Abstract
Artificial intelligence (AI) has permeated various sectors, including the pharmaceutical industry and research, where it has been utilized to efficiently identify new chemical entities with desirable properties. The application of AI algorithms to drug discovery presents both remarkable opportunities and challenges. This review article focuses on the transformative role of AI in medicinal chemistry. We delve into the applications of machine learning and deep learning techniques in drug screening and design, discussing their potential to expedite the early drug discovery process. In particular, we provide a comprehensive overview of the use of AI algorithms in predicting protein structures, drug–target interactions, and molecular properties such as drug toxicity. While AI has accelerated the drug discovery process, data quality issues and technological constraints remain challenges. Nonetheless, new relationships and methods have been unveiled, demonstrating AI’s expanding potential in predicting and understanding drug interactions and properties. For its full potential to be realized, interdisciplinary collaboration is essential. This review underscores AI’s growing influence on the future trajectory of medicinal chemistry and stresses the importance of ongoing synergies between computational and domain experts.
1. Introduction
Artificial intelligence (AI), a field within computer science, focuses on developing methods that empower computers to perform tasks typically associated with human intelligence, such as thinking and learning. AI is having a revolutionary impact on various facets of our lives and spanning across numerous industry sectors, with the pharmaceutical sector experiencing no exception to this transformation [1]. Also, in the medical field, deep learning techniques classify lung cancer with high accuracy, and AI addresses challenges in processing continuous streams of big data from medical IoT devices [2,3]. The emergence of AI has ushered in a new era in drug discovery research, delivering a paradigm shift from traditional trial-and-error-based or hypothesis-driven methods toward more rational and data-driven approaches [1]. The value of AI is immense as it serves as a technology that can significantly reduce the extensive time and financial investments required for the discovery of a new drug.
When used properly, AI technologies can help analyze vast amounts of data, such as genomic, proteomic, and chemical information, to identify potential drug molecules and predict drug efficacy or toxicity [4]. By analyzing complex datasets and identifying hidden patterns, machine learning (ML) or deep learning (DL) algorithms can find novel targets associated with multi-omics data and help search for novel chemical entities with biological activities. They have not only expedited the identification of potential drug candidates but have also proven invaluable in the process of drug repurposing [5]. AI can predict potential new uses for existing drugs, a breakthrough that has the potential to accelerate the drug development process and reduce associated costs [5]. This capability is particularly significant in addressing urgent medical needs, as repurposing existing drugs can bypass lengthy and costly phases of preclinical testing and safety evaluation. Moreover, AI has emerged as a key tool for personalized medicine by aiding the development of drugs that are tailored to individual patients’ genetic profiles. In the future, the demand for AI in drug discovery is expected to grow as the technology becomes more advanced and many more data become available [1].
In the realm of medicinal chemistry, AI has shown promising results in the discovery of new chemical scaffolds with therapeutic potential. It has the capacity to scrutinize vast chemical spaces and extract meaningful patterns, thereby significantly reducing the time required for identifying potential drug candidates [4,6]. ML/DL algorithms can be trained to predict the biological activities, pharmacokinetic properties, and also toxicity profiles of molecules [7]. In addition, the current DL methods can generate novel molecular structures that match desired therapeutic profiles [8,9]. Building on the past decade’s remarkable advances, AI is now being harnessed to automate the process of drug design. Molecular docking, the method that predicts the interaction between a small molecule and a protein, has traditionally been a computationally intensive task. Today, AI is being utilized to predict the likelihood of molecular binding, its strength, and the most energetically favorable position, thereby automating this critical process. In addition, it can be utilized to optimize the chemical structures of drug candidates for enhanced efficacy and reduced toxicity.
While the promise of AI in medicinal chemistry is profound, the integration of AI into drug discovery pipelines presents ongoing challenges [4,10]. Issues related to the quality and availability of data, interpretability of AI models, and regulatory considerations persist [4]. However, as we navigate the ongoing digital transformation, it becomes increasingly evident that AI-based approaches hold immense potential to revolutionize drug discovery and reshape the field of medicinal chemistry. By leveraging AI’s capabilities and addressing the associated challenges, we can harness the power of this technology to accelerate the discovery of safe and effective molecules, ultimately revolutionizing the drug discovery process.
In this article, we provide a comprehensive review of the state-of-the-art technologies that employ AI in medicinal chemistry. We explore the current advancements and future prospects in this rapidly evolving field, shedding light on the transformative role of AI and its potential impact on drug discovery. The main purpose of this article is not only to outline the breakthroughs AI has facilitated but also to critically evaluate where it falls short or poses new challenges. By considering both the promises and pitfalls of AI in this domain, we aim to offer a balanced perspective that will guide future endeavors. Through a holistic understanding of the state of AI in drug discovery, we aspire to foster a foundation for its more robust and insightful application in the efficient and innovative drug discovery.
2. AI/ML Algorithms and Bio Big Data Utilized in Drug Discovery Research
Bio big data encompass a wide range of data types, such as genomic, proteomic, and transcriptomic data, collected from various sources such as high-throughput experiments and clinical studies, providing invaluable insights for drug discovery [6]. AI/ML algorithms, which are computational methods that allow computers to learn from data and recognize patterns, help researchers navigate these vast amounts of bio big data and identify potential drug candidates more effectively and accurately, revolutionizing the drug discovery process [4,11]. In order to optimally exploit AI and ML strategies within the context of drug discovery, it is essential to grasp the fundamental principles that underpin a range of machine learning methodologies. These methodologies, including supervised, unsupervised, and reinforcement learning, are employed to address a diverse array of research challenges within this domain.
2.1. Overview of ML Algorithms
AI is a broad field that encompasses various computational techniques, enabling machines to mimic human-like intelligence capabilities, such as learning, reasoning, and problem-solving. ML is a subset of AI specifically focusing on the development of algorithms that learn, adapt, and perform tasks through data processing and analysis [12]. By identifying patterns, making predictions, and refining algorithms based on input data, ML allows machines to improve their prediction performance and decision-making capabilities autonomously over time. ML algorithms can be broadly categorized into three types: supervised learning, unsupervised learning, and reinforcement learning.
2.1.1. Supervised Learning
Supervised learning is a type of machine learning where algorithms are trained using labeled data, meaning each input data sample is paired with the appropriate or correct output [12]. The algorithm uses these input–output pairs to learn a model that can make accurate predictions for new, unseen data. Supervised learning algorithms, such as support vector machine (SVM), support vector regression (SVR), naïve Bayes, tree-based, and random forest (RF), can identify potential drug candidates by analyzing large datasets and identifying patterns and relationships that humans may not easily detect (Figure 1) [13,14].
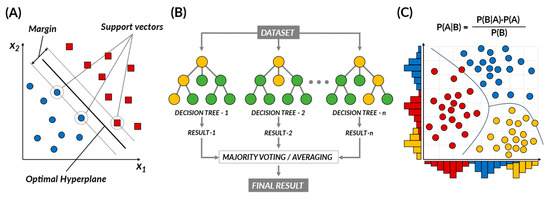
Figure 1.
Representative supervised learning algorithms. (A) Support vector machine. (B) Random forest. (C) Naïve Bayes.
- (1)
- Support Vector Machine (SVM)
The support vector machine (SVM) is a powerful tool, rooted in the principle of structural risk minimization, and is capable of classifying data, identifying outliers, and performing regression analysis. The core of the SVM methodology is the identification of an optimal decision boundary (i.e., hyperplane) that best separates data points across different classes [15]. This hyperplane is constructed by maximizing the margin, which represents the distance between the decision boundary and the closest training samples, also referred to as support vectors. In drug discovery, SVM is primarily employed to predict the biological activity of compounds or to classify molecular properties. One of the key strengths of SVM in such tasks is its ability to handle high-dimensional data and detect complex patterns, particularly within large and noisy datasets [16,17,18]. This makes SVM one of the top performers in predicting chemical and biological properties. However, it is crucial to note that SVM’s performance can be sensitive to the selection of the kernel function and its parameters [19]. Additionally, when dealing with imbalanced datasets, where one class significantly outnumbers the other, SVM may require additional processing steps to balance the data before application [19].
SVMs have established themselves as a significant tool in drug discovery due to their superior ability to analyze complex cheminformatics data. Their use extends to various tasks: they help in virtual screening processes [20,21], predicting drug–target interactions [22,23], and identifying new drug targets [24,25]. They are instrumental in predicting drug similarity in the quantitative structure–activity relationship (QSAR) domain, where the assumption is that structurally similar compounds will exhibit similar drug activities [23,26]. Furthermore, SVMs are employed to forecast activity cliffs, pairs of structurally similar compounds with a significant activity difference towards a specific target, thereby contributing to our understanding of critical drug–target interactions and aiding in new drug development [16,17,18].
- (2)
- Naïve Bayes
The naïve Bayes algorithm is a probabilistic machine learning model rooted in Bayes’ theorem, a principle in probability theory that describes how to update the probabilities of hypotheses when given evidence [12,14]. Specifically, Bayes’ theorem is a mathematical principle that provides a way to update the probabilities of our previous hypotheses based on new evidence [14]. In other words, when new information is given, it helps us decide how to apply it to a hypothesis. The naïve Bayes algorithm takes this principle and applies it with a “naïve” assumption of conditional independence between features [27]. Essentially, it considers each data attribute as independent, thus, simplifying multivariate problems into separate univariate issues. This strong assumption simplifies computations and enables the handling of high-dimensional data with ease. In practice, naïve Bayes has been widely adopted in various fields, such as document analysis, spam detection, and cheminformatics, particularly in drug discovery and drug–target interaction prediction [14]. It is noted for its robustness and versatility. However, despite its simplicity and speed, naïve Bayes has its limitations. It assumes that the attributes in the dataset are entirely independent, which might not accurately reflect the actual dependencies present in the data. Moreover, while naïve Bayes performs reasonably well as a classifier, it is known to be a less reliable probability estimator, so its output probabilities should be interpreted cautiously.
In drug design, the naïve Bayes algorithm has been widely applied, helping predict the biological activities of compounds, assisting in the early selection of promising candidates, and estimating results before laboratory experiments [7,28]. It can predict protein–protein [29] and drug–drug interactions [30], which is vital for understanding cellular pathways and managing polypharmacy, where patients take multiple drugs. This algorithm can also anticipate drug–target interactions, facilitating drug repurposing and side effect prediction [31,32,33,34]. Lastly, it can classify compounds into specific categories quickly, although it operates on the assumption of feature independence, which may not always hold [27,35].
- (3)
- Random Forest (RF)
Tree-based ML algorithms use decision trees (DTs) to predict target values based on observed features. DTs are flowchart-like structures where each internal node represents a feature, branches represent decision rules, and leaf nodes indicate outcomes, allowing for classification and regression tasks. However, single decision trees are prone to overfitting and struggle to generalize to new data. To overcome this limitation, ensemble methods, such as random forest (RF), prove to be particularly beneficial [12,13] The RF algorithm creates an ensemble of DTs, each built on a different sample of the data [14]. Each split in these DTs is determined from a different subset of features, leading to decorrelation between the trees. This strategy combats the overfitting problem often encountered with single DTs. By aggregating results from numerous, ideally uncorrelated, DTs, RF leverages the power of ensemble learning, enhancing its predictive power and stability. RF provides benefits in early drug discovery, including enhanced feature selection and predictive ability in QSAR analysis, making it useful for handling large, high-dimensional datasets in virtual screening [36]. However, to manage overfitting risks, careful data partitioning, model complexity control, and cross-validation are necessary. Analyzing feature importance can improve interpretability [12].
Building on these strengths, RF has been integrated into various stages of drug development, such as predicting chemical and drug properties, protein-related predictions, conducting virtual screening and docking studies, drug response prediction, polypharmacology research, and drug side effects prediction. Specifically, RF has proven helpful in QSAR modeling to correlate a drug’s chemical structure with its biological activity, estimating key parameters such as drug solubility and solvent density [36]. In protein-related predictions, RF assists in determining protein pKa values and protein–protein affinity, as well as identifying protein function and type, which is vital in target-based drug design [37]. Additionally, RF models facilitate efficient virtual screening of compound libraries to predict potential binding with target proteins, making them indispensable in integrated virtual screening and docking studies, including peptide docking studies [38].
2.1.2. Unsupervised Learning
Unsupervised learning is a method that trains a machine in the absence of any correct answers. It traverses unlabeled data, striving to decipher latent patterns or structures devoid of pre-defined output [5]. In this case, the learning process often involves grouping vast amounts of data based on similar characteristics, a process known as clustering. Even though the correct answer for the input value is not known, unsupervised learning can be used to uncover hidden patterns or features within the data, making it a powerful strategy for clustering and dimensionality reduction. Unsupervised learning algorithms, encompassing hidden Markov models (HMMs), growing self-organizing maps (GSOMs), k-means clustering, principal component analysis (PCA), autoencoders, and t-SNE, exhibit the capacity to cluster similar molecules, unearth novel molecular scaffolds, or reveal previously unknown correlations between biological entities (Figure 2).
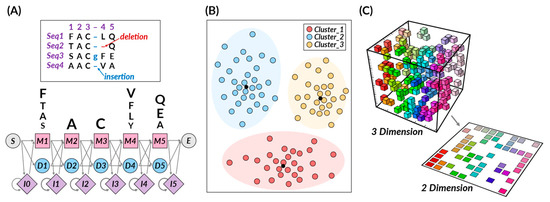
Figure 2.
Representative unsupervised learning algorithms. (A) Hidden Markov models. (B) K-means clustering. (C) T-distributed stochastic neighbor embedding.
- (1)
- Hidden Markov Models (HMMs)
Hidden Markov models (HMMs) are probabilistic models developed to work with sequential data. These models rely on a set of unobserved, hidden states and the probability of observable outputs that each state generates [39]. In a specific state, an outcome or observation can be produced according to an associated probability distribution, making HMMs an instrumental tool for various applications. HMMs excel at processing sequential data, modeling temporal dependencies, and managing missing and noisy data, making them robust against overfitting due to their probabilistic nature [40]. However, since HMMs rely on the Markov assumption, asserting that future states depend only on the current state, it might not always be a realistic presumption for many real-world scenarios.
HMMs have proven to be an invaluable tool in drug discovery, primarily due to their prowess in analyzing sequential biological data. They find extensive applications in various pivotal tasks. HMMs are critical in protein homology detection, efficiently identifying and classifying protein families within sequences [41,42]. This ability is vital in discovering new proteins that serve as potential targets for novel drugs. Further, HMMs play a significant role in protein sequence analysis, a critical process in understanding the function of a protein and selecting it as a target in the early stages of drug development [43,44]. Through augmenting sequence analysis, HMMs reveal more accurate and profound insights, thereby enhancing the outcomes. Lastly, HMMs are instrumental in predicting protein structures and 3D modeling. Accurately predicting and modeling complex protein structures is a crucial part of drug discovery, as it assists in predicting the efficacy and binding characteristics of a potential drug [45].
- (2)
- K-means Clustering
K-means clustering is a powerful tool, grounded in the partitioning principle, adept at classifying data into distinct ‘k’ clusters. A centroid, the mean of all data points within that cluster, characterizes each cluster. This algorithm aims to assign each data point to the nearest cluster, creating homogeneous groups of similar data points [1]. In drug discovery, k-means clustering is primarily used to define proper molecular descriptors, compute the similarities between compound samples, and group compound features based on computed similarities. A key strength of k-means in drug development is its ability to handle high-dimensional data and discern complex patterns, particularly within large and noisy datasets. This capability makes k-means one of the foremost performers in predicting chemical and biological properties. However, it is essential to note that k-means performance can be sensitive to the initial selection of centroids and the predetermined number of clusters. Furthermore, when dealing with imbalanced datasets, k-means may require additional pre-processing steps to balance the data before application. Despite these challenges, the simplicity, scalability, and flexibility of the k-means algorithm make it a vital tool in drug discovery.
Building upon the basic principles, k-means clustering finds extensive use in drug discovery, primarily due to its ability to handle multidimensional data. It helps define molecular descriptors, numerical entities that represent a compound’s physicochemical properties, thereby aiding in predicting its behavior [46,47]. The technique is also proficient in calculating similarities between compound samples, revealing relationships among compounds, and selecting potential drug candidates [48,49]. In addition, k-means clustering is used for clustering compound properties and selecting protein structures based on similarities [46,48,50]. Such grouping helps analyze a drug’s effect, and by identifying similar protein conformations, it enhances the performance of ensemble docking [51,52].
- (3)
- T-Distributed Stochastic Neighbor Embedding (t-SNE)
T-distributed stochastic neighbor embedding (t-SNE) is a technique that simplifies high-dimensional data into a more digestible, low-dimensional form while preserving the relative similarities of data points [1]. In short, t-SNE evaluates the similarity of data points in a high-dimensional space, giving higher probabilities to those more similar [53,54]. It maps these points to a lower-dimensional space, aiming to keep these similarities intact [1,53]. The ultimate goal is to create an easier-to-understand visualization while respecting the original data structure. A key advantage of t-SNE is its unique ability to maintain local and global high-dimensional data structures, unveiling patterns other reduction techniques such as PCA might overlook [54]. While t-SNE is effective for visualizing data, it does have limitations. It requires calculating pairwise similarities for all data points, which can be computationally demanding for large datasets [55]. Also, it often struggles to identify relevant clusters at varying scales and is sensitive to hyperparameters, necessitating careful tuning.
As a result, t-SNE plays a central role in drug design, particularly in compound clustering, drug target exploration, molecular representation, and drug design. t-SNE enables a comprehensive understanding and analysis of complex biological data and compound similarity by visualizing high-dimensional data in low-dimensional space. In particular, t-SNE assists in predicting the behavior of compounds through molecular descriptors, which are unique physicochemical characteristics, ultimately playing a crucial role in selecting potential drug candidates [56]. Moreover, t-SNE is employed to visualize biological data, aiding in the understanding of the relationship between drugs and their targets [57,58]. This can help discover new drug targets or identify new uses for existing drugs. Lastly, in visualizing complex biological data such as protein structures and gene expression profiles in lower dimensions, t-SNE enhances the technical aspects of molecular representation and drug design [59]. Due to its versatility and efficiency, t-SNE is expected to continue to play an essential role in shaping drug development strategies.
2.1.3. Reinforcement Learning
Reinforcement learning, a unique branch of machine learning, fine-tunes decision-making strategies through rewards or penalties for each action. Akin to learning via trial and error, a reward is given when the desired result is achieved, and the machine is trained to maximize this reward. If supervised learning and unsupervised learning proceed in a given static environment with the provided data, reinforcement learning, on the other hand, includes the process of collecting data in a dynamic environment. Reinforcement learning algorithms such as Q-learning and Monte Carlo tree search (MCTS) started to be used to revolutionize processes such as molecular docking, de novo drug design, and drug property optimization. These algorithms navigate molecular configurations, assist in constructing novel drug molecules, and balance objectives such as efficacy and side effects to produce promising drug candidates. It can aid the discovery of effective drug molecules and novel therapeutic strategies in a more creative, innovative way.
For example, Q-learning, a specific application of reinforcement learning, optimizes decisions through an intricate balance of ‘exploration’ and ‘exploitation,’ thus, enabling the model to continue learning while maximizing rewards [60]. Despite potential challenges such as computational intensity and the risk of suboptimal results if not properly balanced, Q-learning proves its worth by aiding in the discovery and optimization of molecular structures and compound characteristics [61]. It particularly excels in multi-property optimization, relationship identification among compounds, and the exploration of molecular space to identify promising candidates.
Further augmenting the capabilities of reinforcement learning in drug discovery is MCTS, another critical tool that enhances decision-making by deftly balancing ‘exploration’ and ‘exploitation’ [62]. Despite its computationally intensive nature and the challenge of striking the right balance, MCTS is indispensable due to its proficiency in navigating vast and complex molecular landscapes. It not only assists in the discovery and design of potential drug candidates but can also customize drugs to bind to specific targets [63,64,65]. MCTS particularly shines in retrosynthetic planning by offering a systematic approach to deconstructing complex organic molecules, thereby streamlining the planning of synthetic routes. By exploring a multitude of synthetic pathways, it helps chemists plan and execute synthesis more efficiently [66]. Additionally, MCTS enhances data mining in drug discovery, unearthing hidden patterns and structures in vast datasets.
2.2. Deep Learning Method
A pivotal advancement in the field of AI was the introduction of deep learning (DL), a subset of ML algorithms, designed to mimic the information-processing mechanism of the human brain. The human brain contains approximately 100 billion neurons, the cells that make up the nervous system. These neurons are intricately connected in multiple layers through a structure called synapses, transmitting signals by exchanging electrochemical signals. This structure of the human nervous system inspired the creation of artificial neurons, leading to the concept of ‘perceptrons’, which marked the beginning of artificial neural networks (ANN). A multilayer perceptron (MLP) is a type of neural network that possesses multiple layers, known as hidden layers, situated between the input and output layers. When there are two or more hidden layers, the term ‘deep’ is used, highlighting the use of consecutive layers.
In DL algorithms, predicted outcomes are generated through multiple layers using the input data, and this prediction is then compared to the actual value to calculate the difference (Figure 3). To reduce this difference, the weights of the previous layers are adjusted in a process called back-propagation. This process is repeatedly performed to continually refine the model. Examples of popular deep learning algorithms include convolutional neural networks (CNNs), often used in image processing tasks; recurrent neural networks (RNNs), particularly effective for sequence data such as time series or natural language; and deep belief networks (DBNs), which utilize unsupervised learning with generative models. Other examples are autoencoders for creating compact representations and generative adversarial networks (GANs) for generating new data that resemble the input data. These diverse algorithms reflect the breadth and depth of deep learning’s potential applications.
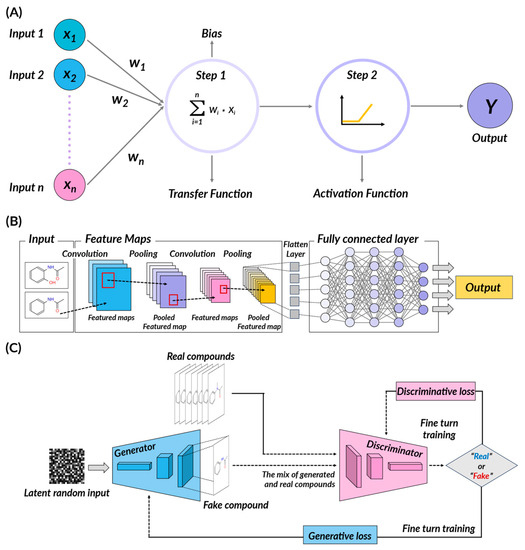
Figure 3.
Schematic illustration of deep learning methods. (A) Simplified representation of perceptron, a unit component of the artificial neural network (ANN), receiving multiple values as inputs and outputting a single value based on transfer and activation function. (B) Schematic diagram of convolutional neural network (CNN). (C) Schematic diagram of generative adversarial network (GAN).
- (1)
- Convolutional Neural Networks (CNNs)
Convolutional neural networks (CNNs) utilize small filters in several layers to detect patterns within data. Initially, CNNs extract simple features from the data and then combine them to extract more complex features. CNNs can extract useful features from images of molecular structures in drug discovery. CNNs have the primary advantage of effectively learning complex features from visual data. However, they require large amounts of data and significant computational resources, which can make model results difficult to interpret. Leveraging their fundamental capabilities, CNNs have found expansive usage in drug design, primarily for their proficiency in handling complex, multidimensional data.
In the field of molecular structure analysis, a CNN aids in deriving purposeful features or ‘descriptors’ that represent a compound’s physicochemical properties. For instance, BindScope employs deep convolutional neural networks to classify and visualize compounds on a large scale, based on their activity or inactivity in structure-based drug discovery [67]. Likewise, the use of artificial intelligence (AI) and machine learning technologies, such as a graph convolutional neural network (Graph-CNN), has facilitated the investigation and implementation of diverse molecular representations in drug discovery screening for identifying and predicting inhibitors of SARS-CoV-2 3CLpro [68]. These descriptors allow for accurate prediction of a compound’s behavior, while CNN excels at assessing similarities between different molecular structures, revealing intricate relationships among compounds. For example, in the ligand-based virtual screening approach, the L3D-PLS model employs CNN to extract crucial interaction features from grids surrounding aligned ligands, outperforming traditional methods in the lead optimization of small datasets [69]. In another application, the CAT–CPI model combines CNN with transformers to improve the prediction of compound-protein interactions, accelerating drug development [70]. Additionally, the FRSite method uses a faster R-CNN-based approach to accurately predict protein binding sites, introducing multi-source 3D data and RPN-3D networks to simultaneously predict the center and size of the binding site [71].
Moreover, CNNs are used to group molecular structures based on similarities, which is crucial for comprehending a drug’s impact and improving the performance of ensemble docking by identifying comparable molecular conformations. For instance, a recent study developed a deep learning model for compound classification using a distributed representation of compounds based on the SMILES notation [72]. Using this representation in a convolutional neural network (CNN), the model could process various compound types while obtaining low-dimensional representations of input features and outperforming standard methods in discriminating compound structures, including identified and unidentified motifs. This approach highlights CNNs’ adaptability and effectiveness in medicinal chemistry and enables a more nuanced understanding of the characteristics of compounds and potential drug interactions. Furthermore, in the context of large-scale data mining in drug design, CNNs assist in revealing patterns, correlations, and structures within enormous datasets.
- (2)
- RecurrentNeural Networks (RNNs)
Recurrent neural networks (RNNs) are a type of neural network designed to process continuous information. Over time, patterns are learned by these networks, which makes them suitable for natural language processing and time-series data analysis. In the case of drug development, RNNs can be beneficial in learning the amino acid sequence of a protein and predicting its impact on a specific illness. The primary benefit of RNNs lies in their ability to understand sequence information. However, as the sequence grows longer, they often fail to retain the initial information, which is a significant drawback.
They aid in constructing innovative drug molecules through various methods, enhancing the field of medicinal chemistry. For instance, combining stack-augmented recurrent neural networks with multi-objective reward-weighted sums in reinforcement learning optimizes the efficient drug design process, proposing a novel way to generate molecules with desired molecular characteristics [73]. By utilizing RNN models, the development of new derivatives of metronidazole and the synthesis and validation of compounds that inhibit bacterial strains such as E. coli, P. aeruginosa, B. subtilis, and S. aureus is explained [74]. The application of memory-augmented techniques using RNN-based architectures such as neural Turing machine (NTM) and differentiable neural computer (DNC) in creating new small molecules, analyzing their performance against simple RNNs, and assessing their validity, novelty, and attribute bias in de novo drug design are also explored [63]. These strategies translate chemical attributes into sequences such as simplified molecular-input line-entry system (SMILES), thus, assisting in forecasting novel potential drug candidates. Moreover, the effectiveness of neural networks, including RNNs, in forecasting drug–target interactions has been recognized. For example, a new deep learning approach using graph neural networks based on 3D structural information has been proposed to predict drug–target interactions [75]. The scope of RNN usage further spans the synthesis and testing of drug efficacy, offering a shift from traditional methods to a more data-centric approach, yielding more precise predictions and superior drug candidates.
- (3)
- Deep Belief Networks (DBNs)
A deep belief network (DBN) is a type of deep neural network structure that employs multiple restricted Boltzmann machines (RBMs) to stack layers of neurons. DBNs are utilized in drug development as a powerful tool to comprehend complex molecular properties. These learned properties are instrumental in the synthesis of potential drug candidates. DBNs exhibit strength in their capability to learn in an unsupervised manner, which enables them to discern intricate patterns within the input data. However, the down side associated with DBNs is that the learning process is intricate and a demands substantial amount of data and computational resources.
Drawing upon foundational concepts, DBNs demonstrate vast potential in drug discovery, particularly due to their capability to model complex, non-linear relationships in multi-dimensional data. They help to define molecular features and accurately predict the biological activity of novel compounds, thereby aiding in the identification of potential new drug candidates. For instance, AI-driven natural language processing and machine learning algorithms have been applied to explore challenges and opportunities in natural product (NPs) drug discovery, with specific AI approaches developed to identify biologically active natural products and capture the molecular ‘patterns’ of these privileged structures [76]. Notably, DBNs augment data mining in drug discovery, aiding in deciphering patterns, correlations, and structures within large datasets. For example, the application of a deep belief network (DBN) with a dropout mechanism to overcome the overfitting problem associated with small sample sizes has introduced a rapid and non-destructive drug identification method using near-infrared spectroscopy [77].
- (4)
- Autoencoders
An autocoder comprises an encoder that compresses input data and a decoder that restores compressed data. In drug discovery, an autoencoder compresses complex molecular properties. These properties are then used to synthesize new drug candidates. Autoencoders have the advantage of learning unsupervised and effectively compressing important characteristics of input data. The disadvantages of autoencoders include their sensitivity to noise in data that contains noise.
Autoencoders have emerged as a powerful tool in drug design, offering promising applications in predicting drug–target binding affinity and generating novel compounds. Specifically, in drug design and synthesis, techniques such as variational autoencoders can be instrumental in engineering novel molecules by decoding latent spaces to generate valid, novel molecular structures that could serve as potential drugs. For example, the problem of generating invalid molecular structures in automated chemical design can be alleviated by recasting it as a constrained Bayesian optimization problem within the latent space of a variational autoencoder, thereby significantly enhancing the validity of the generated molecules [78]. By utilizing their ability to capture and compress high-dimensional data, autoencoders can effectively extract latent features from chemical structures and learn representations that capture the underlying relationships between drugs and their targets. This enables accurate prediction of binding affinities and facilitates the identification of potential drug candidates. For instance, a deep-unsupervised-learning-based method called AutoDTI++ has been proposed to enhance the performance of drug–target interaction (DTI) predictions [79]. Furthermore, autoencoders excel in predicting drug–protein interactions by learning the intricate patterns and dependencies between chemical compounds and protein structures. By encoding the molecular features of drugs and proteins into a lower-dimensional space, autoencoders can effectively capture complex interactions and predict the likelihood of binding events. This capability enhances the performance of virtual screening methods and enables efficient exploration of the vast chemical space. Specifically, a deep learning framework that combines variational autoencoders and attention mechanisms, using CNNs to extract local features, has been proposed to obtain crucial information about drugs and proteins and improve drug–protein interaction (DPI) predictions [80].
- (5)
- Generative Adversarial Networks (GANs)
Generative adversarial networks (GANs) comprise a generator and a discriminator, two neural networks that learn from their interactions with each other. GANs can create new data that closely resemble real-world data. In drug development, GANs can produce new drug candidates that resemble real-world drugs. GANs have the advantage of being generative models that can learn from unsupervised data, but the disadvantage is their unstable learning process that makes it challenging to maintain a balance between the generator and discriminator.
Drawing upon their foundational principles, generative adversarial networks (GANs) have seen substantial application in drug discovery, primarily owing to their ability to learn complex data distributions. They play a crucial role in de novo molecular design, wherein deep generative models can effectively learn from existing data and generate novel molecules, addressing the inefficiencies and time-consuming aspects of traditional methods. In particular, recent developments in deep generative models for de novo molecular design have been reviewed, categorizing these models into two types, examining their strengths and weaknesses, and identifying current challenges [9]. Moreover, a new technique utilizing a deep learning GAN called “DNMG” has been proposed to integrate the 3D information of molecules and effectively predict and explore drug properties and binding affinities for new drug design [81].
GANs are also pivotal in the generation and analysis of high-content images in drug discovery assays. Specifically, a computer-based framework that employs three variations of GANs has been proposed for the automatic analysis of large-scale image data generated from drug tests. Among them, the DCGAN, in particular, has been applied to create realistic synthetic images that can be used to study the effects of drugs on cells and bacteria [82]. By learning data distribution, GANs can create synthetic images, enhancing the automatic analysis of voluminous image data generated in drug screening processes. Moreover, they prove instrumental in predicting drug–drug interactions (DDIs) by learning the patterns within large-scale data, leading to an understanding of unforeseen interactions. Specifically, a novel deep learning model known as DGANDDI utilizes two GAN architectures to deeply explore complementary knowledge between drug attributes and DDI network topology [83]. Another significant utilization of GANs is in predicting drug–target interactions, where they can learn the potential interactions between drugs and their respective protein targets, thereby directing the process of drug discovery. In particular, a novel approach using GANs implements a semi-supervised learning method that leverages both labeled and unlabeled data to predict the binding affinity between drugs and targets [84].
2.3. Performance Metrics
Performance metrics are tools for evaluating a model’s efficacy, with different metrics utilized for classification and regression tasks. Accuracy, precision, recall, and the F1 score are commonly used to assess how accurately the model categorizes data. On the other hand, for regression, the mean squared error (MSE), mean absolute error (MAE), and root mean squared error (RMSE) measure the discrepancy between predicted and actual values. These metrics play a crucial role in understanding and enhancing model performance.
- (1)
- Metrics for Classification Models
Classification models are supervised learning algorithms that categorize given data into one of the predefined classes. Key terminologies for evaluating the performance of the model include true positive (TP), true negative (TN), false positive (FP), and false negative (FN). TP represents the number of positive cases correctly detected by the model, making it an important metric for evaluating model performance. TN represents the number of negative cases correctly ignored by the model, which is especially crucial when wanting to avoid incorrect positive predictions. FP is when the model incorrectly predicts a data point that is actually negative as positive. Such predictions can give users incorrect information or unnecessary alerts. FN is when the model incorrectly predicts a data point that is actually positive as negative. FNs are missed positive cases by the model, which can have severe consequences. For instance, in a medical diagnostic model detecting diseases, an FN could result in a patient with the disease not receiving a diagnosis. Such terminology forms the basis for calculating performance metrics of classification models, which are essential to accurately determine the strengths, weaknesses, and areas of improvement of the model. Based on these terms, several performance metrics are calculated. These metrics reflect various aspects of classification model performance such as accuracy, sensitivity, and specificity, and their importance can vary depending on the specific application area [85].
The accuracy (ACC) indicates the proportion of data correctly classified by the model among its predictions. It is useful when the class distribution of the data is uniform. If one class greatly outnumbers the other, it can be challenging to fully assess the model’s performance based solely on accuracy.
The precision (PREC) denotes the ratio of data points that are actually positive among those the model classified as positive. This metric becomes important when the cost of incorrect positive predictions is high.
The recall (REC) or sensitivity represents the ratio of data points the model predicted as positive among the actual positive data points. It is critical when the cost of incorrect negative predictions is high.
The F1 score is the harmonic mean of precision and recall, indicating a balance between the two metrics. It is especially useful when one class’s sample size is much smaller than the other.
The specificity (SPEC) illustrates the ratio of actual negative class data points correctly predicted as negative by the model.
All these metrics fall within the [0, 1] range, where 1 indicates perfect prediction based on the metric, and 0 indicates an entirely incorrect prediction. Each performance metric emphasizes different aspects of the model, so the importance of a particular metric can increase depending on the specific application or problem. Therefore, it is crucial to consider these metrics comprehensively when evaluating and optimizing model performance.
- (2)
- Metrics for Regression Models
A regression model is an algorithm that models the relationship between one or more independent variables and a continuous dependent variable. Such models are used to predict a continuous output value for given input variables. Various metrics are employed to assess the performance of a regression model by measuring the difference between the predicted and actual values.
The mean squared error (MSE) is the average of the squared differences between predicted and actual values. Due to its squaring of prediction errors, large errors can significantly inflate this value.
The mean absolute error (MAE) is the average of the absolute differences between predicted and actual values. It calculates the absolute error for each prediction and then averages those values.
The root mean squared error (RMSE) is the positive square root of MSE. RMSE provides an interpretation of the prediction error size in the original unit. While it gives greater weight to large errors, RMSE can be more interpretable as it represents the actual size of the prediction error.
The R-squared, also known as the coefficient of determination, is a metric that illustrates the predictive power of a regression model. It indicates how well the model explains the variability in the data.
These metrics play a crucial role in evaluating the performance of a model, emphasizing different aspects. Therefore, when assessing and optimizing the performance of a regression model, it is essential to consider these metrics comprehensively.
2.4. Databases in Drug Research
The effectiveness of ML or DL methods is predominantly contingent upon the availability of substantial, accurate, and reliable data. As technological advances continue to make data generation faster and more affordable, the volume of chemical, biological, and medical data has grown exponentially. There is a continuous effort to centralize these datasets and make them publicly available for research around the globe. This crucial information can be procured from many public databases, which serve as rich repositories of information that are indispensable for drug discovery. These databases encapsulate a wide array of knowledge about the efficacy of various drugs, potential side effects, the nature of drug targets, and intricate chemical structures. However, it is essential to understand that not all databases are equivalent in terms of the type and depth of information they contain. Each database is unique, housing specific sets of data. Therefore, gaining a robust understanding of the nature of the information these databases provide, and how to utilize it effectively, is essential for researchers. Table 1 summarizes the publicly available databases that contain various aspects of data utilized in drug discovery.

Table 1.
Databases utilized in drug discovery research.
3. AI in Structural Biology
Structural biology, encompassing the study of the three-dimensional (3D) structures of biological macromolecules, is essential for understanding the precise mechanisms underlying living organisms. Experimental techniques such as X-ray crystallography, nuclear magnetic resonance (NMR) spectroscopy, and cryo-electron microscopy (cryo-EM) have been employed to elucidate numerous structures. However, these experimental methods often require extensive labor, time, and financial resources, posing limitations for specific molecules or complexes. AI has advanced structural biology by providing computational approaches to overcome these experimental limitations [118,119]. AI techniques, especially DL methods, and data-driven modeling, have been applied by extracting meaningful patterns and features from vast and complex biological sequence and structural data. AI offers innovative tools for researchers, enabling protein structure prediction, protein design, and more, and continues to evolve alongside advancements in hardware and algorithm.
3.1. Protein Folding and Its Prediction
Protein folding, a highly intricate process, plays a fundamental role in determining the functional properties of biological macromolecules. Despite the limited number of around 20 amino acids, the varying structures and functions depending on their arrangements are of significant research interest in life sciences. Therefore, structural studies on protein folding are essential for understanding biological mechanisms and developing therapeutic strategies for diseases. However, observing the actual structure of proteins requires substantial resources and time. Hence, researchers attempt to predict protein folding and 3D structures using genetic information.
Despite the high demand for protein structure prediction, it remains challenging, and achieving perfect prediction is still elusive. Even minor variations in protein sequences can lead to drastic changes in overall structure and, in some cases, result in loss of function. On the other hand, certain amino acids share similar chemical properties, which can lead to minimal structural differences in some mutations. Additionally, despite the astronomical range of conformational possibilities resulting from the rotations of amino acids in flexible polypeptide chains, most small proteins fold spontaneously on a millisecond or even microsecond timescale. In order to address these Levinthal’s paradoxes, scientists have conducted extensive research in this area [120,121]. Researchers have attempted to predict structures using computational thermodynamic hypotheses, but this method has not yielded perfect results. For instance, predicting conformations of a protein within a biological system is challenging because even slight inaccuracies in the computation of the significant free energy difference between folded and unfolded states can lead to incorrect predictions. Therefore, recent arguments suggest that instead of traditional thermodynamic hypotheses, the non-equilibrium and active nature of proteins within the biological context requires modeling using fluctuating free-energy landscapes [122].
To overcome these limitations, various advancements and refinements have been made in the field of homology modeling techniques. Homology modeling, also known as comparative modeling, predicts the structure of a target protein using experimentally determined structures of homologous proteins as templates. Homology modeling contributes to understanding protein structure and function, aiding hypothesis generation and experimental design. This approach leverages the principle that proteins with similar sequences adopt similar structures. Key steps include selecting a suitable template with high sequence similarity, aligning the target sequence with the template, generating a model, and model optimization followed by validation [123]. The model accuracy largely depends on sequence similarity, and adopting strategies such as utilizing multiple templates, along with implementing processes such as energy minimization and loop modeling, can significantly enhance this precision.
3.2. Biomolecular Structure Prediction by Computational Methods
As research methods and technologies have advanced, protein structure and sequencing data accumulation has been accelerating. The Protein Data Bank (PDB), established in 1971, serves as the primary archive housing the largest collection of protein 3D structures [124]. As of 2022, the number of experimentally determined 3D protein structures has exceeded 200,000, with the pace of data accumulation steadily increasing [125]. Additionally, the UniProt Knowledgebase (UniProtKB) [109] provides information on protein sequences including functional annotations and currently holds over 220 million sequences as of 2023. This database also encompasses protein structure visualization data, including predicted structures by AI (i.e., AlphaFold, which is described in the following section).
MODELLER [126], which started development in 1993, is a representative program that can generate homology models using these accumulated databases. When researchers provide the sequence of the desired protein and structures of similar sequences, the program automatically creates models that satisfy spatial restraints using comparative protein structure modeling [127]. The SWISS-MODEL server, another tool in the field, provides an automated web service for generating homology models, with the ProMod3 modeling engine at its core [128]. This program utilizes QMEANDisCo [129] for model quality estimation and has demonstrated excellence through the CAMEO project, showcasing its effectiveness [130]. I-Tasser [131] has garnered significant attention due to an outstanding performance. I-Tasser combines various methods, including threading, ab initio modeling, and structure assembly, to generate models [132]. It utilizes a hierarchical approach, starting with the identification of template structures through threading, followed by fragment assembly simulations and refinement. The program has demonstrated competitive performance in several international structure prediction competitions, such as critical assessment of protein structure prediction (CASP) [133].
Collaborative efforts such as CASP are underway, providing a valuable platform for evaluating AI-based biomolecule structure prediction research [134,135]. Various research teams engage in structure prediction by categorizing protein sequences into cases amenable to template-based modeling (TBM) or those requiring free modeling (FM) and then evaluating their predictions. In 2018, DeepMind’s AlphaFold participated in CASP13 and introduced a novel approach by enhancing the traditional fragment assembly technique using deep learning (DL) methods. AlphaFold utilizes a deep residual convolutional neural network (CNN) to effectively capture intricate patterns within the protein data (Figure 4). The neural network undergoes training to make predictions about protein structures, and through gradient descent, it minimizes the potential energy to stabilize and achieve accurate structure prediction [136]. AlphaFold2 emerged during CASP14, introducing a novel neural network block called Evoformer [119,135]. Evoformer enhances the accuracy of structure prediction by facilitating the exchange of information within the multiple sequence alignment (MSA) and learning the relationships between sequences. It captures intricate spatial relationships, thereby improving the prediction of protein structures. trRosetta is also an approach that incorporates CNN into the existing RosettaFold framework [137]. By utilizing CNN, it directly predicts inter-residue distances and torsion angles from protein sequences and MSAs. These predictions are then integrated with the fragment assembly approach of RosettaFold to generate protein structure models.
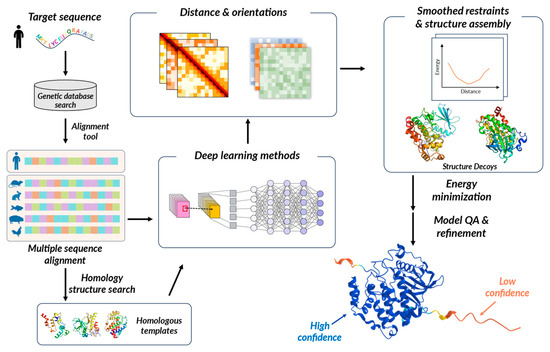
Figure 4.
Workflow of DL-based protein structure prediction.
3.3. Advancements in Protein Structural Research through AI
With the successful application of AI in predicting the structures of individual proteins, there has been a growing interest in applying AI to a broader range of structural biology research. One such area is the prediction of protein complex structures, which is expected to make significant contributions to the study of host–pathogen interactions [118]. DeepMind has released AlphaFold-Multimer to predict protein complex structures [138,139]. However, the coevolution within a protein and between proteins exhibits distinct patterns, posing limitations to complex prediction using MSA-based methods. Moreover, there are challenges in accurately predicting heterodimeric complexes compared to homodimeric ones, and the accuracy tends to decrease as the number of chains increases. In response to these issues, efforts have been made to address them by introducing ESMFold [140], a language model-based approach, aimed at improving the prediction accuracy.
The epigenetic dimension of protein structure (EDPS) represents another important objective that needs to be addressed [141]. Currently, neural network (NN)-based modeling algorithms face significant limitations in epigenetic protein structure prediction. This is particularly evident in membrane proteins [142], where template-based modeling (TBM) demonstrates higher accuracy than NN-based modeling, owing to the incorporation of lipid bilayer template data. This observation suggests that specific lipid species in the membrane environment may influence protein structure formation [143,144]. Comparative studies on G-protein-coupled receptor (GPCR) structure prediction supports this notion [145], as TBM exhibits relatively higher accuracy in loop regions, possibly due to the influence of the surrounding environment. Consequently, further research is required to explore additional methods that can improve the accuracy of EDPS prediction, especially for membrane proteins, even in the absence of suitable templates.
The integration of AI in protein structural research has shown notable advancements and potential in this field. While some challenges persist, particularly in predicting complex interactions and navigating the epigenetic aspects of protein structure, AI’s adaptability and the ongoing refinement of its methodologies have shown promise in overcoming these hurdles. Tools such as AlphaFold or ESMFold represent the innovative solutions developed to address existing limitations. However, the journey towards the comprehensive and accurate prediction of protein structures, including complex and epigenetically influenced structures, is ongoing. As AI technologies continue to evolve, it is anticipated that they will play an increasingly critical role in unfolding the mysteries of protein structures, thereby revolutionizing our approach to structure-based drug discovery.
4. AI in Medicinal Chemistry or Cheminformatics
AI has also revolutionized medicinal chemistry and cheminformatics by providing innovative tools and approaches for drug discovery, such as deep generative models for molecular design, as well as the prediction of drug–target interactions or drug toxicity [47]. AI-driven approaches enable the exploration of vast chemical space, leading to the discovery of novel compounds with therapeutic potential. They can also facilitate drug repurposing by analyzing large-scale data to identify connections between drugs and diseases, expanding the possibilities for addressing unmet medical needs. Regarding chemical structures, AI methods generally use molecular fingerprints as input data. They are trained to find patterns in these fingerprints that correlate with the properties of interest, such as biological activity or physicochemical properties. This process is crucial in various cheminformatics tasks such as virtual screening, QSAR modeling, and de novo drug design. Thus, the relationship between molecular fingerprints and AI methods in cheminformatics is one of symbiosis, with each enabling the other’s functionality.
4.1. Molecular Fingerprints
Molecular fingerprints are binary or count-based representations that encode the chemical structure into a format that computational models can readily process. They can be seen as a bit string (usually several hundreds or thousands of bits long), where each bit represents the presence or absence of a particular substructure or property within the molecule [146]. Capturing different aspects of the molecular structure, such as the presence of certain functional groups, topological features, or 3D properties, it can transform these complex structures into simplified, standardized vectors [147]. The primary use of the molecular fingerprint is for creating a simplified digital representation of complex molecules, which allows efficient comparisons and searches in large molecular databases. It can be also used as a particular form of molecular descriptor that is commonly applied to a wide range of tasks, from virtual screening to prediction model building [148].
Molecular fingerprints can be categorized into the following classes [149,150,151]: (1) substructure keys-based fingerprints that generate bit strings depending on the presence or absence of substructures in the compound, (2) topological fingerprints that focus on topological routes and represent all possible connectivity routes between atoms, (3) circular fingerprints that are also related to atom paths similar to topological fingerprints but represent the connectivity of atoms not in a linear way but in a radius way, illustrating the atom’s environment [152], and (4) pharmacophore fingerprints that encode atoms by the pharmacophoric functional groups and atom bonds by several distance ranges [153]. Some fingerprints cannot be in these classes. SMIfp [154], which is calculated from SMILES, is a good example of that exception. Figure 5 and Table 2 summarizes the representative fingerprints commonly used in drug discovery research.
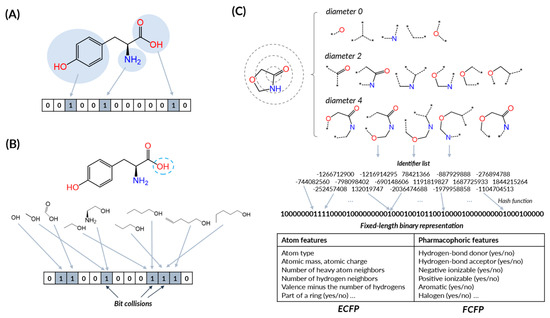
Figure 5.
Schematic representation of molecular fingerprints. (A) Substructure key-based method. (B) Topological or path-based method. (C) Circular methods.
Fingerprints can be used to calculate molecular similarity or distance and help identify molecules with similar structures or properties, clustering molecules into groups, or predict the properties or activities of new molecules based on their similarity to known ones. As a tool in machine learning, molecular fingerprints can be used as input features for models predicting the properties or activities of molecules. In this context, the model learns to associate certain patterns in the fingerprint with the property or activity being predicted. The major drawback of molecular fingerprints is that they generally ignore biological context and do not contain 3D information. Various approaches have been proposed to overcome these limitations. For example, atom pair 3D fingerprints are developed from 2D atom pair fingerprints to contain 3D information [155]. Protein–ligand interaction fingerprints (PLIF) encode information about protein–ligand interactions, such as hydrogen bonds, ionic interactions, and surface contacts according to the residues [156].

Table 2.
Various types of molecular fingerprints.
Table 2.
Various types of molecular fingerprints.
| Category | Molecular Fingerprint | Description | Ref. |
|---|---|---|---|
| Substructure key-based | Molecular ACCess system (MACCS) keys |
| [157] |
| PubChem fingerprint |
| [158] | |
| Topological | Daylight fingerprint |
| [150] |
| Atom pairs2D fingerprints (APFP) |
| [159] | |
| Circular | Extended-connectivity fingerprints (ECFP) |
| [152] |
| Molprint2D |
| [160] | |
| Pharmacophoric | Functional-class fingerprints (FCFP) |
| [152] |
| SMILES-based | SMIfp |
| [154] |
4.2. Deep Generative Model for Molecular Design
A deep generative model is a type of neural network trained on existing data with a high-dimensional probability distribution, which it then uses to create new samples from that distribution [10]. In drug discovery research, deep generative models can be utilized in the area of de novo molecular design, a computational methodology that generates novel molecules with desired properties [8,161,162]. While QSAR models can be used to predict the biological activities of unknown chemicals based on their structures, which are derived from other chemicals with various known biological activities, de novo design is employed to generate novel chemical structures with desired pharmacological properties, using structure–bioactivity data as a basis [161]. They are trained on large databases of known molecules to learn the underlying patterns and structures in the data. This learned knowledge is then leveraged to generate new, unseen molecules that are likely to be physicochemically promising or biologically active.
Popular DL algorithms, such as recurrent neural networks (RNNs), variational autoencoders (VAEs), and generative adversarial networks (GANs), have been applied to de novo molecular design. RNNs are commonly used for data sequence modeling and generation and sequence-to-sequence mapping. In de novo design, they can also be used for analyzing molecular sequence data, such as SMILES [162]. Once the RNNs are trained by target sequences, they can generate new sequences that follow the conditional probability distributions learned from the training set [162]. VAEs can represent high-dimensional complex data by learning a low-dimensional latent space in an unsupervised manner [163]. When the VAE model is applied to de novo design, the encoder converts the molecule to a latent vector representation, and the decoder generates a novel chemical space from the latent vector representation [164]. GANs contain a generator and a discriminator. The generator creates new data based on input data, and the discriminator identifies whether the data are real or generated. During the training, these two components compete against each other, and when the discriminator cannot distinguish between generated data and real input data, the training ends [164]. In this process, GANs can generate novel molecules by using patterns and structures in a pre-existing training dataset.
Deep generative models, despite facing certain challenges, have shown remarkable potential for de novo design in early discovery stages. These models can be utilized to generate novel and diverse chemical scaffolds with desirable predicted values. However, to fully realize their potential, further research is needed to address key issues, such as lack of interpretability. Nonetheless, the early successes of these models have opened up exciting avenues for innovation and creativity that were previously unattainable.
4.3. Prediction of Drug–Target Interaction (DTI)
Drugs control our body’s physiological activities to exert therapeutic effects, which are achieved through the interaction between the drug and its target protein, known as drug–target interaction (DTI) [165]. A drug, which serves as a ligand, binds to the pocket in a protein, often referred to as a binding site, inducing changes in physiological activity. These pockets can vary in size and depth, and ligand binding can alter the protein structure, impacting its function through molecular interactions such as ionic bonds, van der Waals interactions, and hydrogen bonds [166,167,168]. They can prevent the protein from interacting with endogenous molecules or cause changes in its activity [165]. This activity can be affected not only by the ligand itself but also by water molecules, metal ion coordination, and various other circumstances [166].
Traditionally, the drug discovery approach has been based on the “one molecule–one target–one disease” paradigm, where the drug produces therapeutic effects by regulating its target. In this approach, it is necessary to test whether a particular protein could be a specific drug target for treatment [169]. However, a single target is not exclusively associated with one disease, and the onset of complex diseases may involve multiple factors. Depending on the circumstances, it may be necessary to intervene in multiple areas along the pathologic mechanism for effective treatment [170]. From this perspective, the importance of DTI research is increasingly recognized, especially regarding side effects, drug repositioning, and drug resistance [171].
The power of AI is apparent in this target identification and virtual screening [172]. ML algorithms, especially during the virtual screening process, follow an approach that differs from conventional structure-based virtual screening (SBVS) or ligand-based virtual screening (LBVS) [35]. They generate statistical models to anticipate the conditions of undiscovered ligands–proteins based on the recognized configurations of protein–ligand compounds and physicochemical characteristics [166]. The research carried out on the three aspects of ML-based DTIs, namely, prediction of existing ligand binding sites, binding affinity, and binding poses, is ultimately aimed at bringing us closer to more efficient drug discovery [170]. Table 3 summarizes a recent application of AI in DTI prediction.

Table 3.
AI-based methodologies to predict DTIs.
4.4. Toxicity Prediction
The significant increase in chemical usage has intensified the need for reliable toxicity prediction, leading to the establishment of the Tox21 program in 2008, a collaborative endeavor undertaken by the U.S. governments, such as the Environmental Protection Agency (EPA). Further strengthening this initiative, the U.S. Food and Drug Administration (FDA) became a part of the consortium in 2010. This program employs high-throughput screening (HTS), an in vitro assay, to scrutinize the biochemical activity of various substances. This methodology has the advantage of not only reducing the time and cost associated with toxicity testing but also mitigating ethical concerns [187,188]. The culmination of these efforts resulted in the creation of the Tox21 10K library, which subsequently fostered the inception of the Tox21 data challenge, an initiative designed to enhance the precision of predictions about holistic human responses utilizing computational methodologies [189]. In parallel, EPA’s Toxicity Forecaster (ToxCast) program, instituted in 2007, assesses materials that could be harmful to human health and the environment via bioactivity profiling [190]. Distinguished from Tox21, ToxCast covers a broader chemical space and grapples with less specific mechanisms of action. Implementing methods analogous to HTS, it endeavors to categorize chemicals and advocate for the appropriate regulation of environmental pollutants [191]. These initiatives persist as critical contributions to the progression of toxicity prediction models.
These valuable Tox21 and ToxCast datasets have been monumental in driving advancements in the field of ADME/Tox prediction. Compounds that enter the human body commonly undergo absorption, distribution, metabolism, and excretion (ADME), with some leading to toxicity [192]. Efforts to decode these processes from a pharmacokinetic/pharmacodynamics (PK/PD) perspective have been put forth but given their interactions with numerous human body structures such as membranes, proteins, and the intra/extracellular environment, the ADME–Tox (ADMET) processes are viewed as multifactorial and intricate. Factors such as the compound’s solubility, membrane permeability, consumed concentration, and partition coefficient play a significant role in the absorption process [193,194]. AI algorithms can be trained on the Tox21/ToxCast datasets to predict the potentially toxic effects of a new compound, which are critical aspects of the drug discovery process [195]. Here, we describe recent applications in three representative endpoints, i.e., hepatotoxicity, cardiotoxicity, and carcinogenicity.
- (1)
- Prediction of Hepatotoxicity
Given the crucial role the liver plays in drug metabolism, assessing the potential for drug-induced liver injury (DILI) is vital for safety reasons [196,197]. Hepatotoxicity, in particular, is a significant issue in drug development, leading to a significant number of drugs being withdrawn due to DILI or not being launched at all [198,199]. Safety concerns mean that only about 31.8% of potential drug candidates progress from preclinical testing to clinical trials [200]. Even among the drugs that are successfully launched, 58% of FDA drugs approved in 2020 and 2021 show signs of hepatotoxicity [201]. Traditional methods of DILI evaluation, such as in vitro and in vivo studies, are both costly and time-consuming [202]. In response to this, there has been a shift towards developing computational methods that can predict DILI quickly and accurately using AI methods.
- (2)
- Prediction of Cardiotoxicity
Cardiotoxicity is a paramount concern in the creation of innovative pharmaceuticals [203]. In line with the International Conference of Harmonization’s guideline (S7B), it is obligatory for all emergent drugs to undergo a pre-clinical examination of their potential to inhibit hERG activities before they are considered for regulatory appraisals [31,204]. The hERG channel, alternatively known as Ether-à-go-go (EAG) proteins, constitutes potassium channels that are manifested in diverse brain areas, endocrine cells, muscles, and the heart [205,206]. These channels play an indispensable role in cardiac function by facilitating the heart’s electrical activity [205]. The blockade of these channels by small molecules can precipitate QT interval prolongation, which may culminate in lethal cardiotoxicity [207,208]. Consequently, it is essential that drug candidates demonstrate minimal hERG inhibition to circumvent such deleterious effects [209].
- (3)
- Prediction of Mutagenicity and Carcinogenicity
Mutagenicity and carcinogenicity are key considerations in the risk assessment of chemicals and pharmaceuticals [210,211]. Mutagenicity refers to a substance’s ability to cause genetic mutations, potentially leading to various disorders, including cancer, while carcinogenicity is a compound’s potential to cause cancer [212,213,214]. Given the correlation between these two and the global burden of cancer, it is vital to evaluate mutagenicity and carcinogenicity [215,216,217]. Challenges in this task include mutagenicity, inconsistencies in Ames test results, false positives and negatives, and reproducibility issues among labs [218,219]. The 1995 ICH guidelines provided a structure for carcinogenicity studies, yet these studies require two years, approximately $1.1 million, and about 500 rodents, making it a laborious and costly process [220]. As such, in silico predictive methods are gaining popularity, with several proposals suggesting the use of ML approaches to increase efficiency [221]. Table 4 summarizes the additional case studies for predicting various toxicity.

Table 4.
AI-based methodologies to predict various toxicity.
5. Conclusions and Future Perspectives
In conclusion, the integration of AI in drug discovery represents a significant paradigm shift in medicinal chemistry, rather than just a technological addition. AI unlocks insights from complex datasets that were previously unreachable, and its value in data-driven drug discovery is poised to become increasingly prominent. However, alongside the high expectations for AI’s potential, it is vital to exercise caution. AI models heavily rely on large amounts of high-quality data, making access to diverse and sufficient data crucial for accurate learning and prediction. In the context of drug discovery, these data encompass information about known compounds, biological processes, disease mechanisms, clinical data, patient adverse events, and more. Due to the intricate nature of biological processes, the multitude of variables impacting drug action, and individual variations, many aspects cannot be interpreted or applied in isolation. This complexity presents challenges for achieving full automation, often requiring domain knowledge-based optimization and expert-guided manual curation. Efforts to address these limitations of AI applications have gained momentum across multiple domains. A significant emphasis is now on gathering or digitalizing diverse datasets, ensuring that AI tools represent a comprehensive spectrum of the data. To overcome data deficiency, researchers are using pre-trained models and fine-tuning them on specific, smaller datasets. This approach makes AI applications more adaptable and data efficient. There is also a concerted effort to bolster the robustness of AI, certifying that it performs reliably on previously unseen data.
Drug discovery and development, especially within the life-critical industry, necessitate human involvement for real-world experimental validation and clinical trials, extending beyond virtual simulations alone. Incorporating AI into these fields amplifies some of the ethical considerations, especially around data privacy, transparency, or potential bias. Furthermore, using AI on electronic medical records could risk patient privacy breaches. Nevertheless, AI technology can greatly enhance the efficiency of experiments for researchers, particularly in computationally intensive tasks or in identifying intricate patterns that might evade human observation. The optimal scenario is one where humans and AI technology collaborate, each leveraging their respective strengths. Addressing technical challenges, ensuring data robustness, validating AI models, and considering ethical implications requires a continuous collaborative approach involving academia, industry, and regulatory agencies.
Author Contributions
Conceptualization, Y.L.; investigation, R.H., H.Y., G.K., H.L. and Y.L.; resources, Y.L.; writing—original draft preparation, R.H., H.Y., G.K., H.L. and Y.L.; writing—review and editing, R.H., H.Y. and Y.L.; funding acquisition, Y.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Research Foundation of Korea (grant numbers 2021M3E5E3080529 and 2022R1C1C1007409) to Y.L. and the Chung-Ang University Graduate Research Scholarship in 2023 to R.H.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data sharing is not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Chen, W.; Liu, X.; Zhang, S.; Chen, S. Artificial intelligence for drug discovery: Resources, methods, and applications. Mol. Ther. Nucl. Acids 2023, 31, 691–702. [Google Scholar] [CrossRef] [PubMed]
- Cifci, M.A. A Deep Learning-Based Framework for Uncertainty Quantification in Medical Imaging Using the DropWeak Technique: An Empirical Study with Baresnet. Diagnostics 2023, 13, 800. [Google Scholar] [CrossRef] [PubMed]
- Wong, J.H.; Zhang, Q.X. Deep Learning of Sparse Patterns in Medical IoT for Efficient Big Data Harnessing. IEEE Access 2023, 11, 25856–25864. [Google Scholar] [CrossRef]
- Alya, A.A. Artificial intelligence in drug design: Algorithms, applications, challenges and ethics. Future Drug Discov. 2021, 3, FDD59. [Google Scholar] [CrossRef]
- Gupta, R.; Srivastava, D.; Sahu, M.; Tiwari, S.; Ambasta, R.K.; Kumar, P. Artificial intelligence to deep learning: Machine intelligence approach for drug discovery. Mol. Divers 2021, 25, 1315–1360. [Google Scholar] [CrossRef]
- Zhu, H. Big Data and Artificial Intelligence Modeling for Drug Discovery. Annu. Rev. Pharmacol. 2020, 60, 573–589. [Google Scholar] [CrossRef]
- Hu, Y.; Lu, Y.; Wang, S.; Zhang, M.Y.; Qu, X.S.; Niu, B. Application of Machine Learning Approaches for the Design and Study of Anticancer Drugs. Curr. Drug Targets 2019, 20, 488–500. [Google Scholar] [CrossRef]
- Tong, X.C.; Liu, X.H.; Tan, X.Q.; Li, X.T.; Jiang, J.X.; Xiong, Z.P.; Xu, T.Y.; Jiang, H.L.; Qiao, N.; Zheng, M.Y. Generative Models for De Novo Drug Design. J. Med. Chem. 2021, 64, 14011–14027. [Google Scholar] [CrossRef]
- Cheng, Y.; Gong, Y.S.; Liu, Y.S.; Song, B.S.; Zou, Q. Molecular design in drug discovery: A comprehensive review of deep generative models. Brief Bioinform. 2021, 22, bbab344. [Google Scholar] [CrossRef]
- Xue, D.Y.; Gong, Y.K.; Yang, Z.Y.; Chuai, G.H.; Qu, S.; Shen, A.Z.; Yu, J.; Liu, Q. Advances and challenges in deep generative models for de novo molecule generation. Wires Comput. Mol. Sci. 2019, 9, e1395. [Google Scholar] [CrossRef]
- Vemula, D.; Jayasurya, P.; Sushmitha, V.; Kumar, Y.N.; Bhandari, V. CADD, AI and ML in drug discovery: A comprehensive review. Eur. J. Pharm. Sci. 2023, 181, 106324. [Google Scholar] [CrossRef] [PubMed]
- Cerchia, C.; Lavecchia, A. New avenues in artificial-intelligence-assisted drug discovery. Drug Discov. Today 2023, 28, 103516. [Google Scholar] [CrossRef] [PubMed]
- Dara, S.; Dhamercherla, S.; Jadav, S.S.; Babu, C.H.M.; Ahsan, M.J. Machine Learning in Drug Discovery: A Review. Artif. Intell. Rev. 2022, 55, 1947–1999. [Google Scholar] [CrossRef] [PubMed]
- Priya, S.; Tripathi, G.; Singh, D.B.; Jain, P.; Kumar, A. Machine learning approaches and their applications in drug discovery and design. Chem. Biol. Drug Des. 2022, 100, 136–153. [Google Scholar] [CrossRef]
- Guedes, I.A.; Barreto, A.M.S.; Marinho, D.; Krempser, E.; Kuenemann, M.A.; Sperandio, O.; Dardenne, L.E.; Miteva, M.A. New machine learning and physics-based scoring functions for drug discovery. Sci. Rep. 2021, 11, 3198. [Google Scholar] [CrossRef]
- Tamura, S.; Miyao, T.; Bajorath, J. Large-scale prediction of activity cliffs using machine and deep learning methods of increasing complexity. J. Cheminform. 2023, 15, 4. [Google Scholar] [CrossRef]
- Stumpfe, D.; Hu, H.B.; Bajorath, J. Advances in exploring activity cliffs. J. Comput. Aid. Mol. Des. 2020, 34, 929–942. [Google Scholar] [CrossRef]
- Heikamp, K.; Hu, X.Y.; Yan, A.X.; Bajorath, J. Prediction of Activity Cliffs Using Support Vector Machines. J. Chem. Inf. Model 2012, 52, 2354–2365. [Google Scholar] [CrossRef]
- Rodriguez-Perez, R.; Bajorath, J. Evolution of Support Vector Machine and Regression Modeling in Chemoinformatics and Drug Discovery. J. Comput. Aid. Mol. Des. 2022, 36, 355–362. [Google Scholar] [CrossRef]
- Warszycki, D.; Struski, L.; Smieja, M.; Kafel, R.; Kurczab, R. Pharmacoprint: A Combination of a Pharmacophore Fingerprint and Artificial Intelligence as a Tool for Computer-Aided Drug Design. J. Chem. Inf. Model 2021, 61, 5054–5065. [Google Scholar] [CrossRef]
- Jayaraj, P.B.; Jain, S. Ligand based virtual screening using SVM on GPU. Comput. Biol. Chem. 2019, 83, 107143. [Google Scholar] [CrossRef] [PubMed]
- Ogura, K.; Sato, T.; Tuki, H.; Honma, T. Support Vector Machine model for hERG inhibitory activities based on the integrated hERG database using descriptor selection by NSGA-II. Sci. Rep. 2019, 9, 12220. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Perez, R.; Vogt, M.; Bajorath, J. Support Vector Machine Classification and Regression Prioritize Different Structural Features for Binary Compound Activity and Potency Value Prediction. ACS Omega 2017, 2, 6371–6379. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Cruz, N.; Medina-Franco, J.L. Epigenetic Target Profiler: A Web Server to Predict Epigenetic Targets of Small Molecules. J. Chem. Inf. Model 2021, 61, 1550–1554. [Google Scholar] [CrossRef]
- Tong, Z.; Zhou, Y.; Wang, J. Identifying potential drug targets in hepatocellular carcinoma based on network analysis and one-class support vector machine. Sci. Rep. 2019, 9, 10442. [Google Scholar] [CrossRef]
- Kwon, S.; Bae, H.; Jo, J.; Yoon, S. Comprehensive ensemble in QSAR prediction for drug discovery. BMC Bioinform. 2019, 20, 521. [Google Scholar] [CrossRef]
- Hou, T.L.; Bian, Y.M.; McGuire, T.; Xie, X.Q. Integrated Multi-Class Classification and Prediction of GPCR Allosteric Modulators by Machine Learning Intelligence. Biomolecules 2021, 11, 870. [Google Scholar] [CrossRef]
- Kaiser, T.M.; Burger, P.B.; Butch, C.J.; Pelly, S.C.; Liotta, D.C. A Machine Learning Approach for Predicting HIV Reverse Transcriptase Mutation Susceptibility of Biologically Active Compounds. J. Chem. Inf. Model 2018, 58, 1544–1552. [Google Scholar] [CrossRef]
- Hu, J.; Zhou, L.W.; Li, B.; Zhang, X.L.; Chen, N.S. Improve hot region prediction by analyzing different machine learning algorithms. BMC Bioinform. 2021, 22, 522. [Google Scholar] [CrossRef]
- Celebi, R.; Uyar, H.; Yasar, E.; Gumus, O.; Dikenelli, O.; Dumontier, M. Evaluation of knowledge graph embedding approaches for drug-drug interaction prediction in realistic settings. BMC Bioinform. 2019, 20, 726. [Google Scholar] [CrossRef]
- Cai, C.P.; Guo, P.F.; Zhou, Y.D.; Zhou, J.W.; Wang, Q.; Zhang, F.X.; Fang, J.S.; Cheng, F.X. Deep Learning-Based Prediction of Drug-Induced Cardiotoxicity. J. Chem. Inf. Model 2019, 59, 1073–1084. [Google Scholar] [CrossRef] [PubMed]
- Madhukar, N.S.; Khade, P.K.; Huang, L.D.; Gayvert, K.; Galletti, G.; Stogniew, M.; Allen, J.E.; Giannakakou, P.; Elemento, O. A Bayesian machine learning approach for drug target identification using diverse data types. Nat. Commun. 2019, 10, 5221. [Google Scholar] [CrossRef]
- Saha, S.; Chatterjee, P.; Halder, A.K.; Nasipuri, M.; Basu, S.; Plewczynski, D. ML-DTD: Machine Learning-Based Drug Target Discovery for the Potential Treatment of COVID-19. Vaccines 2022, 10, 1643. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.K.A.; Malim, N.H.A.H. Comparative Studies on Resampling Techniques in Machine Learning and Deep Learning Models for Drug-Target Interaction Prediction. Molecules 2023, 28, 1663. [Google Scholar] [CrossRef] [PubMed]
- Carpenter, K.A.; Huang, X.D. Machine Learning-based Virtual Screening and Its Applications to Alzheimer’s Drug Discovery: A Review. Curr. Pharm. Des. 2018, 24, 3347–3358. [Google Scholar] [CrossRef] [PubMed]
- Zakharov, A.V.; Varlamova, E.V.; Lagunin, A.A.; Dmitriev, A.V.; Muratov, E.N.; Fourches, D.; Kuz’min, V.E.; Poroikov, V.V.; Tropsha, A.; Nicklaus, M.C. QSAR Modeling and Prediction of Drug-Drug Interactions. Mol. Pharm. 2016, 13, 545–556. [Google Scholar] [CrossRef] [PubMed]
- Chen, A.Y.; Lee, J.; Damjanovic, A.; Brooks, B.R. Protein pK(a) Prediction by Tree-Based Machine Learning. J. Chem. Theory Comput. 2022, 18, 2673–2686. [Google Scholar] [CrossRef]
- Cooper, K.; Baddeley, C.; French, B.; Gibson, K.; Golden, J.; Lee, T.; Pierre, S.; Weiss, B.; Yang, J. Novel Development of Predictive Feature Fingerprints to Identify Chemistry-Based Features for the Effective Drug Design of SARS-CoV-2 Target Antagonists and Inhibitors Using Machine Learning. ACS Omega 2021, 6, 4857–4877. [Google Scholar] [CrossRef]
- Brekkan, A.; Jonsson, S.; Karlsson, M.O.; Plan, E.L. Handling underlying discrete variables with bivariate mixed hidden Markov models in NONMEM. J. Pharmacokinet. Pharmacodyn. 2019, 46, 591–604. [Google Scholar] [CrossRef]
- Tamposis, I.A.; Tsirigos, K.D.; Theodoropoulou, M.C.; Kontou, P.I.; Bagos, P.G. Semi-supervised learning of Hidden Markov Models for biological sequence analysis. Bioinformatics 2019, 35, 2208–2215. [Google Scholar] [CrossRef]
- Steinegger, M.; Meier, M.; Mirdita, M.; Vohringer, H.; Haunsberger, S.J.; Soding, J. HH-suite3 for fast remote homology detection and deep protein annotation. BMC Bioinform. 2019, 20, 473. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, N.P.; Nute, M.; Mirarab, S.; Warnow, T. HIPPI: Highly accurate protein family classification with ensembles of HMMs. BMC Genom. 2016, 17, 765. [Google Scholar] [CrossRef] [PubMed]
- Li, J.F.; Lee, J.Y.; Liao, L. A new algorithm to train hidden Markov models for biological sequences with partial labels. BMC Bioinform. 2021, 22, 162. [Google Scholar] [CrossRef] [PubMed]
- Tamposis, I.A.; Tsirigos, K.D.; Theodoropoulou, M.C.; Kontou, P.I.; Tsaousis, G.N.; Sarantopoulou, D.; Litou, Z.I.; Bagos, P.G. JUCHMME: A Java Utility for Class Hidden Markov Models and Extensions for biological sequence analysis. Bioinformatics 2019, 35, 5309–5312. [Google Scholar] [CrossRef]
- Kaur, H.; Lynn, A.M. Mapping the FtsQBL divisome components in bacterial NTD pathogens as potential drug targets. Front. Genet. 2023, 13, 1010870. [Google Scholar] [CrossRef]
- Gupta, A.; Zhou, H.X. Machine Learning-Enabled Pipeline for Large-Scale Virtual Drug Screening. J. Chem. Inf. Model 2021, 61, 4236–4244. [Google Scholar] [CrossRef]
- David, L.; Thakkar, A.; Mercado, R.; Engkvist, O. Molecular representations in AI-driven drug discovery: A review and practical guide. J. Cheminform. 2020, 12, 56. [Google Scholar] [CrossRef]
- Madugula, S.S.; John, L.; Nagamani, S.; Gaur, A.S.; Poroikov, V.V.; Sastry, G.N. Molecular descriptor analysis of approved drugs using unsupervised learning for drug repurposing. Comput. Biol. Med. 2021, 138, 104856. [Google Scholar] [CrossRef]
- Huang, L.; Luo, H.M.; Li, S.N.; Wu, F.X.; Wang, J.X. Drug-drug similarity measure and its applications. Brief Bioinform. 2021, 22, bbaa265. [Google Scholar] [CrossRef]
- Nedyalkova, M.; Simeonov, V. Partitioning Pattern of Natural Products Based on Molecular Properties Descriptors Representing Drug-Likeness. Symmetry 2021, 13, 546. [Google Scholar] [CrossRef]
- McKay, K.; Hamilton, N.B.; Remington, J.M.; Schneebeli, S.T.; Li, J.N. Essential Dynamics Ensemble Docking for Structure-Based GPCR Drug Discovery. Front. Mol. Biosci. 2022, 9, 879212. [Google Scholar] [CrossRef]
- Chandak, T.; Mayginnes, J.P.; Mayes, H.; Wong, C.F. Using machine learning to improve ensemble docking for drug discovery. Proteins 2020, 88, 1263–1270. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.L.; Li, Q.; Sun, J.; Tan, S.H.; Tang, Y.H.; Zhao, M.M.; Li, Y.Y.; Cao, X.; Zhao, J.C.; Yang, J.K. Potential drug discovery for COVID-19 treatment targeting Cathepsin L using a deep learning-based strategy. Comput. Struct. Biotechnol. J. 2022, 20, 2442–2454. [Google Scholar] [CrossRef] [PubMed]
- Andronov, M.; Fedorov, M.V.; Sosnin, S. Exploring Chemical Reaction Space with Reaction Difference Fingerprints and Parametric t-SNE. ACS Omega 2021, 6, 30743–30751. [Google Scholar] [CrossRef]
- Thomas, M.; Smith, R.T.; O’Boyle, N.M.; de Graaf, C.; Bender, A. Comparison of structure- and ligand-based scoring functions for deep generative models: A GPCR case study. J. Cheminform. 2021, 13, 39. [Google Scholar] [CrossRef]
- Barnard, T.; Hagan, H.; Tseng, S.; Sosso, G.C. Less may be more: An informed reflection on molecular descriptors for drug design and discovery. Mol. Syst. Des. Eng. 2020, 5, 317–329. [Google Scholar] [CrossRef]
- Liu, G.N.; Singha, M.; Pu, L.M.; Neupane, P.; Feinstein, J.; Wu, H.C.; Ramanujam, J.; Brylinski, M. GraphDTI: A robust deep learning predictor of drug-target interactions from multiple heterogeneous data. J. Cheminform. 2021, 13, 58. [Google Scholar] [CrossRef]
- Xu, X.L.; Xie, Z.M.; Yang, Z.Y.; Li, D.F.; Xu, X.M. A t-SNE Based Classification Approach to Compositional Microbiome Data. Front. Genet. 2020, 11, 620143. [Google Scholar] [CrossRef]
- Karagiannaki, I.; Gourlia, K.; Lagani, V.; Pantazis, Y.; Tsamardinos, I. Learning biologically-interpretable latent representations for gene expression data. Mach. Learn. 2022, 1–31. [Google Scholar] [CrossRef]
- Zhang, L.P.; Tang, L.; Zhang, S.L.; Wang, Z.Z.; Shen, X.H.; Zhang, Z.Q. A Self-Adaptive Reinforcement-Exploration Q-Learning Algorithm. Symmetry 2021, 13, 1057. [Google Scholar] [CrossRef]
- Tang, B.W.; He, F.M.; Liu, D.P.; He, F.; Wu, T.; Fang, M.J.; Niu, Z.M.; Wu, Z.; Xu, D. AI-Aided Design of Novel Targeted Covalent Inhibitors against SARS-CoV-2. Biomolecules 2022, 12, 746. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.X.; Qian, Y.J.; Gao, H.Y.; Coley, C.W.; Mo, Y.M.; Barzilay, R.; Jensen, K.F. Towards efficient discovery of green synthetic pathways with Monte Carlo tree search and reinforcement learning. Chem. Sci. 2020, 11, 10959–10972. [Google Scholar] [CrossRef] [PubMed]
- Lee, G.; Jang, G.H.; Kang, H.Y.; Song, G. Predicting aptamer sequences that interact with target proteins using an aptamer-protein interaction classifier and a Monte Carlo tree search approach. PLoS ONE 2021, 16, e0253760. [Google Scholar] [CrossRef] [PubMed]
- Yoshizawa, T.; Ishida, S.; Sato, T.; Ohta, M.; Honma, T.; Terayama, K. Selective Inhibitor Design for Kinase Homologs Using Multiobjective Monte Carlo Tree Search. J. Chem. Inf. Model 2022, 62, 5351–5360. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.B.; Pei, J.F.; Lai, L.H. Structure-based de novo drug design using 3D deep generative models. Chem. Sci. 2021, 12, 13664–13675. [Google Scholar] [CrossRef] [PubMed]
- Genheden, S.; Thakkar, A.; Chadimova, V.; Reymond, J.L.; Engkvist, O.; Bjerrum, E. AiZynthFinder: A fast, robust and flexible open-source software for retrosynthetic planning. J. Cheminform. 2020, 12, 70. [Google Scholar] [CrossRef]
- Skalic, M.; Martinez-Rosell, G.; Jimenez, J.; De Fabritiis, G. PlayMolecule BindScope: Large scale CNN-based virtual screening on the web. Bioinformatics 2019, 35, 1237–1238. [Google Scholar] [CrossRef]
- Haneczok, J.; Delijewski, M. Machine learning enabled identification of potential SARS-CoV-2 3CLpro inhibitors based on fixed molecular fingerprints and Graph-CNN neural representations. J. Biomed. Inform. 2021, 119, 103821. [Google Scholar] [CrossRef]
- Huo, X.; Xu, J.; Xu, M.; Chen, H. An improved 3D quantitative structure-activity relationships (QSAR) of molecules with CNN-based partial least squares model. Artif. Intell. Life Sci. 2023, 3, 100065. [Google Scholar] [CrossRef]
- Qian, Y.; Wu, J.; Zhang, Q. CAT-CPI: Combining CNN and transformer to learn compound image features for predicting compound-protein interactions. Front. Mol. Biosci. 2022, 9, 963912. [Google Scholar] [CrossRef]
- Jiang, M.J.; Wei, Z.Q.; Zhang, S.G.; Wang, S.; Wang, X.F.; Li, Z. FRSite: Protein drug binding site prediction based on faster R-CNN. J. Mol. Graph. Model. 2019, 93, 107454. [Google Scholar] [CrossRef] [PubMed]
- Hirohara, M.; Saito, Y.; Koda, Y.; Sato, K.; Sakakibara, Y. Convolutional neural network based on SMILES representation of compounds for detecting chemical motif. BMC Bioinform. 2018, 19, 526. [Google Scholar] [CrossRef] [PubMed]
- Hu, P.W.; Zou, J.P.; Yu, J.L.; Shi, S.P. De novo drug design based on Stack-RNN with multi-objective reward-weighted sum and reinforcement learning. J. Mol. Model. 2023, 29, 121. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.N.; Yang, L.J.; Ding, N.; Li, G.W.; Cai, J.J.; An, X.L.; Wang, Z.J.; Qin, J.; Niu, Y.Z. Recurrent neural network (RNN) model accelerates the development of antibacterial metronidazole derivatives. RSC Adv. 2022, 12, 22893–22901. [Google Scholar] [CrossRef]
- Lim, J.; Ryu, S.; Park, K.; Choe, Y.J.; Ham, J.; Kim, W.Y. Predicting Drug-Target Interaction Using a Novel Graph Neural Network with 3D Structure-Embedded Graph Representation. J. Chem. Inf. Model. 2019, 59, 3981–3988. [Google Scholar] [CrossRef]
- Saldivar-Gonzalez, F.I.; Aldas-Bulos, V.D.; Medina-Franco, J.L.; Plisson, F. Natural product drug discovery in the artificial intelligence era. Chem. Sci. 2022, 13, 1526–1546. [Google Scholar] [CrossRef]
- Yang, H.; Hu, B.; Pan, X.; Yan, S.; Feng, Y.; Zhang, X.; Yin, L.; Hu, C. Deep belief network-based drug identification using near infrared spectroscopy. J. Innov. Opt. Health Sci. 2017, 10, 1630011. [Google Scholar] [CrossRef]
- Griffiths, R.R.; Hernandez-Lobato, J.M. Constrained Bayesian optimization for automatic chemical design using variational autoencoders. Chem. Sci. 2020, 11, 577–586. [Google Scholar] [CrossRef]
- Sajadi, S.Z.; Chahooki, M.A.Z.; Gharaghani, S.; Abbasi, K. AutoDTI plus plus: Deep unsupervised learning for DTI prediction by autoencoders. BMC Bioinform. 2021, 22, 204. [Google Scholar] [CrossRef]
- Zhang, Y.; Hu, Y.Q.; Li, H.H.; Liu, X.Y. Drug-protein interaction prediction via variational autoencoders and attention mechanisms. Front. Genet. 2022, 13, 1032779. [Google Scholar] [CrossRef]
- Song, T.; Ren, Y.Q.; Wang, S.; Han, P.F.; Wang, L.L.; Li, X.; Rodriguez-Paton, A. DNMG: Deep molecular generative model by fusion of 3D information for de novo drug design. Methods 2023, 211, 10–22. [Google Scholar] [CrossRef]
- Hussain, S.; Anees, A.; Das, A.; Nguyen, B.P.; Marzuki, M.; Lin, S.P.; Wright, G.; Singhal, A. High-content image generation for drug discovery using generative adversarial networks. Neural Netw. 2020, 132, 353–363. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Li, K.; Shi, J. DGANDDI: Double Generative Adversarial Networks for Drug-Drug Interaction Prediction. IEEE/ACM Trans. Comput. Biol. Bioinform. 2023, 20, 1854–1863. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.L.; Wang, J.J.; Pang, L.; Liu, Y.; Zhang, J. GANsDTA: Predicting Drug-Target Binding Affinity Using GANs. Front. Genet. 2020, 10, 1243. [Google Scholar] [CrossRef] [PubMed]
- Hicks, S.A.; Strumke, I.; Thambawita, V.; Hammou, M.; Riegler, M.A.; Halvorsen, P.; Parasa, S. On evaluation metrics for medical applications of artificial intelligence. Sci. Rep. 2022, 12, 5979. [Google Scholar] [CrossRef]
- Kim, S.; Chen, J.; Cheng, T.J.; Gindulyte, A.; He, J.; He, S.Q.; Li, Q.L.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem 2023 update. Nucleic Acids Res. 2022, 51, D1373–D1380. [Google Scholar] [CrossRef]
- Mendez, D.; Gaulton, A.; Bento, A.P.; Chambers, J.; De Veij, M.; Felix, E.; Magarinos, M.P.; Mosquera, J.F.; Mutowo, P.; Nowotka, M.; et al. ChEMBL: Towards direct deposition of bioassay data. Nucleic Acids Res. 2019, 47, D930–D940. [Google Scholar] [CrossRef]
- Irwin, J.J.; Tang, K.G.; Young, J.; Dandarchuluun, C.; Wong, B.R.; Khurelbaatar, M.; Moroz, Y.S.; Mayfield, J.; Sayle, R.A. ZINC20-A Free Ultralarge-Scale Chemical Database for Ligand Discovery. J. Chem. Inf. Model. 2020, 60, 6065–6073. [Google Scholar] [CrossRef]
- Pence, H.E.; Williams, A. ChemSpider: An Online Chemical Information Resource. J. Chem. Educ. 2010, 87, 1123–1124. [Google Scholar] [CrossRef]
- Wishart, D.S.; Feunang, Y.D.; Guo, A.C.; Lo, E.J.; Marcu, A.; Grant, J.R.; Sajed, T.; Johnson, D.; Li, C.; Sayeeda, Z.; et al. DrugBank 5.0: A major update to the DrugBank database for 2018. Nucleic Acids Res. 2018, 46, D1074–D1082. [Google Scholar] [CrossRef]
- Avram, S.; Wilson, T.B.; Curpan, R.; Halip, L.; Borota, A.; Bora, A.; Bologa, C.G.; Holmes, J.; Knockel, J.; Yang, J.J.; et al. DrugCentral 2023 extends human clinical data and integrates veterinary drugs. Nucleic Acids Res. 2022, 51, D1276–D1287. [Google Scholar] [CrossRef] [PubMed]
- Drugs@FDA: FDA-Approved Drugs. Available online: https://www.accessdata.fda.gov/scripts/cder/daf/ (accessed on 22 July 2023).
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Karp, P.D.; Billington, R.; Caspi, R.; Fulcher, C.A.; Latendresse, M.; Kothari, A.; Keseler, I.M.; Krummenacker, M.; Midford, P.E.; Ong, Q.; et al. The BioCyc collection of microbial genomes and metabolic pathways. Brief Bioinform. 2019, 20, 1085–1093. [Google Scholar] [CrossRef] [PubMed]
- Gillespie, M.; Jassal, B.; Stephan, R.; Milacic, M.; Rothfels, K.; Senff-Ribeiro, A.; Griss, J.; Sevilla, C.; Matthews, L.; Gong, C.Q.; et al. The reactome pathway knowledgebase 2022. Nucleic Acids Res. 2022, 50, D687–D692. [Google Scholar] [CrossRef]
- Wishart, D.S.; Guo, A.C.; Oler, E.; Wang, F.; Anjum, A.; Peters, H.; Dizon, R.; Sayeeda, Z.; Tian, S.Y.; Lee, B.L.; et al. HMDB 5.0: The Human Metabolome Database for 2022. Nucleic Acids Res. 2022, 50, D622–D631. [Google Scholar] [CrossRef]
- Orchard, S.; Ammari, M.; Aranda, B.; Breuza, L.; Briganti, L.; Broackes-Carter, F.; Campbell, N.H.; Chavali, G.; Chen, C.; del-Toro, N.; et al. The MIntAct project-IntAct as a common curation platform for 11 molecular interaction databases. Nucleic Acids Res. 2014, 42, D358–D363. [Google Scholar] [CrossRef]
- Oughtred, R.; Rust, J.; Chang, C.; Breitkreutz, B.J.; Stark, C.; Willems, A.; Boucher, L.; Leung, G.; Kolas, N.; Zhang, F.; et al. The BioGRID database: A comprehensive biomedical resource of curated protein, genetic, and chemical interactions. Protein Sci. 2021, 30, 187–200. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2022, 51, D638–D646. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Santos, A.; von Mering, C.; Jensen, L.J.; Bork, P.; Kuhn, M. STITCH 5: Augmenting protein-chemical interaction networks with tissue and affinity data. Nucleic Acids Res. 2016, 44, D380–D384. [Google Scholar] [CrossRef]
- Gilson, M.K.; Liu, T.Q.; Baitaluk, M.; Nicola, G.; Hwang, L.; Chong, J. BindingDB in 2015: A public database for medicinal chemistry, computational chemistry and systems pharmacology. Nucleic Acids Res. 2016, 44, D1045–D1053. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhang, Y.T.; Lian, X.C.; Li, F.C.; Wang, C.X.; Zhu, F.; Qiu, Y.Q.; Chen, Y.Z. Therapeutic target database update 2022: Facilitating drug discovery with enriched comparative data of targeted agents. Nucleic Acids Res. 2022, 50, D1398–D1407. [Google Scholar] [CrossRef]
- Harding, S.D.; Armstrong, J.F.; Faccenda, E.; Southan, C.; Alexander, S.P.H.; Davenport, A.P.; Pawson, A.J.; Spedding, M.; Davies, J.A.; NC-IUPHAR. The IUPHAR/BPS guide to PHARMACOLOGY in 2022: Curating pharmacology for COVID-19, malaria and antibacterials. Nucleic Acids Res. 2022, 50, D1282–D1294. [Google Scholar] [CrossRef] [PubMed]
- Freshour, S.L.; Kiwala, S.; Cotto, K.C.; Coffman, A.C.; McMichael, J.F.; Song, J.J.; Griffith, M.; Griffith, O.L.; Wagner, A.H. Integration of the Drug-Gene Interaction Database (DGIdb 4.0) with open crowdsource efforts. Nucleic Acids Res. 2021, 49, D1144–D1151. [Google Scholar] [CrossRef] [PubMed]
- Davis, A.P.; Wiegers, T.C.; Johnson, R.J.; Sciaky, D.; Wiegers, J.; Mattingly, C.J. Comparative Toxicogenomics Database (CTD): Update 2023. Nucleic Acids Res. 2022, 51, D1257–D1262. [Google Scholar] [CrossRef] [PubMed]
- Ganter, B.; Snyder, R.D.; Halbert, D.N.; Lee, M.D. Toxicogenomics in drug discovery and development: Mechanistic analysis of compound/class-dependent effects using the DrugMatrix® database. Pharmacogenomics 2006, 7, 1025–1044. [Google Scholar] [CrossRef] [PubMed]
- OECD eChemPortal. Available online: https://www.echemportal.org/echemportal/ (accessed on 25 July 2023).
- Kuhn, M.; Letunic, I.; Jensen, L.J.; Bork, P. The SIDER database of drugs and side effects. Nucleic Acids Res. 2016, 44, D1075–D1079. [Google Scholar] [CrossRef] [PubMed]
- Bateman, A.; Martin, M.J.; Orchard, S.; Magrane, M.; Ahmad, S.; Alpi, E.; Bowler-Barnett, E.H.; Britto, R.; Cukura, A.; Denny, P.; et al. UniProt: The Universal Protein Knowledgebase in 2023. Nucleic Acids Res. 2022, 51, D523–D531. [Google Scholar] [CrossRef]
- Paysan-Lafosse, T.; Blum, M.; Chuguransky, S.; Grego, T.; Pinto, B.L.; Salazar, G.A.; Bileschi, M.L.; Bork, P.; Bridge, A.; Colwell, L.; et al. InterPro in 2022. Nucleic Acids Res. 2022, 51, D418–D427. [Google Scholar] [CrossRef]
- Benson, D.A.; Cavanaugh, M.; Clark, K.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Sayers, E.W. GenBank. Nucleic Acids Res. 2012, 41, D36–D42. [Google Scholar] [CrossRef]
- Burley, S.K.; Bhikadiya, C.; Bi, C.X.; Bittrich, S.; Chao, H.Y.; Chen, L.; Craig, P.A.; Crichlow, G.V.; Dalenberg, K.; Duarte, J.M.; et al. RCSB Protein Data Bank (RCSB.org): Delivery of experimentally-determined PDB structures alongside one million computed structure models of proteins from artificial intelligence/machine learning. Nucleic Acids Res. 2022, 51, D488–D508. [Google Scholar] [CrossRef]
- Feng, Z.K.; Chen, L.; Maddula, H.; Akcan, O.; Oughtred, R.; Berman, H.M.; Westbrook, J. Ligand Depot: A data warehouse for ligands bound to macromolecules. Bioinformatics 2004, 20, 2153–2155. [Google Scholar] [CrossRef] [PubMed]
- Keenan, A.B.; Jenkins, S.L.; Jagodnik, K.M.; Koplev, S.; He, E.; Torre, D.; Wang, Z.C.; Dohlman, A.B.; Silverstein, M.C.; Lachmann, A.; et al. The Library of Integrated Network-Based Cellular Signatures NIH Program: System-Level Cataloging of Human Cells Response to Perturbations. Cell Syst. 2018, 6, 13–24. [Google Scholar] [CrossRef]
- Chang, A.; Jeske, L.; Ulbrich, S.; Hofmann, J.; Koblitz, J.; Schomburg, I.; Neumann-Schaal, M.; Jahn, D.; Schomburg, D. BRENDA, the ELIXIR core data resource in 2021: New developments and updates. Nucleic Acids Res. 2021, 49, D498–D508. [Google Scholar] [CrossRef] [PubMed]
- Sorokina, M.; Merseburger, P.; Rajan, K.; Yirik, M.A.; Steinbeck, C. COCONUT online: Collection of Open Natural Products database. J. Cheminform. 2021, 13, 2. [Google Scholar] [CrossRef]
- Landaburu, L.U.; Berenstein, A.J.; Videla, S.; Maru, P.; Shanmugam, D.; Chernomoretz, A.; Aguero, F. TDR Targets 6: Driving drug discovery for human pathogens through intensive chemogenomic data integration. Nucleic Acids Res. 2020, 48, D992–D1005. [Google Scholar] [CrossRef]
- Bryant, P. Deep learning for protein complex structure prediction. Curr. Opin. Struct. Biol. 2023, 79, 102529. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Zidek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Ivankov, D.N.; Finkelstein, A.V. Solution of Levinthal’s Paradox and a Physical Theory of Protein Folding Times. Biomolecules 2020, 10, 250. [Google Scholar] [CrossRef]
- Rose, G.D. Protein folding—Seeing is deceiving. Protein Sci. 2021, 30, 1606–1616. [Google Scholar] [CrossRef]
- Sorokina, I.; Mushegian, A.R.; Koonin, E.V. Is Protein Folding a Thermodynamically Unfavorable, Active, Energy-Dependent Process? Int. J. Mol. Sci. 2022, 23, 521. [Google Scholar] [CrossRef]
- Muhammed, M.T.; Aki-Yalcin, E. Homology modeling in drug discovery: Overview, current applications, and future perspectives. Chem. Biol. Drug Des. 2019, 93, 12–20. [Google Scholar] [CrossRef] [PubMed]
- Burley, S.K.; Berman, H.M.; Duarte, J.M.; Feng, Z.K.; Flatt, J.W.; Hudson, B.P.; Lowe, R.; Peisach, E.; Piehl, D.W.; Rose, Y.; et al. Protein Data Bank: A Comprehensive Review of 3D Structure Holdings and Worldwide Utilization by Researchers, Educators, and Students. Biomolecules 2022, 12, 1425. [Google Scholar] [CrossRef] [PubMed]
- Burley, S.K.; Bhikadiya, C.; Bi, C.X.; Bittrich, S.; Chen, L.; Crichlow, G.V.; Duarte, J.M.; Dutta, S.; Fayazi, M.; Feng, Z.K.; et al. RCSB Protein Data Bank: Celebrating 50 years of the PDB with new tools for understanding and visualizing biological macromolecules in 3D. Protein Sci. 2022, 31, 187–208. [Google Scholar] [CrossRef]
- Sali, A.; Blundell, T.L. Comparative Protein Modeling by Satisfaction of Spatial Restraints. J. Mol. Biol. 1993, 234, 779–815. [Google Scholar] [CrossRef]
- Webb, B.; Sali, A. Protein Structure Modeling with MODELLER. Methods Mol. Biol. 2021, 2199, 239–255. [Google Scholar] [CrossRef]
- Studer, G.; Tauriello, G.; Bienert, S.; Biasini, M.; Johner, N.; Schwede, T. ProMod3—A versatile homology modelling toolbox. PLoS Comput. Biol. 2021, 17, e1008667. [Google Scholar] [CrossRef]
- Studer, G.; Rempfer, C.; Waterhouse, A.M.; Gumienny, R.; Haas, J.; Schwede, T. QMEANDisCo-distance constraints applied on model quality estimation. Bioinformatics 2020, 36, 1765–1771. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Zhang, Y. I-TASSER server for protein 3D structure prediction. BMC Bioinform. 2008, 9, 40. [Google Scholar] [CrossRef] [PubMed]
- Roy, A.; Kucukural, A.; Zhang, Y. I-TASSER: A unified platform for automated protein structure and function prediction. Nat. Protoc. 2010, 5, 725–738. [Google Scholar] [CrossRef]
- Kryshtafovych, A.; Monastyrskyy, B.; Fidelis, K.; Moult, J.; Schwede, T.; Tramontano, A. Evaluation of the template-based modeling in CASP12. Proteins 2018, 86, 321–334. [Google Scholar] [CrossRef]
- Zheng, W.; Li, Y.; Zhang, C.X.; Zhou, X.G.; Pearce, R.; Bell, E.W.; Huang, X.Q.; Zhang, Y. Protein structure prediction using deep learning distance and hydrogen-bonding restraints in CASP14. Proteins 2021, 89, 1734–1751. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Zidek, A.; Potapenko, A.; et al. Applying and improving AlphaFold at CASP14. Proteins 2021, 89, 1711–1721. [Google Scholar] [CrossRef]
- Senior, A.W.; Evans, R.; Jumper, J.; Kirkpatrick, J.; Sifre, L.; Green, T.; Qin, C.L.; Zidek, A.; Nelson, A.W.R.; Bridgland, A.; et al. Improved protein structure prediction using potentials from deep learning. Nature 2020, 577, 706–710. [Google Scholar] [CrossRef] [PubMed]
- Anishchenko, I.; Baek, M.; Park, H.; Hiranuma, N.; Kim, D.E.; Dauparas, J.; Mansoor, S.; Humphreys, I.R.; Baker, D. Protein tertiary structure prediction and refinement using deep learning and Rosetta in CASP14. Proteins 2021, 89, 1722–1733. [Google Scholar] [CrossRef] [PubMed]
- Evans, R.; O’Neill, M.; Pritzel, A.; Antropova, N.; Senior, A.; Green, T.; Žídek, A.; Bates, R.; Blackwell, S.; Yim, J.; et al. Protein complex prediction with AlphaFold-Multimer. bioRxiv 2022. bioRxiv:10.1101/2021.10.04.463034. [Google Scholar]
- Gao, M.; An, D.N.; Parks, J.M.; Skolnick, J. AF2Complex predicts direct physical interactions in multimeric proteins with deep learning. Nat. Commun. 2022, 13, 1744. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.M.; Akin, H.; Rao, R.S.; Hie, B.; Zhu, Z.K.; Lu, W.T.; Smetanin, N.; Verkuil, R.; Kabeli, O.; Shmueli, Y.; et al. Evolutionary-scale prediction of atomic-level protein structure with a language model. Science 2023, 379, 1123–1130. [Google Scholar] [CrossRef]
- Azzaz, F.; Yahi, N.; Chahinian, H.; Fantini, J. The Epigenetic Dimension of Protein Structure Is an Intrinsic Weakness of the AlphaFold Program. Biomolecules 2022, 12, 1527. [Google Scholar] [CrossRef]
- Tourlet, S.; Radjasandirane, R.; Diharce, J.; de Brevern, A.G. AlphaFold2 Update and Perspectives. BioMedInformatics 2023, 3, 378–390. [Google Scholar] [CrossRef]
- Sciacca, M.F.; Lolicato, F.; Tempra, C.; Scollo, F.; Sahoo, B.R.; Watson, M.D.; Garcia-Vinuales, S.; Milardi, D.; Raudino, A.; Lee, J.C.; et al. Lipid-Chaperone Hypothesis: A Common Molecular Mechanism of Membrane Disruption by Intrinsically Disordered Proteins. ACS Chem. Neurosci. 2020, 11, 4336–4350. [Google Scholar] [CrossRef] [PubMed]
- Fantini, J. How sphingolipids bind and shape proteins: Molecular basis of lipid-protein interactions in lipid shells, rafts and related biomembrane domains. Cell. Mol. Life Sci. 2003, 60, 1027–1032. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.; Su, B.H.; Tseng, Y.J. Comparative studies of AlphaFold, RoseTTAFold and Modeller: A case study involving the use of G-protein-coupled receptors. Brief Bioinform. 2022, 23, bbac308. [Google Scholar] [CrossRef]
- Tovar, A.; Eckert, H.; Bajorath, J. Comparison of 2D fingerprint methods for multiple-template similarity searching on compound activity classes of increasing structural diversity. ChemMedChem 2007, 2, 208–217. [Google Scholar] [CrossRef] [PubMed]
- Carracedo-Reboredo, P.; Linares-Blanco, J.; Rodriguez-Fernandez, N.; Cedron, F.; Novoa, F.J.; Carballal, A.; Maojo, V.; Pazos, A.; Fernandez-Lozano, C. A review on machine learning approaches and trends in drug discovery. Comput. Struct. Biotech. J. 2021, 19, 4538–4558. [Google Scholar] [CrossRef]
- Ding, Y.; Chen, M.C.; Guo, C.; Zhang, P.; Wang, J.W. Molecular fingerprint-based machine learning assisted QSAR model development for prediction of ionic liquid properties. J. Mol. Liq. 2021, 326, 115212. [Google Scholar] [CrossRef]
- Riniker, S.; Landrum, G.A. Open-source platform to benchmark fingerprints for ligand-based virtual screening. J. Cheminform. 2013, 5, 26. [Google Scholar] [CrossRef]
- Cereto-Massague, A.; Ojeda, M.J.; Valls, C.; Mulero, M.; Garcia-Vallve, S.; Pujadas, G. Molecular fingerprint similarity search in virtual screening. Methods 2015, 71, 58–63. [Google Scholar] [CrossRef]
- Gao, K.F.; Nguyen, D.D.; Sresht, V.; Mathiowetz, A.M.; Tu, M.H.; Wei, G.W. Are 2D fingerprints still valuable for drug discovery? Phys. Chem. Chem. Phys. 2020, 22, 8373–8390. [Google Scholar] [CrossRef]
- Rogers, D.; Hahn, M. Extended-Connectivity Fingerprints. J. Chem. Inf. Model. 2010, 50, 742–754. [Google Scholar] [CrossRef]
- McGregor, M.J.; Muskal, S.M. Pharmacophore fingerprinting. 1. Application to QSAR and focused library design. J. Chem. Inf. Comput. Sci. 1999, 39, 569–574. [Google Scholar] [CrossRef]
- Schwartz, J.; Awale, M.; Reymond, J.L. SMIfp (SMILES fingerprint) Chemical Space for Virtual Screening and Visualization of Large Databases of Organic Molecules. J. Chem. Inf. Model. 2013, 53, 1979–1989. [Google Scholar] [CrossRef] [PubMed]
- Awale, M.; Jin, X.; Reymond, J.L. Stereoselective virtual screening of the ZINC database using atom pair 3D-fingerprints. J. Cheminform. 2015, 7, 3. [Google Scholar] [CrossRef] [PubMed]
- Da, C.; Kireev, D. Structural Protein-Ligand Interaction Fingerprints (SPLIF) for Structure-Based Virtual Screening: Method and Benchmark Study. J. Chem. Inf. Model. 2014, 54, 2555–2561. [Google Scholar] [CrossRef] [PubMed]
- Durant, J.L.; Leland, B.A.; Henry, D.R.; Nourse, J.G. Reoptimization of MDL keys for use in drug discovery. J. Chem. Inf. Comput. Sci. 2002, 42, 1273–1280. [Google Scholar] [CrossRef]
- PubChem Substructure Fingerprint. Available online: https://ftp.ncbi.nlm.nih.gov/pubchem/specifications/pubchem_fingerprints.pdf (accessed on 22 July 2023).
- Carhart, R.E.; Smith, D.H.; Venkataraghavan, R. Atom Pairs as Molecular-Features in Structure Activity Studies—Definition and Applications. J. Chem. Inf. Comput. Sci. 1985, 25, 64–73. [Google Scholar] [CrossRef]
- Bender, A.; Mussa, H.Y.; Glen, R.C.; Reiling, S. Similarity searching of chemical databases using atom environment descriptors (MOLPRINT 2D): Evaluation of performance. J. Chem. Inf. Comput. Sci. 2004, 44, 1708–1718. [Google Scholar] [CrossRef]
- Schneider, P.; Schneider, G. De Novo Design at the Edge of Chaos. J. Med. Chem. 2016, 59, 4077–4086. [Google Scholar] [CrossRef]
- Mouchlis, V.D.; Afantitis, A.; Serra, A.; Fratello, M.; Papadiamantis, A.G.; Aidinis, V.; Lynch, I.; Greco, D.; Melagraki, G. Advances in De Novo Drug Design: From Conventional to Machine Learning Methods. Int. J. Mol. Sci. 2021, 22, 1676. [Google Scholar] [CrossRef]
- Girin, L.; Leglaive, S.; Bie, X.Y.; Diard, J.; Hueber, T.; Alameda-Pineda, X. Dynamical Variational Autoencoders: A Comprehensive Review. Found. Trends Mach. Learn. 2021, 15, 1–175. [Google Scholar] [CrossRef]
- Prykhodko, O.; Johansson, S.V.; Kotsias, P.C.; Arus-Pous, J.; Bjerrum, E.J.; Engkvist, O.; Chen, H.M. A de novo molecular generation method using latent vector based generative adversarial network. J. Cheminform. 2019, 11, 74. [Google Scholar] [CrossRef] [PubMed]
- Sachdev, K.; Gupta, M.K. A comprehensive review of feature based methods for drug target interaction prediction. J. Biomed. Inform. 2019, 93, 103159. [Google Scholar] [CrossRef]
- Dhakal, A.; McKay, C.; Tanner, J.J.; Cheng, J.L. Artificial intelligence in the prediction of protein-ligand interactions: Recent advances and future directions. Brief Bioinform. 2022, 23, bbab476. [Google Scholar] [CrossRef] [PubMed]
- Bissantz, C.; Kuhn, B.; Stahl, M. A Medicinal Chemist’s Guide to Molecular Interactions. J. Med. Chem. 2010, 53, 6241. [Google Scholar] [CrossRef]
- Chen, D.L.; Oezguen, N.; Urvil, P.; Ferguson, C.; Dann, S.M.; Savidge, T.C. Regulation of protein-ligand binding affinity by hydrogen bond pairing. Sci. Adv. 2016, 2, e1501240. [Google Scholar] [CrossRef] [PubMed]
- Anusuya, S.; Kesherwani, M.; Priya, K.V.; Vimala, A.; Shanmugam, G.; Velmurugan, D.; Gromiha, M.M. Drug-Target Interactions: Prediction Methods and Applications. Curr. Protein Pept. Sci. 2018, 19, 537–561. [Google Scholar] [CrossRef]
- Chen, X.; Yan, C.C.; Zhang, X.T.; Zhang, X.; Dai, F.; Yin, J.; Zhang, Y.D. Drug-target interaction prediction: Databases, web servers and computational models. Brief Bioinform. 2016, 17, 696–712. [Google Scholar] [CrossRef]
- Bagherian, M.; Sabeti, E.; Wang, K.; Sartor, M.A.; Nikolovska-Coleska, Z.; Najarian, K. Machine learning approaches and databases for prediction of drug-target interaction: A survey paper. Brief Bioinform. 2021, 22, 247–269. [Google Scholar] [CrossRef]
- Xu, L.; Ru, X.Q.; Song, R. Application of Machine Learning for Drug-Target Interaction Prediction. Front. Genet. 2021, 12, 680117. [Google Scholar] [CrossRef]
- Yamanishi, Y.; Araki, M.; Gutteridge, A.; Honda, W.; Kanehisa, M. Prediction of drug-target interaction networks from the integration of chemical and genomic spaces. Bioinformatics 2008, 24, I232–I240. [Google Scholar] [CrossRef]
- Thafar, M.A.; Olayan, R.S.; Ashoor, H.; Albaradei, S.; Bajic, V.B.; Gao, X.; Gojobori, T.; Essack, M. DTiGEMS plus: Drug-target interaction prediction using graph embedding, graph mining, and similarity-based techniques. J. Cheminform. 2020, 12, 44. [Google Scholar] [CrossRef]
- Mysinger, M.M.; Carchia, M.; Irwin, J.J.; Shoichet, B.K. Directory of Useful Decoys, Enhanced (DUD-E): Better Ligands and Decoys for Better Benchmarking. J. Med. Chem. 2012, 55, 6582–6594. [Google Scholar] [CrossRef]
- Liu, H.; Sun, J.J.; Guan, J.H.; Zheng, J.; Zhou, S.G. Improving compound-protein interaction prediction by building up highly credible negative samples. Bioinformatics 2015, 31, 221–229. [Google Scholar] [CrossRef]
- Tsubaki, M.; Tomii, K.; Sese, J. Compound-protein interaction prediction with end-to-end learning of Neural Netw. for graphs and sequences. Bioinformatics 2019, 35, 309–318. [Google Scholar] [CrossRef]
- Wang, S.Y.; Shan, P.; Zhao, Y.L.; Zuo, L. GanDTI: A multi-task neural network for drug-target interaction prediction. Comput. Biol. Chem. 2021, 92, 107476. [Google Scholar] [CrossRef]
- Hecker, N.; Ahmed, J.; Eichborn, J.; Dunkel, M.; Macha, K.; Eckert, A.; Gilson, M.K.; Bourne, P.E.; Preissner, R. SuperTarget goes quantitative: Update on drug-target interactions. Nucleic Acids Res. 2012, 40, D1113–D1117. [Google Scholar] [CrossRef]
- Ding, Y.J.; Tang, J.J.; Guo, F.; Zou, Q. Identification of drug-target interactions via multiple kernel-based triple collaborative matrix factorization. Brief Bioinform. 2022, 23, bbab582. [Google Scholar] [CrossRef] [PubMed]
- Zitnik, M.; Sosic, R.; Leskovec, J. BioSNAP Datasets: Stanford Biomedical Network Dataset Collection. Available online: http://snap.stanford.edu/biodata/ (accessed on 25 July 2023).
- Davis, M.I.; Hunt, J.P.; Herrgard, S.; Ciceri, P.; Wodicka, L.M.; Pallares, G.; Hocker, M.; Treiber, D.K.; Zarrinkar, P.P. Comprehensive analysis of kinase inhibitor selectivity. Nat. Biotechnol. 2011, 29, 1046–1051. [Google Scholar] [CrossRef] [PubMed]
- Song, T.; Zhang, X.D.; Ding, M.; Rodriguez-Paton, A.; Wang, S.D.; Wang, G. DeepFusion: A deep learning based multi-scale feature fusion method for predicting drug-target interactions. Methods 2022, 204, 269–277. [Google Scholar] [CrossRef] [PubMed]
- Yazdani-Jahromi, M.; Yousefi, N.; Tayebi, A.; Kolanthai, E.; Neal, C.J.; Seal, S.; Garibay, O.O. AttentionSiteDTI: An interpretable graph-based model for drug-target interaction prediction using NLP sentence-level relation classification. Brief Bioinform. 2022, 23, bbac272. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Zhang, Z.Q.; Guan, J.H.; Zhou, S.G. Effective drug-target interaction prediction with mutual interaction neural network. Bioinformatics 2022, 38, 3582–3589. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.; Zhu, C.; Zhong, F.; Liu, L. MDTips: A multimodal-data-based drug-target interaction prediction system fusing knowledge, gene expression profile, and structural data. Bioinformatics 2023, 39, btad411. [Google Scholar] [CrossRef] [PubMed]
- Richard, A.M.; Huang, R.L.; Waidyanatha, S.; Shinn, P.; Collins, B.J.; Thillainadarajah, I.; Grulke, C.M.; Williams, A.J.; Lougee, R.R.; Judson, R.S.; et al. The Tox21 10K Compound Library: Collaborative Chemistry Advancing Toxicology. Chem. Res. Toxicol. 2021, 34, 189–216. [Google Scholar] [CrossRef] [PubMed]
- Thomas, R.S.; Paules, R.S.; Simeonov, A.; Fitzpatrick, S.C.; Crofton, K.M.; Casey, W.M.; Mendrick, D.L. The US Federal Tox21 Program: A Strategic and Operational Plan for Continued Leadership. Altex 2018, 35, 163–168. [Google Scholar] [CrossRef] [PubMed]
- Mayr, A.; Klambauer, G.; Unterthiner, T.; Hochreiter, S. DeepTox: Toxicity Prediction using Deep Learning. Front. Environ. Sci. 2016, 3, 80. [Google Scholar] [CrossRef]
- Richard, A.M.; Judson, R.S.; Houck, K.A.; Grulke, C.M.; Volarath, P.; Thillainadarajah, I.; Yang, C.H.; Rathman, J.; Martin, M.T.; Wambaugh, J.F.; et al. ToxCast Chemical Landscape: Paving the Road to 21st Century Toxicology. Chem. Res. Toxicol. 2016, 29, 1225–1251. [Google Scholar] [CrossRef]
- Dix, D.J.; Houck, K.A.; Martin, M.T.; Richard, A.M.; Setzer, R.W.; Kavlock, R.J. The ToxCast program for prioritizing toxicity testing of environmental chemicals. Toxicol. Sci. 2007, 95, 5–12. [Google Scholar] [CrossRef]
- Duran-Iturbide, N.A.; Diaz-Eufracio, B.I.; Medina-Franco, J.L. In Silico ADME/Tox Profiling of Natural Products: A Focus on BIOFACQUIM. ACS Omega 2020, 5, 16076–16084. [Google Scholar] [CrossRef]
- Negus, S.S.; Banks, M.L. Pharmacokinetic-Pharmacodynamic (PKPD) Analysis with Drug Discrimination. Curr. Top Behav. Neurosci. 2018, 39, 245–259. [Google Scholar] [CrossRef]
- Maltarollo, V.G.; Gertrudes, J.C.; Oliveira, P.R.; Honorio, K.M. Applying machine learning techniques for ADME-Tox prediction: A review. Expert Opin. Drug Met. 2015, 11, 259–271. [Google Scholar] [CrossRef]
- Wenzel, J.; Matter, H.; Schmidt, F. Predictive Multitask Deep Neural Network Models for ADME-Tox Properties: Learning from Large Data Sets. J. Chem. Inf. Model. 2019, 59, 1253–1268. [Google Scholar] [CrossRef] [PubMed]
- Almazroo, O.A.; Miah, M.K.; Venkataramanan, R. Drug Metabolism in the Liver. Clin. Liver Dis. 2017, 21, 1–20. [Google Scholar] [CrossRef]
- Xu, Z.Y.; Kang, Q.J.; Yu, Z.H.; Tian, L.C.; Zhang, J.X.; Wang, T. Research on the Species Difference of the Hepatotoxicity of Medicine Based on Transcriptome. Front. Pharmacol. 2021, 12, 647084. [Google Scholar] [CrossRef] [PubMed]
- Bjornsson, E.S. Drug-induced liver injury: An overview over the most critical compounds. Arch. Toxicol. 2015, 89, 327–334. [Google Scholar] [CrossRef] [PubMed]
- Walker, P.A.; Ryder, S.; Lavado, A.; Dilworth, C.; Riley, R.J. The evolution of strategies to minimise the risk of human drug-induced liver injury (DILI) in drug discovery and development. Arch. Toxicol. 2020, 94, 2559–2585. [Google Scholar] [CrossRef]
- Takebe, T.; Imai, R.; Ono, S. The Current Status of Drug Discovery and Development as Originated in United States Academia: The Influence of Industrial and Academic Collaboration on Drug Discovery and Development. Clin. Transl. Sci. 2018, 11, 597–606. [Google Scholar] [CrossRef]
- Clinton, J.W.; Kiparizoska, S.; Aggarwal, S.; Woo, S.; Davis, W.; Lewis, J.H. Drug-Induced Liver Injury: Highlights and Controversies in the Recent Literature. Drug Saf. 2021, 44, 1125–1149. [Google Scholar] [CrossRef]
- Ai, H.X.; Chen, W.; Zhang, L.; Huang, L.C.; Yin, Z.M.; Hu, H.; Zhao, Q.; Zhao, J.; Liu, H.S. Predicting Drug-Induced Liver Injury Using Ensemble Learning Methods and Molecular Fingerprints. Toxicol. Sci. 2018, 165, 100–107. [Google Scholar] [CrossRef]
- Li, M.Y.; Peng, L.M.; Chen, X.P. Pharmacogenomics in drug-induced cardiotoxicity: Current status and the future. Front. Cardiovasc. Med. 2022, 9, 966261. [Google Scholar] [CrossRef]
- Food and Drug Administration. International Conference on Harmonisation; guidance on S7B Nonclinical Evaluation of the Potential for Delayed Ventricular Repolarization (QT Interval Prolongation) by Human Pharmaceuticals. Fed. Regist. 2005, 70, 61133–61134. [Google Scholar]
- Lamothe, S.M.; Guo, J.; Li, W.T.; Yang, T.H.; Zhang, S.T. The Human Ether-a-go-go-related Gene (hERG) Potassium Channel Represents an Unusual Target for Protease-mediated Damage. J. Biol. Chem. 2016, 291, 20387–20401. [Google Scholar] [CrossRef] [PubMed]
- Babcock, J.J.; Li, M. hERG channel function: Beyond long QT. Acta Pharmacol. Sin. 2013, 34, 329–335. [Google Scholar] [CrossRef] [PubMed]
- De Bruin, M.L.; Pettersson, M.; Meyboom, R.H.B.; Hoes, A.W.; Leufkens, H.G.M. Anti-HERG activity and the risk of drug-induced arrhythmias and sudden death. Eur. Heart J. 2005, 26, 590–597. [Google Scholar] [CrossRef]
- Thomas, D.; Karle, C.A.; Kiehn, J. The cardiac hERG/I-Kr potassium channel as pharmacological target: Structure, function, regulation, and clinical applications. Curr. Pharm. Des. 2006, 12, 2271–2283. [Google Scholar] [CrossRef]
- Stergiopoulos, C.; Tsopelas, F.; Valko, K. Prediction of hERG inhibition of drug discovery compounds using biomimetic HPLC measurements. ADMET DMPK 2021, 9, 191–207. [Google Scholar] [CrossRef] [PubMed]
- Honma, M. An assessment of mutagenicity of chemical substances by (quantitative) structure-activity relationship. Genes Environ. 2020, 42, 23. [Google Scholar] [CrossRef]
- Zhang, L.; Ai, H.X.; Chen, W.; Yin, Z.M.; Hu, H.; Zhu, J.F.; Zhao, J.; Zhao, Q.; Liu, H.S. CarcinoPred-EL: Novel models for predicting the carcinogenicity of chemicals using molecular fingerprints and ensemble learning methods. Sci. Rep. 2017, 7, 2118. [Google Scholar] [CrossRef]
- Basu, A.K. DNA Damage, Mutagenesis and Cancer. Int. J. Mol. Sci. 2018, 19, 970. [Google Scholar] [CrossRef]
- Drevon, C.; Piccoli, C.; Montesano, R. Mutagenicity Assays of Estrogenic Hormones in Mammalian Cells. Mutat. Res. 1981, 89, 83–90. [Google Scholar] [CrossRef]
- Ferguson, L.R. Chronic inflammation and mutagenesis. Mutat. Res. Fund. Mol. Mech. Mutagenes. 2010, 690, 3–11. [Google Scholar] [CrossRef]
- Barnes, J.L.; Zubair, M.; John, K.; Poirier, M.C.; Martin, F.L. Carcinogens and DNA damage. Biochem. Soc. T 2018, 46, 1213–1224. [Google Scholar] [CrossRef] [PubMed]
- Fradkin, P.; Young, A.; Atanackovic, L.; Frey, B.; Lee, L.J.; Wang, B. A graph neural network approach for molecule carcinogenicity prediction. Bioinformatics 2022, 38, i84–i91. [Google Scholar] [CrossRef] [PubMed]
- Bartsch, H.; Tomatis, L. Comparison between Carcinogenicity and Mutagenicity Based on Chemicals Evaluated in the IARC Monographs. Environ. Health Persp. 1983, 47, 305–317. [Google Scholar] [CrossRef] [PubMed]
- Hughes, J.P.; Rees, S.; Kalindjian, S.B.; Philpott, K.L. Principles of early drug discovery. Brit. J. Pharmacol. 2011, 162, 1239–1249. [Google Scholar] [CrossRef]
- Knuiman, M.W.; Laird, N.M.; Louis, T.A. Inter-Laboratory Variability in Ames Assay Results. Mutat. Res. 1987, 180, 171–182. [Google Scholar] [CrossRef]
- Galloway, S.M. International Regulatory Requirements for Genotoxicity Testing for Pharmaceuticals Used in Human Medicine, and Their Impurities and Metabolites. Environ. Mol. Mutagen. 2017, 58, 296–324. [Google Scholar] [CrossRef]
- Li, T.; Tong, W.D.; Roberts, R.; Liu, Z.C.; Thakkar, S. DeepCarc: Deep learning-powered carcinogenicity prediction using model-level representation. Front. Artif. Intell. 2022, 4, 757780. [Google Scholar] [CrossRef]
- Zaslayskiy, M.; Jegou, S.; Tramel, E.W.; Wainrib, G. ToxicBlend: Virtual screening of toxic compound with ensemble predictors. Comput. Toxicol. 2019, 10, 81–88. [Google Scholar] [CrossRef]
- Wu, Z.Q.; Ramsundar, B.; Feinberg, E.N.; Gomes, J.; Geniesse, C.; Pappu, A.S.; Leswing, K.; Pande, V. MoleculeNet: A benchmark for molecular machine learning. Chem. Sci. 2018, 9, 513–530. [Google Scholar] [CrossRef]
- Registry of Toxic Effects of Chemical Substances (RTECS). Available online: https://www.3ds.com/ko/products-services/biovia/ (accessed on 25 July 2023).
- Sharma, B.; Chenthamarakshan, V.; Dhurandhar, A.; Pereira, S.; Hendler, J.A.; Dordick, J.S.; Das, P. Accurate clinical toxicity prediction using multi-task deep neural nets and contrastive molecular explanations. Sci. Rep. 2023, 13, 4908. [Google Scholar] [CrossRef]
- Gold, L.S.; Manley, N.B.; Slone, T.H.; Rohrbach, L.; Garfinkel, G.B. Supplement to the Carcinogenic Potency Database (CPDB): Results of animal bioassays published in the general literature through 1997 and by the National Toxicology Program in 1997–1998. Toxicol. Sci. 2005, 85, 747–808. [Google Scholar] [CrossRef] [PubMed]
- Veith, H.; Southall, N.; Huang, R.L.; James, T.; Fayne, D.; Artemenko, N.; Shen, M.; Inglese, J.; Austin, C.P.; Lloyd, D.G.; et al. Comprehensive characterization of cytochrome P450 isozyme selectivity across chemical libraries. Nat. Biotechnol. 2009, 27, 1050–1055. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.B.; Lou, C.F.; Sun, L.X.; Li, J.; Cai, Y.C.; Wang, Z.; Li, W.H.; Liu, G.X.; Tang, Y. admetSAR 2.0: Web-service for prediction and optimization of chemical ADMET properties. Bioinformatics 2019, 35, 1067–1069. [Google Scholar] [CrossRef]
- Wei, Y.; Li, S.S.; Li, Z.L.; Wan, Z.W.; Lin, J.P. Interpretable-ADMET: A web service for ADMET prediction and optimization based on deep neural representation. Bioinformatics 2022, 38, 2863–2871. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.Z.; Yan, Z.Y.; Huang, Y.Y.; Liu, L.H.; He, D.L.; Wang, W.; Fang, X.M.; Zhang, X.N.; Wang, F.; Wu, H.; et al. HelixADMET: A robust and endpoint extensible ADMET system incorporating self-supervised knowledge transfer. Bioinformatics 2022, 38, 3444–3453. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.J.; Suzuki, A.; Thakkar, S.; Yu, K.; Hu, C.C.; Tong, W.D. DILIrank: The largest reference drug list ranked by the risk for developing drug-induced liver injury in humans. Drug Discov. Today 2016, 21, 648–653. [Google Scholar] [CrossRef]
- Liu, A.; Walter, M.; Wright, P.; Bartosik, A.; Dolciami, D.; Elbasir, A.; Yang, H.B.; Bender, A. Prediction and mechanistic analysis of drug-induced liver injury (DILI) based on chemical structure. Biol. Direct 2021, 16, 6. [Google Scholar] [CrossRef]
- Thakkar, S.; Li, T.; Liu, Z.C.; Wu, L.H.; Roberts, R.; Tong, W.D. Drug-induced liver injury severity and toxicity (DILIst): Binary classification of 1279 drugs by human hepatotoxicity. Drug Discov. Today 2020, 25, 201–208. [Google Scholar] [CrossRef]
- TDC Benckmark Dataset. Available online: https://tdcommons.ai/single_pred_tasks/tox/#dili-drug-induced-liver-injury (accessed on 25 July 2023).
- Lim, S.; Kim, Y.; Gu, J.; Lee, S.; Shin, W.; Kim, S. Supervised chemical graph mining improves drug-induced liver injury prediction. iScience 2023, 26, 105677. [Google Scholar] [CrossRef]
- Kadioglu, O.; Klauck, S.M.; Fleischer, E.; Shan, L.T.; Efferth, T. Selection of safe artemisinin derivatives using a Mach. Learn.-based cardiotoxicity platform and in vitro and in vivo validation. Arch. Toxicol. 2021, 95, 2485–2495. [Google Scholar] [CrossRef]
- Kazius, J.; McGuire, R.; Bursi, R. Derivation and validation of toxicophores for mutagenicity prediction. J. Med. Chem. 2005, 48, 312–320. [Google Scholar] [CrossRef] [PubMed]
- Hansen, K.; Mika, S.; Schroeter, T.; Sutter, A.; ter Laak, A.; Steger-Hartmann, T.; Heinrich, N.; Muller, K.R. Benchmark Data Set for in Silico Prediction of Ames Mutagenicity. J. Chem. Inf. Model. 2009, 49, 2077–2081. [Google Scholar] [CrossRef] [PubMed]
- Mattocks, A.R. Chemistry and Toxicology of Pyrrolizidine Alkaloids; Academic Press: Cambridge, MA, USA, 1986. [Google Scholar]
- European Food Safety Authority (EFSA) Dataset. Available online: https://data.europa.eu/data/datasets/database-pesticide-genotoxicity-endpoints?locale=data (accessed on 25 July 2023).
- Helma, C.; Schoning, V.; Drewe, J.; Boss, P. A Comparison of Nine Machine Learning Mutagenicity Models and Their Application for Predicting Pyrrolizidine Alkaloids. Front. Pharmacol. 2021, 12, 708050. [Google Scholar] [CrossRef] [PubMed]
- Inventory of Hazardous Chemicals. Available online: https://www.mem.gov.cn/fw/cxfw/ (accessed on 25 July 2023).
- The Globally Harmonized System of Classification and Labeling of Chemicals (GHS). Available online: https://unece.org/# (accessed on 25 July 2023).
- Hao, N.; Sun, P.X.; Zhao, W.J.; Li, X.X. Application of a developed triple-classification machine learning model for carcinogenic prediction of hazardous organic chemicals to the US, EU, and WHO based on Chinese database. Ecotoxicol. Environ. Safe 2023, 255, 114806. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).