Tumor Suppressor MicroRNAs in Clinical and Preclinical Trials for Neurological Disorders
Abstract
1. Introduction
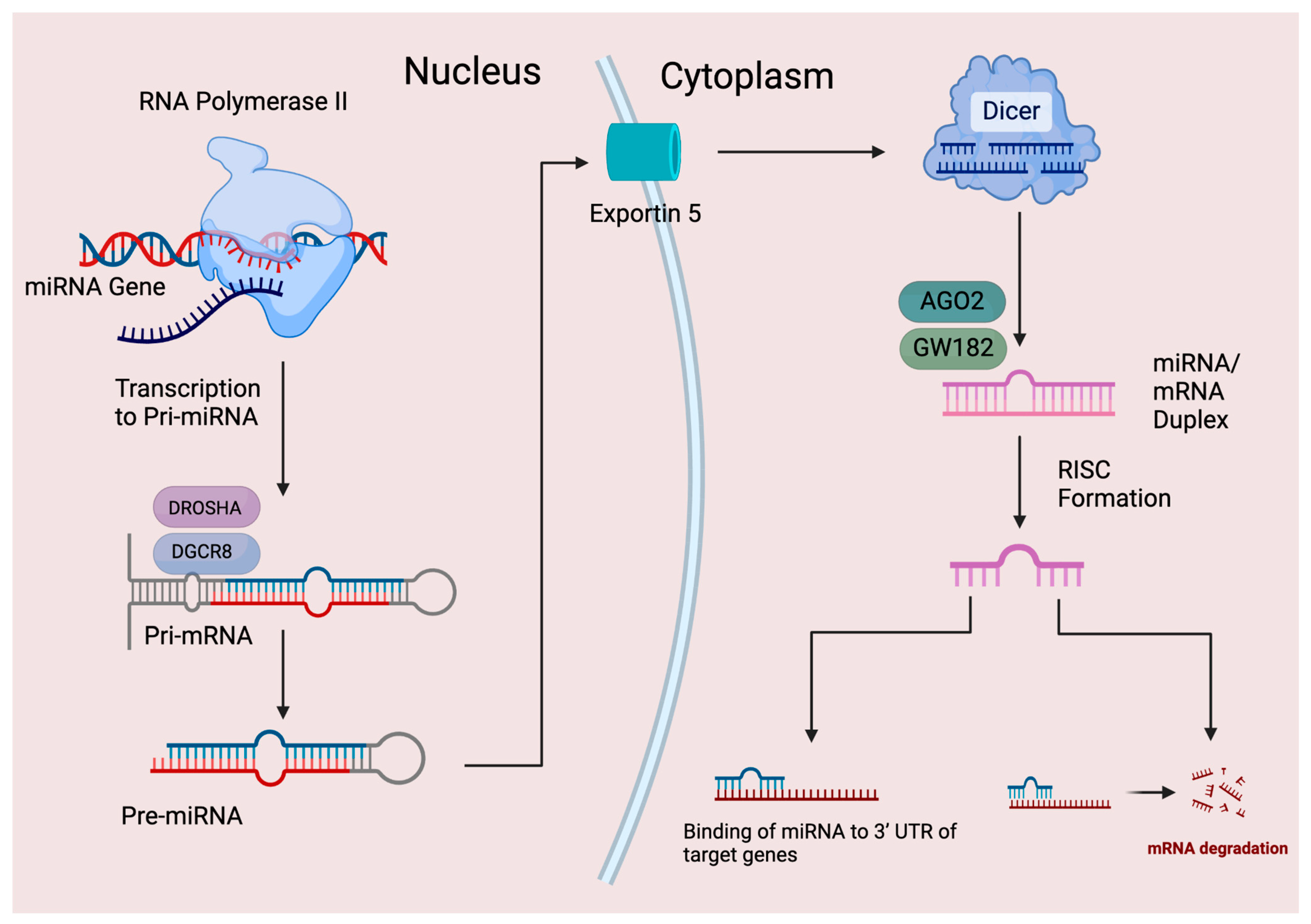
2. MicroRNA History and Biogenesis
3. MicroRNA Biomarkers in Diseases
3.1. MicroRNA Dysregulation and Biomarkers in Cancers
3.2. MicroRNA Dysregulation and Biomarkers in Acute Neurological Disorders:
3.3. MicroRNA Dysregulation and Biomarkers in Neurodegenerative Disorders
4. MiRNAs Consistently Upregulated or Downregulated in Both Cancers and Neurological Disorders
5. Clinical Trials of miR drugs for Neurological Disorders
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Liu, D.Z.; Ander, B.P. Cell cycle inhibition without disruption of neurogenesis is a strategy for treatment of aberrant cell cycle diseases: An update. Sci. World J. 2012, 2012, 491737. [Google Scholar] [CrossRef] [PubMed]
- Lui, A.; Vanleuven, J.; Perekopskiy, D.; Liu, D.; Xu, D.; Alzayat, O.; Elgokhy, T.; Do, T.; Gann, M.; Martin, R.; et al. FDA-Approved Kinase Inhibitors in Preclinical and Clinical Trials for Neurological Disorders. Pharmaceuticals 2022, 15, 1546. [Google Scholar] [CrossRef] [PubMed]
- Lui, A.; Alzayat, O.; Do, T.; Perekopskiy, D.; Gann, M.; Elgokhy, T.S.; Gao, J.; Liu, D. Multi-targeted anti-inflammatory drugs for the treatment of neurological disorders. Neural Regen. Res. 2023, 18, 805–806. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, S.; Selvan, S.T.; Archunan, G.; Gulyas, B.; Padmanabhan, P. MicroRNAs -the next generation therapeutic targets in human diseases. Theranostics 2013, 3, 930–942. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, M.F. Drug target miRNAs: Chances and challenges. Trends Biotechnol. 2014, 32, 578–585. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, C.; Sharma, A.R.; Sharma, G.; Doss, C.G.P.; Lee, S.S. Therapeutic miRNA and siRNA: Moving from Bench to Clinic as Next Generation Medicine. Mol. Ther. Nucleic Acids 2017, 8, 132–143. [Google Scholar] [CrossRef] [PubMed]
- Martin, C.G.; Kim, H.; Yun, S.; Livingston, W.; Fetta, J.; Mysliwiec, V.; Baxter, T.; Gill, J.M. Circulating miRNA associated with posttraumatic stress disorder in a cohort of military combat veterans. Psychiatry Res. 2017, 251, 261–265. [Google Scholar] [CrossRef] [PubMed]
- Redell, J.B.; Moore, A.N.; Ward, N.H., 3rd; Hergenroeder, G.W.; Dash, P.K. Human traumatic brain injury alters plasma microRNA levels. J. Neurotrauma 2010, 27, 2147–2156. [Google Scholar] [CrossRef]
- Hannon, G.J. RNA interference. Nature 2002, 418, 244–251. [Google Scholar] [CrossRef]
- Reynolds, A.; Leake, D.; Boese, Q.; Scaringe, S.; Marshall, W.S.; Khvorova, A. Rational siRNA design for RNA interference. Nat. Biotechnol. 2004, 22, 326–330. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef]
- Jakymiw, A.; Pauley, K.M.; Li, S.; Ikeda, K.; Lian, S.; Eystathioy, T.; Satoh, M.; Fritzler, M.J.; Chan, E.K. The role of GW/P-bodies in RNA processing and silencing. J. Cell Sci. 2007, 120, 1317–1323. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Fan, J.; Belasco, J.G. MicroRNAs direct rapid deadenylation of mRNA. Proc. Natl. Acad. Sci. USA 2006, 103, 4034–4039. [Google Scholar] [CrossRef] [PubMed]
- Lim, L.P.; Lau, N.C.; Garrett-Engele, P.; Grimson, A.; Schelter, J.M.; Castle, J.; Bartel, D.P.; Linsley, P.S.; Johnson, J.M. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 2005, 433, 769–773. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef] [PubMed]
- Wightman, B.; Ha, I.; Ruvkun, G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 1993, 75, 855–862. [Google Scholar] [CrossRef] [PubMed]
- Horvitz, H.R.; Sulston, J.E. Isolation and genetic characterization of cell-lineage mutants of the nematode Caenorhabditis elegans. Genetics 1980, 96, 435–454. [Google Scholar] [CrossRef] [PubMed]
- Chalfie, M.; Horvitz, H.R.; Sulston, J.E. Mutations that lead to reiterations in the cell lineages of C. elegans. Cell 1981, 24, 59–69. [Google Scholar] [CrossRef]
- Lee, R.; Feinbaum, R.; Ambros, V. A short history of a short RNA. Cell 2004, 116, S89–S92. [Google Scholar] [CrossRef]
- Almeida, M.I.; Reis, R.M.; Calin, G.A. MicroRNA history: Discovery, recent applications, and next frontiers. Mutat. Res. 2011, 717, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Reinhart, B.J.; Slack, F.J.; Basson, M.; Pasquinelli, A.E.; Bettinger, J.C.; Rougvie, A.E.; Horvitz, H.R.; Ruvkun, G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 2000, 403, 901–906. [Google Scholar] [CrossRef]
- Pasquinelli, A.E.; Reinhart, B.J.; Slack, F.; Martindale, M.Q.; Kuroda, M.I.; Maller, B.; Hayward, D.C.; Ball, E.E.; Degnan, B.; Müller, P.; et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature 2000, 408, 86–89. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef]
- Fire, A.; Xu, S.; Montgomery, M.K.; Kostas, S.A.; Driver, S.E.; Mello, C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998, 391, 806–811. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, A.J.; Baulcombe, D.C. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 1999, 286, 950–952. [Google Scholar] [CrossRef] [PubMed]
- Zamore, P.D.; Tuschl, T.; Sharp, P.A.; Bartel, D.P. RNAi: Double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 2000, 101, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Hammond, S.M.; Bernstein, E.; Beach, D.; Hannon, G.J. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 2000, 404, 293–296. [Google Scholar] [CrossRef] [PubMed]
- Hutvágner, G.; McLachlan, J.; Pasquinelli, A.E.; Bálint, E.; Tuschl, T.; Zamore, P.D. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 2001, 293, 834–838. [Google Scholar] [CrossRef]
- Grishok, A.; Pasquinelli, A.E.; Conte, D.; Li, N.; Parrish, S.; Ha, I.; Baillie, D.L.; Fire, A.; Ruvkun, G.; Mello, C.C. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 2001, 106, 23–34. [Google Scholar] [CrossRef]
- Pong, S.K.; Gullerova, M. Noncanonical functions of microRNA pathway enzymes—Drosha, DGCR8, Dicer and Ago proteins. FEBS Lett. 2018, 592, 2973–2986. [Google Scholar] [CrossRef]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef]
- Broughton, J.P.; Lovci, M.T.; Huang, J.L.; Yeo, G.W.; Pasquinelli, A.E. Pairing beyond the Seed Supports MicroRNA Targeting Specificity. Mol. Cell 2016, 64, 320–333. [Google Scholar] [CrossRef] [PubMed]
- Vasudevan, S. Posttranscriptional upregulation by microRNAs. Wiley Interdiscip. Rev. RNA 2012, 3, 311–330. [Google Scholar] [CrossRef]
- Makarova, J.A.; Shkurnikov, M.U.; Wicklein, D.; Lange, T.; Samatov, T.R.; Turchinovich, A.A.; Tonevitsky, A.G. Intracellular and extracellular microRNA: An update on localization and biological role. Prog. Histochem. Cytochem. 2016, 51, 33–49. [Google Scholar] [CrossRef]
- Hayes, J.; Peruzzi, P.P.; Lawler, S. MicroRNAs in cancer: Biomarkers, functions and therapy. Trends Mol. Med. 2014, 20, 460–469. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chen, J.; Sen, S. MicroRNA as Biomarkers and Diagnostics. J. Cell. Physiol. 2016, 231, 25–30. [Google Scholar] [CrossRef]
- Huang, W. MicroRNAs: Biomarkers, Diagnostics, and Therapeutics. Methods Mol. Biol. 2017, 1617, 57–67. [Google Scholar] [CrossRef] [PubMed]
- Tüfekci, K.U.; Oner, M.G.; Meuwissen, R.L.; Genç, S. The role of microRNAs in human diseases. Methods Mol. Biol. 2014, 1107, 33–50. [Google Scholar] [CrossRef]
- Paul, P.; Chakraborty, A.; Sarkar, D.; Langthasa, M.; Rahman, M.; Bari, M.; Singha, R.S.; Malakar, A.K.; Chakraborty, S. Interplay between miRNAs and human diseases. J. Cell. Physiol. 2018, 233, 2007–2018. [Google Scholar] [CrossRef]
- Mori, M.A.; Ludwig, R.G.; Garcia-Martin, R.; Brandão, B.B.; Kahn, C.R. Extracellular miRNAs: From Biomarkers to Mediators of Physiology and Disease. Cell Metab. 2019, 30, 656–673. [Google Scholar] [CrossRef] [PubMed]
- Kakimoto, Y.; Tanaka, M.; Kamiguchi, H.; Ochiai, E.; Osawa, M. MicroRNA Stability in FFPE Tissue Samples: Dependence on GC Content. PLoS ONE 2016, 11, e0163125. [Google Scholar] [CrossRef]
- Glinge, C.; Clauss, S.; Boddum, K.; Jabbari, R.; Jabbari, J.; Risgaard, B.; Tomsits, P.; Hildebrand, B.; Kääb, S.; Wakili, R.; et al. Stability of Circulating Blood-Based MicroRNAs—Pre-Analytic Methodological Considerations. PLoS ONE 2017, 12, e0167969. [Google Scholar] [CrossRef] [PubMed]
- Condrat, C.E.; Thompson, D.C.; Barbu, M.G.; Bugnar, O.L.; Boboc, A.; Cretoiu, D.; Suciu, N.; Cretoiu, S.M.; Voinea, S.C. miRNAs as Biomarkers in Disease: Latest Findings Regarding Their Role in Diagnosis and Prognosis. Cells 2020, 9, 276. [Google Scholar] [CrossRef]
- Buhagiar, A.; Seria, E.; Borg, M.; Borg, J.; Ayers, D. Overview of microRNAs as liquid biopsy biomarkers for colorectal cancer sub-type profiling and chemoresistance. Cancer Drug Resist. 2021, 4, 934–945. [Google Scholar] [CrossRef]
- Hutter, K.; Rülicke, T.; Szabo, T.G.; Andersen, L.; Villunger, A.; Herzog, S. The miR-15a/16-1 and miR-15b/16-2 clusters regulate early B cell development by limiting IL-7 receptor expression. Front. Immunol. 2022, 13, 967914. [Google Scholar] [CrossRef]
- Peng, Y.; Croce, C.M. The role of MicroRNAs in human cancer. Signal Transduct. Target. Ther. 2016, 1, 15004. [Google Scholar] [CrossRef]
- Ratnasari, N.; Lestari, P.; Renovaldi, D.; Raditya Ningsih, J.; Qoriansas, N.; Wardana, T.; Hakim, S.; Signa Aini Gumilas, N.; Indrarti, F.; Triwikatmani, C.; et al. Potential plasma biomarkers: miRNA-29c, miRNA-21, and miRNA-155 in clinical progression of Hepatocellular Carcinoma patients. PLoS ONE 2022, 17, e0263298. [Google Scholar] [CrossRef]
- Oura, K.; Morishita, A.; Masaki, T. Molecular and Functional Roles of MicroRNAs in the Progression of Hepatocellular Carcinoma-A Review. Int. J. Mol. Sci. 2020, 21, 8362. [Google Scholar] [CrossRef] [PubMed]
- Chu, R.; Mo, G.; Duan, Z.; Huang, M.; Chang, J.; Li, X.; Liu, P. miRNAs affect the development of hepatocellular carcinoma via dysregulation of their biogenesis and expression. Cell Commun. Signal. 2014, 12, 45. [Google Scholar] [CrossRef]
- Mo, W.Y.; Cao, S.Q. MiR-29a-3p: A potential biomarker and therapeutic target in colorectal cancer. Clin. Transl. Oncol. 2023, 25, 563–577. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.Y.; Zhang, Q.; Wang, D.D.; Yan, W.; Sha, H.H.; Zhao, J.H.; Yang, S.J.; Zhang, H.D.; Hou, J.C.; Xu, H.Z.; et al. MiR-29a: A potential therapeutic target and promising biomarker in tumors. Biosci. Rep. 2018, 38, BSR20171265. [Google Scholar] [CrossRef] [PubMed]
- Bonci, D.; De Maria, R. miR-15/miR-16 loss, miR-21 upregulation, or deregulation of their target genes predicts poor prognosis in prostate cancer patients. Mol. Cell Oncol. 2016, 3, e1109744. [Google Scholar] [CrossRef] [PubMed]
- Arrighetti, N.; Beretta, G.L. miRNAs as Therapeutic Tools and Biomarkers for Prostate Cancer. Pharmaceutics 2021, 13, 380. [Google Scholar] [CrossRef] [PubMed]
- Jin, W.; Chen, F.; Wang, K.; Song, Y.; Fei, X.; Wu, B. miR-15a/miR-16 cluster inhibits invasion of prostate cancer cells by suppressing TGF-β signaling pathway. Biomed. Pharmacother. 2018, 104, 637–644. [Google Scholar] [CrossRef] [PubMed]
- He, B.; Zhao, Z.; Cai, Q.; Zhang, Y.; Zhang, P.; Shi, S.; Xie, H.; Peng, X.; Yin, W.; Tao, Y.; et al. miRNA-based biomarkers, therapies, and resistance in Cancer. Int. J. Biol. Sci. 2020, 16, 2628–2647. [Google Scholar] [CrossRef] [PubMed]
- Parra-Medina, R.; Lopez-Kleine, L.; Ramirez-Clavijo, S.; Payan-Gomez, C. Identification of candidate miRNAs in early-onset and late-onset prostate cancer by network analysis. Sci. Rep. 2020, 10, 12345. [Google Scholar] [CrossRef] [PubMed]
- Jickling, G.C.; Ander, B.P.; Zhan, X.; Noblett, D.; Stamova, B.; Liu, D. microRNA expression in peripheral blood cells following acute ischemic stroke and their predicted gene targets. PLoS ONE 2014, 9, e99283. [Google Scholar] [CrossRef]
- Liu, D.Z.; Tian, Y.; Ander, B.P.; Xu, H.; Stamova, B.S.; Zhan, X.; Turner, R.J.; Jickling, G.; Sharp, F.R. Brain and blood microRNA expression profiling of ischemic stroke, intracerebral hemorrhage, and kainate seizures. J. Cereb. Blood Flow Metab. 2010, 30, 92–101. [Google Scholar] [CrossRef]
- Jeyaseelan, K.; Lim, K.Y.; Armugam, A. MicroRNA expression in the blood and brain of rats subjected to transient focal ischemia by middle cerebral artery occlusion. Stroke 2008, 39, 959–966. [Google Scholar] [CrossRef]
- Lv, B.; Cheng, X.; Sharp, F.R.; Ander, B.P.; Liu, D.Z. MicroRNA-122 Mimic Improves Stroke Outcomes and Indirectly Inhibits NOS2 After Middle Cerebral Artery Occlusion in Rats. Front. Neurosci. 2018, 12, 767. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.Z. Methods (of MicroRNA Therapeutics) for Treating Brain Injury. WO Patent 2019165267A1, 2019. Available online: https://patents.google.com/patent/WO2019165267A1/en (accessed on 29 August 2019).
- Liu, D.Z.; Jickling, G.C.; Ander, B.P.; Hull, H.; Zhan, X.; Cox, C.; Shroff, N.; Dykstra-Aiello, C.; Stamova, B.; Sharp, F.R. Elevating microRNA-122 in blood improves outcomes after temporary middle cerebral artery occlusion in rats. J. Cereb. Blood Flow Metab. 2016, 36, 1374–1383. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Hou, X.; Ren, G.; Zhang, Y.; Cheng, H. Dynamic changes in miR-124 levels in patients with acute cerebral infarction. Int. J. Neurosci. 2019, 129, 649–653. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Feng, Z.; Du, L.; Huang, Y.; Ge, J.; Deng, Y.; Mei, Z. The Potential Role of MicroRNA-124 in Cerebral Ischemia Injury. Int. J. Mol. Sci. 2019, 21, 120. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Li, Z.; He, T.; Qu, M.; Jiang, L.; Li, W.; Shi, X.; Pan, J.; Zhang, L.; Wang, Y.; et al. M2 microglia-derived exosomes protect the mouse brain from ischemia-reperfusion injury via exosomal miR-124. Theranostics 2019, 9, 2910–2923. [Google Scholar] [CrossRef]
- Bernstein, D.L.; Zuluaga-Ramirez, V.; Gajghate, S.; Reichenbach, N.L.; Polyak, B.; Persidsky, Y.; Rom, S. miR-98 reduces endothelial dysfunction by protecting blood-brain barrier (BBB) and improves neurological outcomes in mouse ischemia/reperfusion stroke model. J. Cereb. Blood Flow Metab. 2020, 40, 1953–1965. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Ma, Q.; Li, Y.; Li, B.; Zhang, L. Inhibition of microRNA-210 suppresses pro-inflammatory response and reduces acute brain injury of ischemic stroke in mice. Exp. Neurol. 2018, 300, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Sun, P.; Zhang, K.; Hassan, S.H.; Zhang, X.; Tang, X.; Pu, H.; Stetler, R.A.; Chen, J.; Yin, K.J. Endothelium-Targeted Deletion of microRNA-15a/16-1 Promotes Poststroke Angiogenesis and Improves Long-Term Neurological Recovery. Circ. Res. 2020, 126, 1040–1057. [Google Scholar] [CrossRef]
- Du, K.; Zhao, C.; Wang, L.; Wang, Y.; Zhang, K.Z.; Shen, X.Y.; Sun, H.X.; Gao, W.; Lu, X. MiR-191 inhibit angiogenesis after acute ischemic stroke targeting VEZF1. Aging 2019, 11, 2762–2786. [Google Scholar] [CrossRef]
- Ghai, V.; Fallen, S.; Baxter, D.; Scherler, K.; Kim, T.K.; Zhou, Y.; Meabon, J.S.; Logsdon, A.F.; Banks, W.A.; Schindler, A.G.; et al. Alterations in Plasma microRNA and Protein Levels in War Veterans with Chronic Mild Traumatic Brain Injury. J. Neurotrauma 2020, 37, 1418–1430. [Google Scholar] [CrossRef]
- Di Pietro, V.; Ragusa, M.; Davies, D.; Su, Z.; Hazeldine, J.; Lazzarino, G.; Hill, L.J.; Crombie, N.; Foster, M.; Purrello, M.; et al. MicroRNAs as Novel Biomarkers for the Diagnosis and Prognosis of Mild and Severe Traumatic Brain Injury. J. Neurotrauma 2017, 34, 1948–1956. [Google Scholar] [CrossRef] [PubMed]
- Xiao, X.; Jiang, Y.; Liang, W.; Wang, Y.; Cao, S.; Yan, H.; Gao, L.; Zhang, L. miR-212-5p attenuates ferroptotic neuronal death after traumatic brain injury by targeting Ptgs2. Mol. Brain 2019, 12, 78. [Google Scholar] [CrossRef] [PubMed]
- Ge, X.; Guo, M.; Hu, T.; Li, W.; Huang, S.; Yin, Z.; Li, Y.; Chen, F.; Zhu, L.; Kang, C.; et al. Increased Microglial Exosomal miR-124-3p Alleviates Neurodegeneration and Improves Cognitive Outcome after rmTBI. Mol. Ther. 2020, 28, 503–522. [Google Scholar] [CrossRef] [PubMed]
- Long, X.; Yao, X.; Jiang, Q.; Yang, Y.; He, X.; Tian, W.; Zhao, K.; Zhang, H. Astrocyte-derived exosomes enriched with miR-873a-5p inhibit neuroinflammation via microglia phenotype modulation after traumatic brain injury. J. Neuroinflammation 2020, 17, 89. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Ge, X.; Yu, J.; Han, Z.; Yin, Z.; Li, Y.; Chen, F.; Wang, H.; Zhang, J.; Lei, P. Increased miR-124-3p in microglial exosomes following traumatic brain injury inhibits neuronal inflammation and contributes to neurite outgrowth via their transfer into neurons. FASEB J. 2018, 32, 512–528. [Google Scholar] [CrossRef] [PubMed]
- Brennan, G.P.; Henshall, D.C. microRNAs in the pathophysiology of epilepsy. Neurosci. Lett. 2018, 667, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.L.; Plotkin, J.L.; Venø, M.T.; von Schimmelmann, M.; Feinberg, P.; Mann, S.; Handler, A.; Kjems, J.; Surmeier, D.J.; O’Carroll, D.; et al. MicroRNA-128 governs neuronal excitability and motor behavior in mice. Science 2013, 342, 1254–1258. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Li, Z.; Zhang, Y.; Wang, G.; Wei, M.; Hu, Y.; Ma, S.; Jiang, Y.; Che, N.; Wang, X.; et al. Targeting of microRNA-199a-5p protects against pilocarpine-induced status epilepticus and seizure damage via SIRT1-p53 cascade. Epilepsia 2016, 57, 706–716. [Google Scholar] [CrossRef]
- Ma, Y. The Challenge of microRNA as a Biomarker of Epilepsy. Curr. Neuropharmacol. 2018, 16, 37–42. [Google Scholar] [CrossRef]
- Ren, L.; Zhu, R.; Li, X. Silencing miR-181a produces neuroprotection against hippocampus neuron cell apoptosis post-status epilepticus in a rat model and in children with temporal lobe epilepsy. Genet. Mol. Res. 2016, 15, 15017798. [Google Scholar] [CrossRef]
- Wang, W.; Wang, X.; Chen, L.; Zhang, Y.; Xu, Z.; Liu, J.; Jiang, G.; Li, J.; Zhang, X.; Wang, K.; et al. The microRNA miR-124 suppresses seizure activity and regulates CREB1 activity. Expert Rev. Mol. Med. 2016, 18, e4. [Google Scholar] [CrossRef] [PubMed]
- Jimenez-Mateos, E.M.; Engel, T.; Merino-Serrais, P.; Fernaud-Espinosa, I.; Rodriguez-Alvarez, N.; Reynolds, J.; Reschke, C.R.; Conroy, R.M.; McKiernan, R.C.; deFelipe, J.; et al. Antagomirs targeting microRNA-134 increase hippocampal pyramidal neuron spine volume in vivo and protect against pilocarpine-induced status epilepticus. Brain Struct. Funct. 2015, 220, 2387–2399. [Google Scholar] [CrossRef] [PubMed]
- Jimenez-Mateos, E.M.; Engel, T.; Merino-Serrais, P.; McKiernan, R.C.; Tanaka, K.; Mouri, G.; Sano, T.; O’Tuathaigh, C.; Waddington, J.L.; Prenter, S.; et al. Silencing microRNA-134 produces neuroprotective and prolonged seizure-suppressive effects. Nat. Med. 2012, 18, 1087–1094. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Xiang, W.; Yanhui, L.; Ruofei, L.; Yunhe, M.; Jiewen, L.; Qing, M. Dysregulation of microRNA-128 expression in WHO grades 2 glioma is associated with glioma-associated epilepsy: Down-regulation of miR-128 induces glioma-associated seizure. Epilepsy Res. 2016, 127, 6–11. [Google Scholar] [CrossRef] [PubMed]
- Cai, Z.; Li, S.; Li, S.; Song, F.; Zhang, Z.; Qi, G.; Li, T.; Qiu, J.; Wan, J.; Sui, H.; et al. Antagonist Targeting microRNA-155 Protects against Lithium-Pilocarpine-Induced Status Epilepticus in C57BL/6 Mice by Activating Brain-Derived Neurotrophic Factor. Front. Pharmacol. 2016, 7, 129. [Google Scholar] [CrossRef]
- Wilson, D.M., 3rd; Cookson, M.R.; Van Den Bosch, L.; Zetterberg, H.; Holtzman, D.M.; Dewachter, I. Hallmarks of neurodegenerative diseases. Cell 2023, 186, 693–714. [Google Scholar] [CrossRef] [PubMed]
- Kritsilis, M.; Rizou, S.V.; Koutsoudaki, P.N.; Evangelou, K.; Gorgoulis, V.G.; Papadopoulos, D. Ageing, Cellular Senescence and Neurodegenerative Disease. Int. J. Mol. Sci. 2018, 19, 2937. [Google Scholar] [CrossRef]
- Johnson, I.P. Age-related neurodegenerative disease research needs aging models. Front. Aging Neurosci. 2015, 7, 168. [Google Scholar] [CrossRef]
- Tzeplaeff, L.; Wilfling, S.; Requardt, M.V.; Herdick, M. Current State and Future Directions in the Therapy of ALS. Cells 2023, 12, 1523. [Google Scholar] [CrossRef]
- Ferguson, M.W.; Kennedy, C.J.; Palpagama, T.H.; Waldvogel, H.J.; Faull, R.L.M.; Kwakowsky, A. Current and Possible Future Therapeutic Options for Huntington’s Disease. J. Cent. Nerv. Syst. Dis. 2022, 14, 11795735221092517. [Google Scholar] [CrossRef]
- Copani, A.; Condorelli, F.; Caruso, A.; Vancheri, C.; Sala, A.; Giuffrida Stella, A.M.; Canonico, P.L.; Nicoletti, F.; Sortino, M.A. Mitotic signaling by beta-amyloid causes neuronal death. FASEB J. 1999, 13, 2225–2234. [Google Scholar] [CrossRef] [PubMed]
- Williamson, R.; Scales, T.; Clark, B.R.; Gibb, G.; Reynolds, C.H.; Kellie, S.; Bird, I.N.; Varndell, I.M.; Sheppard, P.W.; Everall, I.; et al. Rapid tyrosine phosphorylation of neuronal proteins including tau and focal adhesion kinase in response to amyloid-beta peptide exposure: Involvement of Src family protein kinases. J. Neurosci. 2002, 22, 10–20. [Google Scholar] [CrossRef] [PubMed]
- Angelucci, F.; Cechova, K.; Valis, M.; Kuca, K.; Zhang, B.; Hort, J. MicroRNAs in Alzheimer’s Disease: Diagnostic Markers or Therapeutic Agents? Front. Pharmacol. 2019, 10, 665. [Google Scholar] [CrossRef] [PubMed]
- Hébert, S.S.; De Strooper, B. Alterations of the microRNA network cause neurodegenerative disease. Trends Neurosci. 2009, 32, 199–206. [Google Scholar] [CrossRef] [PubMed]
- Vilardo, E.; Barbato, C.; Ciotti, M.; Cogoni, C.; Ruberti, F. MicroRNA-101 regulates amyloid precursor protein expression in hippocampal neurons. J. Biol. Chem. 2010, 285, 18344–18351. [Google Scholar] [CrossRef] [PubMed]
- Delay, C.; Calon, F.; Mathews, P.; Hébert, S.S. Alzheimer-specific variants in the 3’UTR of Amyloid precursor protein affect microRNA function. Mol. Neurodegener. 2011, 6, 70. [Google Scholar] [CrossRef] [PubMed]
- Hebert, S.S.; Horre, K.; Nicolai, L.; Papadopoulou, A.S.; Mandemakers, W.; Silahtaroglu, A.N.; Kauppinen, S.; Delacourte, A.; De Strooper, B. Loss of microRNA cluster miR-29a/b-1 in sporadic Alzheimer’s disease correlates with increased BACE1/beta-secretase expression. Proc. Natl. Acad. Sci. USA 2008, 105, 6415–6420. [Google Scholar] [CrossRef]
- Smith, P.; Al Hashimi, A.; Girard, J.; Delay, C.; Hébert, S.S. In Vivo regulation of amyloid precursor protein neuronal splicing by microRNAs. J. Neurochem. 2011, 116, 240–247. [Google Scholar] [CrossRef] [PubMed]
- Fang, M.; Wang, J.; Zhang, X.; Geng, Y.; Hu, Z.; Rudd, J.A.; Ling, S.; Chen, W.; Han, S. The miR-124 regulates the expression of BACE1/β-secretase correlated with cell death in Alzheimer’s disease. Toxicol. Lett. 2012, 209, 94–105. [Google Scholar] [CrossRef]
- Zong, Y.; Wang, H.; Dong, W.; Quan, X.; Zhu, H.; Xu, Y.; Huang, L.; Ma, C.; Qin, C. miR-29c regulates BACE1 protein expression. Brain Res. 2011, 1395, 108–115. [Google Scholar] [CrossRef]
- Zhu, H.C.; Wang, L.M.; Wang, M.; Song, B.; Tan, S.; Teng, J.F.; Duan, D.X. MicroRNA-195 downregulates Alzheimer’s disease amyloid-beta production by targeting BACE1. Brain Res. Bull. 2012, 88, 596–601. [Google Scholar] [CrossRef]
- Nakano, M.; Kubota, K.; Hashizume, S.; Kobayashi, E.; Chikenji, T.S.; Saito, Y.; Fujimiya, M. An enriched environment prevents cognitive impairment in an Alzheimer’s disease model by enhancing the secretion of exosomal microRNA-146a from the choroid plexus. Brain Behav. Immun. Health 2020, 9, 100149. [Google Scholar] [CrossRef]
- Huang, P.; Xu, M.; He, Y.X. MicroRNA-146a regulates the expression of the Aβ1-42 protein in Alzheimer’s disease. Eur. Rev. Med. Pharmacol. Sci. 2021, 25, 2003–2012. [Google Scholar] [CrossRef]
- Ma, Y.; Ye, J.; Zhao, L.; Pan, D. MicroRNA-146a inhibition promotes total neurite outgrowth and suppresses cell apoptosis, inflammation, and STAT1/MYC pathway in PC12 and cortical neuron cellular Alzheimer’s disease models. Braz. J. Med. Biol. Res. 2021, 54, e9665. [Google Scholar] [CrossRef]
- Liang, C.; Zou, T.; Zhang, M.; Fan, W.; Zhang, T.; Jiang, Y.; Cai, Y.; Chen, F.; Chen, X.; Sun, Y.; et al. MicroRNA-146a switches microglial phenotypes to resist the pathological processes and cognitive degradation of Alzheimer’s disease. Theranostics 2021, 11, 4103–4121. [Google Scholar] [CrossRef]
- Fu, X.; Liu, J.; Xie, J.; Chen, G.; Zhang, H.; Meng, F.; Wu, M.; Li, Q.; Liu, Y.; Wang, W.; et al. Identification of potential therapeutic and diagnostic characteristics of Alzheimer disease by targeting the miR-132-3p/FOXO3a-PPM1F axis in APP/PS1 mice. Brain Res. 2022, 1790, 147983. [Google Scholar] [CrossRef]
- Walgrave, H.; Balusu, S.; Snoeck, S.; Vanden Eynden, E.; Craessaerts, K.; Thrupp, N.; Wolfs, L.; Horré, K.; Fourne, Y.; Ronisz, A.; et al. Restoring miR-132 expression rescues adult hippocampal neurogenesis and memory deficits in Alzheimer’s disease. Cell Stem Cell 2021, 28, 1805–1821.e8. [Google Scholar] [CrossRef]
- Qu, J.; Xiong, X.; Hujie, G.; Ren, J.; Yan, L.; Ma, L. MicroRNA-132-3p alleviates neuron apoptosis and impairments of learning and memory abilities in Alzheimer’s disease by downregulation of HNRNPU stabilized BACE1. Cell Cycle 2021, 20, 2309–2320. [Google Scholar] [CrossRef]
- Liang, Y.; Wang, L. Inflamma-MicroRNAs in Alzheimer’s Disease: From Disease Pathogenesis to Therapeutic Potentials. Front. Cell Neurosci. 2021, 15, 785433. [Google Scholar] [CrossRef]
- Zhang, X.; Ma, W.; Xue, W.; Wang, Y.; Chen, P.; Li, Q.; Li, Y.Y.; Hu, X.; Zhao, Y.; Zhou, H. miR-181a plays the tumor-suppressor role in chronic myeloid leukemia CD34+ cells partially via SERPINE1. Cell. Mol. Life Sci. 2023, 81, 10. [Google Scholar] [CrossRef]
- Viera, G.M.; Salomao, K.B.; de Sousa, G.R.; Baroni, M.; Delsin, L.E.A.; Pezuk, J.A.; Brassesco, M.S. miRNA signatures in childhood sarcomas and their clinical implications. Clin. Transl. Oncol. 2019, 21, 1583–1623. [Google Scholar] [CrossRef]
- Hermansen, S.K.; Kristensen, B.W. MicroRNA biomarkers in glioblastoma. J. Neurooncol. 2013, 114, 13–23. [Google Scholar] [CrossRef]
- Rodriguez-Ortiz, C.J.; Prieto, G.A.; Martini, A.C.; Forner, S.; Trujillo-Estrada, L.; LaFerla, F.M.; Baglietto-Vargas, D.; Cotman, C.W.; Kitazawa, M. miR-181a negatively modulates synaptic plasticity in hippocampal cultures and its inhibition rescues memory deficits in a mouse model of Alzheimer’s disease. Aging Cell 2020, 19, e13118. [Google Scholar] [CrossRef]
- Sanuki, R.; Yamamura, T. Tumor Suppressive Effects of miR-124 and Its Function in Neuronal Development. Int. J. Mol. Sci. 2021, 22, 5919. [Google Scholar] [CrossRef]
- Kohama, I.; Kosaka, N.; Chikuda, H.; Ochiya, T. An Insight into the Roles of MicroRNAs and Exosomes in Sarcoma. Cancers 2019, 11, 428. [Google Scholar] [CrossRef]
- Yaghoubi, N.; Zahedi Avval, F.; Khazaei, M.; Aghaee-Bakhtiari, S.H. MicroRNAs as potential investigative and predictive biomarkers in colorectal cancer. Cell Signal. 2021, 80, 109910. [Google Scholar] [CrossRef]
- Hou, T.Y.; Zhou, Y.; Zhu, L.S.; Wang, X.; Pang, P.; Wang, D.Q.; Liuyang, Z.Y.; Man, H.; Lu, Y.; Zhu, L.Q.; et al. Correcting abnormalities in miR-124/PTPN1 signaling rescues tau pathology in Alzheimer’s disease. J. Neurochem. 2020, 154, 441–457. [Google Scholar] [CrossRef]
- Saiki, S.; Sato, S.; Hattori, N. Molecular pathogenesis of Parkinson’s disease: Update. J. Neurol. Neurosurg. Psychiatry 2012, 83, 430–436. [Google Scholar] [CrossRef]
- Junn, E.; Lee, K.W.; Jeong, B.S.; Chan, T.W.; Im, J.Y.; Mouradian, M.M. Repression of alpha-synuclein expression and toxicity by microRNA-7. Proc. Natl. Acad. Sci. USA 2009, 106, 13052–13057. [Google Scholar] [CrossRef]
- Galter, D.; Westerlund, M.; Carmine, A.; Lindqvist, E.; Sydow, O.; Olson, L. LRRK2 expression linked to dopamine-innervated areas. Ann. Neurol. 2006, 59, 714–719. [Google Scholar] [CrossRef]
- Cho, H.J.; Liu, G.; Jin, S.M.; Parisiadou, L.; Xie, C.; Yu, J.; Sun, L.; Ma, B.; Ding, J.; Vancraenenbroeck, R.; et al. MicroRNA-205 regulates the expression of Parkinson’s disease-related leucine-rich repeat kinase 2 protein. Hum. Mol. Genet. 2013, 22, 608–620. [Google Scholar] [CrossRef]
- Chen, Y.; Gao, C.; Sun, Q.; Pan, H.; Huang, P.; Ding, J.; Chen, S. MicroRNA-4639 Is a Regulator of DJ-1 Expression and a Potential Early Diagnostic Marker for Parkinson’s Disease. Front. Aging Neurosci. 2017, 9, 232. [Google Scholar] [CrossRef]
- Saraiva, C.; Paiva, J.; Santos, T.; Ferreira, L.; Bernardino, L. MicroRNA-124 loaded nanoparticles enhance brain repair in Parkinson’s disease. J. Control. Release 2016, 235, 291–305. [Google Scholar] [CrossRef]
- Tufekci, K.U.; Meuwissen, R.; Genc, S.; Genc, K. Inflammation in Parkinson’s disease. Adv. Protein Chem. Struct. Biol. 2012, 88, 69–132. [Google Scholar] [CrossRef]
- Ling, Z.; Fan, G.; Yao, D.; Zhao, J.; Zhou, Y.; Feng, J.; Zhou, G.; Chen, Y. MicroRNA-150 functions as a tumor suppressor and sensitizes osteosarcoma to doxorubicin-induced apoptosis by targeting RUNX2. Exp. Ther. Med. 2020, 19, 481–488. [Google Scholar] [CrossRef]
- Lv, T.; Jiang, L.; Kong, L.; Yang, J. MicroRNA-29c-3p acts as a tumor suppressor gene and inhibits tumor progression in hepatocellular carcinoma by targeting TRIM31. Oncol. Rep. 2020, 43, 953–964. [Google Scholar] [CrossRef]
- Li, H.; Yu, L.; Li, M.; Chen, X.; Tian, Q.; Jiang, Y.; Li, N. MicroRNA-150 serves as a diagnostic biomarker and is involved in the inflammatory pathogenesis of Parkinson’s disease. Mol. Genet. Genom. Med. 2020, 8, e1189. [Google Scholar] [CrossRef]
- Wang, R.; Li, Q.; He, Y.; Yang, Y.; Ma, Q.; Li, C. miR-29c-3p inhibits microglial NLRP3 inflammasome activation by targeting NFAT5 in Parkinson’s disease. Genes Cells 2020, 25, 364–374. [Google Scholar] [CrossRef]
- Ghafouri-Fard, S.; Gholipour, M.; Taheri, M. MicroRNA Signature in Melanoma: Biomarkers and Therapeutic Targets. Front. Oncol. 2021, 11, 608987. [Google Scholar] [CrossRef]
- Hu, Y.B.; Zhang, Y.F.; Wang, H.; Ren, R.J.; Cui, H.L.; Huang, W.Y.; Cheng, Q.; Chen, H.Z.; Wang, G. miR-425 deficiency promotes necroptosis and dopaminergic neurodegeneration in Parkinson’s disease. Cell Death Dis. 2019, 10, 589. [Google Scholar] [CrossRef]
- Ricci, C.; Marzocchi, C.; Battistini, S. MicroRNAs as Biomarkers in Amyotrophic Lateral Sclerosis. Cells 2018, 7, 219. [Google Scholar] [CrossRef]
- Shioya, M.; Obayashi, S.; Tabunoki, H.; Arima, K.; Saito, Y.; Ishida, T.; Satoh, J. Aberrant microRNA expression in the brains of neurodegenerative diseases: miR-29a decreased in Alzheimer disease brains targets neurone navigator 3. Neuropathol. Appl. Neurobiol. 2010, 36, 320–330. [Google Scholar] [CrossRef]
- Cattaneo, E.; Zuccato, C.; Tartari, M. Normal huntingtin function: An alternative approach to Huntington’s disease. Nat. Rev. Neurosci. 2005, 6, 919–930. [Google Scholar] [CrossRef]
- Zuccato, C.; Tartari, M.; Crotti, A.; Goffredo, D.; Valenza, M.; Conti, L.; Cataudella, T.; Leavitt, B.R.; Hayden, M.R.; Timmusk, T.; et al. Huntingtin interacts with REST/NRSF to modulate the transcription of NRSE-controlled neuronal genes. Nat. Genet. 2003, 35, 76–83. [Google Scholar] [CrossRef]
- Johnson, R.; Zuccato, C.; Belyaev, N.D.; Guest, D.J.; Cattaneo, E.; Buckley, N.J. A microRNA-based gene dysregulation pathway in Huntington’s disease. Neurobiol. Dis. 2008, 29, 438–445. [Google Scholar] [CrossRef]
- Martí, E.; Pantano, L.; Bañez-Coronel, M.; Llorens, F.; Miñones-Moyano, E.; Porta, S.; Sumoy, L.; Ferrer, I.; Estivill, X. A myriad of miRNA variants in control and Huntington’s disease brain regions detected by massively parallel sequencing. Nucleic Acids Res. 2010, 38, 7219–7235. [Google Scholar] [CrossRef]
- Lee, S.T.; Chu, K.; Im, W.S.; Yoon, H.J.; Im, J.Y.; Park, J.E.; Park, K.H.; Jung, K.H.; Lee, S.K.; Kim, M.; et al. Altered microRNA regulation in Huntington’s disease models. Exp. Neurol. 2011, 227, 172–179. [Google Scholar] [CrossRef]
- Fukuoka, M.; Takahashi, M.; Fujita, H.; Chiyo, T.; Popiel, H.A.; Watanabe, S.; Furuya, H.; Murata, M.; Wada, K.; Okada, T.; et al. Supplemental Treatment for Huntington’s Disease with miR-132 that Is Deficient in Huntington’s Disease Brain. Mol. Ther. Nucleic Acids 2018, 11, 79–90. [Google Scholar] [CrossRef]
- Rafat, M.; Moraghebi, M.; Afsa, M.; Malekzadeh, K. The outstanding role of miR-132-3p in carcinogenesis of solid tumors. Hum. Cell 2021, 34, 1051–1065. [Google Scholar] [CrossRef]
- Li, Y.; Zu, L.; Wang, Y.; Wang, M.; Chen, P.; Zhou, Q. miR-132 inhibits lung cancer cell migration and invasion by targeting SOX4. J. Thorac. Dis. 2015, 7, 1563–1569. [Google Scholar] [CrossRef]
- Otmani, K.; Lewalle, P. Tumor Suppressor miRNA in Cancer Cells and the Tumor Microenvironment: Mechanism of Deregulation and Clinical Implications. Front. Oncol. 2021, 11, 708765. [Google Scholar] [CrossRef]
- Feng, J.; Hu, S.; Liu, K.; Sun, G.; Zhang, Y. The Role of MicroRNA in the Regulation of Tumor Epithelial-Mesenchymal Transition. Cells 2022, 11, 1981. [Google Scholar] [CrossRef]
- Elnaggar, G.N.; El-Hifnawi, N.M.; Ismail, A.; Yahia, M.; Elshimy, R.A.A. Micro RNA-148a Targets Bcl-2 in Patients with Non-Small Cell Lung Cancer. Asian Pac. J. Cancer Prev. 2021, 22, 1949–1955. [Google Scholar] [CrossRef]
- Peng, Y.; Zhang, X.; Feng, X.; Fan, X.; Jin, Z. The crosstalk between microRNAs and the Wnt/β-catenin signaling pathway in cancer. Oncotarget 2017, 8, 14089–14106. [Google Scholar] [CrossRef]
- Ashrafizadeh, M.; Ang, H.L.; Moghadam, E.R.; Mohammadi, S.; Zarrin, V.; Hushmandi, K.; Samarghandian, S.; Zarrabi, A.; Najafi, M.; Mohammadinejad, R.; et al. MicroRNAs and Their Influence on the ZEB Family: Mechanistic Aspects and Therapeutic Applications in Cancer Therapy. Biomolecules 2020, 10, 1040. [Google Scholar] [CrossRef]
- Davalos, V.; Moutinho, C.; Villanueva, A.; Boque, R.; Silva, P.; Carneiro, F.; Esteller, M. Dynamic epigenetic regulation of the microRNA-200 family mediates epithelial and mesenchymal transitions in human tumorigenesis. Oncogene 2012, 31, 2062–2074. [Google Scholar] [CrossRef]
- Sun, F.; Li, S.G.; Zhang, H.W.; Hua, F.W.; Sun, G.Z.; Huang, Z. MiRNA-411 attenuates inflammatory damage and apoptosis following spinal cord injury. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 491–498. [Google Scholar] [CrossRef]
- Rodrigues, F.B.; Wild, E.J. Huntington’s Disease Clinical Trials Corner: April 2020. J. Huntingtons Dis. 2020, 9, 185–197. [Google Scholar] [CrossRef]
- Zhao, S.; Li, J.; Zhang, G.; Wang, Q.; Wu, C.; Zhang, Q.; Wang, H.; Sun, P.; Xiang, R.; Yang, S. Exosomal miR-451a Functions as a Tumor Suppressor in Hepatocellular Carcinoma by Targeting LPIN1. Cell Physiol. Biochem. 2019, 53, 19–35. [Google Scholar] [CrossRef]
- Sun, H.; Jiang, P. MicroRNA-451a acts as tumor suppressor in cutaneous basal cell carcinoma. Mol. Genet. Genom. Med. 2018, 6, 1001–1009. [Google Scholar] [CrossRef]
- Tan, J.; Li, C.; Ren, L.; Zhu, X.; Hua, F.; Fu, Y. miR-451a suppresses papillary thyroid cancer cell proliferation and invasion and facilitates apoptosis through targeting DCBLD2 and AKT1. Mol. Cell Probes 2022, 66, 101863. [Google Scholar] [CrossRef]
- Keskin, S.; Brouwers, C.C.; Sogorb-Gonzalez, M.; Martier, R.; Depla, J.A.; Vallès, A.; van Deventer, S.J.; Konstantinova, P.; Evers, M.M. AAV5-miHTT Lowers Huntingtin mRNA and Protein without Off-Target Effects in Patient-Derived Neuronal Cultures and Astrocytes. Mol. Ther. Methods Clin. Dev. 2019, 15, 275–284. [Google Scholar] [CrossRef]
- Miniarikova, J.; Zimmer, V.; Martier, R.; Brouwers, C.C.; Pythoud, C.; Richetin, K.; Rey, M.; Lubelski, J.; Evers, M.M.; van Deventer, S.J.; et al. AAV5-miHTT gene therapy demonstrates suppression of mutant huntingtin aggregation and neuronal dysfunction in a rat model of Huntington’s disease. Gene Ther. 2017, 24, 630–639. [Google Scholar] [CrossRef]
- Rupaimoole, R.; Slack, F.J. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017, 16, 203–222. [Google Scholar] [CrossRef]
- Winkle, M.; El-Daly, S.M.; Fabbri, M.; Calin, G.A. Noncoding RNA therapeutics—Challenges and potential solutions. Nat. Rev. Drug Discov. 2021, 20, 629–651. [Google Scholar] [CrossRef]
- Laganà, A.; Acunzo, M.; Romano, G.; Pulvirenti, A.; Veneziano, D.; Cascione, L.; Giugno, R.; Gasparini, P.; Shasha, D.; Ferro, A.; et al. miR-Synth: A computational resource for the design of multi-site multi-target synthetic miRNAs. Nucleic Acids Res. 2014, 42, 5416–5425. [Google Scholar] [CrossRef]
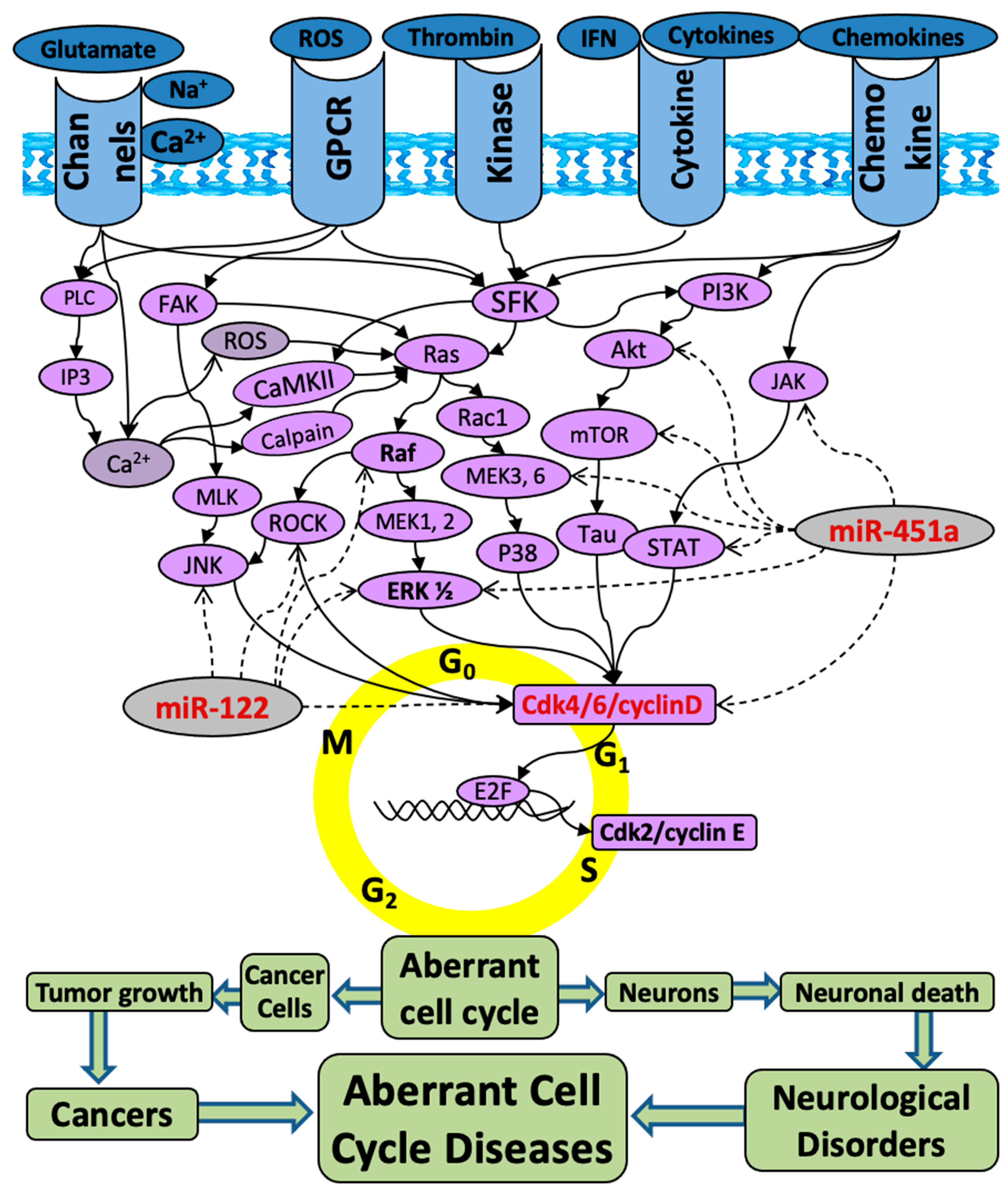
| (a) | ||
| miRNAs Upregulated in Cancers Only | miRNAs Upregulated in Both Cancers and Neurological Disorders | miRNAs Upregulated in Neurological Disorders Only |
| miR-BART7, miR-BART13, miR-2, miR-3, miR-3P, miR-9-1, miR-9-2, miR-9-3, miR-15b-5p, miR-16-1-3p, miR-16-2, miR-16-2-3p, miR-18, miR-19b, miR-19b-3p, miR-20, miR-20a-3p, miR-20b-3p, miR-21-3p, miR-24-2, miR-24-3p, miR-25a, miR-25-5p, miR-27, miR-27a, miR-27-3p, miR-28, miR-28-3p, miR-29a-3p, miR-29c-3p, miR-30b, miR-30c-2, miR-30e-3p, miR-31-5p, miR-32-5p, miR-33, miR-33b-3p, miR-45, miR-48, miR-59, miR-81a, miR-92a-3p, miR-92b, miR-93-5p, miR-95-3p, miR-96-3p, miR-96-5p, miR-98-5p, miR-106a-3633, miR-106b-25, miR-125b-1-3p, miR-126a, miR-129-1, miR-129-2, miR-130, miR-130a-3p, miR-131a, miR-132-5p, miR-135, miR-135a, miR-135b, miR-138-1-3p, miR-138-2-3p, miR-138b, miR-141b, miR-145b, miR-146-5p, miR-146b-3p, miR-146b-5p, miR-148b, miR-151, miR-151-3p, miR-151a, miR-151a-3p, miR-152-3p, miR-159, miR-159a, miR-181a, miR-181a-2-3p, miR-181b-5p, miR-185-5p, miR-187-5p, miR-189-5p, miR-190, miR-194-5p, miR-196, miR-196b, miR-197-5p, miR-199-5p, miR-199-s, miR-203a, miR-203b, miR-208a, miR-208a-3p, miR-210, miR-210-3p, miR-215-5p, miR-218-1, miR-221/222 cluster, miR-222-3p, miR-222-5p, miR-223-3p, miR-244, miR-275, miR-300, miR-301, miR-301b, miR-301b-3p, miR-302-367 cluster, miR-302-3p, miR-302b-3p, miR-320e, miR-325, miR-339-5p, miR-361, miR-365a, miR-365b, miR-367, miR-369-3p, miR-370-3p, miR-371, miR-371a-3p, miR-371a-5p, miR-373-3p, miR-373-5p, miR-374a, miR-378a-5p, miR-378d, miR-409, miR-411, miR-412, miR-412-3p, miR-421, miR-423-3p, miR-423-5p, miR-449, miR-450a, miR-450b-5p, miR-452-5p, miR-466, miR-483, miR-483-5p, miR-485-3p, miR-485-5, miR-493-3p, miR-498, miR-500, miR-500a, miR-502-3p, miR-503-5p, miR-505-3p, miR-506-514 cluster, miR-512-5p, miR-513-1, miR-513-2, miR-514, miR-516a-3p, miR-517, miR-517b-3p, miR-518a, miR-518c, miR-519a, miR-520c-3p, miR-520f, miR-520g, miR-520h, miR-548a, miR-548ah, miR-548ar, miR-548b, miR-551b-3p, miR-551b-5p, miR-556-5p, miR-574-5p, miR-582-3p, miR-584-5p, miR-589, miR-592, miR-615-3p, miR-616, miR-618, miR-626, miR-628, miR-628-5p, miR-629-5p, miR-633, miR-635, miR-640, miR-642a-5p, miR-642b-3p, miR-645, miR-647, miR-657, miR-6741-3p, miR-675-3p, miR-711, miR-764, miR-767, miR-769, miR-857, miR-877-3p, miR-886-3p, miR-886-5p, miR-933, miR-938, miR-939, miR-1185-2-3p, miR-1234, miR-1247, miR-1248, miR-1254, miR-1269, miR-1273g-3p, miR-1275, miR-1293, miR-1303, miR-1307, miR-1323, miR-1537, miR-1537-3p, miR-1825, miR-1908, miR-1908-5p, miR-3065, miR-3131, miR-3136, miR-3141, miR-3144-3p, miR-3147, miR-3151, miR-3153, miR-3162, miR-3176, miR-3177-3p, miR-3189, miR-3200-5p, miR-3201, miR-3613-3p, miR-3917, miR-3976, miR-4267, miR-4270, miR-4283, miR-4289, miR-4418, miR-4429, miR-4465, miR-4484, miR-4644, miR-4652, miR-4664-3p, miR-4665-5p, miR-4709, miR-4764-3p, miR-5001-5p, miR-5100, miR-5191-3p, miR-5694, miR-6796-3p, miR-6826, miR-6852, miR-6875, miR-7641, miR-7702, let-5p, let-7b, let-7b-3p, let-7c-5p, let-7d-5p, let-7f-2, let-7f-5p, let-7g | miR-18a-5p, miR-18b-5p, miR-19a-3p, miR-20a-5p, miR-22-5p, miR-105-5p, miR-126-5p, miR-181a-5p, miR-223-5p, miR-323b-3p, miR-372-3p, miR-373, miR-455-3p, miR-488, miR-549, miR-582-5p, miR-585, miR-595, miR-627-5p, miR-671-5p, miR-762, miR-877-5p, miR-941, miR-1225-5p, miR-1249, miR-3613-5p, miR-4317, miR-4435, miR-iR-4293, let-7e-5p, mIR-17-92 cluster | miR-3p-57664, miR-7-1-3p, miR-7b-5p, miR-9-3p, miR-9a-3p, miR-10, miR-10a, miR-15a, miR-17-5p, miR-18b, miR-21a-5p, miR-25, miR-26a-2, miR-26b-3p, miR-27a-5p, miR-27b, miR-27b-5p, miR-30a-3p, miR-30a-5p, miR-30c-2-3p, miR-30d, miR-30e-5p, miR-32, miR-32-3p, miR-93a-5p, miR-99a, miR-99a-5p, miR-99b, miR-99b-5p, miR-101-3p, miR-101b-3p, miR-106-5p, miR-124a, miR-124-2, miR-124-3, miR-125a-5p, miR-125b-2, miR-128b, miR-130b, miR-133, miR-133a-1, miR-133a-2-3p, miR-134-5, miR-135b-3p, miR-136, miR-136-5p, miR-139, miR-142, miR-142a-5p, miR-145, miR-146b, miR-150, miR-152, miR-154-5p, miR-155-5p, miR-181a-3p, miR-181b-1-3p, miR-181c-3p, miR-184, miR-186-3p, miR-187, miR-191, miR-192-3p, miR-192-5p, miR-193b-5p, miR-194, miR-194-3p, miR-195, miR-195-3p, miR-196a, miR-199a, miR-199b, miR-200b, miR-200c, miR-203, miR-203-3p, miR-204-5p, miR-206, miR-208a-5p, miR-220c, miR-290, miR-211, miR-211-5p, miR-219a.2-3p, miR-219a-5p, miR-292-5p, miR-298, miR-300-3p, miR-302d-3p, miR-322, miR-323a-5p, miR-328-5p, miR-330-3p, miR-339, miR-339-3p, miR-340-3p, miR-345-3p, miR-362, miR-362-3p, miR-363-3p, miR-365a, miR-375, miR-376b-3p, miR-378c, miR-381, miR-382-3p, miR-424, miR-424-5p, miR-425, miR-433-3p, miR-434-3p, miR-451-5p, miR-454-3p, miR-455-5p, miR-486, miR-488-3p, miR-490-3p, miR-494, miR-495-3p, miR-499, miR-499a-3p, miR-499a-5p, miR-500a-3p, miR-505-5p, miR-509-3-5p, miR-513a-5p, miR-518f-3p, miR-519, miR-520d-3p, miR-520f-3p, miR-525-5p, miR-532, miR-532-3p, miR-545, miR-548at-5p, miR-548d, miR-550, miR-552, miR-553, miR-572, miR-579, miR-579-3p, miR-590-5p, miR-601, miR-602, miR-603, miR-611, miR-613, miR-615-5p, miR-617, miR-623, miR-627, miR-628-3p, miR-629-3p, miR-637, miR-638, miR-641, miR-656-3p, miR-659, miR-660, miR-660-5p, miR-663a, miR-665, miR-668, miR-671, miR-671-3p, miR-675, miR-682, miR-686, miR-762, miR-765, miR-874, miR-883a-3p, miR-883b-3p, miR-891, miR-920, miR-923 miR-942, miR-943, miR-1183, miR-1184, miR-1185-1-3p, miR-1202, miR-1224, miR-1246, miR-1249, miR-1255b, miR-1261, miR-1274a, miR-1274b, miR-1277-3p, miR-1285, miR-1285-5p, miR-1289, miR-1290, miR-1291, miR-1321, miR-1843a-5p, miR-2861, miR-3190-3p, miR-3195, miR-3196, miR-3615, miR-3646, miR-3928-5p, miR-3939, miR-4306, miR-4433b-3p, miR-4449, miR-4521, miR-4649-5p, miR-4669, miR-4674, miR-4747-3p, miR-5001-3p, miR-5010-3p, miR-6096-5p, miR-6716-3p, miR-6736-3p, miR-6740-3p, miR-6747-3p, miR-6750-5p, miR-6753-3p, miR-6754-3p, miR-6761-3p, miR-6762-3p, miR-6777-3p, miR-6778-3p, miR-6787-3p, miR-6836-3p, miR-6867-5p, miR-6875-3p, miR-8082, miR-B19527a, let-7-c-3p let-7e, let-7g-3p, miR-PC-5p-12969, miR-23-27-24 cluster |
| (b) | ||
| miRNAs Downregulated in Cancers Only | miRNAs Downregulated in Both Cancers and Neurological Disorders | miRNAs Downregulated in Neurological Disorders Only |
| miR-1-3p, miR-6-3p, miR-10, miR-15-5p, miR-15/16 cluster, miR-15a-3p, miR-16-1, miR-16.1, miR-23a-3p, miR-24-1-5p, miR-26, miR-26a-5p, miR-27a-5p, miR-29b, miR-29b-1-5p, miR-29c-5p, miR-30, miR-30b-3p, miR-30b-5p, miR-30c-2-3p, miR-33a-5p, miR-34b-3p, miR-34b-5p, miR-92, miR-92a cluster, miR-93-3p, miR-99-3b, miR-99b-5p, miR-103b, miR-104-5p, miR-122a, miR-124-5p, miR-126b, miR-127-3p, miR-128-1, miR-129-3p, miR-129b-5p, miR-130b-3p, miR-130b-5p, miR-132-5p, miR-133, miR-133a, miR-133a-3p, miR-134-5p, miR-135-5p, miR-136, miR-136-5p, miR-137, miR-138-5p, miR-145, miR-147, miR-147b, miR-148a-3p, miR-149-5p, miR-152, miR-153, miR-153-3p, miR-181a-3p, miR-181d-5p, miR-188, miR-193-3p, miR-193-5p, miR-193a, miR-196b-5p, miR-199-3p, miR-216-5p, miR-216a, miR-216a-3p, miR-216b, miR-217, miR-218-5p, miR-219a-2-3p, miR-220, miR-220a, miR-220b, miR-269-3p, miR-296-3p, miR-299, miR-302, miR-302a, miR-302b, miR-302c, miR-320a-3p, miR-323, miR-323-3p, miR-323-5p, miR-328, miR-328-3p, miR-330-3p, miR-330-3p-3p, miR-330-5p, miR-333-3p, miR-335-5p, miR-336, miR-337, miR-339-3p, miR-342, miR-362, miR-363, miR-365b-3p, miR-365b-5p, miR-374c, miR-374c-5p, miR-376, miR-376a-3p, miR-381-3p, miR-382-5p, miR-383-5p, miR-384, miR-385, miR-397a, miR-397b, miR-397c, miR-411-5p, miR-424-3p, miR-429-5p, miR-432, miR-433-3p, miR-448, miR-449b, miR-449c, miR-451, miR-451a.1, miR-454-3p, miR-485-5p, miR-486-3p, miR-487b, miR-488-3p, miR-488-5p, miR-489, miR-489-3p, miR-490-3p, miR-491, miR-491-3p, miR-491-5p, miR-497-5p, miR-506-3p, miR-508-3p, miR-509-5p, miR-509-3-5p, miR-511-5p, miR-513, miR-513c-5p, miR-514a-3p, miR-516b, miR-519, miR-520a-3p, miR-524-5p, miR-526b, miR-526b-5p, miR-532-5p, miR-539, miR-544, miR-545, miR-548b-5p, miR-548c-3p, miR-551b, miR-564, miR-573, miR-574, miR-579-3p, miR-582, miR-584, miR-590-3p, miR-593-3p, miR-596, miR-599, miR-601, miR-605, miR-610, miR-612, miR-613, miR-622, miR-625, miR-625-5p, miR-627, miR-628-3p, miR-632, miR-634, miR-636, miR-637, miR-639, miR-650, miR-652-5p, miR-654-5p, miR-655, miR-656, miR-661, miR-665, miR-708, miR-708-5p, miR-744, miR-744-5p, miR-760, miR-768-3p, miR-770-5p, miR-802, miR-874, miR-877, miR-886-3p, miR-887, miR-891a, miR-892b, miR-936, miR-937, miR-937-5p, miR-942, miR-1179, miR-1180-3p, miR-1182, miR-1203, miR-1205, miR-1226, miR-1227, miR-1228-5p, miR-1231, miR-1250, miR-1257, miR-1260, miR-1266, miR-1271, miR-1274a, miR-1280, miR-1283, miR-1287, miR-1296, miR-1301-3p, miR-1302, miR-1321, miR-1587, miR-1913, miR-2682-3p, miR-2861, miR-3065-5p, miR-3156-3p, miR-3170, miR-3185, miR-3196, miR-3612, miR-3614, miR-3651, miR-3662, miR-3666, miR-3714, miR-3928, miR-3960, miR-4259, miR-4282, miR-4324, miR-4430, miR-4458, miR-4467, miR-4478, miR-4485-5p, miR-4488, miR-4501, miR-4633-5p, miR-4635, miR-4647, miR-4706, miR-4730, miR-4731, miR-5096, miR-6086, miR-6503, miR-6510, miR-6511b-5p, miR-6887-5p, let-7a | miR-28-5p, miR-148-3p, miR-199b-3p, miR-208b, miR-219-5p, miR-320b, miR-323a-3p, miR-330, miR-374b, miR-449b-5p, miR-493-5p, miR-509-3p, miR-511, miR-575, miR-1297, miR-4487, let-7i-5p | miR-2-5p, miR-7, miR-7c-5p, miR-7g, miR-9a-5p, miR-15, miR-15b, miR-15b-3p, miR-15b-5p, miR-16-2-3p, miR-16-5p, miR-17-3p, miR-18a, miR-19b-5p, miR-22-5p, miR-23b, miR-23b-3p, miR-26a, miR-26a-1, miR-26a-2-5p, miR-26b-5p, miR-27a-3p, miR-29, miR-30c, miR-31-5p, miR-33b-5p, miR-92a-5p, miR-92b, miR-96, miR-96-5p, miR-98, miR-98-5p, miR-101, miR-103, miR-106a-5, miR-106b-3p, miR-112, miR-122, miR-125, miR-125a, miR-125a-3p, miR-125b-1-3p, miR-126-3p, miR-128-2-5p, miR-130a, miR-130-b-5p, miR-134-3p, miR-139-3p, miR-140, miR-146-5p, miR-146a-3p, miR-146b-5p, miR-148b, miR-148b-3p, miR-148b-5p, miR-149, miR-150-3p, miR-151-3p, miR-151b, miR-161, miR-181-3p, miR-181-5p, miR-182, miR-182-5p, miR-183, miR-183-5p, miR-186, miR-186-5p, miR-187-3p, miR-187-5p, miR-190, miR-192, miR-193a-5p, miR-193b, miR-199a-5p, miR-203a-3p, miR-203b-5p, miR-204, miR-208a, miR-208b-3p, miR-212, miR-212-3p, miR-219, miR-221-5p, miR-298-5p, miR-299-5p, miR-301, miR-320, miR-320a, miR-320c, miR-320e, miR-325-5p, miR-331-3p, miR-335-3p, miR-337-5p, miR-340, miR-340-5p, miR-342-3p, miR-351-3p, miR-352, miR-362-5p, miR-370-3p, miR-374a, miR-374b-5p, miR-379, miR-380, miR-423-3p, miR-423-5p, miR-425-p, miR-433, miR-483-3p, miR-485, miR-486-1, miR-487a, miR-487b-3p, miR-493, miR-500, miR-501-5p, miR-502, miR-502-3p, miR-502-5p, miR-519e, miR-543, miR-545-3p, miR-574-3p, miR-576-5p, miR-598, miR-598-3p, miR-605-5p, miR-626, miR-629, miR-652, miR-669c-5p, miR-744, miR-744-3p, miR-764-3p, miR-767, miR-769, miR-769-5p, miR-873-3p, miR-877-3p, miR-885-3p, miR-885-5p, miR-886-5p, miR-1185-2-3p, miR-1224-5p, miR-1233-5p, miR-1234-3p, miR-1237-5p, miR-1249-3p, miR-1259, miR-1294, miR-1299, miR-1303-3p, miR-1306-3p, miR-1825, miR-1909-3p, miR-1915-3p, miR-3552, miR-3613-3p, miR-3916, miR-3935, miR-4299, miR-4322, miR-4422, miR-4429, miR-4448, miR-4530, miR-4745-5p, miR-4772-3p, miR-6131, miR-6236, miR-6805-5p, miR-6781-5p, miR-7046-3p, miR-7660-3p, miR-7665-5p, miR-7674-5p, miR-8071, let-7a-5p, let-7c, let-7g |
| Clinical Trial NCT Number | Aims of Clinical Trial | Phase | Start and End Date | Status | Results |
|---|---|---|---|---|---|
| NCT04120493 | Establish safety and proof-of-concept of AMT-130 in patients with early-stage HD. | Interventional (Phase I/II) | 6 September 2019–June 2019 (estimated) | Recruiting | None |
| NCT05243017 | Examine safety and efficacy of AMT-130 in patients with HD. | Interventional (Phase I/II) | 7 October 2021–7 October 2027 (estimated) | Recruiting | None |
| NCT06100276 | Examine safety, tolerability, and efficacy of AMT-162 in patients with ALS. | Interventional (phase 1/II) | 1 Febuary 2024–27 Febuary 2032 (estimated) | Not Recruiting Yet | None |
| NCT06063850 | Examine safety, tolerability, and efficacy of AMT-260 in patients with unilateral refractory mesial temporal lobe epilepsy. | Interventional (phase I/II) | 17 November 2023–30 November 2026 (estimated) | Recruiting | None |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lui, A.; Do, T.; Alzayat, O.; Yu, N.; Phyu, S.; Santuya, H.J.; Liang, B.; Kailash, V.; Liu, D.; Inslicht, S.S.; et al. Tumor Suppressor MicroRNAs in Clinical and Preclinical Trials for Neurological Disorders. Pharmaceuticals 2024, 17, 426. https://doi.org/10.3390/ph17040426
Lui A, Do T, Alzayat O, Yu N, Phyu S, Santuya HJ, Liang B, Kailash V, Liu D, Inslicht SS, et al. Tumor Suppressor MicroRNAs in Clinical and Preclinical Trials for Neurological Disorders. Pharmaceuticals. 2024; 17(4):426. https://doi.org/10.3390/ph17040426
Chicago/Turabian StyleLui, Austin, Timothy Do, Omar Alzayat, Nina Yu, Su Phyu, Hillary Joy Santuya, Benjamin Liang, Vidur Kailash, Dewey Liu, Sabra S. Inslicht, and et al. 2024. "Tumor Suppressor MicroRNAs in Clinical and Preclinical Trials for Neurological Disorders" Pharmaceuticals 17, no. 4: 426. https://doi.org/10.3390/ph17040426
APA StyleLui, A., Do, T., Alzayat, O., Yu, N., Phyu, S., Santuya, H. J., Liang, B., Kailash, V., Liu, D., Inslicht, S. S., Shahlaie, K., & Liu, D. (2024). Tumor Suppressor MicroRNAs in Clinical and Preclinical Trials for Neurological Disorders. Pharmaceuticals, 17(4), 426. https://doi.org/10.3390/ph17040426







