Selective Delivery to Cardiac Muscle Cells Using Cell-Specific Aptamers
Abstract
:1. Introduction
2. Results
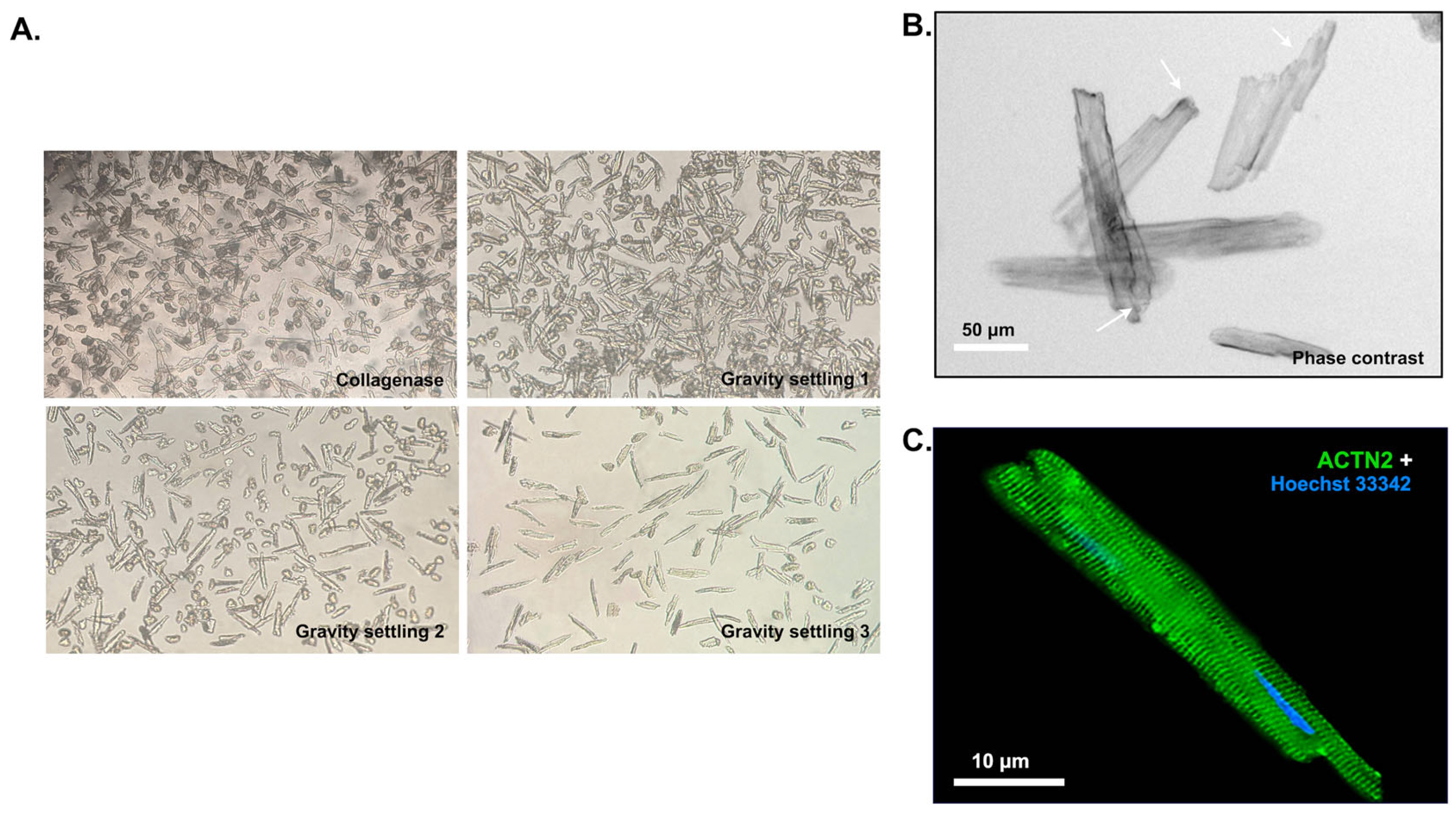
2.1. In Vivo SELEX for Cardiomyocyte Selection
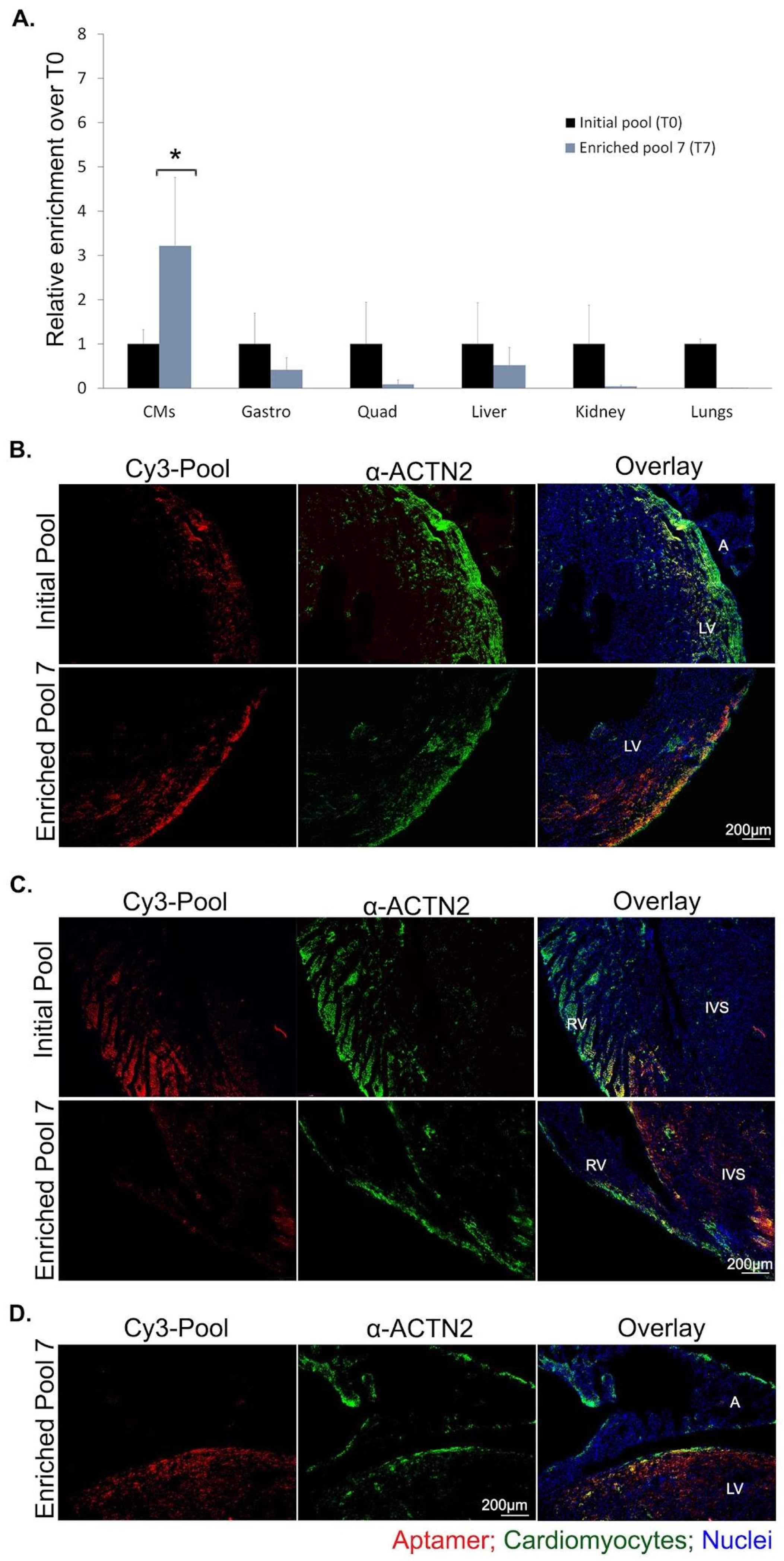
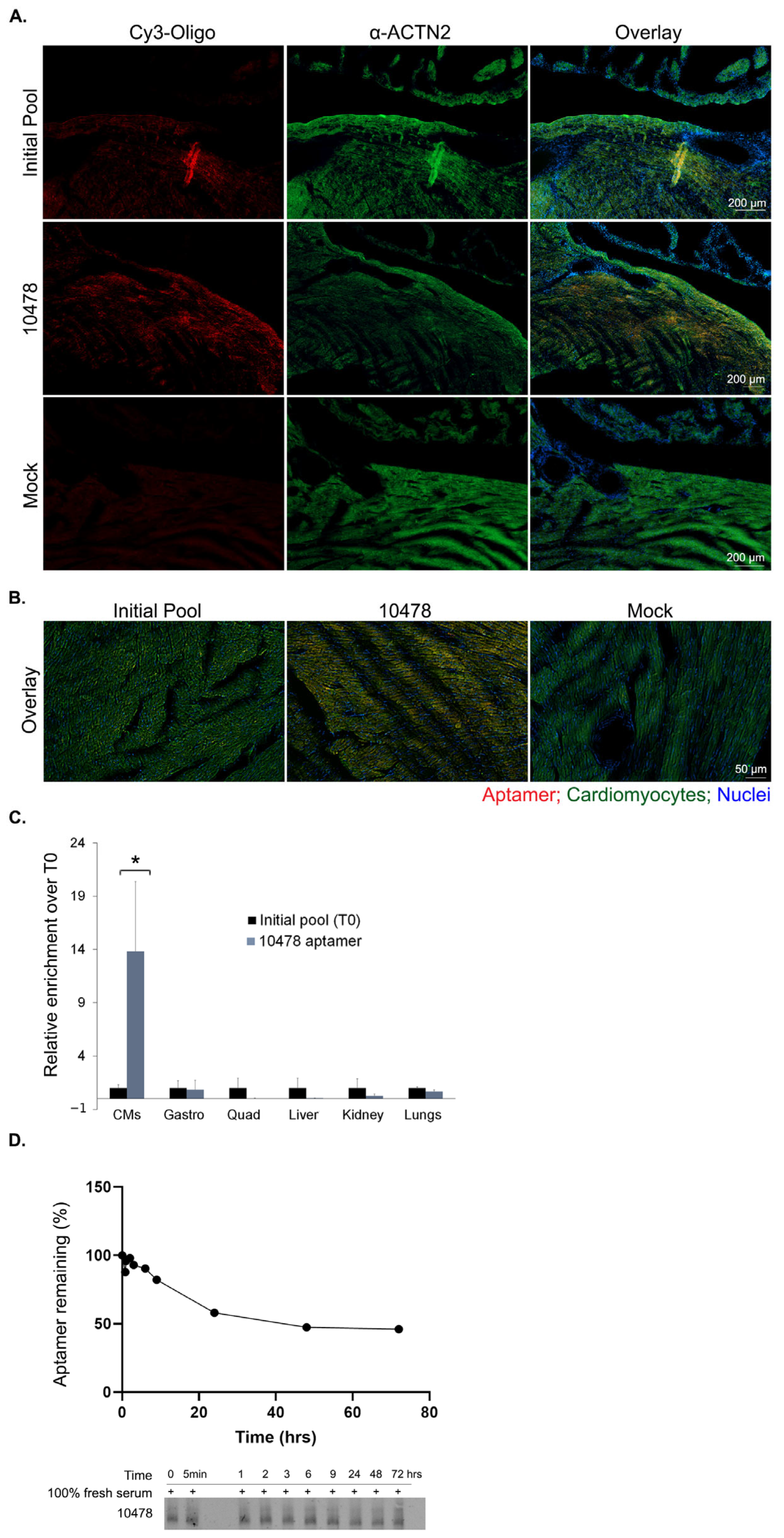
2.2. Enriched Aptamer Pool Selectivity for Cardiomyocytes
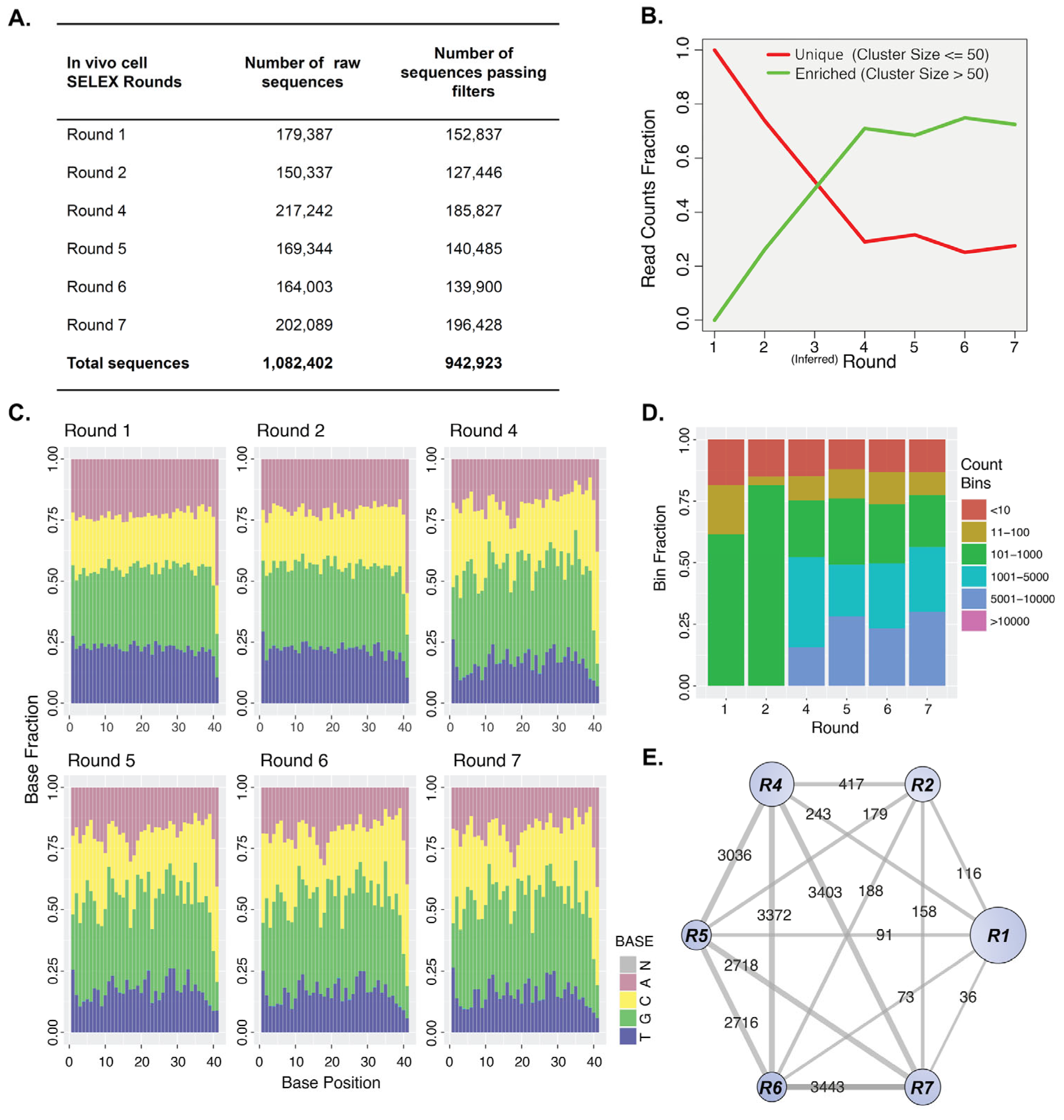
2.3. Sequencing Analysis of Aptamer Enrichment
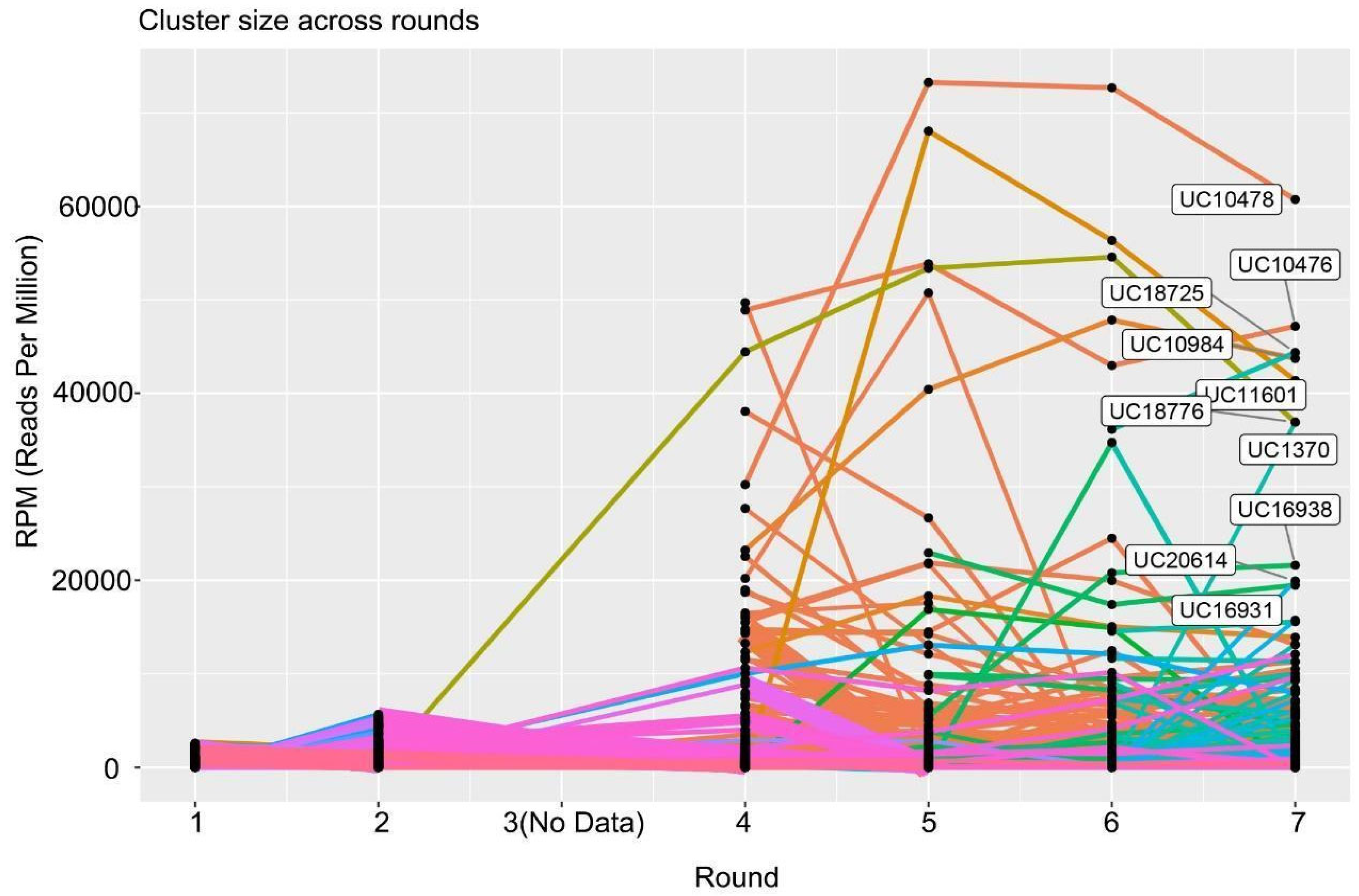
2.4. Identification of Candidate Aptamer Clusters
2.5. Secondary NGS Analysis for Diversity Confirmation
2.6. 2′F-Py RNA Aptamer 10478 Shows Selective Localization in CMs
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Oligonucleotides
4.3. Aptamer Library Preparation
4.4. In Vivo SELEX
4.5. PCR Cycle Optimization
4.6. Next Generation Sequencing for Aptamer Enrichment
4.7. Bioinformatics Analysis of NGS Data
4.8. RT-qPCR Quantification
4.9. Immunocytochemistry
4.10. Tissue Harvesting and Preparation
4.11. Tissue Processing and (Immuno)Staining
4.12. Fluorescence Microscopy
4.13. Serum Stability Assay
4.14. Secondary Structure Prediction
4.15. Statistical Analysis
4.16. Terms and Definitions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| UCID | Unique cluster identification number assigned to the NGS data, abbreviated UC# that allows the identification of the different clusters/families of sequences in the NGS data following SELEX. |
| Clusters | Groups or families of biological sequences that share similarities in their primary sequence. Sequence-clustering algorithms attempt to group the sequences that are somewhat related and the most represented sequence serves as the seed sequence of the cluster. |
| Unique sequences | Any sequence in the NGS data that presents with ≤50 read counts. |
| Enriched sequences | Any sequence in the NGS data that presents with >50 read counts. |
| Number of raw sequences | The total number of reads going off the sequencer and into data analysis. |
| Read count | The number of reads/sequences going off the sequencer or that align to a reference sequence (e.g., to the seed sequence of a cluster). |
| Read count fraction | This is the proportion of the sum of the read counts (sequences) in each class (enriched or unique) over the total number of reads in the dataset, per round. Total read in the dataset = 1. |
| Bin fraction | Bin fraction denotes the proportion of each bin over the total population of reads on the dataset. Total reads = 1. The fraction of read counts was derived by binning the sequences with respect to the read counts of each across 6 bands (≤10, 11–100, 101–1000, 1001–5000, 5001–10,000, >10,000). |
| Base fraction | Nucleotide (base) fractions per position, in a given round, were obtained by calculating the frequency of each base per position over the total number of reads. Total number of reads = 1. |
| Reads per million (RPM) | The read counts per cluster were divided by the “per million” scaling factor. This normalizes for sequencing depth, giving the reads per million. The “per million” scaling factor is derived by counting the total reads in a sample (i.e., SELEX round) and then dividing that number by 1,000,000. |
| Technical duplicate | Technical replicates are repeated measurements of the same sample that demonstrate the variability of the protocol. Technical replicates are important because they address the reproducibility of the assay or technique. In this study, a technical duplicate is used in RT-qPCR assay to ensure the validity of the method (i.e., the pipetting technique). It is the same cDNA pipetted into multiple wells, thus Ct values with little variability should be obtained. The mean value from these replicates is then used as a representative value for each biological sample in subsequent data analyses. |
| Biological replicate | Biological replicates are parallel measurements of biologically distinct samples that capture random biological variation, which can be a subject of study or a source of noise itself. Biological replicates address how widely your experimental results can be generalized. Unless otherwise stated, three biologically distinct samples (n = 3) were used in each experiment (biological triplicate). |
Appendix B
| SELEX Round | Ranking | UCID | Cluster ID | Cluster Counts | Top Seq Counts | SequenceID | Random Region Sequence |
|---|---|---|---|---|---|---|---|
| 1 | 1 | UC145 | Cluster 144 | 396 | 345 | >2-345-2257.31_2 | AGGGTAAGCCTTTCCATCGGGTCGACTTCGGATTGCATCG |
| 2 | UC156 | Cluster 155 | 394 | 346 | >1-346-2263.85_1 | TGTGAGTGATTACGCTCTGTGCGTATGGGGACAGTTCCGC | |
| 3 | UC538 | Cluster 537 | 386 | 333 | >3-333-2178.79_3 | AAAGTCTACAGGTGAAAGGCGTCACCGCGAGGCGAGCGTT | |
| 4 | UC124 | Cluster 123 | 384 | 326 | >5-326-2132.99_5 | CGGTGCACTGGCATGCTGGACCGGAGGTCAGGACGGTCGG | |
| 5 | UC17 | Cluster 16 | 381 | 323 | >6-323-2113.36_6 | TGGCCCGCTACTCCGCGGTCTATACTAGTATTCCGTAACA | |
| 6 | UC45 | Cluster 44 | 378 | 323 | >6-323-2113.36_7 | ACTGTGTCGATCAGGTAAACGACACTTGCGGCCTGCTATA | |
| 7 | UC88 | Cluster 87 | 374 | 321 | >8-321-2100.28_8 | TACCCCATAATAGGCCTTGTAGGATCGTAGACGTTACGTC | |
| 8 | UC504 | Cluster 503 | 374 | 314 | >13-314-2054.48_13 | TACTTGACAACACTAGTGATAGCAGAATCGCGAGACCGCA | |
| 9 | UC400 | Cluster 399 | 371 | 327 | >4-327-2139.53_4 | GTGGACGAGCCGGGCATGGTCGAGTGTGAAGGGAGCCGCG | |
| 10 | UC605 | Cluster 604 | 370 | 321 | >8-321-2100.28_9 | CCGGCGACTCTCGCGAACAGCTTCCCATCCGCATTTGTGG | |
| 2 | 1 | UC3215 | Cluster 141 | 723 | 635 | >1-635-4884.31_1 | TGCCGCAGGGTGTGGATTGAATTGACGGTGAGACGCGCAC |
| 2 | UC7082 | Cluster 35 | 718 | 631 | >2-631-4853.55_2 | CGGTGGACGTGTAGCGGGAATCCGCGGCAAACACAGAGCT | |
| 3 | UC7117 | Cluster 75 | 717 | 615 | >5-615-4730.48_5 | GGCCCAACGTGGTTGGGGTCAACACGCGGGATTCGGGGTT | |
| 4 | UC7087 | Cluster 40 | 705 | 609 | >7-609-4684.33_7 | ATAGCGTCCGGCTAGGCTTTCTCGGTGCGCAGCGGAGACA | |
| 5 | UC7091 | Cluster 45 | 701 | 617 | >4-617-4745.86_4 | GCACTTGCAGCGCGGTGTACGCTAACGCCTGGGCCGGTGA | |
| 6 | UC7171 | Cluster 135 | 690 | 622 | >3-622-4784.32_3 | TCTGTGCACGGCATCCGCTTAGAGTGTCCGGTCGGACATC | |
| 7 | UC3458 | Cluster 269 | 689 | 610 | >6-610-4692.02_6 | AGGGCGGGTCGCGGGCCTGGTGATTGGACGGAGGCTGGCC | |
| 8 | UC7116 | Cluster 74 | 683 | 606 | >8-606-4661.25_8 | CGATGCCCTGTGGTCGGTCGCCCGGCAGGGCTGTGCAGTT | |
| 9 | UC7053 | Cluster 2 | 677 | 578 | >15-578-4445.88_15 | CGGTTCCGAGCGTTGGTGGAGGACGCGGGTAGGCGGACGT | |
| 10 | UC7189 | Cluster 155 | 677 | 585 | >11-585-4499.72_11 | TGAGCCTGCGCGCGGGGGGAGGCGGCGGAGGACCAGTAGT | |
| 3 | N/A—No back up sample for NGS analysis | ||||||
| 4 | 1 | UC10474 | Cluster 2 | 9235 | 8057 | >1-8057-43057.24_1 | CGACGGACAAGGCTCACCGTGGCCATGTGAGCTCGGGCGC |
| 2 | UC10476 | Cluster 5 | 9086 | 8034 | >2-8034-42934.33_2 | TGCGACGTGGGCGCGTCATGCTGCGCGGTGCTGTGCACGC | |
| 3 | UC1370 | Cluster 3 | 8256 | 7138 | >3-7138-38146.03_3 | ACGGGCGCCCGTGCATAAGGTGCGGCGGGCTGACGTGTCG | |
| 4 | UC10479 | Cluster 8 | 7072 | 5973 | >4-5973-31920.18_4 | CACGCGGCGGCCGTGAATGGTCACGGAGGCGAGCTGTGCC | |
| 5 | UC10478 | Cluster 7 | 5617 | 4911 | >5-4911-26244.77_5 | TGCAGGTGCATGTGGGATCACGCGCGGTTAGGTCGCCGCG | |
| 6 | UC10483 | Cluster 12 | 5143 | 4463 | >6-4463-23850.62_6 | TCACGGGCGTGGCGGGCGACGAGCCACGGAGCGGGGTTGC | |
| 7 | UC10984 | Cluster 547 | 4316 | 3932 | >7-3932-21012.92_7 | ATGTCACGAACGAGGGCGTGCTCGCGTGGTGCGGAGGCA | |
| 8 | UC10488 | Cluster 17 | 4190 | 3664 | >8-3664-19580.70_8 | GTGCGCCACAGGTGTTACGGTGGTGCATCCGTGGGCTGCG | |
| 9 | UC10481 | Cluster 10 | 3752 | 3309 | >9-3309-17683.56_9 | CGGAGCCACCGGCGCGTGGGTGCGGGTGCGGCCACCAGCA | |
| 10 | UC10487 | Cluster 16 | 3528 | 3109 | >10-3109-16614.74_10 | TGACGGCCCTGCAAGGAGGGCTAGGATGTCGCTGTTGCGC | |
| 5 | 1 | UC10478 | Cluster 4 | 10293 | 8302 | >1-8302-56908.23_1 | TGCAGGTGCATGTGGGATCACGCGCGGTTAGGTCGCCGCG |
| 2 | UC11601 | Cluster 5 | 9563 | 8251 | >2-8251-56558.64_2 | CGACGGACAAGGCTCACCGTGGCCATGTGAGCTCGGGCGC | |
| 3 | UC10476 | Cluster 9 | 7566 | 6655 | >3-6655-45618.44_3 | TGCGACGTGGGCGCGTCATGCTGCGCGGTGCTGTGCACGC | |
| 4 | UC1370 | Cluster 7 | 7499 | 6515 | >4-6515-44658.77_4 | ACGGGCGCCCGTGCATAAGGTGCGGCGGGCTGACGTGTCG | |
| 5 | UC10481 | Cluster 8 | 7128 | 6262 | >5-6262-42924.52_5 | CGGAGCCACCGGCGCGTGGGTGCGGGTGCGGCCACCAGCA | |
| 6 | UC10984 | Cluster 428 | 5679 | 5206 | >6-5206-35685.89_6 | ATGTCACGAACGAGGGCGTGCTCGCGTGGTGCGGAGGCA | |
| 7 | UC10479 | Cluster 10 | 3747 | 3314 | >7-3314-22716.68_7 | CACGCGGCGGCCGTGAATGGTCACGGAGGCGAGCTGTGCC | |
| 8 | UC16931 | Cluster 12 | 3223 | 2828 | >8-2828-19385.27_8 | TGGGGGCTCAGTGACGGCGCGTCGTCGTTGAGCAGCGGCA | |
| 9 | UC10484 | Cluster 11 | 3070 | 2545 | >9-2545-17445.37_9 | CGCGGCCCCGGTAGTGTGGCTGGAGGGGTTGTTGTCGACA | |
| 10 | UC10494 | Cluster 2 | 3057 | 2508 | >10-2508-17191.74_10 | CGTGGGACGGCCGGCGTAGGGTCGGCAGCGAGTGGCGCGC | |
| 6 | 1 | UC10478 | Cluster 3 | 10172 | 8763 | >1-8763-61923.64_1 | TGCAGGTGCATGTGGGATCACGCGCGGTTAGGTCGCCGCG |
| 2 | UC11601 | Cluster 4 | 7883 | 6702 | >2-6702-47359.61_2 | CGACGGACAAGGCTCACCGTGGCCATGTGAGCTCGGGCGC | |
| 3 | UC1370 | Cluster 1 | 7636 | 6472 | >3-6472-45734.31_3 | ACGGGCGCCCGTGCATAAGGTGCGGCGGGCTGACGTGTCG | |
| 4 | UC10984 | Cluster 311 | 6694 | 6016 | >4-6016-42512.00_4 | ATGTCACGAACGAGGGCGTGCTCGCGTGGTGCGGAGGCA | |
| 5 | UC10476 | Cluster 7 | 6010 | 5149 | >5-5149-36385.35_5 | TGCGACGTGGGCGCGTCATGCTGCGCGGTGCTGTGCACGC | |
| 6 | UC18725 | Cluster 10 | 5056 | 4306 | >6-4306-30428.30_6 | CGGAGCCACCGGCGCGTGGGTGCGGGTGCGGCCACCAGCA | |
| 7 | UC17023 | Cluster 8 | 4858 | 4173 | >8-4173-29488.46_8 | CACGCGGCGGCCGTGAATGGTCACGGAGGCGAGCTGTGCC | |
| 8 | UC18722 | Cluster 5 | 4856 | 4182 | >7-4182-29552.06_7 | TCACGGTGGGATGACTGAAGGTCTGGTGCGACCGGGGCGC | |
| 9 | UC10489 | Cluster 17 | 3427 | 2949 | >9-2949-20839.07_9 | GGCGCGCCAGTCGCTCCGAGGGAGGGTGCGACGGTGCGTC | |
| 10 | UC16938 | Cluster 15 | 2911 | 1953 | >12-1953-13800.85_12 | CACGGCAACTGTGAGGCAAAAACGCCTTTGGCCCGGCGCT | |
| 7 | 1 | UC10478 | Cluster 10 | 10716 | 9067 | >1-9067-52008.17_1 | TGCAGGTGCATGTGGGATCACGCGCGGTTAGGTCGCCGCG |
| 2 | UC10476 | Cluster 3 | 8321 | 6970 | >2-6970-39979.81_2 | TGCGACGTGGGCGCGTCATGCTGCGCGGTGCTGTGCACGC | |
| 3 | UC18725 | Cluster 8 | 7826 | 6729 | >4-6729-38597.44_4 | CGGAGCCACCGGCGCGTGGGTGCGGGTGCGGCCACCAGCA | |
| 4 | UC10984 | Cluster 263 | 7715 | 6915 | >3-6915-39664.33_3 | ATGTCACGAACGAGGGCGTGCTCGCGTGGTGCGGAGGCA | |
| 5 | UC11601 | Cluster 2 | 7300 | 6184 | >5-6184-35471.33_5 | CGACGGACAAGGCTCACCGTGGCCATGTGAGCTCGGGCGC | |
| 6 | UC1370 | Cluster 11 | 6511 | 5386 | >7-5386-30894.01_7 | ACGGGCGCCCGTGCATAAGGTGCGGCGGGCTGACGTGTCG | |
| 7 | UC18776 | Cluster 11 | 6511 | 5386 | >7-5386-30894.01_7 | ACGGGCGCCCGTGCATAAGGTGCGGCGGGCTGACGTGTCG | |
| 8 | UC16938 | Cluster 35 | 3812 | 2578 | >12-2578-14787.37_12 | CACGGCAACTGTGAGGCAAAAACGCCTTTGGCCCGGCGCT | |
| 9 | UC20614 | Cluster 5 | 3516 | 3017 | >10-3017-17305.46_10 | CTGCCGGCGGTTGGGCCCTGGGCGGGCCAGCGGATGTCGC | |
| 10 | UC16931 | Cluster 14 | 3437 | 2860 | >11-2860-16404.91_11 | TGGGGGCTCAGTGACGGCGCGTCGTCGTTGAGCAGCGGCA | |
References
- Zhang, Y.; Lai, B.S.; Juhas, M. Recent Advances in Aptamer Discovery and Applications. Molecules 2019, 24, 941. [Google Scholar] [CrossRef]
- Byun, J. Recent Progress and Opportunities for Nucleic Acid Aptamers. Life 2021, 11, 193. [Google Scholar] [CrossRef]
- Cheng, C.; Chen, Y.H.; Lennox, K.A.; Behlke, M.A.; Davidson, B.L. In vivo SELEX for Identification of Brain-penetrating Aptamers. Mol. Ther. Nucleic Acids 2013, 2, e67. [Google Scholar] [CrossRef]
- Vorobyeva, M.A.; Dymova, M.A.; Novopashina, D.S.; Kuligina, E.V.; Timoshenko, V.V.; Kolesnikov, I.A.; Taskaev, S.Y.; Richter, V.A.; Venyaminova, A.G. Tumor Cell-Specific 2′-Fluoro RNA Aptamer Conjugated with Closo-Dodecaborate as A Potential Agent for Boron Neutron Capture Therapy. Int. J. Mol. Sci. 2021, 22, 7326. [Google Scholar] [CrossRef]
- Maio, G.; Enweronye, O.; Zumrut, H.E.; Batool, S.; Van, N.; Mallikaratchy, P. Systematic optimization and modification of a DNA aptamer with 2’-O-methyl RNA analogues. ChemistrySelect 2017, 2, 2335–2340. [Google Scholar] [CrossRef]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef]
- Philippou, S.; Mastroyiannopoulos, N.P.; Makrides, N.; Lederer, C.W.; Kleanthous, M.; Phylactou, L.A. Selection and Identification of Skeletal-Muscle-Targeted RNA Aptamers. Mol. Ther. Nucleic Acids 2018, 10, 199–214. [Google Scholar] [CrossRef]
- Thiel, W.H.; Bair, T.; Peek, A.S.; Liu, X.; Dassie, J.; Stockdale, K.R.; Behlke, M.A.; Miller, F.J.; Giangrande, P.H. Rapid Identification of Cell-Specific, Internalizing RNA Aptamers with Bioinformatics Analyses of a Cell-Based Aptamer Selection. PLoS ONE 2012, 7, e43836. [Google Scholar] [CrossRef]
- Thiel, K.W.; Hernandez, L.I.; Dassie, J.P.; Thiel, W.H.; Liu, X.; Stockdale, K.R.; Rothman, A.M.; Hernandez, F.J.; McNamara, J.O.; Giangrande, P.H. Delivery of chemo-sensitizing siRNAs to HER2+-breast cancer cells using RNA aptamers. Nucleic Acids Res. 2012, 40, 6319–6337. [Google Scholar] [CrossRef]
- Daei, P.; Ramezanpour, M.; Khanaki, K.; Tabarzad, M.; Nikokar, I.; Ch, M.H.; Elmi, A. Aptamer-based Targeted Delivery of miRNA let-7d to Gastric Cancer Cells as a Novel Anti-Tumor Therapeutic Agent. Iran J. Pharm. Res. 2018, 17, 1537–1549. [Google Scholar]
- Porciani, D.; Cardwell, L.N.; Tawiah, K.D.; Alam, K.K.; Lange, M.J.; Daniels, M.A.; Burke, D.H. Modular cell-internalizing aptamer nanostructure enables targeted delivery of large functional RNAs in cancer cell lines. Nat. Commun. 2018, 9, 2283. [Google Scholar] [CrossRef]
- Di Leandro, L.; Giansanti, F.; Mei, S.; Ponziani, S.; Colasante, M.; Ardini, M.; Angelucci, F.; Pitari, G.; D’angelo, M.; Cimini, A.; et al. Aptamer-Driven Toxin Gene Delivery in U87 Model Glioblastoma Cells. Front. Pharmacol. 2021, 12, 588306. [Google Scholar] [CrossRef]
- Fu, Z.; Xiang, J. Aptamer-Functionalized Nanoparticles in Targeted Delivery and Cancer Therapy. Int. J. Mol. Sci. 2020, 21, 9123. [Google Scholar] [CrossRef]
- Cadinoiu, A.N.; Rata, D.M.; Atanase, L.I.; Daraba, O.M.; Gherghel, D.; Vochita, G.; Popa, M. Aptamer-Functionalized Liposomes as a Potential Treatment for Basal Cell Carcinoma. Polymers 2019, 11, 1515. [Google Scholar] [CrossRef]
- Liang, X.; Wang, Y.; Shi, H.; Dong, M.; Han, H.; Li, Q. Nucleolin-Targeting AS1411 Aptamer-Modified Micelle for the Co-Delivery of Doxorubicin and miR-519c to Improve the Therapeutic Efficacy in Hepatocellular Carcinoma Treatment. Int. J. Nanomed. 2021, 16, 2569–2584. [Google Scholar] [CrossRef]
- Jafari, R.; Zolbanin, N.M.; Majidi, J.; Atyabi, F.; Yousefi, M.; Jadidi-Niaragh, F.; Aghebati-Maleki, L.; Shanehbandi, D.; Zangbar, M.-S.S.; Rafatpanah, H.; et al. Anti-Mucin1 Aptamer-Conjugated Chitosan Nanoparticles for Targeted Co-Delivery of Docetaxel and IGF-1R siRNA to SKBR3 Metastatic Breast Cancer Cells. Iran Biomed. J. 2019, 23, 21–33. [Google Scholar] [CrossRef]
- Mi, J.; Liu, Y.; Rabbani, Z.N.; Yang, Z.; Urban, J.H.; Sullenger, B.A.; Clary, B.M. In vivo selection of tumor-targeting RNA motifs. Nat. Chem. Biol. 2010, 6, 22–24. [Google Scholar] [CrossRef]
- Mi, J.; Ray, P.; Liu, J.; Kuan, C.-T.; Xu, J.; Hsu, D.; Sullenger, B.A.; White, R.R.; Clary, B.M. In Vivo Selection Against Human Colorectal Cancer Xenografts Identifies an Aptamer That Targets RNA Helicase Protein DHX9. Mol. Ther. Nucleic Acids 2016, 5, e315. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, Y.; Yang, H.; Qin, M.; Ding, X.; Liu, R.; Jiang, Y. In Vivo SELEX of an Inhibitory NSCLC-Specific RNA Aptamer from PEGylated RNA Library. Mol. Ther. Nucleic Acids 2018, 10, 187–198. [Google Scholar] [CrossRef]
- Chen, L.; He, W.; Jiang, H.; Wu, L.; Xiong, W.; Li, B.; Zhou, Z.; Qian, Y. In vivo SELEX of bone targeting aptamer in prostate cancer bone metastasis model. Int. J. Nanomed. 2019, 14, 149–159. [Google Scholar] [CrossRef]
- Zhou, J.; Rossi, J. Aptamers as targeted therapeutics: Current potential and challenges. Nat. Rev. Drug Discov. 2017, 16, 181–202. [Google Scholar] [CrossRef]
- Ackers-Johnson, M.; Li, P.Y.; Holmes, A.P.; O’Brien, S.-M.; Pavlovic, D.; Foo, R.S. A Simplified, Langendorff-Free Method for Concomitant Isolation of Viable Cardiac Myocytes and Nonmyocytes From the Adult Mouse Heart. Circ. Res. 2016, 119, 909–920. [Google Scholar] [CrossRef]
- Goyenvalle, A.; Griffith, G.; Babbs, A.; El Andaloussi, S.; Ezzat, K.; Avril, A.; Dugovic, B.; Chaussenot, R.; Ferry, A.; Voit, T.; et al. Functional correction in mouse models of muscular dystrophy using exon-skipping tricyclo-DNA oligomers. Nat. Med. 2015, 21, 270–275. [Google Scholar] [CrossRef]
- Chernikov, I.V.; Vlassov, V.V.; Chernolovskaya, E.L. Current Development of siRNA Bioconjugates: From Research to the Clinic. Front. Pharmacol. 2019, 10, 444. [Google Scholar] [CrossRef]
- Michel, N.A.; Ljubojevic-Holzer, S.; Bugger, H.; Zirlik, A. Cellular Heterogeneity of the Heart. Front. Cardiovasc. Med. 2022, 9, 868466. [Google Scholar] [CrossRef]
- Desjardins, C.A.; Yao, M.; Hall, J.; O’donnell, E.; Venkatesan, R.; Spring, S.; Wen, A.; Hsia, N.; Shen, P.; Russo, R.; et al. Enhanced exon skipping and prolonged dystrophin restoration achieved by TfR1-targeted delivery of antisense oligonucleotide using FORCE conjugation in mdx mice. Nucleic Acids Res. 2022, 50, 11401–11414. [Google Scholar] [CrossRef]
- Aupy, P.; Zarrouki, F.; Sandro, Q.; Gastaldi, C.; Buclez, P.-O.; Mamchaoui, K.; Garcia, L.; Vaillend, C.; Goyenvalle, A. Long-Term Efficacy of AAV9-U7snRNA-Mediated Exon 51 Skipping in mdx52 Mice. Mol. Ther. Methods Clin. Dev. 2020, 17, 1037–1047. [Google Scholar] [CrossRef]
- Negishi, Y.; Ishii, Y.; Nirasawa, K.; Sasaki, E.; Endo-Takahashi, Y.; Suzuki, R.; Maruyama, K. PMO Delivery System Using Bubble Liposomes and Ultrasound Exposure for Duchenne Muscular Dystrophy Treatment. Methods Mol. Biol. 2018, 1687, 185–192. [Google Scholar] [CrossRef]
- Wang, M.; Wu, B.; Tucker, J.D.; Shah, S.N.; Lu, P.; Lu, Q. Triazine-cored polymeric vectors for antisense oligonucleotide delivery in vitro and in vivo. J. Nanobiotechnol. 2020, 18, 34. [Google Scholar] [CrossRef]
- Moretti, A.; Fonteyne, L.; Giesert, F.; Hoppmann, P.; Meier, A.B.; Bozoglu, T.; Baehr, A.; Schneider, C.M.; Sinnecker, D.; Klett, K.; et al. Somatic gene editing ameliorates skeletal and cardiac muscle failure in pig and human models of Duchenne muscular dystrophy. Nat. Med. 2020, 26, 207–214. [Google Scholar] [CrossRef]
- Tsoumpra, M.K.; Fukumoto, S.; Matsumoto, T.; Takeda, S.; Wood, M.J.A.; Aoki, Y. Peptide-conjugate antisense based splice-correction for Duchenne muscular dystrophy and other neuromuscular diseases. EBioMedicine 2019, 45, 630–645. [Google Scholar] [CrossRef]
- Gan, L.; Wu, L.C.; Wood, J.A.; Yao, M.; Treleaven, C.M.; Estrella, N.L.; Wentworth, B.M.; Hanson, G.J.; Passini, M.A. A cell-penetrating peptide enhances delivery and efficacy of phosphorodiamidate morpholino oligomers in mdx mice. Mol. Ther. Nucleic Acids 2022, 30, 17–27. [Google Scholar] [CrossRef]
- Frazier, K.S. Antisense Oligonucleotide Therapies: The Promise and the Challenges from a Toxicologic Pathologist’s Perspective. Toxicol. Pathol. 2015, 43, 78–89. [Google Scholar] [CrossRef]
- Sutermaster, B.A.; Darling, E.M. Considerations for high-yield, high-throughput cell enrichment: Fluorescence versus magnetic sorting. Sci. Rep. 2019, 9, 227. [Google Scholar] [CrossRef]
- Gartz, M.; Beatka, M.; Prom, M.J.; Strande, J.L.; Lawlor, M.W. Cardiomyocyte-produced miR-339-5p mediates pathology in Duchenne muscular dystrophy cardiomyopathy. Hum. Mol. Genet. 2021, 30, 2347–2361. [Google Scholar] [CrossRef]
- Civit, L.; Theodorou, I.; Frey, F.; Weber, H.; Lingnau, A.; Gröber, C.; Blank, M.; Dambrune, C.; Stunden, J.; Beyer, M.; et al. Targeting hormone refractory prostate cancer by in vivo selected DNA libraries in an orthotopic xenograft mouse model. Sci. Rep. 2019, 9, 4976. [Google Scholar] [CrossRef]
- Jauset-Rubio, M.; Botero, M.L.; Skouridou, V.; Aktas, G.B.; Svobodova, M.; Bashammakh, A.S.; El-Shahawi, M.S.; Alyoubi, A.O.; O’sullivan, C.K. One-Pot SELEX: Identification of Specific Aptamers against Diverse Steroid Targets in One Selection. ACS Omega 2019, 4, 20188–20196. [Google Scholar] [CrossRef]
- Kolm, C.; Cervenka, I.; Aschl, U.J.; Baumann, N.; Jakwerth, S.; Krska, R.; Mach, R.L.; Sommer, R.; DeRosa, M.C.; Kirschner, A.K.T.; et al. DNA aptamers against bacterial cells can be efficiently selected by a SELEX process using state-of-the art qPCR and ultra-deep sequencing. Sci. Rep. 2020, 10, 20917. [Google Scholar] [CrossRef]
- Wu, M.; Wang, Y.; Wang, Y.; Zhang, M.; Luo, Y.; Tang, J.; Wang, Z.; Wang, D.; Hao, L.; Wang, Z.; et al. Paclitaxel-loaded and A10-3.2 aptamer-targeted poly(lactide-co-glycolic acid) nanobubbles for ultrasound imaging and therapy of prostate cancer. Int. J. Nanomed. 2017, 12, 5313–5330. [Google Scholar] [CrossRef]
- Biscans, A.; Coles, A.; Haraszti, R.; Echeverria, D.; Hassler, M.; Osborn, M.; Khvorova, A. Diverse lipid conjugates for functional extra-hepatic siRNA delivery in vivo. Nucleic Acids Res. 2019, 47, 1082–1096. [Google Scholar] [CrossRef]
- Chen, Z.; Liu, H.; Jain, A.; Zhang, L.; Liu, C.; Cheng, K. Discovery of Aptamer Ligands for Hepatic Stellate Cells Using SELEX. Theranostics 2017, 7, 2982–2995. [Google Scholar] [CrossRef]
- Wang, T.; Gantier, M.P.; Xiang, D.; Bean, A.G.; Bruce, M.; Zhou, S.-F.; Khasraw, M.; Ward, A.; Wang, L.; Wei, M.Q.; et al. EpCAM Aptamer-mediated Survivin Silencing Sensitized Cancer Stem Cells to Doxorubicin in a Breast Cancer Model. Theranostics 2015, 5, 1456–1472. [Google Scholar] [CrossRef]
- Leng, L.; Dong, X.; Gao, X.; Ran, N.; Geng, M.; Zuo, B.; Wu, Y.; Li, W.; Yan, H.; Han, G.; et al. Exosome-mediated improvement in membrane integrity and muscle function in dystrophic mice. Mol. Therapy 2021, 29, 1459–1470. [Google Scholar] [CrossRef]
- Da Pieve, C.; Blackshaw, E.; Missailidis, S.; Perkins, A.C. PEGylation and Biodistribution of an anti-MUC1 Aptamer in MCF-7 Tumor-Bearing Mice. Bioconjug. Chem. 2012, 23, 1377–1381. [Google Scholar] [CrossRef]
- Biscans, A.; Caiazzi, J.; McHugh, N.; Hariharan, V.; Muhuri, M.; Khvorova, A. Docosanoic acid conjugation to siRNA enables functional and safe delivery to skeletal and cardiac muscles. Mol. Ther. 2021, 29, 1382–1394. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef]
- Li, W.; Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef]
- Morgan, M.; Anders, S.; Lawrence, M.; Aboyoun, P.; Pagès, H.; Gentleman, R. ShortRead: A bioconductor package for input, quality assessment and exploration of high-throughput sequence data. Bioinformatics 2009, 25, 2607–2608. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Kratschmer, C.; Levy, M. Effect of Chemical Modifications on Aptamer Stability in Serum. Nucleic Acid Ther. 2017, 27, 335–344. [Google Scholar] [CrossRef]
- Friedman, N.; Cai, L.; Xie, X.S. Linking stochastic dynamics to population distribution: An analytical framework of gene expression. Phys. Rev. Lett. 2006, 97, 168302. [Google Scholar] [CrossRef]

| Gender | Male |
| Age | 8 weeks old |
| Average yield | 5.53 × 105 ± 3.79 × 104 |
| Average viability | 81.7 ± 7.64 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Philippou, S.; Mastroyiannopoulos, N.P.; Tomazou, M.; Oulas, A.; Ackers-Johnson, M.; Foo, R.S.; Spyrou, G.M.; Phylactou, L.A. Selective Delivery to Cardiac Muscle Cells Using Cell-Specific Aptamers. Pharmaceuticals 2023, 16, 1264. https://doi.org/10.3390/ph16091264
Philippou S, Mastroyiannopoulos NP, Tomazou M, Oulas A, Ackers-Johnson M, Foo RS, Spyrou GM, Phylactou LA. Selective Delivery to Cardiac Muscle Cells Using Cell-Specific Aptamers. Pharmaceuticals. 2023; 16(9):1264. https://doi.org/10.3390/ph16091264
Chicago/Turabian StylePhilippou, Styliana, Nikolaos P. Mastroyiannopoulos, Marios Tomazou, Anastasios Oulas, Matthew Ackers-Johnson, Roger S. Foo, George M. Spyrou, and Leonidas A. Phylactou. 2023. "Selective Delivery to Cardiac Muscle Cells Using Cell-Specific Aptamers" Pharmaceuticals 16, no. 9: 1264. https://doi.org/10.3390/ph16091264
APA StylePhilippou, S., Mastroyiannopoulos, N. P., Tomazou, M., Oulas, A., Ackers-Johnson, M., Foo, R. S., Spyrou, G. M., & Phylactou, L. A. (2023). Selective Delivery to Cardiac Muscle Cells Using Cell-Specific Aptamers. Pharmaceuticals, 16(9), 1264. https://doi.org/10.3390/ph16091264








