The Designed Pore-Forming Antimicrobial Peptide C14R Combines Excellent Activity against the Major Opportunistic Human Pathogen Pseudomonas aeruginosa with Low Cytotoxicity
Abstract
1. Introduction
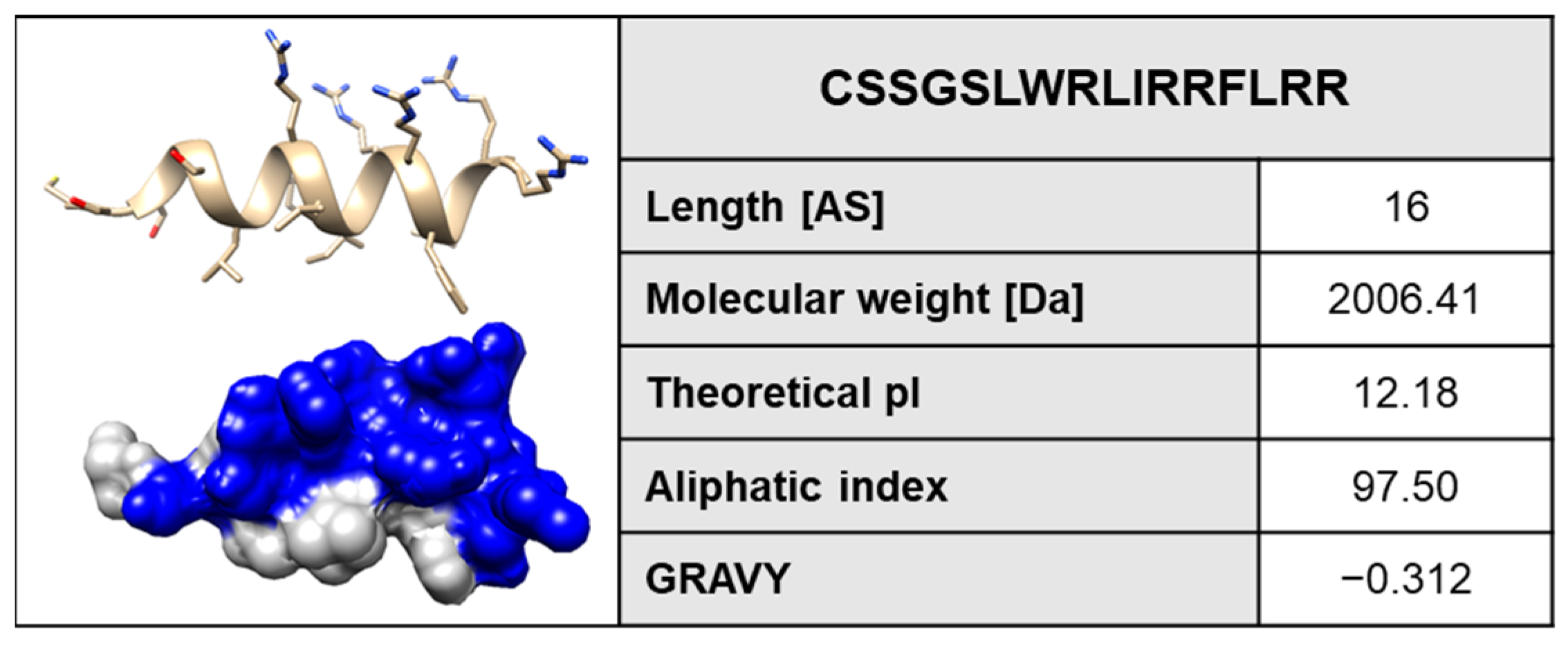
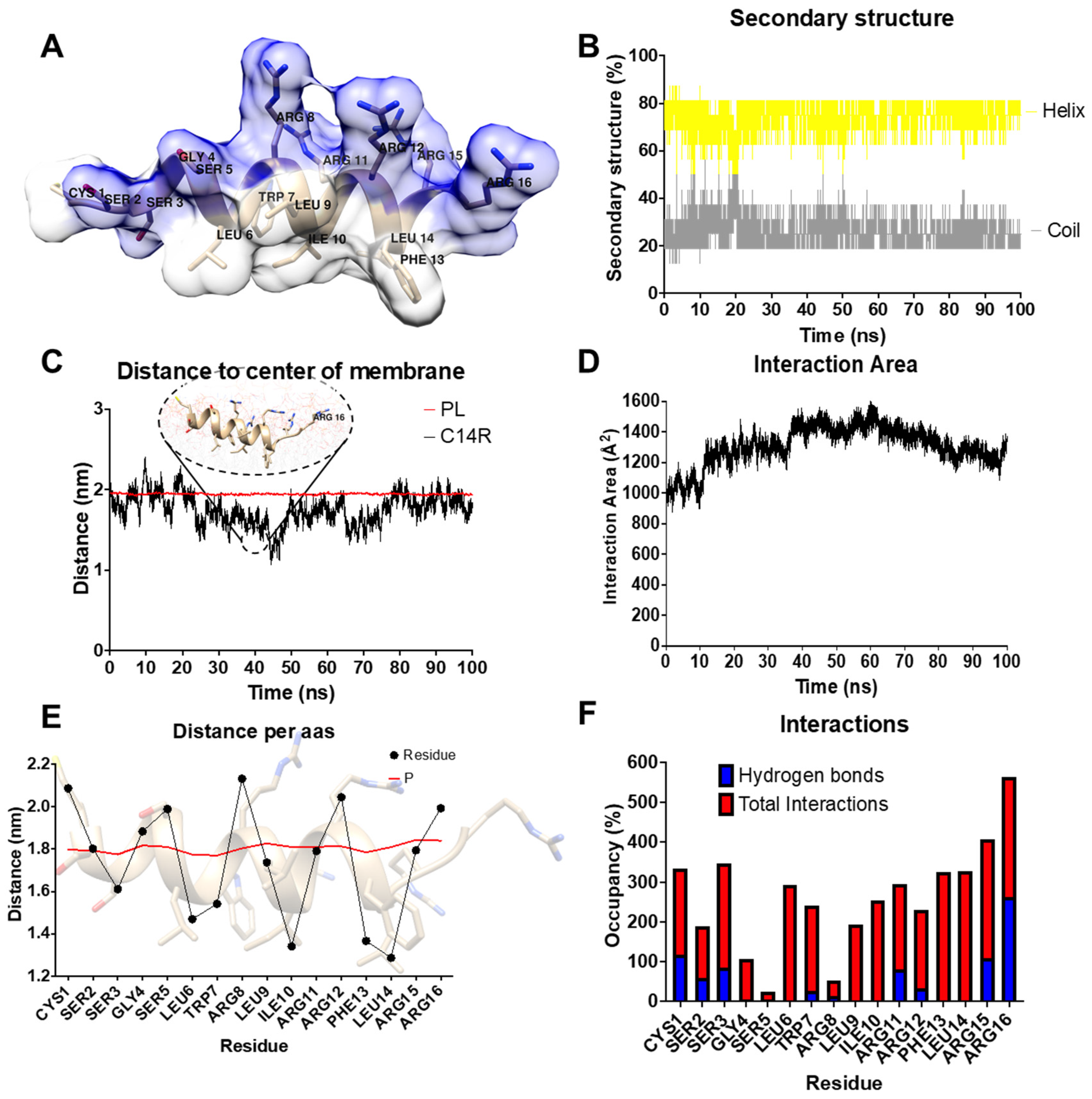
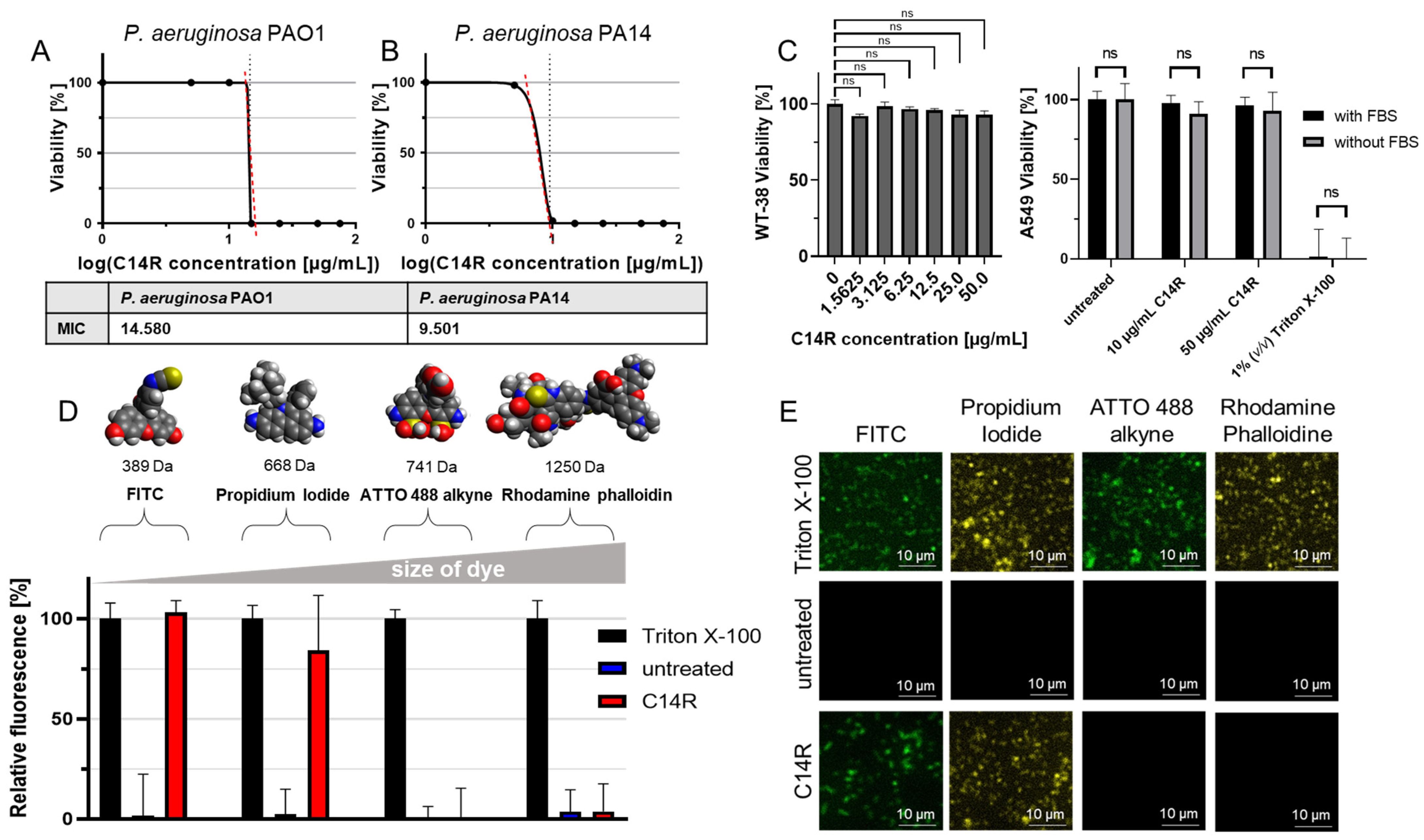
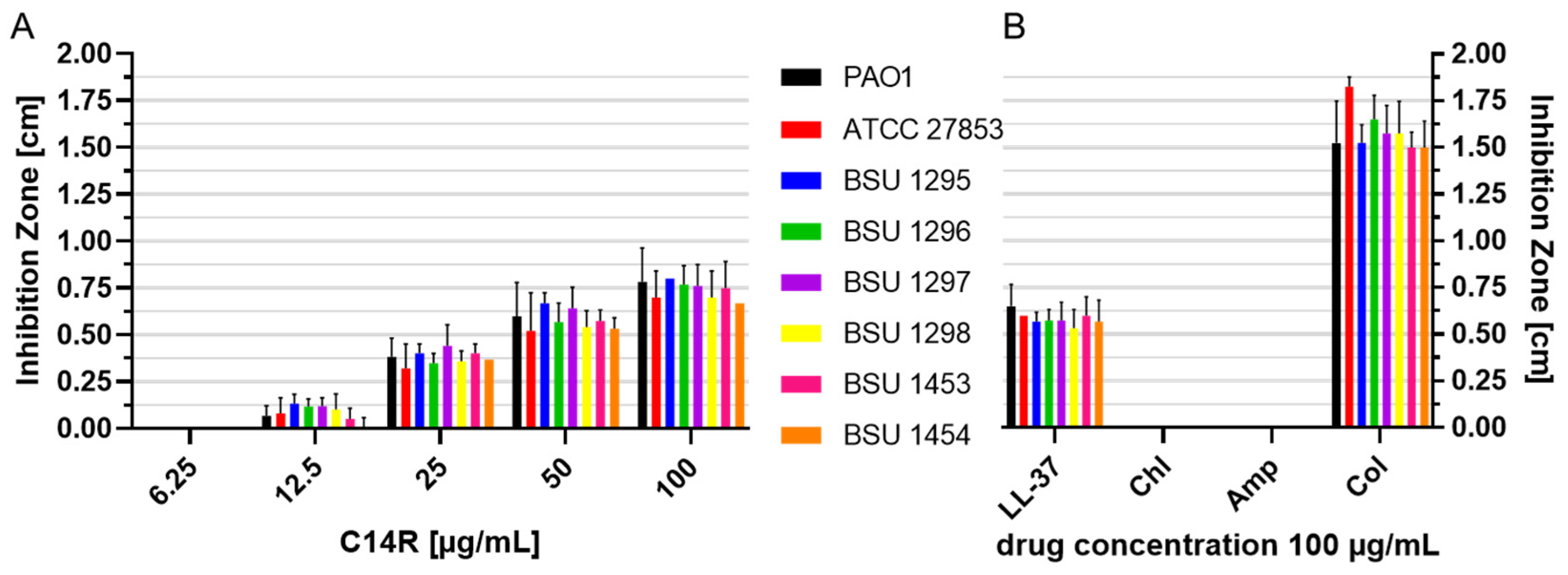
2. Results
3. Discussion
4. Materials and Methods
4.1. Molecular Dynamics Simulations
4.2. Peptide Synthesis
4.3. Cultivation of Pseudomonas Species
4.4. Viability Testing and Quantification
4.5. Cytotoxicity Testing
4.6. C14R Permeabilization Assay
4.7. Overlay Radial Diffusion Assay
4.8. Hemolysis Assay
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, Research, and Development of New Antibiotics: The WHO Priority List of Antibiotic-Resistant Bacteria and Tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef] [PubMed]
- Zasloff, M. Antimicrobial Peptides of Multicellular Organisms. Nature 2002, 415, 389–395. [Google Scholar] [CrossRef] [PubMed]
- Peters, B.M.; Shirtliff, M.E.; Jabra-Rizk, M.A. Antimicrobial Peptides: Primeval Molecules or Future Drugs? PLoS Pathog. 2010, 6, e1001067. [Google Scholar] [CrossRef] [PubMed]
- Radek, K.; Gallo, R. Antimicrobial Peptides: Natural Effectors of the Innate Immune System. Semin. Immunopathol. 2007, 29, 27–43. [Google Scholar] [CrossRef] [PubMed]
- Bahar, A.A.; Ren, D. Antimicrobial Peptides. Pharmaceuticals 2013, 6, 1543–1575. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Zhang, M.; Lai, R.; Zhang, Z. Chemical Modifications to Increase the Therapeutic Potential of Antimicrobial Peptides. Peptides 2021, 146, 170666. [Google Scholar] [CrossRef] [PubMed]
- Magana, M.; Pushpanathan, M.; Santos, A.L.; Leanse, L.; Fernandez, M.; Ioannidis, A.; Giulianotti, M.A.; Apidianakis, Y.; Bradfute, S.; Ferguson, A.L.; et al. The Value of Antimicrobial Peptides in the Age of Resistance. Lancet Infect. Dis. 2020, 20, e216–e230. [Google Scholar] [CrossRef]
- López-Abarrategui, C.; McBeth, C.; Mandal, S.M.; Sun, Z.J.; Heffron, G.; Alba-Menéndez, A.; Migliolo, L.; Reyes-Acosta, O.; García-Villarino, M.; Nolasco, D.O.; et al. Cm-P5: An Antifungal Hydrophilic Peptide Derived from the Coastal Mollusk Cenchritis Muricatus (Gastropoda: Littorinidae). FASEB J. 2015, 29, 3315–3325. [Google Scholar] [CrossRef]
- Kubiczek, D.; Raber, H.; Gonzalez-García, M.; Morales-Vicente, F.; Staendker, L.; Otero-Gonzalez, A.J.; Rosenau, F. Derivates of the Antifungal Peptide CM-P5 Inhibit Development of Candida Auris Biofilms in Vitro. Antibiotics 2020, 9, 363. [Google Scholar] [CrossRef]
- Vicente, F.E.M.; González-Garcia, M.; Diaz Pico, E.; Moreno-Castillo, E.; Garay, H.E.; Rosi, P.E.; Jimenez, A.M.; Campos-Delgado, J.A.; Rivera, D.G.; Chinea, G.; et al. Design of a Helical-Stabilized, Cyclic, and Nontoxic Analogue of the Peptide Cm-P5 with Improved Antifungal Activity. ACS Omega 2019, 4, 19081–19095. [Google Scholar] [CrossRef]
- Rodriguez, A.; Martell-Huguet, E.M.; González-García, M.; Alpízar-Pedraza, D.; Alba, A.; Vazquez, A.A.; Grieshober, M.; Spellerberg, B.; Stenger, S.; Münch, J.; et al. Identification and Characterization of Three New Antimicrobial Peptides from the Marine Mollusk Nerita Versicolor (Gmelin, 1791). Int. J. Mol. Sci. 2023, 24, 3852. [Google Scholar] [CrossRef] [PubMed]
- García, M.G.; Rodríguez, A.; Alba, A.; Vázquez, A.A.; Vicente, F.E.M.; Pérez-Erviti, J.; Spellerberg, B.; Stenger, S.; Grieshober, M.; Conzelmann, C.; et al. New Antibacterial Peptides from the Freshwater Mollusk Pomacea Poeyana (Pilsbry, 1927). Biomolecules 2020, 10, 1473. [Google Scholar] [CrossRef]
- Raber, H.F.; Sejfijaj, J.; Kissmann, A.K.; Wittgens, A.; Gonzalez-Garcia, M.; Alba, A.; Vázquez, A.A.; Vicente, F.E.M.; Erviti, J.P.; Kubiczek, D.; et al. Antimicrobial Peptides Pom-1 and Pom-2 from Pomacea Poeyana Are Active against Candida Auris, c. Parapsilosis and c. Albicans Biofilms. Pathogens 2021, 10, 496. [Google Scholar] [CrossRef] [PubMed]
- Amann, V.; Kissmann, A.-K.; Mildenberger, V.; Krebs, I.; Perez-Erviti, J.A.; Martell-Huguet, E.M.; Otero-Gonzalez, A.J.; Morales-Vicente, F.; Rodríguez-Castaño, G.P.; Firacative, C.; et al. Cm-P5 Peptide Dimers Inhibit Biofilms of Candida Albicans Clinical Isolates, C. Parapsilosis and Fluconazole-Resistant Mutants of C. Auris. Int. J. Mol. Sci. 2023, 24, 9788. [Google Scholar] [CrossRef] [PubMed]
- Jenssen, H.; Hamill, P.; Hancock, R.E.W. Peptide Antimicrobial Agents. Clin. Microbiol. Rev. 2006, 19, 491–511. [Google Scholar] [CrossRef] [PubMed]
- Browne, K.; Chakraborty, S.; Chen, R.; Willcox, M.D.; Black, D.S.; Walsh, W.R.; Kumar, N. A New Era of Antibiotics: The Clinical Potential of Antimicrobial Peptides. Int. J. Mol. Sci. 2020, 21, 7047. [Google Scholar] [CrossRef] [PubMed]
- Tossi, A.; Sandri, L.; Giangaspero, A. Amphipathic, α-Helical Antimicrobial Peptides. Pept. Sci. 2004, 55, 4–30. [Google Scholar] [CrossRef]
- Shai, Y. Mode of Action of Membrane Active Antimicrobial Peptides. Pept. Sci. 2004, 66, 236–248. [Google Scholar] [CrossRef]
- Jean-François, F.; Elezgaray, J.; Berson, P.; Vacher, P.; Dufourc, E.J. Pore Formation Induced by an Antimicrobial Peptide: Electrostatic Effects. Biophys. J. 2008, 95, 5748–5756. [Google Scholar] [CrossRef]
- Brogden, K.A. Antimicrobial Peptides: Pore Formers or Metabolic Inhibitors in Bacteria? Nat. Rev. Microbiol. 2005, 3, 238. [Google Scholar] [CrossRef]
- Li, W.; Separovic, F.; O’Brien-Simpson, N.M.; Wade, J.D. Chemically Modified and Conjugated Antimicrobial Peptides against Superbugs. Chem. Soc. Rev. 2021, 50, 4932–4973. [Google Scholar] [CrossRef] [PubMed]
- Torres, M.D.T.; Sothiselvam, S.; Lu, T.K.; de la Fuente-Nunez, C. Peptide Design Principles for Antimicrobial Applications. J. Mol. Biol. 2019, 431, 3547–3567. [Google Scholar] [CrossRef] [PubMed]
- Hilpert, K.; Volkmer-Engert, R.; Walter, T.; Hancock, R.E.W. High-Throughput Generation of Small Antibacterial Peptides with Improved Activity. Nat. Biotechnol. 2005, 23, 1008–1012. [Google Scholar] [CrossRef] [PubMed]
- Chan, D.I.; Prenner, E.J.; Vogel, H.J. Tryptophan- and Arginine-Rich Antimicrobial Peptides: Structures and Mechanisms of Action. Biochim. Et. Biophys. Acta (BBA)—Biomembr. 2006, 1758, 1184–1202. [Google Scholar] [CrossRef] [PubMed]
- Torcato, I.M.; Huang, Y.-H.; Franquelim, H.G.; Gaspar, D.; Craik, D.J.; Castanho, M.A.R.B.; Troeira Henriques, S. Design and Characterization of Novel Antimicrobial Peptides, R-BP100 and RW-BP100, with Activity against Gram-Negative and Gram-Positive Bacteria. Biochim. Et. Biophys. Acta (BBA)—Biomembr. 2013, 1828, 944–955. [Google Scholar] [CrossRef]
- Badosa, E.; Ferre, R.; Planas, M.; Feliu, L.; Besalú, E.; Cabrefiga, J.; Bardají, E.; Montesinos, E. A Library of Linear Undecapeptides with Bactericidal Activity against Phytopathogenic Bacteria. Peptides 2007, 28, 2276–2285. [Google Scholar] [CrossRef]
- McDonald, E.F.; Jones, T.; Plate, L.; Meiler, J.; Gulsevin, A. Benchmarking AlphaFold2 on Peptide Structure Prediction. Structure 2023, 31, 111–119.e2. [Google Scholar] [CrossRef]
- Rettie, S.A.; Campbell, K.V.; Bera, A.K.; Kang, A.; Kozlov, S.; De La Cruz, J.; Adebomi, V.; Zhou, G.; DiMaio, F.; Ovchinnikov, S.; et al. Cyclic Peptide Structure Prediction and Design Using AlphaFold. bioRxiv 2023, 26, 2023.02.25.529956. [Google Scholar] [CrossRef]
- Gleason, N.J.; Vostrikov, V.V.; Greathouse, D.V.; Grant, C.V.; Opella, S.J.; Koeppe, R.E. Tyrosine Replacing Tryptophan as an Anchor in GWALP Peptides. Biochemistry 2012, 51, 2044–2053. [Google Scholar] [CrossRef]
- He, J.; Baldini, R.L.; Déziel, E.; Saucier, M.; Zhang, Q.; Liberati, N.T.; Lee, D.; Urbach, J.; Goodman, H.M.; Rahme, L.G. The Broad Host Range Pathogen Pseudomonas Aeruginosa Strain PA14 Carries Two Pathogenicity Islands Harboring Plant and Animal Virulence Genes. Proc. Natl. Acad. Sci. USA 2004, 101, 2530–2535. [Google Scholar] [CrossRef]
- Brogden, N.K.; Brogden, K.A. Will New Generations of Modified Antimicrobial Peptides Improve Their Potential as Pharmaceuticals? Int. J. Antimicrob. Agents 2011, 38, 217–225. [Google Scholar] [CrossRef] [PubMed]
- Jung, O.; Smeets, R.; Hartjen, P.; Schnettler, R.; Feyerabend, F.; Klein, M.; Wegner, N.; Walther, F.; Stangier, D.; Henningsen, A.; et al. Improved In Vitro Test Procedure for Full Assessment of the Cytocompatibility of Degradable Magnesium Based on ISO 10993-5/-12. Int. J. Mol. Sci. 2019, 20, 255. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Narayana, J.L.; Mishra, B.; Zhang, Y.; Wang, F.; Wang, C.; Zarena, D.; Lushnikova, T.; Wang, X. Design of Antimicrobial Peptides: Progress Made with Human Cathelicidin LL-37. In Antimicrobial Peptides. Advances in Experimental Medicine and Biology; Springer: Singapore, 2019; pp. 215–240. [Google Scholar]
- Huang, Y.; Huang, J.; Chen, Y. Alpha-Helical Cationic Antimicrobial Peptides: Relationships of Structure and Function. Protein Cell 2010, 1, 143–152. [Google Scholar] [CrossRef] [PubMed]
- Ambrosio, R.L.; Rosselló, C.A.; Casares, D.; Palmieri, G.; Anastasio, A.; Escribá, P.V. The Antimicrobial Peptide 1018-K6 Interacts Distinctly with Eukaryotic and Bacterial Membranes, the Basis of Its Specificity and Bactericidal Activity. Int. J. Mol. Sci. 2022, 23, 12392. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, V.; Feio, M.J.; Bastos, M. Role of Lipids in the Interaction of Antimicrobial Peptides with Membranes. Prog. Lipid Res. 2012, 51, 149–177. [Google Scholar] [CrossRef] [PubMed]
- Huan, Y.; Kong, Q.; Mou, H.; Yi, H. Antimicrobial Peptides: Classification, Design, Application and Research Progress in Multiple Fields. Front. Microbiol. 2020, 11, 582779. [Google Scholar] [CrossRef] [PubMed]
- Deleu, M.; Crowet, J.-M.; Nasir, M.N.; Lins, L. Complementary Biophysical Tools to Investigate Lipid Specificity in the Interaction between Bioactive Molecules and the Plasma Membrane: A Review. Biochim. Et. Biophys. Acta (BBA)—Biomembr. 2014, 1838, 3171–3190. [Google Scholar] [CrossRef]
- Plotnikova, J.M.; Rahme, L.G.; Ausubel, F.M. Pathogenesis of the Human Opportunistic Pathogen Pseudomonas Aeruginosa PA14 in Arabidopsis. Plant Physiol. 2000, 124, 1766–1774. [Google Scholar] [CrossRef]
- Lépine, F.; Déziel, E.; Milot, S.; Rahme, L.G. A Stable Isotope Dilution Assay for the Quantification of the Pseudomonas Quinolone Signal in Pseudomonas Aeruginosa Cultures. Biochim. Et. Biophys. Acta (BBA)—Gen. Subj. 2003, 1622, 36–41. [Google Scholar] [CrossRef]
- Tan, M.-W.; Rahme, L.G.; Sternberg, J.A.; Tompkins, R.G.; Ausubel, F.M. Pseudomonas Aeruginosa Killing of Caenorhabditis Elegans Used to Identify, P. Aeruginosa Virulence Factors. Proc. Natl. Acad. Sci. USA 1999, 96, 2408–2413. [Google Scholar] [CrossRef]
- Greco, I.; Molchanova, N.; Holmedal, E.; Jenssen, H.; Hummel, B.D.; Watts, J.L.; Håkansson, J.; Hansen, P.R.; Svenson, J. Correlation between Hemolytic Activity, Cytotoxicity and Systemic in Vivo Toxicity of Synthetic Antimicrobial Peptides. Sci. Rep. 2020, 10, 13206. [Google Scholar] [CrossRef] [PubMed]
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.-C.; Williams, K.L.; Appel, R.D.; Hochstrasser, D.F. Protein Identification and Analysis Tools in the ExPASy Server. In 2-D Proteome Analysis Protocols; Humana Press: Totowa, NJ, USA, 1999; pp. 531–552. [Google Scholar]
- Jo, S.; Kim, T.; Im, W. Automated Builder and Database of Protein/Membrane Complexes for Molecular Dynamics Simulations. PLoS ONE 2007, 2, e880. [Google Scholar] [CrossRef] [PubMed]
- Jo, S.; Kim, T.; Iyer, V.G.; Im, W. CHARMM-GUI: A Web-based Graphical User Interface for CHARMM. J. Comput. Chem. 2008, 29, 1859–1865. [Google Scholar] [CrossRef] [PubMed]
- Jo, S.; Lim, J.B.; Klauda, J.B.; Im, W. CHARMM-GUI Membrane Builder for Mixed Bilayers and Its Application to Yeast Membranes. Biophys. J. 2009, 97, 50–58. [Google Scholar] [CrossRef] [PubMed]
- Wu, E.L.; Cheng, X.; Jo, S.; Rui, H.; Song, K.C.; Dávila-Contreras, E.M.; Qi, Y.; Lee, J.; Monje-Galvan, V.; Venable, R.M.; et al. CHARMM-GUI Membrane Builder toward Realistic Biological Membrane Simulations. J. Comput. Chem. 2014, 35, 1997–2004. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Phillips, J.C.; Braun, R.; Wang, W.; Gumbart, J.; Tajkhorshid, E.; Villa, E.; Chipot, C.; Skeel, R.D.; Kalé, L.; Schulten, K. Scalable Molecular Dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. [Google Scholar] [CrossRef]
- Vanommeslaeghe, K.; Hatcher, E.; Acharya, C.; Kundu, S.; Zhong, S.; Shim, J.; Darian, E.; Guvench, O.; Lopes, P.; Vorobyov, I.; et al. CHARMM General Force Field: A Force Field for Drug-like Molecules Compatible with the CHARMM All-atom Additive Biological Force Fields. J. Comput. Chem. 2010, 31, 671–690. [Google Scholar] [CrossRef]
- Best, R.B.; Zhu, X.; Shim, J.; Lopes, P.E.M.; Mittal, J.; Feig, M.; MacKerell, A.D. Optimization of the Additive CHARMM All-Atom Protein Force Field Targeting Improved Sampling of the Backbone ϕ, ψ and Side-Chain χ1 and χ2 Dihedral Angles. J. Chem. Theory Comput. 2012, 8, 3257–3273. [Google Scholar] [CrossRef]
- Klauda, J.B.; Venable, R.M.; Freites, J.A.; O’Connor, J.W.; Tobias, D.J.; Mondragon-Ramirez, C.; Vorobyov, I.; MacKerell, A.D.; Pastor, R.W. Update of the CHARMM All-Atom Additive Force Field for Lipids: Validation on Six Lipid Types. J. Phys. Chem. B 2010, 114, 7830–7843. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of Simple Potential Functions for Simulating Liquid Water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Cuendet, M.A.; van Gunsteren, W.F. On the Calculation of Velocity-Dependent Properties in Molecular Dynamics Simulations Using the Leapfrog Integration Algorithm. J. Chem. Phys. 2007, 127, 184102. [Google Scholar] [CrossRef] [PubMed]
- Darden, T.; York, D.; Pedersen, L. Particle Mesh Ewald: An N⋅log(N) Method for Ewald Sums in Large Systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef]
- Davidchack, R.L.; Handel, R.; Tretyakov, M.V. Langevin Thermostat for Rigid Body Dynamics. J. Chem. Phys. 2009, 130, 234101. [Google Scholar] [CrossRef] [PubMed]
- Feller, S.E.; Zhang, Y.; Pastor, R.W.; Brooks, B.R. Constant Pressure Molecular Dynamics Simulation: The Langevin Piston Method. J. Chem. Phys. 1995, 103, 4613–4621. [Google Scholar] [CrossRef]
- Stover, C.K.; Pham, X.Q.; Erwin, A.L.; Mizoguchi, S.D.; Warrener, P.; Hickey, M.J.; Brinkman, F.S.L.; Hufnagle, W.O.; Kowalik, D.J.; Lagrou, M.; et al. Complete Genome Sequence of Pseudomonas Aeruginosa PAO1, an Opportunistic Pathogen. Nature 2000, 406, 959–964. [Google Scholar] [CrossRef]
- CLSI M27-A3: Reference Method for Broth Dilution Antifungal Susceptibily Testing of Yeasts; CLSI: Berwyn, PA, USA, 2008; Volume 28.
- Fai, P.B.; Grant, A. A Rapid Resazurin Bioassay for Assessing the Toxicity of Fungicides. Chemosphere 2009, 74, 1165–1170. [Google Scholar] [CrossRef]
- Mosmann, T. Rapid Colorimetric Assay for Cellular Growth and Survival: Application to Proliferation and Cytotoxicity Assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
- Lieber, M.; Todaro, G.; Smith, B.; Szakal, A.; Nelson-Rees, W. A Continuous Tumor-cell Line from a Human Lung Carcinoma with Properties of Type II Alveolar Epithelial Cells. Int. J. Cancer 1976, 17, 62–70. [Google Scholar] [CrossRef]
- Vogel, V.; Bauer, R.; Mauerer, S.; Schiffelholz, N.; Haupt, C.; Seibold, G.M.; Fändrich, M.; Walther, P.; Spellerberg, B. Angicin, a Novel Bacteriocin of Streptococcus Anginosus. Sci. Rep. 2021, 11, 24377. [Google Scholar] [CrossRef]
- Groß, R.; Bauer, R.; Krüger, F.; Rücker-Braun, E.; Olari, L.-R.; Ständker, L.; Preising, N.; Rodríguez, A.A.; Conzelmann, C.; Gerbl, F.; et al. A Placenta Derived C-Terminal Fragment of β-Hemoglobin with Combined Antibacterial and Antiviral Activity. Front. Microbiol. 2020, 11, 508. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mildenberger, V.; Alpízar-Pedraza, D.; Martell-Huguet, E.M.; Krämer, M.; Bolotnikov, G.; Otero-Gonzalez, A.J.; Weil, T.; Rodriguez-Alfonso, A.; Preising, N.; Ständker, L.; et al. The Designed Pore-Forming Antimicrobial Peptide C14R Combines Excellent Activity against the Major Opportunistic Human Pathogen Pseudomonas aeruginosa with Low Cytotoxicity. Pharmaceuticals 2024, 17, 83. https://doi.org/10.3390/ph17010083
Mildenberger V, Alpízar-Pedraza D, Martell-Huguet EM, Krämer M, Bolotnikov G, Otero-Gonzalez AJ, Weil T, Rodriguez-Alfonso A, Preising N, Ständker L, et al. The Designed Pore-Forming Antimicrobial Peptide C14R Combines Excellent Activity against the Major Opportunistic Human Pathogen Pseudomonas aeruginosa with Low Cytotoxicity. Pharmaceuticals. 2024; 17(1):83. https://doi.org/10.3390/ph17010083
Chicago/Turabian StyleMildenberger, Vanessa, Daniel Alpízar-Pedraza, Ernesto M. Martell-Huguet, Markus Krämer, Grigory Bolotnikov, Anselmo J. Otero-Gonzalez, Tanja Weil, Armando Rodriguez-Alfonso, Nico Preising, Ludger Ständker, and et al. 2024. "The Designed Pore-Forming Antimicrobial Peptide C14R Combines Excellent Activity against the Major Opportunistic Human Pathogen Pseudomonas aeruginosa with Low Cytotoxicity" Pharmaceuticals 17, no. 1: 83. https://doi.org/10.3390/ph17010083
APA StyleMildenberger, V., Alpízar-Pedraza, D., Martell-Huguet, E. M., Krämer, M., Bolotnikov, G., Otero-Gonzalez, A. J., Weil, T., Rodriguez-Alfonso, A., Preising, N., Ständker, L., Vogel, V., Spellerberg, B., Kissmann, A.-K., & Rosenau, F. (2024). The Designed Pore-Forming Antimicrobial Peptide C14R Combines Excellent Activity against the Major Opportunistic Human Pathogen Pseudomonas aeruginosa with Low Cytotoxicity. Pharmaceuticals, 17(1), 83. https://doi.org/10.3390/ph17010083







