Abstract
Antimicrobial resistance (AMR) due to the prevalence of multidrug-resistant (MDR) pathogens is rapidly increasing worldwide, and the identification of new antimicrobial agents with innovative mechanisms of action is urgently required. Medicinal plants that have been utilised for centuries with minor side effects may hold great promise as sources of effective antimicrobial products. The free-living nematode Caenorhabditis elegans (C. elegans) is an excellent live infection model for the discovery and development of new antimicrobial compounds. However, while C. elegans has widely been utilised to explore the effectiveness and toxicity of synthetic antibiotics, it has not been used to a comparable extent for the analysis of natural products. By screening the PubMed database, we identified articles reporting the use of the C. elegans model for the identification of natural products endowed with antibacterial and antifungal potential, and we critically analysed their results. The studies discussed here provide important information regarding “in vivo” antimicrobial effectiveness and toxicity of natural products, as evaluated prior to testing in conventional vertebrate models, thereby supporting the relevance of C. elegans as a highly proficient model for their identification and functional assessment. However, their critical evaluation also underlines that the characterisation of active phytochemicals and of their chemical structure, and the unravelling of their mechanisms of action represent decisive challenges for future research in this area.
1. Introduction
Antimicrobial resistance (AMR) occurs when pathogens such as bacteria, fungi, viruses, and parasites evolve in ways rendering current treatments, including antibiotics, ineffective. Despite efforts and initiatives promoted by the World Health Organisation (WHO), the spiralling increase in AMR remains one of the top 10 health issues worldwide [1,2,3]. While the overprescription and misapplication of antimicrobials represent major factors underlying the emergence of multidrug-resistant (MDR) pathogens [4], worldwide trade, migration, and travel have accelerated their spread [5].
A recent study reported that while globally, in 2019, 4.95 million deaths could be considered as associated with AMR, 1.27 million of them could be directly attributed to infections by MDR pathogens [6]. If no action is taken, the WHO has warned that this figure could reach 10 million deaths per year by 2050 [6], thus surpassing cancer, diabetes, and heart disease as the leading cause of death worldwide [7]. Moreover, the unavailability of effective therapies, leading to prolonged hospital stays and work absenteeism, could increasingly burden health systems’ economies due to higher treatment costs [4].
In 2019, three infectious diseases were associated with AMR [6] worldwide. They include lower respiratory infections, linked to >1.5 million deaths, followed by bloodstream and intra-abdominal infections [6]. Importantly, six pathogens, namely, Escherichia coli, Staphylococcus aureus, Klebsiella pneumoniae, Streptococcus pneumoniae, Acinetobacter baumannii, and Pseudomonas aeruginosa, were responsible for a total of 929,000 deaths attributable to AMR worldwide [6], with E coli being the leading cause [6]. In particular, Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species, collectively defined as ESKAPE pathogens, have developed resistance to several classes of antibiotics, allowing them to “escape” the bacteriostatic or bactericidal effects of current treatments [8]. These pathogens represent leading causes of nosocomial infections and are associated with high mortality rates, prolonged hospital stays, and increased healthcare costs [9,10,11,12]. New antimicrobials targeting ESKAPE and other pathogens are urgently needed [13].
Caenorhabditis elegans (C. elegans) has widely been used in the past two decades as a model organism for drug discovery [14,15] and to explore “in vivo” mechanisms of action of previously identified compounds. Most importantly, it has also been proficiently utilised for the identification of novel antimicrobials, with a major focus on libraries of synthetic compounds [16,17].
Plant-based natural products have been used for centuries as sources of antimicrobial compounds, and they may provide safe, alternative antimicrobial treatments. Considering the role of C. elegans in the discovery of novel drugs and in the unravelling of their molecular mechanisms, here we highlight the importance of using this model in the study of plant-derived antibacterial and antifungal compounds and review previous studies in this research area.
2. Current Issues with Antimicrobial Drug Discovery
Current antimicrobial drug discovery rates fail to meet the urgent demand for novel compounds to treat life-threatening infections caused by MDR pathogens. Limited returns on investment and scarce funding are some of the factors contributing to insufficient antimicrobial drug development, which requires approximately 10 years from preclinical trials until market authorisation according to WHO estimations [2]. Indeed, between 2020 and 2021, only one antimicrobial compound has been approved by the US Food and Drug Administration (FDA) and the European Medicines Agency (EMA) [18]. Moreover, compounds lacking cross-resistance, with novel mechanisms of action or molecular targets, or those belonging to new classes of drugs are relatively rare. In 2022, WHO reported that out of 32 prioritised new antimicrobial compounds in clinical trials, only 6 could be considered innovative [2].
Initial stages of antimicrobial drug screening typically utilise in vitro methods focusing on direct bacteriostatic or bactericidal effects. However, the use of these methods in the screening of compounds with novel mechanisms of action presents several limitations. Culturing of the defined microorganisms in viable conditions may be difficult. The high concentrations of compounds used for in vitro screening may prove toxic or may have poor pharmacokinetic properties in later stages of development [19,20]. Moreover, some compounds are unable to cross the MDR-barrier of Gram-negative bacteria [21] or are ineffective when tested in vivo [19,22,23].
Overall, current in vitro assays appear to be inadequate for the discovery of novel antimicrobials [24], and more robust in vivo multicellular models are necessary.
3. Advantages of C. elegans as a Model for the Screening of Novel Compounds
To address these issues, in the last decade, antimicrobial drug discovery technology has moved towards the use of whole animal models, such as zebrafish and C. elegans, for the screening of new compounds and the identification of their mechanisms of action.
C. elegans, a bacteria-eating organism, can be infected and killed by a variety of human pathogens, including Gram-negative bacteria such as Pseudomonas aeruginosa and Salmonella enterica; Gram-positive bacteria such as Staphylococcus aureus, Streptococcus pneumoniae, and Enterococcus faecalis; and fungi such as those of the Candida species [16,17,24,25].
To screen for novel antimicrobial compounds, the C. elegans natural food source, E. coli OP50, is replaced by a pathogenic bacterium, e.g., Pseudomonas aeruginosa. Survival assays are performed either on agar or in liquid media [16,25,26,27]. Initially, worms are incubated with pathogenic organisms to establish infection before transferring them into assay plates (12-, 24-, 96-, or 384-well plates) that are subsequently added with different concentrations of novel compounds. The number of live and dead worms can be counted visually under the microscope or, in automated assays, by using a cell-permeable dye, identifying dead worms, and image analysis software [17,28] (Figure 1). Notably, automated assays with 10–20 worms per well in 96- or 384-well plates allow for the screening of large numbers of compounds [29].
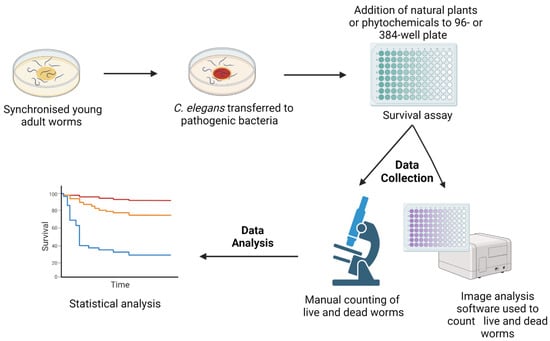
Figure 1.
A schematic diagram of the survival assays used for the C. elegans-based screening of novel compounds with antimicrobial activity. Synchronised C. elegans are generated either by egg laying or the bleaching method. Worms are grown in E. coli OP50 plates for 2–3 days at 20 °C until their adulthood. Subsequently, they are transferred to plates seeded with pathogenic microbes. Following an incubation period, worms are transferred to 12-, 24-, 96-, or 384-well assay plates containing different concentrations of antimicrobial compounds. Numbers of live and dead worms can be scored manually under the microscope or by imaging software using a cell permeable dye identifying dead nematodes. The image was created with BioRender.com.
The main advantage of the live animal model is that it not only allows for the assessment of direct antimicrobial effects of defined agents with novel mechanisms of action or molecular targets, but it also addresses their potential role as immune modulators [30], indirectly contributing to fighting microbial virulence and increasing the likelihood of host survival. Moreover, the use of a whole live animal model provides valuable information on systemic toxicity of specific compounds in the early stages of drug discovery.
Importantly, the small size and short lifespan of C. elegans convincingly support its suitability as a model organism. Indeed, C. elegans is a 1 mm long, free-living, transparent nematode that develops from a hatched egg to adulthood in 3 days at 20 °C. Once they reach adulthood, wild-type worms live for three additional weeks. The short lifespan and quick generation time accelerate the antimicrobial discovery process and critically favour its automation.
C. elegans was the first multicellular organism whose genome was fully sequenced (C. elegans Sequencing Consortium 1998). Most notably, C. elegans is a self-fertilised organism, and all offspring are genetically identical, thus providing a distinct advantage compared to fruit flies and murine models [31]. Indeed, inbreeding of mutant strains to clarify mechanisms of action of novel compounds can be easily performed starting from a highly homogeneous genetic background.
Thus, C. elegans qualifies as an excellent model organism, bridging the gap between in vitro assays and the complexities of vertebrate models.
4. Natural Plants as Sources of Novel Antimicrobial Agents
Natural plants have long been used in traditional medicine [32,33]. In particular, many plant species have been utilised to treat microbial infections, and scientists have capitalised on traditional knowledge to screen natural products with potential antimicrobial activity and to attempt to decipher their mechanisms of action [32].
The extraction of novel antimicrobial agents from natural plants is cost-effective compared to synthetic chemistry methods, since plants are usually easily available and purification processes are relatively inexpensive [33]. This is particularly important in developing countries where a variety of infectious diseases are endemic. Natural plant extracts may provide a more accessible and affordable treatment for some of them.
Notably, natural plants offer a range of secondary metabolites, including phenols, flavonoids, sulphur compounds, phenolic glycosides, saponins, unsaturated lactones, cryogenics, and/or glucosinolates, thereby representing rich sources of novel compounds with antimicrobial properties [34,35,36]. These metabolites are usually not necessary for plant survival but can offer protection against predators and pathogens [37]. Indeed, many of them have already been shown to display antimicrobial activity [34,38,39]. Their diversity suggests that a large number of plant species can be explored as potential sources of antimicrobial activity [37,39,40,41].
Importantly, antimicrobial compounds extracted from natural plants have been reported to be characterised by fewer side effects compared to synthetic agents [42,43], possibly due to the complex mixture of different metabolites present in natural plants, which may have synergistic effects or counteract the toxicity of individual components. While this complexity could hinder the standardisation of natural plant extracts, it provides a strong incentive for an updated analysis of interactions occurring between different metabolites, also addressing their antioxidant, anti-inflammatory, and anticancer properties [44,45].
5. Methodology
All previous works on the use of Caenorhabditis elegans as an in vivo model for the discovery and development of natural plant-based antimicrobial compounds were collected from the PubMed database, examined, and reported in the present review. A total of 203 peer-reviewed papers published in the English language were included in this review. The following search terms were used: “Natural products, Caenorhabditis elegans, antibacterial activity”, overall yielding 41 entries; and “Natural products, Caenorhabditis elegans, antifungal activity”, overall yielding 10 entries. One entry shared both terms. Reports published until April 2023 were considered. Entries focusing on anthelminthic activity were excluded. Within this database, our analysis focused on a critical evaluation of natural products active against MDR pathogens and tested in the C. elegans model.
6. Natural Products Active against Bacterial Infection
In vitro and in vivo antibacterial activities of natural plant products, as tested using the C. elegans model, are illustrated in Table 1.

Table 1.
Antibacterial activity of natural plant products in the C. elegans in vivo model. * MoA: mechanism of action; ** N.D.: not determined.
Staphylococcus aureus (S. aureus), a Gram-positive bacterium, frequently implicated in various community- and hospital-acquired infections, has developed resistance against penicillin and semisynthetic methicillin. The ability to produce potent toxins and enzymes and form biofilms makes these bacteria highly feared pathogens. Methicillin-resistant S. aureus (MRSA) has become widespread in all settings, and new strains continue to emerge and develop higher resistance to the few available antibiotics [75].
A liquid-based assay used to screen novel compounds has shown that natural extracts from Nypa fruticans (mangrove palm), Swietenia macrophylla (big leaf mahogany), Curcuma longa (turmeric), Eurycoma longifolia (longjack), Orthosiphon stamineus (cat’s whiskers), and Silybum eburneum (milk thistle) increase the lifespan of S. aureus-infected C. elegans by 2.8-fold [46]. N. fruticans husks and C. longa inhibit S. aureus growth at concentrations <200 μg/mL. Furthermore, extracts from N. fruticans roots, S. macrophylla seeds, and O. stamineus leaves protect worms from MRSA, and reduced colonisation of the nematode intestines by S. aureus was observed following treatment with N. fruticans root and O. stamineus leaf extracts. These results suggest that products from these plants, historically used for their analgesic, anti-inflammatory, and antioxidative effects in Southeast Asia, could promote host immunity and/or decrease bacterial virulence [46].
In another study, a herbal formulation used to treat wound infections and derived mainly from leaves and bark of plants such as Azadirachta indica (Neem), Acacia nilotica (Babul), Ocimum sanctum (Tulsi), and Annona squamosa (sugar apple) showed promising activity upon repeated exposure of C. elegans to S. aureus. This formulation, containing the Curcuma longa (turmeric) rhizome and seed oil of Ricinus communis (castor oil), inhibits bacterial growth and production of staphyloxanthin, a quorum-sensing (QS)-regulated pigment, at concentrations ≥0.025% v/v. In addition, it prevents biofilm formation and significantly modifies the bacterial transcriptome by targeting genes associated with haemolysis, virulence, enzyme activity, basic cellular processes, quorum-sensing, and transcriptional regulation [51].
P. aeruginosa is another opportunistically resistant bacterium posing major healthcare challenges, due to a range of virulence factors [76], with intrinsic and acquired mechanisms including low outer membrane permeability, multidrug efflux pumps, and enzymes. Moreover, P. aeruginosa demonstrates adaptive resistance through persistent biofilm formation in infected tissues [77].
Secondary metabolites from endophytic fungi of Alangium, Angelica sinensis (female ginseng), Bupleurum chinense, Herba plantaginis (plantain), Schisandra chinensis (magnolia vine fruit), Herba menthae (mint), and Stephania japonica (snake vine), used in traditional Chinese medicine (TCM), were tested in a liquid-based, slow-killing assay with C. elegans infected with MDR P. aeruginosa. This study identified 36 extracts prolonging C. elegans survival, and 4 with antimicrobial activity against P. aeruginosa. The 18s rRNA amplicon identified the fungal strains producing these four extracts with in vitro and in vivo activity as Alternaria sp. from Angelica sinensis; Herba plantaginis and Phoma exigua from Alangium sp.; and Aspergillus sydowii from Plantago depressa [54].
In another study, polysaccharide extracts from Sophora moorcroftiana seeds, a medicinal shrub endemic to China, were shown to improve the lifespan of C. elegans as well as its reproductive capacity upon P. aeruginosa infection, and they demonstrated specific antibacterial activity [55]. Both studies suggest that these natural compounds may enhance host immunity and/or reduce bacterial virulence.
Swietenia macrophylla (S. macrophylla), a source of limonoids (modified terpenoids), has shown antimicrobial activity against MRSA [46]. Moreover, S. macrophylla seed extracts significantly improve the survival of P. aeruginosa-infected C. elegans. Notably, seed ethyl acetate and methanol extracts do not inhibit P. aeruginosa growth. However, S. macrophylla enhances the expression of the immune modulator gene lys-7, which is known to be suppressed by the QS mechanisms of P. aeruginosa [56,57].
Modulation of the QS mechanisms by natural products is further supported by studies highlighting the antibacterial effects of CBO. In vitro studies have shown that CBO inhibits elastases A and B and regulates signalling molecules of P. aeruginosa. Importantly, it also decreases the expression and production of other virulence factors, such as chitinase, pyocyanin, exopolysaccharide, and biofilm formation, at subinhibitory concentrations (<3.2%). Treatment of P. aeruginosa-infected C. elegans with 1.6% v/v clove oil improves worm survival in vivo [58,59]. Similar effects were observed by using Murraya koenigii (curry tree), a medicinal plant with anti-infective and antioxidant properties [60].
Marine plants, such as the macroalgae Chondrus crispus (C. crispus), a red seaweed, are rich sources of compounds with immunomodulatory abilities [78]. A water extract of C. crispus has been shown to improve the survival of P. aeruginosa-infected C. elegans at a 500 µg/mL concentration. These effects result from both host immune-system activation through the pmk-1, daf-2/daf-16, and skn-1 pathways, and QS and virulence gene suppression [61]. Furthermore, an extract of Ascophyllum nodosum, a brown seaweed, promotes the survival of C. elegans through similar mechanisms and by significantly reducing biofilm formation by P. aeruginosa [62].
Ayurveda, an ancient holistic approach used for disease prevention and treatment, has also been extensively studied in the context of innovative drug discovery. For instance, TF, a formulation of the three plants Phyllanthus emblica (Indian gooseberry), Terminalia bellerica (Bibhitaka), and Terminalia chebula (Chebulic Myrobalan), is commonly used in Ayurvedic medicine to improve general health and to treat a variety of diseases [79].
C. elegans has been used as an in vivo model to test TF antibacterial activity against S. aureus, P. aeruginosa, S. pyogenes, S. marcescens, and C. violaceum [63]. All pathogens except S. pyogenes showed reduced virulence (18–45%) towards the nematode host following pretreatment with TF. Moreover, C. elegans pretreated with TF also showed an increased survival rate (14–41%) following infection with these five pathogens. In addition, TF is effective in worms infected with S. marcescens, C. violaceum, or S. aureus. However, TF is ineffective in worms exposed to a mixed population of S. aureus and P. aeruginosa. TF antibacterial activity can be attributed to a combination of reduced QS pigment production in Gram-negative bacteria, virulent enzyme inhibition in S. aureus, and upregulation of the host immune defence accompanied by decreased biofilm formation by S. marcescens and S. aureus. In a similar study, bacteria pretreated with TF leaf extracts showed decreased virulence associated with QS modifications [79].
Psoralea corylifolia (Babchi), a herb used in traditional Chinese medicine and Ayurveda to treat skin diseases [80], has also been studied for its QS-dampening constituents. Its seed extracts contain bakuchiol, a terpenophenol that inhibits in vitro biofilm formation by P. aeruginosa, A. hydrophila, C. violaceum, S. marcescens, and L. monocytogenes.
To study their effects in vivo, C. elegans infected with P. aeruginosa were treated with 1000 µg/mL P. corylifolia seed extract. Worms survived longer than untreated nematodes [64]. While increased survival was also observed in a similar study testing Mangifera indica (mango) leaf extracts [65], underlying in vivo mechanisms remain to be investigated.
Streptococcus pyogenes (S. pyogenes), a Gram-positive, β-haemolytic streptococcus, can cause highly contagious infections with serious complications, such as rheumatic fever. Although first-line treatment with penicillin is still effective, some resistant strains are emerging [81]. The bioflavonoid fukugiside from Garcinia travancorica (mangosteen) has shown promising concentration-dependent antibacterial properties against S. pyogenes in vitro by downregulating various virulence genes and reducing its ability to evade phagocytosis. Fukugiside significantly improves the survival rate of C. elegans infected with S. pyogenes without displaying any toxicity [66]. Betulin, a triterpenoid present in the bark of birch trees, has also shown similar effects in vivo [82]. Interestingly, in an innovative antimicrobial approach, extracts of Tripterygium wilfordii (Thunder God Vine) were able to photosensitise S. pyogenes and S. aureus for the treatment of skin infections. Accordingly, the combination of T. wilfordii extract and antimicrobial photodynamic therapy improved the lifespan of C. elegans infected with S. pyogenes [67].
Among pathogens that conventional antimicrobials cannot eradicate [83], shigatoxin-producing enterohemorrhagic E coli 0157:H7 causes bloody diarrhoea, which can only be managed supportively. Within this context, the C. elegans model was used to test the antibacterial properties of clove oil, known to downregulate various virulence factors, including QS molecules, and biofilm formation [58,59]. Indeed, eugenol, the major constituent of clove oil, displays high efficacy against E coli 0157:H7 and prolongs nematode survival [68].
Another gut pathogen, Salmonella typhimurium, is susceptible to B. oleracea var. Botrytis (cauliflower) byproduct extracts. Studies in cauliflower-treated C. elegans have shown a dose-dependent reduction in Salmonella typhimurium virulence factors, significantly reduced colonisation, and prolonged nematode survival [69,70].
In addition to flavonoids, phytoestrogens such as honokiol and magnolol, derived from magnolia species, have demonstrated antibacterial activity [71]. The survival of Vibrio cholerae-infected C. elegans can be significantly prolonged by treatment with these polyphenols. In vitro studies suggest that these compounds decrease host inflammatory responses to this potent diarrhoeal pathogen [74].
Burkholderia pseudomallei, a tropical pathogen that causes serious melioidosis, shows reduced pathogenicity in vivo when infected C. elegans are treated with curcumin (turmeric). Further investigations have shown that this effect relies on alterations of the bacterial transcriptome, resulting in the attenuation of virulence factors [72].
7. Natural Products Active against Fungal Infection
The C. elegans model has also successfully been used for the discovery of antifungal natural products (Table 2).

Table 2.
Antifungal activity of natural plant products in the C. elegans in vivo model. * MoA: mechanism of action; ** N.D.: not determined.
Candida albicans (C. albicans), causing opportunistic infections, has represented a major target for the discovery of natural antifungal products. The limited array of antifungal drugs and the intrinsic ability of C. albicans to quickly develop tolerance highlight the significance of the C. elegans infection model in this regard [89].
A total of 2560 natural products were screened to identify compounds favouring the survival of C. albicans-infected nematodes, and 12 triterpenoid-based saponins significantly prolonged it. Six of these compounds also inhibited in vitro growth of the yeast, including its resistant strains, and two prevented biofilm formation [87].
Saponins are plant glycosides known in traditional medicine for their antitumorigenic, antimicrobial, and anti-inflammatory properties. Although they can be toxic at high concentrations due to their haemolytic properties [90], they have shown promising potential as enhancers of photosensitiser uptake in combination with photodynamic treatments [87].
Gallic acid and gallates are secondary metabolites present in various plants and are known for their antioxidant and anti-inflammatory properties [91]. In vitro studies support their antifungal effects against Candida spp., Cryptococcus spp., Paracoccidioides spp., and Histoplasma capsulatum [92]. The in vivo model of C. albicans–C. elegans indicates that they significantly increase the nematode lifespan at >1 µg/mL concentrations, although their toxicity is also increased at high concentrations [84].
Another hydrophobic polyphenol, thymol, also prolongs the survival of C. albicans-infected worms and shows antibiofilm activity in vitro. This compound, extracted from thyme, also prevents dysregulation of the host p38 MAPK signalling pathway, playing a key role in the immune response [93].
An investigative study identified 15 out of 1266 tested compounds that promoted the longevity of infected worms and prevented the fungal yeast–hyphal transition. They included plant-based compounds, caffeic acid phenethyl ester (CAPE), and lapachol [85]. CAPE is a polyphenolic ester naturally occurring in plants, and its bioactive form can be extracted from honeybee hive propolis. Its mechanism of action has been proposed to rely on yeast gene suppression and immunomodulation [94]. On the other hand, lapachol, a naphthoquinone derived from the bark of Tabebuia avellaneda (lapacho tree), is used in traditional Amazonian medicine as an antibacterial and antiparasitic agent [45]. Both compounds were shown to inhibit C. albicans biofilm formation in vitro, although only CAPE demonstrated antifungal effects in a murine model [85].
Fungal filamentation and biofilm formation are key virulence factors accounting for C. albicans pathogenicity by allowing microbes to colonise mucosal surfaces and medical devices, and to invade human tissues. Moreover, the biofilm matrix renders fungi resistant to immune defences and drugs, leading to intractable infections [95]. The non-toxic naturally occurring antifungals magnolol and honokiol significantly suppress in vitro yeast cell adhesion, hyphal growth, and biofilm formation at 4–32 μg/mL concentrations by downregulating virulence genes, including those of the Ras1-cAMP-Efg1 pathway. Importantly, both honokiol and magnolol significantly prolong the lifespan of C. albicans-infected worms by preventing colonisation [86]. In a similar study, tetrandrine (TET), an alkaloid obtained from the roots of Stephania tetrandra, a medicinal herb known as Fang Ji used in Chinese herbal medicine for joint diseases, inhibited the yeast-to-hyphae transition and biofilm formation in vitro by downregulating the expression of genes from the Ras/cAMP pathway, responsible for QS and biofilm formation [96]. Accordingly, TET enhanced the lifespan of infected nematodes at the same concentration range as magnolol and honokiol [88].
Major issues related to the use of antifungal agents are associated with their adverse effects and drug-resistance development, since treatment requires long-term administration due to frequent relapses [97]. Therefore, natural plants may represent a safe and effective alternative and C. elegans may provide an early detection tool to identify compounds with selective toxicity against fungi and/or unravel alternative mechanisms of action inhibiting fungal infection.
8. Conclusions
Microbial diseases have long been treated with plant extracts. The growing issue of antibiotic resistance has prompted research into natural substances as cutting-edge antimicrobial agents. The discovery of natural substances with antibacterial and antifungal characteristics has greatly benefited from using C. elegans as an in vivo model. The C. elegans model has been used to investigate simultaneously the toxicity and antimicrobial efficacy of plant-based products using survival assays. Furthermore, the interactions between plant-based products and host–pathogen interactions have been studied to delineate the mechanisms of action involved, such as quorum-sensing inhibition and host immune-induction activity. Moreover, C. elegans mutant strains and molecular techniques have been utilised to decipher the signalling pathways involved in the antimicrobial efficacy of plant-based products. However, issues such as poorly standardised plant extract preparations and inadequate data on safety and toxicity still exist.
Most importantly, as clearly emerging from our study, for a large number of natural products, mechanisms of action are still poorly defined. Moreover, the nature and chemical structure of active compounds and secondary metabolites are frequently unclear. Addressing these issues will represent a major research challenge in the near future, and the use of the C. elegans model may powerfully contribute to this effort by accelerating the identification of effective phytochemicals and by more reliably delineating their mechanisms of action.
On the other hand, since converting plant products into innovative antimicrobial drugs requires major funding to pinpoint active ingredients and standardise production and distribution processes, the use of C. elegans may lessen the financial risk involved and help promote investment in this area.
Author Contributions
Conceptualization, S.H.O.Z. and H.K.A.; methodology, S.H.O.Z. and H.K.A.; software, writing—original draft preparation, S.H.O.Z. and J.S.B.; writing—review and editing, S.H.O.Z., F.N.H., R.A.S. and H.K.A.; visualization, S.H.O.Z.; supervision. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We thank Giulio C. Spagnoli, National Research Council, Institute of Translational Pharmacology, Rome, Italy, for valuable improvement and discussions of the manuscript draft.
Conflicts of Interest
The authors declare no conflict of interest.
References
- WHO. Global Action Plan on Antimicrobial Resistance. Available online: https://www.who.int/publications-detail-redirect/9789241509763 (accessed on 4 April 2023).
- WHO. World Health Statistics. 2022. Available online: https://www.who.int/news/item/20-05-2022-world-health-statistics-2022 (accessed on 4 April 2023).
- Willemsen, A.; Reid, S.; Assefa, Y. A review of national action plans on antimicrobial resistance: Strengths and weaknesses. Antimicrob. Resist. Infect. Control 2022, 11, 90. [Google Scholar] [CrossRef]
- Zinner, S.H. The search for new antimicrobials: Why we need new options. Expert Rev. Anti Infect. Ther. 2005, 3, 907–913. [Google Scholar] [CrossRef] [PubMed]
- MacPherson, D.W.; Gushulak, B.D.; Baine, W.B.; Bala, S.; Gubbins, P.O.; Holtom, P.; Segarra-Newnham, M. Population mobility, globalization, and antimicrobial drug resistance. Emerg. Infect. Dis. 2009, 15, 1727–1732. [Google Scholar] [CrossRef]
- Antimicrobial Resistance Collaborators. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef] [PubMed]
- Interagency Coordination Group on Antimicrobial Resistance. No time to wait: Securing the future from drug-resistant infections. In Report to the Secretary General of the United Nations; World Health Organisation: Geneva, Switzerland, 2019. [Google Scholar]
- Boucher, H.W.; Talbot, G.H.; Bradley, J.S.; Edwards, J.E.; Gilbert, D.; Rice, L.B.; Scheld, M.; Spellberg, B.; Bartlett, J. Bad bugs, no drugs: No ESKAPE! An update from the Infectious Diseases Society of America. Clin. Infect. Dis. 2009, 48, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Peleg, A.Y.; Hooper, D.C. Hospital-acquired infections due to gram-negative bacteria. N. Engl. J. Med. 2010, 362, 1804–1813. [Google Scholar] [CrossRef]
- Founou, R.C.; Founou, L.L.; Essack, S.Y. Clinical and economic impact of antibiotic resistance in developing countries: A systematic review and meta-analysis. PLoS ONE 2017, 12, e0189621. [Google Scholar] [CrossRef] [PubMed]
- Rice, L.B. Federal funding for the study of antimicrobial resistance in nosocomial pathogens: No ESKAPE. J. Infect. Dis. 2008, 197, 1079–1081. [Google Scholar] [CrossRef]
- Navidinia, M. The clinical importance of emerging ESKAPE pathogens in nosocomial infections. Arch. Adv. Biosci. 2016, 7, 43–57. [Google Scholar] [CrossRef]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Link, E.M.; Hardiman, G.; Sluder, A.E.; Johnson, C.D.; Liu, L.X. Therapeutic target discovery using Caenorhabditis elegans. Pharmacogenomics 2000, 1, 203–217. [Google Scholar] [CrossRef]
- Jones, A.K.; Buckingham, S.D.; Sattelle, D.B. Chemistry-to-gene screens in Caenorhabditis elegans. Nat. Rev. Drug Discov. 2005, 4, 321–330. [Google Scholar] [CrossRef]
- Moy, T.I.; Ball, A.R.; Anklesaria, Z.; Casadei, G.; Lewis, K.; Ausubel, F.M. Identification of novel antimicrobials using a live-animal infection model. Proc. Natl. Acad. Sci. USA 2006, 103, 10414–10419. [Google Scholar] [CrossRef]
- Moy, T.I.; Conery, A.L.; Larkins-Ford, J.; Wu, G.; Mazitschek, R.; Casadei, G.; Lewis, K.; Carpenter, A.E.; Ausubel, F.M. High-throughput screen for novel antimicrobials using a whole animal infection model. ACS Chem. Biol. 2009, 4, 527–533. [Google Scholar] [CrossRef]
- Gigante, V.; Sati, H.; Beyer, P. Recent advances and challenges in antibacterial drug development. ADMET DMPK 2022, 10, 147–151. [Google Scholar] [CrossRef] [PubMed]
- Lipinski, C.; Hopkins, A. Navigating chemical space for biology and medicine. Nature 2004, 432, 855–861. [Google Scholar] [CrossRef]
- Projan, S.J.; Shlaes, D.M. Antibacterial drug discovery: Is it all downhill from here? Clin. Microbiol. Infect. 2004, 10 (Suppl. S4), 18–22. [Google Scholar] [CrossRef] [PubMed]
- Li, X.Z.; Nikaido, H. Efflux-mediated drug resistance in bacteria. Drugs 2004, 64, 159–204. [Google Scholar] [CrossRef] [PubMed]
- Horrobin, D.F. Modern biomedical research: An internally self-consistent universe with little contact with medical reality? Nat. Rev. Drug Discov. 2003, 2, 151–154. [Google Scholar] [CrossRef] [PubMed]
- Williams, M. A return to the fundamentals of drug discovery? Curr. Opin. Investig. Drugs 2004, 5, 29–33. [Google Scholar] [PubMed]
- Silver, L.L. Challenges of antibacterial discovery. Clin. Microbiol. Rev. 2011, 24, 71–109. [Google Scholar] [CrossRef] [PubMed]
- Adonizio, A.; Leal, S.M.; Ausubel, F.M.; Mathee, K. Attenuation of Pseudomonas aeruginosa virulence by medicinal plants in a Caenorhabditis elegans model system. J. Med. Microbiol. 2008, 57, 809–813. [Google Scholar] [CrossRef] [PubMed]
- Burns, A.R.; Kwok, T.C.; Howard, A.; Houston, E.; Johanson, K.; Chan, A.; Cutler, S.R.; McCourt, P.; Roy, P.J. High-throughput screening of small molecules for bioactivity and target identification in Caenorhabditis elegans. Nat. Protoc. 2006, 1, 1906–1914. [Google Scholar] [CrossRef]
- Tampakakis, E.; Okoli, I.; Mylonakis, E. A C. elegans-based, whole animal, in vivo screen for the identification of antifungal compounds. Nat. Protoc. 2008, 3, 1925–1931. [Google Scholar] [CrossRef] [PubMed]
- Gosai, S.J.; Kwak, J.H.; Luke, C.J.; Long, O.S.; King, D.E.; Kovatch, K.J.; Johnston, P.A.; Shun, T.Y.; Lazo, J.S.; Perlmutter, D.H.; et al. Automated high-content live animal drug screening using C. elegans expressing the aggregation prone serpin alpha1-antitrypsin Z. PLoS ONE 2010, 5, e15460. [Google Scholar] [CrossRef]
- Giunti, S.; Andersen, N.; Rayes, D.; De Rosa, M.J. Drug discovery: Insights from the invertebrate Caenorhabditis elegans. Pharmacol. Res. Perspect. 2021, 9, e00721. [Google Scholar] [CrossRef] [PubMed]
- Peterson, N.D.; Pukkila-Worley, R. Caenorhabditis elegans in high-throughput screens for anti-infective compounds. Curr. Opin. Immunol. 2018, 54, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Phillips, P.C. Self-fertilization sweeps up variation in the worm genome. Nat. Genet. 2012, 44, 237–238. [Google Scholar] [CrossRef] [PubMed]
- Newman, D.J.; Cragg, G.M. Natural Products as Sources of New Drugs over the Nearly Four Decades from 01/1981 to 09/2019. J. Nat. Prod. 2020, 83, 770–803. [Google Scholar] [CrossRef] [PubMed]
- Fabricant, D.S.; Farnsworth, N.R. The value of plants used in traditional medicine for drug discovery. Environ. Health Perspect. 2001, 109 (Suppl. S1), 69–75. [Google Scholar] [CrossRef]
- Cowan, M.M. Plant products as antimicrobial agents. Clin. Microbiol. Rev. 1999, 12, 564–582. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Garibay, F.; Téllez-Valdez, O.; Moreno-Torres, G.; Calderón, J.S. Flavonoids from Tephrosia major. A New Prenyl-β-hydroxychalcone. Z. Für Naturforschung C 2002, 57, 579–583. [Google Scholar] [CrossRef] [PubMed]
- Bennett, R.N.; Wallsgrove, R.M. Secondary metabolites in plant defence mechanisms. N. Phytol. 1994, 127, 617–633. [Google Scholar] [CrossRef]
- Savoia, D. Plant-derived antimicrobial compounds: Alternatives to antibiotics. Future Microbiol. 2012, 7, 979–990. [Google Scholar] [CrossRef] [PubMed]
- Mohammadi, A.; Nazari, H.; Imani, S.; Amrollahi, H. Antifungal activities and chemical composition of some medicinal plants. J. Mycol. Med. 2014, 24, e1–e8. [Google Scholar] [CrossRef] [PubMed]
- Cappiello, F.; Loffredo, M.R.; Del Plato, C.; Cammarone, S.; Casciaro, B.; Quaglio, D.; Mangoni, M.L.; Botta, B.; Ghirga, F. The Revaluation of Plant-Derived Terpenes to Fight Antibiotic-Resistant Infections. Antibiotics 2020, 9, 325. [Google Scholar] [CrossRef]
- Shin, J.; Prabhakaran, V.S.; Kim, K.S. The multi-faceted potential of plant-derived metabolites as antimicrobial agents against multidrug-resistant pathogens. Microb Pathog. 2018, 116, 209–214. [Google Scholar] [CrossRef]
- Othman, L.; Sleiman, A.; Abdel-Massih, R.M. Antimicrobial Activity of Polyphenols and Alkaloids in Middle Eastern Plants. Front. Microbiol. 2019, 10, 911. [Google Scholar] [CrossRef]
- Lewis, K.; Ausubel, F.M. Prospects for plant-derived antibacterials. Nat. Biotechnol. 2006, 24, 1504–1507. [Google Scholar] [CrossRef]
- Ruddaraju, L.K.; Pammi, S.V.N.; Guntuku, G.S.; Padavala, V.S.; Kolapalli, V.R.M. A review on anti-bacterials to combat resistance: From ancient era of plants and metals to present and future perspectives of green nano technological combinations. Asian J. Pharm. Sci. 2020, 15, 42–59. [Google Scholar] [CrossRef]
- Rodriguez-Yoldi, M.J. Anti-Inflammatory and Antioxidant Properties of Plant Extracts. Antioxidants 2021, 10, 921. [Google Scholar] [CrossRef]
- Cragg, G.; Newman, D. Natural products and drug discovery and development: A history of success and continuing promise for the future. Planta Med. 2014, 80, IL1. [Google Scholar] [CrossRef]
- Kong, C.; Yehye, W.A.; Abd Rahman, N.; Tan, M.W.; Nathan, S. Discovery of potential anti-infectives against Staphylococcus aureus using a Caenorhabditis elegans infection model. BMC Complement Altern. Med. 2014, 14, 4. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Abd-Rashid, B.A.; Ismail, Z.; Ismail, R.; Mak, J.W.; Pook, P.C.; Er, H.M.; Ong, C.E. In vitro effects of active constituents and extracts of Orthosiphon stamineus on the activities of three major human cDNA-expressed cytochrome P450 enzymes. Chem. Biol. Interact. 2011, 190, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Yam, M.F.; Mohamed, E.A.; Ang, L.F.; Pei, L.; Darwis, Y.; Mahmud, R.; Asmawi, M.Z.; Basir, R.; Ahmad, M. A simple isocratic HPLC method for the simultaneous determination of sinensetin, eupatorin, and 3’-hydroxy-5,6,7,4’-tetramethoxyflavone in Orthosiphon stamineus extracts. J. Acupunct. Meridian Stud. 2012, 5, 176–182. [Google Scholar] [CrossRef] [PubMed]
- Alexpandi, R.; Prasanth, M.I.; Ravi, A.V.; Balamurugan, K.; Durgadevi, R.; Srinivasan, R.; De Mesquita, J.F.; Pandian, S.K. Protective effect of neglected plant Diplocyclos palmatus on quorum sensing mediated infection of Serratia marcescens and UV-A induced photoaging in model Caenorhabditis elegans. J. Photochem. Photobiol. B 2019, 201, 111637. [Google Scholar] [CrossRef]
- Kim, Y.; Lee, J.W.; Kang, S.G.; Oh, S.; Griffiths, M.W. Bifidobacterium spp. influences the production of autoinducer-2 and biofilm formation by Escherichia coli O157:H7. Anaerobe 2012, 18, 539–545. [Google Scholar] [CrossRef]
- Patel, P.; Joshi, C.; Kothari, V. Anti-Pathogenic Efficacy and Molecular Targets of a Polyherbal Wound- Care Formulation (Herboheal) against Staphylococcus aureus. Infect. Disord. Drug Targets 2019, 19, 193–206. [Google Scholar] [CrossRef]
- Lee, K.M.; Lim, J.; Nam, S.; Yoon, M.Y.; Kwon, Y.K.; Jung, B.Y.; Park, Y.; Park, S.; Yoon, S.S. Inhibitory effects of broccoli extract on Escherichia coli O157:H7 quorum sensing and in vivo virulence. FEMS Microbiol. Lett. 2011, 321, 67–74. [Google Scholar] [CrossRef]
- Choi, E.J.; Kim, H.I.; Kim, J.A.; Jun, S.Y.; Kang, S.H.; Park, D.J.; Son, S.J.; Kim, Y.; Shin, O.S. The herbal-derived honokiol and magnolol enhances immune response to infection with methicillin-sensitive Staphylococcus aureus (MSSA) and methicillin-resistant S. aureus (MRSA). Appl. Microbiol. Biotechnol. 2015, 99, 4387–4396. [Google Scholar] [CrossRef]
- Zhou, Y.M.; Shao, L.; Li, J.A.; Han, L.Z.; Cai, W.J.; Zhu, C.B.; Chen, D.J. An efficient and novel screening model for assessing the bioactivity of extracts against multidrug-resistant Pseudomonas aeruginosa using Caenorhabditis elegans. Biosci. Biotechnol. Biochem. 2011, 75, 1746–1751. [Google Scholar] [CrossRef][Green Version]
- Zhang, Y.; Mi, D.Y.; Wang, J.; Luo, Y.P.; Yang, X.; Dong, S.; Ma, X.M.; Dong, K.Z. Constituent and effects of polysaccharides isolated from Sophora moorcroftiana seeds on lifespan, reproduction, stress resistance, and antimicrobial capacity in Caenorhabditis elegans. Chin. J. Nat. Med. 2018, 16, 252–260. [Google Scholar] [CrossRef]
- Dharmalingam, K.; Tan, B.K.; Mahmud, M.Z.; Sedek, S.A.; Majid, M.I.; Kuah, M.K.; Sulaiman, S.F.; Ooi, K.L.; Khan, N.A.; Muhammad, T.S.; et al. Swietenia macrophylla extract promotes the ability of Caenorhabditis elegans to survive Pseudomonas aeruginosa infection. J. Ethnopharmacol. 2012, 139, 657–663. [Google Scholar] [CrossRef]
- Evans, E.A.; Kawli, T.; Tan, M.W. Pseudomonas aeruginosa suppresses host immunity by activating the DAF-2 insulin-like signaling pathway in Caenorhabditis elegans. PLoS Pathog. 2008, 4, e1000175. [Google Scholar] [CrossRef]
- Haripriyan, J.; Omanakuttan, A.; Menon, N.D.; Vanuopadath, M.; Nair, S.S.; Corriden, R.; Nair, B.G.; Nizet, V.; Kumar, G.B. Clove Bud Oil Modulates Pathogenicity Phenotypes of the Opportunistic Human Pathogen Pseudomonas aeruginosa. Sci. Rep. 2018, 8, 3437. [Google Scholar] [CrossRef] [PubMed]
- Husain, F.M.; Ahmad, I.; Asif, M.; Tahseen, Q. Influence of clove oil on certain quorum-sensing-regulated functions and biofilm of Pseudomonas aeruginosa and Aeromonas hydrophila. J. Biosci. 2013, 38, 835–844. [Google Scholar] [CrossRef] [PubMed]
- Ganesh, P.S.; Rai, R.V. Inhibition of quorum-sensing-controlled virulence factors of Pseudomonas aeruginosa by Murraya koenigii essential oil: A study in a Caenorhabditis elegans infectious model. J. Med. Microbiol. 2016, 65, 1528–1535. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Hafting, J.; Critchley, A.T.; Banskota, A.H.; Prithiviraj, B. Components of the cultivated red seaweed Chondrus crispus enhance the immune response of Caenorhabditis elegans to Pseudomonas aeruginosa through the pmk-1, daf-2/daf-16, and skn-1 pathways. Appl. Environ. Microbiol. 2013, 79, 7343–7350. [Google Scholar] [CrossRef] [PubMed]
- Kandasamy, S.; Khan, W.; Evans, F.; Critchley, A.T.; Prithiviraj, B. Tasco(R): A product of Ascophyllum nodosum enhances immune response of Caenorhabditis elegans against Pseudomonas aeruginosa infection. Mar. Drugs 2012, 10, 84–105. [Google Scholar] [CrossRef]
- Patel, H.; Patel, F.; Jani, V.; Jha, N.; Ansari, A.; Paliwal, B.; Rathod, B.; Patel, D.; Patel, P.; Kothari, V. Anti-pathogenic potential of a classical ayurvedic Triphala formulation. F1000Res 2019, 8, 1126. [Google Scholar] [CrossRef]
- Husain, F.M.; Ahmad, I.; Khan, F.I.; Al-Shabib, N.A.; Baig, M.H.; Hussain, A.; Rehman, M.T.; Alajmi, M.F.; Lobb, K.A. Seed Extract of Psoralea corylifolia and Its Constituent Bakuchiol Impairs AHL-Based Quorum Sensing and Biofilm Formation in Food- and Human-Related Pathogens. Front. Cell. Infect. Microbiol. 2018, 8, 351. [Google Scholar] [CrossRef] [PubMed]
- Husain, F.M.; Ahmad, I.; Al-Thubiani, A.S.; Abulreesh, H.H.; AlHazza, I.M.; Aqil, F. Leaf Extracts of Mangifera indica L. Inhibit Quorum Sensing—Regulated Production of Virulence Factors and Biofilm in Test Bacteria. Front. Microbiol. 2017, 8, 727. [Google Scholar] [CrossRef]
- Nandu, T.G.; Subramenium, G.A.; Shiburaj, S.; Viszwapriya, D.; Iyer, P.M.; Balamurugan, K.; Rameshkumar, K.B.; Karutha Pandian, S. Fukugiside, a biflavonoid from Garcinia travancorica inhibits biofilm formation of Streptococcus pyogenes and its associated virulence factors. J. Med. Microbiol. 2018, 67, 1391–1401. [Google Scholar] [CrossRef]
- Alam, S.T.; Hwang, H.; Son, J.D.; Nguyen, U.T.T.; Park, J.S.; Kwon, H.C.; Kwon, J.; Kang, K. Natural photosensitizers from Tripterygium wilfordii and their antimicrobial photodynamic therapeutic effects in a Caenorhabditis elegans model. J. Photochem. Photobiol. B 2021, 218, 112184. [Google Scholar] [CrossRef]
- Kim, Y.G.; Lee, J.H.; Gwon, G.; Kim, S.I.; Park, J.G.; Lee, J. Essential Oils and Eugenols Inhibit Biofilm Formation and the Virulence of Escherichia coli O157:H7. Sci. Rep. 2016, 6, 36377. [Google Scholar] [CrossRef]
- Ibáñez-Peinado, D.; Pina-Pérez, C.; García-Carrión, G.; Martínez, A.; Rodrigo, D. In vivo Antimicrobial Activity Assessment of a Cauliflower By-Product Extract Against Salmonella Typhimurium. Front. Sustain. Food Syst. 2020, 4, 8. [Google Scholar] [CrossRef]
- Sanz-Puig, M.; Lazaro, E.; Armero, C.; Alvares, D.; Martinez, A.; Rodrigo, D.S. Typhimurium virulence changes caused by exposure to different non-thermal preservation treatments using C. elegans. Int. J. Food Microbiol. 2017, 262, 49–54. [Google Scholar] [CrossRef] [PubMed]
- Jacobo-Salcedo Mdel, R.; Gonzalez-Espindola, L.A.; Alonso-Castro, A.J.; Gonzalez-Martinez Mdel, R.; Dominguez, F.; Garcia-Carranca, A. Antimicrobial activity and cytotoxic effects of Magnolia dealbata and its active compounds. Nat. Prod. Commun. 2011, 6, 1121–1124. [Google Scholar] [CrossRef]
- Eng, S.A.; Nathan, S. Curcumin rescues Caenorhabditis elegans from a Burkholderia pseudomallei infection. Front. Microbiol. 2015, 6, 290. [Google Scholar] [CrossRef][Green Version]
- Yang, J.; Chen, Z.; Ching, P.; Shi, Q.; Li, X. An integrated microfluidic platform for evaluating in vivo antimicrobial activity of natural compounds using a whole-animal infection model. Lab. Chip. 2013, 13, 3373–3382. [Google Scholar] [CrossRef]
- Kim, H.-I.; Kim, J.-A.; Choi, E.-J.; Harris, J.B.; Jeong, S.-Y.; Son, S.-J.; Kim, Y.; Shin, O.S. In vitro and in vivo antimicrobial efficacy of natural plant-derived compounds against Vibrio cholerae of O1 El Tor Inaba serotype. Biosci. Biotechnol. Biochem. 2015, 79, 475–483. [Google Scholar] [CrossRef] [PubMed]
- Lakhundi, S.; Zhang, K. Methicillin-Resistant Staphylococcus aureus: Molecular Characterization, Evolution, and Epidemiology. Clin. Microbiol. Rev. 2018, 31, 10–1128. [Google Scholar] [CrossRef]
- Silby, M.W.; Winstanley, C.; Godfrey, S.A.C.; Levy, S.B.; Jackson, R.W. Pseudomonas genomes: Diverse and adaptable. FEMS Microbiol. Rev. 2011, 35, 652–680. [Google Scholar] [CrossRef] [PubMed]
- Pang, Z.; Raudonis, R.; Glick, B.R.; Lin, T.J.; Cheng, Z. Antibiotic resistance in Pseudomonas aeruginosa: Mechanisms and alternative therapeutic strategies. Biotechnol. Adv. 2019, 37, 177–192. [Google Scholar] [CrossRef]
- Holdt, S.L.; Kraan, S. Bioactive compounds in seaweed: Functional food applications and legislation. J. Appl. Phycol. 2011, 23, 543–597. [Google Scholar] [CrossRef]
- Bag, A.; Bhattacharyya, S.K.; Pal, N.K. Antibacterial potential of hydroalcoholic extracts of triphala components against multidrug-resistant uropathogenic bacteria—A preliminary report. Indian J. Exp. Biol. 2013, 51, 709–714. [Google Scholar]
- Khushboo, P.S.; Jadhav, V.M.; Kadam, V.J.; Sathe, N.S. Psoralea corylifolia Linn.-“Kushtanashini”. Pharmacogn. Rev. 2010, 4, 69–76. [Google Scholar] [CrossRef]
- Kanwal, S.; Vaitla, P. Streptococcus Pyogenes; StatPearls: Treasure Island, FL, USA, 2023. [Google Scholar]
- Viszwapriya, D.; Subramenium, G.A.; Prithika, U.; Balamurugan, K.; Pandian, S.K. Betulin inhibits virulence and biofilm of Streptococcus pyogenes by suppressing ropB core regulon, sagA and dltA. Pathog. Dis. 2016, 74, ftw088. [Google Scholar] [CrossRef] [PubMed]
- Vaou, N.; Stavropoulou, E.; Voidarou, C.; Tsigalou, C.; Bezirtzoglou, E. Towards Advances in Medicinal Plant Antimicrobial Activity: A Review Study on Challenges and Future Perspectives. Microorganisms 2021, 9, 2041. [Google Scholar] [CrossRef]
- Singulani, J.L.; Scorzoni, L.; Gomes, P.C.; Nazare, A.C.; Polaquini, C.R.; Regasini, L.O.; Fusco-Almeida, A.M.; Mendes-Giannini, M.J.S. Activity of gallic acid and its ester derivatives in Caenorhabditis elegans and zebrafish (Danio rerio) models. Future Med. Chem. 2017, 9, 1863–1872. [Google Scholar] [CrossRef]
- Breger, J.; Fuchs, B.B.; Aperis, G.; Moy, T.I.; Ausubel, F.M.; Mylonakis, E. Antifungal chemical compounds identified using a C. elegans pathogenicity assay. PLoS Pathog. 2007, 3, e18. [Google Scholar] [CrossRef]
- Sun, L.; Liao, K.; Wang, D. Effects of Magnolol and Honokiol on Adhesion, Yeast-Hyphal Transition, and Formation of Biofilm by Candida albicans. PLoS ONE 2015, 10, e0117695. [Google Scholar] [CrossRef]
- Coleman, J.J.; Okoli, I.; Tegos, G.P.; Holson, E.B.; Wagner, F.F.; Hamblin, M.R.; Mylonakis, E. Characterization of plant-derived saponin natural products against Candida albicans. ACS Chem. Biol. 2010, 5, 321–332. [Google Scholar] [CrossRef]
- Zhao, L.X.; Li, D.D.; Hu, D.D.; Hu, G.H.; Yan, L.; Wang, Y.; Jiang, Y.Y. Effect of Tetrandrine against Candida albicans Biofilms. PLoS ONE. 2013, 8, e79671. [Google Scholar] [CrossRef]
- Costa-de-Oliveira, S.; Rodrigues, A.G. Candida albicans Antifungal Resistance and Tolerance in Bloodstream Infections: The Triad Yeast-Host-Antifungal. Microorganisms 2020, 8, 154. [Google Scholar] [CrossRef]
- Podolak, I.; Galanty, A.; Sobolewska, D. Saponins as cytotoxic agents: A review. Phytochem. Rev. 2010, 9, 425–474. [Google Scholar] [CrossRef] [PubMed]
- Rosas, E.C.; Correa, L.B.; das Graças Henriques, M. Chapter 28—Antiinflammatory Properties of Schinus terebinthifolius and Its Use in Arthritic Conditions. In Bioactive Food as Dietary Interventions for Arthritis and Related Inflammatory Diseases, 2nd ed.; Watson, R.R., Preedy, V.R., Watson, R.R., Preedy, V.R., Eds.; Academic Press: Cambridge, MA, USA, 2019; pp. 489–505. [Google Scholar]
- De Paula, E.S.A.C.; Costa-Orlandi, C.B.; Gullo, F.P.; Sangalli-Leite, F.; de Oliveira, H.C.; da Silva Jde, F.; Scorzoni, L.; Pitangui Nde, S.; Rossi, S.A.; Benaducci, T.; et al. Antifungal Activity of Decyl Gallate against Several Species of Pathogenic Fungi. Evid. Based Complement. Altern. Med. 2014, 2014, 506273. [Google Scholar] [CrossRef] [PubMed]
- Shu, C.; Sun, L.; Zhang, W. Thymol has antifungal activity against Candida albicans during infection and maintains the innate immune response required for function of the p38 MAPK signaling pathway in Caenorhabditis elegans. Immunol. Res. 2016, 64, 1013–1024. [Google Scholar] [CrossRef] [PubMed]
- Murtaza, G.; Karim, S.; Akram, M.R.; Khan, S.A.; Azhar, S.; Mumtaz, A.; Bin Asad, M.H. Caffeic acid phenethyl ester and therapeutic potentials. Biomed. Res. Int. 2014, 2014, 145342. [Google Scholar] [CrossRef]
- Gulati, M.; Nobile, C.J. Candida albicans biofilms: Development, regulation, and molecular mechanisms. Microbes Infect. 2016, 18, 310–321. [Google Scholar] [CrossRef]
- Ahmad Khan, M.S.; Altaf, M.M.; Sajid, M. Chapter 14—Insights of Phyto-Compounds as Antipathogenic Agents: Controlling Strategies for Inhibiting Biofilms and Quorum Sensing in Candida albicans. In New Look to Phytomedicine; Ahmad Khan, M.S., Ahmad, I., Chattopadhyay, D., Eds.; Academic Press: Cambridge, MA, USA, 2019; pp. 367–389. [Google Scholar]
- Benson, J.M.; Nahata, M.C. Clinical use of systemic antifungal agents. Clin. Pharm. 1988, 7, 424–438. [Google Scholar] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).