Unraveling the Radioprotective Mechanisms of UV-Resistant Bacillus subtilis ASM-1 Extracted Compounds through Molecular Docking
Abstract
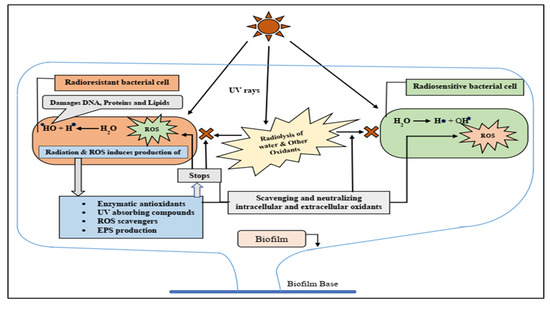
1. Introduction
2. Results
2.1. Isolation and Screening for Radioresistant Bacteria
2.2. Characterization of Strain ASM-1
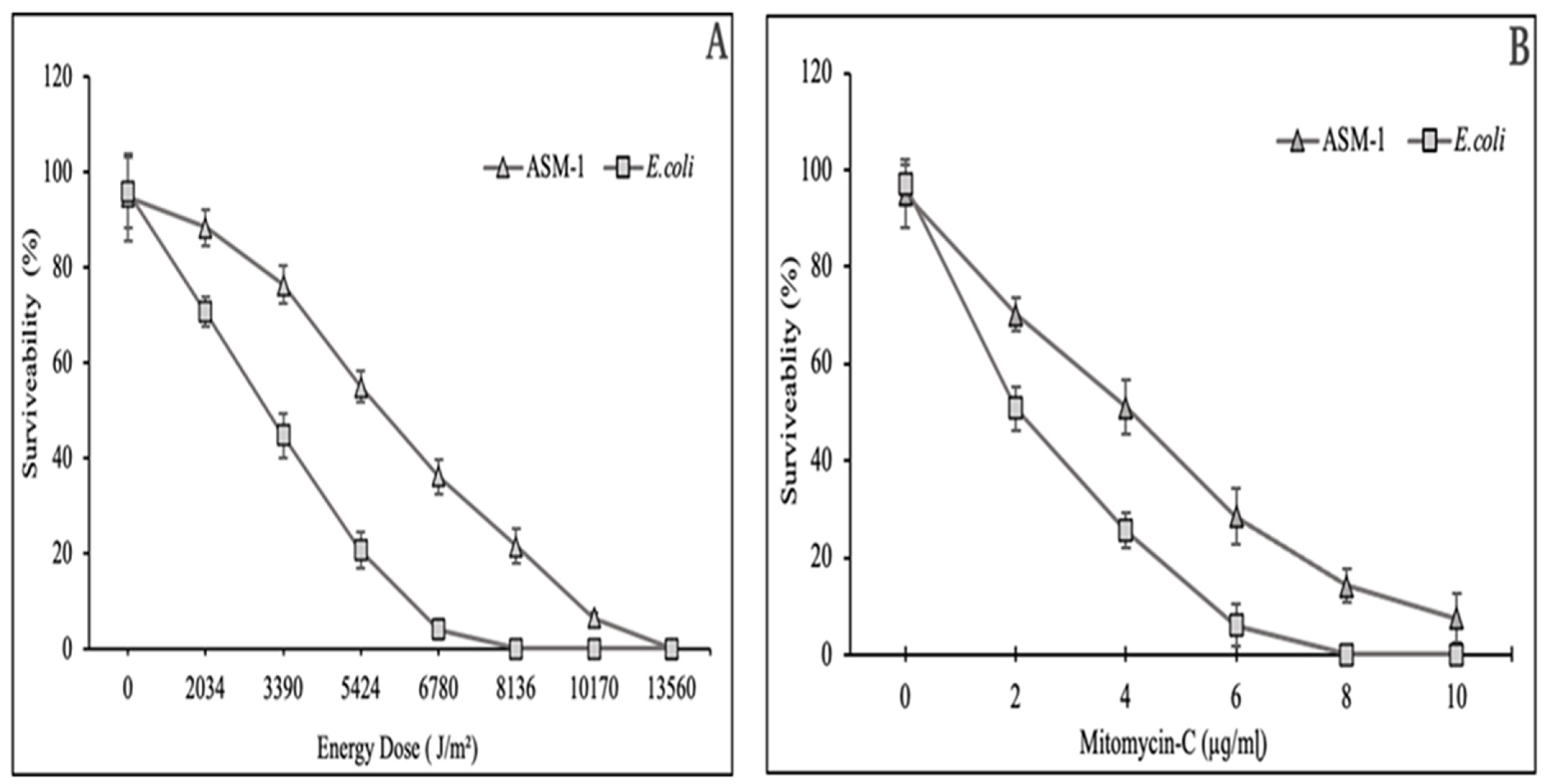
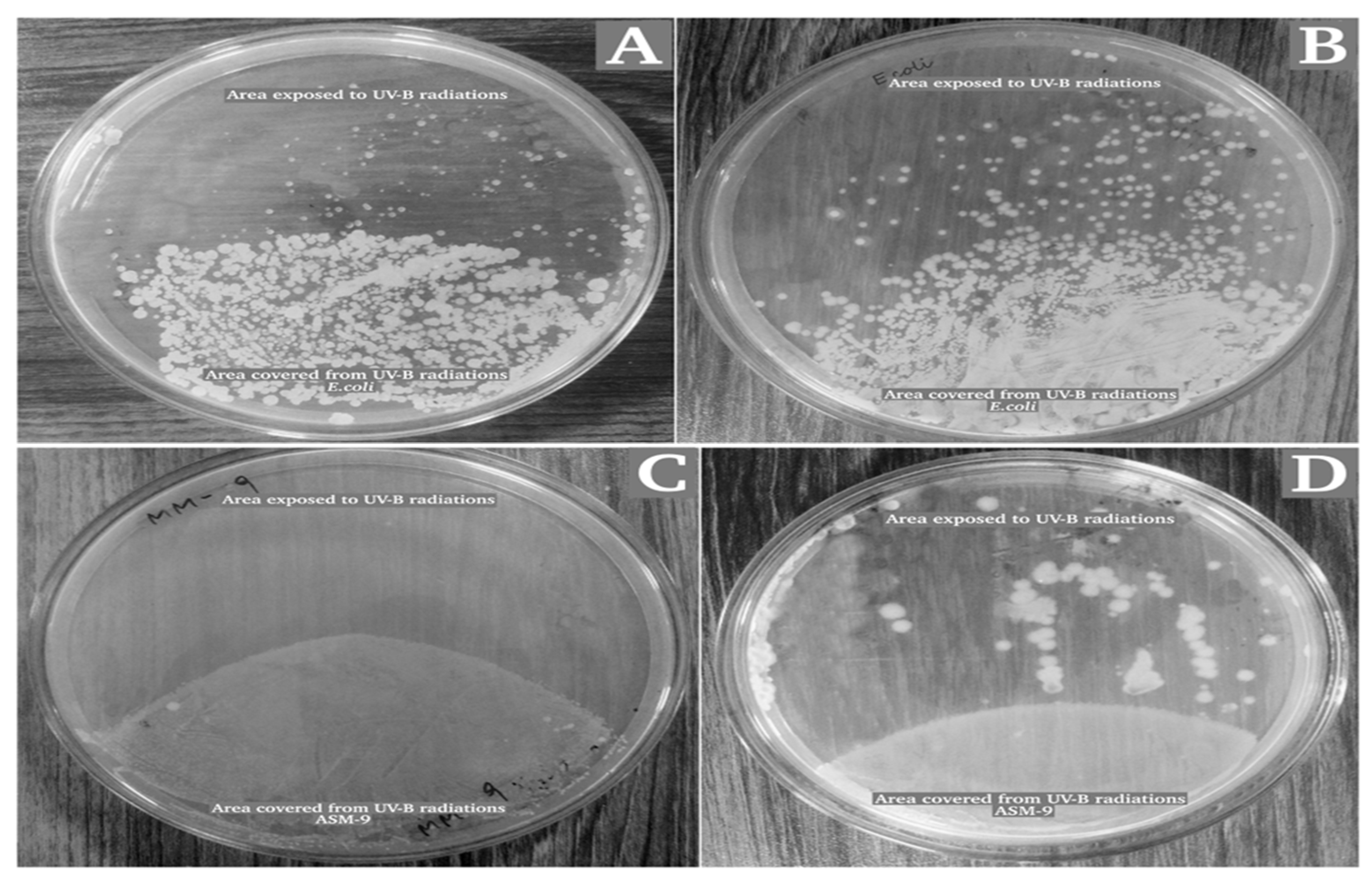
2.3. Survival Rate of Bacillus subtilis Strain ASM-1 under UVB Radiation and Mitomycin-C
2.4. Purification and Selection of the Extracellular Crude Extracts
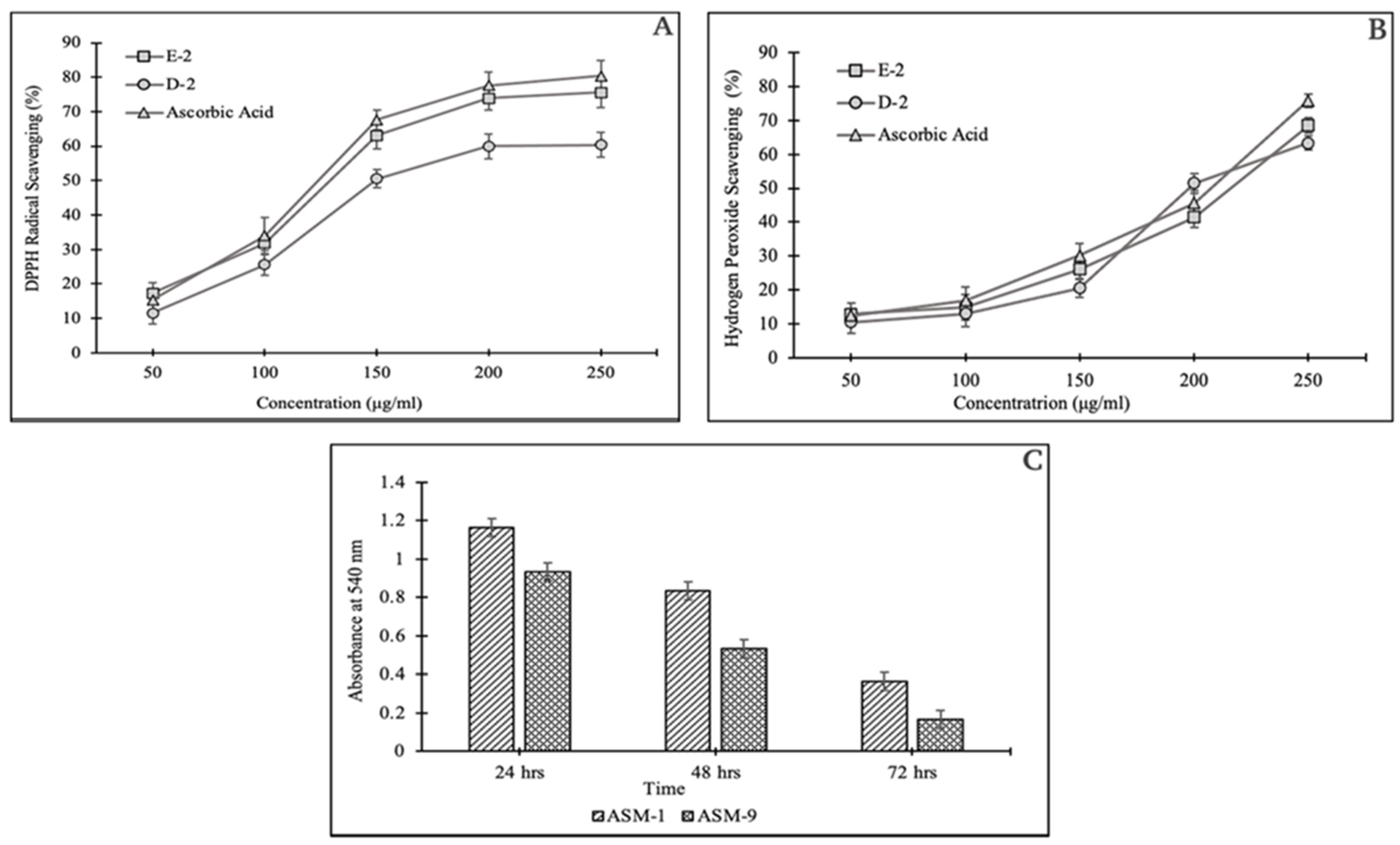
2.5. In Vitro Evaluation for the Antioxidant Potential of the Selected Fractions
2.5.1. DPPH Assay
2.5.2. Hydrogen Peroxide Scavenging Assay
2.5.3. Cytotoxic and Antibacterial Potential
2.5.4. Biofilm Detection Assay
2.5.5. GC–MS Analysis of the Selected Fractions
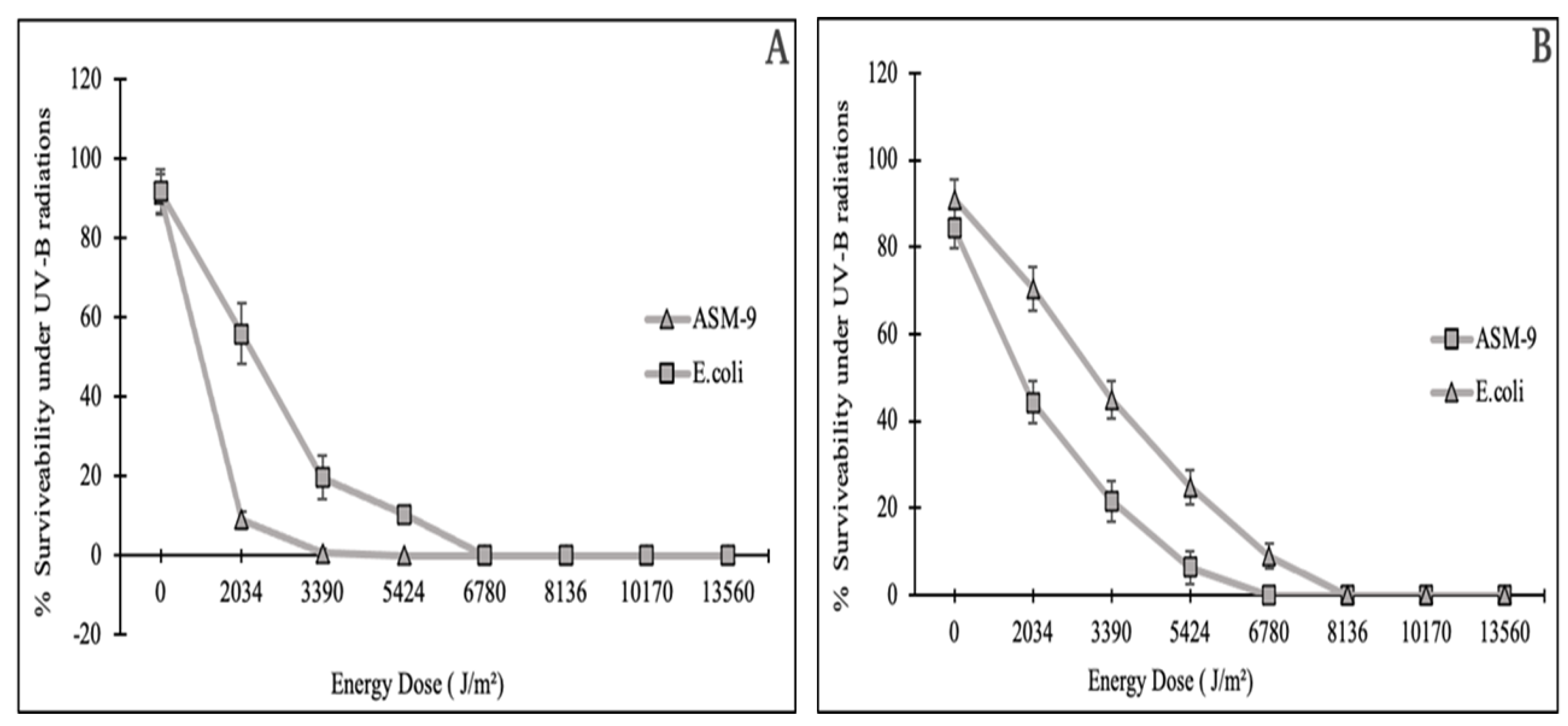
2.6. Radioprotective Activity of ASM-1 Extracts on Selected Radiosensitive Strains
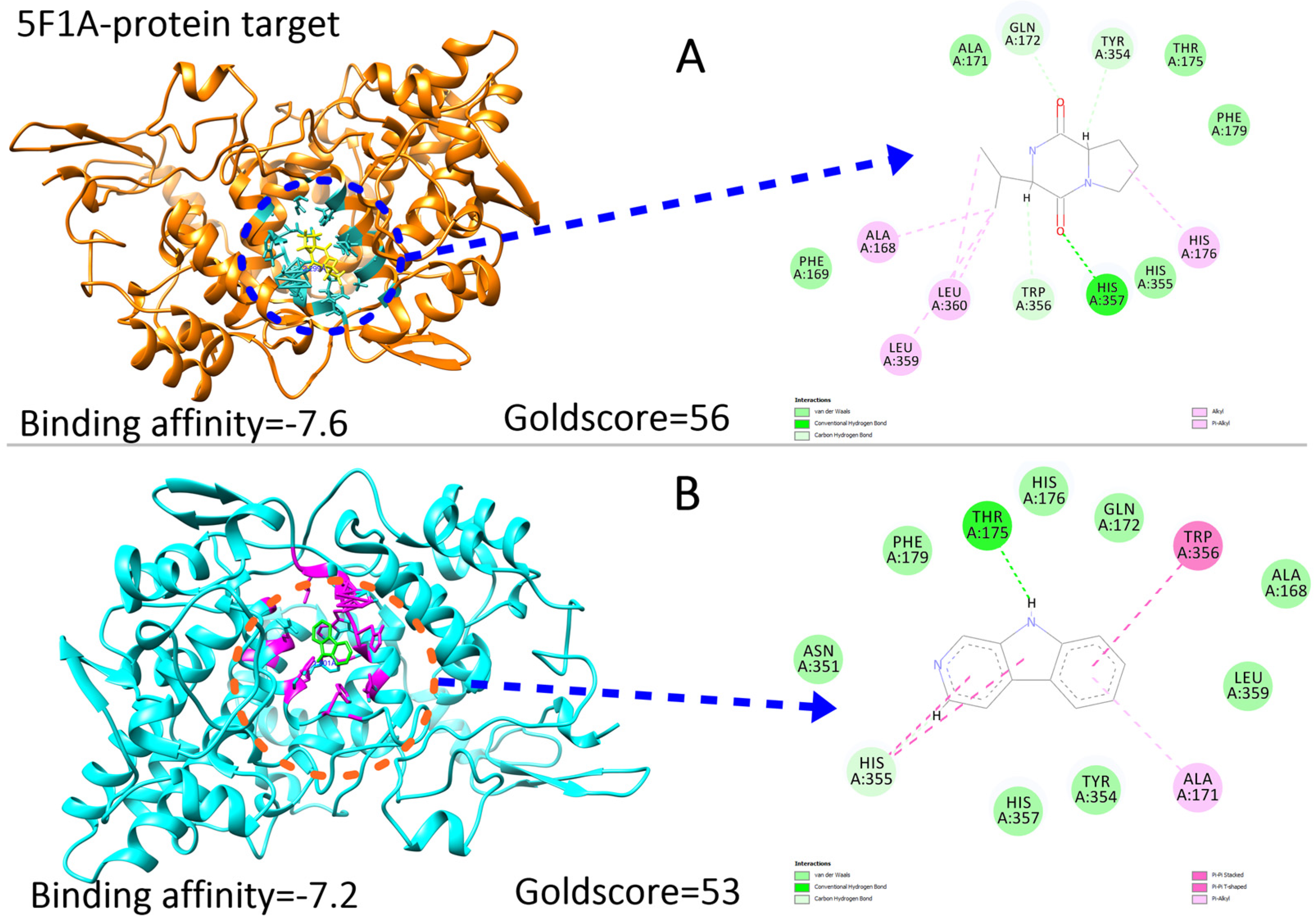
2.7. Ligand Preparations and Molecular Docking
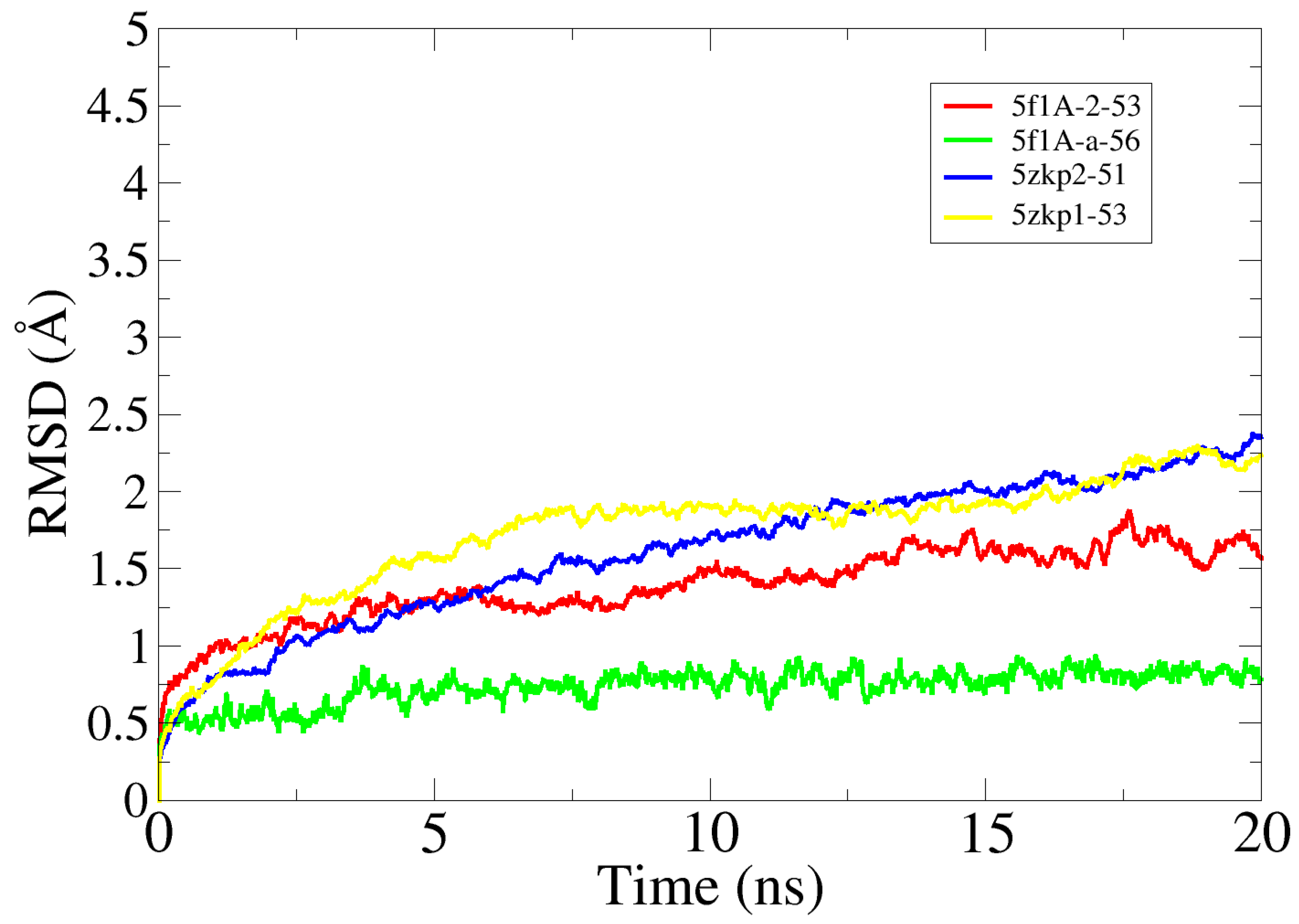
2.8. Molecular Dynamics Simulation Analysis
3. Discussion
4. Materials and Methods
4.1. Isolation and Screening of Radioresistant Bacteria
4.2. Identification of Radioresistant Bacterial Strain ASM-1
4.3. Survival Curve of Strain ASM-1 at UVB and Oxidative Stress
4.4. Extraction of Extracellular Bioactive Compounds from Bacillus subtilis ASM-1 Strain
4.5. Partial Purification of Extracellular Bioactive Compounds from B. subtilis ASM-1
4.6. In Vitro Bioassays of Purified Fractions from B. subtilis ASM-1
4.6.1. DPPH Radical Scavenging Assay
4.6.2. Hydrogen Peroxide Scavenging Assay
4.6.3. Cytotoxic and Antibacterial Potential of ASM-1 Fractions
4.6.4. Biofilm Detection through Microtiter Plate Method
4.6.5. Gas Chromatography–Mass Spectrometry Analysis for ASM-1 Fractions
4.7. In Vitro Analysis of ASM-1 Extracts in Protecting Radiosensitive Microorganisms under UVB Radiation
4.8. Ligand Preparation
4.9. Molecular Docking
4.10. Molecular Dynamics Simulations
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Giddings, L.-A.; Newman, D. Bioactive Compounds from Terrestrial Extremophiles; Springer International Publishing: Berlin/Heidelberg, Germany, 2015; ISBN 978-3-319-13259-4. [Google Scholar]
- Nocker, A.; Shah, M.; Dannenmann, B.; Schulze-Osthoff, K.; Wingender, J.; Probst, A.J. Assessment of UV-C-Induced Water Disinfection by Differential PCR-Based Quantification of Bacterial DNA Damage. J. Microbiol. Methods 2018, 149, 89–95. [Google Scholar] [CrossRef]
- Sakudo, A.; Toyokawa, Y.; Nakamura, T.; Yagyu, Y.; Imanishi, Y. Nitrogen Gas Plasma Treatment of Bacterial Spores Induces Oxidative Stress that Damages the Genomic DNA. Mol. Med. Rep. 2017, 15, 396–402. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.Z.-H.; Luo, X.-S.; Liu, M.; Huang, Q. Diversity of Ionizing Radiation-Resistant Bacteria Obtained from the Taklimakan Desert. J. Basic Microbiol. 2015, 55, 135–140. [Google Scholar] [CrossRef]
- Becker, J.; Wittmann, C. Microbial Production of Extremolytes—High-Value Active Ingredients for Nutrition, Health Care, and Well-Being. Curr. Opin. Biotechnol. 2020, 65, 118–128. [Google Scholar] [CrossRef]
- Latifi, A.; Ruiz, M.; Zhang, C.-C. Oxidative Stress in Cyanobacteria. FEMS Microbiol. Rev. 2009, 33, 258–278. [Google Scholar] [CrossRef] [PubMed]
- Chaudhri, A.; Nadeem, M.; Rahman, A.; Alam, T.; Sajjad, W.; Hasan, F.; Badshah, M.; Khan, S.; Rehman, F.; Shah, A. Antioxidative and Radioprotective Properties of Glycosylated Flavonoid, Xanthorhamnin from Radio-Resistant Bacterium Bacillus Indicus Strain TMC-6. Curr. Microbiol. 2020, 77, 1245–1253. [Google Scholar] [CrossRef]
- Shuryak, I.; Matrosova, V.Y.; Gaidamakova, E.K.; Tkavc, R.; Grichenko, O.; Klimenkova, P.; Volpe, R.P.; Daly, M.J. Microbial Cells Can Cooperate to Resist High-Level Chronic Ionizing Radiation. PLoS ONE 2017, 12, e0189261. [Google Scholar] [CrossRef]
- Verdonk, M.L.; Cole, J.C.; Hartshorn, M.J.; Murray, C.W.; Taylor, R.D. Improved Protein–Ligand Docking Using GOLD. Proteins Struct. Funct. Bioinform. 2003, 52, 609–623. [Google Scholar] [CrossRef]
- Ahmad, S.; Abbasi, H.W.; Shahid, S.; Gul, S.; Abbasi, S.W. Molecular Docking, Simulation and MM-PBSA Studies of Nigella Sativa Compounds: A Computational Quest to Identify Potential Natural Antiviral for COVID-19 Treatment. J. Biomol. Struct. Dyn. 2020, 39, 4225–4233. [Google Scholar] [CrossRef]
- Riesenman, P.J.; Nicholson, W.L. Role of the Spore Coat Layers in Bacillus Subtilis Spore Resistance to Hydrogen Peroxide, Artificial UV-C, UV-B, and Solar UV Radiation. Appl. Environ. Microbiol. 2000, 66, 620–626. [Google Scholar] [CrossRef] [PubMed]
- Pailliè-Jiménez, M.E.; Stincone, P.; Brandelli, A. Natural Pigments of Microbial Origin. Front. Sustain. Food Syst. 2020, 4, 590439. [Google Scholar] [CrossRef]
- Nayak, T.; Sengupta, I.; Dhal, P.K. A New Era of Radiation Resistance Bacteria in Bioremediation and Production of Bioactive Compounds with Therapeutic Potential and Other Aspects: An in-Perspective Review. J. Environ. Radioact. 2021, 237, 106696. [Google Scholar] [CrossRef] [PubMed]
- Laroussi, M.; Leipold, F. Evaluation of the Roles of Reactive Species, Heat, and UV Radiation in the Inactivation of Bacterial Cells by Air Plasmas at Atmospheric Pressure. Int. J. Mass Spectrom. 2004, 233, 81–86. [Google Scholar] [CrossRef]
- Célariès, B.; Rima, G.; Court, L.; Lion, C.; Laval, J.D. Radioprotective Activity and Synthesis of Siladithioacetals and Germadithioacetals Derived from N-Substituted Naphthylethylimidazoline. Met.-Based Drugs 2001, 8, 199–210. [Google Scholar] [CrossRef]
- Brand-Williams, W.; Cuvelier, M.E.; Berset, C. Use of a Free Radical Method to Evaluate Antioxidant Activity. LWT Food Sci. Technol. 1995, 28, 25–30. [Google Scholar] [CrossRef]
- Kadaikunnan, S.; Rejiniemon, T.S.; Khaled, J.M.; Alharbi, N.S.; Mothana, R. In-Vitro Antibacterial, Antifungal, Antioxidant and Functional Properties of Bacillus amyloliquefaciens. Ann. Clin. Microbiol. Antimicrob. 2015, 14, 9. [Google Scholar] [CrossRef]
- Lemon, K.P.; Earl, A.M.; Vlamakis, H.C.; Aguilar, C.; Kolter, R. Biofilm Development with an Emphasis on Bacillus subtilis. Curr. Top. Microbiol. Immunol. 2008, 322, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Ghorab, M.M.; Ragab, F.A.; Alqasoumi, S.I.; Alafeefy, A.M.; Aboulmagd, S.A. Synthesis of Some New Pyrazolo[3,4-d]Pyrimidine Derivatives of Expected Anticancer and Radioprotective Activity. Eur. J. Med. Chem. 2010, 45, 171–178. [Google Scholar] [CrossRef] [PubMed]
- Sverdlov, R.L.; Brinkevich, S.D.; Shadyro, O.I. Effects of Tryptophan Derivatives and β-Carboline Alkaloids on Radiation- and Peroxide-Induced Transformations of Ethanol. Radiat. Phys. Chem. 2014, 98, 77–85. [Google Scholar] [CrossRef]
- Liu, M.-H.; Zhang, Q.; Zhang, Y.-H.; Lu, X.-Y.; Fu, W.-M.; He, J.-Y. Chemical Analysis of Dietary Constituents in Rosa Roxburghii and Rosa Sterilis Fruits. Molecules 2016, 21, 1204. [Google Scholar] [CrossRef]
- Elamin, M.M.; Abdelrahim, N.A.; Elhag, D.E.A.; Joseph, M.R.P.; Hamid, M.E. Bioactive Pyrrole-Pyrazine Derivative from a Novel Bacillus Species and Review of the Literature. Afr. J. Pharm. Pharmacol. 2021, 15, 138–151. [Google Scholar] [CrossRef]
- Gao, H.; Li, P.; Xu, X.; Zeng, Q.; Guan, W. Research on Volatile Organic Compounds From Bacillus subtilis CF-3: Biocontrol Effects on Fruit Fungal Pathogens and Dynamic Changes During Fermentation. Front. Microbiol. 2018, 9, 456. [Google Scholar] [CrossRef]
- Sajjad, W.; Ahmad, S.; Aziz, I.; Azam, S.S.; Hasan, F.; Shah, A.A. Antiproliferative, Antioxidant and Binding Mechanism Analysis of Prodigiosin from Newly Isolated Radio-Resistant Streptomyces Sp. Strain WMA-LM31. Mol. Biol. Rep. 2018, 45, 1787–1798. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Chen, X.; Jiang, Y.; Jiang, C.; Li, Q.; Chen, X.; Jiang, Y.; Jiang, C. Cultural, Physiological, and Biochemical Identification of Actinobacteria. In Actinobacteria-Basics and Biotechnological Applications; IntechOpen: London, UK, 2016; ISBN 978-953-51-2248-7. [Google Scholar]
- Wright, M.H.; Adelskov, J.; Greene, A.C. Bacterial DNA Extraction Using Individual Enzymes and Phenol/Chloroform Separation. J. Microbiol. Biol. Educ. 2017, 18, 10–1128. [Google Scholar] [CrossRef]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S Ribosomal DNA Amplification for Phylogenetic Study. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Mattimore, V.; Battista, J.R. Radioresistance of Deinococcus Radiodurans: Functions Necessary to Survive Ionizing Radiation Are Also Necessary to Survive Prolonged Desiccation. J. Bacteriol. 1996, 178, 633–637. [Google Scholar] [CrossRef] [PubMed]
- Kedare, S.B.; Singh, R.P. Genesis and Development of DPPH Method of Antioxidant Assay. J. Food Sci. Technol. 2011, 48, 412–422. [Google Scholar] [CrossRef]
- Elmastaş, M.; Gülçin, İ.; Işildak, Ö.; Küfrevioğlu, Ö.İ.; İbaoğlu, K.; Aboul-Enein, H.Y. Radical Scavenging Activity and Antioxidant Capacity of Bay Leaf Extracts. J. Iran. Chem. Soc. 2006, 3, 258–266. [Google Scholar] [CrossRef]
- Zhang, Y.; Mu, J.; Han, J.; Gu, X. An Improved Brine Shrimp Larvae Lethality Microwell Test Method. Toxicol. Mech. Methods 2012, 22, 23–30. [Google Scholar] [CrossRef]
- Christensen, G.D.; Simpson, W.A.; Younger, J.J.; Baddour, L.M.; Barrett, F.F.; Melton, D.M.; Beachey, E.H. Adherence of Coagulase-Negative Staphylococci to Plastic Tissue Culture Plates: A Quantitative Model for the Adherence of Staphylococci to Medical Devices. J. Clin. Microbiol. 1985, 22, 996–1006. [Google Scholar] [CrossRef] [PubMed]
- Tahir, H.A.S.; Gu, Q.; Wu, H.; Raza, W.; Hanif, A.; Wu, L.; Colman, M.V.; Gao, X. Plant Growth Promotion by Volatile Organic Compounds Produced by Bacillus subtilis SYST2. Front. Microbiol. 2017, 8, 171. [Google Scholar] [CrossRef]
- Dallakyan, S.; Olson, A.J. Small-Molecule Library Screening by Docking with PyRx. In Chemical Biology; Springer: Berlin/Heidelberg, Germany, 2015; pp. 243–250. [Google Scholar]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Case, D.A.; Betz, R.M.; Cerutti, D.S.; Cheatham, T.; Darden, T.; Duke, R.E.; Giese, T.J.; Gohlke, H.; Götz, A.W.; Homeyer, N.; et al. Amber 16, University of California, San Francisco; University of California: San Francisco, CA, USA, 2016. [Google Scholar] [CrossRef]
- Sprenger, K.G.; Jaeger, V.W.; Pfaendtner, J. The General AMBER Force Field (GAFF) Can Accurately Predict Thermodynamic and Transport Properties of Many Ionic Liquids. J. Phys. Chem. B 2015, 119, 5882–5895. [Google Scholar] [CrossRef] [PubMed]
- Case, D.A.; Babin, V.; Berryman, J.T.; Betz, R.M.; Cai, Q.; Cerutti, D.S.; Cheatham III, T.E.; Darden, T.A.; Duke, R.E.; Gohlke, H.; et al. The FF14SB Force Field. Amber 2014, 14, 29–31. [Google Scholar]
- Izaguirre, J.A.; Catarello, D.P.; Wozniak, J.M.; Skeel, R.D. Langevin Stabilization of Molecular Dynamics. J. Chem. Phys. 2001, 114, 2090–2098. [Google Scholar] [CrossRef]
- Price, D.J.; Brooks, C.L. A Modified TIP3P Water Potential for Simulation with Ewald Summation. J. Chem. Phys. 2004, 121, 10096–10103. [Google Scholar] [CrossRef]
- Roe, D.R.; Cheatham, T.E. PTRAJ and CPPTRAJ: Software for Processing and Analysis of Molecular Dynamics Trajectory Data. J. Chem. Theory Comput. 2013, 9, 3084–3095. [Google Scholar] [CrossRef]



| Exposure Time | Radiant Exposure in (J/m2) He = Ee × t (Sec) | UV Resistant Isolates |
|---|---|---|
| 3 min | 2.034 × 103 J/m2 | * TMC 1 to TMC 18 ** ASM 1 to ASM 8 |
| 5 min | 3.390 × 103 J/m2 | TMC 2, TMC 7, TMC 6, TMC 9, TMC 11, TMC 12, TMC15, ASM 2, ASM 1, ASM 3, ASM 5, TMM 6, ASM 8 |
| 8 min | 5.424 × 103 J/m2 | TMC 2, TMC 7, TMC 6, TMC 9, TMC 12, TMC15, ASM 1, ASM 5, ASM 6, ASM 8 |
| 10 min | 6.780 × 103 J/m2 | ASM-1, ASM-6, ASM-8 |
| 12 min | 8.136 × 103 J/m2 | ASM-1 |
| Fractions | Retention Time | Content (%) | Chemical Compounds | Molecular Formula | Molecular Weight | Class |
|---|---|---|---|---|---|---|
| E-2 | 3.083 | 1.03 | Pyrrolo [1,2-a] pyrazine -1,4-dione, hexahydro-3-(2-methyl propyl) | C11H18N2O2 | 210 | Pyrrolopyrazine |
| 4.146 | 0.14 | 9H-Pyrido [3,4-b] indole | C11H8N2 | 168 | β-carboline alkaloid | |
| 4.123 | 0.56 | Pyrrolo [1,2a] pyrazine-1,4-dione, hexahydro -3-(phenylmethyl) | C7H10N2O2 | 154 | Pyrrolopyrazine | |
| 7.521 | 0.77 | n-Nonadecanol-1 | C19H40O | 284 | Alcohol | |
| 11.614 | 0.21 | Dodecane | C12H26 | 170 | Alkane | |
| 15.291 | 1.91 | Phenol,2,2′-methylenebis [6-(1,1-dimethylethyl)-4-methyl | C23H32O2 | 340 | Phenol | |
| D-2 | 3.412 | 1.09 | Benzamide | C7H7NO | 121 | Amide |
| 4.243 | 0.28 | 1,2-benzenedicarboxylic acid, monobutyl ester | C12H14O4 | 222 | Dicarboxylic acid Butyl ester | |
| 5.461 | 0.32 | 1,2-Benzenedicarboxylic acid, diisooctyl ester | C24H38O4 | 390 | Dicarboxylic acid Octyl ester | |
| 11.065 | 0.37 | (S)-6-Hydroxyheptanoic acid | C7H14O2 | 130 | Fatty acid | |
| 11.534 | 0.04 | 1-Octanol | C8H18O | 130 | Alcohol |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rahman, A.U.; Ali, A.; Ahmad, F.; Ahmad, S.; Alharbi, M.; Alasmari, A.F.; Fayyaz, A.; Rana, Q.u.a.; Khan, S.; Hasan, F.; et al. Unraveling the Radioprotective Mechanisms of UV-Resistant Bacillus subtilis ASM-1 Extracted Compounds through Molecular Docking. Pharmaceuticals 2023, 16, 1139. https://doi.org/10.3390/ph16081139
Rahman AU, Ali A, Ahmad F, Ahmad S, Alharbi M, Alasmari AF, Fayyaz A, Rana Qua, Khan S, Hasan F, et al. Unraveling the Radioprotective Mechanisms of UV-Resistant Bacillus subtilis ASM-1 Extracted Compounds through Molecular Docking. Pharmaceuticals. 2023; 16(8):1139. https://doi.org/10.3390/ph16081139
Chicago/Turabian StyleRahman, Asim Ur, Aftab Ali, Faisal Ahmad, Sajjad Ahmad, Metab Alharbi, Abdullah F. Alasmari, Amna Fayyaz, Qurrat ul ain Rana, Samiullah Khan, Fariha Hasan, and et al. 2023. "Unraveling the Radioprotective Mechanisms of UV-Resistant Bacillus subtilis ASM-1 Extracted Compounds through Molecular Docking" Pharmaceuticals 16, no. 8: 1139. https://doi.org/10.3390/ph16081139
APA StyleRahman, A. U., Ali, A., Ahmad, F., Ahmad, S., Alharbi, M., Alasmari, A. F., Fayyaz, A., Rana, Q. u. a., Khan, S., Hasan, F., Badshah, M., & Shah, A. A. (2023). Unraveling the Radioprotective Mechanisms of UV-Resistant Bacillus subtilis ASM-1 Extracted Compounds through Molecular Docking. Pharmaceuticals, 16(8), 1139. https://doi.org/10.3390/ph16081139