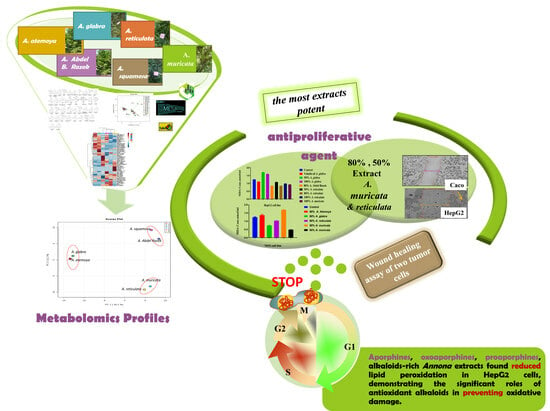
Comprehensive Tools of Alkaloid/Volatile Compounds–Metabolomics and DNA Profiles: Bioassay-Role-Guided Differentiation Process of Six Annona sp. Grown in Egypt as Anticancer Therapy
Abstract
1. Introduction
2. Results
2.1. Extraction of Plant Material and Isolation of DNA, Followed by Fingerprinting Analysis
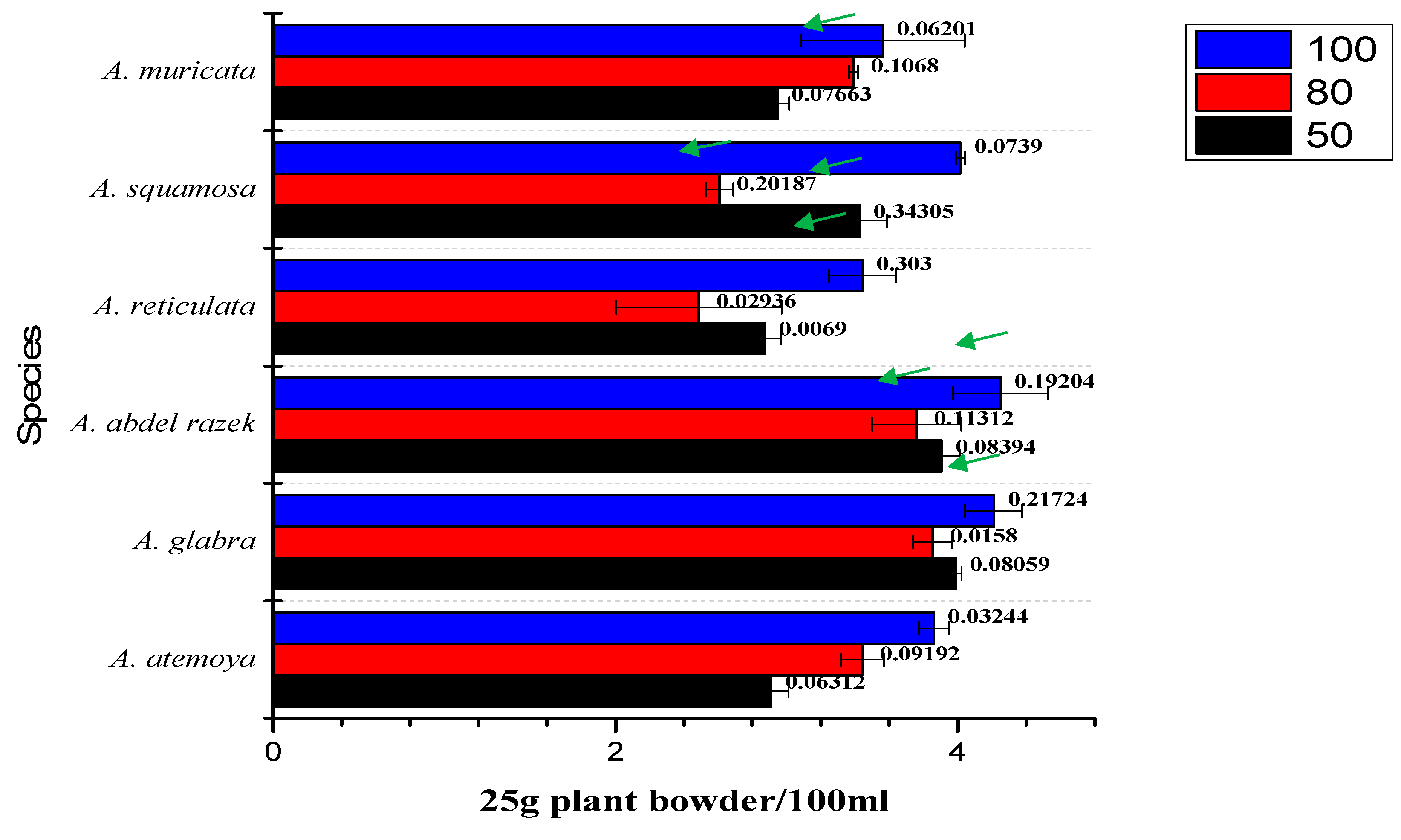
2.2. Bioassay-Guided Differentiation Process for Six Annona Species and Isolated Total Alkaloids
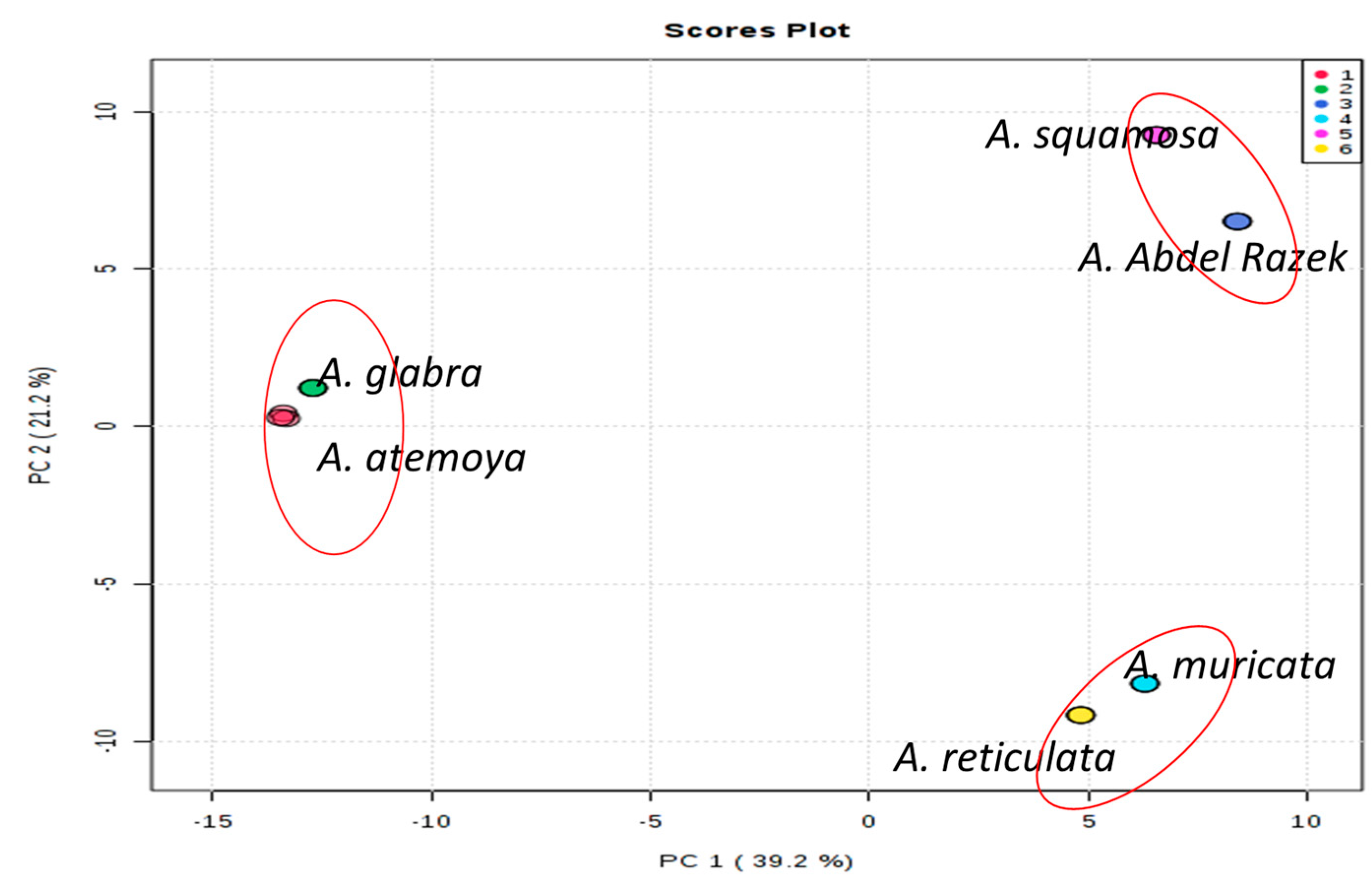
2.3. Taxonomic—DNA Fingerprinting for Six Annona Species
2.4. Volatile Oils of Annona sp. and Chemical Characterization Using GC-MS
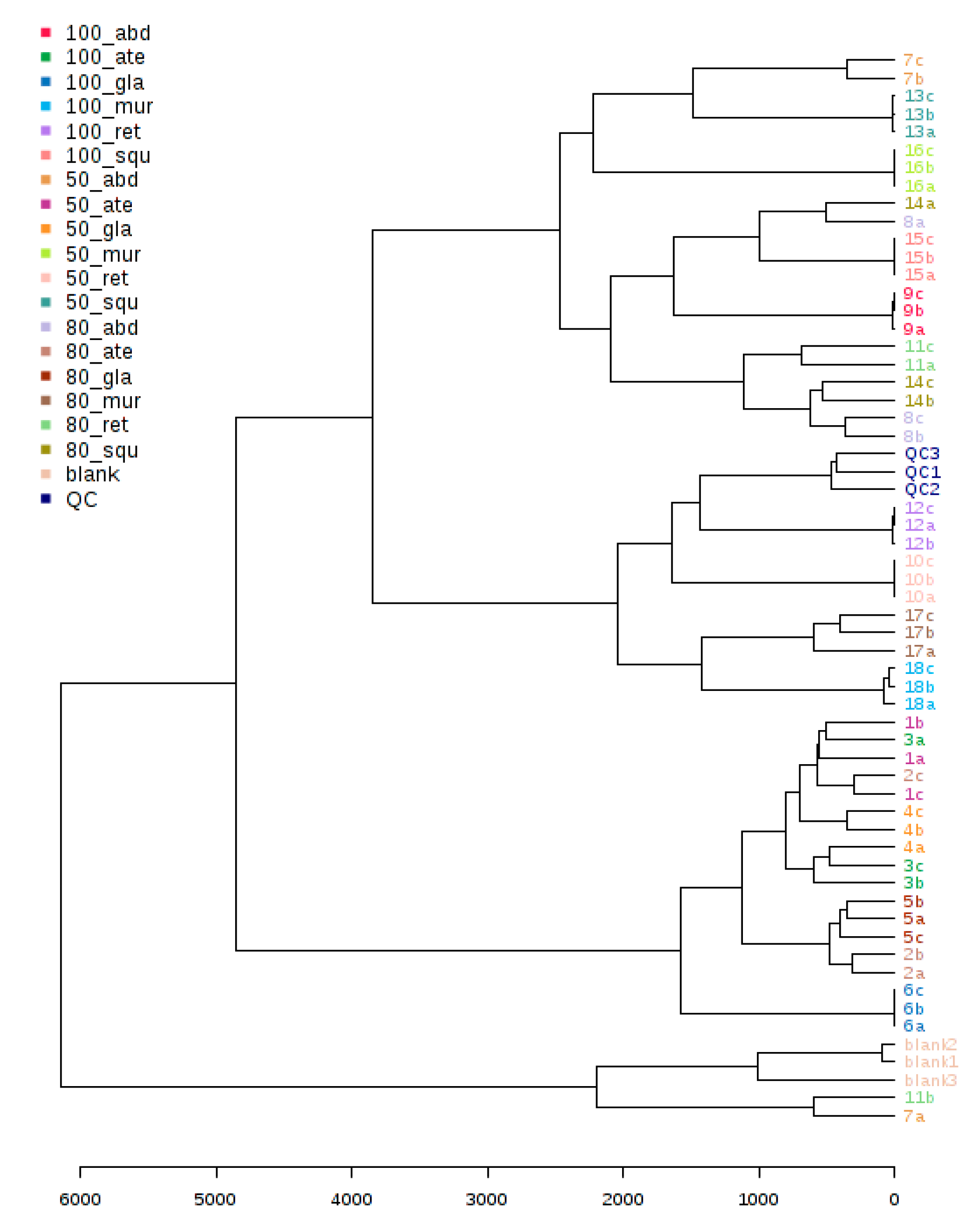
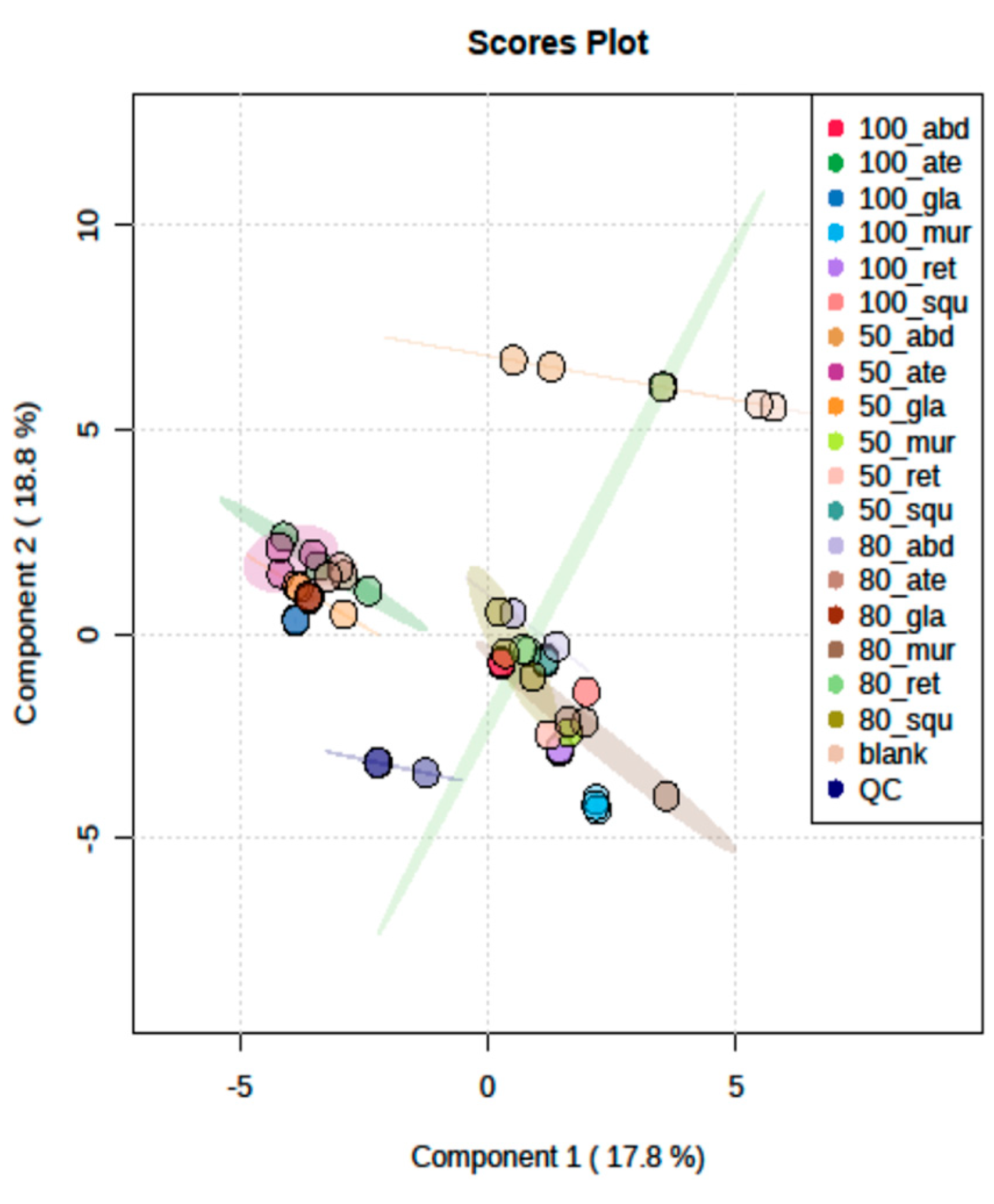
2.5. LC/MSMS Profiles of Six Annona sp. with 18 Differentiation Extracts
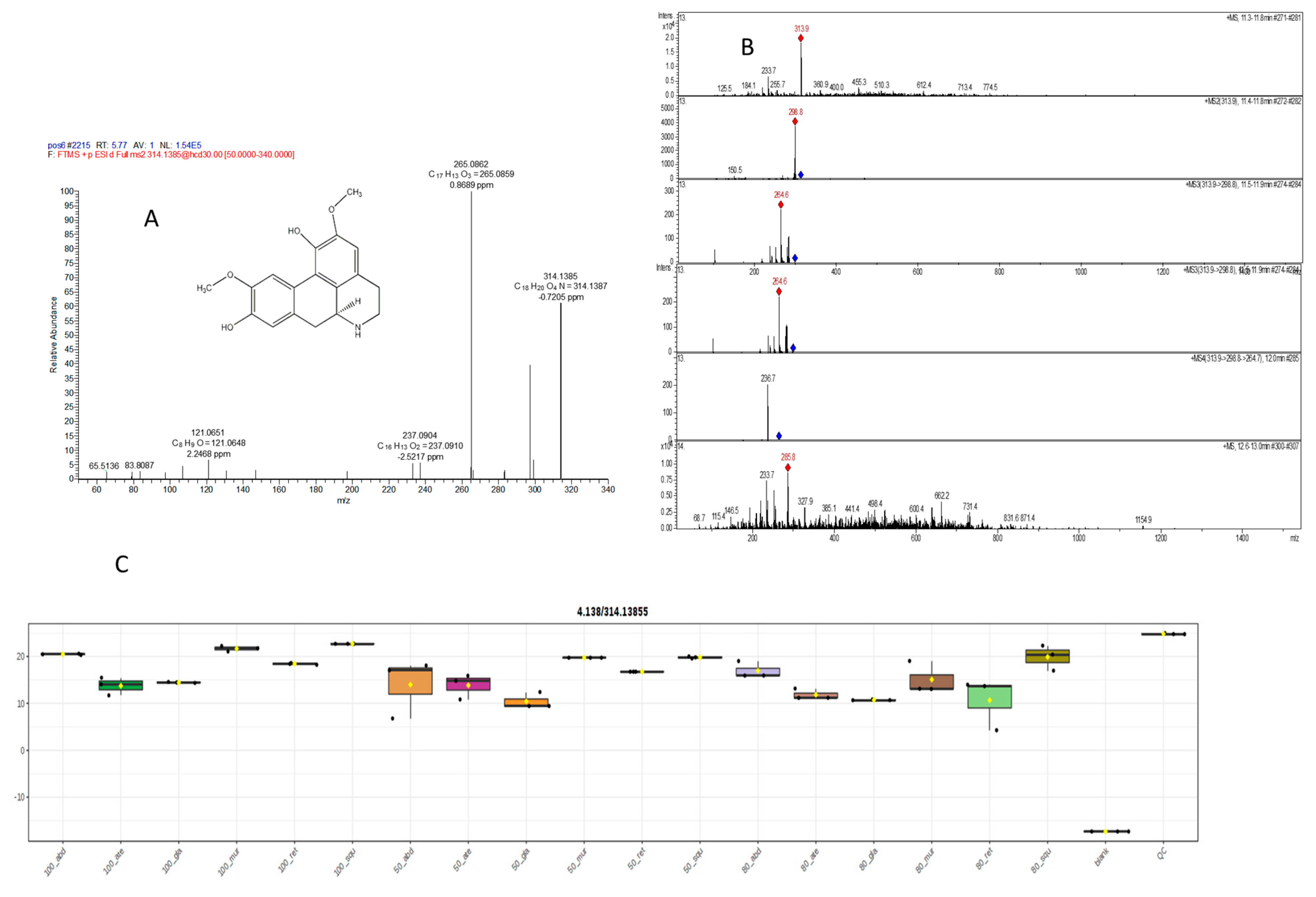
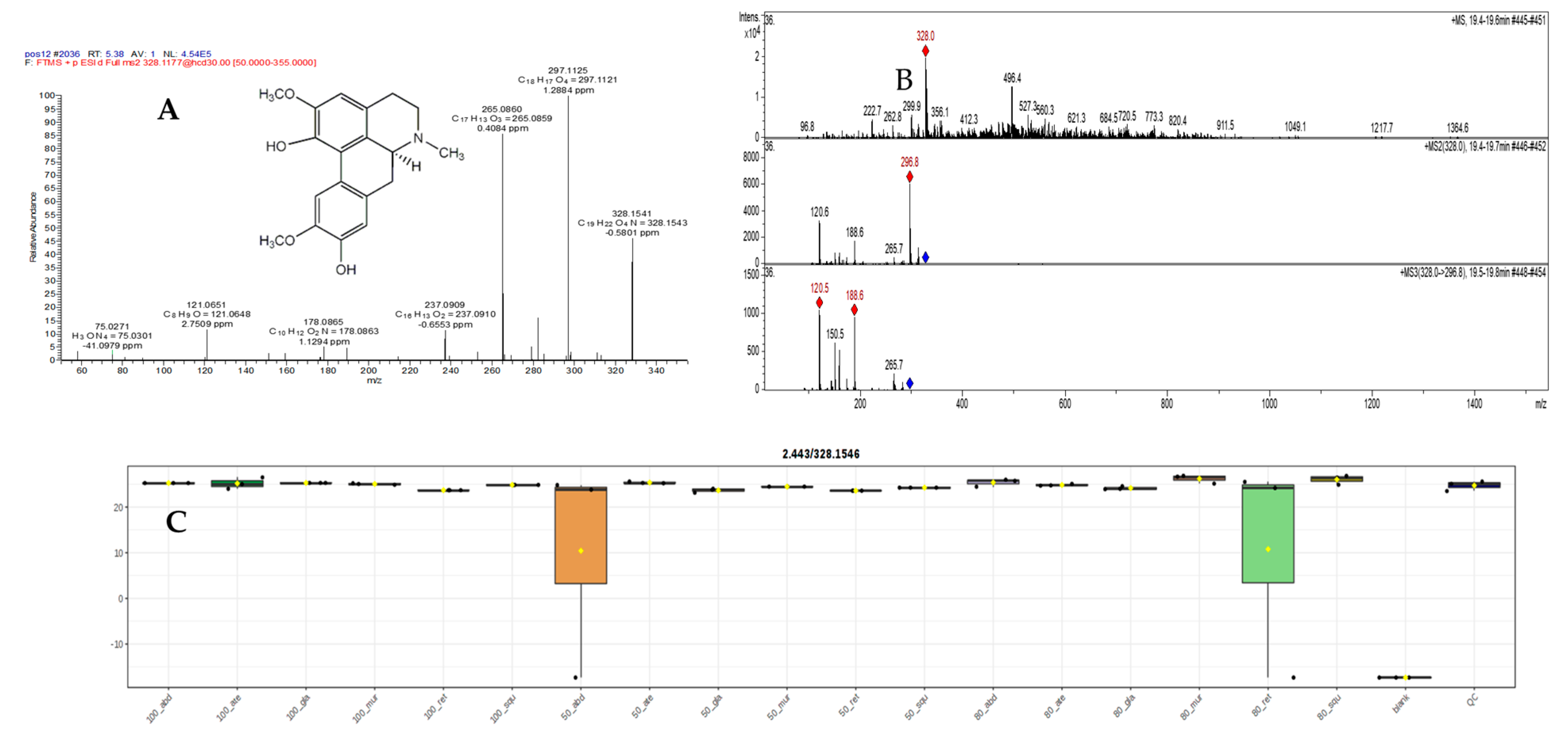
2.5.1. Mass Spectral Analysis of Norisoboldinedemethyl
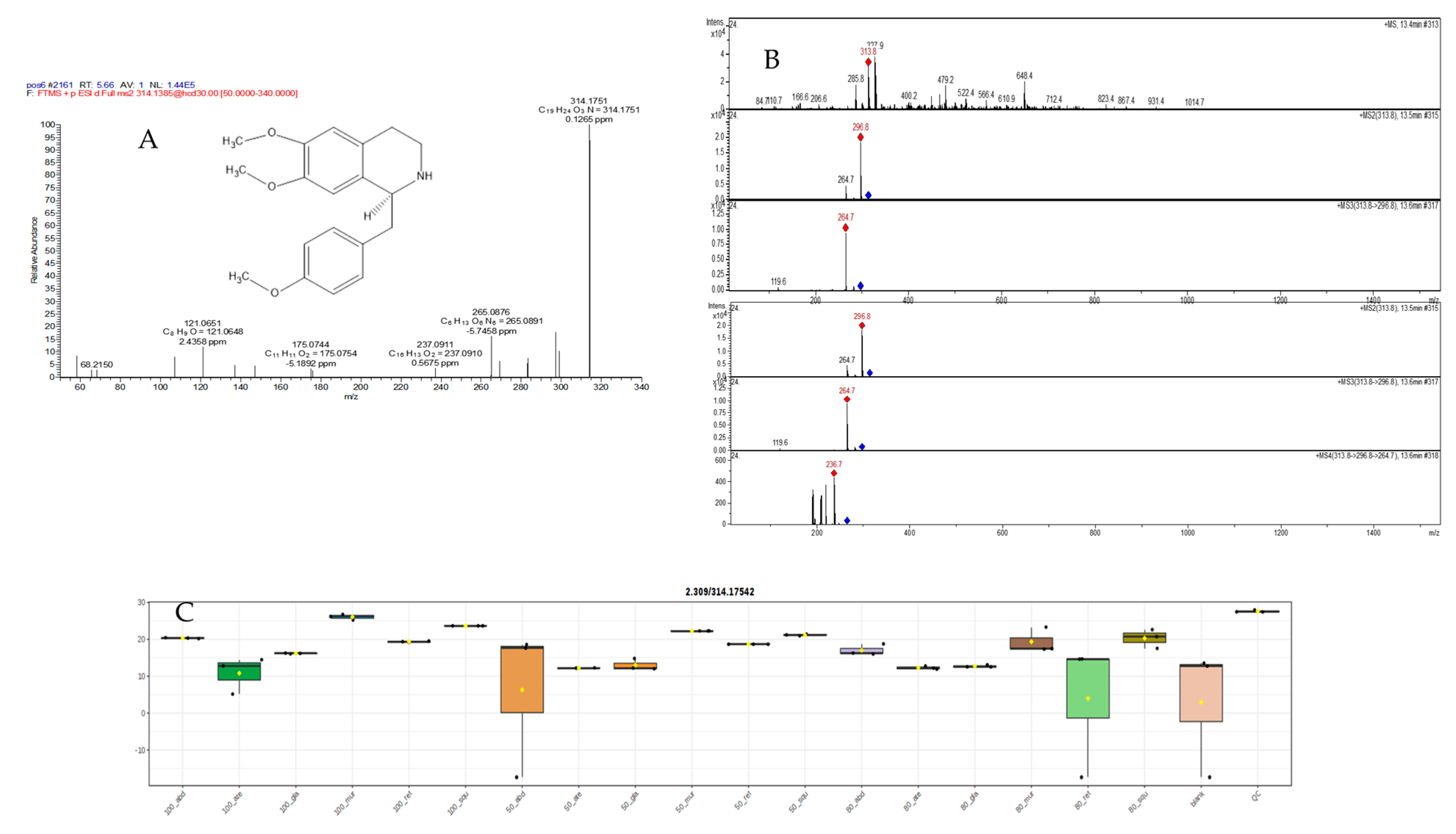
2.5.2. Mass Spectral Analysis of N,N-Dimethylcoclaurie
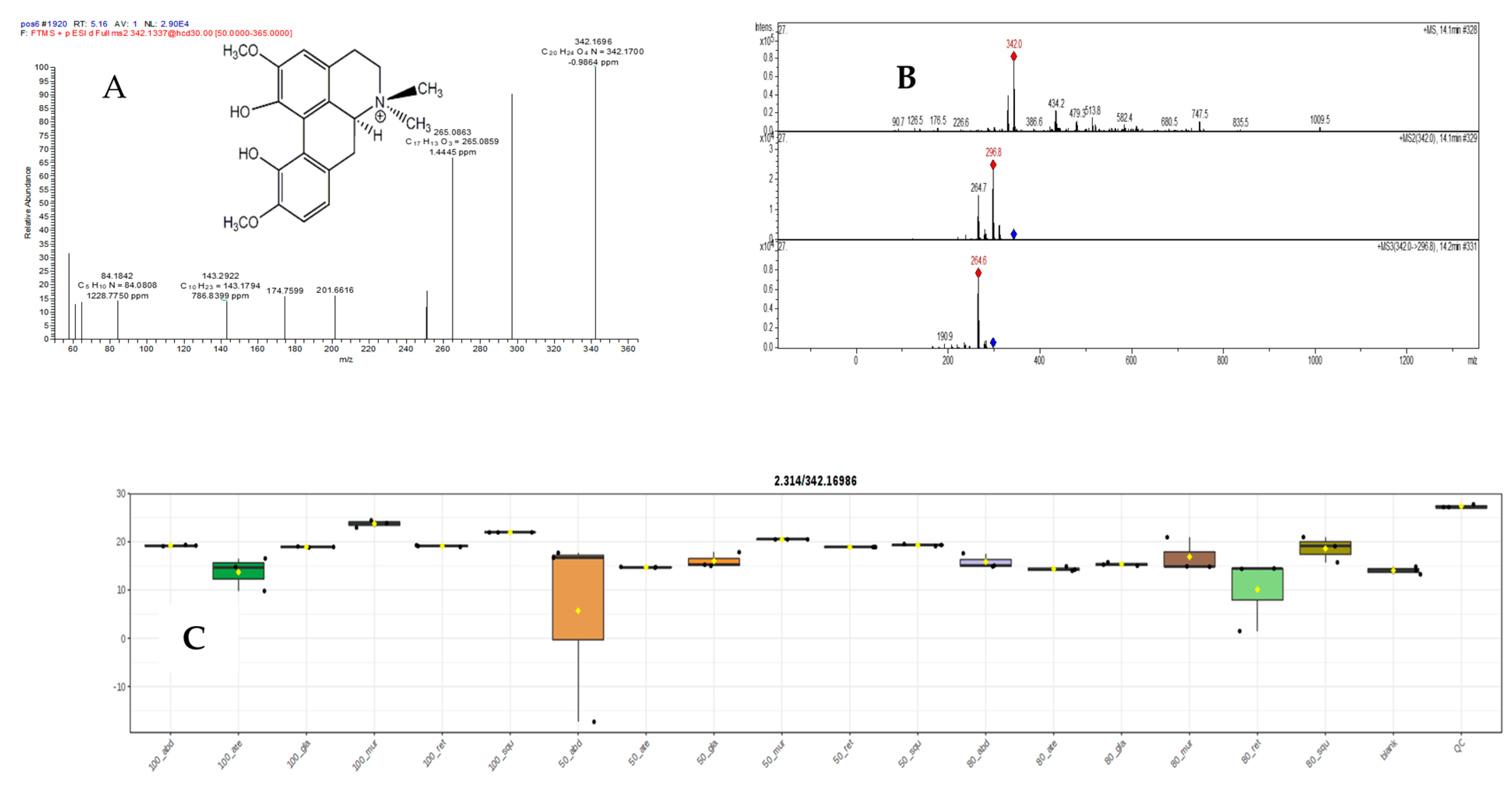
2.5.3. Mass Spectral Analysis of Magnoflorine
2.5.4. Mass Spectral Analysis of Isoboldine
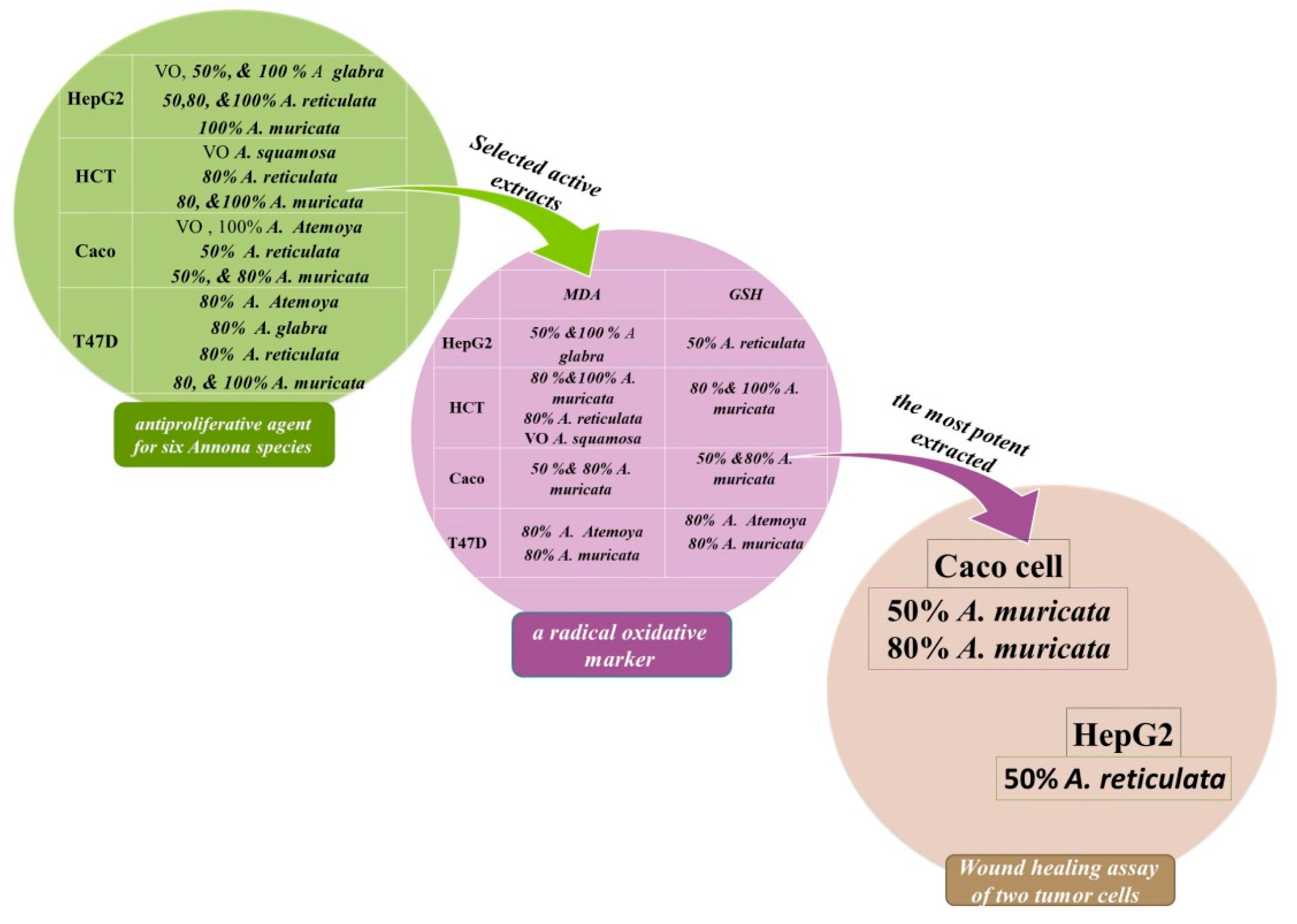
2.6. Antiproliferative Agent for Six Annona Species Grown in Egypt
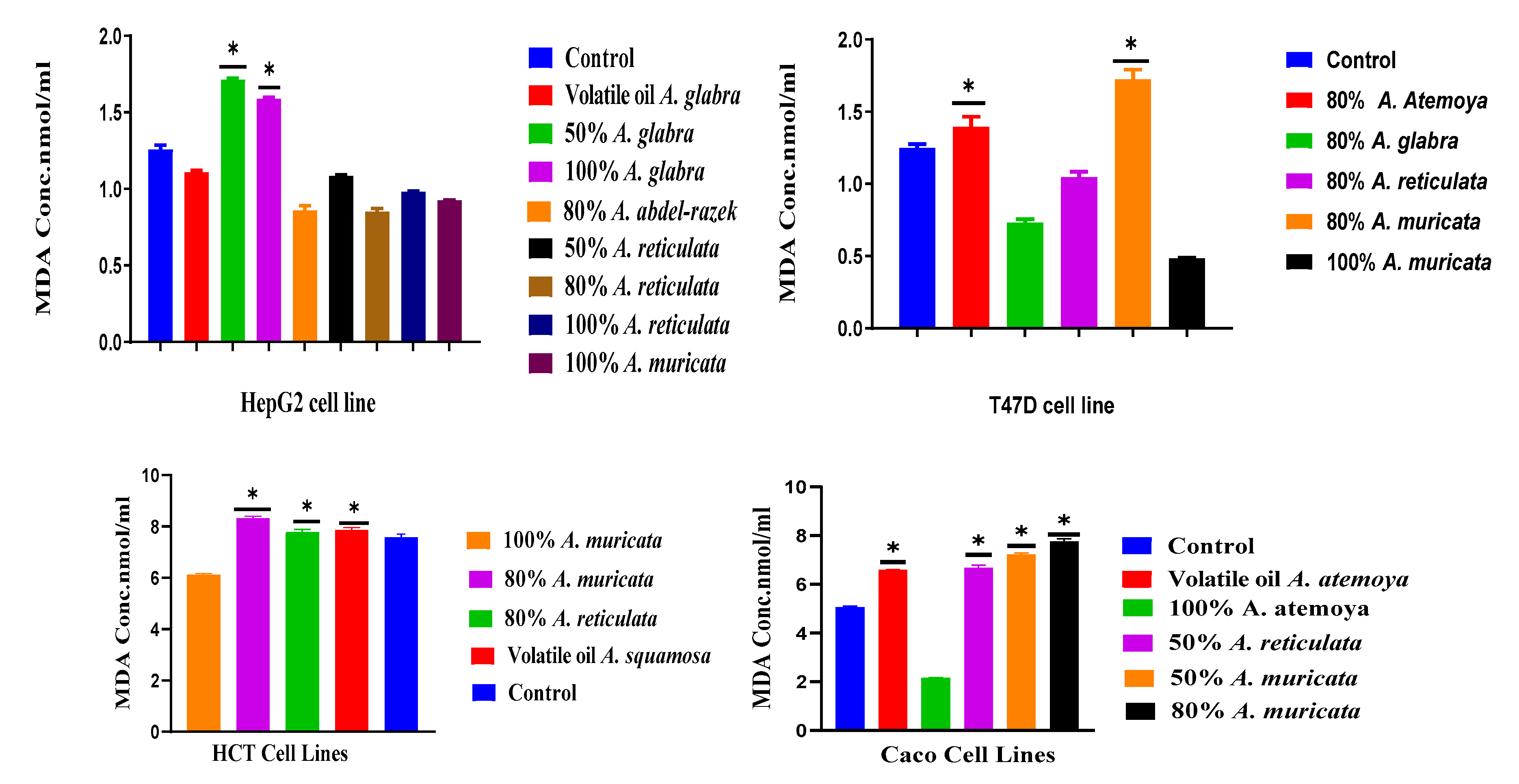
2.6.1. Determination of Total Lipid Peroxide Content (Measured as Malonaldialdehyde)
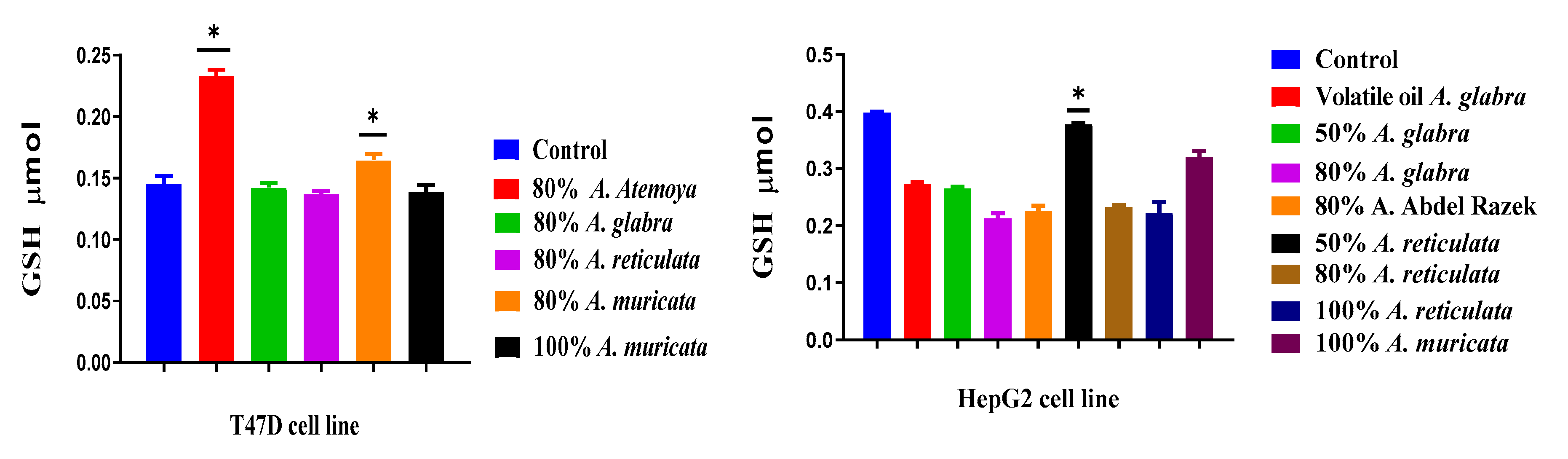
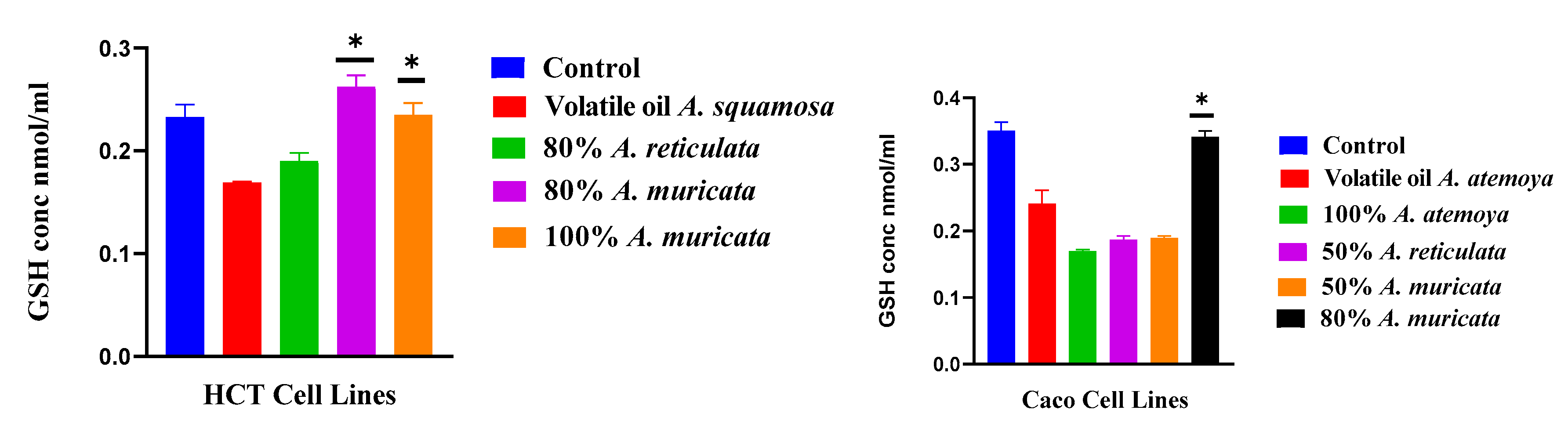
2.6.2. Determination of Reduced Glutathione Content
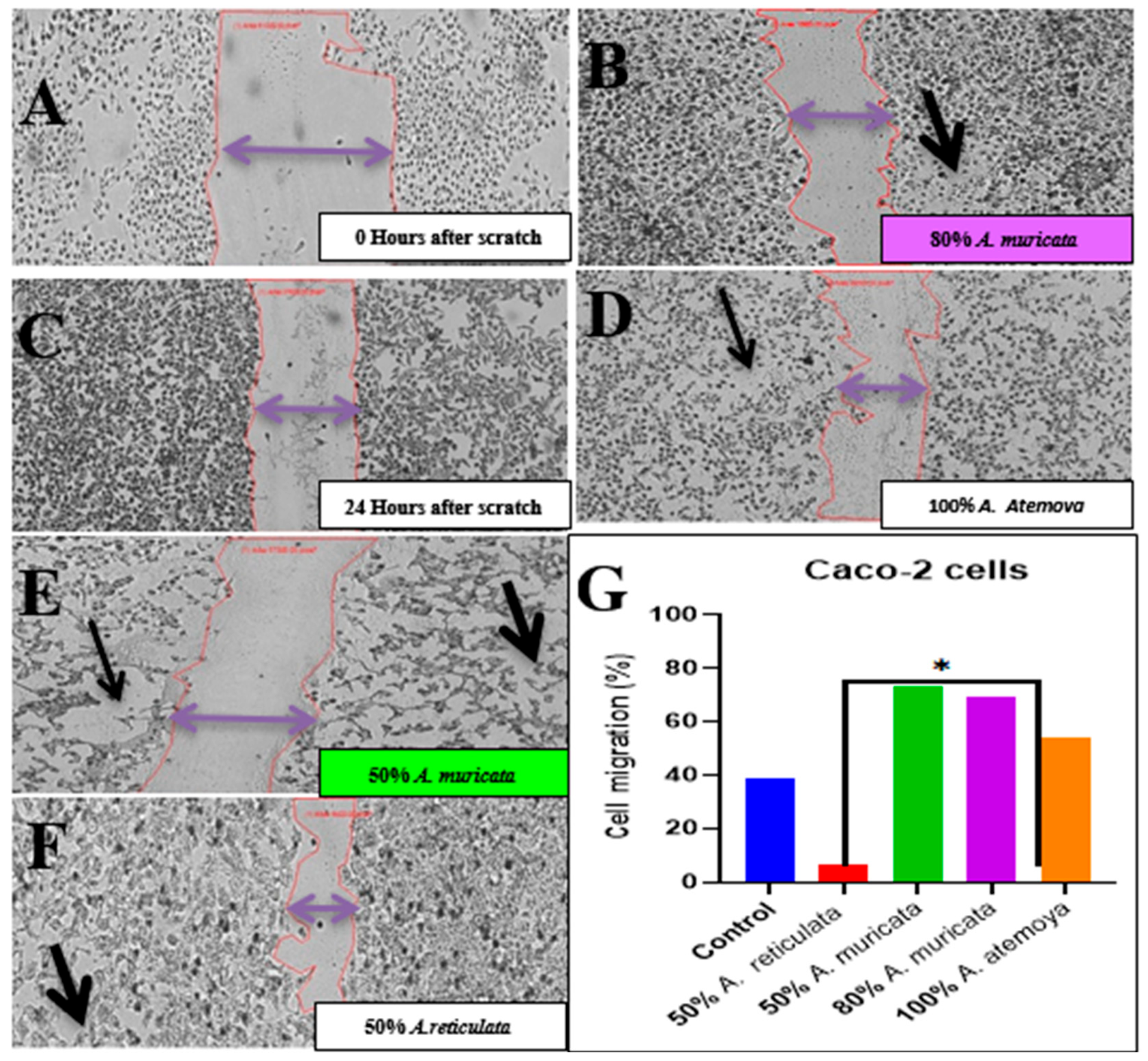
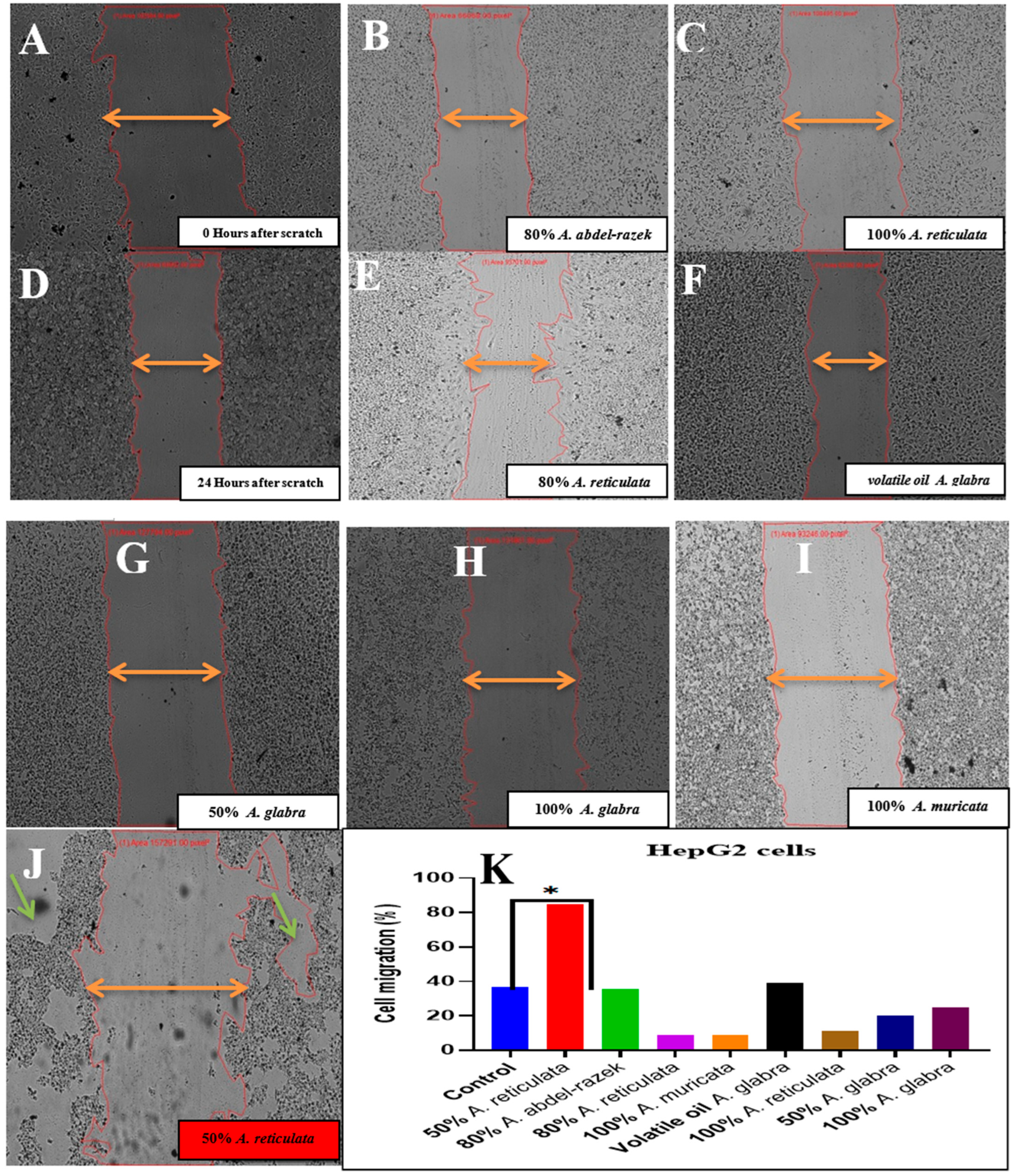
2.6.3. Wound Healing Assay In Vitro of Two Tumor Cells: Inhibition of Cell Migration by Selected Cytotoxic Extracts
3. Discussion
3.1. Optimization of Extraction Method of Total Alkaloids
3.2. Metabolomics and Fingerprint Profiles of Cultivated Annona Species
3.3. Quantification of Malondialdehyde as a Biomarker of Oxidative Stress in Human Hepatoma HepG2 and T47D Cell Cultures
3.4. The Frequency of Non-Enzymatic Antioxidants of GSH in Liver Cancer (HepG2) and Breast Cancer (T47D) as Tumor Grade Will Be Determined
4. Material and Methods
4.1. Collection of Plant Material and Isolation of DNA
4.2. A Bioassay-Guided Differentiation Method Was Conducted for Six Annona Species and the Isolated Total Alkaloids
4.3. GC/MS Analysis (Determination and Identification)
4.4. LC/MSMS Analysis
- The dissolution of 10 mg from each of the eighteen extracts was performed in 1 milliliter of methanol, followed by filtration using a sarangi filter. Regarding the extract that had been diluted, dilution was repeated thrice.
- The LC/MS-ion-trap Esquire was equipped with ESI and operated in positive ion mode. The scan range was 100–3000 m/z and scan resolution was 13,000 m/z/s). A nebulizer, gas flow of 30 psi, 9.1 L/min, and temperature of 310 °C were used. The skimmer was set at -10.0 V [83].
- The LCMSMS system consisted of a Q-Exactive hybrid MS/MS quadrupole-Orbitrap mass spectrometer and UPLC (Waters, Milford, CT, USA). The separation of chromatographic components employed solvents (A:B) such as water acidified with 0.1% formic acid and acetonitrile, with a mobile phase flow rate of 0.3 mL/min. This was accomplished through a gradient program consisting of the following steps: from 0 to 15 min, the composition changed from 50% A to 50% B; from 15 to 22 min, the composition changed to 98% B and remained at this level for 22 min; from 22 to 23 min, the composition changed back to 95% A until 27 min, at which point the system returned to its initial conditions and was re-equilibrated for 3 min [83]. Our approach involved the processing of data obtained from mass spectrometric fragmentations of 74 alkaloid metabolites using ion mobility tandem mass spectrometry. This allowed us to generate a comprehensive fragmentation pattern, as well as retention time and MS/MS information.
- The MS-DIAL 4.60 tool, which utilizes the MSP format for the purpose of filtering noisy spectra through a fundamental spectral similarity computation, offers enhanced and standardized untargeted metabolomics by exporting the four portions of the imported raw MS data to a common output format (abf). The identification of the distinct compounds involved the utilization of precise molecular masses (within a range of less than 5 ppm), mass spectra, retention periods, internet databases (ChEBI, Metlin, PubChem, and KNApSAck, ChemSpider), as well as literature data [84,85].
4.5. Model of Antiproliferation for Six Annona sp.
4.5.1. Estimation of Potential Cytotoxicity of Extracts on Cell Lines Using Sulphorhodamine-B (SRB) Assay
Estimation of Total Lipid Peroxide Content (Measured as Malonaldialdehyde)
Estimation of Reduced Glutathione Content
4.5.2. Wound Healing Assay
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Goding Sauer, A.; Fedewa, S.A.; Butterly, L.F.; Anderson, J.C.; Cercek, A.; Smith, R.A.; Jemal, A. Colorectal cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 145–164. [Google Scholar] [CrossRef] [PubMed]
- Jacobo-Herrera, N.; Pérez-Plasencia, C.; Castro-Torres, V.A.; Martínez-Vázquez, M.; González-Esquinca, A.R.; Zentella-Dehesa, A. Selective acetogenins and their potential as anticancer agents. Front. Pharmacol. 2019, 10, 783. [Google Scholar] [CrossRef] [PubMed]
- Sultana, S.; Asif, H.M.; Nazar, H.M.I.; Akhtar, N.; Rehman, J.U.; Rehman, R.U. Medicinal plants combating against cancer-a green anticancer approach. Asian Pac. J. Cancer Prev. 2014, 15, 4385–4394. [Google Scholar] [CrossRef]
- Newman, D.J.; Cragg, G.M. Natural products as sources of new drugs from 1981 to 2014. J. Nat. Prod. 2016, 79, 629–661. [Google Scholar] [CrossRef]
- Da Rocha, A.B.; Lopes, R.M.; Schwartsmann, G. Natural products in anticancer therapy. Curr. Opin. Pharmacol. 2001, 1, 364–369. [Google Scholar] [CrossRef]
- Nobili, S.; Lippi, D.; Witort, E.; Donnini, M.; Bausi, L.; Mini, E.; Capaccioli, S. Natural compounds for cancer treatment and prevention. Pharmacol. Res. 2009, 59, 365–378. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Decker, C.C.; Zechner, L.; Krstin, S.; Wink, M. In vitro wound healing of tumor cells: Inhibition of cell migration by selected cytotoxic alkaloids. BMC Pharmacol. Toxicol. 2019, 20, 1–12. [Google Scholar] [CrossRef]
- Zayed, M.M.; Rozan, M.A.; Ziena, H.M. Chemical composition, bioactive and phenolic compounds in three varieties of Annona fruit grown in Egypt. Alex. Sci. Exch. J. 2022, 43, 199–207. [Google Scholar] [CrossRef]
- Haggag, W.M.; Nofal, M. Improving the biological control of botryodiplodia disease on some Annona cultivars using single or multi-bioagents in Egypt. Biol. Control 2006, 38, 341–349. [Google Scholar] [CrossRef]
- Mohammed, M.A.; Hamed, M.A.; El-Gengaihia, S.E.; Eneinc, A.M.A.; Ahmedc, O.K.; Hassana, E.M. In vitro screening of Annona cherimola leaves and bark for their antioxidant activity and in vivo assessment as protective agents against gastric ulcer in rats. Plant Arch. 2020, 20, 2658–2668. [Google Scholar]
- Moghadamtousi, S.Z.; Fadaeinasab, M.; Nikzad, S.; Mohan, G.; Ali, H.M.; Kadir, H.A. Annona muricata (Annonaceae): A review of its traditional uses, isolated acetogenins and biological activities. Int. J. Mol. Sci. 2015, 16, 15625–15658. [Google Scholar] [CrossRef] [PubMed]
- El-Gengaihi, S.E.; Aboul-Enein, A.M.; Mohammed, M.A. Antiproliferative effect and chemical constituents of Annona species. Plant Arch. 2020, 20, 2650–2657. [Google Scholar]
- Al Kazman, B.S.; Harnett, J.E.; Hanrahan, J.R. Traditional uses, phytochemistry and pharmacological activities of Annonacae. Molecules 2022, 27, 3462. [Google Scholar] [CrossRef] [PubMed]
- Fofana, S.; Keita, A.; Balde, S.; Ziyaev, R.; Aripova, S. Alkaloids from leaves of Annona muricata. Chem. Nat. Compd. 2012, 48, 714. [Google Scholar] [CrossRef]
- Pandey, N.; Barve, D. Phytochemical and pharmacological review on Annona squamosa linn. Int. J. Res. Pharm. Biomed. Sci. 2011, 2, 1404–1412. [Google Scholar]
- Khan, N.; Kumar, N.; Ballal, A.; Datta, D.; Belle, V.S. Unveiling antioxidant and anti-cancer potentials of characterized Annona reticulata leaf extract in 1, 2-dimethylhydrazine-induced colorectal cancer in wistar rats. J. Ayurveda Integr. Med. 2021, 12, 579–589. [Google Scholar] [CrossRef] [PubMed]
- Mannino, G.; Gentile, C.; Porcu, A.; Agliassa, C.; Caradonna, F.; Bertea, C.M. Chemical profile and biological activity of cherimoya (Annona cherimola mill.) and atemoya (Annona atemoya) leaves. Molecules 2020, 25, 2612. [Google Scholar] [CrossRef]
- Mohammed, M.A.; Hamed, M.A.; El-Gengaihi, S.E.; Enein, A.M.A.; Kachlicki, P.; Hassan, E.M. Profiling of secondary metabolites and DNA typing of three different Annona cultivars grown in Egypt. Metabolomics 2022, 18, 49. [Google Scholar] [CrossRef]
- Xiao, Y.; Wang, Y.-K.; Xiao, X.-R.; Zhao, Q.; Huang, J.-F.; Zhu, W.-F.; Li, F. Metabolic profiling of coumarins by the combination of uplc-ms-based metabolomics and multiple mass defect filter. Xenobiotica 2020, 50, 1076–1089. [Google Scholar] [CrossRef]
- Doyle, J.J.; Doyle, J.L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Collard, B.C.; Mackill, D.J. Start codon targeted (scot) polymorphism: A simple, novel DNA marker technique for generating gene-targeted markers in plants. Plant Mol. Biol. Report. 2009, 27, 86–93. [Google Scholar] [CrossRef]
- Patti, G.J.; Yanes, O.; Siuzdak, G. Metabolomics: The apogee of the omics trilogy. Nat. Rev. Mol. Cell Biol. 2012, 13, 263–269. [Google Scholar] [CrossRef]
- Xia, J.; Mandal, R.; Sinelnikov, I.V.; Broadhurst, D.; Wishart, D.S. Metaboanalyst 2.0—A comprehensive server for metabolomic data analysis. Nucleic Acids Res. 2012, 40, W127–W133. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.C.; Chang, F.R.; Chao, Y.C.; Teng, C.M. Antiplatelet and vasorelaxing actions of aporphinoids from Cassytha filiformis. Phytother. Res. Int. J. Devoted Pharmacol. Toxicol. Eval. Nat. Prod. Deriv. 1998, 12, S39–S41. [Google Scholar] [CrossRef]
- Martínez-Vázquez, M.; Estrada-Reyes, R. Secondary metabolismin in Annonaceae: Potencial source of drugs. Rev. Bras. Frutic. 2014, 36, 141–146. [Google Scholar] [CrossRef][Green Version]
- Chang, F.; Chen, K.; Ko, F.; Teng, C.; Wu, Y.-C. Bioactive alkaloids from Annona reticulata. Chin. Pharm. J. 1995, 47, 483–491. [Google Scholar]
- Chang, F.-R.; Wei, J.-L.; Teng, C.-m.; Wu, Y.-C. Two new 7-dehydroaporphine alkaloids and antiplatelet action aporphines from the leaves of Annona purpurea. Phytochemistry 1998, 49, 2015–2018. [Google Scholar] [CrossRef] [PubMed]
- Guinaudeau, H.; Leboeuf, M.; Cavé, A. Aporphine alkaloids. II. J. Nat. Prod. 1979, 42, 325–360. [Google Scholar] [CrossRef]
- Si, D.Y.; Zhao, S.X.; Deng, J.Z. A 4, 5-dioxoaporphine from the aerial parts of Stephania tetrandra. J. Nat. Prod. 1992, 55, 828–829. [Google Scholar] [CrossRef]
- Moriyasu, M.; Ichimaru, M.; Nishiyama, Y.; Kato, A. Isolation of alkaloids from plant materials by the combination of ion-pair extraction and preparative ion-pair hplc using sodium perchlorate-1-magnoliae cortex. Pharm. J. 1994, 48, 282–286. [Google Scholar]
- Glasby, J.S.P. Encyclopedia of the Alkaloids; Springer: Boston, MA, USA, 1975. [Google Scholar] [CrossRef]
- Kuo, R.-Y.; Chang, F.-R.; Chen, C.-Y.; Teng, C.-M.; Yen, H.-F.; Wu, Y.-C. Antiplatelet activity of n-methoxycarbonyl aporphines from Rollinia mucosa. Phytochemistry 2001, 57, 421–425. [Google Scholar] [CrossRef]
- Simeon, S.; Rios, J.; Villar, A. Alkaloids from Annona cherimolia (mill.) stem bark. Plantes Med. Phytother. 1989, 23, 159–161. [Google Scholar]
- Chen, K.-S.; Wu, Y.-C.; Teng, C.-M.; Ko, F.-N.; Wu, T.-S. Bioactive alkaloids from illigera luzonensis. J. Nat. Prod. 1997, 60, 645–647. [Google Scholar] [CrossRef]
- Chang, F.-R.; Wei, J.-L.; Teng, C.-M.; Wu, Y.-C. Antiplatelet aggregation constituents from Annona purpurea. J. Nat. Prod. 1998, 61, 1457–1461. [Google Scholar] [CrossRef] [PubMed]
- Contreras, M.d.M.; Bribi, N.; Gómez-Caravaca, A.M.; Gálvez, J.; Segura-Carretero, A. Alkaloids profiling of Fumaria capreolata by analytical platforms based on the hyphenation of gas chromatography and liquid chromatography with quadrupole-time-of-flight mass spectrometry. Int. J. Anal. Chem. 2017, 2017, 5178729. [Google Scholar] [CrossRef]
- Nishiyama, Y.; Moriyasu, M.; Ichimaru, M.; Iwasa, K.; Kato, A.; Mathenge, S.G.; Mutiso, P.B.C.; Juma, F.D. Quaternary isoquinoline alkaloids from Xylopia parviflora. Phytochemistry 2004, 65, 939–944. [Google Scholar] [CrossRef]
- Bhakuni, D.S.; Jain, S. Alkaloids of Cocculus laurifolius dc. Tetrahedron 1980, 36, 3107–3114. [Google Scholar] [CrossRef]
- Bentley, K.W. Β-phenylethylamines and the isoquinoline alkaloids. Nat. Prod. Rep. 2002, 19, 332–356. [Google Scholar] [CrossRef]
- Uprety, H.; Bhakuni, D.; Dhar, M. Aporphine alkaloids of litsea sebifera, l. Wightiana and Actinodaphne obovata. Phytochemistry 1972, 11, 3057–3059. [Google Scholar] [CrossRef]
- Chang, F.R.; Chen, C.Y.; Hsieh, T.J.; Cho, C.P.; Wu, Y.C. Chemical constituents from Annona glabra iii. J. Chin. Chem. Soc. 2000, 47, 913–920. [Google Scholar] [CrossRef]
- Singla, D.; Sharma, A.; Kaur, J.; Panwar, B.; Raghava, G.P. Biadb: A curated database of benzylisoquinoline alkaloids. BMC Pharmacol. 2010, 10, 4. [Google Scholar] [CrossRef]
- Wu, F.-E.; Gu, Z.-M.; Zeng, L.; Zhao, G.-X.; Zhang, Y.; McLaughlin, J.L.; Sastrodihardjo, S. Two new cytotoxic monotetrahydrofuran Annonaceous acetogenins, annomuricins a and b, from the leaves of Annona muricata. J. Nat. Prod. 1995, 58, 830–836. [Google Scholar] [CrossRef] [PubMed]
- Pachaly, P.; Adnan, A.; Will, G. Nmr-assignments of n-acylaporphine alkaloids from tinospora crispa. Planta Medica 1992, 58, 184–187. [Google Scholar] [CrossRef] [PubMed]
- Saito, M.; Alvarenga, M. Alkaloids from Annona cacans. Fitoterapia-Milano 1994, 65, 87. [Google Scholar]
- Baxter, H.; Harborne, J.; Moss, G. Phytochemical Dictionary: A Handbook of Bioactive Compounds from Plants, 2nd ed.; Taylor and Francis Ltd.: London, UK, 1999. [Google Scholar]
- Chen, C.Y.; Chang, F.R.; Wu, Y.C. The constituents from the stems of Annona cherimola. J. Chin. Chem. Soc. 1997, 44, 313–319. [Google Scholar] [CrossRef]
- Kunitomo, J.; Oshikata, M.; Akasu, M. The alkaloids of Stephania cepharantha hayata cultivated in japan (ii) (author’s transl). Yakugaku Zasshi J. Pharm. Soc. Jpn. 1981, 101, 951. [Google Scholar] [CrossRef] [PubMed]
- Achenbach, H.; Frey, D.; Waibel, R. 6a, 7-dehydro-2-hydroxy-4, 5-dioxonoraporphine and other alkaloids from monocyclanthus vignei: 13c-nmr studies on 4, 5-dioxoaporphines. J. Nat. Prod. 1991, 54, 1331–1336. [Google Scholar] [CrossRef]
- Fleischer, T.C.; Waigh, R.D.; Waterman, P.G. Pogostol o-methyl ether and artabotrol: Two novel sesquiterpenes from the stem bark of Artabotrys stenopetalus. J. Nat. Prod. 1997, 60, 1054–1056. [Google Scholar] [CrossRef]
- Lan, Y.-H.; Chang, F.-R.; Yu, J.-H.; Yang, Y.-L.; Chang, Y.-L.; Lee, S.-J.; Wu, Y.-C. Cytotoxic styrylpyrones from goniothalamus a muyon. J. Nat. Prod. 2003, 66, 487–490. [Google Scholar] [CrossRef]
- Áiqbal Choudhary, M. Diterpenoid and steroidal alkaloids. Nat. Prod. Rep. 1999, 16, 619–635. [Google Scholar]
- Lavault, M.; Bruneton, J.; Cavé, A.; Chan, K.C.; Deverre, J.R.; Sevenet, T.; Guinaudeau, H. Alcaloïdes bisbenzylisoquinoléiques de albertisia cf. A. papuana. Can. J. Chem. 1987, 65, 343–347. [Google Scholar] [CrossRef]
- Wijeratne, E.K.; Hatanaka, Y.; Kikuchi, T.; Tezuka, Y.; Gunatilaka, A.L. A dioxoaporphine and other alkaloids of two Annonaceous plants of sri lanka. Phytochemistry 1996, 42, 1703–1706. [Google Scholar] [CrossRef]
- Puri, B.; Hall, A. Phytochemical Dictionary: A Handbook of Bioactive Compounds from Plants; CRC Press: Boca Raton, FL, USA, 1998. [Google Scholar]
- Nishiyama, Y.; Moriyasu, M.; Ichimaru, M.; Iwasa, K.; Kato, A.; Mathenge, S.G.; Mutiso, P.B.C.; Juma, F.D. Secondary and tertiary isoquinoline alkaloids from Xylopia parviflora. Phytochemistry 2006, 67, 2671–2675. [Google Scholar] [CrossRef]
- Chen, C.-Y.; Chang, F.-R.; Pan, W.-B.; Wu, Y.-C. Four alkaloids from Annona cherimola. Phytochemistry 2001, 56, 753–757. [Google Scholar] [CrossRef]
- Rabêlo, S.V.; de Souza Araújo, C.; de Oliveira Costa, V.C.; Tavares, J.F.; da Silva, M.S.; Barbosa Filho, J.M.; da Silva Almeida, J.R.G. Genus Annona L. (Annonaceae): A review. In Nutraceuticals and Functional Foods; Nova Science Publishers: New York, NY, USA, 2013; p. 41. [Google Scholar]
- Yang, Y.L.; Chang, F.R.; Wu, Y.C. Annosqualine: A novel alkaloid from the stems of Annona squamosa. Helv. Chim. Acta 2004, 87, 1392–1399. [Google Scholar] [CrossRef]
- Chang, F.-R.; Chen, C.-Y.; Wu, P.-H.; Kuo, R.-Y.; Chang, Y.-C.; Wu, Y.-C. New alkaloids from Annona purpurea. J. Nat. Prod. 2000, 63, 746–748. [Google Scholar] [CrossRef]
- Tsugawa, H.; Nakabayashi, R.; Mori, T.; Yamada, Y.; Takahashi, M.; Rai, A.; Sugiyama, R.; Yamamoto, H.; Nakaya, T.; Yamazaki, M. A cheminformatics approach to characterize metabolomes in stable-isotope-labeled organisms. Nat. Methods 2019, 16, 295–298. [Google Scholar] [CrossRef] [PubMed]
- Yuan, T.; Zhang, C.-R.; Yang, S.-P.; Yin, S.; Wu, W.-B.; Dong, L.; Yue, J.-M. Sesquiterpenoids and phenylpropanoids from Chloranthus serratus. J. Nat. Prod. 2008, 71, 2021–2025. [Google Scholar] [CrossRef]
- Sun, W.; Sheng, J. Handbook of Natural Active Constituents; Chinese Medicinal Science and Technology Press: Beijing, China, 1998; Volume 166. [Google Scholar]
- Leboeuf, M.; Legueut, C.; Cavé, A.; Desconclois, J.; Forgacs, P.; Jacquemin, H. Alcaloïdes des Annonacées xxix: Alcaloïdes de l’Annona muricata L. Planta Medica 1981, 42, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Chia, Y.-C.; Chang, F.-R.; Teng, C.-M.; Wu, Y.-C. Aristolactams and dioxoaporphines from Fissistigma balansae and Fissistigma oldhamii. J. Nat. Prod. 2000, 63, 1160–1163. [Google Scholar] [CrossRef] [PubMed]
- Yoon, M.-A.; Jeong, T.-S.; Park, D.-S.; Xu, M.-Z.; Oh, H.-W.; Song, K.-B.; Lee, W.S.; Park, H.-Y. Antioxidant effects of quinoline alkaloids and 2, 4-di-tert-butylphenol isolated from Scolopendra subspinipes. Biol. Pharm. Bull. 2006, 29, 735–739. [Google Scholar] [CrossRef] [PubMed]
- Cortes, D.; Torrero, M.Y.; Pilar, D.O.M.; Luz, C.M.; Cavé, A.; Hadi, A. Norstephalagine and atherospermidine: Two smooth muscle relaxant aporphines from Artabotrys maingayi. J. Nat. Prod. 1990, 53, 503. [Google Scholar] [CrossRef]
- Hébert, J.; Gravel, D. O-nitrophenylethylene glycol: A photosensitive protecting group for aldehydes and ketones. Can. J. Chem. 1974, 52, 187–189. [Google Scholar] [CrossRef]
- Chen, J.-Z.; Chou, G.-X.; Wang, C.-H.; Yang, L.; Bligh, S.A.; Wang, Z.-T. Characterization of new metabolites from in vivo biotransformation of norisoboldine by liquid chromatography/mass spectrometry and nmr spectroscopy. J. Pharm. Biomed. Anal. 2010, 52, 687–693. [Google Scholar] [CrossRef] [PubMed]
- Aguilar-Hernández, G.; Zepeda-Vallejo, L.G.; García-Magaña, M.d.L.; Vivar-Vera, M.d.l.Á.; Pérez-Larios, A.; Girón-Pérez, M.I.; Coria-Tellez, A.V.; Rodríguez-Aguayo, C.; Montalvo-González, E. Extraction of alkaloids using ultrasound from pulp and by-products of soursop fruit (Annona muricata L.). Appl. Sci. 2020, 10, 4869. [Google Scholar] [CrossRef]
- Ellaithy, A.; Abdel-khalek, A.; Mohammed, M. The potency of ricinine biopesticide from Ricinus communis leaves as an alternative host for mass rearing process of tetranychus urticae and two predatory Phytoseiid mites. Egypt. J. Chem. 2022, 65, 535–549. [Google Scholar]
- Gavamukulya, Y.; Wamunyokoli, F.; El-Shemy, H.A. Annona muricata: Is the natural therapy to most disease conditions including cancer growing in our backyard? A systematic review of its research history and future prospects. Asian Pac. J. Trop. Med. 2017, 10, 835–848. [Google Scholar] [CrossRef]
- Paulo, M.d.Q.; Barbosa-Filho, J.; Lima, E.O.; Maia, R.F.; de Cassia, R.; Barbosa, B.; Kaplan, M.A.C. Antimicrobial activity of benzylisoquinoline alkaloids from Annona salzmanii dc. J. Ethnopharmacol. 1992, 36, 39–41. [Google Scholar] [CrossRef]
- Stevigny, C.; Bailly, C.; Quetin-Leclercq, J. Cytotoxic and antitumor potentialities of aporphinoid alkaloids. Curr. Med. Chem.-Anti-Cancer Agents 2005, 5, 173–182. [Google Scholar] [CrossRef]
- Rodríguez-Arce, E.; Cancino, P.; Arias-Calderón, M.; Silva-Matus, P.; Saldías, M. Oxoisoaporphines and aporphines: Versatile molecules with anticancer effects. Molecules 2020, 25, 108. [Google Scholar] [CrossRef]
- El-Gengaihi, S.E.; Hamed, M.A.; Khalaf-Allah, A.E.-R.M.; Mohammed, M.A. Golden berry juice attenuates the severity of hepatorenal injury. J. Diet. Suppl. 2013, 10, 357–369. [Google Scholar] [CrossRef] [PubMed]
- Carocho, M.; Ferreira, I.C. A review on antioxidants, prooxidants and related controversy: Natural and synthetic compounds, screening and analysis methodologies and future perspectives. Food Chem. Toxicol. 2013, 51, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Mansour, H.H.; Elkady, A.A.; Elrefaei, A.H.; Hafez, H.F. Radioprotective, antioxidant and antitumor efficacy of Annona muricata L. Leaf extract. IJBB 2018, 55, 205–214. [Google Scholar]
- Khalaf-Allah, A.E.-R.M.; El-Gengaihi, S.E.; Hamed, M.A.; Zahran, H.G.; Mohammed, M.A. Chemical composition of golden berry leaves against hepato-renal fibrosis. J. Diet. Suppl. 2016, 13, 378–392. [Google Scholar] [CrossRef]
- Dice, L.R. Measures of the amount of ecologic association between species. Ecology 1945, 26, 297–302. [Google Scholar] [CrossRef]
- Williams, J.G.; Kubelik, A.R.; Livak, K.J.; Rafalski, J.A.; Tingey, S.V. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990, 18, 6531–6535. [Google Scholar] [CrossRef] [PubMed]
- Günther, K. Über die taxonomische gliederung und die geographische verbreitung der insektenordnung der phasmatodea. Contrib. Entomol. 1953, 3, 541–563. [Google Scholar]
- Mohammed, M.A.; Attia, H.N.; El-Gengaihi, S.E.; Maklad, Y.A.; Ahmed, K.A.; Kachlicki, P. Comprehensive metabolomic, lipidomic and pathological profiles of baobab (Adansonia digitata) fruit pulp extracts in diabetic rats. J. Pharm. Biomed. Anal. 2021, 201, 114139. [Google Scholar] [CrossRef]
- Tsugawa, H.; Ikeda, K.; Takahashi, M.; Satoh, A.; Mori, Y.; Uchino, H.; Okahashi, N.; Yamada, Y.; Tada, I.; Bonini, P. Ms-dial 4: Accelerating lipidomics using an ms/ms, ccs, and retention time atlas. bioRxiv 2020. [Google Scholar] [CrossRef]
- Mohammed, M.A.; Ibrahim, B.M.; Abdel-Latif, Y.; Hassan, A.H.; El Raey, M.A.; Hassan, E.M.; El-Gengaihi, S.E. Pharmacological and metabolomic profiles of Musa acuminata wastes as a new potential source of anti-ulcerative colitis agents. Sci. Rep. 2022, 12, 10595. [Google Scholar] [CrossRef]
- Skehan, P.; Storeng, R.; Scudiero, D.; Monks, A.; McMahon, J.; Vistica, D.; Warren, J.T.; Bokesch, H.; Kenney, S.; Boyd, M.R. New colorimetric cytotoxicity assay for anticancer-drug screening. JNCI J. Natl. Cancer Inst. 1990, 82, 1107–1112. [Google Scholar] [CrossRef] [PubMed]
- Moustafa, G.O.; Shalaby, A.; Naglah, A.M.; Mounier, M.M.; El-Sayed, H.; Anwar, M.M.; Nossier, E.S. Synthesis, characterization, in vitro anticancer potentiality, and antimicrobial activities of novel peptide–glycyrrhetinic-acid-based derivatives. Molecules 2021, 26, 4573. [Google Scholar] [CrossRef] [PubMed]
- Draper, H.; Hadley, M. [43] Malondialdehyde determination as index of lipid peroxidation. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 1990; Volume 186, pp. 421–431. [Google Scholar]
- Ellman, G.L. Tissue sulfhydryl groups. Arch. Biochem. Biophys. 1959, 82, 70–77. [Google Scholar] [CrossRef]
- Tam, J.C.W.; Lau, K.M.; Liu, C.L.; To, M.H.; Kwok, H.F.; Lai, K.K.; Lau, C.P.; Ko, C.H.; Leung, P.C.; Fung, K.P. The in vivo and in vitro diabetic wound healing effects of a 2-herb formula and its mechanisms of action. J. Ethnopharmacol. 2011, 134, 831–838. [Google Scholar] [CrossRef]
- Mohammed, M.A. Fighting cytokine storm and immunomodulatory deficiency: By using natural products therapy up to now. Front. Pharmacol. 2023, 14, 1111329. [Google Scholar] [CrossRef] [PubMed]

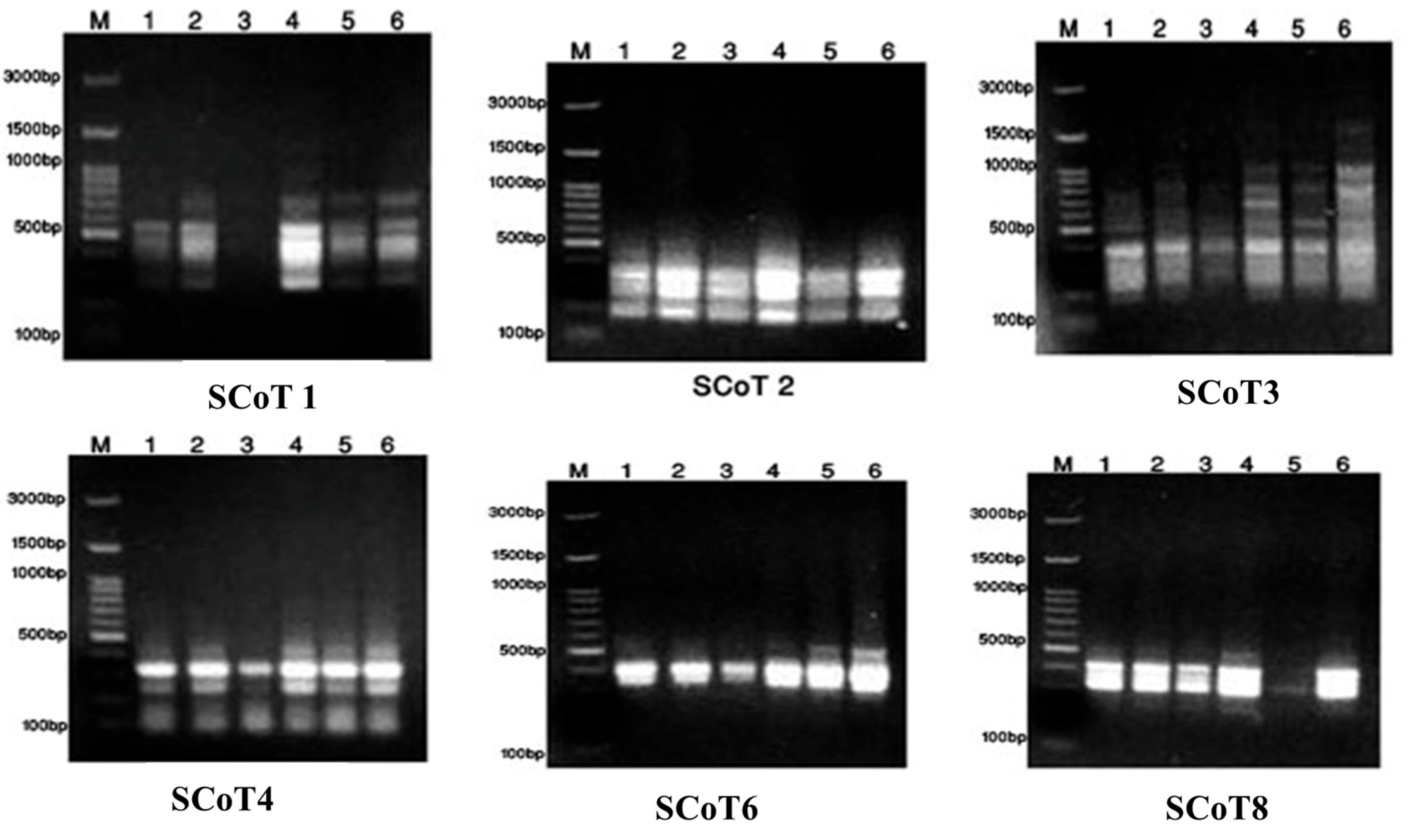
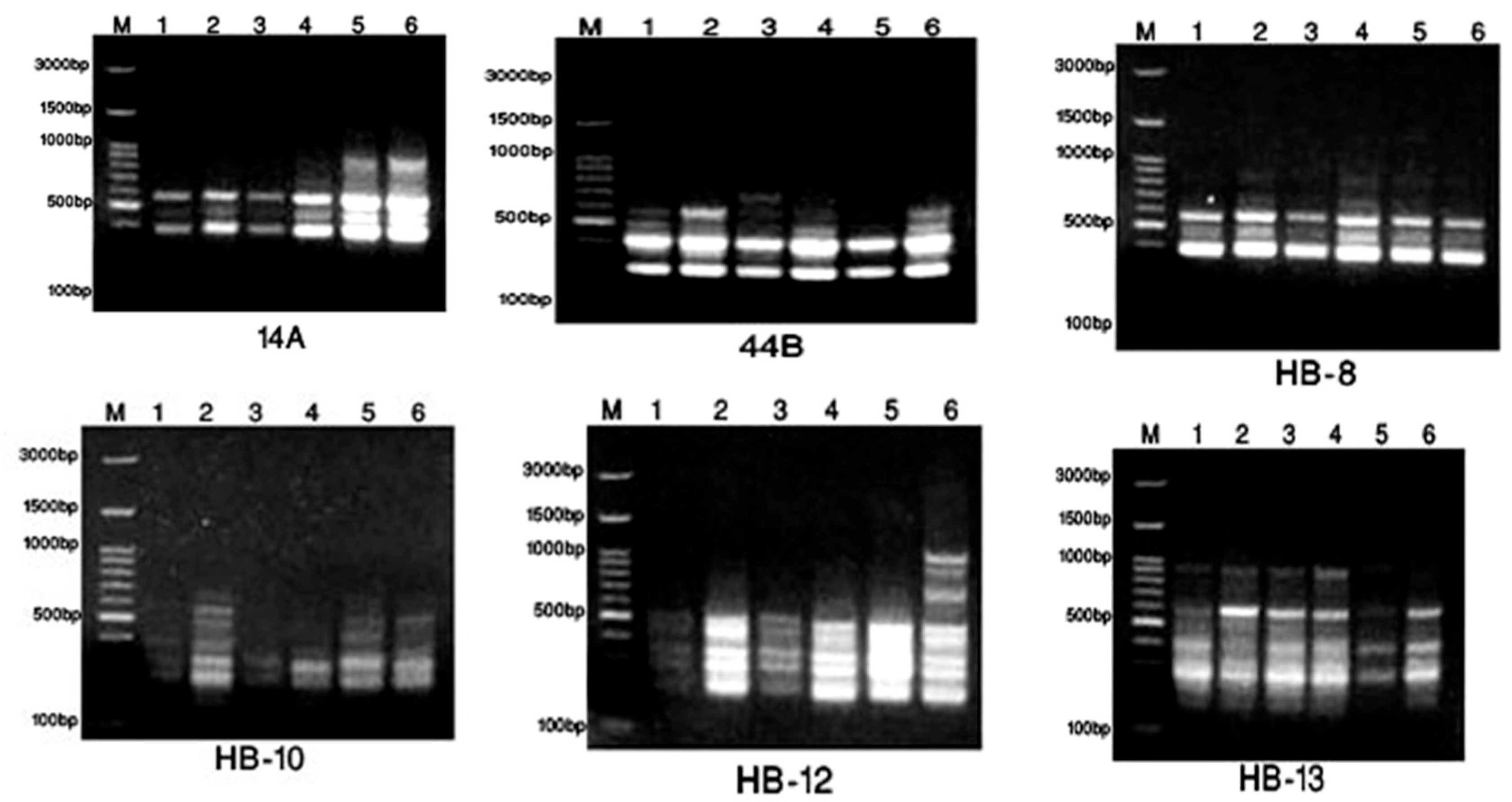
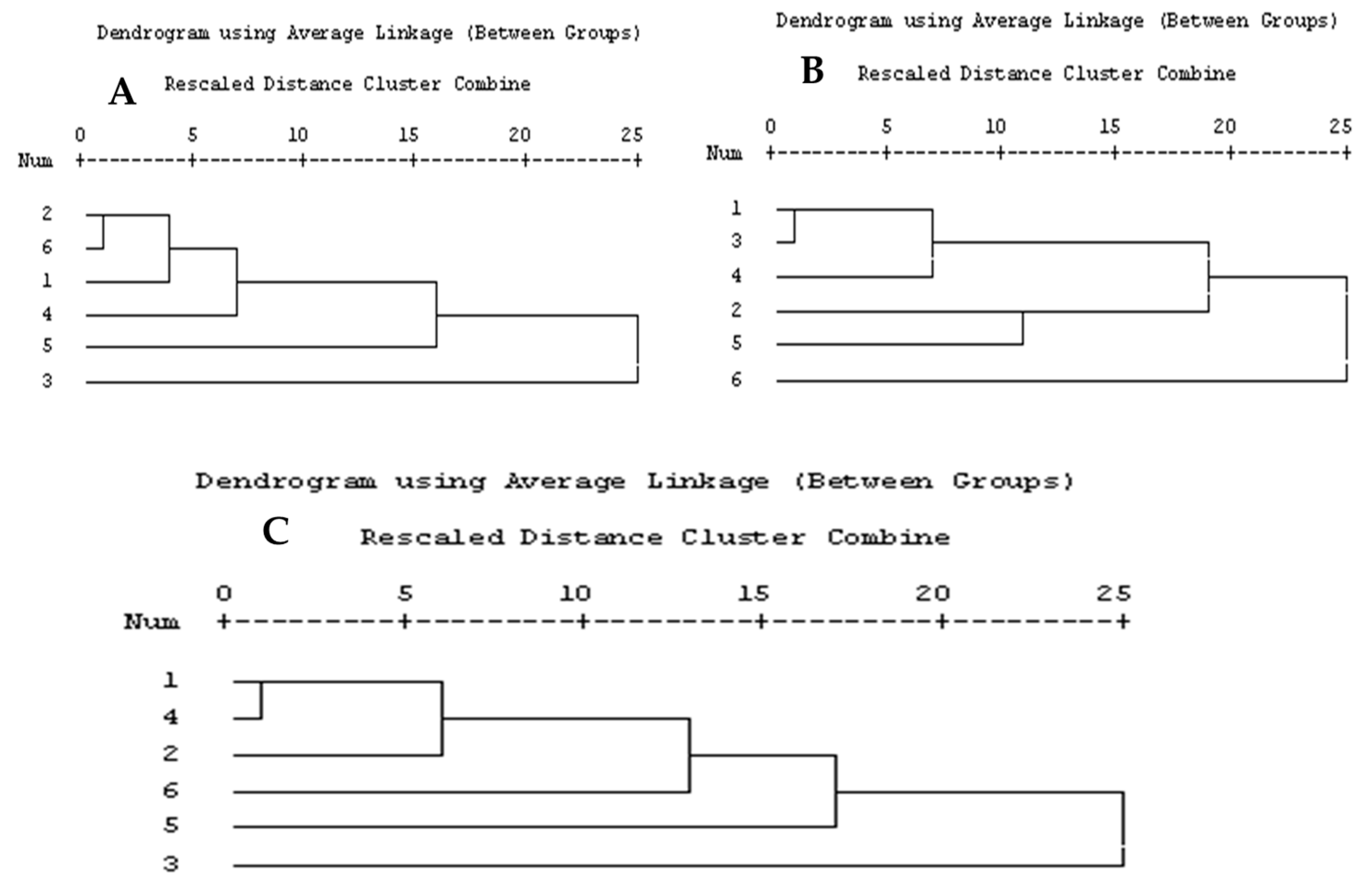
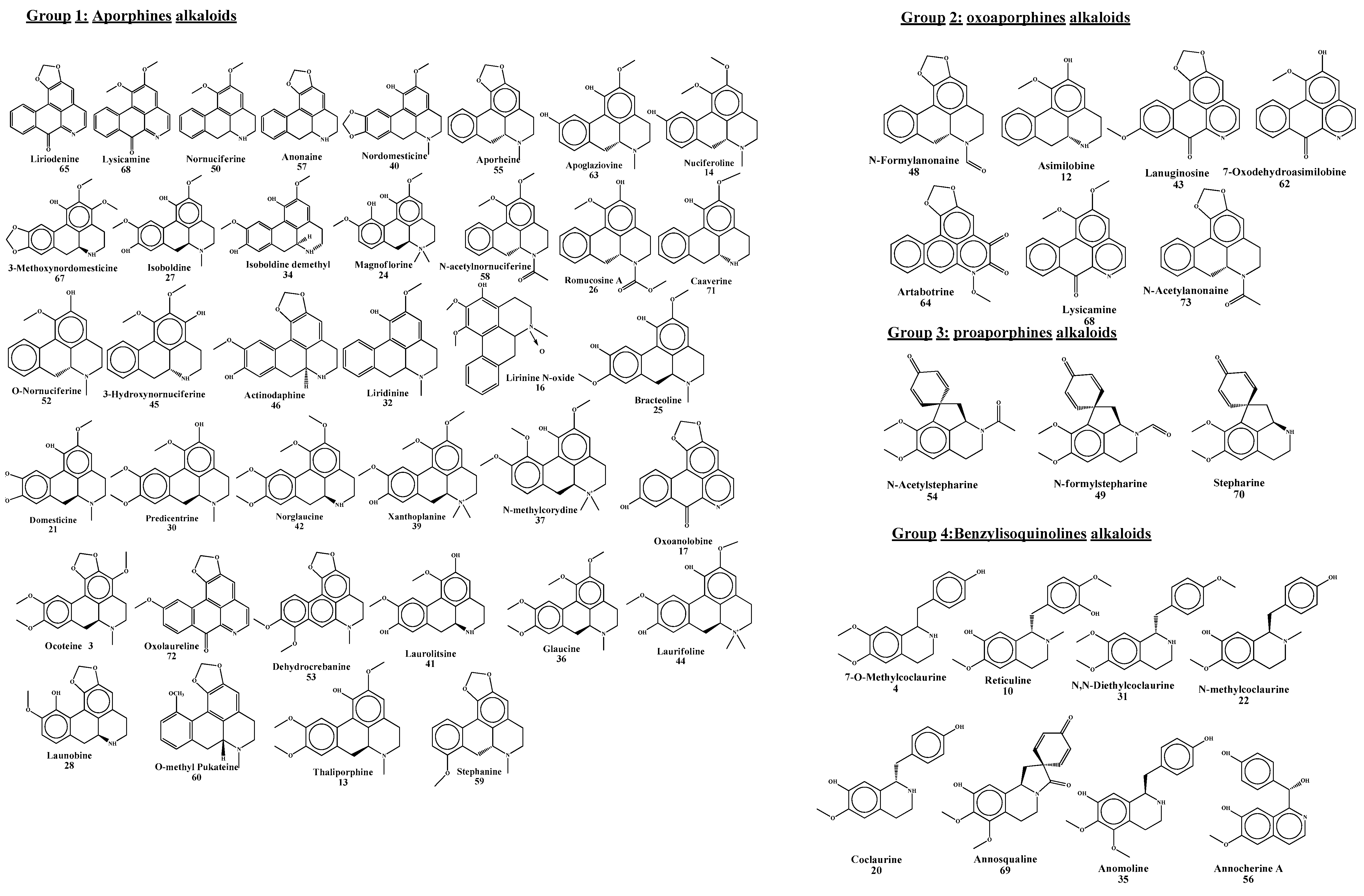
| No. | Primer | ISSR Sequence | Primer | SCoT Sequence |
|---|---|---|---|---|
| 1 | A-14 | 5′ CTC-TCT-CTC-TCT-CTC-TTG 3′ | SCoT 1 | 5′ ACG-ACA-TGG-CGA-CCA-CGC 3′ |
| 2 | B-44 | 5′ CTC-TCT-CTC-TCT-CTC-TGC 3′ | SCoT 2 | 5′ ACC-ATG-GCT-ACC-ACC-GGC 3′ |
| 3 | HB-8 | 5′ GAG-AGA-GAG-AGA-GG 3′ | SCoT 3 | 5′ ACG-ACA-TGG-CGA-CCC-ACA 3′ |
| 4 | HB-10 | 5′ GAG-AGA-GAG-AGA-CC 3′ | SCoT 4 | 5′ ACC-ATG-GCT-ACC-ACC-GCA 3′ |
| 5 | HB-12 | 5′ CAC-CAC-CAC-GC 3′ | SCoT 6 | 5′ CAA-TGG-CTA-CCA-CTA-CAG 3′ |
| 6 | HB-13 | 5′ GAG-GAG-GAG-C 3′ | SCoT 8 | 5′ ACA-ATG GCT-ACC-ACT-ACC 3′ |
| Peak No. | RT | Identified Constituents | Chemical Formula | MS (M/e) | % Components in Annona sp. | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| m/z | No. of Scans | Main Significant Fragments | Base Peak | A. atemoya | A. glabra | A. abdel-razek | A. reticulata | A. squamosa | A. muricata | ||||
| 1 | 11.333 | α-Thujene | C10H16 | 136 | 17 | 121, 105, 93, 77, 65, 53 | 93 | 0 | 0 | 0 | 1.89 | 0.2 | 0 |
| 2 | 11.641 | α-Pinene | C10H16 | 136 | 17 | 121, 93, 77, 67 | 93 | 1.31 | 4.85 | 8.03 | 1.85 | 4.94 | 9.1 |
| 3 | 12.178 | Camphene | C10H16 | 136 | 17 | 121, 107, 93, 79 | 93 | 0.35 | 1.57 | 0.35 | 0.19 | 1.91 | 0.16 |
| 4 | 12.959 | β-Pinene | C10H16 | 136 | 19 | 121, 107, 93, 69, 53 | 93 | 0 | 0.4 | 8.43 | 1.47 | 1.62 | 9.45 |
| 5 | 13.088 | Sabinene | C10H16 | 136 | 19 | 121, 105, 93, 77, 69, 53 | 93 | 0 | 0 | 0.27 | 11.9 | 0.78 | 2 |
| 6 | 13.694 | β-Myrcene | C10H16 | 136 | 16 | 121, 93, 69, 41 | 93 | 3.06 | 6.36 | 1.11 | 1.49 | 0.27 | 3.17 |
| 7 | 14.201 | α-Phellandrene | C10H16 | 136. | 16 | 121, 93, 77 | 93 | 2.28 | 5.96 | 0.41 | 0 | 0.13 | |
| 8 | 14.906 | β-Cymene | C10H14 | 134 | 17 | 119, 103, 91, 77, 51 | 119 | 1.04 | 1.08 | 0 | 0.29 | 0 | 0 |
| 9 | 15.034 | Eucalyptol | C10H18O | 154 | 17 | 139, 121, 108, 93, 81, 71, 55 | 81 | 0 | 0 | 0 | 0 | 0 | 2.41 |
| 10 | 15.099 | D-Limonene | C10H16 | 136 | 17 | 121, 107, 93, 79, 68, 53 | 93, 68 | 2.42 | 3.41 | 3.27 | 1.45 | 3.25 | 3.08 |
| 11 | 15.792 | β-Ocimene | C10H16 | 136 | 16 | 105, 93, 79, 53 | 93 | 2.95 | 8.61 | 2.52 | 0.39 | 0.24 | 0.7 |
| 12 | 16.171 | γ-Terpinene | C10H16 | 136 | 18 | 121, 105, 93, 77, 65 | 93 | 0 | 0 | 0 | 1.68 | 0.18 | 0.15 |
| 13 | 16.526 | Iso-β-terpineol | C10H18O | 154 | 20 | 121, 93, 71, 55 | 71 | 0 | 0 | 0 | 0.13 | 0 | 0 |
| 14 | 17.261 | α-Terpinolene | C10H16 | 136 | 16 | 121, 105, 93, 79, 67, 53 | 93 | 0 | 0 | 0 | 0.75 | 0.18 | |
| 15 | 17.704 | Linalool | C10H18O | 154 | 21 | 136, 121, 93, 71, 55 | 71 | 0.4 | 0.51 | 0.72 | 0.52 | 0.2 | 0.33 |
| 16 | 18.514 | Trans-p-2-Menthen-1-ol | C10H18O | 154 | 21 | 139, 111, 93, 79, 55, 43 | 43 | 0 | 0 | 0 | 0.17 | 0 | 0 |
| 17 | 20.828 | Trans-3(10)-Caren-2-ol | C10H16O | 152 | 20 | 137, 119, 109.000, 95, 81, 69, 41 | 109 | 0 | 0.27 | 0 | 0 | 0 | 0 |
| 18 | 20.548 | Terpinen-4-ol | C10H18O | 154 | 18 | 125, 93, 71, 55 | 71 | 0 | 0 | 0 | 3.08 | 0.41 | 0.24 |
| 19 | 20.956 | α-Terpineol | C10H18O | 154 | 18 | 121, 107, 93, 81, 59, 43, 31 | 59 | 0 | 0.23 | 0 | 0.19 | 0 | 0.15 |
| 20 | 21.592 | Acetic acid, octyl ester | C10H20O2 | 172 | 17 | 152, 112, 70, 43 | 43 | 0.56 | 0 | 0 | 0 | 0 | 0 |
| 21 | 22.285 | Cis-3-Hexenyl isovalerate | C11H20O2 | 184 | 16 | 152, 103, 67, 57 | 67 | 0 | 0 | 0 | 0 | 0 | 0.11 |
| 22 | 24.226 | Bornyl acetate | C12H20O2 | 172 | 17 | 154, 121, 108, 95, 80, 55 | 95 | 0.35 | 0.52 | 0 | 0 | 0.9 | 0.13 |
| 23 | 25.963 | δ-Elemene | C15H24 | 204 | 16 | 189, 161, 136, 121, 93, 77, 55, 41 | 121 | 0 | 0 | 21.4 | 4.33 | 5.22 | 6.99 |
| 24 | 26.33 | α-Cubebene | C15H24 | 204 | 16 | 189, 161, 133, 105, 91, 81, 55 | 105 | 0 | 0 | 0 | 0.21 | 0.27 | 0.25 |
| 25 | 27.222 | α-Copaene | C15H24 | 204 | 16 | 189, 161, 133, 119, 93, 81, 55 | 161, 119 | 1.52 | 0.89 | 0.58 | 1.88 | 1.04 | 3.06 |
| 26 | 27.414 | γ-Muurolene | C15H24 | 204 | 20 | 190, 161, 147.1100, 133, 105, 91, 69, 55 | 161 | 0 | 0.48 | 0 | 0 | 0 | 0 |
| 27 | 27.484 | Bicyclo [5.2.0]nonane, 4-methylene-2,8,8-trimethyl-2-vinyl- | C15H24 | 204 | 25 | 189, 161, 133, 105, 93, 81, 67, 53 | 81 | 0 | 0 | 0 | 1.42 | 0 | 1.21 |
| 28 | 27.718 | β-elemene | C15H24 | 204 | 24 | 189, 175, 161, 147, 119, 107, 93, 67, 55 | 161, 93 | 2.4 | 1.75 | 8.9 | 11.21 | 4.96 | 9.72 |
| 29 | 28.009 | (+)-α-Himachalene | C15H24 | 204 | 20 | 189, 175, 161, 119, 93, 55 | 93 | 0 | 0.4 | 0 | 0 | 0 | 0 |
| 30 | 28.027 | α-Gurjunene | C15H24 | 204 | 13 | 189, 178, 161, 147, 133, 119, 105, 79, 55 | 105 | 0 | 0 | 0 | 0.15 | 1.43 | 0.15 |
| 31 | 28.242 | Cis-Caryophyllene | C15H24 | 204 | 28 | 189, 165, 117, 132, 119, 119, 91, 55 | 105 | 0.33 | 4.1 | 0.88 | 0 | 10.77 | 1.4 |
| 32 | 28.749 | Caryophyllene | C15H24 | 204 | 29 | 189, 161, 133, 120, 105, 93, 79, 69, 55 | 133 | 28.09 | 19.59 | 5.37 | 11.66 | 16.63 | 6.22 |
| 33 | 28.936 | Β-cubebene | C15H24 | 204 | 27 | 189, 161, 133, 105, 79 | 161 | 0.91 | 0.43 | 1.05 | 1.21 | 0.55 | 0.86 |
| 34 | 29.047 | Trans-α-Bergamotene | C15H24 | 204 | 26 | 189, 161, 133, 119, 107, 93 | 119, 93 | 4.2 | 2.39 | 5.15 | 0.83 | 0 | 0.33 |
| 35 | 29.425 | 7-epi-α-Cadinene | C15H24 | 204 | 29 | 189, 161, 147, 133, 119, 105, 91, 79, 55 | 161, 91 | 0.36 | 0 | 0.4 | 0.37 | 0 | 0 |
| 36 | 29.659 | Cis-β-Farnesene | C15H24 | 204 | 16 | 161, 147, 133, 105, 93, 69 | 69 | 0.45 | 0.2 | 0 | 0.17 | 0 | 0 |
| 37 | 29.787 | Humulene | C15H24 | 204 | 22 | 175, 147, 121, 107, 93, 80 | 93 | 5.22 | 3.84 | 1.49 | 2.46 | 5 | 1.75 |
| 38 | 30.545 | γ-Muurolene | C15H24 | 204 | 17 | 189, 161, 147, 133, 105, 79, 55 | 161 | 0.3 | 0.56 | 0.46 | 1.58 | 2.08 | 1 |
| 39 | 30.795 | Germacrene D | C15H24 | 204 | 26 | 177, 161, 147, 133, 119, 105, 79, 55 | 161 | 9.5 | 4.98 | 13.06 | 10.74 | 4.47 | 6.32 |
| 40 | 30.993 | β-Selinene | C15H24 | 204 | 19 | 189, 161, 147, 133, 105, 79 | 105, 161 | 0 | 0 | 0.28 | 0 | 0.26 | 0.51 |
| 41 | 31.081 | δ-Selinene | C15H24 | 204 | 20 | 189, 175, 161, 147, 133, 91, 55 | 189, 161 | 0 | 0 | 0.29 | 0 | 0 | 11.12 |
| 42 | 31.355 | γ-Elemene | C15H24 | 204 | 35 | 161, 121, 93, 41 | 121, 93 | 0 | 0 | 2.3 | 0 | 2.79 | 0 |
| 43 | 31.366 | (+)-γ-Gurjunene | C15H24 | 204 | 20 | 189, 179, 161, 133, 121, 93, 79, 67 | 121, 93 | 0 | 0 | 0 | 6.95 | 0 | 0 |
| 44 | 31.413 | α-Muurolene | C15H24 | 204 | 22 | 189, 161, 133, 119, 105, 91, 79, 55 | 105 | 0.96 | 0.33 | 0 | 0 | 0.77 | 0 |
| 45 | 31.578 | α-Farnesene | C15H24 | 204 | 16 | 189, 161, 133, 107, 93, 69, 55 | 93 | 0.82 | 0.38 | 0 | 0 | 0 | 0 |
| 46 | 31.675 | β-Bisabolene | C15H24 | 204 | 20 | 189, 161, 134, 107, 93, 79, 69 | 93, 69 | 1.38 | 0.73 | 0 | 0 | 0 | 0.27 |
| 47 | 32.165 | Cubedol | C15H26O | 224 | 29 | 207, 189, 161, 145, 91, 79, 67.0700, 55 | 161 | 0 | 0 | 0 | 0 | 0.43 | 0 |
| 48 | 32.008 | Trans-γ-cadinene | C15H24 | 204 | 22 | 191.1200, 161.1000, 133.1000, 105.1000, 79.0900 | 161 | 0.36 | 0.64 | 0.34 | 0.34 | 2.27 | 0.96 |
| 49 | 31.669 | β-Bisabolene | C15H24 | 204 | 25 | 189, 161, 134.1200, 105, 93, 79, 69 | 161 | 0 | 0 | 0 | 0.93 | 0 | 0 |
| 50 | 32.147 | Cubedol | C15H26O | 224 | 33 | 207, 189, 161, 135, 79, 55 | 161 | 0 | 0 | 0 | 0.53 | 0.38 | 0.34 |
| 51 | 34.514 | (-)-Globulol | C15H26O | 224 | 33 | 204, 189, 177.1200, 161, 122, 105, 81, 55 | 43 | 0 | 0 | 0 | 0 | 0 | 0.14 |
| 52 | 32.346 | δ-Cadinene | C15H24 | 204 | 24 | 189, 161, 119, 91, 69 | 161 | 2.19 | 2.17 | 0.73 | 1.51 | 2.48 | 1.81 |
| 53 | 32.678 | α-Patchoulene (1.alpha.,3a.alpha.,7.alpha.,8a.beta.)- | C15H24 | 204 | 24 | 189, 161, 119, 93, 79 | 93 | 1.84 | 1.03 | 0 | 0.16 | 0 | 0 |
| 54 | 33.628 | Elemol | C15H26O | 224 | 39 | 189, 161, 135, 107, 79, 59 | 93 | 0 | 0 | 2.45 | 0.33 | 0.67 | 0 |
| 55 | 34.153 | Nerolidol | C15H26O | 222 | 32 | 189, 161, 136, 107, 93, 69, 55 | 69 | 1.03 | 1.11 | 0.34 | 0.75 | 0.53 | 0.29 |
| 56 | 34.823 | Squalene | C30H50 | 381 | 38 | 207, 161, 95, 81, 69, 53 | 69 | 0 | 0 | 0 | 0 | 0 | 0.6 |
| 57 | 35.138 | (−)-Spathulenol | C15H24O | 220 | 37 | 220, 205, 187, 159, 119, 105, 91, 79, 43 | 43 | 4.65 | 0.35 | 0 | 0.58 | 0.84 | 0.53 |
| 58 | 35.365 | Caryophyllene oxide | C15H24O | 220 | 36 | 205, 161, 135, 121, 109, 79, 69, 43 | 43 | 4.15 | 3.68 | 0.28 | 1.19 | 1.77 | 0 |
| 59 | 35.767 | Veridiflorol | C15H26O | 222 | 31 | 204, 177, 149.1100, 135, 121, 107, 81, 43 | 43 | 0.4 | 0.47 | 0 | 0.5 | 0.89 | 0 |
| 60 | 36.624 | β-Ionone | C13H20O | 192 | 22 | 205, 177, 161, 121, 91, 55 | 177 | 0 | 0 | 0 | 0 | 0.28 | 0 |
| 61 | 36.233 | β-Eudesmol | C15H26O | 222 | 222 | 204, 177, 164, 149, 133, 107, 81, 59 | 149 | 0 | 0 | 0 | 0 | 0 | 0.27 |
| 62 | 37.09 | Germacrene D-4-ol | C15H26O | 222 | 20 | 207, 161, 93 | 93 | 0 | 0 | 3.71 | 0.54 | 0 | 0 |
| 63 | 37.265 | β-Guaiene | C15H24 | 204 | 35 | 179, 145, 119, 105, 79, 55 | 119 | 0 | 0 | 0.8 | 0 | 0.87 | 0 |
| 64 | 37.318 | Cubenol | C15H26O | 222 | 30 | 204, 179, 161, 119, 105 | 119 | 0.49 | 1.22 | 0.37 | 0.9 | 0 | 0.28 |
| 65 | 37.428 | Iso-spathulenol | C15H24O | 220 | 36 | 204, 177, 162, 133, 119, 105, 91, 79, 55 | 119, 91 | 0 | 0 | 1.39 | 0 | 0.77 | 0 |
| 66 | 37.941 | Tau-Cadinol | C15H26O | 222 | 50 | 204, 189, 161, 121, 95 | 161 | 3.33 | 0.52 | 1.52 | 2.19 | 8.27 | 3.45 |
| 67 | 38.116 | Torreyol | C15H26O | 222 | 22 | 204, 189, 161,136, 119, 79 | 161 | 0.57 | 3.7 | 0 | 1.64 | 0.7 | 0.13 |
| 68 | 38.244 | Cis-α-Bisabolene | C15H24 | 204 | 38 | 136, 93 | 93 | 1.26 | 0.48 | 0 | 0 | 0 | 0 |
| 69 | 38.361 | Tau-Muurolol | C15H26O | 222 | 37 | 204, 161, 134, 105, 81 | 161 | 0 | 0 | 0.64 | 0.25 | 0.38 | 0 |
| 70 | 38.495 | α-Cadinol | C15H26O | 222 | 28 | 204, 189, 161, 137, 121, 95, 81, 55 | 95 | 3.66 | 0.96 | 0.94 | 0 | 3.83 | 2.61 |
| 71 | 40.716 | Alloaromadendrene oxide-(1) | C15H24O | 220 | 23 | 177, 135, 107, 69 | 69 | 0.34 | 1.94 | 0 | 0 | 0 | 0 |
| 72 | 40.926 | Bergamotol, Z-.alpha.-trans- | C15H24O | 220 | 77 | 187, 119, 93 | 93 | 1.69 | 1.23 | 0 | 0 | 0 | 0 |
| 73 | 43.024 | Farnesol, acetate | C17H28O2 | 253 | 22 | 189, 136, 93, 69 | 69 | 0.45 | 0.7 | 0 | 0 | 0 | 0.37 |
| 74 | 48.293 | Neoisolongifolene, 8-bromo- | C15H23Br | 282 | 40 | 225, 203, 175, 91, 41 | 203 | 0.32 | 1 | 0.23 | 0.39 | 2.56 | 0 |
| 75 | 49.208 | Chlorpyrifos | C9H11Cl3NO3PS | 313 | 24 | 313, 257, 196, 170, 124, 96 | 96 | 0 | 0 | 0 | 0.12 | 0 | 0 |
| 76 | 50.146 | Geranyllinalool | C20H34O | 313 | 50 | 203, 135, 69 | 69 | 0.41 | 0.43 | 0.54 | 0 | 0.91 | 0.67 |
| Total Identification | 98.3 | 96.45 | 100.59 | 95.87 | 99.97 | 95.1 | |||||||
| Un-identification compound | 2.02 | 3.55 | −0.59 | 4.13 | 0.03 | 4.9 | |||||||
| Oxygenated compounds | 22.48 | 17.84 | 13.7 | 13.61 | 23.03 | 13.05 | |||||||
| Non-oxygenated compounds | 75.82 | 78.61 | 86.89 | 82.26 | 76.94 | 82.05 | |||||||
| Different Extract | Volatile Oils of Annona sp. | Different Alcoholic Extracts of Annona sp. | ||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A. Atemoya | A. glabra | A. abdel-razek | A. reticulata | A. squamosa | A. muricata | A. Atemoya | A. glabra | A. abdel-razek | A. reticulata | A. squamosa | A. muricata | |||||||||||||
| IC50 (μg/mL) | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | ||||||
| 50% | 80% | 100% | 50% | 80% | 100% | 50% | 80% | 100% | 50% | 80% | 100% | 50% | 80% | 100% | 50% | 80% | 100% | |||||||
| HepG2 Liver Cell | 32 | 18 | >100 | >100 | 50 | >100 | 80 | 36.5 | 43 | 14.5 | 77 | 13 | >100 | 10 | 41 | 12.5 | 10 | 13.5 | 56 | 50 | >100 | 29 | 31 | 10.5 |
| MCF7 Breast Cell | 77 | 26.5 | 81.5 | >100 | 42.5 | >100 | 8.5 | 85 | 9 | 19 | 42.5 | 8.5 | 99 | 28 | 83.5 | 36.5 | 29.5 | 10.5 | 11 | >100 | >100 | 10 | 10.5 | 9.5 |
| HCT Colon Cell | 73 | >100 | >100 | >100 | 35.5 | 54 | >100 | >100 | >100 | >100 | >100 | >100 | >100 | >100 | >100 | >100 | 48 | 68 | >100 | >100 | >100 | >100 | 36.5 | 24 |
| Caco Colon Cell | 22 | >100 | 99 | >100 | 49.5 | 33 | 44 | 49.5 | 24.5 | 93 | >100 | 50 | >100 | >100 | >100 | 12 | 47 | 85.5 | >100 | >100 | >100 | 21 | 15 | 68 |
| T47D Breast Cell | >100 | >100 | >100 | >100 | >100 | >100 | >100 | 18.5 | 63.5 | >100 | 23 | >100 | >100 | >100 | >100 | >100 | 40.5 | 53 | >100 | >100 | >100 | 37.5 | 38 | 43 |
| No. | RT | Tentative Identification | Chemical Formula | High-Resolution MS Data | ∆ppm | Annona sp. | ID 1, 2, and 3 | Ref. | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| [M + H]+ m/z Measured and Calculated | Major Fragments | A. Atemoya | A. glabra | A. abdel-razek | A. reticulata | A. aquamosa | A. muricata | |||||||||||||||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | |||||||||
| Group 1: Aporphine | ||||||||||||||||||||||||||
| 3 | 2.21 | Ocoteine C Thalicmine | C21H23NO4 | 370.1720, 370.1721 | 208.1178, 185.9153, 148.7571, 109.2159, 74.0610 | −0.0224 | * | 1 | [24] | |||||||||||||||||
| 6 | 2.28 | Apomorphine B | C17H17O2N | 268.1326, 268.1040 | 251.1066, 219.0805, 191.0805, 136.0620, 97.0295 | −4.303 | * | 2 | [25] | |||||||||||||||||
| 12 | 3.39 | Asimilobine A | C17H17O2N | 268.1341, 268.1333 | 251.1067, 236.0836, 219.0806, 191.0856, 163.0761, 163.0761 | 3.3151 | * | 1 | [26] | |||||||||||||||||
| 13 | 3.85 | Thaliporphine B | C20H24NO4+ | 342.1703, 342.1700 | 297.1121, 265.0858, 237.0905, 178.0861, 123.0440, 58.0661 | 0.7974 | 1 | [27] | ||||||||||||||||||
| 14 | 4.08 | Nuciferoline C | C19H21NO3 | 312.1592, 312.1594 | 259.0969, 265.0862, 205.4610, 171.2698, 144.4292, 104.0781, 76.6994 | 0.6723 | * | 1 | [28] | |||||||||||||||||
| 16 | 4.34 | Lirinine N-oxide C | C19H21O4N | 328.1544, 328.1543 | 297.1124, 283.0966, 265.0868, 178.0864, 116.0532 | 0.709 | * | - | [28] | |||||||||||||||||
| 19 | 4.81 | N-acetyl-3-methoxynornantenine C | C22H23NO6 | 398.1598, 398.1598 | 377.8857, 298.1073, 176.0712, 155.0394, 65.0490 | −0.1366 | * | - | [28] | |||||||||||||||||
| 21 | 5.02 | Domesticine C | C19H19NO4 | 326.1385, 326.1387 | 295.0963, 265.0859, 244.4052, 225.6086, 128.8098, 118.4798, 78.7120, 66.3822 | 0.694 | * | 1 | [29] | |||||||||||||||||
| 24 | 5.16 | Magnoflorine B | C20H24NO4+ | 342.1696, 342.1700 | 297.1120, 265.0863, 251.0703, 201.6616, 174.7599, 143.2922, 84.1842 | −0.9864 | * | 1 | [30] | |||||||||||||||||
| 25 | 5.19 | Bracteoline C | C19H21NO4 | 328.1542, 328.1543 | 297.1128, 265.0860, 237.0910, 178.0865, 121.0652, 75.0269 | 0.3941 | 1 | [31] | ||||||||||||||||||
| 26 | 5.24 | Romucosine A B | C19 H19O4N | 326.1389, 326.1387 | 295.0965, 265.0859, 237.0911, 193.3112, 183.8072 | 0.5224 | * | * | * | * | 1 | [32] | ||||||||||||||
| 27 | 5.38 | Isoboldine B | C19H21NO4 | 328.1541, 238.1543 | 297.1125, 285.1139, 265.0860, 237.060, 237.0909, 178.0865, 121.0651, 58.0660 | −0.5801 | * | 1 | [33] | |||||||||||||||||
| 28 | 5.48 | Launobine C | C18H17O4 N | 312.1228, 312.1230 | 297.1003, 263.1003, 263.0699, 242.381, 64.7326 | −0.6449 | * | 1 | [34] | |||||||||||||||||
| 30 | 5.62 | Predicentrine B | C20H24NO4+ | 342.1701, 342.1700 | 311.1278, 296.1034, 279.1015, 264.0779, 248.0827, 178.0862, 58.0660 | 0.3515 | * | 1 | [35] | |||||||||||||||||
| 32 | 5.69 | Liridinine C | C19H21NO3 | 312.1593, 321.1594 | 280.0738, 263.0703, 235.0760, 205.0648, 118.0079 | −0.379 | * | - | [28] | |||||||||||||||||
| 34 | 5.77 | Derivative of isoboldinedemethyl C | C18H19NO4 | 314.1385, 314.1387 | 283.1325, 265.0862, 237.0904, 197.1719, 147.051, 121.061 | −0.7207 | * | - | [36] | |||||||||||||||||
| 36 | 5.8 | Glaucine A O,O-dimethylisoboldine | C21H26NO4 | 356.1856, 356.1856 | 325.1433, 311.1281, 294.1255, 279.1017, 237.0919, 213.5202, 93.0936 | 0.0754 | * | 1 | [26] | |||||||||||||||||
| 37 | 5.8 | N-methylcorydine B | C21H26NO4 | 356.1850, 356.1856 | 311.1278, 279.1016, 264.0788, 164.0421, 147.0438, 85.2029 | −1.8747 | * | 1 | [37] | |||||||||||||||||
| 38 | 5.85 | Nuciferine B | C19H21O2N | 296.1646, 296.1645 | 278.1177, 251.1067, 219.0807, 145.4770, 109.4571, 88.9068, 58.0662 | 0.3591 | * | 1 | [33] | |||||||||||||||||
| 39 | 5.86 | Xanthoplanine B | C21H26NO4 | 356.1859, 356.1856 | 311.1275, 279.1022, 264.5139, 206.1176, 186.3449, 137.7469, 70.8474 | 0.867 | * | 1 | [37] | |||||||||||||||||
| 40 | 5.90 | Nordomesticine A | C18H17 O4N | 312.1226, 312.1230 | 294.1129, 259.5082, 159.2854, 106.766 | −1.3293 | 1 | [37] | ||||||||||||||||||
| 41 | 5.96 | Laurolitsine B | C18H19NO4 | 314.1383, 314.1387 | 297.1125, 265.0860, 237.0921, 147.0439, 121.0653, 92.6976 | −1.1091 | * | 1 | [38] | |||||||||||||||||
| 42 | 5.98 | Norglaucine A | C20H24NO4+ | 342.1695, 342.1700 | 311.1279, 296.1046, 279.1016, 265.0851, 248.032, 149.1094, 75.1861 | −1.5215 | 1 | [39] | ||||||||||||||||||
| 43 | 6.03 | Asimilobine A | C17H17NO2 | 368.1331, 268.1332 | 251.1066, 219.805, 191.0854, 116.0090 | -0.5546 | * | 1 | [26] | |||||||||||||||||
| 46 | 6.57 | (-)-Actinodaphine C | C18H17O4N | 312.1235, 312.1230 | 295.0964, 265.0857, 237.0913, 200.9195, 177.3906, 158.8117, 106.9259 | 1.4084 | 3 | [40] | ||||||||||||||||||
| 47 | 6.77 | Xylopine A | C18H17O3N | 296.1284, 296.1281 | 265.0861, 188.0704, 125.3992, 90.0052 | 0.9034 | * | 1 | [28] | |||||||||||||||||
| 44 | 6.07 | Laurifoline C | C20H24NO4+ | 342.1705, 342.1700 | 311.1267, 297.1129, 265.0868, 178.0860, 64.9947, 58.0660 | 1.4217 | * | 1 | [38] | |||||||||||||||||
| 48 | 6.87 | N-formylanonaine A (-)-N-formylanonaine | C18H15O3N | 294.1124, 294.1125 | 263.0702, 236.1060, 127.6403, 114.6490 | −0.2504 | * | * | 1 | [41] | ||||||||||||||||
| 50 | 6.94 | Nornuciferine ASanjoinineia Daechualkaloid E | C18H19O2N | 282.1480, 282.1489 | 265.1222, 250.0989, 243.1039, 164.0425, 129.0027 | −2.8872 | * | 1 | [41] | |||||||||||||||||
| 52 | 7.17 | N-methylasimilobine B O-nornuciferine | C18H19NO2 | 282.1247, 282.1237 | 265.1223, 250.0986, 234.1042, 186.6200, 118.7022, 806066 | 3.4064 | 1 | [42] | ||||||||||||||||||
| 55 | 7.33 | Roemerine B (-)-Aporheine | C18H17NO2 | 280.1340, 280.1332 | 249.0910, 234.1497, 207.0805, 150.0269, 117.1255, 77.8964 | 2.7374 | * | 1 | [43] | |||||||||||||||||
| 57 | 7.91 | Anonaine A | C17H15O2N | 266.1175, 266.1176 | 249.0910, 219.0805, 191.0854, 174.0443 | −0.2353 | * | * | 1 | [41] | ||||||||||||||||
| 58 | 7.96 | N-acetylnornuciferine C | C20H21NO3 | 324.1596, 324.1594 | 309.1001, 171.3005, 111.1272 | 0.4823 | 1 | [44] | ||||||||||||||||||
| 59 | 8 | Stephanine B | C19H19NO3 | 310.1438, 310.1438 | 279.1014, 249.010, 194.0215, 75.1965 | 0.093 | * | * | 1 | [45] | ||||||||||||||||
| 60 | 8.06 | O-methyl pukateine C | C19H20NO3 | 310.1426, 310.1438 | 279.1014, 249.0904, 85.4723 62.2940 | −3.6462 | * | 3 | [28] | |||||||||||||||||
| 61 | 8.85 | N-formylstepharine C | C19H19NO4 | 326.1377, 326.1387 | 309.1119, 279.1015, 266.0927, 129.4605, 102.2371, 91.6374 | −3.0333 | * | 1 | [28] | |||||||||||||||||
| 63 | 9 | Apoglaziovine A | C18H19NO3 | 298.1442, 298.1438 | 281.1166, 249.0910, 221.1166, 194.1585, 127.0428, 64.0436 | 1.325 | * | 1 | [28] | |||||||||||||||||
| 67 | 10.88 | 3-Methoxynordomesticine C | C19H19O5N | 342.1336, 342.1336 | 292.8240, 279.1023, 265.0850, 136.3306, 93.0376 | 0.004 | * | * | * | 1 | [28] | |||||||||||||||
| 71 | 12 | Caaverine B | C17H17O2N | 268.1312, 268.1332 | 235.0067, 148.0763, 131.0494, 121.0652, 107.0496, 59.0501 | 0.5546 | * | 1 | [46] | |||||||||||||||||
| Group 2: Oxoaporphine alkaloids | ||||||||||||||||||||||||||
| 1 | 1.64 | Annonbraine A | C19 H11O4N | 318.0385, 318.0397 | 290.0431, 242.0245, 218.9947, 137.8421, 90.9476 | −1.219 | * | * | * | - | [41] | |||||||||||||||
| 17 | 4.44 | Oxoanolobine B | C17H19NO4 | 302.1479, 302.1387 | 253.0852, 225.0901, 191.0707, 159.0440, 121.0653, 107.0497, 69.7030 | −2.6683 | * | 1 | [47] | |||||||||||||||||
| 53 | 7.24 | Dehydrocrebanine C | C20H19NO4 | 338.1385, 338.1387 | 323.1148, 177.0544, 91.6380, 61.0048 | −0.4889 | * | 1 | [48] | |||||||||||||||||
| 62 | 8.67 | 7-Oxodehydroasimilobine B | C17H11NO3 | 278.0813, 278.0812 | 263.0581, 236.9391, 213.9227, 149.0237, 105.1089 | 0.3544 | * | 1 | [49] | |||||||||||||||||
| 64 | 9.58 | Artabotrine B | C18H11O5N | 322.0708, 322.0710 | 278.40501, 164.5077, 157.0504, 110.9065, 71.1918 | −0.7101 | * | 1 | [50] | |||||||||||||||||
| 65 | 10.03 | Liriodenine A | C17H9O3N | 276.0654, 276.0655 | 249.9080, 226.9840, 208.8835, 127.9462, 110.9124, 64.1885 | −0.4367 | * | 1 | [51] | |||||||||||||||||
| 66 | 10.3 | Lanuginosine A | C18H11NO4 | 306.0760, 306.0761 | 274.2528, 230.1798, 166.1937, 140.2873, 70.0659 | −0.412 | * | 1 | [42] | |||||||||||||||||
| 68 | 10.9 | Lysicamine A (Oxonuciferine) | C18H13NO3 | 292.0965, 292.0968 | 277.0733, 248.0707, 218.1884, 163.0764, 128.8807, 81.0039 | −1.0021 | * | * | 1 | [41] | ||||||||||||||||
| 72 | 12.84 | Oxolaureline C 10-Methoxyliriodenine | C18H10NO4 | 306.0764, 306.0761 | 267.3079, 240.9942, 131.1421, 102.4868, 78.4019 | 0.9839 | * | 2 | [28] | |||||||||||||||||
| 73 | 15.13 | N-acetylbongaridine C (-)-N-acetylanonaine | C19H17O3N | 308.1284, 308.1281 | 249.0909, 219.0807, 156.2953, 134.3902, 86.0606 | 0.8682 | * | 1 | [52] | |||||||||||||||||
| Group 3: Proaporphine alkaloids | ||||||||||||||||||||||||||
| 49 | 6.88 | (-)-N-formylstepharine C | C19H19NO4 | 326.1382, 326.1387 | 296.0966, 265.0860, 237.0910, 171.9669, 69.8721 | −1.4426 | 1 | [53] | ||||||||||||||||||
| 54 | 7.27 | N-acetylstepharine C | C20H21NO4 | 340.1540, 340.1543 | 328.6469, 297.4160, 218.9519, 160.7969, 129.9211, 92.8186, 71.1164 | −1.0083 | 1 | [54] | ||||||||||||||||||
| 70 | 11.7 | Stepharine C | C18H19NO3 | 298.1447, 298.1438 | 221.9773, 177.0549, 145.0285, 105.0706, 69.1952 | 3.0651 | * | 1 | [41] | |||||||||||||||||
| Group 4: Benzylisoquinoline and isoquinoline alkaloids | ||||||||||||||||||||||||||
| 4 | 2.22 | 7-O-methylcoclaurine C | C18H21NO3 | 300.1595, 300.1594 | 283.1328, 269.1170, 223.1120, 192.1016, 176.0754, 137.0598, 121.0651, 107.0496, 58.0661 | 0.2159 | * | 1 | [30] | |||||||||||||||||
| 10 | 2.35 | Reticuline A | C19H23O4N | 330.1697, 330.1700 | 192.1019, 175.0754, 137.0597 | 0.08374 | * | * | * | 1 | [55] | |||||||||||||||
| 20 | 5.01 | Coclaurine B | C17H19NO3 | 300.1594, 300.1594 | 283.1328, 269.1171, 237.0916, 175.0754, 137.0597, 121.0652, 89.0605 | 0.0126 | * | 1 | [30] | |||||||||||||||||
| 22 | 5.1 | N-methylcoclaurine B | C18H21O3N | 300.1594, 300.1541 | 283.1328, 269.1171, 237.0916, 175.0754, 137.0597, 121.0652, 89.005 | 0.0126 | * | * | 1 | [56] | ||||||||||||||||
| 31 | 5.66 | N,N-dimethylcoclaurine C | C19H24NO3+ | 314.1751, 314.1751 | 297.1111, 265.0876, 237.0911, 175.0744, 147.0446, 121.0651 | 0.1265 | * | - | [36] | |||||||||||||||||
| 56 | 7.49 | Annocherine A A | C17H15NO4 | 298.1076, 298.1074 | 280.0967, 192.0660, 168.9584, 129.7724, 60.6217 | 0.6373 | 1 | [57] | ||||||||||||||||||
| 35 | 5.78 | Anomoline B | C18H21NO4 | 316.1540, 316.1543 | 267.1016, 239.1066, 191.0703, 159.0440, 121.0652 | -0.9883 | 1 | [58] | ||||||||||||||||||
| 69 | 11.03 | Annosqualine A | C19H19NO5 | 342.1337, 342.1336, | 324.1225, 265.0876, 222.0770, 189.0428, 84.9932 | 0.2873 | * | 1 | [59] | |||||||||||||||||
| Other compounds | ||||||||||||||||||||||||||
| 5 | 2.33 | Pallidine B Morphinandienone | C19H21NO4 | 328.1536, 328.1543 | 247.1118, 256.0858, 237.0908, 203.5709 | −2.2541 | 1 | [60] | ||||||||||||||||||
| 7 | 2.28 | Nicotinamide 2 Nicotine alkaloid | C6H6N2O | 123.055 | 96.0555, 80.0498, 67.0552 | −0.1804 | * | [61] | ||||||||||||||||||
| 8 | 2.28 | Chlorantene B C Sesquiterpenoid alkaloids | C15H19O5N | 294.1323, 294.1336 | 276.1443, 230.1388, 212.1283, 173.9608, 132.1021, 97.0292, 86.0972 | 0.1276 | * | * | * | 1 | [62] | |||||||||||||||
| 2 | 1.99 | 6-(Alpha-D-glucosaminyl)-1D-myo-inositol | C12H24O10N | 342.1402, 342.1403 | 306.1182, 283.1075, 282.1075, 240.0867, 204.0874, 162.0761, 127.0393 | 0.3943 | * | * | - | |||||||||||||||||
| 9 | 2.37 | Isoleucine C Amino acids | C6H12NO2 | 132.1021, 132.1019 | 114.0553, 97.0289, 86.0972, 68.0504, 58.0661 | 1.1019 | * | 1 | [61] | |||||||||||||||||
| 11 | 2.39 | Sarracine C Pylrrolidine | C18H27O5N | 338.1972, 338.1975 | 285.1080, 254.1135, 237.0869, 211.1084, 159.0766, 114.0553 | 1.001 | * | [63] | ||||||||||||||||||
| 15 | 4.29 | Phenylacetaldehyde | C8H8O | 121.0651, 121.0647 | 103.0547, 93.0706, 53.0397 | −0.3087 | * | - | - | |||||||||||||||||
| 18 | 4.45 | Coreximine A Protoberberine | C19H21O4N | 328.1545, 328.1543 | 313.1313, 265.0865, 235.8464, 192.1019, 178.0864, 151.0754, 117.0734, 93.0376, 58.0660 | 0.4428 | 1 | [64] | ||||||||||||||||||
| 23 | 5.15 | Piperolactam C B Aristolactamalkaloids | C18H15O4N | 310.1073, 310.1074 | 279.0889, 165.6389, 101.8022, 70.9469, 63.6019 | −0.2731 | * | * | 1 | [65] | ||||||||||||||||
| 29 | 5.55 | Scopoletin C | C10H9O4 | 193.0498, 193.0495 | 165.0547, 133.0285, 147.0444 | 1.3956 | 1 | [66] | ||||||||||||||||||
| 33 | 5.73 | Unknown | C20H31O6N | 382.2227, 382.2224 | 273.1493, 191.0694, 161.5542, 132.7013, 65.1538 | 0.8729 | * | - | - | |||||||||||||||||
| 45 | 6.36 | 3-Hydroxynornuciferine B | C18H19NO3 | 298.1434, 298.1438 | 281.1170, 249.0911, 221.0973, 160.1834, 94.6420, 59.5692 | −1.234 | * | 1 | [67] | |||||||||||||||||
| 51 | 6.96 | Unknown | C22H17ON2 | 326.1411, 326.1414 | 309.1119, 294.0886, 278.0939, 265.0858, 251.1050 | −0.9582 | - | - | ||||||||||||||||||
| 74 | 16.27 | Benzophenone-3 C | C14 H12O3 | 229.0856, 229.0859 | 151.0391, 105.0340, 54.08002 | −0.7861 | * | 2 | [68] | |||||||||||||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mohammed, M.A.; Elzefzafy, N.; El-Khadragy, M.F.; Alzahrani, A.; Yehia, H.M.; Kachlicki, P. Comprehensive Tools of Alkaloid/Volatile Compounds–Metabolomics and DNA Profiles: Bioassay-Role-Guided Differentiation Process of Six Annona sp. Grown in Egypt as Anticancer Therapy. Pharmaceuticals 2024, 17, 103. https://doi.org/10.3390/ph17010103
Mohammed MA, Elzefzafy N, El-Khadragy MF, Alzahrani A, Yehia HM, Kachlicki P. Comprehensive Tools of Alkaloid/Volatile Compounds–Metabolomics and DNA Profiles: Bioassay-Role-Guided Differentiation Process of Six Annona sp. Grown in Egypt as Anticancer Therapy. Pharmaceuticals. 2024; 17(1):103. https://doi.org/10.3390/ph17010103
Chicago/Turabian StyleMohammed, Mona A., Nahla Elzefzafy, Manal F. El-Khadragy, Abdulhakeem Alzahrani, Hany Mohamed Yehia, and Piotr Kachlicki. 2024. "Comprehensive Tools of Alkaloid/Volatile Compounds–Metabolomics and DNA Profiles: Bioassay-Role-Guided Differentiation Process of Six Annona sp. Grown in Egypt as Anticancer Therapy" Pharmaceuticals 17, no. 1: 103. https://doi.org/10.3390/ph17010103
APA StyleMohammed, M. A., Elzefzafy, N., El-Khadragy, M. F., Alzahrani, A., Yehia, H. M., & Kachlicki, P. (2024). Comprehensive Tools of Alkaloid/Volatile Compounds–Metabolomics and DNA Profiles: Bioassay-Role-Guided Differentiation Process of Six Annona sp. Grown in Egypt as Anticancer Therapy. Pharmaceuticals, 17(1), 103. https://doi.org/10.3390/ph17010103