Approaches, Strategies and Procedures for Identifying Anti-Inflammatory Drug Lead Molecules from Natural Products
Abstract
1. Introduction
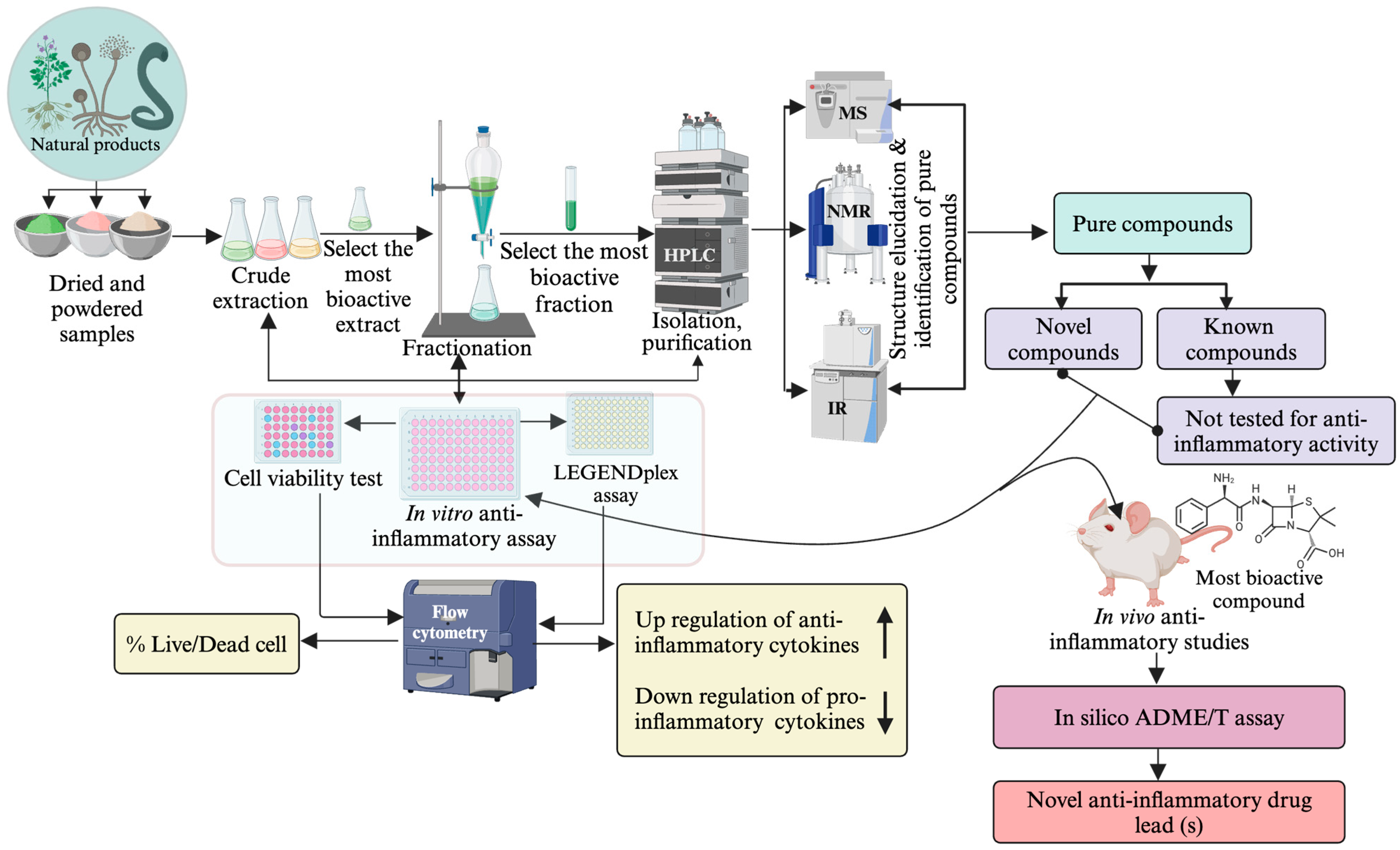
2. Approaches and Techniques for the Biodiscovery of Anti-Inflammatory Drug Leads
2.1. Approaches
2.1.1. Biorational Approach
Ethnopharmacology Approach
The Ecological Approach
2.1.2. Chemorational Approach
2.1.3. Random Approach/Find-and-Grind Approach
2.2. Recent Advances in Anti-Inflammatory Drug Discovery Approaches
2.2.1. Computer-Aided Drug Discovery
2.2.2. Artificial Intelligence (AI) in Drug Discovery
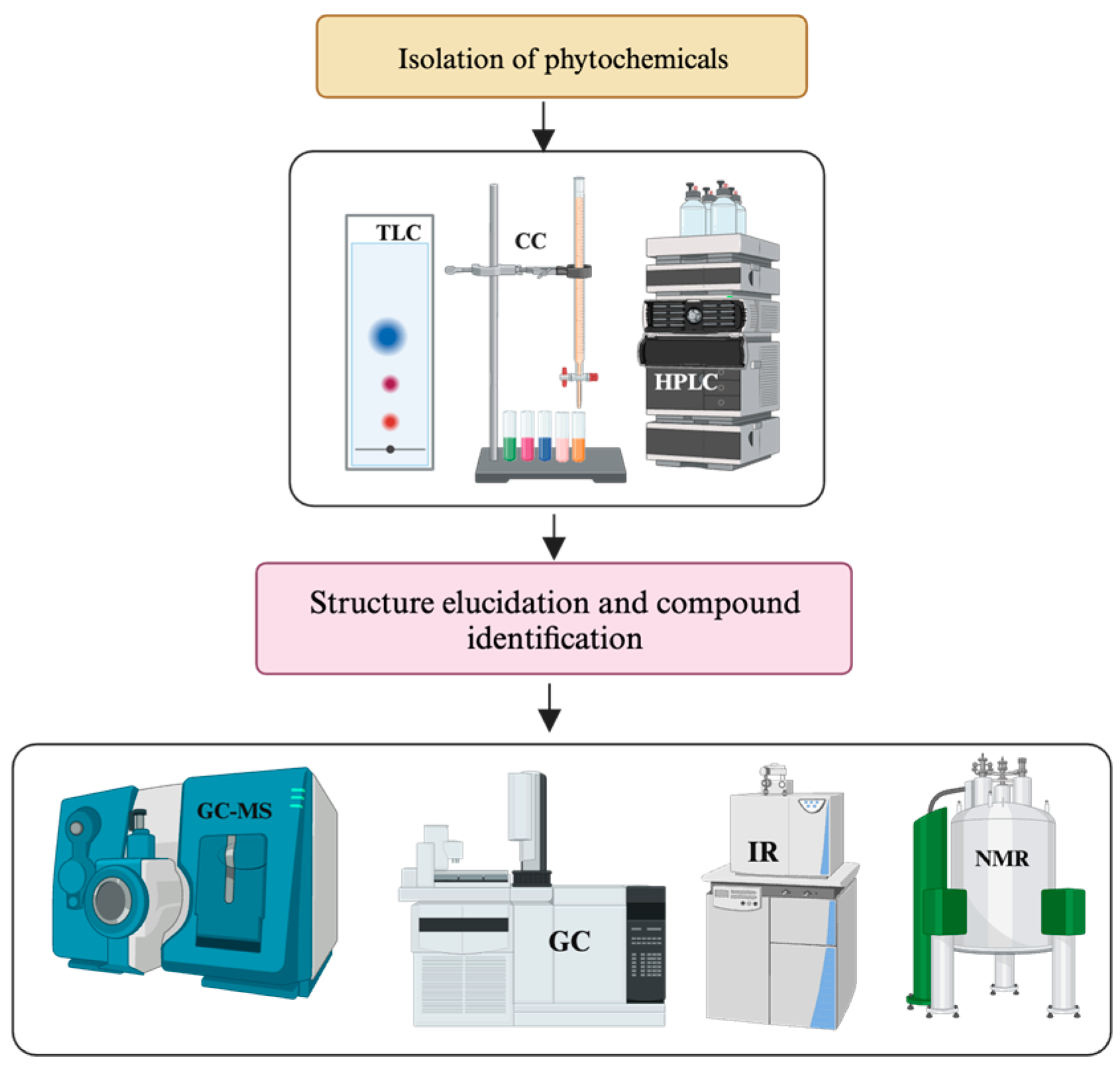
2.3. Methods for Extracting, Purifying, and Determining the Structures of Anti-Inflammatory SMs from NPs
2.3.1. Extraction and Fractionation
2.3.2. Phytochemical Screening
2.3.3. Isolation of Anti-Inflammatory SMs (Bioactivity-Guided)
Thin Layer Chromatography/Preparative TLC
Column Chromatography
High-Pressure Liquid Chromatography (HPLC)
Ultra-Performance Liquid Chromatography (UPLC)
2.3.4. Identification of SMs and Structure Elucidation of Novel Molecules
Nuclear Magnetic Resonance (NMR) Spectroscopy
Liquid Chromatography-Mass Spectrometry (LC-MS/LC-MS/MS)
UV-Visible Spectroscopy
Infrared Spectroscopy (IR)
High-Resolution Mass Spectrometry (HRMS)
3. Screening Isolated Compounds for Anti-Inflammatory Properties
3.1. In Vitro Assays for Anti-Inflammatory Screening of SMs and Crude Extracts
3.1.1. Human Leukemia Monocytic Cell Line (THP-1)
3.1.2. Caco-2 Cell Line
3.1.3. Human Colorectal Adenocarcinoma Cell Line (HT29)
3.1.4. The murine Macrophage Cell Line (RAW 264.7)
3.1.5. Peripheral Blood Mononuclear Cells (PBMCs) Assay
3.2. In-Vivo Experimental Models for Anti-Inflammatory Screening
3.2.1. 2 2,4,6-Trinitrobenzene Sulfonic Acid (TNBS)-Induced Colitis Model
3.2.2. Dextran Sodium Sulfate (DSS)-Induced Colitis Model
3.2.3. Oxazolone ((4-Ethoxylmethylene-2-Phenyloxazol-5-One)-Induced Colitis Model (OC))
3.2.4. Winnie Mouse Model of Colitis
3.2.5. T Cell Adoptive Transfer Model
4. Conclusions and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ramos, A.E.F.; Evanno, L.; Poupon, E.; Champy, P.; Beniddir, M.A. Natural products targeting strategies involving molecular networking: Different manners, one goal. Nat. Prod. Rep. 2019, 36, 960–980. [Google Scholar] [CrossRef] [PubMed]
- Butler, M.S.; Robertson, A.A.; Cooper, M.A. Natural product and natural product derived drugs in clinical trials. Nat. Prod. Rep. 2014, 31, 1612–1661. [Google Scholar] [CrossRef] [PubMed]
- Mishra, B.B.; Tiwari, V.K. Natural products: An evolving role in future drug discovery. Eur. J. Med. 2011, 46, 4769–4807. [Google Scholar] [CrossRef] [PubMed]
- Butler, M.S. The role of natural product chemistry in drug discovery. J. Nat. Prod. 2004, 67, 2141–2153. [Google Scholar] [CrossRef] [PubMed]
- Marderosian, A.; Beutler, J. The Review of Natural Products; Facts and Comparisons: St. Louis, MO, USA, 2002. [Google Scholar]
- Varela, M.L.; Mogildea, M.; Moreno, I.; Lopes, A. Acute inflammation and metabolism. Inflammation 2018, 41, 1115–1127. [Google Scholar] [CrossRef] [PubMed]
- Serhan, C.N.; Ward, P.A.; Gilroy, D.W. Fundamentals of Inflammation; Cambridge University Press: Cambridge, UK, 2010. [Google Scholar]
- Roy, S.; Bagchi, D.; Raychaudhuri, S.P. Chronic Inflammation: Molecular Pathophysiology, Nutritional and Therapeutic Interventions; CRC Press: Boca Raton, FL, USA, 2012. [Google Scholar]
- Olajide, O.A.; Sarker, S.D. Anti-inflammatory natural products. Annu. Rep. Med. Chem. 2020, 55, 153–177. [Google Scholar]
- Schoultz, I.; Söderholm, J.D.; McKay, D.M. Is metabolic stress a common denominator in inflammatory bowel disease? Inflamm. Bowel Dis. 2011, 17, 2008–2018. [Google Scholar] [CrossRef]
- Horiuchi, T.; Mitoma, H.; Harashima, S.-I.; Tsukamoto, H.; Shimoda, T. Transmembrane TNF-α: Structure, function and interaction with anti-TNF agents. Rheumatology 2010, 49, 1215–1228. [Google Scholar] [CrossRef]
- Yeshi, K.; Turpin, G.; Jamtsho, T.; Wangchuk, P. Indigenous uses, phytochemical analysis, and anti-inflammatory properties of Australian tropical medicinal plants. Molecules 2022, 27, 3849. [Google Scholar] [CrossRef]
- Newman, D.J.; Cragg, G.M. Natural products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019. Nat. Prod. Rep. 2020, 83, 770–803. [Google Scholar] [CrossRef]
- de la Torre, B.G.; Albericio, F. The pharmaceutical industry in 2019: An analysis of FDA drug approvals from the perspective of molecules. Molecules 2020, 25, 745. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Song, J.; Kong, L.; Yuan, T.; Li, W.; Zhang, W.; Hou, B.; Lu, Y.; Du, G. The strategies and techniques of drug discovery from natural products. Pharmacol. Ther. 2020, 216, 107686. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-F. Historical story on natural medicinal chemistry: The most preeminent natural organic chemist of 20th century-Robert Burns Woodward. Chin. Tradit. Herb. Drugs 2017, 24, 1484–1498. [Google Scholar]
- Li, Z.; Chen, K.; Rose, P.; Zhu, Y.Z. Natural products in drug discovery and development: Synthesis and medicinal perspective of leonurine. Front. Chem. 2022, 10, 1036329. [Google Scholar] [CrossRef] [PubMed]
- Mak, K.-K.; Pichika, M.R. Artificial intelligence in drug development: Present status and future prospects. Drug Discov. Today 2019, 24, 773–780. [Google Scholar] [CrossRef] [PubMed]
- Paul, D.; Sanap, G.; Shenoy, S.; Kalyane, D.; Kalia, K.; Tekade, R.K. Artificial intelligence in drug discovery and development. Drug Discov. Today 2021, 26, 80–93. [Google Scholar] [CrossRef]
- Harvey, A.L. Medicines from nature: Are natural products still relevant to drug discovery? Trends Pharmacol. Sci. 1999, 20, 196–198. [Google Scholar] [CrossRef]
- Strohl, W.R. The role of natural products in a modern drug discovery program. Drug Discov. Today 2000, 5, 39–41. [Google Scholar] [CrossRef]
- Fabricant, D.S.; Farnsworth, N.R. The value of plants used in traditional medicine for drug discovery. Environ. Health Perspect. 2001, 109 (Suppl. S1), 69–75. [Google Scholar] [CrossRef]
- Huie, C.W. A review of modern sample-preparation techniques for the extraction and analysis of medicinal plants. Anal. Bioanal. Chem. 2002, 373, 23–30. [Google Scholar] [CrossRef]
- Wang, B.; Ma, L.; Tao, X.; Lipsky, P. Triptolide, an active component of the Chinese herbal remedy Tripterygium wilfordii Hook F, inhibits production of nitric oxide by decreasing inducible nitric oxide synthase gene transcription. Arthritis Rheumatol. 2004, 50, 2995–3003. [Google Scholar] [CrossRef] [PubMed]
- Peyrin-Biroulet, L.; Loftus, E.V., Jr.; Colombel, J.F.; Sandborn, W.J. The natural history of adult Crohn’s disease in population-based cohorts. Am. J. Gastroenterol. 2010, 105, 289–297. [Google Scholar] [CrossRef] [PubMed]
- Joo, Y.-E. Natural product-derived drugs for the treatment of inflammatory bowel diseases. Intest. Res. 2014, 12, 103–109. [Google Scholar] [CrossRef] [PubMed]
- Na, S.Y.; Moon, W. Perspectives on Current and Novel Treatments for Inflammatory Bowel Disease. Gut Liver 2019, 13, 604–616. [Google Scholar] [CrossRef]
- Busingye, D.; Pollack, A.; Chidwick, K. Prevalence of inflammatory bowel disease in the Australian general practice population: A cross-sectional study. PLoS ONE 2021, 16, e0252458. [Google Scholar] [CrossRef]
- Tammam, M.A.; Daskalaki, M.G.; Tsoureas, N.; Kolliniati, O.; Mahdy, A.; Kampranis, S.C.; Tsatsanis, C.; Roussis, V.; Ioannou, E. Secondary Metabolites with Anti-Inflammatory Activity from Laurencia majuscula Collected in the Red Sea. Mar. Drugs 2023, 21, 79. [Google Scholar] [CrossRef]
- Zhou, Y.; Wang, D.; Yan, W. Treatment effects of natural products on inflammatory bowel disease in vivo and their mechanisms: Based on animal experiments. Nutrients 2023, 15, 1031. [Google Scholar] [CrossRef]
- Dzobo, K. The role of natural products as sources of therapeutic agents for innovative drug discovery. J. Compr. Pharm. 2022, 408–422. [Google Scholar] [CrossRef]
- Nasim, N.; Sandeep, I.S.; Mohanty, S. Plant-derived natural products for drug discovery: Current approaches and prospects. Nucleus 2022, 65, 399–411. [Google Scholar] [CrossRef]
- Wangchuk, P.; Apte, S.H.; Smout, M.J.; Groves, P.L.; Loukas, A.; Doolan, D.L. Defined small molecules produced by Himalayan medicinal plants display Immunomodulatory properties. Int. J. Mol. Sci. 2018, 19, 3490. [Google Scholar] [CrossRef]
- Wangchuk, P.; Loukas, A. Techniques and Technologies for the Biodiscovery of Novel Small Molecule Drug Lead Compounds from Natural Products. In Natural Products and Drug Discovery; Elsevier: Amsterdam, The Netherlands, 2018; pp. 435–465. [Google Scholar]
- Anwar, N.; Teo, Y.K.; Tan, J.B.L. The Role of Plant Metabolites in Drug Discovery: Current Challenges and Future Perspectives. In Natural Bio-active Compounds; Springer: Berlin/Heidelberg, Germany, 2019; Volume 2, pp. 25–51. [Google Scholar] [CrossRef]
- Heinrich, M. 3.12—Ethnopharmacology and Drug Discovery. In Comprehensive Natural Products II; Liu, H.-W., Mander, L., Eds.; Elsevier: Oxford, UK, 2010; pp. 351–381. [Google Scholar]
- Gempo, N.; Yeshi, K.; Jamtsho, T.; Jamtsho, L.; Samten; Wangchuk, P. Development of quality control parameters for two Bhutanese medicinal plants (Aster flaccidus Bunge and Aster diplostephioides (DC.) Benth. ex C.B.Clarke) using traditional and modern pharmacognostical platforms. Heliyon 2024, 10, e24969. [Google Scholar] [CrossRef] [PubMed]
- Wangchuk, P.; Samten; Jamtsho, T. Phytopharmaceutical properties and quality assessment of two Himalayan medicinal plants, Meconopsis horridula and Meconopsis simplicifolia. J. Herb. Med. 2023, 38, 100628. [Google Scholar] [CrossRef]
- Thompson, J.B.; Hawkins, J.A. Ethnopharmacological disease classification and bioprospecting: The diversity of plant drugs used to treat cancer. bioRxiv 2023. [Google Scholar] [CrossRef]
- Saha, D.; Sarankar, S.K. Critical review on potentials of ethnopharmacological, ethnomedicinal and traditional practices of Madhuca longifolia (J. Koenig Ex L.) JF Macbr.(Family: Sapotaceae). Pharm. Sci. 2023, 21, 30–36. [Google Scholar] [CrossRef]
- Pirintsos, S.; Panagiotopoulos, A.; Bariotakis, M.; Daskalakis, V.; Lionis, C.; Sourvinos, G.; Karakasiliotis, I.; Kampa, M.; Castanas, E. From Traditional Ethnopharmacology to Modern Natural Drug Discovery: A Methodology Discussion and Specific Examples. Molecules 2022, 27, 4060. [Google Scholar] [CrossRef] [PubMed]
- Arya, A.; Kumar, S. Medicinal Plants: Ethnomedicine, Pharmacognosy and Therapeutic Values, 1st ed.; Sharma, G.M.A., Kumar, S., Eds.; Anu Book Publisher and Distributors: New Delhi, India, 2022; p. 23. [Google Scholar]
- Tang, W.; Zuo, J.-P. Immunosuppressant discovery from Tripterygium wilfordii Hook f: The novel triptolide analog (5R)-5-hydroxytriptolide (LLDT-8). Acta Pharmacol. Sin. 2012, 33, 1112–1118. [Google Scholar] [CrossRef] [PubMed]
- Kotha, R.R.; Luthria, D.L. Curcumin: Biological, pharmaceutical, nutraceutical, and analytical aspects. Molecules 2019, 24, 2930. [Google Scholar] [CrossRef]
- Vecchi Brumatti, L.; Marcuzzi, A.; Tricarico, P.M.; Zanin, V.; Girardelli, M.; Bianco, A.M. Curcumin and inflammatory bowel disease: Potential and limits of innovative treatments. Molecules 2014, 19, 21127–21153. [Google Scholar] [CrossRef]
- Süntar, I. Importance of ethnopharmacological studies in drug discovery: Role of medicinal plants. Phytochem. Rev. 2020, 19, 1199–1209. [Google Scholar] [CrossRef]
- Kumar, P.; Mathew, S.; Gamage, R.; Bodkin, F.; Doyle, K.; Rossetti, I.; Wagnon, I.; Zhou, X.; Raju, R.; Gyengesi, E. From the Bush to the Brain: Preclinical Stages of Ethnobotanical Anti-Inflammatory and Neuroprotective Drug Discovery—An Australian Example. Int. J. Mol. Sci. 2023, 24, 11086. [Google Scholar] [CrossRef]
- Shilling, A.J. The Chemical Ecology and Drug Discovery Potential of the Antarctic Red Alga Plocamium cartilagineum and the Antarctic Sponge Dendrilla membranosa. Ph.D. Thesis, University of South Florida, Tampa, FL, USA, 2019. [Google Scholar]
- Adegboye, O.; Field, M.A.; Kupz, A.; Pai, S.; Sharma, D.; Smout, M.J.; Wangchuk, P.; Wong, Y.; Loiseau, C. Natural-Product-Based Solutions for Tropical Infectious Diseases. Clin. Microbiol. Rev. 2021, 34, e0034820. [Google Scholar] [CrossRef]
- Atanasov, A.G.; Waltenberger, B.; Pferschy-Wenzig, E.-M.; Linder, T.; Wawrosch, C.; Uhrin, P.; Temml, V.; Wang, L.; Schwaiger, S.; Heiss, E.H. Discovery and resupply of pharmacologically active plant-derived natural products: A review. Biotechnol. Adv. 2015, 33, 1582–1614. [Google Scholar] [CrossRef]
- Coley, P.D.; Kursor, T.A. Anti-Herbivore Defenses of Young Tropical Leaves: Physiological Constraints and Ecological Trade-offs. In Tropical Forest Plant Ecophysiology; Springer: Berlin/Heidelberg, Germany, 1996; pp. 305–336. [Google Scholar]
- Coley, P.D.; Heller, M.V.; Aizprua, R.; Araúz, B.; Flores, N.; Correa, M.; Gupta, M.; Solis, P.N.; Ortega-Barría, E.; Romero, L.I.; et al. Using Ecological Criteria to Design Plant Collection Strategies for Drug Discovery. Front. Ecol. Environ. 2003, 1, 421–428. [Google Scholar] [CrossRef]
- Yeshi, K.; Ruscher, R.; Miles, K.; Crayn, D.; Liddell, M.; Wangchuk, P. Antioxidant and Anti-Inflammatory Activities of Endemic Plants of the Australian Wet Tropics. Plants 2022, 11, 2519. [Google Scholar] [CrossRef] [PubMed]
- Ritmejerytė, E.; Ryan, R.Y.M.; Byatt, B.J.; Peck, Y.; Yeshi, K.; Daly, N.L.; Zhao, G.; Crayn, D.; Loukas, A.; Pyne, S.G.; et al. Anti-inflammatory properties of novel galloyl glucosides isolated from the Australian tropical plant Uromyrtus metrosideros. Chem.-Biol. Interact. 2022, 368, 110124. [Google Scholar] [CrossRef] [PubMed]
- Trefzer, R.; Elpeleg, O.; Gabrusskaya, T.; Stepensky, P.; Mor-Shaked, H.; Grosse, R.; Brandt, D.T. Characterization of a L136P mutation in Formin-like 2 (FMNL2) from a patient with chronic inflammatory bowel disease. PLoS ONE 2021, 16, e0252428. [Google Scholar] [CrossRef] [PubMed]
- Wangchuk, P.; Yeshi, K.; Loukas, A. Metabolomics and lipidomics studies of parasitic helminths: Molecular diversity and identification levels achieved by using different characterisation tools. Metabolomics 2023, 19, 63. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Sarkar, T.; Pati, S.; Kari, Z.A.; Edinur, H.A.; Chakraborty, R. Novel bioactive compounds from marine sources as a tool for functional food development. Front. Mar. Sci. 2022, 9, 832957. [Google Scholar] [CrossRef]
- Beattie, A.J.; Hay, M.; Magnusson, B.; de Nys, R.; Smeathers, J.; Vincent, J.F. Ecology and bioprospecting. Austral Ecol. 2011, 36, 341–356. [Google Scholar] [CrossRef] [PubMed]
- Cordell, G.A.; Quinn-Beattie, M.L.; Farnsworth, N.R. The potential of alkaloids in drug discovery. Phytother. Res. 2001, 15, 183–205. [Google Scholar] [CrossRef]
- Pelletier, S.W. Chemistry of the Alkaloids; Pelletier, S.W., Ed.; Van Nostrand Reinhold: New York, NY, USA, 1970. [Google Scholar]
- Katiyar, C.; Gupta, A.; Kanjilal, S.; Katiyar, S. Drug discovery from plant sources: An integrated approach. Ayu 2012, 33, 10–19. [Google Scholar] [CrossRef]
- Sadybekov, A.V.; Katritch, V. Computational approaches streamlining drug discovery. Nature 2023, 616, 673–685. [Google Scholar] [CrossRef]
- Sun, D.; Gao, W.; Hu, H.; Zhou, S. Why 90% of clinical drug development fails and how to improve it? Acta Pharm. Sin. 2022, 12, 3049–3062. [Google Scholar] [CrossRef]
- Powrie, F.; Leach, M.W.; Mauze, S.; Menon, S.; Caddle, L.B.; Coffman, R.L. Inhibition of Thl responses prevents inflammatory bowel disease in scid mice reconstituted with CD45RBhi CD4+ T cells. Immunity 1994, 1, 553–562. [Google Scholar] [CrossRef] [PubMed]
- Johnson, T.O.; Akinsanmi, A.O.; Ejembi, S.A.; Adeyemi, O.E.; Oche, J.-R.; Johnson, G.I.; Adegboyega, A.E. Modern drug discovery for inflammatory bowel disease: The role of computational methods. World J. Gastroenterol. 2023, 29, 310–331. [Google Scholar] [CrossRef] [PubMed]
- Baig, M.H.; Ahmad, K.; Rabbani, G.; Danishuddin, M.; Choi, I. Computer aided drug design and its application to the development of potential drugs for neurodegenerative disorders. Curr. Neuropharmacol. 2018, 16, 740–748. [Google Scholar] [CrossRef] [PubMed]
- Sliwoski, G.; Kothiwale, S.; Meiler, J.; Edward, W.; Lowe, J. Computational Methods in Drug Discovery. Pharmacol. Rev. 2014, 66, 334–395. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, L.G.; Dos Santos, R.N.; Oliva, G.; Andricopulo, A.D. Molecular docking and structure-based drug design strategies. Molecules 2015, 20, 13384–13421. [Google Scholar] [CrossRef] [PubMed]
- Geronikaki, A.A.; Lagunin, A.A.; Hadjipavlou-Litina, D.I.; Eleftheriou, P.T.; Filimonov, D.A.; Poroikov, V.V.; Alam, I.; Saxena, A.K. Computer-aided discovery of anti-inflammatory thiazolidinones with dual cyclooxygenase/lipoxygenase inhibition. J. Med. Chem. 2008, 51, 1601–1609. [Google Scholar] [CrossRef] [PubMed]
- Liaras, K.; Fesatidou, M.; Geronikaki, A. Thiazoles and thiazolidinones as COX/LOX inhibitors. Molecules 2018, 23, 685. [Google Scholar] [CrossRef]
- Jukič, M.; Bren, U. Machine learning in antibacterial drug design. Front. Pharmacol. 2022, 13, 864412. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.; Srivastava, D.; Sahu, M.; Tiwari, S.; Ambasta, R.K.; Kumar, P. Artificial intelligence to deep learning: Machine intelligence approach for drug discovery. Mol. Divers. 2021, 25, 1315–1360. [Google Scholar] [CrossRef]
- Najmi, A.; Javed, S.A.; Al Bratty, M.; Alhazmi, H.A. Modern Approaches in the Discovery and Development of Plant-Based Natural Products and Their Analogues as Potential Therapeutic Agents. Molecules 2022, 27, 349. [Google Scholar] [CrossRef] [PubMed]
- Cragg, G.M.; Newman, D.J. Natural products: A continuing source of novel drug leads. Biochim. Biophys. Acta Gen. Subj. 2013, 1830, 3670–3695. [Google Scholar] [CrossRef]
- Madhukar, N.S.; Khade, P.K.; Huang, L.; Gayvert, K.; Galletti, G.; Stogniew, M.; Allen, J.E.; Giannakakou, P.; Elemento, O. A Bayesian machine learning approach for drug target identification using diverse data types. Nat. Commun. 2019, 10, 5221. [Google Scholar] [CrossRef]
- Bruderer, S.; Paruzzo, F.; Bolliger, C. Deep Learning-Based Phase and Baseline Correction of 1D 1H NMR Spectra. Public Bruker White Paper. 2021. Available online: https://www.bruker.com/en/products-and-solutions/mr/nmr-software/topspin.html (accessed on 16 July 2023).
- Hou, T.; Zhang, W.; Xia, K.; Qiao, X.; Xu, X. ADME evaluation in drug discovery. 5. Correlation of Caco-2 permeation with simple molecular properties. J. Chem. Inf. Comput. 2004, 44, 1585–1600. [Google Scholar] [CrossRef]
- Sakiyama, Y. The use of machine learning and nonlinear statistical tools for ADME prediction. Expert Opin. Drug Metab. Toxicol. 2009, 5, 149–169. [Google Scholar] [CrossRef]
- Trotter, M.W.; Holden, S.B. Support vector machines for ADME property classification. Qsar Comb. Sci. 2003, 22, 533–548. [Google Scholar] [CrossRef]
- Tao, L.; Zhang, P.; Qin, C.; Chen, S.Y.; Zhang, C.; Chen, Z.; Zhu, F.; Yang, S.Y.; Wei, Y.Q.; Chen, Y.Z. Recent progresses in the exploration of machine learning methods as in-silico ADME prediction tools. Adv. Drug Deliv. Rev. 2015, 86, 83–100. [Google Scholar] [CrossRef]
- Selvaraj, C.; Chandra, I.; Singh, S.K. Artificial intelligence and machine learning approaches for drug design: Challenges and opportunities for the pharmaceutical industries. Mol. Divers. 2022, 26, 1893–1913. [Google Scholar] [CrossRef]
- Howarth, A.; Ermanis, K.; Goodman, J.M. DP4-AI automated NMR data analysis: Straight from spectrometer to structure. Chem. Sci. 2020, 11, 4351–4359. [Google Scholar] [CrossRef]
- Huber, F.; van der Burg, S.; van der Hooft, J.J.; Ridder, L. MS2DeepScore: A novel deep learning similarity measure to compare tandem mass spectra. J. Cheminform. 2021, 13, 84. [Google Scholar] [CrossRef]
- Newman, D.J.; Cragg, G.M. Natural products as sources of new drugs from 1981 to 2014. J. Nat. Prod. 2016, 79, 629–661. [Google Scholar] [CrossRef]
- Premarathna, A.D.; Tuvikene, R.; Somasiri, M.; De Silva, M.; Adhikari, R.; Ranahewa, T.; Wijesundara, R.; Wijesekera, S.; Dissanayake, I.; Wangchuk, P. A novel therapeutic effect of mannitol-rich extract from the brown seaweed Sargassum ilicifolium using in vitro and in vivo models. BMC Complement. Med. Ther. 2023, 23, 26. [Google Scholar]
- Wangchuk, P.; Anderson, D.; Yeshi, K.; Loukas, A. Identification of small molecules of the infective stage of human hookworm using LCMS-based metabolomics and lipidomics protocols. ACS Infect. Dis. 2021, 7, 3264–3276. [Google Scholar] [CrossRef] [PubMed]
- Wangchuk, P.; Samten. GC-FID coupled GC-MS analysis of the essential oil and the recorded biological activities of Meconopsis simplicifolia. J. Biol. Act. Prod. Nat. 2015, 5, 365–372. [Google Scholar]
- Zhang, Q.-W.; Lin, L.-G.; Ye, W.-C. Techniques for extraction and isolation of natural products: A comprehensive review. Chin. Med. 2018, 13, 20. [Google Scholar] [CrossRef] [PubMed]
- Osorio-Tobón, J.F. Recent advances and comparisons of conventional and alternative extraction techniques of phenolic compounds. J. Food Sci. Technol. 2020, 57, 4299–4315. [Google Scholar] [CrossRef]
- Liu, S.-C.; Lin, J.-T.; Wang, C.-K.; Chen, H.-Y.; Yang, D.-J. Antioxidant properties of various solvent extracts from lychee (Litchi chinenesis Sonn.) flowers. Food Chem. 2009, 114, 577–581. [Google Scholar] [CrossRef]
- Abubakar, A.R.; Haque, M. Preparation of medicinal plants: Basic extraction and fractionation procedures for experimental purposes. J. Pharm. Bioallied Sci. 2020, 12, 1–10. [Google Scholar] [CrossRef]
- El Maaiden, E.; Bouzroud, S.; Nasser, B.; Moustaid, K.; El Mouttaqi, A.; Ibourki, M.; Boukcim, H.; Hirich, A.; Kouisni, L.; El Kharrassi, Y. A Comparative Study between Conventional and Advanced Extraction Techniques: Pharmaceutical and Cosmetic Properties of Plant Extracts. Molecules 2022, 27, 2074. [Google Scholar] [CrossRef]
- Cesari, I.; Hoerlé, M.; Simoes-Pires, C.; Grisoli, P.; Queiroz, E.F.; Dacarro, C.; Marcourt, L.; Moundipa, P.F.; Carrupt, P.A.; Cuendet, M.; et al. Anti-inflammatory, antimicrobial and antioxidant activities of Diospyros bipindensis (Gürke) extracts and its main constituents. J. Ethnopharmacol. 2013, 146, 264–270. [Google Scholar] [CrossRef] [PubMed]
- Brusotti, G.; Cesari, I.; Frassà, G.; Grisoli, P.; Dacarro, C.; Caccialanza, G. Antimicrobial properties of stem bark extracts from Phyllanthus muellerianus (Kuntze) Excell. J. Ethnopharmacol. 2011, 135, 797–800. [Google Scholar] [CrossRef] [PubMed]
- Cos, P.; Vlietinck, A.J.; Berghe, D.V.; Maes, L. Anti-infective potential of natural products: How to develop a stronger in vitro ‘proof-of-concept’. J. Ethnopharmacol. 2006, 106, 290–302. [Google Scholar] [CrossRef] [PubMed]
- Pintać, D.; Majkić, T.; Torović, L.; Orčić, D.; Beara, I.; Simin, N.; Mimica–Dukić, N.; Lesjak, M. Solvent selection for efficient extraction of bioactive compounds from grape pomace. Ind. Crops Prod. 2018, 111, 379–390. [Google Scholar] [CrossRef]
- Sasidharan, S.; Chen, Y.; Saravanan, D.; Sundram, K.; Latha, L.Y. Extraction, isolation and characterization of bioactive compounds from plants’ extracts. Afr. J. Tradit. Complement. Altern. Med. 2011, 8, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Albuquerque, B.R.; Prieto, M.; Barreiro, M.F.; Rodrigues, A.; Curran, T.P.; Barros, L.; Ferreira, I.C. Catechin-based extract optimization obtained from Arbutus unedo L. fruits using maceration/microwave/ultrasound extraction techniques. Ind. Crops Prod. 2017, 95, 404–415. [Google Scholar] [CrossRef]
- Jovanović, A.A.; Đorđević, V.B.; Zdunić, G.M.; Pljevljakušić, D.S.; Šavikin, K.P.; Gođevac, D.M.; Bugarski, B.M. Optimization of the extraction process of polyphenols from Thymus serpyllum L. herb using maceration, heat-and ultrasound-assisted techniques. Sep. Purif. Technol. 2017, 179, 369–380. [Google Scholar] [CrossRef]
- Chanivet, M.; Durán-Guerrero, E.; Rodríguez-Dodero, M.d.C.; Barroso, C.G.; Castro, R. Application of accelerating energies to the maceration of sherry vinegar with citrus fruits. J. Sci. Food Agric. 2021, 101, 2235–2246. [Google Scholar] [CrossRef]
- Wang, W.-Y.; Qu, H.-B.; Gong, X.-C. Research progress on percolation extraction process of traditional Chinese medicines. Chin. Med. J. 2020, 45, 1039–1046. [Google Scholar]
- Shejawale, D.D.; Murugesh, C.; Rastogi, N.; Subramanian, R. Effect of feed particle size and solvent flow rate on soybean oil extraction in a percolation type extractor. J. Food Sci. Technol. 2022, 59, 4723–4730. [Google Scholar] [CrossRef]
- Zhang, M.; Zhao, J.; Dai, X.; Li, X. Extraction and Analysis of Chemical Compositions of Natural Products and Plants. Separations 2023, 10, 598. [Google Scholar] [CrossRef]
- Ahmad, R.; Ahmad, N.; Riaz, M.; Al-tarouti, M.; Aloufi, F.; AlDarwish, A.; Alalaq, B.; Alhanfoush, B.; Khan, Z. Optimization of extraction and quantification technique for phenolics content of garlic (Allium sativum): An application for comparative phytochemical evaluation based on cultivar origin. Biomed. Chromatogr. 2020, 34, e4942. [Google Scholar] [CrossRef]
- Sevindik, O.; Kelebek, H.; Rombolà, A.D.; Selli, S. Grape seed oil volatiles and odour activity values: A comparison with Turkish and Italian cultivars and extraction methods. J. Food Sci. Technol. 2022, 59, 1968–1981. [Google Scholar] [CrossRef]
- Wei, M.-C.; Xiao, J.; Yang, Y.-C. Extraction of α-humulene-enriched oil from clove using ultrasound-assisted supercritical carbon dioxide extraction and studies of its fictitious solubility. Food Chem. 2016, 210, 172–181. [Google Scholar] [CrossRef]
- Yang, Y.-C.; Wei, M.-C. Development and characterization of a green procedure for apigenin extraction from Scutellaria barbata D. Don. Food Chem. 2018, 252, 381–389. [Google Scholar] [CrossRef]
- Alara, O.R.; Abdurahman, N.H.; Ukaegbu, C.I. Soxhlet extraction of phenolic compounds from Vernonia cinerea leaves and its antioxidant activity. J. Appl. Res. Med. Aromat. Plants 2018, 11, 12–17. [Google Scholar] [CrossRef]
- Jones, W.P.; Kinghorn, A.D. Extraction of Plant Secondary Metabolites. In Natural Products Isolation. Methods in Molecular Biology; Sarker, S., Nahar, L., Eds.; Humana Press: Totowa, NJ, USA, 2012; Volume 864, pp. 341–366. [Google Scholar]
- Bagade, S.B.; Patil, M. Recent advances in microwave assisted extraction of bioactive compounds from complex herbal samples: A review. Crit. Rev. Anal. Chem. 2021, 51, 138–149. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.; Xiong, X.; Huang, G. Ultrasound-assisted extraction and characteristics of maize polysaccharides from different sites. Ultrason. Sonochem. 2023, 95, 106416. [Google Scholar] [CrossRef] [PubMed]
- Achat, S.; Tomao, V.; Madani, K.; Chibane, M.; Elmaataoui, M.; Dangles, O.; Chemat, F. Direct enrichment of olive oil in oleuropein by ultrasound-assisted maceration at laboratory and pilot plant scale. Ultrason. Sonochem. 2012, 19, 777–786. [Google Scholar] [CrossRef]
- Hu, W.; Zhao, Y.; Yang, Y.; Zhang, H.; Ding, C.; Hu, C.; Zhou, L.; Zhang, Z.; Yuan, S.; Chen, Y. Microwave-assisted extraction, physicochemical characterization and bioactivity of polysaccharides from Camptotheca acuminata fruits. Int. J. Biol. Macromol. 2019, 133, 127–136. [Google Scholar] [CrossRef]
- Donelian, A.; Carlson, L.; Lopes, T.; Machado, R. Comparison of extraction of patchouli (Pogostemon cablin) essential oil with supercritical CO2 and by steam distillation. J. Supercrit. Fluids. 2009, 48, 15–20. [Google Scholar] [CrossRef]
- Zhang, H.; Huang, T.; Liao, X.; Zhou, Y.; Chen, S.; Chen, J.; Xiong, W. Extraction of Camphor Tree Essential Oil by Steam Distillation and Supercritical CO2 Extraction. Molecules 2022, 27, 5385. [Google Scholar] [CrossRef]
- Conte, R.; Gullich, L.M.; Bilibio, D.; Zanella, O.; Bender, J.P.; Carniel, N.; Priamo, W.L. Pressurized liquid extraction and chemical characterization of safflower oil: A comparison between methods. Food Chem. 2016, 213, 425–430. [Google Scholar] [CrossRef] [PubMed]
- Çam, M.; Hışıl, Y. Pressurised water extraction of polyphenols from pomegranate peels. Food Chem. 2010, 123, 878–885. [Google Scholar] [CrossRef]
- Ma, X.; Zheng, C.; Han, L.; Xie, B.; Jia, J.; Cao, Z.; Li, Y.; Chen, Y. Synergistic therapeutic actions of herbal ingredients and their mechanisms from molecular interaction and network perspectives. Drug Discov. Today 2009, 14, 579–588. [Google Scholar] [CrossRef] [PubMed]
- Caesar, L.K.; Cech, N.B. Synergy and antagonism in natural product extracts: When 1 + 1 does not equal 2. Nat. Prod. Rep. 2019, 36, 869–888. [Google Scholar] [CrossRef] [PubMed]
- Yadav, B.; Jogawat, A.; Rahman, M.S.; Narayan, O.P. Secondary metabolites in the drought stress tolerance of crop plants: A review. Gene Rep. 2021, 23, 101040. [Google Scholar] [CrossRef]
- Li, Y.; Kong, D.; Fu, Y.; Sussman, M.R.; Wu, H. The effect of developmental and environmental factors on secondary metabolites in medicinal plants. Plant Physiol. Biochem. 2020, 148, 80–89. [Google Scholar] [CrossRef]
- Chew, Y.L.; Ling Chan, E.W.; Tan, P.L.; Lim, Y.Y.; Stanslas, J.; Goh, J.K. Assessment of phytochemical content, polyphenolic composition, antioxidant and antibacterial activities of Leguminosae medicinal plants in Peninsular Malaysia. BMC Complement. Altern. Med. 2011, 11, 12. [Google Scholar] [CrossRef]
- Khalil, N.; Sperotto, J.; Manfron, M. Antiinflammatory activity and acute toxicity of Dodonaea viscosa. Fitoterapia 2006, 77, 478–480. [Google Scholar] [CrossRef] [PubMed]
- Salinas-Sánchez, D.O.; Herrera-Ruiz, M.; Pérez, S.; Jiménez-Ferrer, E.; Zamilpa, A. Anti-inflammatory activity of hautriwaic acid isolated from Dodonaea viscosa leaves. Molecules 2012, 17, 4292–4299. [Google Scholar] [CrossRef] [PubMed]
- Maleki, S.J.; Crespo, J.F.; Cabanillas, B. Anti-inflammatory effects of flavonoids. Food Chem. 2019, 299, 125124. [Google Scholar] [CrossRef] [PubMed]
- Moura, A.; Silva, E.; Fraga, M.; Wanderley, A.; Afiatpour, P.; Maia, M. Antiinflammatory and chronic toxicity study of the leaves of Ageratum conyzoides L. in rats. Phytomedicine 2005, 12, 138–142. [Google Scholar] [CrossRef]
- de Mello, S.V.G.V.; da Rosa, J.S.; Facchin, B.M.; Luz, A.B.G.; Vicente, G.; Faqueti, L.G.; Rosa, D.W.; Biavatti, M.W.; Fröde, T.S. Beneficial effect of Ageratum conyzoides Linn (Asteraceae) upon inflammatory response induced by carrageenan into the mice pleural cavity. J. Ethnopharmacol. 2016, 194, 337–347. [Google Scholar] [CrossRef]
- Peng, J.; Zheng, T.T.; Li, X.; Liang, Y.; Wang, L.J.; Huang, Y.C.; Xiao, H.T. Plant-Derived Alkaloids: The Promising Disease-Modifying Agents for Inflammatory Bowel Disease. Front. Pharmacol. 2019, 10, 351. [Google Scholar] [CrossRef]
- Wangchuk, P.; Keller, P.A.; Pyne, S.G.; Taweechotipatr, M.; Tonsomboon, A.; Rattanajak, R.; Kamchonwongpaisan, S. Evaluation of an ethnopharmacologically selected Bhutanese medicinal plants for their major classes of phytochemicals and biological activities. J. Ethnopharmacol. 2011, 137, 730–742. [Google Scholar] [CrossRef]
- Nortjie, E.; Basitere, M.; Moyo, D.; Nyamukamba, P. Extraction Methods, Quantitative and Qualitative Phytochemical Screening of Medicinal Plants for Antimicrobial Textiles: A Review. Plants 2022, 11, 2011. [Google Scholar] [CrossRef]
- Raju, R.; Singh, A.; Bodkin, F.; Münch, G. Costatamins A–C, new 4-phenylcoumarins with anti-inflammatory activity from the Australian woodland tree Angophora costata (Myrtaceae). Fitoterapia 2019, 133, 171–174. [Google Scholar] [CrossRef]
- Zhang, C.; Deng, J.; Liu, D.; Tuo, X.; Yu, Y.; Yang, H.; Wang, N. Nuciferine inhibits proinflammatory cytokines via the PPARs in LPS-induced RAW264.7 cells. Molecules 2018, 23, 2723. [Google Scholar] [CrossRef]
- Fua, Y.-H. Structurally diverse indole alkaloids from Ochrosia elliptica. Heterocycles 2017, 94, 743–749. [Google Scholar] [CrossRef]
- Raju, R.; Gunawardena, D.; Ahktar, M.A.; Low, M.; Reddell, P.; Munch, G. Anti-Inflammatory Chemical Profiling of the Australian Rainforest Tree Alphitonia petriei (Rhamnaceae). Molecules 2016, 21, 1521. [Google Scholar] [CrossRef]
- Xue, P.-H.; Zhang, N.; Liu, D.; Zhang, Q.-R.; Duan, J.-S.; Yu, Y.-Q.; Li, J.-Y.; Cao, S.-J.; Zhao, F.; Kang, N. Cytotoxic and anti-inflammatory sesquiterpenes from the whole plants of Centipeda minima. J. Nat. Prod. 2021, 84, 247–258. [Google Scholar] [CrossRef] [PubMed]
- Jing, W.; Xiaolan, C.; Yu, C.; Feng, Q.; Haifeng, Y. Pharmacological effects and mechanisms of tannic acid. Biomed. Pharmacother. 2022, 154, 113561. [Google Scholar] [CrossRef]
- Srisook, K.; Srisook, E.; Nachaiyo, W.; Chan-In, M.; Thongbai, J.; Wongyoo, K.; Chawsuanthong, S.; Wannasri, K.; Intasuwan, S.; Watcharanawee, K. Bioassay-guided isolation and mechanistic action of anti-inflammatory agents from Clerodendrum inerme leaves. J. Ethnopharmacol. 2015, 165, 94–102. [Google Scholar] [CrossRef]
- Tang, J.; Diao, P.; Shu, X.; Li, L.; Xiong, L. Quercetin and quercitrin attenuates the inflammatory response and oxidative stress in LPS-induced RAW264.7 cells: In Vitro assessment and a theoretical model. Biomed Res. Int. 2019, 2019, 7039802. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Huo, X.; Liu, W.; Li, K.; Sun, Z.; Tian, J. Apigenin exhibits anti-inflammatory effects in LPS-stimulated BV2 microglia through activating GSK3β/Nrf2 signaling pathway. Immunopharmacol. Immunotoxicol. 2020, 42, 9–16. [Google Scholar] [CrossRef]
- Pongprayoon, U.; Baeckström, P.; Jacobsson, U.; Lindström, M.; Bohlin, L. Antispasmodic activity of β-damascenone and E-phytol isolated from Ipomoea pes-caprae. Planta Medica 1992, 58, 19–21. [Google Scholar] [CrossRef]
- Van, Q.T.T.; Vien, L.T.; Hanh, T.T.H.; Huong, P.T.T.; Cuong, N.T.; Thao, N.P.; Thuan, N.H.; Dang, N.H.; Thanh, N.V.; Cuong, N.X. Acylated flavonoid glycosides from Barringtonia racemosa. Nat. Prod. Res. 2020, 34, 1276–1281. [Google Scholar] [CrossRef]
- Legault, J.; Perron, T.; Mshvildadze, V.; Girard-Lalancette, K.; Perron, S.; Laprise, C.; Sirois, P.; Pichette, A. Antioxidant and anti-inflammatory activities of quercetin 7-O-β-D-glucopyranoside from the leaves of Brasenia schreberi. J. Med. Food 2011, 14, 1127–1134. [Google Scholar] [CrossRef]
- Beena, P.; Rajesh, K.; Arul, B. Preliminary phytochemical screening of Cicer arietinum in folklore medicine for hepatoprotection. J. Innov. Pharm. Biol. Sci. 2016, 3, 153–159. [Google Scholar]
- Pandey, A.; Tripathi, S. Concept of standardization, extraction and pre phytochemical screening strategies for herbal drug. J. Pharmacogn. Phytochem. 2014, 2, 115–119. [Google Scholar]
- Wallis, T.E. Textbook of Pharmacognosy; CBS: Hounslow, UK, 2005. [Google Scholar]
- Trease, G.E.; Evans, W.C. Pharmacognosy; Baillere Tindall: London, UK, 1983. [Google Scholar]
- Poole, C.F. Sample preparation for planar chromatography. J. Sep. Sci. 2023, 46, 2300071. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, R.; Parambil, J.V. Evolution of extraction technique for the separation of bioactive compounds from Aegle marmelos. Sep. Sci. Technol. 2023, 58, 667–681. [Google Scholar] [CrossRef]
- Chittasupho, C.; Chaobankrang, K.; Sarawungkad, A.; Samee, W.; Singh, S.; Hemsuwimon, K.; Okonogi, S.; Kheawfu, K.; Kiattisin, K.; Chaiyana, W. Antioxidant, Anti-Inflammatory and Attenuating Intracellular Reactive Oxygen Species Activities of Nicotiana tabacum var. Virginia Leaf Extract Phytosomes and Shape Memory Gel Formulation. Gels 2023, 9, 78. [Google Scholar] [CrossRef] [PubMed]
- Smith, H.; Doyle, S.; Murphy, R. Target directed identification of natural bioactive compounds from filamentous fungi. Food Chem. 2023, 405, 134743. [Google Scholar] [CrossRef]
- Zhang, Z.; Pang, X.; Xuewu, D.; Ji, Z.; Jiang, Y. Role of peroxidase in anthocyanin degradation in litchi fruit pericarp. Food Chem. 2005, 90, 47–52. [Google Scholar] [CrossRef]
- Qu, B.; Liu, Y.; Shen, A.; Guo, Z.; Yu, L.; Liu, D.; Huang, F.; Peng, T.; Liang, X. Combining multidimensional chromatography-mass spectrometry and feature-based molecular networking methods for the systematic characterization of compounds in the supercritical fluid extract of Tripterygium wilfordii Hook F. Analyst 2023, 148, 61–73. [Google Scholar] [CrossRef]
- Peiris, D.; Fernando, D.; Senadeera, S.; Ranaweera, C. Phytochemical Screening for Medicinal Plants: Guide for Extraction Methods. Asian J. Plant Sci. 2023, 11, 13–34. [Google Scholar] [CrossRef]
- Schendzielorz, M.; Schmidt, T.; Puchalla, N.; Csuk, R.; Kramell, A.E. TLC and HPTLC-APCI-MS for the rapid discrimination of plant resins frequently used for lacquers and varnishes by artists and conservators. Phytochem. Anal. 2023, 35, 64–76. [Google Scholar] [CrossRef]
- Wangchuk, P. Plant alkaloids: Classification, Isolation and Drug Development. In Medicinal Plants: Chemistry, Pharmacology and Therapeutic Applications; Swamy, M., Patra, J.K., Rudramurthy, G.R., Eds.; Routledge: London, UK, 2019; pp. 131–137. [Google Scholar]
- Hahn-Deinstrop, E. Applied Thin-Layer Chromatography: Best Practice and Avoidance of Mistakes; John Wiley & Sons: Hoboken, NJ, USA, 2007. [Google Scholar]
- Ingle, K.P.; Deshmukh, A.G.; Padole, D.A.; Dudhare, M.S.; Moharil, M.P.; Khelurkar, V.C. Phytochemicals: Extraction methods, identification and detection of bioactive compounds from plant extracts. J. Pharmacogn. Phytochem. 2017, 6, 32–36. [Google Scholar]
- van Beek, T.A.; Tetala, K.K.; Koleva, I.I.; Dapkevicius, A.; Exarchou, V.; Jeurissen, S.M.; Claassen, F.W.; van der Klift, E.J. Recent developments in the rapid analysis of plants and tracking their bioactive constituents. Phytochem. Rev. 2009, 8, 387–399. [Google Scholar] [CrossRef]
- Zhang, J.; Zhou, Z.; Yang, J.; Zhang, W.; Bai, Y.; Liu, H. Thin layer chromatography/plasma assisted multiwavelength laser desorption ionization mass spectrometry for facile separation and selective identification of low molecular weight compounds. Anal. Chem. 2012, 84, 1496–1503. [Google Scholar] [CrossRef]
- Bucar, F.; Wube, A.; Schmid, M. Natural product isolation–how to get from biological material to pure compounds. Nat. Prod. Rep. 2013, 30, 525–545. [Google Scholar] [CrossRef] [PubMed]
- Ben Salah, H.; Allouche, N. Plant-Based Chemicals Extraction and Isolation. In Plant Based “Green Chemistry 2.0” Moving from Evolutionary to Revolutionary; Springer: Singapore, 2019; pp. 89–117. [Google Scholar]
- Albayrak, G.; Demir, S.; Kose, F.A.; Baykan, S. New coumarin glycosides from endemic Prangos heyniae H. Duman & MF Watson. Nat. Prod. Res. 2023, 37, 227–239. [Google Scholar]
- Todorova, G.; Lazarova, I.; Mikhova, B.; Kostova, I. Anthraquinone, naphthalene, and naphthoquinone components of Asphodeline lutea. Chem. Nat. Compd. 2010, 46, 322–323. [Google Scholar] [CrossRef]
- Park, J.E.; Lee, T.H.; Ham, S.L.; Subedi, L.; Hong, S.M.; Kim, S.Y.; Choi, S.U.; Kim, C.S.; Lee, K.R. Anticancer and Anti-Neuroinflammatory Constituents Isolated from the Roots of Wasabia japonica. Antioxidants 2022, 11, 482. [Google Scholar] [CrossRef]
- McChesney, J.D.; Rodenburg, D.L. Preparative chromatography and natural products discovery. Curr. Opin. Biotechnol. 2014, 25, 111–113. [Google Scholar] [CrossRef]
- Jandera, P. Stationary and mobile phases in hydrophilic interaction chromatography: A review. Anal. Chim. Acta 2011, 692, 1–25. [Google Scholar] [CrossRef]
- Hsu, Y.-C.; Ou, S.-M.; Zhuang, K.-R.; Kuo, A.-L.; Li, W.-J.; Huang, C.-Y.; Lin, C.-H.; Chen, J.-J.; Fu, S.-L. Hypericum sampsonii exhibits anti-inflammatory activity in a lipopolysaccharide-induced sepsis mouse model. J. Tradit. Complement. Med. 2023, 13, 379–388. [Google Scholar] [CrossRef]
- Wu, Q.; Hou, X.; Lv, H.; Li, H.; Zhao, L.; Qiu, H. Synthesis of octadecylamine-derived carbon dots and application in reversed phase/hydrophilic interaction liquid chromatography. J. Chromatogr. A. 2021, 1656, 462548. [Google Scholar] [CrossRef] [PubMed]
- Yuan, C.; Dang, J.; Han, Y.; Liu, C.; Yu, S.; Lv, Y.; Cui, Y.; Wang, Z.; Li, G. Preparative isolation of maltol glycoside from Dianthus superbus and its anti-inflammatory activity in vitro. RSC Adv. 2022, 12, 5031–5041. [Google Scholar] [CrossRef]
- Uckoo, R.M.; Jayaprakasha, G.; Patil, B.S. Chromatographic techniques for the separation of polymethoxyflavones from citrus. In Emerging Trends in Dietary Components for Preventing and Combating Disease; ACS Publications: Washington, DC, USA, 2012; pp. 3–15. [Google Scholar]
- Maciejewska-Turska, M.; Pecio, Ł.; Zgórka, G. Isolation of Mirificin and Other Bioactive Isoflavone Glycosides from the Kudzu Root Lyophilisate Using Centrifugal Partition and Flash Chromatographic Techniques. Molecules 2022, 27, 6227. [Google Scholar] [CrossRef]
- Mahato, N.; Sinha, M.; Sharma, K.; Koteswararao, R.; Cho, M.H. Modern Extraction and Purification Techniques for Obtaining High Purity Food-Grade Bioactive Compounds and Value-Added Co-Products from Citrus Wastes. Foods 2019, 8, 523. [Google Scholar] [CrossRef] [PubMed]
- Todorova, V.; Ivanov, K.; Karcheva-Bahchevanska, D.; Ivanova, S. Development and Validation of High-Performance Liquid Chromatography for Identification and Quantification of Phytoecdysteroids Ecdysterone and Turkesterone in Dietary Supplements. Processes 2023, 11, 1786. [Google Scholar] [CrossRef]
- Srivastava, N.; Singh, A.; Kumari, P.; Nishad, J.H.; Gautam, V.S.; Yadav, M.; Bharti, R.; Kumar, D.; Kharwar, R.N. Advances in extraction technologies: Isolation and purification of bioactive compounds from biological materials. In Natural Bioactive Compounds; Elsevier: Amsterdam, The Netherlands, 2021; pp. 409–433. [Google Scholar]
- El-Maraghy, C.M. Implementation of green chemistry to develop HPLC/UV and HPTLC methods for the quality control of Fluconazole in presence of two official impurities in drug substance and pharmaceutical formulations. Sustain. Chem. Pharm. 2023, 33, 101124. [Google Scholar] [CrossRef]
- Shin, D.H.; Priefer, R. A Review of the Efficacy of LC-MS. In Quantitative and Qualitative Determination Technologies of Counterfeit Drugs; Priefer, R., Ed.; CRC Press: Boca Raton, FL, USA, 2023. [Google Scholar]
- Bhat, M.H.; Fayaz, M.; Kumar, A.; Dar, A.A.; Jain, A.K. Chromatographic method for determination of the amino acid content in Dioscorea bulbifera L. Tubers by RP-HPLC. Pharm. Sci. 2019, 25, 65–69. [Google Scholar] [CrossRef]
- Dar, A.A.; Sangwan, P.L.; Singh, N.; Kumar, A. Method validation and simultaneous quantification of five triterpenoids from Codonopsis ovata by high-performance thin-layer chromatography. JPC 2019, 32, 251–256. [Google Scholar] [CrossRef]
- Wani, A.A.; Dar, A.A.; Jan, I.; Sofi, K.A.; Sofi, J.A.; Dar, I.H. Dissipation, risk assessment, half-life period and method validation of carbendazim and triazophos in green pea by high-performance liquid chromatography. Sep. Sci. Plus 2019, 2, 284–290. [Google Scholar] [CrossRef]
- Aierken, K.; Li, J.; Xu, N.; Wu, T.; Zang, D.; Aisa, H.A. Chemical constituents of Rumex dentatus L. and their antimicrobial and anti-inflammatory activities. Phytochemistry 2023, 205, 113509. [Google Scholar] [CrossRef]
- Richardson, S.D. Environmental mass spectrometry: Emerging contaminants and current issues. Anal. Chem. 2012, 84, 747–778. [Google Scholar] [CrossRef]
- Nguyen, D.T.T.; Guillarme, D.; Rudaz, S.; Veuthey, J.L. Fast analysis in liquid chromatography using small particle size and high pressure. J. Sep. Sci. 2006, 29, 1836–1848. [Google Scholar] [CrossRef] [PubMed]
- Swartz, M.E. UPLC™: An introduction and review. J. Liq. Chromatogr. Relat. Technol. 2005, 28, 1253–1263. [Google Scholar] [CrossRef]
- Ramachandra, B. Development of impurity profiling methods using modern analytical techniques. Crit Rev Anal Chem. 2017, 47, 24–36. [Google Scholar] [CrossRef] [PubMed]
- Almeida, C.M.M. Overview of Sample Preparation and Chromatographic Methods to Analysis Pharmaceutical Active Compounds in Waters Matrices. Separations 2021, 8, 16. [Google Scholar] [CrossRef]
- Matos, M.J.; Santana, L.; Uriarte, E.; Abreu, O.A.; Molina, E.; Yordi, E.G. Coumarins—An important class of phytochemicals. Phytochem.-Isol. Characterisation Role Hum. Health 2015, 25, 533–538. [Google Scholar]
- Altemimi, A.; Lakhssassi, N.; Baharlouei, A.; Watson, D.G.; Lightfoot, D.A. Phytochemicals: Extraction, isolation, and identification of bioactive compounds from plant extracts. Plants 2017, 6, 42. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, R.N. Rediscovering natural product biodiversity. Drug Discov. Today 1999, 4, 449–451. [Google Scholar] [CrossRef]
- Hughes, D. New HTS imaging technology deal. Drug Discov. Today 1998, 3, 438–439. [Google Scholar] [CrossRef]
- Hashi, K.; Ohki, S.; Matsumoto, S.; Nishijima, G.; Goto, A.; Deguchi, K.; Yamada, K.; Noguchi, T.; Sakai, S.; Takahashi, M.; et al. Achievement of 1020MHz NMR. J. Magn. Reson. 2015, 256, 30–33. [Google Scholar] [CrossRef]
- Mbayachi, V.B.; Tian, Z.-Y.; Dai, W.-K.; Ayejoto, D.A.; Wang, Z.-M.; Zhang, X.; Khalil, M. Nuclear Magnetic Resonance. In Advanced Diagnostics in Combustion Science; Springer: Berlin/Heidelberg, Germany, 2023; pp. 245–308. [Google Scholar]
- Kemp, W. Organic Spectroscopy; Bloomsbury Publishing: London, UK, 2017. [Google Scholar]
- Hu, G.; Qiu, M. Machine learning-assisted structure annotation of natural products based on MS and NMR data. Nat. Prod. Rep. 2023, 40, 1735–1753. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Pan, H.; Zhai, G.; Luo, Q.; Li, Y.; Fang, C.; Shi, F. Widespread occurrence of in-source fragmentation in the analysis of natural compounds by LC-ESI-MS. Rapid Commun. Mass Spectrom. 2023, 37, e9519. [Google Scholar] [CrossRef] [PubMed]
- da Silva Bezerra, K. Perspective Chapter: High-Performance Liquid Chromatography Coupled to Mass Spectrometry—The Advance in Chemical Analysis. In High Performance Liquid Chromatography-Recent Advances and Applications; IntechOpen: Rijeka, Croatia, 2023. [Google Scholar]
- Wang, H.; Chen, K.; Xue, R.; Turghun, C.; Han, B. Identification of the chemical constituents in cullen corylifolium ethanolic extract by LC-MS/MS and GC-MS. Nat. Prod. Res. 2023, 37, 1392–1396. [Google Scholar] [CrossRef]
- Sashidhara, K.V.; Rosaiah, J.N. Various dereplication strategies using LC-MS for rapid natural product lead identification and drug discovery. Nat. Prod. Commun. 2007, 2, 1934578X0700200218. [Google Scholar] [CrossRef]
- Cançado, G.G.L.; Fiuza, J.A.; de Paiva, N.C.N.; de Carvalho Dhom Lemos, L.; Ricci, N.D.; Gazzinelli-Guimarães, P.H.; Martins, V.G.; Bartholomeu, D.C.; Negrão-Corrêa, D.A.; Carneiro, C.M.; et al. Hookworm products ameliorate dextran sodium sulfate-induced colitis in BALB/c mice. J. Leukoc. Biol. 2011, 17, 2275–2286. [Google Scholar] [CrossRef]
- Pitt, J.J. Principles and applications of liquid chromatography-mass spectrometry in clinical biochemistry. Clin. Biochem. Rev. 2009, 30, 19. [Google Scholar]
- Rahman, M. Application of computational methods in isolation of plant secondary metabolites. In Computational Phytochemistry; Elsevier: Amsterdam, The Netherlands, 2018; pp. 107–139. [Google Scholar]
- Summers, R.W.; Elliott, D.; Urban, J.; Thompson, R.; Weinstock, J. Trichuris suis therapy in Crohn’s disease. Gut 2005, 54, 87–90. [Google Scholar] [CrossRef]
- Yang, F.; Wang, H.; Feng, G.; Zhang, S.; Wang, J.; Cui, L. Rapid Identification of Chemical Constituents in Hericium erinaceus Based on LC-MS/MS Metabolomics. J. Food Qual. 2021, 2021, 5560626. [Google Scholar] [CrossRef]
- Urbano, M.; De Castro, M.D.L.; Pérez, P.M.; García-Olmo, J.; Gomez-Nieto, M.A. Ultraviolet–visible spectroscopy and pattern recognition methods for differentiation and classification of wines. Food Chem. 2006, 97, 166–175. [Google Scholar] [CrossRef]
- Barnes, R.B.; Bonner, L.G. The early history and the methods of infrared spectroscopy. Am. J. Phys. 1936, 4, 181–189. [Google Scholar] [CrossRef]
- Coates, J. Interpretation of Infrared Spectra, a Practical Approach; Wiley: New York, NY, USA, 2000. [Google Scholar]
- Walsh, K.B.; Blasco, J.; Zude-Sasse, M.; Sun, X. Visible-NIR ‘point’spectroscopy in postharvest fruit and vegetable assessment: The science behind three decades of commercial use. Postharvest Biol. Technol. 2020, 168, 111246. [Google Scholar] [CrossRef]
- Johnson, J.B.; Naiker, M. Seeing red: A review of the use of near-infrared spectroscopy (NIRS) in entomology. Appl. Spectrosc. Rev. 2020, 55, 810–839. [Google Scholar] [CrossRef]
- Johnson, J.B.; Walsh, K.B.; Naiker, M.; Ameer, K. The Use of Infrared Spectroscopy for the Quantification of Bioactive Compounds in Food: A Review. Molecules 2023, 28, 3215. [Google Scholar] [CrossRef]
- Alberts, M.; Laino, T.; Vaucher, A.C. Leveraging Infrared Spectroscopy for Automated Structure Elucidation (version 1.0.0). Anal. Chem. 2023. [Google Scholar] [CrossRef]
- Nabeel, O. IR Spectroscopy in Qualitative and Quantitative Analysis. In Infrared Spectroscopy, Marwa, E.-A., Khalid, A.-S., Ahmed, S.E.-S., Eds.; IntechOpen: Rijeka, Croatia, 2022. [Google Scholar]
- Triastuti, A.; Pradana, D.A.; Setiawan, I.D.; Fakhrudin, N.; Himmi, S.K.; Widyarini, S.; Rohman, A. In vivo anti-inflammatory activities of Plantago major extract and fractions and analysis of their phytochemical components using a high-resolution mass spectrometry. Res. Pharm. Sci. 2022, 17, 665–676. [Google Scholar] [CrossRef] [PubMed]
- Kouloura, E.; Danika, E.; Kim, S.; Hoerlé, M.; Cuendet, M.; Halabalaki, M.; Skaltsounis, L.A. Rapid Identification of Coumarins from Micromelum falcatum by UPLC-HRMS/MS and Targeted Isolation of Three New Derivatives. Molecules 2014, 19, 15042–15057. [Google Scholar] [CrossRef]
- Wallace, M.A.G.; McCord, J.P. High-resolution mass spectrometry. In Breathborne Biomarkers and the Human Volatilome; Elsevier: Amsterdam, The Netherlands, 2020; pp. 253–270. [Google Scholar]
- Menger, F.; Gago-Ferrero, P.; Wiberg, K.; Ahrens, L. Wide-scope screening of polar contaminants of concern in water: A critical review of liquid chromatography-high resolution mass spectrometry-based strategies. Trends Environ. Anal. Chem. 2020, 28, e00102. [Google Scholar] [CrossRef]
- Jenny, M.; Klieber, M.; Zaknun, D.; Schroecksnadel, S.; Kurz, K.; Ledochowski, M.; Schennach, H.; Fuchs, D. In Vitro testing for anti-inflammatory properties of compounds employing peripheral blood mononuclear cells freshly isolated from healthy donors. J. Inflamm. Res. 2011, 60, 127–135. [Google Scholar] [CrossRef]
- Ponce de León-Rodríguez, M.d.C.; Guyot, J.-P.; Laurent-Babot, C. Intestinal in vitro cell culture models and their potential to study the effect of food components on intestinal inflammation. Crit. Rev. Food Sci. Nutr. 2019, 59, 3648–3666. [Google Scholar] [CrossRef]
- Cong, L.; Gao, Z.; Zheng, Y.; Ye, T.; Wang, Z.; Wang, P.; Li, M.; Dong, B.; Yang, W.; Li, Q. Electrical stimulation inhibits Val-boroPro-induced pyroptosis in THP-1 macrophages via sirtuin3 activation to promote autophagy and inhibit ROS generation. Aging 2020, 12, 6415. [Google Scholar] [CrossRef]
- Li, H.; Wang, Y.; Song, Y. MicroRNA-26b inhibits the immune response to Mycobacterium tuberculosis (M. tb) infection in THP-1 cells via targeting TGFβ-activated kinase-1 (TAK1), a promoter of the NF-κB pathway. Int. J. Clin. Exp. Pathol. 2018, 11, 1218. [Google Scholar]
- Wang, T.; Liu, W.; Li, C.; Si, G.; Liang, Z.; Yin, J. Mist1 promoted inflammation in colitis model via K+-ATPase NLRP3 inflammasome by SNAI1. Pathol. Res. Pract. 2021, 224, 153511. [Google Scholar] [CrossRef]
- Li, Q.; Cheng, H.; Liu, Y.; Wang, X.; He, F.; Tang, L. Activation of mTORC1 by LSECtin in macrophages directs intestinal repair in inflammatory bowel disease. Cell Death Dis. 2020, 11, 918. [Google Scholar] [CrossRef] [PubMed]
- Park, H.J.; Kim, J.; Saima, F.T.; Rhee, K.-J.; Hwang, S.; Kim, M.Y.; Baik, S.K.; Eom, Y.W.; Kim, H.-S. Adipose-derived stem cells ameliorate colitis by suppression of inflammasome formation and regulation of M1-macrophage population through prostaglandin E2. Biochem. Biophys. Res. Commun. 2018, 498, 988–995. [Google Scholar] [CrossRef] [PubMed]
- Xue, L.; Liu, C.; Ma, H.; Seeram, N.P.; Neto, C.C. Anti-inflammatory activities of cranberry fruit extracts in human THP-1 monocytes are influenced by their phytochemical composition. Food Sci. Technol. 2022, 2, 75–83. [Google Scholar] [CrossRef]
- Bisht, A.; Dickens, M.; Rutherfurd-Markwick, K.; Thota, R.; Mutukumira, A.N.; Singh, H. Chlorogenic Acid Potentiates the Anti-Inflammatory Activity of Curcumin in LPS-Stimulated THP-1 Cells. Nutrients 2020, 12, 2706. [Google Scholar] [CrossRef]
- Zhao, D.R.; Jiang, Y.S.; Sun, J.Y.; Li, H.H.; Sun, X.T.; Zhao, M.M. Amelioration of 4-methylguaiacol on LPS-induced inflammation in THP-1 cells through NF-κB/IκBα/AP-1 and Nrf2/HO-1 signaling pathway. J. Funct. Foods 2019, 55, 95–103. [Google Scholar] [CrossRef]
- Chanput, W.; Mes, J.J.; Wichers, H.J. THP-1 cell line: An in vitro cell model for immune modulation approach. Int. Immunopharmacol. 2014, 23, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Cousins, R.J.; Blanchard, R.K.; Popp, M.P.; Liu, L.; Cao, J.; Moore, J.B.; Green, C.L. A global view of the selectivity of zinc deprivation and excess on genes expressed in human THP-1 mononuclear cells. Proc. Natl. Acad. Sci. USA 2003, 100, 6952–6957. [Google Scholar] [CrossRef]
- Lea, T. Caco-2 Cell Line. In The Impact of Food Bioactives on Health: In Vitro and Ex Vivo Models; Verhoeckx, K., Cotter, P., López-Expósito, I., Kleiveland, C., Lea, T., Mackie, A., Requena, T., Swiatecka, D., Wichers, H., Eds.; Springer: Berlin/Heidelberg, Germany, 2015; pp. 103–111. [Google Scholar]
- Primavera, R.; Palumbo, P.; Celia, C.; Cinque, B.; Carata, E.; Carafa, M.; Paolino, D.; Cifone, M.G.; Di Marzio, L. An insight of in vitro transport of PEGylated non-ionic surfactant vesicles (NSVs) across the intestinal polarized enterocyte monolayers. Eur. J. Pharm. Biopharm. 2018, 127, 432–442. [Google Scholar] [CrossRef]
- Lozoya-Agullo, I.; Araujo, F.; Gonzalez-Alvarez, I.; Merino-Sanjuan, M.; Gonzalez-Alvarez, M.; Bermejo, M.; Sarmento, B. Usefulness of Caco-2/HT29-MTX and Caco-2/HT29-MTX/Raji B coculture models to predict intestinal and colonic permeability compared to Caco-2 monoculture. Mol. Pharm. 2017, 14, 1264–1270. [Google Scholar] [CrossRef] [PubMed]
- Borgonetti, V.; Cocetta, V.; Biagi, M.; Carnevali, I.; Governa, P.; Montopoli, M. Anti-inflammatory activity of a fixed combination of probiotics and herbal extract in an in-vitro model of intestinal inflammation by stimulating Caco-2 cells with LPS-conditioned THP-1 cells medium. Minerva Pediatr. 2022, 74, 511–518. [Google Scholar] [CrossRef] [PubMed]
- Weber, L.; Kuck, K.; Jürgenliemk, G.; Heilmann, J.; Lipowicz, B.; Vissiennon, C. Anti-Inflammatory and Barrier-Stabilising Effects of Myrrh, Coffee Charcoal and Chamomile Flower Extract in a Co-Culture Cell Model of the Intestinal Mucosa. Biomolecules 2020, 10, 1033. [Google Scholar] [CrossRef] [PubMed]
- Andrews, C.; McLean, M.H.; Durum, S.K. Cytokine Tuning of Intestinal Epithelial Function. Front. Immunol. 2018, 9, 1270. [Google Scholar] [CrossRef] [PubMed]
- Akiyama, Y.; Matsumura, N.; Ono, A.; Hayashi, S.; Funaki, S.; Tamura, N.; Kimoto, T.; Jiko, M.; Haruna, Y.; Sarashina, A.; et al. Prediction of Oral Drug Absorption in Rats from In Vitro Data. Pharm. Res. 2023, 40, 359–373. [Google Scholar] [CrossRef]
- Xu, R.; Zhu, H.; Hu, L.; Yu, B.; Zhan, X.; Yuan, Y.; Zhou, P. Characterization of the intestinal absorption of morroniside from Cornus officinalis Sieb. et Zucc via a Caco-2 cell monolayer model. PLoS ONE 2020, 15, e0227844. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, Z.-Q.; Zheng, R.-J.; Jiang, J.-J.; Li, D.-D.; Zhou, W.-W. Paenibacillus exopolysaccharide repairs GI inflammation by suppressing MAPK and NF-κB and restoring lipid production in Caco-2 cell line. J. Funct. Foods 2023, 107, 105709. [Google Scholar] [CrossRef]
- Restivo, I.; Basilicata, M.G.; Giardina, I.C.; Massaro, A.; Pepe, G.; Salviati, E.; Pecoraro, C.; Carbone, D.; Cascioferro, S.; Parrino, B.; et al. A Combination of Polymethoxyflavones from Citrus sinensis and Prenylflavonoids from Humulus lupulus Counteracts IL-1β-Induced Differentiated Caco-2 Cells Dysfunction via a Modulation of NF-κB/Nrf2 Activation. Antioxidants 2023, 12, 1621. [Google Scholar]
- Joshi, A.; Soni, A.; Acharya, S. In vitro models and ex vivo systems used in inflammatory bowel disease. Vitr. Models 2022, 1, 213–227. [Google Scholar] [CrossRef]
- Rajendiran, V.; Natarajan, V.; Devaraj, S.N. Anti-inflammatory activity of Alpinia officinarum hance on rat colon inflammation and tissue damage in DSS induced acute and chronic colitis models. Food Sci. Hum. Wellness 2018, 7, 273–281. [Google Scholar] [CrossRef]
- Jo, A.; Een Kim, C.; Lee, M. Serratane triterpenoids isolated from Lycopodium clavatum by bioactivity-guided fractionation attenuate the production of inflammatory mediators. Bioorg. Chem. 2020, 96, 103632. [Google Scholar] [CrossRef] [PubMed]
- Loganes, C.; Lega, S.; Bramuzzo, M.; Vecchi Brumatti, L.; Piscianz, E.; Valencic, E.; Tommasini, A.; Marcuzzi, A. Curcumin Anti-Apoptotic Action in a Model of Intestinal Epithelial Inflammatory Damage. Nutrients 2017, 9, 578. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.-A.; Kim, D.-K.; Kang, O.-H.; Choi, Y.-A.; Park, H.-J.; Choi, S.-C.; Kim, T.-H.; Yun, K.-J.; Nah, Y.-H.; Lee, Y.-M. Inhibitory effect of luteolin on TNF-α-induced IL-8 production in human colon epithelial cells. Int. Immunopharmacol. 2005, 5, 209–217. [Google Scholar] [CrossRef] [PubMed]
- Raschke, W.; Baird, S.; Ralph, P.; Nakoinz, I. Functional macrophage cell lines transformed by Abelson leukemia virus. Cell 1978, 15, 261–267. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.; Zou, Y.; Peng, J. Oregano Essential Oil Attenuates RAW264.7 Cells from Lipopolysaccharide-Induced Inflammatory Response through Regulating NADPH Oxidase Activation-Driven Oxidative Stress. Molecules 2018, 23, 1857. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhang, L.; Xu, S.; Pan, J.; Zhang, Q.; Lu, R. Surface-layer protein from Lactobacillus acidophilus NCFM inhibits lipopolysaccharide-induced inflammation through MAPK and NF-κB signaling pathways in RAW264. 7 cells. J. Agri. Food Chem. 2018, 66, 7655–7662. [Google Scholar] [CrossRef]
- Gao, R.; Shu, W.; Shen, Y.; Sun, Q.; Bai, F.; Wang, J.; Li, D.; Li, Y.; Jin, W.; Yuan, L. Sturgeon protein-derived peptides exert anti-inflammatory effects in LPS-stimulated RAW264. 7 macrophages via the MAPK pathway. J. Funct. Foods. 2020, 72, 104044. [Google Scholar] [CrossRef]
- Kim, Y.; Lim, H.J.; Jang, H.-J.; Lee, S.; Jung, K.; Lee, S.W.; Lee, S.-J.; Rho, M.-C. Portulaca oleracea extracts and their active compounds ameliorate inflammatory bowel diseases in vitro and in vivo by modulating TNF-α, IL-6 and IL-1β signalling. Food Res. Int. 2018, 106, 335–343. [Google Scholar] [CrossRef]
- Kim, J.E.; Min, S.K.; Hong, J.-M.; Kim, K.H.; Han, S.J.; Yim, J.H.; Park, H.; Kim, I.-C. Anti-inflammatory effects of methanol extracts from the Antarctic lichen, Amandinea sp. in LPS-stimulated raw 264.7 macrophages and zebrafish. Fish Shellfish Immunol. 2020, 107, 301–308. [Google Scholar] [CrossRef]
- Krajewska, J.B.; Długosz, O.; Sałaga, M.; Banach, M.; Fichna, J. Silver nanoparticles based on blackcurrant extract show potent anti-inflammatory effect in vitro and in DSS-induced colitis in mice. Int. J. Pharm. 2020, 585, 119549. [Google Scholar] [CrossRef] [PubMed]
- Zhen, D.; Xuan, T.-Q.; Hu, B.; Bai, X.; Fu, D.-N.; Wang, Y.; Wu, Y.; Yang, J.; Ma, Q. Pteryxin attenuates LPS-induced inflammatory responses and inhibits NLRP3 inflammasome activation in RAW264. 7 cells. J. Ethnopharmacol. 2022, 284, 114753. [Google Scholar] [CrossRef]
- Morgan, R.E.; Ahn, S.; Nzimiro, S.; Fotie, J.; Phelps, M.A.; Cotrill, J.; Yakovich, A.J.; Sackett, D.L.; Dalton, J.T.; Werbovetz, K.A. Inhibitors of tubulin assembly identified through screening a compound library. Chem. Biol. Drug Des. 2008, 72, 513–524. [Google Scholar] [CrossRef]
- Merly, L.; Smith, S.L. Murine RAW 264.7 cell line as an immune target: Are we missing something? Immunopharmacol. Immunotoxicol. 2017, 39, 55–58. [Google Scholar] [CrossRef]
- Elisia, I.; Pae, H.B.; Lam, V.; Cederberg, R.; Hofs, E.; Krystal, G. Comparison of RAW264.7, human whole blood and PBMC assays to screen for immunomodulators. J. Immunol. Methods 2018, 452, 26–31. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Guan, Y.; Yang, R.; Li, J.; Wang, J.; Jia, A.-Q. Anti-inflammatory activity of 3-cinnamoyltribuloside and its metabolomic analysis in LPS-activated RAW 264.7 cells. BMC Complement. Med. Ther. 2020, 20, 329. [Google Scholar] [CrossRef]
- Berghaus, L.J.; Moore, J.N.; Hurley, D.J.; Vandenplas, M.L.; Fortes, B.P.; Wolfert, M.A.; Boons, G.-J. Innate immune responses of primary murine macrophage-lineage cells and RAW 264.7 cells to ligands of Toll-like receptors 2, 3, and 4. Comp. Immunol. Microbiol. Infect. Dis. 2010, 33, 443–454. [Google Scholar] [CrossRef]
- Taciak, B.; Białasek, M.; Braniewska, A.; Sas, Z.; Sawicka, P.; Kiraga, Ł.; Rygiel, T.; Król, M. Evaluation of phenotypic and functional stability of RAW 264.7 cell line through serial passages. PLoS ONE 2018, 13, e0198943. [Google Scholar] [CrossRef]
- Verhoeckx, K.; Cotter, P.; López-Expósito, I.; Kleiveland, C.; Lea, T.; Mackie, A.; Requena, T.; Swiatecka, D.; Wichers, H. The Impact of Food Bioactives on Health: In Vitro and Ex Vivo Models; Springer: Cham, Switzerland, 2015. [Google Scholar]
- Kleiveland, C.R. Peripheral Blood Mononuclear Cells. In The Impact of Food Bioactives on Health: In Vitro and Ex Vivo Models; Springer: Cham, Switzerland, 2015; pp. 161–167. [Google Scholar]
- Yoshimura, T.; Mitsuyama, K.; Sakemi, R.; Takedatsu, H.; Yoshioka, S.; Kuwaki, K.; Mori, A.; Fukunaga, S.; Araki, T.; Morita, M. Evaluation of serum leucine-rich alpha-2 glycoprotein as a new inflammatory biomarker of inflammatory bowel disease. Mediat. Inflamm. 2021, 2021, 8825374. [Google Scholar] [CrossRef] [PubMed]
- Morita, T.; Mitsuyama, K.; Yamasaki, H.; Mori, A.; Yoshimura, T.; Araki, T.; Morita, M.; Tsuruta, K.; Yamasaki, S.; Kuwaki, K. Gene expression of transient receptor potential channels in peripheral blood mononuclear cells of inflammatory bowel disease patients. J. Clin. Med. 2020, 9, 2643. [Google Scholar] [CrossRef]
- Ju, J.K.; Cho, Y.-N.; Park, K.-J.; Kwak, H.D.; Jin, H.-M.; Park, S.-Y.; Kim, H.S.; Kee, S.-J.; Park, Y.-W. Activation, deficiency, and reduced IFN-γ production of mucosal-associated invariant T cells in patients with inflammatory bowel disease. J. Innate Immun. 2020, 12, 422–434. [Google Scholar] [CrossRef]
- Paprocka, R.; Kołodziej, P.; Wiese-Szadkowska, M.; Helmin-Basa, A.; Bogucka-Kocka, A. Evaluation of Anthelmintic and Anti-Inflammatory Activity of 1,2,4-Triazole Derivatives. Molecules 2022, 27, 4488. [Google Scholar] [CrossRef] [PubMed]
- Gahtani, R.M.; Shaikh, A.; Kamli, H. Computational and Preclinical Analysis of 2-(4-Methyl)benzylidene-4,7-dimethyl Indan-1-one (IPX-18): A Novel Arylidene Indanone Small Molecule with Anti-Inflammatory Activity via NF-κB and Nrf2 Signaling. Biomedicines 2023, 11, 716. [Google Scholar] [CrossRef] [PubMed]
- Sedgwick, J.B.; Menon, I.; Gern, J.E.; Busse, W.W. Effects of inflammatory cytokines on the permeability of human lung microvascular endothelial cell monolayers and differential eosinophil transmigration. J. Allergy Clin. Immunol. 2002, 110, 752–756. [Google Scholar] [CrossRef]
- Gabay, C. Interleukin-6 and chronic inflammation. Arthritis Res. Ther. 2006, 8, S3. [Google Scholar] [CrossRef] [PubMed]
- Kayhan, S.; Guzel, A.; Duran, L.; Tutuncu, S.; Guzel, A.; Gunaydın, M.; Salis, O.; Okuyucu, A.; Selcuk, M.Y. Effects of leflunomide on inflamation and fibrosis in bleomycine induced pulmonary fibrosis in wistar albino rats. J. Thorac. Dis. 2013, 5, 641. [Google Scholar] [PubMed]
- Mangan, D.F.; Wahl, S.M. Differential regulation of human monocyte programmed cell death (apoptosis) by chemotactic factors and pro-inflammatory cytokines. J. Immunol. 1991, 147, 3408–3412. [Google Scholar] [CrossRef] [PubMed]
- Rogers, P.D.; Thornton, J.; Barker, K.S.; McDaniel, D.O.; Sacks, G.S.; Swiatlo, E.; McDaniel, L.S. Pneumolysin-dependent and-independent gene expression identified by cDNA microarray analysis of THP-1 human mononuclear cells stimulated by Streptococcus pneumoniae. Infect. Immun. 2003, 71, 2087–2094. [Google Scholar] [CrossRef]
- Facchin, B.M.; dos Reis, G.O.; Vieira, G.N.; Mohr, E.T.B.; da Rosa, J.S.; Kretzer, I.F.; Demarchi, I.G.; Dalmarco, E.M. Inflammatory biomarkers on an LPS-induced RAW 264.7 cell model: A systematic review and meta-analysis. Inflamm. Res. 2022, 71, 741–758. [Google Scholar] [CrossRef]
- Mizoguchi, E.; Low, D.; Ezaki, Y.; Okada, T. Recent updates on the basic mechanisms and pathogenesis of inflammatory bowel diseases in experimental animal models. Intest. Res. 2020, 18, 151–167. [Google Scholar] [CrossRef]
- Baydi, Z.; Limami, Y.; Khalki, L.; Zaid, N.; Naya, A.; Mtairag, E.M.; Oudghiri, M.; Zaid, Y. An Update of Research Animal Models of Inflammatory Bowel Disease. Sci. World J. 2021, 2021, 7479540. [Google Scholar] [CrossRef]
- DeVoss, J.; Diehl, L. Murine Models of Inflammatory Bowel Disease (IBD) Challenges of Modeling Human Disease. Toxicol. Pathol. 2014, 42, 99–110. [Google Scholar] [CrossRef]
- Lee, C.H.; Koh, S.-J.; Radi, Z.A.; Habtezion, A. Animal models of inflammatory bowel disease: Novel experiments for revealing pathogenesis of colitis, fibrosis, and colitis-associated colon cancer. Intest. Res. 2023, 21, 295–305. [Google Scholar] [CrossRef]
- Goyal, N.; Rana, A.; Ahlawat, A.; Bijjem, K.R.V.; Kumar, P. Animal models of inflammatory bowel disease: A review. Inflammopharmacology 2014, 22, 219–233. [Google Scholar] [CrossRef]
- Wirtz, S.; Popp, V.; Kindermann, M.; Gerlach, K.; Weigmann, B.; Fichtner-Feigl, S.; Neurath, M.F. Chemically induced mouse models of acute and chronic intestinal inflammation. Nat. Protoc. 2017, 12, 1295–1309. [Google Scholar] [CrossRef]
- Katsandegwaza, B.; Horsnell, W.; Smith, K. Inflammatory Bowel Disease: A Review of Pre-Clinical Murine Models of Human Disease. Int. J. Mol. Sci. 2022, 23, 9344. [Google Scholar] [CrossRef]
- Fuss, I.J.; Neurath, M.; Boirivant, M.; Klein, J.S.; De La Motte, C.; Strong, S.A.; Fiocchi, C.; Strober, W. Disparate CD4+ lamina propria (LP) lymphokine secretion profiles in inflammatory bowel disease. Crohn’s disease LP cells manifest increased secretion of IFN-gamma, whereas ulcerative colitis LP cells manifest increased secretion of IL-5. J. Immunol. 1996, 157, 1261–1270. [Google Scholar] [CrossRef] [PubMed]
- Antoniou, E.; Margonis, G.A.; Angelou, A.; Pikouli, A.; Argiri, P.; Karavokyros, I.; Papalois, A.; Pikoulis, E. The TNBS-induced colitis animal model: An overview. Ann. Med. Surg. 2016, 11, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Neurath, M.F.; Fuss, I.; Kelsall, B.L.; Stüber, E.; Strober, W. Antibodies to interleukin 12 abrogate established experimental colitis in mice. J. Exp. Med. 1995, 182, 1281–1290. [Google Scholar] [CrossRef]
- Ellrichmann, M.; Wietzke-Braun, P.; Dhar, S.; Nikolaus, S.; Arlt, A.; Bethge, J.; Kuehbacher, T.; Wintermeyer, L.; Balschun, K.; Klapper, W. Endoscopic ultrasound of the colon for the differentiation of Crohn’s disease and ulcerative colitis in comparison with healthy controls. Aliment. Pharmacol. Ther. 2014, 39, 823–833. [Google Scholar] [CrossRef] [PubMed]
- Gambari, R.; Borgatti, M.; Lampronti, I.; Fabbri, E.; Brognara, E.; Bianchi, N.; Piccagli, L.; Yuen, M.C.-W.; Kan, C.-W.; Hau, D.K.-P. Corilagin is a potent inhibitor of NF-kappaB activity and downregulates TNF-alpha induced expression of IL-8 gene in cystic fibrosis IB3-1 cells. Int. Immunopharmacol. 2012, 13, 308–315. [Google Scholar] [CrossRef]
- Lee, I.-A.; Hyun, Y.-J.; Kim, D.-H. Berberine ameliorates TNBS-induced colitis by inhibiting lipid peroxidation, enterobacterial growth and NF-κB activation. Eur. J. Pharmacol. 2010, 648, 162–170. [Google Scholar] [CrossRef]
- Li, C.; Xi, Y.; Li, S.; Zhao, Q.; Cheng, W.; Wang, Z.; Zhong, J.; Niu, X.; Chen, G. Berberine ameliorates TNBS induced colitis by inhibiting inflammatory responses and Th1/Th17 differentiation. Mol. Immunol. 2015, 67, 444–454. [Google Scholar] [CrossRef]
- Jiang, X.; Chen, X.; Dong, R.; Wang, J.; Pan, Y.; Cao, Y. Establishment of a mouse model of inflammatory bowel disease using dextran sulfate sodium. Adv. Clin. Exp. Med. 2023. [Google Scholar] [CrossRef] [PubMed]
- Dou, W.; Zhang, J.; Sun, A.; Zhang, E.; Ding, L.; Mukherjee, S.; Wei, X.; Chou, G.; Wang, Z.-T.; Mani, S. Protective effect of naringenin against experimental colitis via suppression of Toll-like receptor 4/NF-κB signalling. Br. J. Nutr. 2013, 110, 599–608. [Google Scholar] [CrossRef] [PubMed]
- Almutary, A.G.; Alnuqaydan, A.M.; Almatroodi, S.A.; Tambuwala, M.M. Comparative Analysis of the Effect of Different Concentrations of Dextran Sodium Sulfate on the Severity and Extent of Inflammation in Experimental Ulcerative Colitis. Appl. Sci. 2023, 13, 3233. [Google Scholar] [CrossRef]
- Prakash, T.; Janadri, S. Anti-inflammatory effect of wedelolactone on DSS induced colitis in rats: IL-6/STAT3 signaling pathway. J. Ayurveda Integr. Med. 2023, 14, 100544. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.; Wannemuehler, M.; Hostetter, J. Mycobacterium avium paratuberculosis infection augments innate immune responses following intestinal epithelial injury. Exp. Biol. Med. 2014, 239, 436–441. [Google Scholar] [CrossRef] [PubMed]
- Jin, W.; Ai, H.; Huang, Q.; Li, C.; He, X.; Jin, Z.; Zuo, Y. Preclinical evidence of probiotics in ulcerative colitis: A systematic review and network meta-analysis. Front. Pharmacol. 2023, 14, 1187911. [Google Scholar] [CrossRef] [PubMed]
- Algieri, F.; Rodriguez-Nogales, A.; Garrido-Mesa, N.; Zorrilla, P.; Burkard, N.; Pischel, I.; Sievers, H.; Benedek, B.; Feistel, B.; Walbroel, B. Intestinal anti-inflammatory activity of the Serpylli herba extract in experimental models of rodent colitis. J. Crohn's Colitis 2014, 8, 775–788. [Google Scholar] [CrossRef]
- Mukhopadhyay, S.; Aich, P. Comparative severity analysis of colitis in C57BL/6 than BALB/c mice: A novel and rapid model of DSS induced colitis. bioRxiv 2020. [Google Scholar] [CrossRef]
- Heller, F.; Fuss, I.J.; Nieuwenhuis, E.E.; Blumberg, R.S.; Strober, W. Oxazolone colitis, a Th2 colitis model resembling ulcerative colitis, is mediated by IL-13-producing NK-T cells. Immunity 2002, 17, 629–638. [Google Scholar] [CrossRef]
- Yang, J.; Zhao, J.; Nakaguchi, T.; Gregersen, H. Biomechanical changes in oxazolone-induced colitis in BALB/C mice. J. Biomech. 2009, 42, 811–817. [Google Scholar] [CrossRef]
- Wang, X.; Ouyang, Q.; Luo, W.J. Oxazolone-induced murine model of ulcerative colitis. Chin. J. Dig. Dis. 2004, 5, 165–168. [Google Scholar] [CrossRef] [PubMed]
- Strober, W.; Fuss, I.J. Proinflammatory cytokines in the pathogenesis of inflammatory bowel diseases. Gastroenterology 2011, 140, 1756–1767.e51. [Google Scholar] [CrossRef] [PubMed]
- Kasaian, M.T.; Page, K.M.; Fish, S.; Brennan, A.; Cook, T.A.; Moreira, K.; Zhang, M.; Jesson, M.; Marquette, K.; Agostinelli, R. Therapeutic activity of an interleukin-4/interleukin-13 dual antagonist on oxazolone-induced colitis in mice. Immunology 2014, 143, 416–427. [Google Scholar] [CrossRef] [PubMed]
- Engel, M.A.; Khalil, M.; Siklosi, N.; Mueller-Tribbensee, S.M.; Neuhuber, W.L.; Neurath, M.F.; Becker, C.; Reeh, P.W. Opposite effects of substance P and calcitonin gene-related peptide in oxazolone colitis. Dig. Liver Dis. 2012, 44, 24–29. [Google Scholar] [CrossRef] [PubMed]
- Charles, P.C.; Weber, K.S.; Cipriani, B.; Brosnan, C.F. Cytokine, chemokine and chemokine receptor mRNA expression in different strains of normal mice: Implications for establishment of a Th1/Th2 bias. J. Neuroimmunol. 1999, 100, 64–73. [Google Scholar] [CrossRef]
- Brierley, S.M.; Linden, D.R. Neuroplasticity and dysfunction after gastrointestinal inflammation. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 611–627. [Google Scholar] [CrossRef] [PubMed]
- Robinson, A.M.; Rahman, A.A.; Carbone, S.E.; Randall-Demllo, S.; Filippone, R.; Bornstein, J.C.; Eri, R.; Nurgali, K. Alterations of colonic function in the Winnie mouse model of spontaneous chronic colitis. Am. J. Physiol. Gastrointest. Liver Physiol. 2017, 312, G85–G102. [Google Scholar] [CrossRef] [PubMed]
- Buisine, M.; Desreumaux, P.; Leteurtre, E.; Copin, M.; Colombel, J.; Porchet, N.; Aubert, J. Mucin gene expression in intestinal epithelial cells in Crohn’s disease. Gut 2001, 49, 544–551. [Google Scholar] [CrossRef]
- Heazlewood, C.K.; Cook, M.C.; Eri, R.; Price, G.R.; Tauro, S.B.; Taupin, D.; Thornton, D.J.; Png, C.W.; Crockford, T.L.; Cornall, R.J. Aberrant mucin assembly in mice causes endoplasmic reticulum stress and spontaneous inflammation resembling ulcerative colitis. PLoS Med. 2008, 5, e54. [Google Scholar] [CrossRef]
- Robinson, A.M.; Gondalia, S.V.; Karpe, A.V.; Eri, R.; Beale, D.J.; Morrison, P.D.; Palombo, E.A.; Nurgali, K. Fecal Microbiota and Metabolome in a Mouse Model of Spontaneous Chronic Colitis: Relevance to Human Inflammatory Bowel Disease. J. Leukoc. Biol. 2016, 22, 2767–2787. [Google Scholar] [CrossRef]
- Yang, W.; Cong, Y. Exploring colitis through dynamic T cell adoptive transfer models. J. Leukoc. Biol. 2023, 29, 1673–1680. [Google Scholar] [CrossRef]
- Eri, R.; McGuckin, M.A.; Wadley, R. T cell transfer model of colitis: A great tool to assess the contribution of T cells in chronic intestinal inflammation. Methods Mol. Biol. 2012, 844, 261–275. [Google Scholar] [CrossRef]
- Morrissey, P.J.; Charrier, K.; Braddy, S.; Liggitt, D.; Watson, J. CD4+ T cells that express high levels of CD45RB induce wasting disease when transferred into congenic severe combined immunodeficient mice. Disease development is prevented by cotransfer of purified CD4+ T cells. J. Exp. Med. 1993, 178, 237–244. [Google Scholar] [CrossRef]
- Birkeland, M.; Metlay, J.; Sanders, V.; Fernandez-Botran, R.; Vitetta, E.; Steinman, R.; Pure, E. Epitopes on CD45R [T200] molecules define differentiation antigens on murine B and T lymphocytes. J. Mol. Cell. Immunol. 1988, 4, 71–85. [Google Scholar]
- Lee, W.T.; Yin, X.-M.; Vitetta, E.S. Functional and ontogenetic analysis of murine CD45Rhi and CD45Rlo CD4+ T cells. J. Immunol. 1990, 144, 3288–3295. [Google Scholar] [CrossRef] [PubMed]
- Park, K.T.; Ehrlich, O.G.; Allen, J.I.; Meadows, P.; Szigethy, E.M.; Henrichsen, K.; Kim, S.C.; Lawton, R.C.; Murphy, S.M.; Regueiro, M.; et al. The cost of inflammatory bowel disease: An initiative from the crohn’s & colitis foundation. Inflamm. Bowel Dis. 2020, 26, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Powrie, F. T cells in inflammatory bowel disease: Protective and pathogenic roles. Immunity 1995, 3, 171–174. [Google Scholar] [CrossRef][Green Version]
- Sartor, R.B. Animal models of intestinal inflammation. Kirsner’s Inflamm. Bowel Dis. 2004, 6, 120–137. [Google Scholar] [CrossRef]
- Ostanin, D.V.; Pavlick, K.P.; Bharwani, S.; D’Souza, D.; Furr, K.L.; Brown, C.M.; Grisham, M.B. T cell-induced inflammation of the small and large intestine in immunodeficient mice. Am. J. Physiol. Gastrointest. Liver Physiol. 2006, 290, G109–G119. [Google Scholar] [CrossRef] [PubMed]
- Nieuwenhuis, E.E.; Neurath, M.F.; Corazza, N.; Iijima, H.; Trgovcich, J.; Wirtz, S.; Glickman, J.; Bailey, D.; Yoshida, M.; Galle, P.R. Disruption of T helper 2-immune responses in Epstein–Barr virus-induced gene 3-deficient mice. Proc. Natl. Acad. Sci. USA 2002, 99, 16951–16956. [Google Scholar] [CrossRef]
- Iijima, H.; Neurath, M.F.; Nagaishi, T.; Glickman, J.N.; Nieuwenhuis, E.E.; Nakajima, A.; Chen, D.; Fuss, I.J.; Utku, N.; Lewicki, D.N. Specific regulation of T helper cell 1–mediated murine colitis by CEACAM1. J. Exp. Med. 2004, 199, 471–482. [Google Scholar] [CrossRef]
- Li, H.; Song, J.; Niu, G.; Zhang, H.; Guo, J.; Shih, D.Q.; Targan, S.R.; Zhang, X. TL1A blocking ameliorates intestinal fibrosis in the T cell transfer model of chronic colitis in mice. Pathol. Res. Pract. 2018, 214, 217–227. [Google Scholar] [CrossRef]
- Bramhall, M.; Flórez-Vargas, O.; Stevens, R.; Brass, A.; Cruickshank, S. Quality of Methods Reporting in Animal Models of Colitis. Inflamm. Bowel Dis. 2015, 21, 1248–1259. [Google Scholar] [CrossRef] [PubMed]
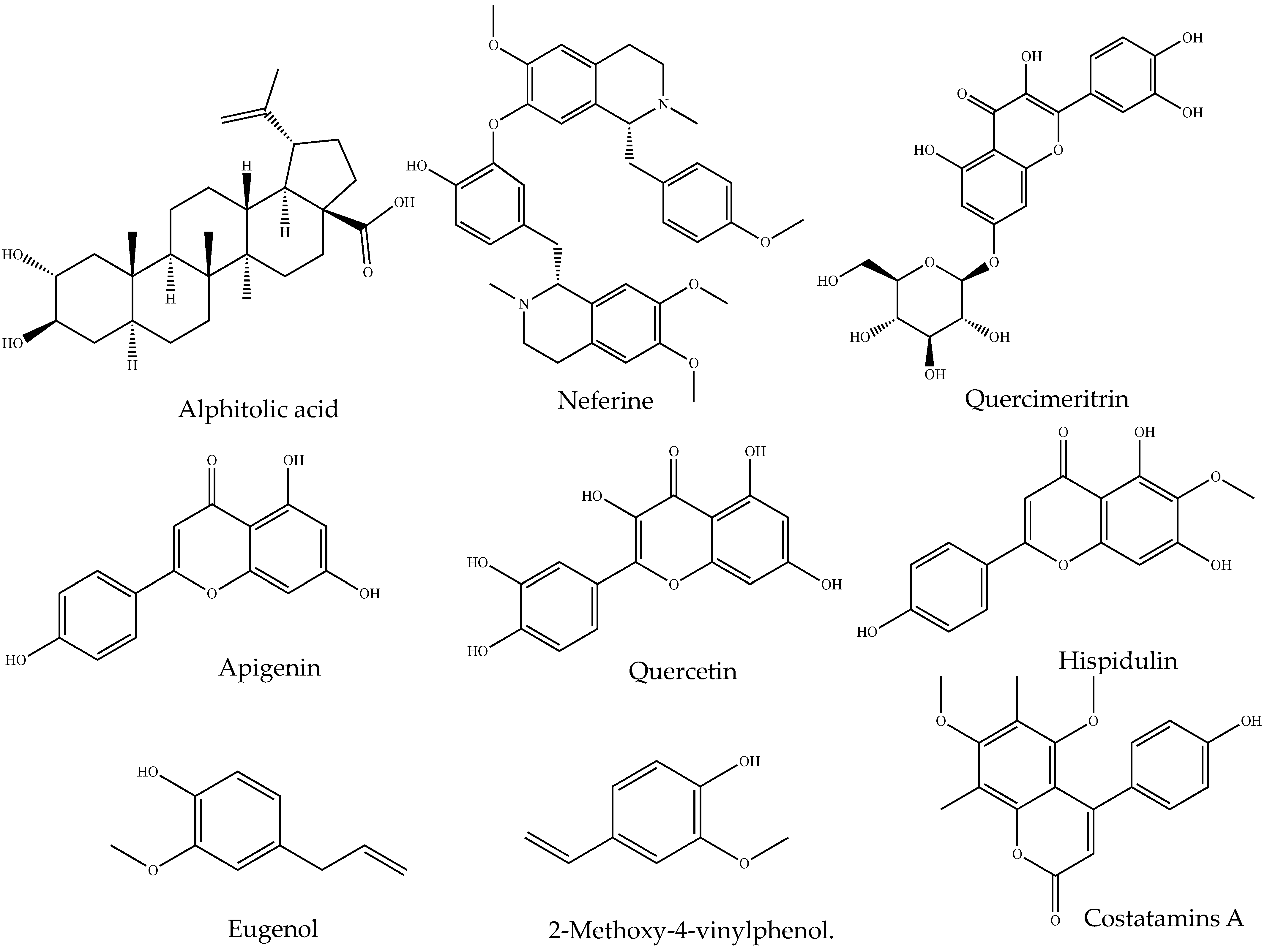
| Source | Class of Organic Compound | Isolated Small Molecules | Anti-Inflammatory Activity |
|---|---|---|---|
| Angophora costata (Gaertn.) Britten | Alkaloid | Costatamins A | Suppressed NO production and decreased TNF secretion in RAW 264.7 cells [131] |
| Nelumbo nucifera Gaertn. | Neferine | Decreased the production of IL-6 and TNF in RAW 264.7 cells activated by lipopolysaccharide activated [132] | |
| Ochrosia elliptica Labill. | 10-Methoxyconolidine | Decreased the level of TNF, IL-6 and NO production in lipopolysaccharide stimulated RAW 264.7 cells [133] | |
| Alphitonia petriei Braid & C.T.White | Terpenoid | alphitolic acid | suppressed the production of NO and TNF in RAW 264.7 cells activated by lipopolysaccharide + IFN-γ [134] |
| Centipeda minima (L.) A.Braun & Asch. | Centiplide A | Suppressed the production of NO in RAW 264.7 cell treated with liposaccharides [135] | |
| Tannins | Tannic acid | HaCaT cells exposed to UVB irradiation when treated with tannic acid, inhibited production of the proinflammatory cytokine IL-18, IL-1, IL-6, TNF, COX-2, and PGE2 and elevate its mRNA expression [136] | |
| Clerodendrum inerme Gaertn. | Flavonoid | Hispidulin | suppressed the production of PGE2 and expression of the expressions of iNOS and COX-2 by blocking NF-κB DNA-binding activity and JNK pathway [137] |
| Nelumbo nucifera Gaertn. | Quercetin | Reduced NO production in lipopolysaccharides treated RAW 264.7 cells [138] | |
| Merremia tridentata (L.) Hallier f. | Apigenin | Inhibit IL-1β, IL-6 and TNF production in lipopolysaccharide induced murine BV2 microglia cells [139] | |
| Ipomoea pes-caprae (L.) R.Br. | Phenolic | Eugenol and 2-Methoxy-4-vinylphenol. | Reduced the synthesis of prostaglandins [140]. |
| Barringtonia racemose (L.) Spreng. | Glycoside | Barringoside I | Exhibited a moderate inhibition NO production in lipopolysaccharide stimulated RAW 264.7 cells [141] |
| Brasenia schreberi J.F.Gmel. | Quercimeritrin | Suppressed the expression iNOS and NO in lipopolysaccharide-stimulated RAW 264.7 cells [142] |
| Types of Tests | Reagent/Chemical Added to Extract | Confirmatory Color Change |
|---|---|---|
| Alkaloid test | ||
| Dragendorff’s test | Potassium bismuth iodide solution (1 mL) | Orange, red precipitate |
| Wagner’s test | Potassium iodide solution (1 mL) | Reddish brown precipitate |
| Mayer’s test | Potassium mercuric iodide solution (1 mL) | Whitish or cream |
| Hager’s test | Saturated ferric solution (1 mL) | Yellow-colored precipitate |
| Steroid test | ||
| Libermann Burchard’s test | Acetic anhydrites + sulfuric acid | Violet to blue-colored ring |
| Terpenoid test | ||
| Copper acetate test | Copper acetate solution (3–4 drops) | Emerald, green color |
| Salkowski’s test | CHCL3 and concentrated H2SO4 (2 and 3 mL respectively) | Reddish brown color |
| Tannins test | ||
| Gelatin’s test | Gelatin solution + sodium chloride (1%) | Appearance of white precipitate |
| Flavonoid test | ||
| Lead acetate test | Lead acetate solution (2–3 drops) | Yellow precipitate |
| Alkaline reagent test | Sodium hydroxide solution (2–3 drops) | Initially yellow color and turns colorless after adding dilute acid |
| Phenolic test | ||
| Ferric chloride test | Ferric chloride (2–3 drops) | Bluish-black color |
| Lead acetate test | Lead acetate (2–3 drops) | Yellow color |
| Gelatine test | Gelatin solution (1%) | White precipitate |
| Mayer’s reagent test (potassium mercuric iodide test) | Mayer’s reagent (1 mL) | white precipitate |
| Anthraquinone test | ||
| Bontrager’s test | Boiled extract (In 10% of HCL for 2–3 min) Add CHCL3 to filtrate (2–3 drops of 10% NH3) Heat mixture (3–4 min) | Pink color |
| Cell Line Assay for IBD | Advantages | Limitations |
|---|---|---|
| THP-1 (Human leukemia monocytic cell line) |
| |
| Caco-2 cell line |
| |
| HT29 (Human colorectal adenocarcinoma cell line) |
|
|
| The murine macrophage cell line (RAW 264.7) |
|
|
| Peripheral blood mononuclear cells (PBMCs) assay |
|
|
| Animal Models | Advantages | Disadvantages |
|---|---|---|
| 2 2,4,6-trinitrobenzene sulfonic acid (TNBS)-induced colitis model |
|
|
| Dextran sodium sulfate (DSS)-induced colitis model |
| |
| Oxazolone ((4-ethoxylmethylene-2-phenyloxazol-5-one)-induced colitis model (OC)) |
| |
| Winnie mouse model of colitis |
|
|
| T cell adoptive transfer model |
|
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jamtsho, T.; Yeshi, K.; Perry, M.J.; Loukas, A.; Wangchuk, P. Approaches, Strategies and Procedures for Identifying Anti-Inflammatory Drug Lead Molecules from Natural Products. Pharmaceuticals 2024, 17, 283. https://doi.org/10.3390/ph17030283
Jamtsho T, Yeshi K, Perry MJ, Loukas A, Wangchuk P. Approaches, Strategies and Procedures for Identifying Anti-Inflammatory Drug Lead Molecules from Natural Products. Pharmaceuticals. 2024; 17(3):283. https://doi.org/10.3390/ph17030283
Chicago/Turabian StyleJamtsho, Tenzin, Karma Yeshi, Matthew J. Perry, Alex Loukas, and Phurpa Wangchuk. 2024. "Approaches, Strategies and Procedures for Identifying Anti-Inflammatory Drug Lead Molecules from Natural Products" Pharmaceuticals 17, no. 3: 283. https://doi.org/10.3390/ph17030283
APA StyleJamtsho, T., Yeshi, K., Perry, M. J., Loukas, A., & Wangchuk, P. (2024). Approaches, Strategies and Procedures for Identifying Anti-Inflammatory Drug Lead Molecules from Natural Products. Pharmaceuticals, 17(3), 283. https://doi.org/10.3390/ph17030283









