Saturated Iso-Type Fatty Acids from the Marine Bacterium Mesoflavibacter zeaxanthinifaciens with Anti-Trypanosomal Potential
Abstract
1. Introduction
2. Results
2.1. Collection of Marine Invertebrates
2.2. Isolation and Identification of Marine Bacteria
2.3. Extraction of Microbial Metabolites and Evaluation of the 50% Inhibitory Concentration (IC50) for T. cruzi Trypomastigotes
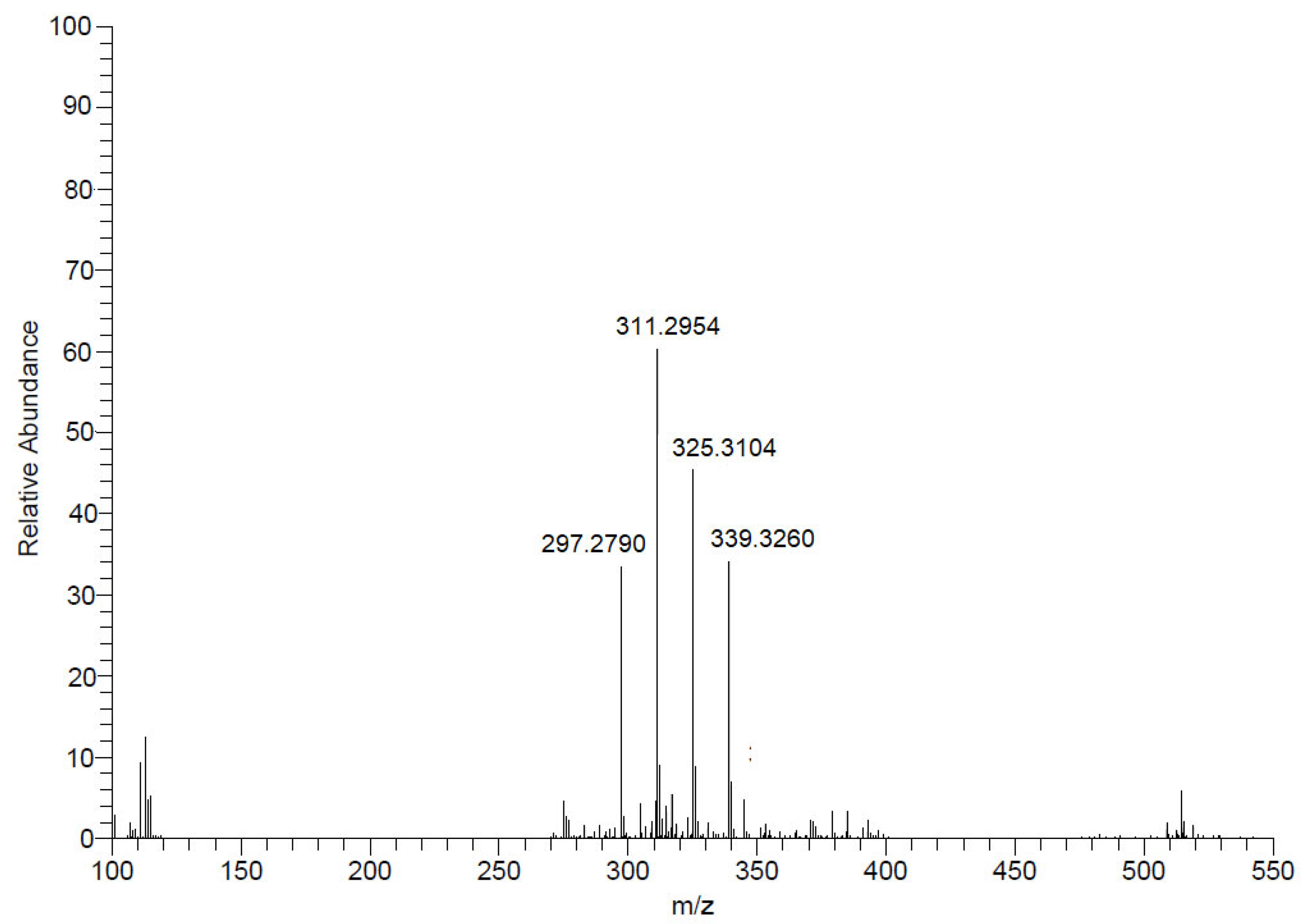
2.4. Fractionation of Mesoflavibacter zeaxanthinifaciens Extract
2.5. Antitrypanosomal Activity of Fraction II of Mesoflavibacter zeaxanthinifaciens EtOAc Extract (FII)
2.6. Hemolytic Activity of Fraction II of Mesoflavibacter zeaxanthinifaciens EtOAc Extract
2.7. Protein Profile of T. cruzi after Treatment with Fraction II of Mesoflavibacter zeaxanthinifaciens EtOAc Extract (FII)
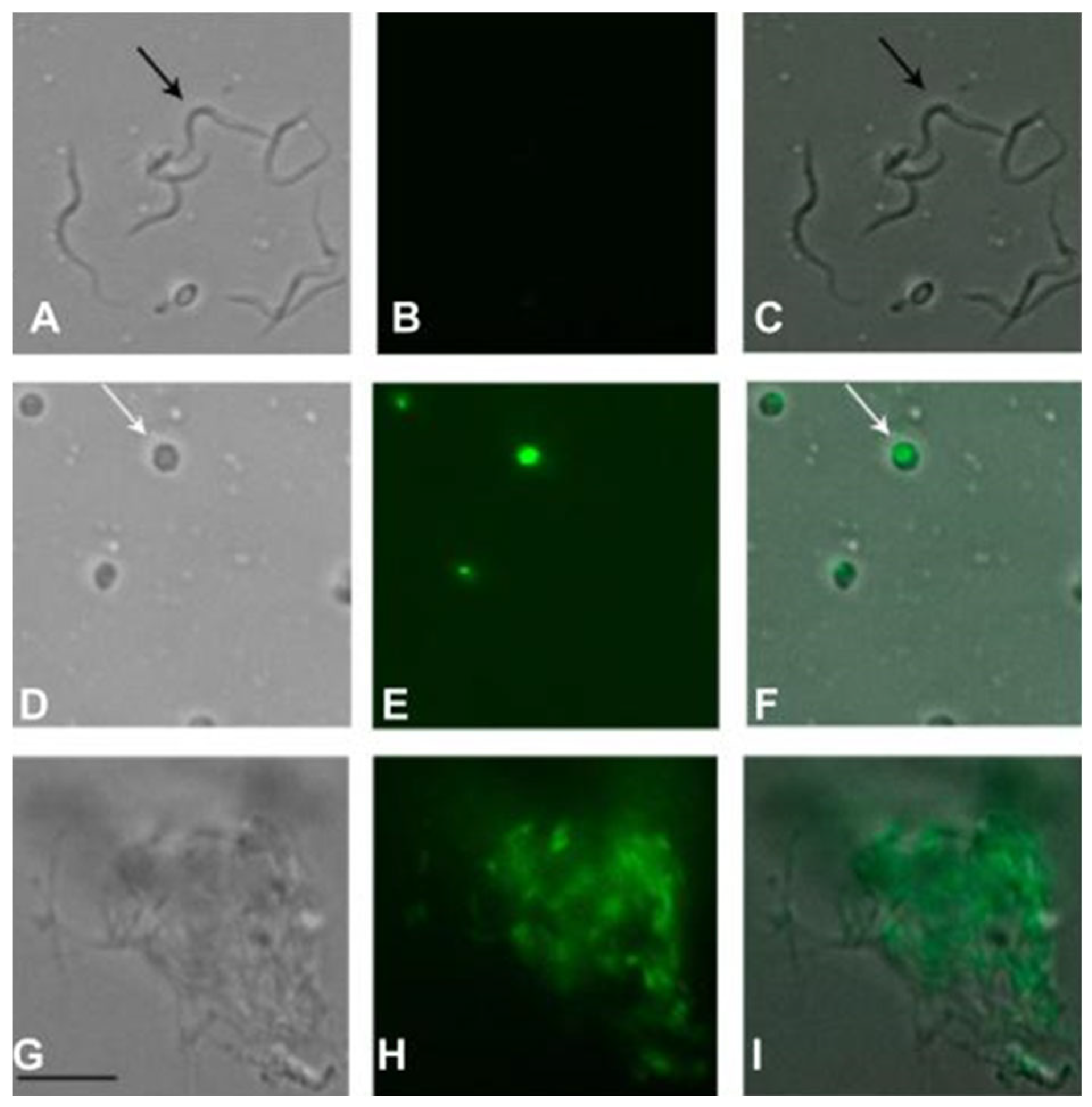
2.8. Plasma Membrane Permeability of Trypomastigotes Treated with Fraction II of Mesoflavibacter zeaxanthinifaciens EtOAc Extract
3. Discussion
4. Materials and Methods
4.1. Mice
4.2. Parasites and Mammalian Cell Maintenance
4.3. Analytical Methods
4.4. Collection of Marine Corals and Sediments
4.5. Isolation and Cultivation of Microorganisms
4.6. Bacteria Identification with Mass Spectrometry (MALDI-ToF/MS)
4.7. Bacterial Identification by Sequencing of the Partial 16S rRNA Gene
4.8. Extraction of Metabolites from Marine Bacteria Grown on Agar and in Broth Medium
4.8.1. Agar
4.8.2. Broth
4.9. Bio-Guided Fractionation of Mesoflavibacter zeaxanthinifaciens Organic Extract
4.10. Evaluation of the Anti-Trypanosoma cruzi Activity
4.11. Cytotoxicity against Mammalian Cells
4.12. Hemolytic Activity
4.13. Evaluation of T. cruzi Trypomastigotes Protein Profile
4.14. Evaluation of Plasma Membrane Permeability
4.15. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Santos, J.D.; Vitorino, I.; Reyes, F.; Vicente, F.; Lage, O.M. From Ocean to Medicine: Pharmaceutical Applications of Metabolites from Marine Bacteria. Antibiotics 2020, 9, 455. [Google Scholar] [CrossRef]
- Atanasov, A.G.; Zotchev, S.B.; Dirsch, V.M.; Orhan, I.E.; Banach, M.; Rollinger, J.M.; Barreca, D.; Weckwerth, W.; Bauer, R.; Bayer, E.A.; et al. Natural Products in Drug Discovery: Advances and Opportunities. Nat. Rev. Drug Discov. 2021, 20, 200–216. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.; Buddie, A.G.; Goss, R.J.M.; Overmann, J.; Lepleux, C.; Brönstrup, M.; Kloareg, B.; Meiners, T.; Brennecke, P.; Ianora, A.; et al. Discovery Pipelines for Marine Resources: An Ocean of Opportunity for Biotechnology? World J. Microbiol. Biotechnol. 2019, 35, 107. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; She, J.; Fu, J.; Wang, J.; Ye, Y.; Yang, B.; Liu, Y.; Zhou, X.; Tao, H. Advances in Natural Products from the Marine-Sponge-Associated Microorganisms with Antimicrobial Activity in the Last Decade. Mar. Drugs 2023, 21, 236. [Google Scholar] [CrossRef] [PubMed]
- Overmann, J.; Lepleux, C. Marine Bacteria and Archaea: Diversity, Adaptations, and Culturability. In The Marine Microbiome: An Untapped Source of Biodiversity and Biotechnological Potential; Springer: Cham, Switzerland, 2016; pp. 21–55. [Google Scholar] [CrossRef]
- Ameen, F.; AlNadhari, S.; Al-Homaidan, A.A. Marine Microorganisms as an Untapped Source of Bioactive Compounds. Saudi J. Biol. Sci. 2021, 28, 224–231. [Google Scholar] [CrossRef] [PubMed]
- Newman, D.J.; Cragg, G.M. Natural Products as Sources of New Drugs over the Nearly Four Decades from 01/1981 to 09/2019. J. Nat. Prod. 2020, 83, 770–803. [Google Scholar] [CrossRef] [PubMed]
- Chan-Bacab, M.J.; Reyes-Estebanez, M.M.; Camacho-Chab, J.C.; Ortega-Morales, B.O. Microorganisms as a Potential Source of Molecules to Control Trypanosomatid Diseases. Molecules 2021, 26, 1388. [Google Scholar] [CrossRef] [PubMed]
- Chagas Disease (Also Known as American Trypanosomiasis). Available online: https://www.who.int/en/news-room/fact-sheets/detail/chagas-disease-(american-trypanosomiasis) (accessed on 5 April 2023).
- Lidani, K.C.F.; Andrade, F.A.; Bavia, L.; Damasceno, F.S.; Beltrame, M.H.; Messias-Reason, I.J.; Sandri, T.L. Chagas Disease: From Discovery to a Worldwide Health Problem. Front. Public Health 2019, 7, 00166. [Google Scholar] [CrossRef] [PubMed]
- Guhl, F.; Ramírez, J.D. Poverty, Migration, and Chagas Disease. Curr. Trop. Med. Rep. 2021, 8, 52–58. [Google Scholar] [CrossRef]
- Guarner, J. Chagas Disease as Example of a Reemerging Parasite. Semin. Diagn. Pathol. 2019, 36, 164–169. [Google Scholar] [CrossRef]
- Morillo, C.A.; Marin-Neto, J.A.; Avezum, A.; Sosa-Estani, S.; Rassi, A.; Rosas, F.; Villena, E.; Quiroz, R.; Bonilla, R.; Britto, C.; et al. Randomized Trial of Benznidazole for Chronic Chagas’ Cardiomyopathy. N. Engl. J. Med. 2015, 373, 1295–1306. [Google Scholar] [CrossRef] [PubMed]
- MarinLit—A Database of the Marine Natural Products Literature. Available online: https://marinlit.rsc.org/ (accessed on 6 March 2024).
- Gerwick, W.H.; Fenner, A.M. Drug Discovery from Marine Microbes. Microb. Ecol. 2013, 65, 800–806. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Luis, S.; Gómez, J.F.; Spadafora, C.; Guzmán, H.M.; Gutiérrez, M. Antitrypanosomal Alkaloids from the Marine Bacterium Bacillus Pumilus. Molecules 2012, 17, 11146–11155. [Google Scholar] [CrossRef] [PubMed]
- Osei, E.; Kwain, S.; Mawuli, G.T.; Anang, A.K.; Owusu, K.B.A.; Camas, M.; Camas, A.S.; Ohashi, M.; Alexandru-Crivac, C.N.; Deng, H.; et al. Paenidigyamycin A, Potent Antiparasitic Imidazole Alkaloid from the Ghanaian Paenibacillus Sp. DE2SH. Mar. Drugs 2019, 17, 9. [Google Scholar] [CrossRef] [PubMed]
- Asker, D.; Beppu, T.; Ueda, K. Mesoflavibacter Zeaxanthinifaciens Gen. Nov., Sp. Nov., a Novel Zeaxanthin-Producing Marine Bacterium of the Family Flavobacteriaceae. Syst. Appl. Microbiol. 2007, 30, 291–296. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.-H.; He, X.; Austin, B. Vibrio Harveyi: A Serious Pathogen of Fish and Invertebrates in Mariculture. Mar. Life Sci. Technol. 2020, 2, 231–245. [Google Scholar] [CrossRef]
- Sampaio, A.; Silva, V.; Poeta, P.; Aonofriesei, F. Vibrio spp.: Life Strategies, Ecology, and Risks in a Changing Environment. Diversity 2022, 14, 97. [Google Scholar] [CrossRef]
- Mansson, M.; Gram, L.; Larsen, T.O. Production of Bioactive Secondary Metabolites by Marine Vibrionaceae. Mar. Drugs 2011, 9, 1440–1468. [Google Scholar] [CrossRef]
- Sottorff, I.; Wiese, J.; Lipfert, M.; Preußke, N.; Sönnichsen, F.D.; Imhoff, J.F. Different Secondary Metabolite Profiles of Phylogenetically Almost Identical Streptomyces Griseus Strains Originating from Geographically Remote Locations. Microorganisms 2019, 7, 166. [Google Scholar] [CrossRef]
- Kaspar, F.; Neubauer, P.; Gimpel, M. Bioactive Secondary Metabolites from Bacillus Subtilis: A Comprehensive Review. J. Nat. Prod. 2019, 82, 2038–2053. [Google Scholar] [CrossRef]
- Zhao, Y.; Mao, X.; Zhang, M.; Yang, W.; Di, H.J.; Ma, L.; Liu, W.; Li, B. The Application of Bacillus Megaterium Alters Soil Microbial Community Composition, Bioavailability of Soil Phosphorus and Potassium, and Cucumber Growth in the Plastic Shed System of North China. Ecosyst. Environ. 2021, 307, 107236. [Google Scholar] [CrossRef]
- Xie, Y.; Peng, Q.; Ji, Y.; Xie, A.; Yang, L.; Mu, S.; Li, Z.; He, T.; Xiao, Y.; Zhao, J.; et al. Isolation and Identification of Antibacterial Bioactive Compounds From Bacillus Megaterium L2. Front. Microbiol. 2021, 12, 645484. [Google Scholar] [CrossRef]
- Cheffi, M.; Maalej, A.; Mahmoudi, A.; Hentati, D.; Marques, A.M.; Sayadi, S.; Chamkha, M. Lipopeptides Production by a Newly Halomonas Venusta Strain: Characterization and Biotechnological Properties. Bioorg. Chem. 2021, 109, 104724. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Quan, C.; Wang, X.; Zhao, P.; Fan, S. Extraction, Purification and Identification of Bacterial Signal Molecules Based on N-Acyl Homoserine Lactones. Microb. Biotechnol. 2011, 4, 479–490. [Google Scholar] [CrossRef] [PubMed]
- Biswas, J.; Jana, S.K.; Mandal, S. Biotechnological Impacts of Halomonas: A Promising Cell Factory for Industrially Relevant Biomolecules. Front. Microbiol. 2022, 14, 2131961. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, Z.; Sun, J.; Xue, C.; Mao, X. Biotechnological Production of Zeaxanthin by Microorganisms. Trends Food Sci. Technol. 2018, 71, 225–234. [Google Scholar] [CrossRef]
- Vila, E.; Hornero-Méndez, D.; Lareo, C.; Saravia, V. Biotechnological Production of Zeaxanthin by an Antarctic Flavobacterium: Evaluation of Culture Conditions. J. Biotechnol. 2020, 319, 54–60. [Google Scholar] [CrossRef]
- Lee, J.H.; Hwang, Y.M.; Baik, K.S.; Choi, K.S.; Ka, J.-O.; Seong, C.N. Mesoflavibacter Aestuarii Sp. Nov., a Zeaxanthin-Producing Marine Bacterium Isolated from Seawater. Int. J. Syst. Evol. Microbiol. 2014, 64 Pt 6, 1932–1937. [Google Scholar] [CrossRef] [PubMed]
- Luiz Franco, O.; Guo, B.; Oves, M.; Ahmar Rauf, M.; Hussain, A.; Qari, H.A.; Aslam Parwaz Khan, A.; Muhammad, P.; Tabish Rehman, M.; Fahad Alajmi, M.; et al. Antibacterial Silver Nanomaterial Synthesis From Mesoflavibacter Zeaxanthinifaciens and Targeting Biofilm Formation. Front. Pharmacol. 2019, 10, 00801. [Google Scholar] [CrossRef]
- English, A.L.; Boufridi, A.; Quinn, R.J.; Kurtböke, D.I. Evaluation of Fermentation Conditions Triggering Increased Antibacterial Activity from a Near-Shore Marine Intertidal Environment-Associated Streptomyces Species. Synth. Syst. Biotechnol. 2017, 2, 28–38. [Google Scholar] [CrossRef]
- Guo, W.; Zhang, Z.; Zhu, T.; Gu, Q.; Li, D. Penicyclones A-E, Antibacterial Polyketides from the Deep-Sea-Derived Fungus Penicillium sp. F23-2. J. Nat. Prod. 2015, 78, 2699–2703. [Google Scholar] [CrossRef] [PubMed]
- Vidar, W.S.; Baumeister, T.U.H.; Caesar, L.K.; Kellogg, J.J.; Todd, D.A.; Linington, R.G.; Kvalheim, O.M.; Cech, N.B. Interaction Metabolomics to Discover Synergists in Natural Product Mixtures. J. Nat. Prod. 2023, 86, 671. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Molina, J.A.; Molina, I. Chagas Disease. Lancet 2018, 391, 82–94. [Google Scholar] [CrossRef] [PubMed]
- Nichols, C.M.; Bowman, J.P.; Guezennec, J. Olleya Marilimosa Gen. Nov., Sp. Nov., an Exopolysaccharide-Producing Marine Bacterium from the Family Flavobacteriaceae, Isolated from the Southern Ocean. Int. J. Syst. Evol. Microbiol. 2005, 55, 1557–1561. [Google Scholar] [CrossRef] [PubMed]
- Londero, V.S.; da Costa-Silva, T.A.; Gomes, K.S.; Ferreira, D.D.; Mesquita, J.T.; Tempone, A.G.; Young, M.C.M.; Jerz, G.; Lago, J.H.G. Acetylenic Fatty Acids from Porcelia Macrocarpa (Annonaceae) against Trypomastigotes of Trypanosoma cruzi: Effect of Octadec-9-Ynoic Acid in Plasma Membrane Electric Potential. Bioorg. Chem. 2018, 78, 307–311. [Google Scholar] [CrossRef] [PubMed]
- Romanelli, M.M.; Amaral, M.; Thevenard, F.; Santa Cruz, L.M.; Regasini, L.O.; Migotto, A.E.; Lago, J.H.G.; Tempone, A.G. Mitochondrial Imbalance of Trypanosoma Cruzi Induced by the Marine Alkaloid 6-Bromo-2′-de- N-Methylaplysinopsin. ACS Omega 2022, 7, 28561–28570. [Google Scholar] [CrossRef]
- Galhardo, T.S.; Ueno, A.K.; Costa-Silva, T.A.; Tempone, A.G.; Carvalho, W.A.; Fischmeister, C.; Bruneau, C.; Mandelli, D.; Lago, J.H.G. New Derivatives from Dehydrodieugenol B and Its Methyl Ether Displayed High Anti-Trypanosoma cruzi Activity and Cause Depolarization of the Plasma Membrane and Collapse the Mitochondrial Membrane Potential. Chem. Biol. Interact. 2022, 366, 110129. [Google Scholar] [CrossRef] [PubMed]
- Efimova, S.S.; Malykhina, A.I.; Ostroumova, O.S. Triggering the Amphotericin B Pore-Forming Activity by Phytochemicals. Membranes 2023, 13, 670. [Google Scholar] [CrossRef]
- Emami, K.; Askari, V.; Ullrich, M.; Mohinudeen, K.; Anil, A.C. Characterization of Bacteria in Ballast Water Using MALDI-TOF Mass Spectrometry. PLoS ONE 2012, 7, 38515. [Google Scholar] [CrossRef]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S Ribosomal DNA Amplification for Phylogenetic Study. J. Bacteriol. 1991, 173, 697. [Google Scholar] [CrossRef]
- Lima, M.L.; Romanelli, M.M.; Borborema, S.E.T.; Johns, D.M.; Migotto, A.E.; Lago, J.H.G. Tempone Antitrypanosomal Activity of Isololiolide Isolated from the Marine Hydroid Macrorhynchia Philippina (Cnidaria, Hydrozoa). Bioorg. Chem. 2019, 89, 103002. [Google Scholar] [CrossRef] [PubMed]
- de Castro Levatti, E.V.; Costa-Silva, T.A.; Morais, T.R.; Fernandes, J.P.S.; Lago, J.H.G.; Tempone, A.G. Lethal Action of Licarin A Derivatives in Leishmania (L.) Infantum: Imbalance of Calcium and Bioenergetic Metabolism. Biochimie 2023, 208, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Londero, V.S.; Rosa, M.E.; Baitello, J.B.; Costa-Silva, T.A.; Monteiro Cruz, L.S.; Tempone, A.G.; Caseli, L.; Henrique Lago, J.G. Barbellatanic Acid, a New Antitrypanosomal Pseudo-Disesquiterpenoid Isolated from Nectandra Barbellata, Displayed Interaction with Protozoan Cell Membrane. BBA-Biomembr. 2023, 1865, 184184. [Google Scholar] [CrossRef] [PubMed]





| Acronym | Origin | Depth (Meters) |
|---|---|---|
| TC | Tubastraea coccinea | 5 |
| MH | Mussismilia hispida | 10 |
| MD | Madracis decactis | 10 |
| SCSB | Sediment of the São Sabastião Channel | 35 |
| SIBUZ | Sediment of Buzios Island | 13 |
| Acronym | Identification | Method |
|---|---|---|
| TC 2.0.2 | Alteromonas macleodi | S |
| TC 2.2 | Vibrio alginolyticus | MS |
| MH 3.0 | Vibrio harveyi | MS |
| MH 3.3 | Vibrio alginolyticus | MS |
| MD 5.0 | Vibrio harveyi | MS |
| SCSB 6.0.2.1 | Shewanella pneumatophori | S |
| SCSB 6.0.2.2 | Mesoflavibacter zeaxanthinifaciens | S |
| SCSB 6.1 | Bacillus megaterium | MS |
| SCSB 6.2 | Vibrio harveyi | MS |
| SIBUZ 7 | Vibrio harveyi | MS |
| SIBUZ 7.2.2 | Halomonas aquamarine | S |
| Acronym | Microorganism | Identity (%) | Query Cover (%) | Acc Number |
|---|---|---|---|---|
| TC 2.0.2 | Alteromonas macleodii | 99.78 | 99 | OP163900 |
| SCSB 6.0.2.1 | Shewanella pneumatophori | 99.72 | 99 | OP163959 |
| SCSB 6.0.2.2 | Mesoflavibacter zeaxanthinifaciens | 100 | 96 | OR479885 |
| SIBUZ 7.2.2 | Halomonas aquamarina | 99.72 | 100 | OP163958 |
| Acronym | Strain | Mass of Metabolites (mg) | IC50 ± SD (μg/mL) |
|---|---|---|---|
| TC 2.0.2 | Alteromonas macleodi | 2.6 | 31.2 ± 2.6 |
| TC 2.2 | Vibrio alginolyticus | 2.1 | 13.9 ± 3.0 |
| MH 3.0 | Vibrio harveyi | 11.4 | 51.3 ± 1.1 |
| MH 3.3 | Vibrio alginolyticus | 8.4 | 25.4 ± 1.2 |
| MD 5.0 | Vibrio harveyi | 3.4 | 18.3 ± 2.6 |
| SCSB 6.0.2.1 | Shewanella pneumatophori | 5.5 | 15.1 ± 3.3 |
| SCSB 6.0.2.2 | Mesoflavibacter zeaxanthinifaciens | 4.8 | 17.9 ± 0.7 |
| SCSB 6.1 | Bacillus megaterium | 2.7 | 59.9 ± 0.1 |
| SCSB 6.2 | Vibrio harveyi | 3.9 | 32.8 ± 3.9 |
| SIBUZ 7 | Vibrio harveyi | 8.9 | 8.0 ± 0.7 |
| SIBUZ 7.2.2 | Halomonas aquamarina | 3.2 | 15.4 ± 0.5 |
| Compound | Trypomastigotes (IC50 ± SD) | Amastigotes (IC50 ± SD) | Cytotoxicity (CC50 ± SD) |
|---|---|---|---|
| Fraction II | 17.7 ± 3.5 μg/mL | 23.8 ± 2.7 μg/mL | >200 μg/mL |
| Benznidazole | 3.6 ± 0.9 μg/mL | 1.4 ± 0.5 μg/mL | 49.4 μg/mL |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santos Ferreira, D.A.; de Castro Levatti, E.V.; Santa Cruz, L.M.; Costa, A.R.; Migotto, Á.E.; Yamada, A.Y.; Camargo, C.H.; Christodoulides, M.; Lago, J.H.G.; Tempone, A.G. Saturated Iso-Type Fatty Acids from the Marine Bacterium Mesoflavibacter zeaxanthinifaciens with Anti-Trypanosomal Potential. Pharmaceuticals 2024, 17, 499. https://doi.org/10.3390/ph17040499
Santos Ferreira DA, de Castro Levatti EV, Santa Cruz LM, Costa AR, Migotto ÁE, Yamada AY, Camargo CH, Christodoulides M, Lago JHG, Tempone AG. Saturated Iso-Type Fatty Acids from the Marine Bacterium Mesoflavibacter zeaxanthinifaciens with Anti-Trypanosomal Potential. Pharmaceuticals. 2024; 17(4):499. https://doi.org/10.3390/ph17040499
Chicago/Turabian StyleSantos Ferreira, Dayana Agnes, Erica Valadares de Castro Levatti, Lucas Monteiro Santa Cruz, Alan Roberto Costa, Álvaro E. Migotto, Amanda Yaeko Yamada, Carlos Henrique Camargo, Myron Christodoulides, João Henrique G. Lago, and Andre Gustavo Tempone. 2024. "Saturated Iso-Type Fatty Acids from the Marine Bacterium Mesoflavibacter zeaxanthinifaciens with Anti-Trypanosomal Potential" Pharmaceuticals 17, no. 4: 499. https://doi.org/10.3390/ph17040499
APA StyleSantos Ferreira, D. A., de Castro Levatti, E. V., Santa Cruz, L. M., Costa, A. R., Migotto, Á. E., Yamada, A. Y., Camargo, C. H., Christodoulides, M., Lago, J. H. G., & Tempone, A. G. (2024). Saturated Iso-Type Fatty Acids from the Marine Bacterium Mesoflavibacter zeaxanthinifaciens with Anti-Trypanosomal Potential. Pharmaceuticals, 17(4), 499. https://doi.org/10.3390/ph17040499







