Rapamycin as a Potential Alternative Drug for Squamous Cell Gingiva Carcinoma (Ca9-22): A Focus on Cell Cycle, Apoptosis and Autophagy Genetic Profile
Abstract
1. Introduction
2. Results
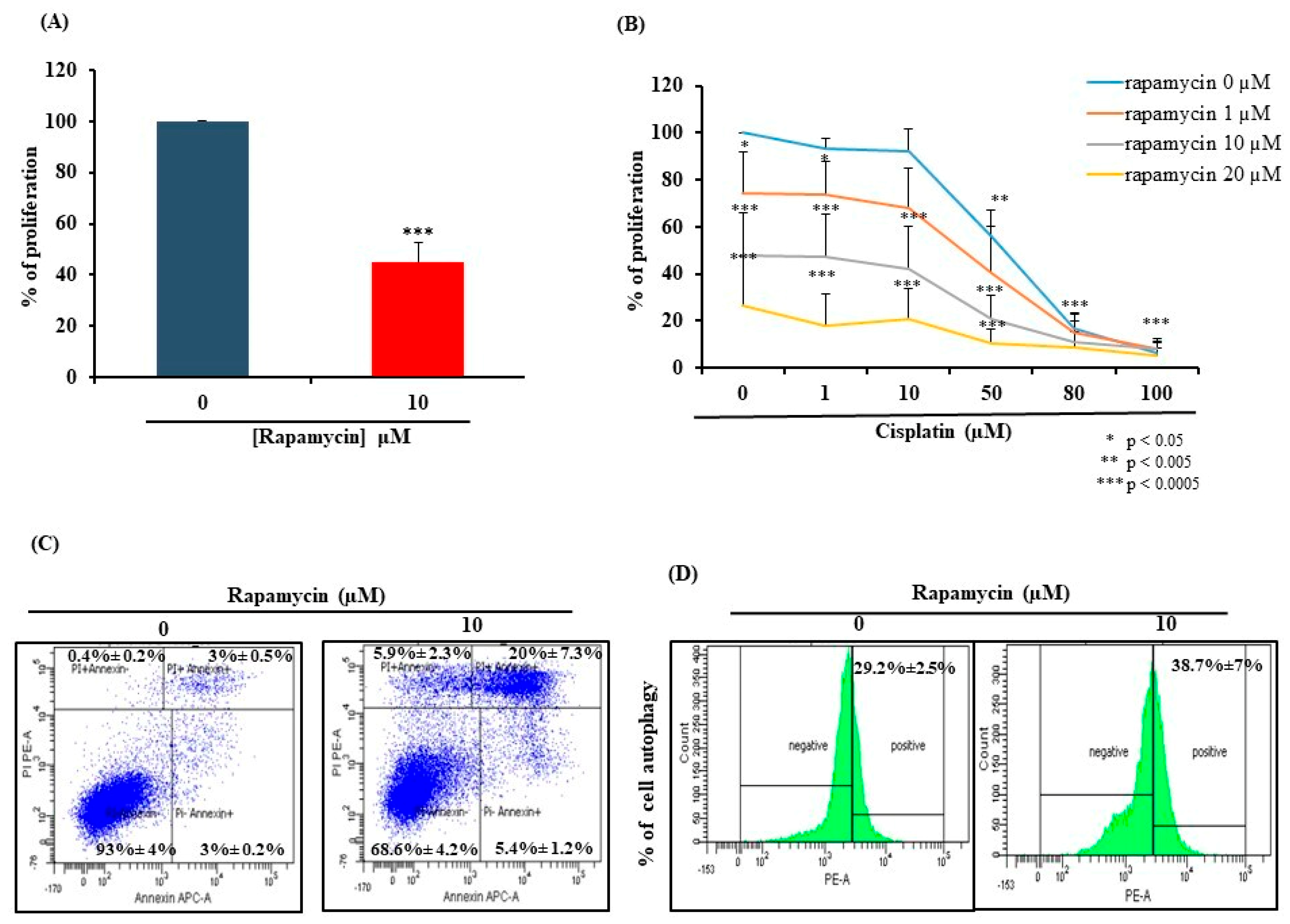
2.1. Rapamycin Exhibits Anti-Oral Cancer Activity through Stimulation of Apoptosis and Autophagy
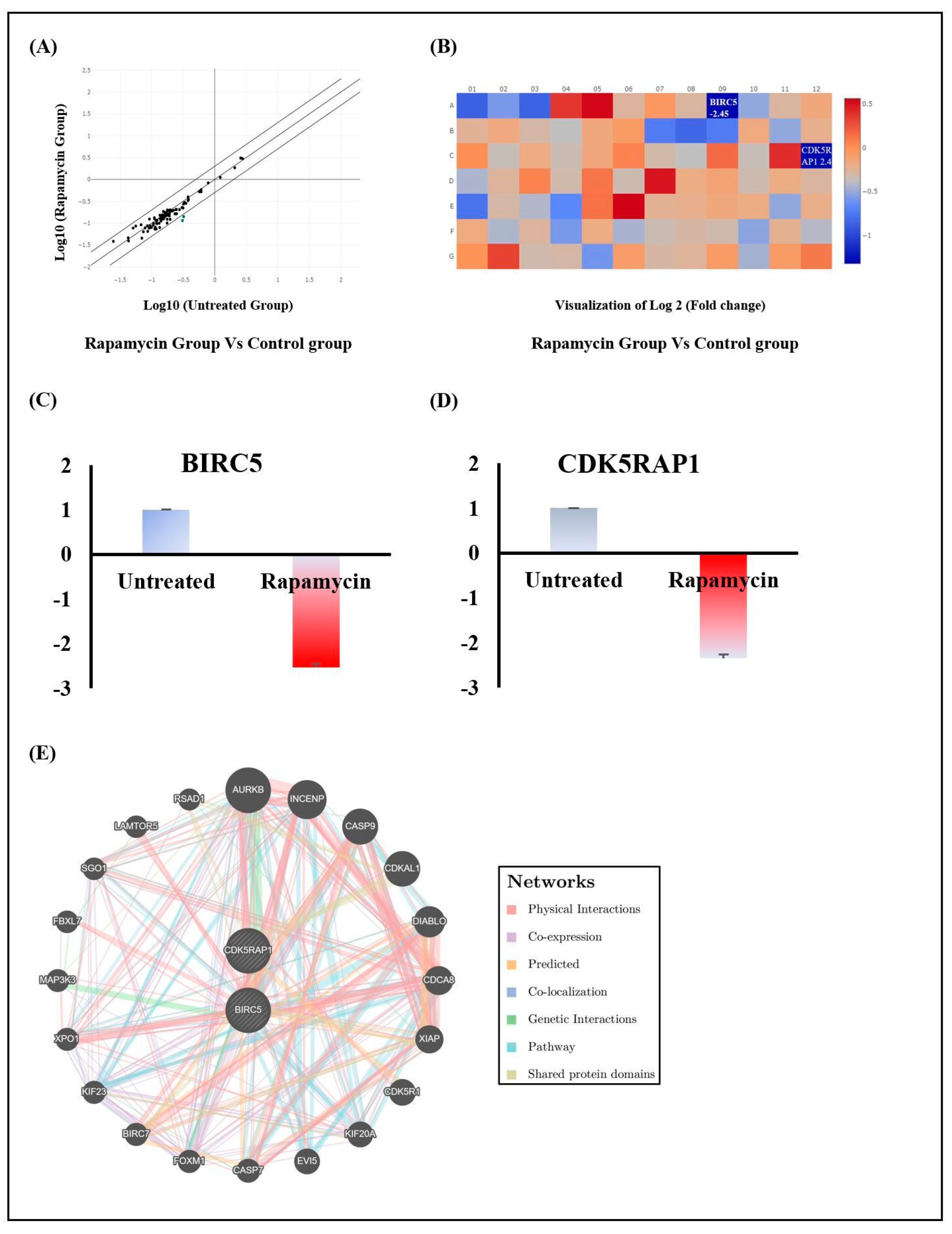
2.2. Rapamycin’s Action Is Mediated through the Repression of Cell Division Modulators BIRC5 and CDK5RAP1
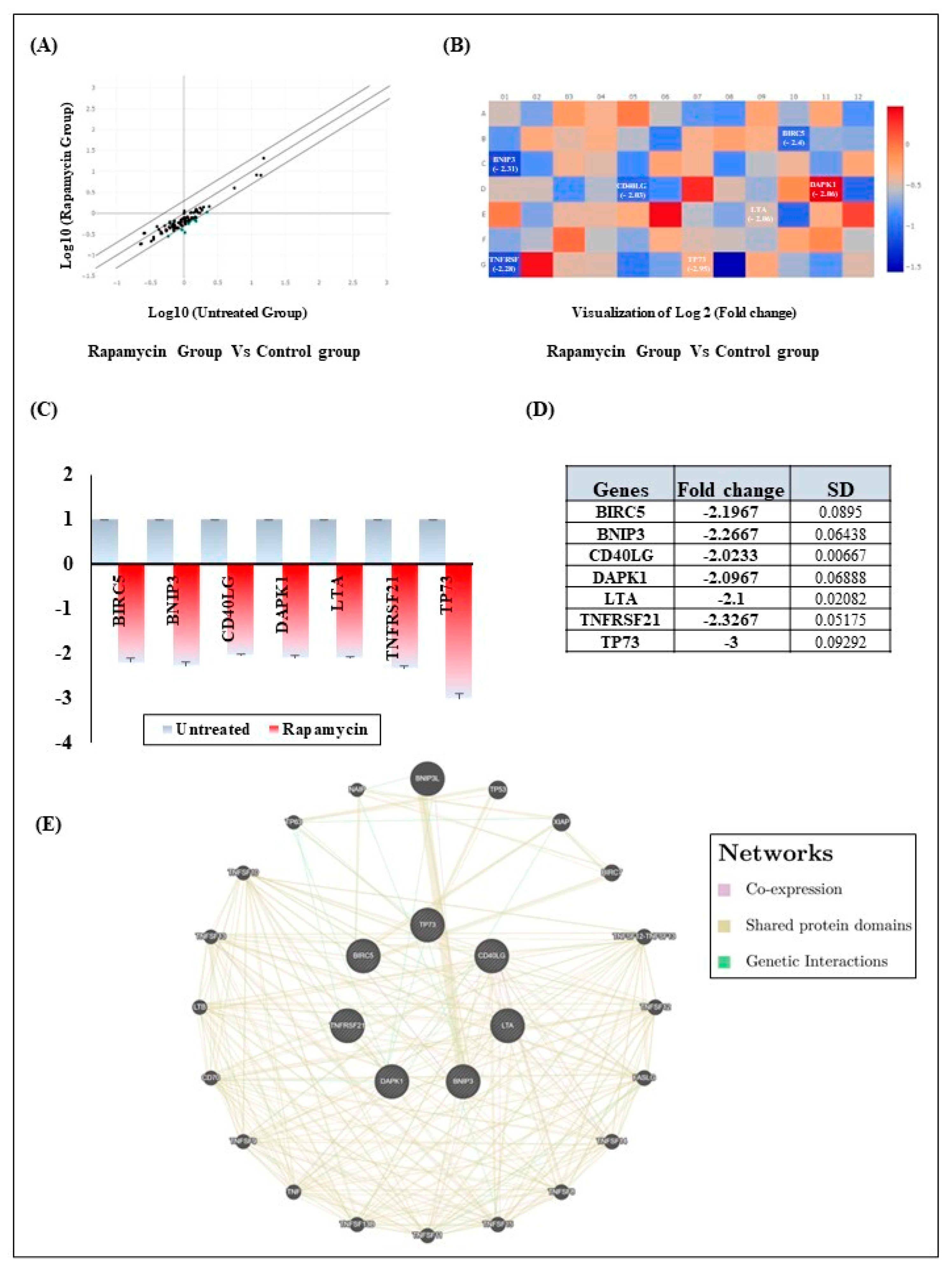
2.3. Rapamycin Induces Gingiva Carcinoma Cell Apoptosis via Downregulation of Intrinsic and Extrinsic Apoptotic Pathways Components
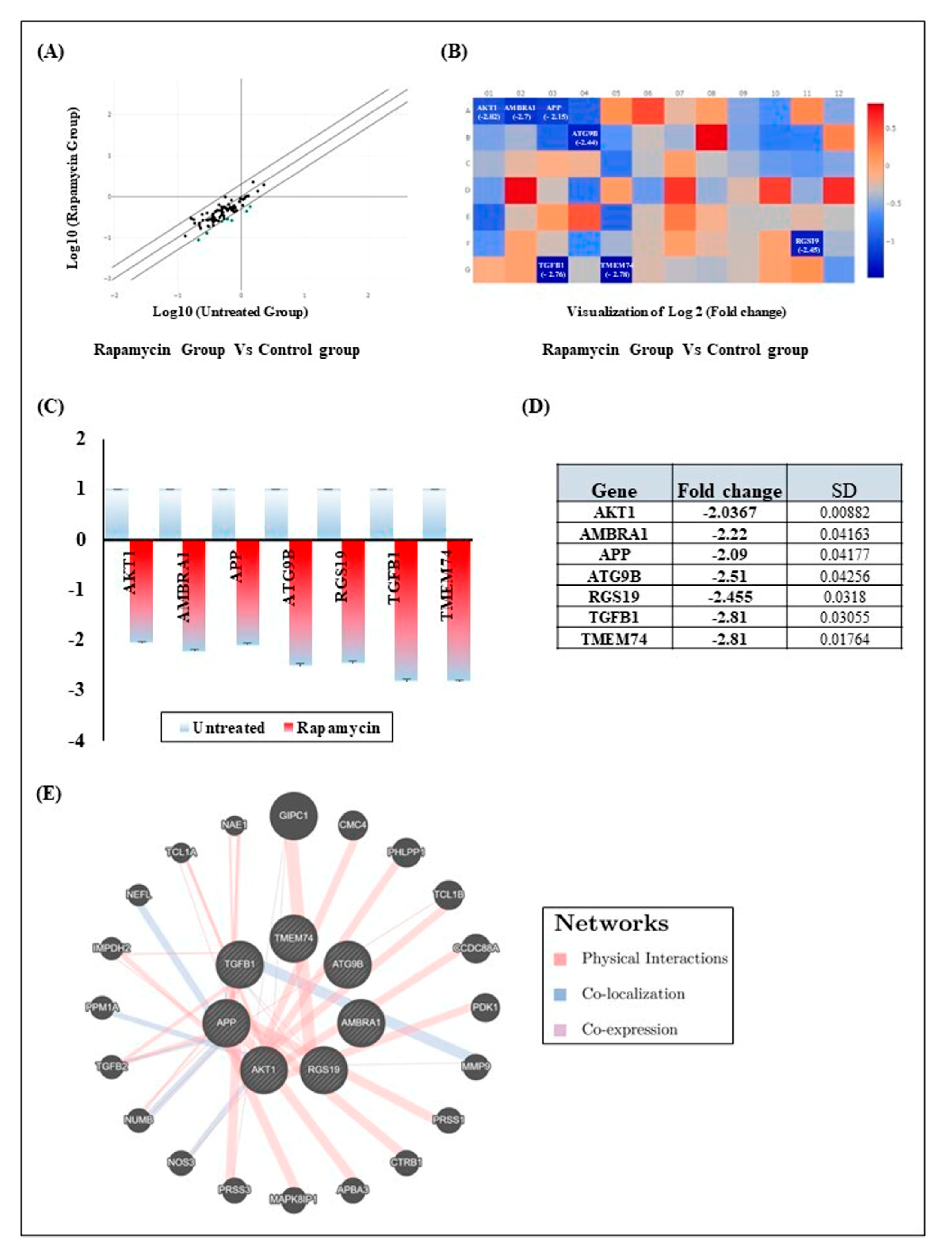
2.4. Rapamycin Treatment Induces Gingiva Carcinoma Cell Autophagy by Inhibition of Seven Autophagy Genes
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Cell Viability Assay
4.3. Cell Apoptosis Assay
4.4. Cell Autophagy Assay
4.5. RNA Extraction and Reverse Transcription
4.6. Gene Expression Using RT2 Profiler PCR Array
4.7. Gene–Gene Interaction Networks
4.8. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. WHO Highlights Oral Health Neglect Affecting Nearly Half of the World’s Population; Global Oral Health Status Report 2022; World Health Organization: Geneva, Switzerland, 2022. [Google Scholar]
- Sarode, G.; Maniyar, N.; Sarode, S.C.; Jafer, M.; Patil, S.; Awan, K.H. Epidemiologic aspects of oral cancer. Dis. Mon. 2020, 66, 100988. [Google Scholar] [CrossRef] [PubMed]
- National Cancer Institute. Side Effects of Cancer Treatment; National Cancer Institute: Bethesda, MD, USA, 2021. [Google Scholar]
- Parmar, A.; Macluskey, M.; Mc Goldrick, N.; Conway, I.D.; Glenny, A.-M.; Clarkson, E.J.; Worthington, H.V.; Chan, K.K. Interventions for the treatment of oral cavity and oropharyngeal cancer: Chemotherapy. Cochrane Database Syst. Rev. 2021, 12, CD006386. [Google Scholar] [PubMed]
- Dasari, S.; Tchounwou, P.B. Cisplatin in cancer therapy: Molecular mechanisms of action. Eur. J. Pharmacol. 2014, 740, 364–378. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Li, Z.; Luo, T.; Shi, H. Targeting the PI3K/AKT/mTOR and RAF/MEK/ERK pathways for cancer therapy. Mol. Biomed. 2022, 3, 47. [Google Scholar] [CrossRef] [PubMed]
- Popova, N.V.; Jücker, M. The Role of mTOR Signaling as a Therapeutic Target in Cancer. Int. J. Mol. Sci. 2021, 22, 1743. [Google Scholar] [CrossRef]
- Alzahrani, A.S. PI3K/Akt/mTOR inhibitors in cancer: At the bench and bedside. Semin. Cancer Biol. 2019, 59, 125–132. [Google Scholar] [CrossRef]
- Harsha, C.; Banik, K.; Ang, H.L.; Girisa, S.; Vikkurthi, R.; Parama, D.; Rana, V.; Shabnam, B.; Khatoon, E.; Kumar, A.P.; et al. Targeting AKT/mTOR in Oral Cancer: Mechanisms and Advances in Clinical Trials. Int. J. Mol. Sci. 2020, 21, 3285. [Google Scholar] [CrossRef] [PubMed]
- Pålsson-McDermott, E.M.; O’Neill, L.A.J. Targeting immunometabolism as an anti-inflammatory strategy. Cell Res. 2020, 30, 300–314. [Google Scholar] [CrossRef]
- Selvarani, R.; Mohammed, S.; Richardson, A. Effect of rapamycin on aging and age-related diseases-past and future. Geroscience 2021, 43, 1135–1158. [Google Scholar] [CrossRef] [PubMed]
- Kuerec, A.H.; Maier, A.B. Why is rapamycin not a rapalog? Gerontology 2023, 69, 657–659. [Google Scholar] [CrossRef]
- Populo, H.; Lopes, J.M.; Soares, P. The mTOR signalling pathway in human cancer. Int. J. Mol. Sci. 2012, 13, 1886–1918. [Google Scholar] [CrossRef] [PubMed]
- Semlali, A.; Papadakos, S.; Contant, C.; Zouaoui, I.; Rouabhia, M. Rapamycin inhibits oral cancer cell growth by promoting oxidative stress and suppressing ERK1/2, NF-kappaB and beta-catenin pathways. Front. Oncol. 2022, 12, 873447. [Google Scholar] [CrossRef]
- Beardmore, V.A.; Ahonen, L.J.; Gorbsky, G.J.; Kallio, M.J. Survivin dynamics increases at centromeres during G2/M phase transition and is regulated by microtubule-attachment and Aurora B kinase activity. J. Cell Sci. 2004, 117, 4033–4042. [Google Scholar] [CrossRef]
- Wang, H.; Wei, L.; Li, C.; Zhou, J.; Li, Z. CDK5RAP1 deficiency induces cell cycle arrest and apoptosis in human breast cancer cell line by the ROS/JNK signaling pathway. Oncol. Rep. 2015, 33, 1089–1096. [Google Scholar] [CrossRef] [PubMed]
- Xiong, J.; Wang, Y.; Gu, Y.; Xue, Y.; Dang, L.; Li, Y. CDK5RAP1 targeting NF-kappaB signaling pathway in human malignant melanoma A375 cell apoptosis. Oncol. Lett. 2018, 15, 4767–4774. [Google Scholar]
- Duronio, R.J.; Xiong, Y. Signaling Pathways that Control Cell Proliferation. Cold Spring Harb. Perspect. Biol. 2013, 5, 4767–4774. [Google Scholar] [CrossRef]
- Pennati, M.; Folini, M.; Zaffaroni, N. Targeting survivin in cancer therapy. Expert Opin. Ther. Targets 2008, 12, 463–476. [Google Scholar] [CrossRef]
- Oparina, N.; Erlandsson, M.C.; Fäldt Beding, A.; Parris, T.; Helou, K.; Karlsson, P.; Einbeigi, Z.; Bokarewa, M.I. Prognostic Significance of BIRC5/Survivin in Breast Cancer: Results from Three Independent Cohorts. Cancers 2021, 13, 2209. [Google Scholar] [CrossRef] [PubMed]
- Li, F. Discovery of survivin inhibitors and beyond: FL118 as a proof of concept. Int. Rev. Cell Mol. Biol. 2013, 305, 217–252. [Google Scholar]
- Du, C.Y.; Fang, M.; Li, Y.; Li, L.; Wang, X. Smac, a mitochondrial protein that promotes cytochrome c-dependent caspase activation by eliminating IAP inhibition. Cell 2000, 102, 33–42. [Google Scholar] [CrossRef]
- Zhang, M.D.; Wu, K.; Wang, M.; Bai, F.; Chen, H. CASP9 As a Prognostic Biomarker and Promising Drug Target Plays a Pivotal Role in Inflammatory Breast Cancer. Int. J. Anal. Chem. 2022, 2022, 1043445. [Google Scholar] [CrossRef]
- Mudde, A.C.A.; Booth, C.; Marsh, R.A. Evolution of Our Understanding of XIAP Deficiency. Front. Pediatr. 2021, 9, 660520. [Google Scholar] [CrossRef] [PubMed]
- Chen, E.R.; He, Y.; Jiang, J.; Yi, J.; Zou, Z.; Song, Q.; Ren, Q.; Lin, Z.; Lu, Y.; Liu, J.; et al. CDCA8 induced by NF-YA promotes hepatocellular carcinoma progression by regulating the MEK/ERK pathway. Exp. Hematol. Oncol. 2023, 12, 9. [Google Scholar] [CrossRef] [PubMed]
- Du, R.; Huang, C.; Liu, K.; Li, X.; Dong, Z. Targeting AURKA in Cancer: Molecular mechanisms and opportunities for Cancer therapy. Mol. Cancer 2021, 20, 15. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, A.; Miligy, I.M.; Toss, M.S.; Green, A.R.; Rakha, E.A. High Inner Centromere Protein (INCENP) expression correlates with aggressive features and predicts poor prognosis in patients with invasive breast cancer. Pathobiology 2023, 90, 377–388. [Google Scholar] [CrossRef]
- Melino, G.; Lu, X.; Gasco, M.; Crook, T.; Knight, R.A. Functional regulation of p73 and p63: Development and cancer. Trends Biochem. Sci. 2003, 28, 663–670. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.L.; Yu, W.; Xiao, H.; Lin, K. BIRC5 is a prognostic biomarker associated with tumor immune cell infiltration. Sci. Rep. 2021, 11, 390. [Google Scholar] [CrossRef] [PubMed]
- Vijayalingam, S.; Pillai, S.G.; Rashmi, R.; Subramanian, T.; Sagartz, J.E.; Chinnadurai, G. Overexpression of BH3-Only Protein BNIP3 Leads to Enhanced Tumor Growth. Genes Cancer 2010, 1, 964–971. [Google Scholar] [CrossRef]
- Burton, T.R.; Gibson, S.B. The role of Bcl-2 family member BNIP3 in cell death and disease: NIPping at the heels of cell death. Cell Death Differ. 2009, 16, 515–523. [Google Scholar] [CrossRef]
- Giatromanolaki, A.; Koukourakis, M.I.; Sowter, H.M.; Sivridis, E.; Gibson, S.; Gatter, K.C.; Harris, A.L. BNIP3 expression is linked with hypoxia-regulated protein expression and with poor prognosis in non-small cell lung cancer. Clin. Cancer Res. 2004, 10, 5566–5571. [Google Scholar] [CrossRef] [PubMed]
- Leo, C.; Horn, L.C.; Höckel, M. Hypoxia and expression of the proapoptotic regulator BNIP3 in cervical cancer. Int. J. Gynecol. Cancer 2006, 16, 1314–1320. [Google Scholar] [CrossRef]
- Sowter, H.M.; Ferguson, M.; Pym, C.; Watson, P.; Fox, S.B.; Han, C.; Harris, A.L. Expression of the cell death genes BNip3 and NIX in ductal carcinoma of the breast: Correlation of BNip3 levels with necrosis and grade. J. Pathol. 2003, 201, 573–580. [Google Scholar] [CrossRef]
- Gorbunova, A.S.; Yapryntseva, M.A.; Denisenko, T.V.; Zhivotovsky, B. BNIP3 in Lung Cancer: To Kill or Rescue? Cancers 2020, 12, 3390. [Google Scholar] [CrossRef] [PubMed]
- Elgueta, R.; Benson, M.J.; De Vries, V.C.; Wasiuk, A.; Guo, Y.; Noelle, R.J. Molecular mechanism and function of CD40/CD40L engagement in the immune system. Immunol. Rev. 2009, 229, 152–172. [Google Scholar] [CrossRef]
- Huang, Y.; Yu, X.; Wang, L.; Zhou, S.; Sun, J.; Feng, N.; Nie, S.; Wu, J.; Gao, F.; Fei, B.; et al. Four Genetic Polymorphisms of Lymphotoxin-Alpha Gene and Cancer Risk: A Systematic Review and Meta-Analysis. PLoS ONE 2013, 8, e82519. [Google Scholar] [CrossRef] [PubMed]
- Bergmann, S.; Pandolfi, P.P. Giving blood: A new role for CD40 in tumorigenesis. J. Exp. Med. 2006, 203, 2409–2412. [Google Scholar] [CrossRef]
- Coussens, L.M.; Werb, Z. Inflammation and cancer. Nature 2002, 420, 860–867. [Google Scholar] [CrossRef] [PubMed]
- Shalapour, S.; Karin, M. Immunity, inflammation, and cancer: An eternal fight between good and evil. J. Clin. Investig. 2015, 125, 3347–3355. [Google Scholar] [CrossRef] [PubMed]
- Pan, H.; Wu, S.; Wang, J.; Zhu, T.; Li, T.; Wan, B.; Liu, B.; Luo, Y.; Ma, X.; Sui, R.; et al. TNFRSF21 mutations cause high myopia. J. Med. Genet. 2019, 56, 671–677. [Google Scholar] [CrossRef]
- Nikolaev, A.; McLaughlin, T.; O’Leary, D.; Tessier-Lavigne, M. N-APP binds DR6 to trigger axon pruning and neuron death via distinct caspases. Nature 2009, 457, 981. [Google Scholar] [CrossRef] [PubMed]
- Movahhed, P.; Saberiyan, M.; Safi, A.; Arshadi, Z.; Kazerouni, F.; Teimori, H. The impact of DAPK1 and mTORC1 signaling association on autophagy in cancer. Mol. Biol. Rep. 2022, 49, 4959–4964. [Google Scholar] [CrossRef] [PubMed]
- Gade, P.; Singh, A.K.; Roy, S.K.; Reddy, S.P.; Kalvakolanu, D.V. Down-regulation of the transcriptional mediator subunit Med1 contributes to the loss of expression of metastasis-associated in human cancers and cancer cells. Int. J. Cancer 2009, 125, 1566–1574. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.L.; Zeng, W.H.; Xia, Y.M. TWEAK/Fn14 signaling in tumors. Tumor Biol. 2017, 39, 1010428317714624. [Google Scholar] [CrossRef] [PubMed]
- Levine, A.J.; Oren, M. The first 30 years of p53: Growing ever more complex. Nat. Rev. Cancer 2009, 9, 749–758. [Google Scholar] [CrossRef]
- Lu, M.; Lin, S.C.; Huang, Y.; Kang, Y.J.; Rich, R.; Lo, Y.C.; Myszka, D.; Han, J.; Wu, H. XIAP induces NF-κB activation via the BIR1/TAB1 interaction and BIR1 dimerization. Mol. Cell 2007, 26, 689–702. [Google Scholar] [CrossRef] [PubMed]
- Schweers, R.L.; Zhang, J.; Randall, M.S.; Loyd, M.R.; Li, W.; Dorsey, F.C.; Kundu, M.; Opferman, J.T.; Cleveland, J.L.; Miller, J.L.; et al. NIX is required for programmed mitochondrial clearance during reticulocyte maturation. Proc. Natl. Acad. Sci. USA 2007, 104, 19500–19505. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.I.; Kiyono, K.; Miyazono, K. Regulation of autophagy by transforming growth factor-beta (TGF-beta) signaling. Autophagy 2010, 6, 645–647. [Google Scholar] [CrossRef]
- Ding, Y.; Kim, S.; Lee, S.Y.; Koo, J.K.; Wang, Z.; Choi, M.E. Autophagy regulates TGF-beta expression and suppresses kidney fibrosis induced by unilateral ureteral obstruction. J. Am. Soc. Nephrol. 2014, 25, 2835–2846. [Google Scholar] [CrossRef] [PubMed]
- Padua, D.; Massague, J. Roles of TGFbeta in metastasis. Cell Res. 2009, 19, 89–102. [Google Scholar] [CrossRef] [PubMed]
- Moustakas, A.; Heldin, C.H. Signaling networks guiding epithelial-mesenchymal transitions during embryogenesis and cancer progression. Cancer Sci. 2007, 98, 1512–1520. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, R.; Shajimoon, A.; Afroz, R.; Gabr, M.; Thomas, W.G.; Little, P.J.; Kamato, D. Akt acts as a switch for GPCR transactivation of the TGF-beta receptor type 1. FEBS J. 2022, 289, 2642–2656. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Marcos, M.; Kietrsunthorn, P.S.; Pavlova, Y.; Adia, M.A.; Ghosh, P.; Farquhar, M.G. Functional characterization of the guanine nucleotide exchange factor (GEF) motif of GIV protein reveals a threshold effect in signaling. Proc. Natl. Acad. Sci. USA 2012, 109, 1961–1966. [Google Scholar] [CrossRef]
- Lin, C.; Ear, J.; Midde, K.; Lopez-Sanchez, I.; Aznar, N.; Garcia-Marcos, M.; Kufareva, I.; Abagyan, R.; Ghosh, P. Structural basis for activation of trimeric Gi proteins by multiple growth factor receptors via GIV/Girdin. Mol. Biol. Cell 2014, 25, 3654–3671. [Google Scholar] [CrossRef] [PubMed]
- Hosokawa, N.; Hara, T.; Kaizuka, T.; Kishi, C.; Takamura, A.; Miura, Y.; Iemura, S.-I.; Natsume, T.; Takehana, K.; Yamada, N.; et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol. Biol. Cell 2009, 20, 1981–1991. [Google Scholar] [CrossRef]
- Sun, Y.Z.; Li, Q.; Zhang, J.; Chen, Z.; He, Q.; Liu, X.; Zhao, N.; Yin, A.; Huang, H.; He, M.; et al. Autophagy regulatory molecule, TMEM74, interacts with BIK and inhibits BIK-induced apoptosis. Cell. Signal. 2017, 36, 34–41. [Google Scholar] [CrossRef]
- Di Bartolomeo, S.; Corazzari, M.; Nazio, F.; Oliverio, S.; Lisi, G.; Antonioli, M.; Pagliarini, V.; Matteoni, S.; Fuoco, C.; Giunta, L.; et al. The dynamic interaction of AMBRA1 with the dynein motor complex regulates mammalian autophagy. J. Cell Biol. 2010, 191, 155–168. [Google Scholar] [CrossRef] [PubMed]
- Orsi, A.; Razi, M.; Dooley, H.C.; Robinson, D.; Weston, A.E.; Collinson, L.M.; Tooze, S.A. Dynamic and transient interactions of Atg9 with autophagosomes, but not membrane integration, are required for autophagy. Mol. Biol. Cell 2012, 23, 1860–1873. [Google Scholar] [CrossRef] [PubMed]
- Young, A.R.J.; Chan, E.Y.W.; Hu, X.W.; Köchl, R.; Crawshaw, S.G.; High, S.; Hailey, D.W.; Lippincott-Schwartz, J.; Tooze, S.A. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J. Cell Sci. 2006, 119, 3888–3900. [Google Scholar] [CrossRef] [PubMed]
- Pagliarini, V.; Wirawan, E.; Romagnoli, A.; Ciccosanti, F.; Lisi, G.; Lippens, S.; Cecconi, F.; Fimia, G.M.; Vandenabeele, P.; Corazzari, M.; et al. Proteolysis of Ambra1 during apoptosis has a role in the inhibition of the autophagic pro-survival response. Cell Death Differ. 2012, 19, 1495–1504. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Chen, Y.; Zhang, J.; Cao, L.; He, M.; Liu, X.; Zhao, N.; Yin, A.; Huang, H.; Wang, L. TMEM74 promotes tumor cell survival by inducing autophagy via interactions with ATG16L1 and ATG9A. Cell Death Dis. 2017, 8, e3031. [Google Scholar] [CrossRef]
- Wang, N.; Tan, H.-Y.; Li, S.; Feng, Y. Atg9b Deficiency Suppresses Autophagy and Potentiates Endoplasmic Reticulum Stress-Associated Hepatocyte Apoptosis in Hepatocarcinogenesis. Theranostics 2017, 7, 2325–2338. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.A.; Rahman, M.S.; Rahman, M.H.; Rasheduzzaman, M.; Mamun-Or-Rashid, A.N.M.; Uddin, M.J.; Rahman, M.R.; Hwang, H.; Pang, M.G.; Rhim, H. Modulatory Effects of Autophagy on APP Processing as a Potential Treatment Target for Alzheimer’s Disease. Biomedicines 2021, 9, 5. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Koo, E.H. The amyloid precursor protein: Beyond amyloid. Mol. Neurodegener. 2006, 1, 5. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Oi, R.; Koizumi, H.; Maeda, I.; Noguchi, A.; Tatsunami, S.; Iwatani, T.; Kawamoto, H.; Tsugawa, K.; Takagi, M. Clinicopathological Significance of TARBP2, APP, and ZNF395 in Breast Cancer. Breast Cancer-Basic Clin. Res. 2016, 10, 211–221. [Google Scholar] [CrossRef]
- Pekarsky, Y.; Hallas, C.; Croce, C.M. The role of TCL1 in human T-cell leukemia. Oncogene 2001, 20, 5638–5643. [Google Scholar] [CrossRef] [PubMed]
- Aznar, N.; Midde, K.K.; Dunkel, Y.; Lopez-Sanchez, I.; Pavlova, Y.; Marivin, A.; Barbazán, J.; Murray, F.; Nitsche, U.; Janssen, K.-P.; et al. Daple is a novel non-receptor GEF required for trimeric G protein activation in Wnt signaling. eLife 2015, 4, e07091. [Google Scholar] [CrossRef] [PubMed]
- Egeblad, M.; Werb, Z. New functions for the matrix metalloproteinases in cancer progression. Nat. Rev. Cancer 2002, 2, 161–174. [Google Scholar] [CrossRef]
- Vihinen, P.; Ala-aho, R.; Kähäri, V.M. Matrix metalloproteinases as therapeutic targets in cancer. Curr. Cancer Drug Targets 2005, 5, 203–220. [Google Scholar] [CrossRef]
- Contant, C.; Rouabhia, M.; Loubaki, L.; Chandad, F.; Semlali, A. Anethole induces anti-oral cancer activity by triggering apoptosis, autophagy and oxidative stress and by modulation of multiple signaling pathways. Sci. Rep. 2021, 11, 13087. [Google Scholar] [CrossRef]
- Semlali, A.; Ajala, I.; Beji, S.; Al-Zharani, M.M.; Rouabhia, M. Synergistic Effect of Anethole and Platinum Drug Cisplatin against Oral Cancer Cell Growth and Migration by Inhibiting MAPKase, Beta-Catenin, and NF-κB Pathways. Pharmaceuticals 2023, 16, 700. [Google Scholar] [CrossRef]
- Semlali, A.; Beji, S.; Ajala, I.; Al-Zharani, M.; Rouabhia, M. Synergistic Effects of New Curcumin Analog (PAC) and Cisplatin on Oral Cancer Therapy. Curr. Issues Mol. Biol. 2023, 45, 5018–5035. [Google Scholar] [CrossRef]
- Lampron, M.C.; Paré, I.; Al-Zharani, M.; Semlali, A.; Loubaki, L. Cannabinoid Mixture Affects the Fate and Functions of B Cells through the Modulation of the Caspase and MAP Kinase Pathways. Cells 2023, 12, 588. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Papadakos, S.; Issa, H.; Alamri, A.; Alamri, A.; Semlali, A. Rapamycin as a Potential Alternative Drug for Squamous Cell Gingiva Carcinoma (Ca9-22): A Focus on Cell Cycle, Apoptosis and Autophagy Genetic Profile. Pharmaceuticals 2024, 17, 131. https://doi.org/10.3390/ph17010131
Papadakos S, Issa H, Alamri A, Alamri A, Semlali A. Rapamycin as a Potential Alternative Drug for Squamous Cell Gingiva Carcinoma (Ca9-22): A Focus on Cell Cycle, Apoptosis and Autophagy Genetic Profile. Pharmaceuticals. 2024; 17(1):131. https://doi.org/10.3390/ph17010131
Chicago/Turabian StylePapadakos, Sofia, Hawraa Issa, Abdulaziz Alamri, Abdullah Alamri, and Abdelhabib Semlali. 2024. "Rapamycin as a Potential Alternative Drug for Squamous Cell Gingiva Carcinoma (Ca9-22): A Focus on Cell Cycle, Apoptosis and Autophagy Genetic Profile" Pharmaceuticals 17, no. 1: 131. https://doi.org/10.3390/ph17010131
APA StylePapadakos, S., Issa, H., Alamri, A., Alamri, A., & Semlali, A. (2024). Rapamycin as a Potential Alternative Drug for Squamous Cell Gingiva Carcinoma (Ca9-22): A Focus on Cell Cycle, Apoptosis and Autophagy Genetic Profile. Pharmaceuticals, 17(1), 131. https://doi.org/10.3390/ph17010131








