone deacetylase HDA-1 in longevity and gene regulation in C. elegans. bioRxiv 2023. [Google Scholar] [CrossRef]
- Ózsvári, B.; Sotgia, F.; Lisanti, M.P. First-in-class candidate therapeutics that target mitochondria and effectively prevent cancer cell metastasis: Mitoriboscins and TPP compounds. Aging 2020, 12, 10162–10179. [Google Scholar] [CrossRef]
- Gao, H.; Yin, J.; Ji, C.; Yu, X.; Xue, J.; Guan, X.; Zhang, S.; Liu, X.; Xing, F. Targeting ubiquitin specific proteases (USPs) in cancer immunotherapy: From basic research to preclinical application. J. Exp. Clin. Cancer Res. 2023, 42, 225. [Google Scholar] [CrossRef] [PubMed]
- Kristie, T.M.; Liang, Y.; Vogel, J.L. Control of alpha-herpesvirus IE gene expression by HCF-1 coupled chromatin modification activities. Biochim. Biophys. Acta 2010, 1799, 257–265. [Google Scholar] [CrossRef] [PubMed]
- Wysocka, J.; Herr, W. The herpes simplex virus VP16-induced complex: The makings of a regulatory switch. Trends Biochem. Sci. 2003, 28, 294–304. [Google Scholar] [CrossRef] [PubMed]
- Daou, S.; Mashtalir, N.; Hammond-Martel, I.; Pak, H.; Yu, H.; Sui, G.; Vogel, J.L.; Kristie, T.M.; Affar, E.B. Crosstalk between O-GlcNAcylation and proteolytic cleavage regulates the host cell factor-1 maturation pathway. Proc. Natl. Acad. Sci. USA 2011, 108, 2747–2752. [Google Scholar] [CrossRef] [PubMed]
- Capotosti, F.; Guernier, S.; Lammers, F.; Waridel, P.; Cai, Y.; Jin, J.; Conaway, J.W.; Conaway, R.C.; Herr, W. O-GlcNAc transferase catalyzes site-specific proteolysis of HCF-1. Cell 2011, 144, 376–388. [Google Scholar] [CrossRef] [PubMed]
- Lazarus, M.B.; Jiang, J.; Kapuria, V.; Bhuiyan, T.; Janetzko, J.; Zandberg, W.F.; Vocadlo, D.J.; Herr, W.; Walker, S. HCF-1 is cleaved in the active site of O-GlcNAc transferase. Science 2013, 342, 1235–1239. [Google Scholar] [CrossRef] [PubMed]
- Bhuiyan, T.; Waridel, P.; Kapuria, V.; Zoete, V.; Herr, W. Distinct OGT-Binding Sites Promote HCF-1 Cleavage. PLoS ONE 2015, 10, e0136636. [Google Scholar] [CrossRef]
- Gabellini, D.; Pedrotti, S. The SUV4-20H Histone Methyltransferases in Health and Disease. Int. J. Mol. Sci. 2022, 23, 4736. [Google Scholar] [CrossRef]
- Sekine, H.; Okazaki, K.; Kato, K.; Alam, M.M.; Shima, H.; Katsuoka, F.; Tsujita, T.; Suzuki, N.; Kobayashi, A.; Igarashi, K.; et al. O-GlcNAcylation Signal Mediates Proteasome Inhibitor Resistance in Cancer Cells by Stabilizing NRF1. Mol. Cell. Biol. 2018, 38, e00252-18. [Google Scholar] [CrossRef]
- Tomlin, F.M.; Gerling-Driessen, U.I.M.; Liu, Y.C.; Flynn, R.A.; Vangala, J.R.; Lentz, C.S.; Clauder-Muenster, S.; Jakob, P.; Mueller, W.F.; Ordoñez-Rueda, D.; et al. Inhibition of NGLY1 Inactivates the Transcription Factor Nrf1 and Potentiates Proteasome Inhibitor Cytotoxicity. ACS Cent. Sci. 2017, 3, 1143–1155. [Google Scholar] [CrossRef]
- Gong, X.; Du, D.; Deng, Y.; Zhou, Y.; Sun, L.; Yuan, S. The structure and regulation of the E3 ubiquitin ligase HUWE1 and its biological functions in cancer. Investig. New Drugs 2020, 38, 515–524. [Google Scholar] [CrossRef] [PubMed]
- Maes, A.; Maes, K.; Vlummens, P.; De Raeve, H.; Devin, J.; Szablewski, V.; De Veirman, K.; Menu, E.; Moreaux, J.; Vanderkerken, K.; et al. Maternal embryonic leucine zipper kinase is a novel target for diffuse large B cell lymphoma and mantle cell lymphoma. Blood Cancer J. 2019, 9, 87. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, B.W.; Xiang, L.; Wu, H.; Alexander, S.A.S.; Zhou, P.; Dai, M.Z.; Wang, X.; Xiong, W.; Zhang, Y.; et al. MLL5 is involved in retinal photoreceptor maturation through facilitating CRX-mediated photoreceptor gene transactivation. iScience 2022, 25, 104058. [Google Scholar] [CrossRef]
- Zhou, P.; Wang, Z.; Yuan, X.; Zhou, C.; Liu, L.; Wan, X.; Zhang, F.; Ding, X.; Wang, C.; Xiong, S.; et al. Mixed lineage leukemia 5 (MLL5) protein regulates cell cycle progression and E2F1-responsive gene expression via association with host cell factor-1 (HCF-1). J. Biol. Chem. 2013, 288, 17532–17543. [Google Scholar] [CrossRef] [PubMed]
- Sebastian, S.; Sreenivas, P.; Sambasivan, R.; Cheedipudi, S.; Kandalla, P.; Pavlath, G.K.; Dhawan, J. MLL5, a trithorax homolog, indirectly regulates H3K4 methylation, represses cyclin A2 expression, and promotes myogenic differentiation. Proc. Natl. Acad. Sci. USA 2009, 106, 4719–4724. [Google Scholar] [CrossRef] [PubMed]
- Mitxitorena, I.; Somma, D.; Mitchell, J.P.; Lepistö, M.; Tyrchan, C.; Smith, E.L.; Kiely, P.A.; Walden, H.; Keeshan, K.; Carmody, R.J. The deubiquitinase USP7 uses a distinct ubiquitin-like domain to deubiquitinate NF-ĸB subunits. J. Biol. Chem. 2020, 295, 11754–11763. [Google Scholar] [CrossRef] [PubMed]
- Pereira-Martins, D.A.; Weinhäuser, I.; Coelho-Silva, J.L.; França-Neto, P.L.; Almeida, L.Y.; Bianco, T.M.; Silva, C.L.; França, R.F.; Traina, F.; Rego, E.M.; et al. MLL5 improves ATRA driven differentiation and promotes xenotransplant engraftment in acute promyelocytic leukemia model. Cell Death Dis. 2021, 12, 371. [Google Scholar] [CrossRef]
- Shi, W.H.; Li, X.; Chang, C.K. [Significance of chromosome 7 abnormalities in myeloid malignancies]. Zhongguo Shi Yan Xue Ye Xue Za Zhi 2014, 22, 1739–1743. [Google Scholar]
- Lemak, A.; Yee, A.; Wu, H.; Yap, D.; Zeng, H.; Dombrovski, L.; Houliston, S.; Aparicio, S.; Arrowsmith, C.H. Solution NMR structure and histone binding of the PHD domain of human MLL5. PLoS ONE 2013, 8, e77020. [Google Scholar] [CrossRef]
- Chen, C.; Chen, Z.; Chio, C.L.; Zhao, Y.; Li, Y.; Liu, Z.; Jin, Z.; Wu, X.; Wei, W.; Zhao, Q.; et al. Higher Expression of WT1 With Lower CD58 Expression may be Biomarkers for Risk Stratification of Patients With Cytogenetically Normal Acute Myeloid Leukemia. Technol. Cancer Res. Treat. 2021, 20, 15330338211052152. [Google Scholar] [CrossRef]
- Zhang, C.C.; Li, Y.; Jiang, C.Y.; Le, Q.M.; Liu, X.; Ma, L.; Wang, F.F. O-GlcNAcylation mediates H2O2-induced apoptosis through regulation of STAT3 and FOXO1. Acta Pharmacol. Sin. 2024. [CrossRef] [PubMed]
- Cai, Y.; Jin, J.; Swanson, S.K.; Cole, M.D.; Choi, S.H.; Florens, L.; Washburn, M.P.; Conaway, J.W.; Conaway, R.C. Subunit composition and substrate specificity of a MOF-containing histone acetyltransferase distinct from the male-specific lethal (MSL) complex. J. Biol. Chem. 2010, 285, 4268–4272. [Google Scholar] [CrossRef] [PubMed]
- Hoe, M.; Nicholas, H.R. Evidence of a MOF histone acetyltransferase-containing NSL complex in C. elegans. Worm 2014, 3, e982967. [Google Scholar] [CrossRef]
- Liang, F.; Li, X.; Shen, X.; Yang, R.; Chen, C. Expression profiles and functional prediction of histone acetyltransferases of the MYST family in kidney renal clear cell carcinoma. BMC Cancer 2023, 23, 586. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Wei, T.; Sun, L.; Wu, T.; Li, F.; Zhao, J.; Chu, J.; Wang, F.; Cai, Y.; Jin, J. The Non-Specific Lethal (NSL) Histone Acetyltransferase Complex Transcriptionally Regulates Yin Yang 1-Mediated Cell Proliferation in Human Cells. Int. J. Mol. Sci. 2022, 23, 3801. [Google Scholar] [CrossRef] [PubMed]
- Neganova, M.E.; Klochkov, S.G.; Aleksandrova, Y.R.; Aliev, G. Histone modifications in epigenetic regulation of cancer: Perspectives and achieved progress. Semin. Cancer Biol. 2022, 83, 452–471. [Google Scholar] [CrossRef] [PubMed]
- He, N.; Ma, D.; Tan, Y.; Liu, M. Upregulation of O-GlcNAc transferase is involved in the pathogenesis of acute myeloid leukemia. Asia Pac. J. Clin. Oncol. 2022, 18, e318–e328. [Google Scholar] [CrossRef]
- Balsollier, C.; Pieters, R.J.; Anderluh, M. Overview of the Assays to Probe O-Linked β-N-Acetylglucosamine Transferase Binding and Activity. Molecules 2021, 26, 1037. [Google Scholar] [CrossRef]
- Aljazi, M.B.; Gao, Y.; Wu, Y.; Mias, G.I.; He, J. Histone H3K36me2-Specific Methyltransferase ASH1L Promotes MLL-AF9-Induced Leukemogenesis. Front. Oncol. 2021, 11, 754093. [Google Scholar] [CrossRef]
- Valerio, D.G.; Xu, H.; Chen, C.W.; Hoshii, T.; Eisold, M.E.; Delaney, C.; Cusan, M.; Deshpande, A.J.; Huang, C.H.; Lujambio, A.; et al. Histone Acetyltransferase Activity of MOF Is Required for MLL-AF9 Leukemogenesis. Cancer Res. 2017, 77, 1753–1762. [Google Scholar] [CrossRef]
- Pessoa Rodrigues, C.; Akhtar, A. Differential H4K16ac levels ensure a balance between quiescence and activation in hematopoietic stem cells. Sci. Adv. 2021, 7, eabi5987. [Google Scholar] [CrossRef] [PubMed]
- Boila, L.D.; Sengupta, A. Evolving insights on histone methylome regulation in human acute myeloid leukemia pathogenesis and targeted therapy. Exp. Hematol. 2020, 92, 19–31. [Google Scholar] [CrossRef] [PubMed]
- Matuleviciute, R.; Cunha, P.P.; Johnson, R.S.; Foskolou, I.P. Oxygen regulation of TET enzymes. FEBS J. 2021, 288, 7143–7161. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, X.; Lu, J. Ten-Eleven-Translocation Genes in Cancer. Cancer Treat. Res. 2023, 190, 363–373. [Google Scholar] [CrossRef] [PubMed]
- He, B.; Zhang, C.; Zhang, X.; Fan, Y.; Zeng, H.; Liu, J.; Meng, H.; Bai, D.; Peng, J.; Zhang, Q.; et al. Tissue-specific 5-hydroxymethylcytosine landscape of the human genome. Nat. Commun. 2021, 12, 4249. [Google Scholar] [CrossRef] [PubMed]
- Joshi, K.; Liu, S.; Breslin, S.J.P.; Zhang, J. Mechanisms that regulate the activities of TET proteins. Cell. Mol. Life Sci. 2022, 79, 363. [Google Scholar] [CrossRef] [PubMed]
- Prasad, R.; Yen, T.J.; Bellacosa, A. Active DNA demethylation-The epigenetic gatekeeper of development, immunity, and cancer. Adv. Genet. 2021, 2, e10033. [Google Scholar] [CrossRef]
- Wallace, L.; Obeng, E.A. Noncoding rules of survival: Epigenetic regulation of normal and malignant hematopoiesis. Front. Mol. Biosci. 2023, 10, 1273046. [Google Scholar] [CrossRef]
- López-Oreja, I.; Gohr, A.; Playa-Albinyana, H.; Giró, A.; Arenas, F.; Higashi, M.; Tripathi, R.; López-Guerra, M.; Irimia, M.; Aymerich, M.; et al. SF3B1 mutation-mediated sensitization to H3B-8800 splicing inhibitor in chronic lymphocytic leukemia. Life Sci. Alliance 2023, 6. [Google Scholar] [CrossRef]
- Playa-Albinyana, H.; Arenas, F.; Royo, R.; Giró, A.; López-Oreja, I.; Aymerich, M.; López-Guerra, M.; Frigola, G.; Beà, S.; Delgado, J.; et al. Chronic lymphocytic leukemia patient-derived xenografts recapitulate clonal evolution to Richter transformation. Leukemia 2023. [Google Scholar] [CrossRef]
- Dey, A.S.; Ayon, N.J.; Bhattacharya, C.; Gutheil, W.G.; Mukherji, M. Positive/negative ion-switching-based LC-MS/MS method for quantification of cytosine derivatives produced by the TET-family 5-methylcytosine dioxygenases. Biol. Methods Protoc. 2020, 5, bpaa019. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, Y.; Wang, C.; Wang, X. TET (Ten-eleven translocation) family proteins: Structure, biological functions and applications. Signal Transduct. Target. Ther. 2023, 8, 297. [Google Scholar] [CrossRef] [PubMed]
- Timmins, M.A.; Wagner, S.D.; Ahearne, M.J. The new biology of PTCL-NOS and AITL: Current status and future clinical impact. Br. J. Haematol. 2020, 189, 54–66. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.W.; Xie, J.; Jiang, J. The Emerging Roles of Protein Interactions with O-GlcNAc Cycling Enzymes in Cancer. Cancers 2022, 14, 5135. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xue, M.; Deng, X.; Dong, L.; Nguyen, L.X.T.; Ren, L.; Han, L.; Li, C.; Xue, J.; Zhao, Z.; et al. TET2-mediated mRNA demethylation regulates leukemia stem cell homing and self-renewal. Cell Stem Cell 2023, 30, 1072–1090.e10. [Google Scholar] [CrossRef] [PubMed]
- Nabil, R.; Hassan, N.M.; Abdellateif, M.S.; Gawdat, R.M.; Elshazly, S.S. The prognostic role of C-KIT, TET1 and TET2 gene expression in Acute Myeloid Leukemia. Mol. Biol. Rep. 2023, 50, 641–653. [Google Scholar] [CrossRef] [PubMed]
- Yao, W.Q.; Wu, F.; Zhang, W.; Chuang, S.S.; Thompson, J.S.; Chen, Z.; Zhang, S.W.; Clipson, A.; Wang, M.; Liu, H.; et al. Angioimmunoblastic T-cell lymphoma contains multiple clonal T-cell populations derived from a common TET2 mutant progenitor cell. J. Pathol. 2020, 250, 346–357. [Google Scholar] [CrossRef]
- Senga, S.S.; Grose, R.P. Hallmarks of cancer-the new testament. Open Biol. 2021, 11, 200358. [Google Scholar] [CrossRef]
- Shekhawat, J.; Gauba, K.; Gupta, S.; Choudhury, B.; Purohit, P.; Sharma, P.; Banerjee, M. Ten-eleven translocase: Key regulator of the methylation landscape in cancer. J. Cancer Res. Clin. Oncol. 2021, 147, 1869–1879. [Google Scholar] [CrossRef]
- Yin, X.; Hu, L.; Xu, Y. Structure and Function of TET Enzymes. Adv. Exp. Med. Biol. 2022, 1389, 239–267. [Google Scholar] [CrossRef]
- Huang, B.; Huang, C.; Zhu, L.; Xie, L.; Wang, Y.; Zhu, N. Exploring the Pharmacological Mechanisms of Tripterygium wilfordii Hook F against Cardiovascular Disease Using Network Pharmacology and Molecular Docking. Biomed. Res. Int. 2021, 2021, 5575621. [Google Scholar] [CrossRef]
- He, R.; Xhabija, B.; Gopi, L.K.; Kurup, J.T.; Xu, Z.; Liu, Z.; Kidder, B.L. H3K4 demethylase KDM5B regulates cancer cell identity and epigenetic plasticity. Oncogene 2022, 41, 2958–2972. [Google Scholar] [CrossRef] [PubMed]
- Zimmerman, S.M.; Lin, P.N.; Souroullas, G.P. Non-canonical functions of EZH2 in cancer. Front. Oncol. 2023, 13, 1233953. [Google Scholar] [CrossRef] [PubMed]
- Shin, H.; Leung, A.; Costello, K.R.; Senapati, P.; Kato, H.; Moore, R.E.; Lee, M.; Lin, D.; Tang, X.; Pirrotte, P.; et al. Inhibition of DNMT1 methyltransferase activity via glucose-regulated O-GlcNAcylation alters the epigenome. Elife 2023, 12, e85595. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Zhao, B.; Li, T.; Dong, H.; Cheng, X.; Gong, W.; Wang, J.; Zhang, J.; Xin, G.; Yu, Y.; et al. Inhibition of O-GlcNAc transferase activates type I interferon-dependent antitumor immunity by bridging cGAS-STING pathway. bioRxiv 2023. [Google Scholar] [CrossRef]
- Ishii, Y.; Kolluri, K.K.; Pennycuick, A.; Zhang, X.; Nigro, E.; Alrifai, D.; Borg, E.; Falzon, M.; Shah, K.; Kumar, N.; et al. BAP1 and YY1 regulate expression of death receptors in malignant pleural mesothelioma. J. Biol. Chem. 2021, 297, 101223. [Google Scholar] [CrossRef] [PubMed]
- Inoue, D.; Fujino, T.; Sheridan, P.; Zhang, Y.Z.; Nagase, R.; Horikawa, S.; Li, Z.; Matsui, H.; Kanai, A.; Saika, M.; et al. A novel ASXL1-OGT axis plays roles in H3K4 methylation and tumor suppression in myeloid malignancies. Leukemia 2018, 32, 1327–1337. [Google Scholar] [CrossRef] [PubMed]
- Chiereghin, C.; Travaglino, E.; Zampini, M.; Saba, E.; Saitta, C.; Riva, E.; Bersanelli, M.; Della Porta, M.G. The Genetics of Myelodysplastic Syndromes: Clinical Relevance. Genes 2021, 12, 1144. [Google Scholar] [CrossRef]
- Jansko-Gadermeir, B.; Leisch, M.; Gassner, F.J.; Zaborsky, N.; Dillinger, T.; Hutter, S.; Risch, A.; Melchardt, T.; Egle, A.; Drost, M.; et al. Myeloid NGS Analyses of Paired Samples from Bone Marrow and Peripheral Blood Yield Concordant Results: A Prospective Cohort Analysis of the AGMT Study Group. Cancers 2023, 15, 2305. [Google Scholar] [CrossRef]
- Boyd, R.J.; Murry, J.B.; Morsberger, L.A.; Klausner, M.; Chen, S.; Gocke, C.D.; McCallion, A.S.; Zou, Y.S. Ring Chromosomes in Hematological Malignancies Are Associated with TP53 Gene Mutations and Characteristic Copy Number Variants. Cancers 2023, 15, 5439. [Google Scholar] [CrossRef]
- Sashida, G. Stem cell regulation and dynamics in myeloid malignancies. Int. J. Hematol. 2023, 117, 789–790. [Google Scholar] [CrossRef] [PubMed]
- Asada, S. Molecular mechanisms by which the mutant ASXL1/BAP1 complex aggravates myeloid leukemia. Rinsho Ketsueki 2020, 61, 392–405. [Google Scholar] [CrossRef] [PubMed]
- Bai, J.; Chen, Z.; Chen, C.; Zhang, M.; Zhang, Y.; Song, J.; Yuan, J.; Jiang, X.; Xing, W.; Yang, J.; et al. Reducing hyperactivated BAP1 attenuates mutant ASXL1-driven myeloid malignancies in human haematopoietic cells. Cancer Lett. 2021, 519, 78–90. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.C.; Agosto-Peña, J. Epigenetic regulation by ASXL1 in myeloid malignancies. Int. J. Hematol. 2023, 117, 791–806. [Google Scholar] [CrossRef] [PubMed]
- Asada, S.; Goyama, S.; Inoue, D.; Shikata, S.; Takeda, R.; Fukushima, T.; Yonezawa, T.; Fujino, T.; Hayashi, Y.; Kawabata, K.C.; et al. Mutant ASXL1 cooperates with BAP1 to promote myeloid leukaemogenesis. Nat. Commun. 2018, 9, 2733. [Google Scholar] [CrossRef] [PubMed]
- Vuelta, E.; Ordoñez, J.L.; Alonso-Pérez, V.; Méndez, L.; Hernández-Carabias, P.; Saldaña, R.; Sevilla, J.; Sebastián, E.; Muntión, S.; Sánchez-Guijo, F.; et al. CRISPR-Cas9 Technology as a Tool to Target Gene Drivers in Cancer: Proof of Concept and New Opportunities to Treat Chronic Myeloid Leukemia. CRISPR J. 2021, 4, 519–535. [Google Scholar] [CrossRef]
- Mannino, M.P.; Hart, G.W. The Beginner’s Guide to O-GlcNAc: From Nutrient Sensitive Pathway Regulation to Its Impact on the Immune System. Front. Immunol. 2022, 13, 828648. [Google Scholar] [CrossRef]
- Woo, S.Y.; Lee, S.Y.; Yu, S.L.; Park, S.J.; Kang, D.; Kim, J.S.; Jeong, I.B.; Kwon, S.J.; Hwang, W.J.; Park, C.R.; et al. MicroRNA-7-5p’s role in the O-GlcNAcylation and cancer metabolism. Noncoding RNA Res. 2020, 5, 201–207. [Google Scholar] [CrossRef]
- Jiang, M.; Xu, B.; Li, X.; Shang, Y.; Chu, Y.; Wang, W.; Chen, D.; Wu, N.; Hu, S.; Zhang, S.; et al. O-GlcNAcylation promotes colorectal cancer metastasis via the miR-101-O-GlcNAc/EZH2 regulatory feedback circuit. Oncogene 2019, 38, 301–316. [Google Scholar] [CrossRef]
- Yu, Z.; He, H.; Jiang, B.; Hu, J. O-GlcNAcylation of CSNK2A1 by OGT is Involved in the Progression of Colorectal Cancer. Mol. Biotechnol. 2024. [Google Scholar] [CrossRef]
- Wang, L.; Feng, Y.; Zhang, C.; Chen, X.; Huang, H.; Li, W.; Zhang, J.; Liu, Y. Upregulation of OGT by Caveolin-1 promotes hepatocellular carcinoma cell migration and invasion. Cell Biol. Int. 2021, 45, 2251–2263. [Google Scholar] [CrossRef] [PubMed]
- Le Minh, G.; Reginato, M.J. Role of O-GlcNAcylation on cancer stem cells: Connecting nutrient sensing to cell plasticity. Adv. Cancer Res. 2023, 157, 195–228. [Google Scholar] [CrossRef] [PubMed]
- Ning, D.; Chen, J.; Du, P.; Liu, Q.; Cheng, Q.; Li, X.; Zhang, B.; Chen, X.; Jiang, L. The crosstalk network of XIST/miR-424-5p/OGT mediates RAF1 glycosylation and participates in the progression of liver cancer. Liver Int. 2021, 41, 1933–1944. [Google Scholar] [CrossRef] [PubMed]
- Lo, W.Y.; Wang, S.J.; Wang, H.J. Non-canonical Interaction Between O-Linked N-Acetylglucosamine Transferase and miR-146a-5p Aggravates High Glucose-Induced Endothelial Inflammation. Front. Physiol. 2020, 11, 1091. [Google Scholar] [CrossRef]
- Dykxhoorn, D.M.; Wang, H.; Da Fonseca Ferreira, A.; Wei, J.; Dong, C. MicroRNA-423-5p Mediates Cocaine-Induced Smooth Muscle Cell Contraction by Targeting Cacna2d2. Int. J. Mol. Sci. 2023, 24, 6584. [Google Scholar] [CrossRef] [PubMed]
- You, Z.; Peng, D.; Cao, Y.; Zhu, Y.; Yin, J.; Zhang, G.; Peng, X. P53 suppresses the progression of hepatocellular carcinoma via miR-15a by decreasing OGT expression and EZH2 stabilization. J. Cell. Mol. Med. 2021, 25, 9168–9182. [Google Scholar] [CrossRef] [PubMed]
- Xia, M.; Wang, S.; Qi, Y.; Long, K.; Li, E.; He, L.; Pan, F.; Guo, Z.; Hu, Z. Corrigendum: Inhibition of O-GlcNAc transferase sensitizes prostate cancer cells to docetaxel. Front. Oncol. 2023, 13, 1257404. [Google Scholar] [CrossRef]
- Huang, W.K.; Shi, H.; Akçakaya, P.; Zeljic, K.; Gangaev, A.; Caramuta, S.; Yeh, C.N.; Bränström, R.; Larsson, C.; Lui, W.O. Imatinib Regulates miR-483-3p and Mitochondrial Respiratory Complexes in Gastrointestinal Stromal Tumors. Int. J. Mol. Sci. 2021, 22, 10600. [Google Scholar] [CrossRef]
- Albuquerque, S.O.; Barros, T.G.; Dias, L.R.S.; Lima, C.; Azevedo, P.; Flores-Junior, L.A.P.; Dos Santos, E.G.; Loponte, H.F.; Pinheiro, S.; Dias, W.B.; et al. Biological evaluation and molecular modeling of peptidomimetic compounds as inhibitors for O-GlcNAc transferase (OGT). Eur. J. Pharm. Sci. 2020, 154, 105510. [Google Scholar] [CrossRef]
- Makwana, V.; Ryan, P.; Malde, A.K.; Anoopkumar-Dukie, S.; Rudrawar, S. Bisubstrate Ether-Linked Uridine-Peptide Conjugates as O-GlcNAc Transferase Inhibitors. ChemMedChem 2021, 16, 477–483. [Google Scholar] [CrossRef]
- Ju Kim, E. O-GlcNAc Transferase: Structural Characteristics, Catalytic Mechanism and Small-Molecule Inhibitors. Chembiochem 2020, 21, 3026–3035. [Google Scholar] [CrossRef]
- Alteen, M.G.; Tan, H.Y.; Vocadlo, D.J. Monitoring and modulating O-GlcNAcylation: Assays and inhibitors of O-GlcNAc processing enzymes. Curr. Opin. Struct. Biol. 2021, 68, 157–165. [Google Scholar] [CrossRef]
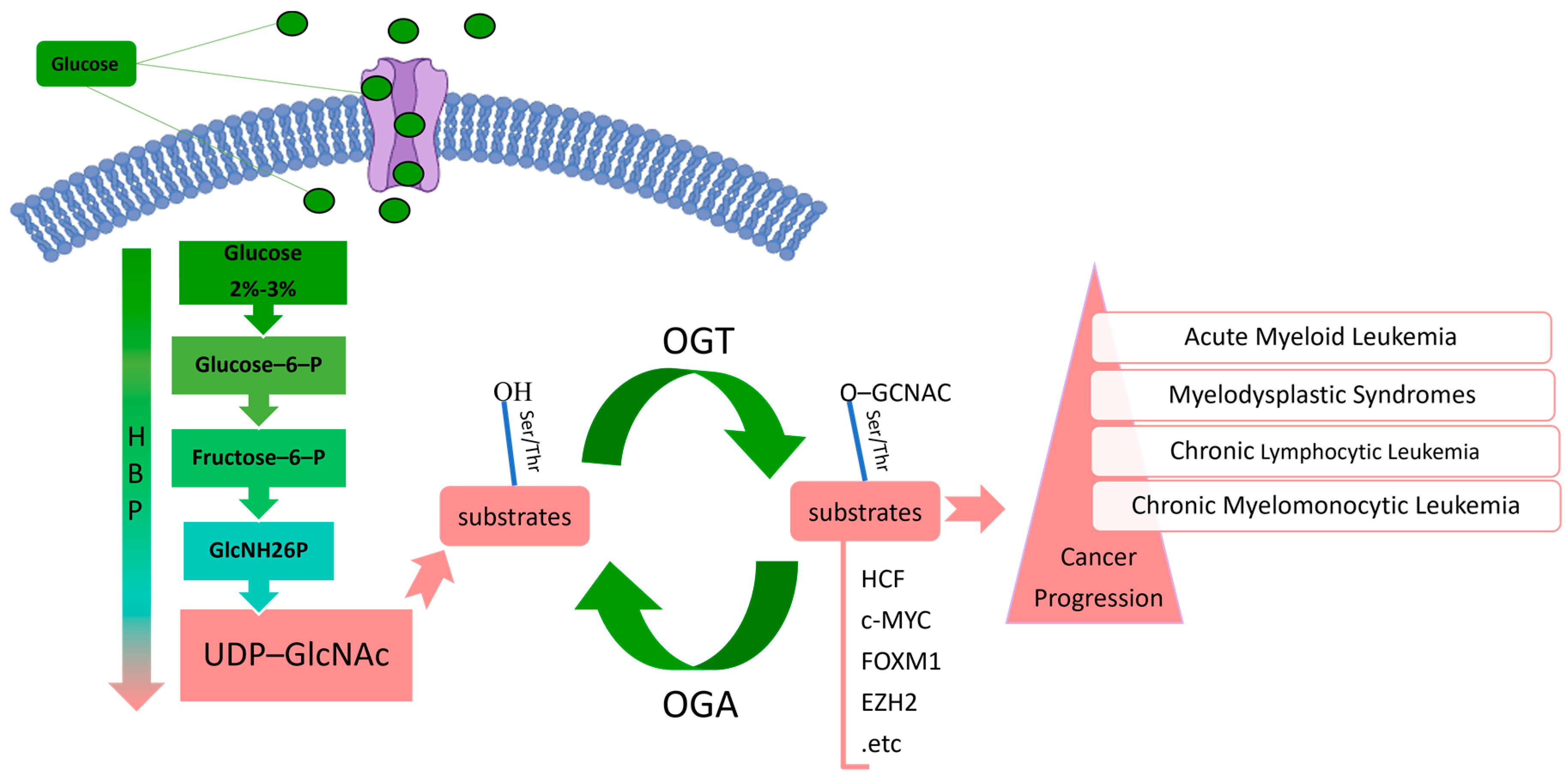
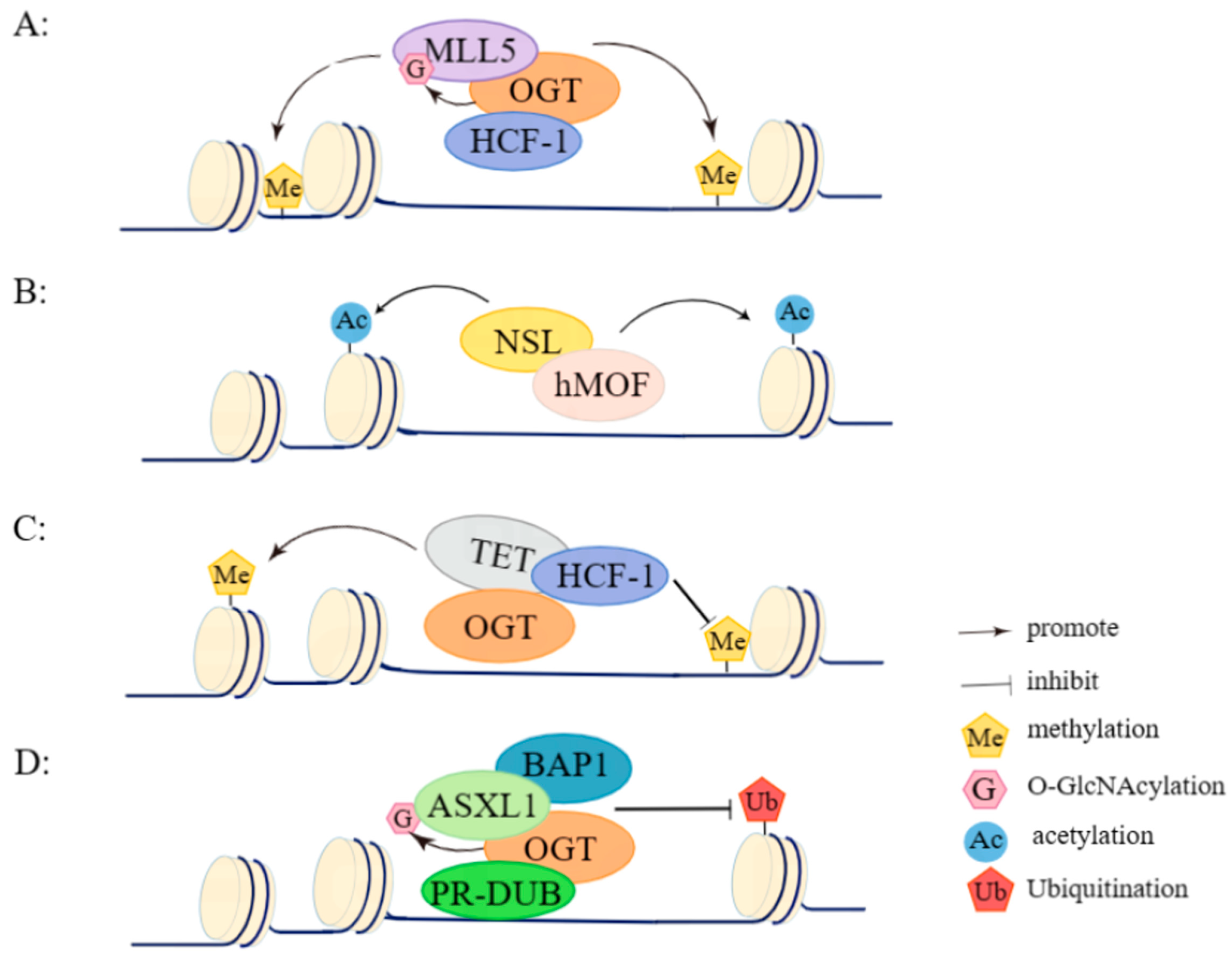
| Binding Protein | Complex | Biological Function(s) | Reference(s) | |
|---|---|---|---|---|
| OGT | HCF-1 | OGT/HCF-1 complex | Regulates the epigenetic modifications and performs related functions by forming temporary complexes in concert with other proteins, such as MYC and MLL5. | [52,53,54,55] |
| MOF, NSL1, NSL2, NSL3, MCRS2, WDR5, OGT, HCF-1 | NSL complex | Catalyzes histone H4K16/K5/K8 acetylation. | [56] | |
| TET. | OGT/TET complex | Recruits OGT to chromatin and subsequently causes the O-GlcNAcylation of histones to regulate gene expression, and OGT-TET2/3 O-GlcNAcylates HCF-1 and regulates SET1/COMPASS-mediated H3K4me3. | [57,58] | |
| HCF-1, BAP1, ASXL2, ASXL3, KDM1B, MBD5/6, MLL3, FOXK1/2 | PR-DUB complex | Catalyzes H2AK119 deubiquitination. | [59,60,61] | |
| OGA | O-GlcNAczyme complex | Regulates signal transduction, glucose metabolism, cell proliferation, and apoptosis, as well as other basic cellular processes. | [62,63,64,65] | |
| HUWE1 | OGT/HUWE1 complex | |||
| Rev-erbα | OGT/Rev-erbα complex |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhuang, S.; Liu, Z.; Wu, J.; Yao, Y.; Li, Z.; Shen, Y.; Yu, B.; Wu, D. Can O-GIcNAc Transferase (OGT) Complex Be Used as a Target for the Treatment of Hematological Malignancies? Pharmaceuticals 2024, 17, 664. https://doi.org/10.3390/ph17060664
Zhuang S, Liu Z, Wu J, Yao Y, Li Z, Shen Y, Yu B, Wu D. Can O-GIcNAc Transferase (OGT) Complex Be Used as a Target for the Treatment of Hematological Malignancies? Pharmaceuticals. 2024; 17(6):664. https://doi.org/10.3390/ph17060664
Chicago/Turabian StyleZhuang, Shiwei, Zhimei Liu, Jinyao Wu, Yudan Yao, Zongyang Li, Yanxiang Shen, Bin Yu, and Donglu Wu. 2024. "Can O-GIcNAc Transferase (OGT) Complex Be Used as a Target for the Treatment of Hematological Malignancies?" Pharmaceuticals 17, no. 6: 664. https://doi.org/10.3390/ph17060664
APA StyleZhuang, S., Liu, Z., Wu, J., Yao, Y., Li, Z., Shen, Y., Yu, B., & Wu, D. (2024). Can O-GIcNAc Transferase (OGT) Complex Be Used as a Target for the Treatment of Hematological Malignancies? Pharmaceuticals, 17(6), 664. https://doi.org/10.3390/ph17060664






