Abstract
Background: Histone deacetylase inhibitors (HDACIs) are a relatively new class of potential drugs for treating cancer. Aim: Discovery of new anticancer agents targeting HDAC. Methods: New uracil and thiouracil derivatives panels were designed and synthesized as HDAC inhibitors. The synthesized compounds were tested against MCF-7, HepG2, and HCT-116. HDAC1 and HDAC4 inhibitory activities of these compounds were tested. The most active member was tested for its potential against cell cycle, apoptosis, caspase-3, and caspase-8. Docking studies were carried out against HDAC1. Results: Compounds 5a, 5b, 5f, 5i, 5k, and 5m exhibited promising cytotoxic activities. HDAC1 and HDAC4 inhibitory activities of these compounds were tested. Regarding the HDAC1 inhibitory activity, compound 5m was the most potent member (IC50 = 0.05 µg/mL) compared to trichostatin A (IC50 = 0.0349 µg/mL). For HDAC4, compound 5m showed superior activity (IC50 = 2.83 µg/mL) than trichostatin A (IC50 = 3.349 µg/mL). Compound 5m showed a high potential to arrest the HCT116 cell cycle at the G0-G1 phase. In addition, it showed an almost 17 times apoptotic effect (37.59%) compared to the control cells (2.17%). Furthermore, Compound 5m showed significant increases in the levels of caspase-3 and caspase-8. Finally, the uracil and thiouracil derivatives showed accepted binding mods against HDAC. Conclusions: Compound 5m has potential anticancer activity targeting HDAC with a significant apoptotic effect.
1. Introduction
Cancer continues to be one of humanity’s most significant public health issues, despite the enormous efforts to combat it [1]. According to the WHO, cancer was the leading cause of death worldwide in 2020, accounting for almost 10 million deaths, or about one in every six [2]. As a result, cancer is viewed as a serious issue for both the economy of nations and the economies of individuals [3,4]. Examples of how the complication of cancer pathology manifests itself include oncogenic mutations, multi-drug resistance, and the activation of compensatory mechanisms [5,6,7]. Finding anticancer options that are more potent and less harmful, based on the various biological and molecular characteristics of cancer pathogenesis, is therefore crucial.
One of the main epigenetic pathways implicated in cancer development is histone acetylation [8]. Histone acetylation is a required precursor to other processes of epigenetic modifications, such as methylation or phosphorylation, and it not only causes genetic alterations on its own [9,10]. Histone acetyltransferases (HAT) and histone deacetylases (HDAC) are two antagonistic categories of enzymes that control the process of histone acetylation. As lysine residues on histone and non-histone proteins are acetylated, heterochromatin is transformed into euchromatin, whereas HDACs play the opposite role by deacetylating chromatin to return it to its more condensed condition [11]
A relatively emerging family of prospective medications for treating hyperproliferative illnesses is histone deacetylase inhibitors (HDACIs) [12,13,14]. These inhibitors bind directly to the HDAC active site and block substrate access, producing an accumulation of acetylated histone [15,16,17]. They can affect differentiation, growth arrest, and/or apoptosis in transformed cell cultures due to their diverse biological activities [18,19]. There is a high demand for novel HDACIs as HDACs have become a key tactic in anticancer drug research [20,21]. Some families of tiny, powerful HDACIs have recently been discovered (Figure 1) [22,23].
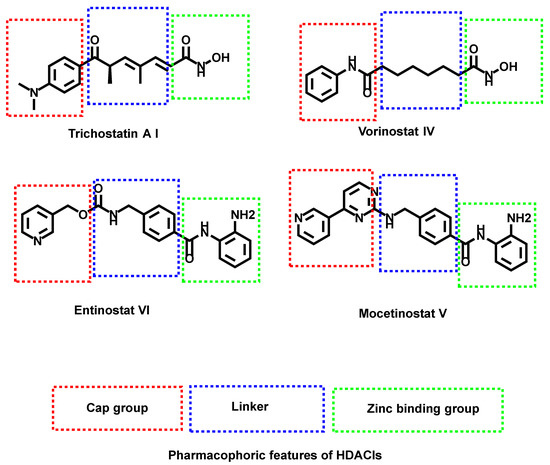
Figure 1.
Some reported HDAC inhibitors and their basic pharmacophoric features.
HDACIs should have a cap group, a spacer, and a functional group as basic pharmacophores [24]. The reported functional groups are hydroxamic acids, carboxylic acids, and phenylene diamines [25].
Besides, uracil and thiouracil moieties are important N-containing heterocycles in medicinal chemistry and drug discovery [26] due to their wide scope of biological activity [26], especially antitumor activities [27]. So, our goal is the design and synthesis of new uracil and thiouracil-containing derivatives targeting HDAC with promising effects against cancer.
Rationale of Molecular Design
Studying the SAR of the HDAC inhibitors class revealed three pharmacophoric features essential for maximal fitting in the active site of HDAC. These features include (i) a zinc-binding region group (ZBG) which can interact with the zinc atom in the active site, (ii) a linker moiety that can occupy the tubular access of the active site, and (iii) a cap group which can occupy the surface recognition motif [28] (Figure 2).
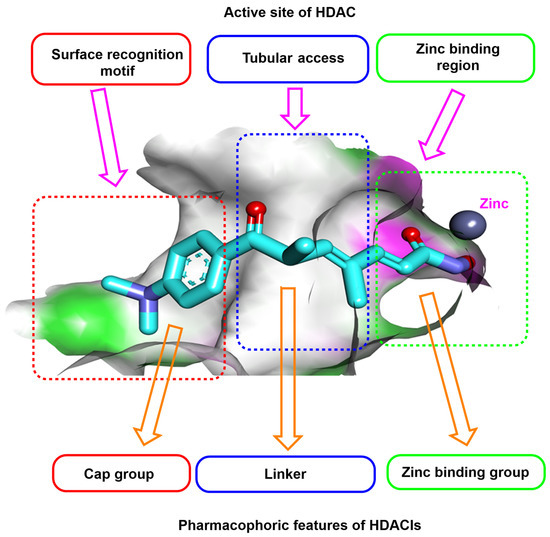
Figure 2.
Schematic diagram of trichostatin A occupying the active site of HDAC and the essential pharmacophoric features of HDAC inhibitors.
In this work, we aimed to synthesize new compounds targeting HDAC. The new compounds were designed to possess the pharmacophoric features of HDAC inhibitors. Many derivatives were applied in this work to reach a good insight into the SAR of the synthesized compounds as potential anticancer agents.
The designed compounds varied in their different pharmacophoric features. For the zinc-binding region, two bioisosters were used. These isosters are the substituted thiouracil (5a–g and 6a–d) and uracil (5h–m and 7a–c) derivatives. Different benzyl derivatives (5a–g and 5i–m) were used as linkers. In one compound (5h), a methylene group was used as a linker. Additional series (6a–d and 7a–c) comprise different cyclic structures as a linker moiety. Regarding the cap group, it was thiouracil (5a–g and 6a–d) and uracil (5h–m and 7a–c) derivatives (Figure 3).
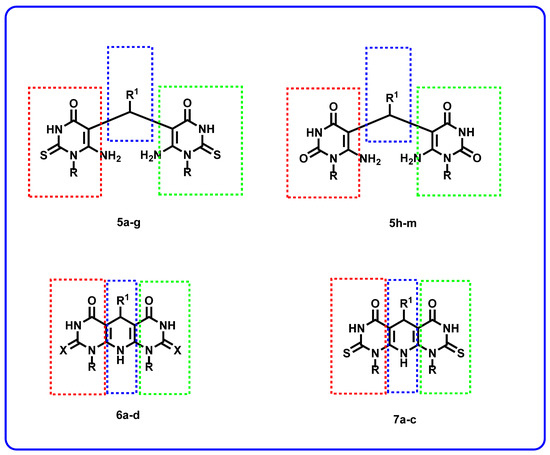
Figure 3.
The target compounds represent the essential pharmacophoric features of HDAC inhibitors.
2. Results and Discussion
2.1. Chemistry
The ability of 6-aminouracil to react with aliphatic or aromatic carbonyl compounds [29,30,31,32,33,34,35] has been amply demonstrated up till now. The search for new biologically active substances has fueled interest in these reactions, as the presence of a uracil moiety that is a pharmacophore in an organic molecule frequently provides the molecule with some kind of biological effect. Significant progress has been made in uracil derivatives in the field of chemistry. Our protocol is directed towards synthesized novel derivatives of 5,5′-(arylmethylene)bis(6-aminouracils) and dipyrimidopyridines. 6-Amino-2-oxo(thioxo)pyrimidine-4-ones 3a,b was used as starting material and prepared according to the reported methods [29,30,31,32,33,34] as shown in Scheme 1.
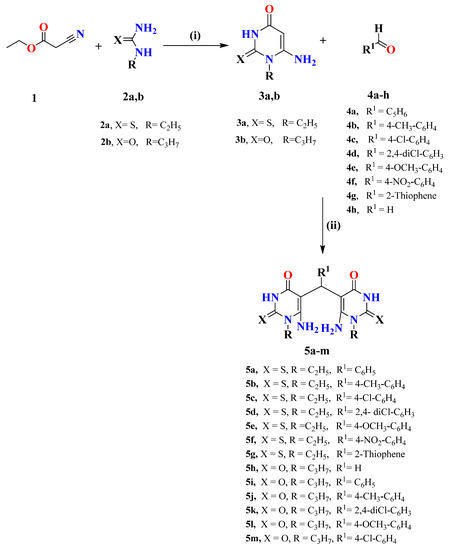
Scheme 1.
Synthesis of compounds 5a–m. Reagents and conditions: (i) C2H5ONa, EtOH, heating under reflux, 8–10 h.; (ii) EtOH, c.HCl, R.T., 2–3 h.
The condensation of appropriate aliphatic or aromatic aldehydes (at the ratio carbonyl compound to aminouracils 1:2) with compounds 3a,b in ethanol in the presence of HCl at room temperature resulting in 5,5′-bisdiaminopyrimidines 5a–m in good yields. The latter compounds undergo intermolecular dehydration, as shown in Figure 4. The desired compounds 5a–m were approved by spectral data 1H-, 13C NMR, IR, and Mass spectra. 1H NMR spectra of compounds 5a–m showed a characteristic singlet signals of CH-5 at the range of δ 5.37–6.05 ppm, as well as broad singlet signals characteristic for the 2NH2-6 group at δ 7.05–7.95 ppm.
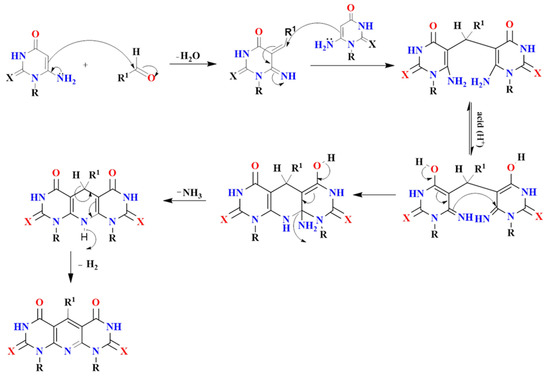
Figure 4.
A plausible reaction mechanism for synthesized compounds 5a–m, 6a–d and 7a–c.
Dipyrimidopyridine derivatives 6a–d were synthesized in good yields from refluxing of compounds 5a, f, h, l with a mixture of AcOH/c.HCl for 2.5 h (Scheme 2). The reaction proceeds through the same idea of the Hantzsch reaction via intramolecular cyclization accompanied by the evolution of NH3 due to the attack of the amino group of one unit to the electrophilic carbon center of C=NH, as illustrated in Figure 4. The novel compounds were revealed by 1H NMR, 13C NMR, IR, and Mass spectra. 1H NMR of compounds 6a–c exhibit the disappearance of the four singlet signal protons characteristic for 2NH2 groups and the appearance of a singlet signal of NH-10 at δ 7.77–7.37 ppm and another singlet signal characteristic for CH-5 at δ 5.46–5.75 ppm. Moreover, compound 6d showed a characteristic singlet signal for NH-10 at δ 7.18 and CH2-5 at δ 3.49 ppm. A characteristic C-5 signal at the δ 80–90 ppm range was noticed in 13C NMR for the mentioned compounds.
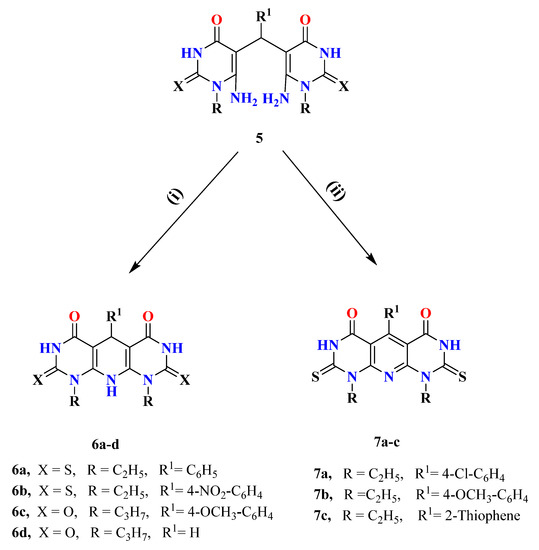
Scheme 2.
Synthesis of compounds 6a–d and 7a–c. Reagents and conditions: (i) g.HAc, c.HCl, heat under reflux, 2.5 h.; (ii) g.HAc, c.HCl, heat under reflux, 4–5 h.
On the other hand, Refluxing compounds 5c, e, g with a mixture of AcOH/c.HCl for 4–5 h take place through intramolecular oxidative cyclization afforded 7a–c (Scheme 2), which is proved by all spectral data. 1H NMR proved, without doubt, the formation of oxidative cyclized compounds 7a–c via the disappearance of the singlet signal of NH-10 at the region of δ 7 ppm as well as the clearance of the spectra from the singlet signal of CH-5 at the range δ 5.50–5.70 ppm. Moreover, the characteristic C-5 signal appeared at the normal deshielded aromatic region in 13C NMR, which proved the complete aromatization of the pyridine ring. A plausible reaction mechanism might be illustrated as follows in Figure 4.
2.2. Biological Testing
2.2.1. In Vitro Cytotoxic Activities
Anti-proliferative effect of the target compounds was assessed against a panel of tumor cell lines, including MCF-7 (human breast cancer cell line), HepG2 (human liver carcinoma cell line), colorectal carcinoma (HCT-116) using MTT assay [36]. Sorafenib was used as a reference drug. From the results presented in Table 1, it is clear that 5a, 5g, and 5f has promising anti-proliferative effect against MCF-7 with IC50 values of 11 ± 1.6, 21 ± 2.2, 9.3 ± 3.4 µM, respectively compared to sorafenib (IC50 = 141 ± 3 µM). In addition, compound 5b showed high activity against HCT-116 cells with an IC50 value of 21 ± 2.4 µM compared to sorafenib (IC50 = 177 ± 0.93 µM). Furthermore, compounds 5i, 5k, and 5m exhibited high cytotoxic effect against HepG2 with IC50 values of 4 ± 1, 5 ± 2, and 3.3 ± 0.56 µM, respectively, compared to sorafenib (IC50 = 17 ± 2.3 µM).

Table 1.
In vitro cytotoxic activities of the target compounds against MCF-7, HepG2, and HCT-116 cell lines.
Some compounds showed moderate activities against MCF-7 as 5c, 5b, 5d, 5e, 5j and 5m with IC50 values of 77 ± 2.3, 55 ± 2.8, 62 ± 2.1, 60 ± 0.49, 71 ± 2, and 52 ± 3.5 µM, respectively. Additionally, compounds 5a, 5c, 5e, 5f, 5g, 5h, and 5i showed moderate cytotoxic activity against HCT-116 cells with IC50 values ranging from 88 ± 2.4 to 97 ± 1.2 µM. On the other hand, the other compounds showed weak activities against the tested cell lines.
2.2.2. Structure-Activity Relationship
The cytotoxicity results show that the synthesized compounds with open chain linkers (5a–m) are more active than those with cyclic linkers (6a–d, and 7a–c). These results may be explained by the flexibility of the open chain linkers, which may give a good chance for flexible orientations in the active pocket of the target enzyme. On the other hand, the cyclic linker may restrict the good fitting with the active site of the receptor.
For the cap group, it was found that uracil derivatives are more active than thiouracil derivatives. These findings may be due to the high chance of the oxygen atom of uracil moiety to form electrostatic attraction at the cap-binding region. On the other hand, the sulfur atom of the thiouracil moiety has less chance to form electrostatic bonds than the uracil moiety. More clarification about the binding pattern of the synthesized compounds was clarified in the docking section.
Depending on the cytotoxicity against MCF-7, we can reach more details about SAR. For the thiouracil derivatives with open-chain linkers (5a–g), it was found that activity decreased upon substitution at the linker region as the order of 4-nitrophenyl > phenyl > thiophene > 4-methylphenyl > 4-methoxyphenyl > 2,4-dichlorophenyl > 4-chlorophenyl.
For the uracil derivatives with open chain linkers (5h–m), it was found that activity decreased upon substitution at the linker region as the order of 4-chlorophenyl > 4-methylphenyl > H > 2,4-dichlorophenyl > phenyl > 4-methoxyphenyl.
For the thiouracil derivatives with cyclic linkers (6a,b), it was found that the substitution phenyl moiety (6a) is more advantageous than the substitution with 4-nitrophenyl moiety (6b).
For the uracil derivatives with cyclic linkers (6c,d), it was found that the unsubstituted cyclic linker (6d) is more advantageous than the substituted one with 4-methoxyphenyl moiety (6c).
Compared to the activity of the compound, the thiouracil derivatives 6c (bearing a propyl moiety at both cap and zinc binding group) and 7b (bearing an ethyl moiety at both cap and zinc binding group) indicated that the substitution with propyl moiety is better for biological activity.
2.2.3. HDAC1 and HDAC4 Inhibitory Assay
HDAC1 plays a critical role in proliferating and senescent cells in culture and young and old tissues in vivo [37]. HDAC1 levels are also essential for regulating apoptosis [38]. Basic and clinical experimental evidence has established that HDAC4 performs various functions [39]. Accordingly, HDAC1 and HDAC4 were selected for testing in this work.
To assess the mechanism of cytotoxicity of the synthesized compounds, HDAC1 and HDAC4 inhibitory activities of the most cytotoxic compounds (5a, 5b, 5f, 5i, 5k, and 5m) were tested. Trichostatin A was used as a reference compound. The results were summarized as IC50 values in Table 2.

Table 2.
Effect of compounds against as HDAC1 and HDAC4.
In general, as appeared in Table 2, the synthesized compounds have higher selectivity towards HDAC1 than HDAC4. These results match the reported behavior of uracil derivatives against HDAC1 [40].
Regarding the HDAC1 inhibitory activity, compound 5m was the most potent member (IC50 = 0.05 µg/mL) compared to trichostatin A (IC50 = 0.035 µg/mL). In addition, compounds 5i and 5k showed strong activities with IC50 values of 0.146 and 0.23 µg/mL, respectively. Furthermore, compounds 5a, 5b, and 5f showed moderate HDAC1 inhibitory activities with IC50 values of 1.34, 1.01, and 1.9 µg/mL, respectively.
For HDAC4 inhibitory activity, compound 5m showed superior activity (IC50 = 2.83 µg/mL) than trichostatin A (IC50 = 3.35 µg/mL). In addition, compounds 5a, 5b, 5f, 5i, and 5k showed moderate HDAC4 inhibitory activities with IC50 values ranging from 5.27 to 8.311 µg/mL.
2.2.4. Cell Cycle Analysis
The effect of the most promising compound, 5m, against the cell cycle was tested in HCT116 cells. The tested cells were subjected to compound 5m with a concentration of 78 µM (IC50 value of compound 5m) after 72 h. As presented in Table 3 and Figure 5, the percent of HCT-116 treated cells increased at the %G0–G1 phase (55.31) compared to its concentration in the control cells (43.82%). On the contrary, the percentage of HCT-116 cells decreased at the S phase from 41.19 to 34.88%. Similarly, it decreased at the G2/M phase from 15.04% to 9.81%. Such findings revealed that compound 5m arrested the HCT-116 cell growth at G0–G1 phase.

Table 3.
Effect of compound 5m on stages of the cell death process in HCT-116 cells after 72 h treatment.
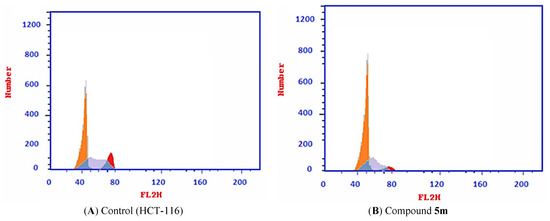
Figure 5.
Flow cytometric analysis of cell cycle phases posts compounds 5m treatment. (A) The representative histograms show the cell cycle distribution of control (HCT-116). (B) HCT-116 cells treated with compound 5m.
2.2.5. Apoptosis Analysis
Compound 5m was tested for apoptotic effect in HCT-116 using Annexin-V/propidium iodide (PI) staining assay. The tested cells were subjected to 78 µM from compound 5m with an incubation time of 72 h. The results revealed that the apoptotic effect of compound 5m was almost 17 times (37.59%) more than observed in control cells (2.17%). The early apoptosis increased from 0.43% to 22.36%. The late apoptosis increased from 0.18 to 13.14% (Table 4 and Supplementary data).

Table 4.
Apoptotic effect of compound 5m on HCT-116 cells after 72 h treatment.
2.2.6. Caspase-3 and Caspase-8 Determination
Due to the potential effect of both caspase-3 and caspase-8 on the apoptosis pathway [41], the effects of the most active candidate 5m on the level of caspase-3 and caspase-8 were tested on HCT-116 72 h. As shown in Table 5, Compound 5m showed significant increases in the levels of caspase-3 and caspase-8 (5- and 2.5-fold, respectively) compared to the control cells. Taking Staurosporine as a positive control, compound 5m showed slightly less activity against the level of caspase-3 and caspase-8, as deduced from Table 5.

Table 5.
Effect of compound 5m on the level of caspase-3 and caspase-8 in HCT-116 cells after 72 h treatment.
2.2.7. Cytotoxicity against Normal Cell Line
The cytotoxicity of the most promising candidate, 5m against normal cells (WI-38), was assessed using an MTT assay. Staurosporine was used as a reference molecule. The results are summarized in Table 1.
The results revealed that compound 5m has very low cytotoxicity against WI-38 cells with an IC50 value of 65.67 µM compared with Staurosporine (IC50 = 51.48 µM). The obtained results indicated that compound 5m is safer than Staurosporine.
2.3. Docking Studies
All the synthesized compounds were docked against the crystal structure of HDAC1 (PDB ID: 1C3R) using MOE2019 software to reach a good insight into their binding pattern. Trichostatin A (The co-crystallized ligand) was utilized as a reference molecule. The binding pattern of some examples is presented below. The binding free energies (∆G) for all the target molecules against HDAC1 are shown in Table 6.

Table 6.
Binding free energies (∆G in Kcal/mol) of the synthesized compounds and trichostatin A against HDAC1.
Trichostatin A exhibited a binding score of −19.11 kcal/mol against HDAC1. The hydroxamic acid group occupied the zinc-binding region forming many hydrogen bonds with Gly140 and Tyr297. Also, the hydroxamic acid group was involved in an electrostatic interaction with zinc ions. Three hydrophobic bonds were formed between the linker chain and Leu265, Phe198, and His170. The surface recognition motif was occupied by the N,N-dimethylaniline moiety (Figure 6).
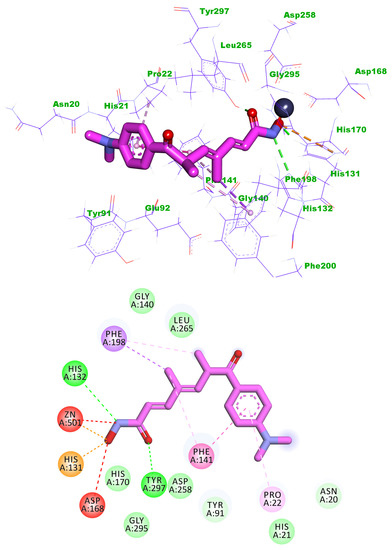
Figure 6.
The Predicted binding mode of Trichostatin A with the active site of HDAC1.
Compound 5b exhibited a binding mode like that of Trichostatin A, with a binding score of −14.13 kcal/mol. One of the two 6-amino-1-ethyl-2-thioxo-2,3-dihydropyrimidin-4(1H)-one moieties occupied the zinc-binding region forming one hydrogen bond withHis131. Such moiety formed three electrostatic bonds with Zn ion, Tyr297, and His132, and two hydrophobic bonds with Phe198 and His132. The chlorobenzene moiety occupied the linker region forming two hydrophobic interactions with Leu265 and Tyr196. The other 6-amino-1-ethyl-2-thioxo-2,3-dihydropyrimidin-4(1H)-one moiety occupied the surface recognition region forming one hydrophobic interaction with Leu265 (Figure 7).
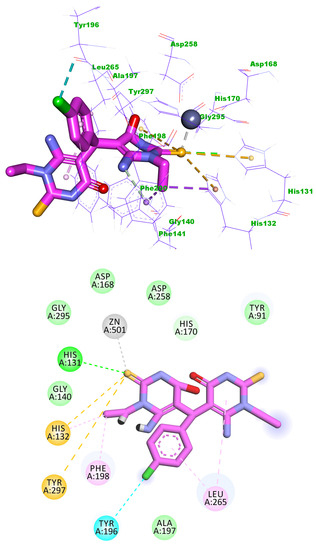
Figure 7.
Predicted binding mode of compound 5b with the active sites of HDAC1.
Compound 5i showed a binding energy of −17.35 kcal/mol. The first 6-amino-1-propylpyrimidine-2,4(1H,3H)-dione moiety occupied the zinc binding region forming two hydrogen bonds with His131 and Asp258. In addition, it formed three electrostatic bonds with Zn ion and His132. Also, it formed two hydrophobic bonds with Cys142 and Leu23. The phenyl moiety occupied the linker region. The other 6-amino-1-propylpyrimidine-2,4(1H,3H)-dione moiety occupied the surface recognition region forming one hydrogen bond and one hydrophobic interaction with Leu265 (Figure 8).
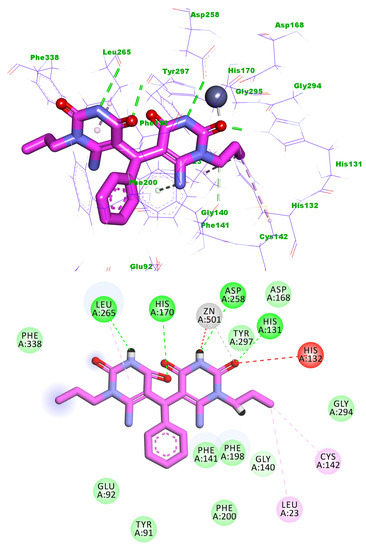
Figure 8.
Predicted binding mode of compound 5i with the active site of HDAC1.
Compound 5k showed a binding energy of −15.36 kcal/mol. One 6-amino-1-propylpyrimidine-2,4(1H,3H)-dione moiety was oriented into the zinc binding region, forming one hydrogen bond with Tyr297. Three electrostatic interactions were formed between the first 6-amino-1-propylpyrimidine-2,4(1H,3H)-dione moiety and Zn ion, His170, and Phe141. Also, it formed three hydrophobic interactions with Phe141, Phe198, and His132. The central tolyl moiety occupied the linker region forming a hydrophobic interaction with Phe200. The other 6-amino-1-propylpyrimidine-2,4(1H,3H)-dione moiety occupied the surface recognition region forming three hydrophobic interactions with Leu265, Pro22, and Phe141 (Figure 9).
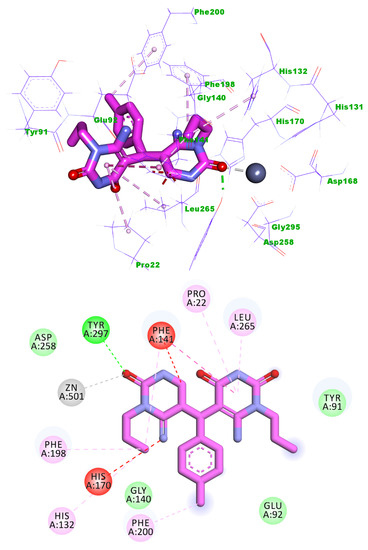
Figure 9.
Predicted binding mode of compound 5k with the active site of HDAC1.
Compound 5m showed a binding energy of −14.99 kcal/mol. The first 6-amino-1-propylpyrimidine-2,4(1H,3H)-dione moiety occupied the zinc binding region forming two hydrogen bonds with Tyr297 and His170. In addition, it formed four electrostatic interactions with Zn ion, Leu265, Gly140, and Phe198. Also, it formed four hydrophobic interactions with Phe141, Phe198, Leu265, and His132. The central 4-chlorophenyl moiety occupied the linker region forming a hydrophobic interaction with Leu265. The other 6-amino-1-propylpyrimidine-2,4(1H,3H)-dione moiety occupied the surface recognition region forming two hydrophobic interactions with Tyr91 and Phe198. In addition, it formed a hydrogen bond with Tyr91 (Figure 10).
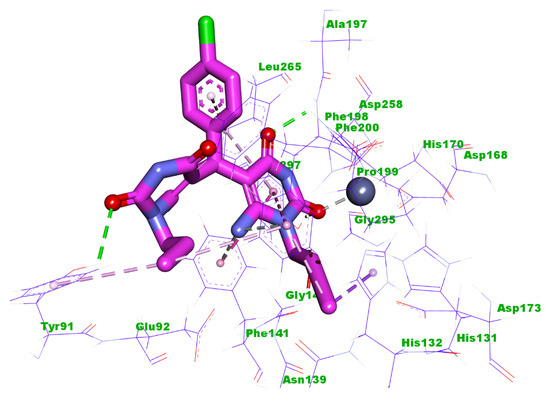
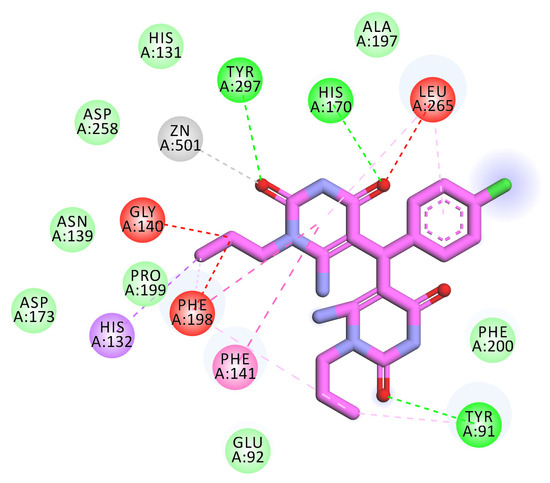
Figure 10.
Predicted binding mode of compound 5m with the active site of HDAC1.
3. Conclusions
Twenty uracil and thiouracil derivatives were synthesized as potential inhibitors for HDAC. These compounds were tested for their cytotoxic effect against MCF-7, HepG2, and HCT-116 cell lines. Some compounds showed promising anti-proliferative activities. 5a, 5b, and 5f, has promising anti-proliferative effect against MCF-7 with IC50 values of 11 ± 1.6, 55 ± 2.8, 9.3 ± 3.4 µM, respectively compared to sorafenib (IC50 = 141 ± 3 µM). In addition, compound 5b showed high activity against HCT-116 cells with an IC50 value of 21 ± 2.4 µM compared to sorafenib (IC50 = 177 ± 0.93 µM). Furthermore, compounds 5i, 5k, and 5m exhibited high cytotoxic effect against HepG2 with IC50 values of 4 ± 1, 5 ± 2, and 3.3 ± 0.56 µM, respectively, compared to sorafenib (IC50 = 17 ± 2.3 µM). SAR study revealed that the synthesized compounds with open chain linkers (5a–m) are more active than that with cyclic linkers (6a–d, and 7a–c). Compound 5m showed an IC50 of 0.05 µg/mL against HDAC1 compared to trichostatin A (IC50 = 0.0349 µg/mL). Furthermore, compound 5m showed superior activity (IC50 = 2.83 µg/mL) than trichostatin A (IC50 = 3.349 µg/mL) against HDAC4. The most promising compound 5m arrested the HCT-116 cell growth at the G0-G1 phase and induced apoptosis by 17-fold compared to the control. In addition, such a compound increased the levels of caspase-3 and caspase-8 in HCT-116 cells. Finally, the docking studies indicated that the synthesized compounds have a binding mode almost like the reference molecule (trichostatin A) against the prospective target (HDAC).
4. Experimental
4.1. Chemistry
4.1.1. General
All advice used in the synthesis and analysis of the new compounds was presented in Supplementary data.
4.1.2. 6-Amino-1-alkyl-2-oxo/thioxo-2,3-dihydropyrimidinones (3a,b)
It was prepared according to the reported method [29,30,31,32,33,34].
4.1.3. 5,5′-(Arylmethylene)bis(6-amino-1-alkyl-2-oxo/thioxo-2,3-dihydropyrimidinones) (5a–m)
A mixture of compounds 6-aminouracil and/or thiouracil (3a and/or 3b) (2.0 mmol) and different appropriate aromatic aldehydes (1.0 mmol) (4a–h) in the presence of conc. hydrochloric acid as a catalyst in absolute ethanol (20 mL) was stirred at room temperature for 2–3 h. The formed precipitate was collected by filtration, washed with methanol, and recrystallized from DMF:ethanol (3:1) afforded the desired compounds (5a–m) in good yields (Scheme 1). The structures of the synthesized compounds were confirmed by 1H and 13C NMR spectroscopy.
5,5′-(Phenylmethylene)bis(6-amino-1-ethyl-2-thioxo-2,3-dihydropyrimidin-4(1H)-one) (5a)
White Solid, (yield: 77%), m.p. > 300 °C; HPLC (99.65%); IR (KBr) cm−1: 3390, 3102 (NH2, NH), 3050 (CH arom.), 2972, 2927 (CH aliph.), 1636 (C=O), 1520 (C=C); 1H NMR (400 MHz, DMSO-d6) δ 12.25 (s, 2H, 2NH, exchangeable with D2O), 7.70 (s, 4H, NH2, exchangeable with D2O), 7.23 (t, J = 7.5 Hz, 2H, arom.), 7.12 (t, J = 7.5 Hz, 1H, arom.), 7.07 (d, J = 7.6 Hz, 2H, arom.), 5.51 (s, 1H, CH-5), 4.62–4.30 (m, 4H, 2CH2), 1.22 (t, J = 6.7 Hz, 6H, 2CH3); 13C NMR (100 MHz, DMSO-d6) δ 174.71, 161.44, 154.62, 138.51, 128.33, 126.89, 125.72, 91.29, 43.64, 34.92, 12.37; MS (70 eV) m/z (%): 430 (M+, 5), 258 (16), 102 (31), 44 (100); Anal. Calcd for C19H22N6O2S2 (430.55): C, 53.00; H, 5.15; N, 19.52; Found: C, 53.19; H, 5.39; N, 19.78.
5,5′-(p-Tolylmethylene)bis(6-amino-1-ethyl-2-thioxo-2,3-dihydropyrimidin-4(1H)-one) (5b)
White solid, (yield: 63%), m.p. = 239–240 °C; HPLC (99.65%); IR (KBr) cm−1; 3376, 3158 (NH2, NH), 3070 (CH arom.), 2976, 2928 (CH aliph.), 1661 (C=O), 1565 (C=C), 833 (p-substituted phenyl); 1H NMR (400 MHz, DMSO-d6) δ 12.42 (s, 2H, NH), 8.04 (d, J = 8.2 Hz, 2H, arom.), 7.34 (m, 2H, arom.), 7.05 (s, 4H, 2NH2), 6.05 (s, 1H, CH-5), 4.58 (q, J = 6.9 Hz, 2H, CH2), 4.30 (q, J=6.8 Hz, 2H, CH2), 2.38 (s, 3H, CH3), 1.34 (t, J = 6.9 Hz, 3H, CH3), 1.13 (t, J = 6.8 Hz, 3H, CH3); 13C NMR (100 MHz, DMSO-d6) δ 172.90, 161.91, 152.27, 140.52, 129.59, 128.67, 128.25, 127.81, 126.58, 94.32, 42.78, 32.93, 21.01, 12.13; MS (70 eV) m/z (%): 444 (M+, 39), 375 (44), 274 (100), 169 (50), 62 (39); Anal. Calcd for C20H24N6O2S2 (444.57): C, 54.03; H, 5.44; N, 18.90; Found: C, 54.21; H, 5.67; N, 19.08.
5,5′-((4-Chlorophenyl)methylene)bis(6-amino-1-ethyl-2-thioxo-2,3-dihydropyrimidin- 4(1H)-one) (5c)
White solid, (yield: 78%), m.p. > 300 °C; IR (KBr) cm−1: 3376, 3153 (NH2, NH), 3050 (CH arom.), 2976, 2928 (CH aliph.), 1661 (C=O), 1565 (C=C), 839 ((p-substituted phenyl); 1H NMR (400 MHz, DMSO-d6) δ 12.28 (s, 2H, 2NH), 7.69 (s, 4H, 2NH2), 7.27 (d, J = 8.5 Hz, 2H, arom.), 7.10 (d, J = 8.5 Hz, 2H, arom.), 5.48 (s, 1H, CH-5), 4.57-4.36 (m, 4H, 2CH2), 1.23 (t, J = 6.7 Hz, 6H, 2CH3); 13C NMR (100 MHz, DMSO-d6) δ 174.29, 164.21, 158.52, 137.26, 129.77, 128.48, 127.71, 87.32, 43.19, 34.08, 11.90; MS (70 eV) m/z (%): 466 (M+2, 4), 464 (M+, 10), 333 (12), 292 (95), 190 (20), 44 (100); Anal. Calcd for C19H21ClN6O2S2 (464.99): C, 49.08; H, 4.55; N, 18.07; Found: C, 49.32; H, 4.69; N, 18.23.
5,5′-((2,4-Dichlorophenyl)methylene)bis(6-amino-1-ethyl-2-thioxo-2,3-dihydro pyrimidin-4(1H)-one) (5d)
White powder (yield: 60%), m.p. = 272–273 °C; IR (KBr) cm−1: 3373, 3145 (NH2, NH), 3065 (CH arom.), 2975, 2931 (CH aliph.), 1663 (C=O), 1572 (C=C), 730 (trisubstituted phenyl); 1H NMR (400 MHz, DMSO-d6) δ 12.29 (s, 2H, 2NH), 7.75 (s, 2H, NH2), 7.46 (d, J = 1.7 Hz, 1H, arom.), 7.33 (dd, J = 8.5, 1.7 Hz, 1H, arom.), 7.28 (d, J = 8.5 Hz, 1H, arom.), 7.16 (s, 2H, NH2), 5.41 (s, 1H, CH-5), 4.45 (m, 4H, 2CH2), 1.22 (t, J = 6.8 Hz, 6H, 2CH3). 13C NMR (100 MHz, DMSO-d6) δ 174.26, 164.18, 153.87, 136.32, 133.15, 131.04, 130.17, 128.90, 126.71, 90.57, 43.24, 33.55, 11.89; MS (70 eV) m/z (%): 503 (M+4, 23), 501 (M+2, 26), 499 (M+, 35), 360 (100), 300 (31), 286 (42), 255 (18); Anal. Calcd for C19H20Cl2N6O2S2 (499.43): C, 45.69; H, 4.04; N, 16.83; Found: C, 45.78; H, 4.21; N, 17.05.
5,5′-((4-Methoxyphenyl)methylene)bis(6-amino-1-ethyl-2-thioxo-2,3-dihydropyrimidin -4(1H)-one) (5e)
White powder (yield: 67%), m.p. = 260–261 °C; 1H NMR (400 MHz, DMSO-d6) δ 12.22 (s, 2H, 2NH), 7.95 (s, 2H, NH2) 7.74 (s, 2H, NH2), 6.96 (d, J = 8.6 Hz, 2H, arom.), 6.79 (d, J = 8.6 Hz, 2H, arom.), 5.45 (s, 1H, CH-5), 4.45 (m, 4H, 2CH2), 3.70 (s, 3H, OCH3), 1.22 (t, J = 6.8 Hz, 6H, 2CH3); 13C NMR (100 MHz, DMSO-d6) δ 174.20, 162.32, 157.06, 153.88, 129.64, 127.45, 113.28, 91.37, 54.94, 43.11, 35.79, 11.90; MS (70 eV) m/z (%):460 (M+, 30), 392 (41), 222 (30), 129 (100), 56 (40); Anal. Calcd for C20H24N6O3S2 (460.57): C, 52.16; H, 5.25; N, 18.25; Found: C, 52.40; H, 5.37; N, 18.49.
5,5′-((4-Nitrophenyl)methylene)bis(6-amino-1-ethyl-2-thioxo-2,3-dihydropyrimidin-4(1H)-one) (5f)
White solid, (yield: 79%) m.p. = 274–275 °C; HPLC (99.00%); IR (KBr) cm−1: 3384, 3137 (NH2, NH), 3060 (CH arom.), 2975, 2931 (CH aliph.), 1662 (C=O), 1555 (C=C), 1505, 1344 (NO2), 849 ((p-substituted phenyl); 1H NMR (400 MHz, DMSO-d6) δ 12.34 (s, 2H, 2NH), 8.10 (d, J = 8.7 Hz, 2H, arom.), 7.69 (s, 4H, 2NH2), 7.38 (d, J = 8.7 Hz, 2H, arom.), 5.58 (s, 1H, CH-5), 4.49 (m, 4H, 2CH2), 1.23 (t, J = 6.8 Hz, 6H, 2CH3); 13C NMR (100 MHz, DMSO-d6) δ 174.42, 162.32, 154.24, 147.27, 145.44, 127.98, 123.02, 90.60, 43.26, 35.79, 11.87; MS (70 eV) m/z (%): 475 (M+, 20), 425 (71), 372 (85), 343 (100), 297 (20) Anal. Calcd for C19H21N7O4S2 (475.54): C, 47.99; H, 4.45; N, 20.62; Found: C, 48.17; H, 4.63; N, 20.89.
5,5′-(Thiophen-2-ylmethylene)bis(6-amino-1-ethyl-2-thioxo-2,3-dihydropyrimidin-4(1H)-one) (5g)
White solid, (yield: 56%) m.p. = 271–272 °C; IR (KBr) cm−1: 3380, 3142 (NH2, NH), 3030 (CH arom.), 2976, 2924 (CH aliph.), 1630 (C=O), 1566 (C=C); 1H NMR (400 MHz, DMSO-d6) δ 12.29 (s, 2H, 2NH), 7.87 (br.s, 4H, 2NH2), 7.27 (d, 1H, arom.), 6.85 (t, 1H, arom.), 6.65 (s, 1H, arom.), 5.64 (s, 1H, CH-5), 4.50 (m, 4H, 2CH2).1.21 (t, J = 6.9 Hz, 6H, 2CH3); 13C NMR (100 MHz, DMSO-d6) δ 174.35, 162.34, 153.85, 143.73, 126.28, 123.79, 123.60, 91.51, 43.11, 18.56, 11.86; MS (70 eV) m/z (%): 436 (M+, 7), 264 (58), 171 (90), 44 (100); Anal. Calcd for C17H20N6O2S3 (436.57): C, 46.77; H, 4.62; N, 19.25; Found: C, 46.98; H, 4.76; N, 19.51.
5,5′-Methylenebis(6-amino-1-propylpyrimidine-2,4(1H,3H)-dione) (5h)
White solid, (yield 49%), m.p. = 256–257 °C; IR (KBr) cm−1: 3342, 3133 (NH2, NH), 2969, 2938 (CH aliph.), 1677 (C=O), 1590 (C=C); 1H NMR (400 MHz, DMSO-d6) δ 11.40 (s, 1H, NH), 10.53 (s, 1H, NH), 7.52 (br. s, 2H, NH2), 7.16 (s, 2H, NH2), 3.64 (m, 4H, 2CH2), 2.66 (s, 2H, CH2), 1.50 (m, 4H, 2 CH2), 0.85 (m, 6H, 2CH3); 13C NMR (100 MHz, DMSO-d6) δ 164.02, 161.53, 150.50, 148.84, 84.97, 80.36, 45.59, 45.23, 20.86, 20.72, 17.75, 11.10, 10.71; MS (70 eV) m/z (%): 350 (M+, 18), 386 (61), 220 (100), 144 (37), 61 (80); Anal. Calcd for C15H22N6O4 (350.38): C, 51.42; H, 6.33; N, 23.99; Found: C, 51.67; H, 6.45; N, 23.75.
5,5′-(Phenylmethylene)bis(6-amino-1-propylpyrimidine-2,4(1H,3H)-dione) (5i)
Pale yellow solid, yield (66%), m.p. = 294–295 °C; HPLC (99.58%); IR (KBr) cm−1: 3388, 3178 (NH2, NH), 3045 (CH arom.), 2969, 2930 (CH aliph.), 1670 (C=O), 1598 (C=C); 1H NMR (400 MHz, DMSO-d6) δ 10.71 (s, 2H, 2NH), 7.73–7.41 (br s, 4H, 2NH2), 7.19 (t, J = 7.6 Hz, 2H, arom.), 7.07 (m, 3H, arom.), 5.45 (s, 1H, CH-5), 3.77 (m, 4H, 2CH2), 1.56 (m, 4H, 2CH2), 0.88 (t, J = 7.4 Hz, 6H, 2CH3); 13C NMR (100 MHz, DMSO-d6) δ 150.08, 139.67, 134.59, 129.49, 129.16, 127.63, 126.44, 124.84, 71.90, 42.78, 34.13, 20.70, 10.76; MS (70 eV) m/z (%): 426 (M+, 39), 256 (42), 215 (43), 106 (100), 40 (17); Anal. Calcd for C21H26N6O4 (426.48): C, 59.14; H, 6.15; N, 19.71; Found: C, 59.36; H, 6.29; N, 19.87.
5,5′-(p-Tolylmethylene)bis(6-amino-1-propylpyrimidine-2,4(1H,3H)-dione) (5j)
Pale yellow solid, (yield 73%), m.p. = 286–287 °C; IR (KBr) cm−1: 3387, 3180 (NH2, NH), 3049 (CH arom.), 2968, 2934 (CH aliph.), 1694 (C=O), 1560 (C=C), 845 (p-substituted phenyl); 1H NMR (400 MHz, DMSO-d6) δ 10.69 (s, 2H, 2NH), 7.67 (br.s, 2H, NH2), 7.39 (br.s, 2H, NH2), 7.00 (d, J = 8.1 Hz, 2H, arom.), 6.92 (d, J = 8.1 Hz, 2H, arom.), 5.40 (s, 1H, CH-5), 3.77 (m, 4H, 2CH2), 2.23 (s, 3H, CH3), 1.55 (m, 4H, 2CH2), 0.88 (t, J = 7.3 Hz, 6H, 2CH3); 13C NMR (100 MHz, DMSO-d6) δ 160.73, 150.17, 145.35, 136.52, 133.64, 129.80, 129.67, 128.33, 126.43, 80.12, 42.84, 33.84, 20.76, 20.54, 10.81; MS (70 eV) m/z (%): 440 (M+, 27), 327 (60), 210 (62), 97 (100), 61 (86); Anal. Calcd for C22H28N6O4 (440.50): C, 59.99; H, 6.41; N, 19.08; Found: C, 60.17; H, 6.53; N, 19.31.
5,5′-((2,4-Dichlorophenyl)methylene)bis(6-amino-1-propylpyrimidine-2,4(1H,3H)-dione) (5k)
White solid, (yield 58%), m.p. 285–286 °C; HPLC (99.53%); IR (KBr) cm−1: 3365, 3176 (NH, NH2), 3049 (CH arom.), 2969, 2939 (CH aliph.), 1692 (C=O), 1568 (C=C), 709 (trisubstituted phenyl); 1H NMR (400 MHz, 6 δ 10.91 (s, 1H, NH), 10.66 (s, 1H, NH), 7.42 (br.s, 2H, NH2), 7.41 (d, J = 2.2 Hz, 1H, arom.), 7.31 (dd, J = 8.5, 2.2 Hz, 1H, arom.), 7.24 (d, J = 8.5 Hz, 1H, arom.), 7.00 (br.s, 2H, NH2), 5.37 (s, 1H, CH-5), 3.86–3.68 (m, 4H, 2CH2), 1.56 (m, 4H, 2CH2), 0.87 (t, 6H, 2CH3); 13C NMR (100 MHz, DMSO) δ 163.73, 154.59, 149.93, 137.95, 133.20, 130.56, 130.19, 128.75, 126.45, 86.26, 42.80, 33.30, 20.67, 10.71; MS (70 eV) m/z (%): 499 (M+4, 12), 497 (M+2, 36), 495 (M+, 13), 345 (100), 257 (57), 135 (66), 59 (66); Anal. Calcd for C21H24 Cl2N6O4 (495.36): C, 50.92; H, 4.88; N, 16.97; Found: C, 51.14; H, 4.95; N, 17.13.
5,5′-((4-Methoxyphenyl)methylene)bis(6-amino-1-propylpyrimidine-2,4(1H,3H)-dione) (5l)
White solid, (yield 64%), m.p. = 289–290 °C; IR (KBr) cm−1: 3376, 3150 (NH, NH2), 3042 (CH arom.), 2963, 2923 (CH aliph.), 1688 (C=O), 1598 (C=C), 843 (p-substituted phenyl); 1H NMR (400 MHz, DMSO-d6) δ 10.78 (s, 1H, NH), 10.62 (s, 1H, NH), 7.87 (d, J = 8.5 Hz, 1H, arom.), 7.65 (br.s, 2H, NH2), 7.37 (br.s, 2H, NH2), 7.13 (d, J = 8.5 Hz, 1H, arom.), 6.94 (d, J = 8.5 Hz, 1H, arom.), 6.76 (d, J = 8.5 Hz, 1H, arom.), 5.39 (s, 1H, CH-5), 3.83 (m, 4H, 2CH2), 3.69 (s, 3H, O-CH3), 1.54 (m, 4H, 2CH2), 0.88 (t, J = 7.2 Hz, 6H, 2CH3); 13C NMR (100 MHz, DMSO-d6) δ 164.70, 157.26, 150.57, 132.28, 131.76, 130.13, 127.89, 114.99, 113.54, 87.41, 56.17, 55.38, 43.24, 33.86, 21.18, 11.23. MS (70 eV) m/z (%): 456 (M+, 42), 306 (86), 231 (88), 191 (100), 57 (36); Anal. Calcd for C22H28N6O5 (456.50): C, 57.88; H, 6.18; N, 18.41; Found: C, 58.09; H, 6.40; N, 18.49.
5,5′-((4-Chlorophenyl)methylene)bis(6-amino-1-propylpyrimidine-2,4(1H,3H)-dione) (5m)
Pale yellow solid, yield (73%), m.p. = 257–258 °C; HPLC (99.71%); IR (KBr) cm−1: 3377, 3189 (NH, NH2), 3045 (CH arom.), 2966, 2930 (CH aliph.), 1666 (C=O), 1565 (C=C), 843 (p-substituted phenyl); 1H NMR (400 MHz, DMSO-d6) δ 10.81 (s, 2H, 2NH), 7.64 (br.s, 4H, 2NH2), 7.24 (d, J = 8.6 Hz, 2H, arom.), 7.07 (d, J = 8.6 Hz, 2H, arom.), 5.42 (s, 1H, CH-5), 3.76 (m, 4H, 2CH2), 1.56 (m, 4H, 2CH2), 0.88 (t, J = 7.3 Hz, 6H, 2CH3); 13C NMR (100 MHz, DMSO-d6) δ 162.38, 150.10, 138.89, 128.50, 127.55, 86.57, 42.88, 35.83, 20.72, 10.80; MS (70 eV) m/z (%): 462 (M+2, 34), 460 (M+, 39), 385 (51), 370 (100), 269 (42), 102 (40); Anal. Calcd for C21H25ClN6O4 (460.92): C, 54.72; H, 5.47; N, 18.23; Found: C, 54.95; H, 5.63; N, 18.50.
4.1.4. 1,9-Dialkyl-2,3,5,8,9,10-hexahydropyrido[2,3-d:6,5-d′]dipyrimidinones (6a–d)
Compounds 5a, 5f, 5h or 5l (0.7 mmol) and glacial acetic acid (5 mL) were heated under reflux in the presence of c. HCl (1 mL) for 2.5 h. After cooling, the reaction mixture was filtered off, washed with ethanol crystallized from DMF and dried in the oven.
1,9-Diethyl-5-phenyl-2,8-dithioxo-2,3,5,8,9,10-hexahydropyrido[2,3-d:6,5-d′]dipyrimidine-4,6(1H,7H)-dione (6a)
White solid, (yield 75%), m.p. = 255–256 °C; 1H NMR (400 MHz, DMSO-d6) δ 12.78 (s, 2H, NH), 7.70 (s, 1H, NH), 7.26 (t, J = 7.5 Hz, 2H, arom.), 7.14 (m, 3H, arom.), 5.46 (s, 1H, CH-5), 4.41–4.31 (m, 4H, 2CH2), 1.25 (t, J = 7.0 Hz, 3H, CH3), 1.19 (t, J = 6.9 Hz, 3H, CH3); MS (70 eV) m/z (%): 413 (M+, 59), 352 (32), 207 (49), 157 (100), 40 (54); Anal. Calcd for C19H19N5O2S2 (413.51): C, 55.19; H, 4.63; N, 16.94; Found: C, 55.42; H, 4.78; N, 16.89.
1,9-Diethyl-5-(4-nitrophenyl)-2,8-dithioxo-2,3,5,8,9,10-hexahydropyrido[2,3-d:6,5-d′]dipyrimidine-4,6(1H,7H)-dione (6b)
Pale yellow solid, (yield: 86%), m.p. = 251–252 °C; IR (KBr) cm−1: 3137 (NH), 3070 (CH arom.), 2977, 2933 (CH aliph.), 1650 (C=O), 1519 (C=C), 1500, 1342 (NO2), 853 (p-substituted phenyl); 1H NMR (400 MHz, DMSO- d6) δ 12.83 (s, 2H, 2NH), 8.17–8.06 (m, 2H, arom.), 7.77 (s, 1H, NH), 7.44 (m, 2H, arom.), 5.57 (s, 1H, CH-5), 4.52–4.35 (m, 4H, 2CH2), 1.25 (t, J = 7.0 Hz, 3H, CH3), 1.19 (t, J = 7.0 Hz, 3H, CH3); 13C NMR (100 MHz, DMSO- d6) δ 173.95, 162.27, 152.16, 147.22, 145.82, 128.00, 123.20, 92.93, 43.75, 34.65, 11.53; MS (70 eV) m/z (%): 458 (M+, 31), 343 (31), 238 (41), 139 (33), 83 (100); Anal. Calcd for C19H18N6O4S2 (458.51): C, 49.77; H, 3.96; N, 18.33; Found: C, 49.98; H, 4.17; N, 18.59.
5-(4-Methoxyphenyl)-1,9-dipropyl-5,10-dihydropyrido[2,3-d:6,5-d′]dipyrimidine-2,4,6,8(1H,3H,7H,9H)-tetraone (6c)
Yellow solid, (yield: 62%), m.p. = 180–181 °C; IR (KBr) cm−1: 3223 (NH), 3091 (CH arom.), 2962, 2937 (CH aliph.), 1647 (C=O), 1543 (C=C), 835 (p-substituted phenyl); 1H NMR (400 MHz, DMSO-d6) δ 11.48 (s, 1H, NH), 11.37 (s, 1H, NH), 8.35–8.27 (m, 3H, arom., NH) 7.08-7.05 (m, 2H, arom.), 5.75 (s, 1H, CH-5), 3.87 (s, 3H, CH3), 3.76-3.71 (m, 4H, 2CH2), 1.57-1.53 (m, 4H, 2CH2), 0.87 (t, J = 7.5 Hz, 6H, 2CH3); 13C NMR (101 MHz, DMSO- d6) δ 163.48, 163.37, 161.42, 161.08, 155.84, 150.38, 150.32, 137.46, 125.25, 125.11, 115.55, 115.50, 113.92, 83.69, 55.69, 42.30, 41.74, 20.82, 11.19; MS (70 eV) m/z (%): 439 (M+, 27), 311 (66), 245(100), 138 (95), 75 (49); Anal. Calcd for C22H25N5O5 (439.47): C, 60.13; 5.73; N, 15.94; Found: C, 59.94; H, 5.91; N, 16.17.
1,9-Dipropyl-5,10-dihydropyrido[2,3-d:6,5-d′]dipyrimidine-2,4,6,8 (1H,3H,7H,9H)-tetraone (6d)
White solid, (yield: 70%), m.p. = 274–275 °C; IR (KBr) cm−1: 3233 (NH), 2969, 2938 (CH aliph.), 1677 (C=O), 1589 (C=C); 1H NMR (400 MHz, DMSO-d6) δ 11.40 (s, 1H, NH), 10.53 (s, 1H, NH), 7.18 (s, 1H, NH), 3.74– 3.60 (m, 4H, 2CH2), 3.49 (s, 2H, CH2), 1.56–1.44 (m, 4H, 2 CH2), 0.86 (t, J = 5.0 Hz, 3H, CH3), 0.83 (t, J = 5.1 Hz, 3H, CH3); 13C NMR (100 MHz, DMSO-d6) δ 161.55, 150.51,150.29, 148.87, 80.37, 45.62, 45.26, 27.70, 20.88, 20.74, 11.11, 10.73; MS (70 eV) m/z (%): 333 (M+, 42), 215 (70), 185 (100), 84 (66); Anal. Calcd for C15H19N5O4 (333.35): C, 54.05; H, 5.75; N, 21.01; Found: C, 54.21; H, 5.89; N, 20.96.
4.1.5. 5Aryl-1,9-Diethyl-2,8-dithioxo-2,3,8,9-tetrahydropyrido[2,3-d:6,5-d′]dipyrimidine-4,6(1H,7H)-diones (7a–c)
A mixture of compound 5c, 5e or 5g (0.7 mmol), glacial acetic acid (5 mL) and c. HCl (1 mL) was heated under reflux for 4–5 h. After cooling, the formed precipitate was collected by filtration, washed with ethanol crystallized from DMF and dried in the oven.
5-(4-Chlorophenyl)-1,9-diethyl-2,8-dithioxo-2,3,8,9-tetrahydropyrido[2,3-d:6,5-d′]dipyrimidine-4,6(1H,7H)-dione (7a)
Yellow solid, (yield: 72%), m.p. = 224–225 °C; IR (KBr) cm−1: 3229 (NH), 3109 (CH arom.), 2976, 2928 (CH aliph.), 1662 (C=O), 1572 (C=C), 834 (p-substituted phenyl); 1H NMR (400 MHz, DMSO-d6) δ 10.01 (s, 2H, 2NH), 7.93 (d, J = 8.5 Hz, 2H, arom.), 7.68 (d, J = 8.5 Hz, 2H, arom.), 4.32–4.22 (m, 2H, CH2), 4.12–4.05 (m, 2H, CH2), 1.30–1.10 (m, 6H, 2CH3); MS (70 eV) m/z (%): 448 (M+2, 6), 446 (M+, 17), 402 (77), 325 (100), 249 (54), 69 (22); Anal. Calcd for C19H16ClN5O2S2 (445.94): C, 51.17; H, 3.62; N, 15.71; Found: C, 51.37; H, 3.76; N, 15.92.
1,9-Diethyl-5-(4-methoxyphenyl)-2,8-dithioxo-2,3,8,9-tetrahydropyrido[2,3-d:6,5-d′]dipyrimidine-4,6(1H,7H)-dione (7b)
Mustard yellow, yield (66%), m.p. = 221–221 °C; IR (KBr) cm−1: 3235 (NH), 3070 (CH arom.), 2988, 2935 (CH aliph.), 1660 (C=O), 1569 (C=C), 843 (p-substituted phenyl); 1H NMR (400 MHz, DMSO-d6) δ 12.47 (s, 1H, NH), 12.40 (s, 1H, NH), 8.43 (d, J = 9.0 Hz, 1H, arom.), 8.31(d, J = 9.0 Hz, 1H, arom.), 7.10–7.07 (m, 2H, arom.), 4.32 (q, J = 6.9 Hz, 4H, 2 CH2), 3.89 (s, 3H, CH3), 1.18 (t, J = 6.9 Hz, 6H, 2CH3); 13C NMR (100 MHz, DMSO-d6) δ 178.64, 178.57, 164.23, 164.10, 162.52, 157.35, 156.82, 156.67, 131.82, 129.66, 127.46, 114.53, 114.14, 113.12, 55.82, 55.70, 54.89, 12.34, 12.26; MS (70 eV) m/z (%): 441 (M+, 18), 355 (41), 235 (100), 113 (28), 50 (39); Anal. Calcd for C20H19N5O3S2 (441.52): C, 54.41; H, 4.34; N, 15.86; Found: C, 54.21; H, 4.51; N, 16.04.
1,9-Diethyl-5-(thiophen-2-yl)-2,8-dithioxo-2,3,8,9-tetrahydropyrido[2,3-d:6,5-d′]dipyrimidine-4,6(1H,7H)-dione (7c)
Brown solid, (yield: 54%), m.p. = 216–217 °C; IR (KBr) cm−1: 3207 (NH), 3050 (CH arom.), 2972, 2926 (CH aliph.), 1657 (C=O), 1568 (C=C); 1H NMR (400 MHz, DMSO-d6) δ 12.50 (s, 2H, 2NH), 8.38 (t, 1H, arom.), 8.27 (t, 1H, arom.), 7.41 (m, 1H, arom.), 4.34 (m, 4H, 2CH2), 1.17 (m, 6H, 2CH3); 13C NMR (100 MHz, DMSO-d6) δ 178.59, 178.55, 161.36, 160.34, 160.21, 159.38, 143.60, 136.87, 128.88, 128.79, 111.98, 100.93, 54.86, 48.17, 14.51, 12.27; MS (70 eV) m/z (%): 417 (M+, 37), 399 (100), 298 (25), 122 (58), 44 (40); Anal. Calcd for C17H15N5O2S3 (417.52): C, 48.90; H, 3.62; N, 16.77; Found: C, 49.08; H, 3.80; N, 16.98.
4.2. Biological Testing
4.2.1. In Vitro Cytotoxic Activity
Anti-proliferative activity activities were tested using MTT assay [36,42,43,44,45] as described in Supplementary Materials.
4.2.2. In vitro HDAC Assay
Compounds 5a, 5b, 5f, 5i, 5k, and 5m were tested for their HDAC inhibitory activities (HDAC1 and HDAC4 subtypes) as described in Supplementary Materials [46,47].
4.2.3. Flow Cytometry Analysis for Cell Cycle
Cell cycle analysis was performed for compound 5m as described in Supplementary Materials [48,49,50].
4.2.4. Flow Cytometry Analysis for Apoptosis
The apoptotic effect of the compound 5m was tested as described in Supplementary data [51,52,53]. Quantitative Real-Time Reverse-Transcriptase PCR (qRT-PCR) technique Using qRT-PCR, the effect of compound 5m on the expression of cleaved caspase-3 and caspase-8 was determined (Supplementary Materials) [54,55,56,57,58].
4.3. Docking Studies
Docking studies were conducted against HDAC1 (http://www.rcsb.org/ accessed on 1 January 2023, PDB code: 1C3R, resolution of 2.00 Å) as described in Supplementary Materials [59].
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ph16070966/s1, S1: chemistry, S2: Methods of biological testing, S3: docking procedure, Figures in S4: the spectral data (1H NMR, 13C NMR, Mass, and IR spectra) of the synthesized compounds, and Figures in S5: the HPLC reports [60].
Author Contributions
Conceptualization, S.E.-K. and M.H. (Mohamed Hagras); methodology, O.R.E., M.A.E.D., M.H. (Maghawry Hegazy), A.A.E.-H., M.M.M. and S.Y.E.; software, I.H.E.; validation, I.H.E. and S.E.-K.; formal analysis M.H. (Maghawry Hegazy), M.A.E.D. and F.A.; investigation, S.E.-K. and M.H. (Mohamed Hagras); resources, S.E.-K. and M.H. (Mohamed Hagras); data curation, S.E.-K. and M.H. (Mohamed Hagras); writing—original draft preparation, S.E.-K., M.A.E.D. and M.H. (Maghawry Hegazy); writing—review and editing, I.H.E. and S.E.-K.; visualization, F.A.; supervision, M.H. (Mohamed Hagras), M.A.E.D. and S.E.-K.; project administration, S.E.-K.; funding acquisition, F.A. All authors have read and agreed to the published version of the manuscript.
Funding
This paper is based on work supported by Science, Technology & Innovation Funding Authority (STDF) under grant number (43229) young researcher.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Sample Availability
Samples of the compounds are available from the authors.
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- WHO. Cancer. Available online: https://www.who.int/health-topics/cancer#tab=tab_1 (accessed on 1 October 2020).
- Badran, M.M.; Abouzid, K.A.M.; Hussein, M.H.M. Synthesis of certain substituted quinoxalines as antimicrobial agents (Part II). Arch. Pharmacal Res. 2003, 26, 107–113. [Google Scholar] [CrossRef]
- World Health Organisation. Cancer—Key Facts. 2018. Available online: https://www.who.int/news-room/fact-sheets/detail/cancer (accessed on 1 March 2023).
- Chabner, B.A.; Roberts, T.G., Jr. Timeline: Chemotherapy and the war on cancer. Nat. Rev. Cancer 2005, 5, 65–72. [Google Scholar] [CrossRef]
- Krauss, G.; Schönbrunner, N.; Cooper, J. Biochemistry of Signal Transduction and Regulation; Wiley Online Library: Weinheim, NJ, USA, 2003; Volume 3. [Google Scholar]
- Nguyen, K.-S.H.; Kobayashi, S.; Costa, D.B. Acquired Resistance to Epidermal Growth Factor Receptor Tyrosine Kinase Inhibitors in Non–Small-Cell Lung Cancers Dependent on the Epidermal Growth Factor Receptor Pathway. Clin. Lung Cancer 2009, 10, 281–289. [Google Scholar] [CrossRef]
- Waldmann, T.; Schneider, R. Targeting histone modifications—Epigenetics in cancer. Curr. Opin. Cell Biol. 2013, 25, 184–189. [Google Scholar] [CrossRef]
- Yang, X.-J.; Seto, E. Lysine Acetylation: Codified Crosstalk with Other Posttranslational Modifications. Mol. Cell 2008, 31, 449–461. [Google Scholar] [CrossRef]
- Yang, X.J.; Grégoire, S. Metabolism, cytoskeleton and cellular signalling in the grip of protein Nϵ-and O-acetylation. EMBO Rep. 2007, 8, 556–562. [Google Scholar] [CrossRef]
- Sun, X.-J.; Man, N.; Tan, Y.; Nimer, S.D.; Wang, L. The Role of Histone Acetyltransferases in Normal and Malignant Hematopoiesis. Front. Oncol. 2015, 5, 108. [Google Scholar] [CrossRef]
- Glauben, R.; Sonnenberg, E.; Zeitz, M.; Siegmund, B. HDAC inhibitors in models of inflammation-related tumorigenesis. Cancer Lett. 2009, 280, 154–159. [Google Scholar] [CrossRef]
- Hanessian, S.; Auzzas, L.; Giannini, G.; Marzi, M.; Cabri, W.; Barbarino, M.; Vesci, L.; Pisano, C. ω-Alkoxy analogues of SAHA (vorinostat) as inhibitors of HDAC: A study of chain-length and stereochemical dependence. Bioorganic Med. Chem. Lett. 2007, 17, 6261–6265. [Google Scholar] [CrossRef]
- Zhu, Y.; Li, H.-F.; Lu, S.; Zheng, Y.-X.; Wu, Z.; Tang, W.-F.; Zhou, X.; Lu, T. Investigation on the isoform selectivity of histone deacetylase inhibitors using chemical feature based pharmacophore and docking approaches. Eur. J. Med. Chem. 2010, 45, 1777–1791. [Google Scholar] [CrossRef]
- Marks, P.A.; Rifkind, R.A.; Richon, V.M.; Breslow, R.; Miller, T.; Kelly, W.K. Histone deacetylases and cancer: Causes and therapies. Nat. Rev. Cancer 2001, 1, 194–202. [Google Scholar] [CrossRef]
- Ekou, L.; Ekou, T.; Opalinski, I.; Gesson, J.P. Histone Deacetylase Inhibitors: Synthesis of Tetrapeptide Analogue SAHA/TPX. E J. Chem. 2011, 8, S79–S84. [Google Scholar] [CrossRef]
- Grozinger, C.M.; Schreiber, S.L. Deacetylase Enzymes: Biological Functions and the Use of Small-Molecule Inhibitors. Chem. Biol. 2002, 9, 3–16. [Google Scholar] [CrossRef]
- Miller, T.A.; Witter, D.J.; Belvedere, S. Histone deacetylase inhibitors. J. Med. Chem. 2003, 46, 5097–5116. [Google Scholar] [CrossRef]
- Yoshida, M.; Horinouchi, S.; Beppu, T. Trichostatin A and trapoxin: Novel chemical probes for the role of histone acetylation in chromatin structure and function. Bioessays 1995, 17, 423–430. [Google Scholar] [CrossRef]
- Methot, J.L.; Chakravarty, P.K.; Chenard, M.; Close, J.; Cruz, J.C.; Dahlberg, W.K.; Fleming, J.; Hamblett, C.L.; Hamill, J.E.; Harrington, P. Exploration of the internal cavity of histone deacetylase (HDAC) with selective HDAC1/HDAC2 inhibitors (SHI-1: 2). Bioorganic Med. Chem. Lett. 2008, 18, 973–978. [Google Scholar] [CrossRef]
- Schäfer, S.; Saunders, L.; Eliseeva, E.; Velena, A.; Jung, M.; Schwienhorst, A.; Strasser, A.; Dickmanns, A.; Ficner, R.; Schlimme, S.; et al. Phenylalanine-containing hydroxamic acids as selective inhibitors of class IIb histone deacetylases (HDACs). Bioorganic Med. Chem. 2008, 16, 2011–2033. [Google Scholar] [CrossRef]
- Islam, N.M.; Kato, T.; Nishino, N.; Kim, H.-J.; Ito, A.; Yoshida, M. Bicyclic peptides as potent inhibitors of histone deacetylases: Optimization of alkyl loop length. Bioorganic Med. Chem. Lett. 2010, 20, 997–999. [Google Scholar] [CrossRef]
- Meinke, P.; Liberator, P. Histone Deacetylase: A Target for Antiproliferative and Antiprotozoal Agents. Curr. Med. Chem. 2001, 8, 211–235. [Google Scholar] [CrossRef]
- Yoshida, M.; Matsuyama, A.; Komatsu, Y.; Nishino, N. From discovery to the coming generation of histone deacetylase inhibitors. Curr. Med. Chem. 2003, 10, 2351–2358. [Google Scholar] [CrossRef] [PubMed]
- Bertrand, P. Inside HDAC with HDAC inhibitors. Eur. J. Med. Chem. 2010, 45, 2095–2116. [Google Scholar] [CrossRef]
- Awad, S.M.; Zohny, Y.M.; Ali, S.A.; Mahgoub, S.; Said, A.M. Design, Synthesis, Molecular Modeling, and Biological Evaluation of Novel Thiouracil Derivatives as Potential Antithyroid Agents. Molecules 2018, 23, 2913. [Google Scholar] [CrossRef]
- El-Naggar, A.M.; Abou-El-Regal, M.M.; El-Metwally, S.A.; Sherbiny, F.F.; Eissa, I.H. Synthesis, characterization and molecular docking studies of thiouracil derivatives as potent thymidylate synthase inhibitors and potential anticancer agents. Mol. Divers. 2017, 21, 967–983. [Google Scholar] [CrossRef]
- Li, Y.; Wang, Y.; Xie, N.; Xu, M.; Qian, P.; Zhao, Y.; Li, S. Design, synthesis and antiproliferative activities of novel benzamides derivatives as HDAC inhibitors. Eur. J. Med. Chem. 2015, 100, 270–276. [Google Scholar] [CrossRef]
- El-Kalyoubi, S.A.; Taher, E.S.; Ibrahim, T.S.; El-Behairy, M.F.; Al-Mahmoudy, A.M.M. Uracil as a Zn-Binding Bioisostere of the Allergic Benzenesulfonamide in the Design of Quinoline–Uracil Hybrids as Anticancer Carbonic Anhydrase Inhibitors. Pharmaceuticals 2022, 15, 494. [Google Scholar] [CrossRef]
- El-Kalyoubi, S.; Agili, F.; Adel, I.; Tantawy, M.A. Novel uracil derivatives depicted potential anticancer agents: In Vitro, molecular docking, and ADME study. Arab. J. Chem. 2022, 15, 103669. [Google Scholar] [CrossRef]
- El-Kalyoubi, S.A.; Ragab, A.; Abu Ali, O.A.; Ammar, Y.A.; Seadawy, M.G.; Ahmed, A.; Fayed, E.A. One-pot synthesis and molecular modeling studies of new bioactive spiro-oxindoles based on uracil derivatives as SARS-CoV-2 inhibitors targeting rna polymerase and spike glycoprotein. Pharmaceuticals 2022, 15, 376. [Google Scholar] [CrossRef]
- El-Kalyoubi, S.; Agili, F.; Zordok, W.A.; El-Sayed, A.S.A. Synthesis, In Silico Prediction and In Vitro Evaluation of Antimicrobial Activity, DFT Calculation and Theoretical Investigation of Novel Xanthines and Uracil Containing Imidazolone Derivatives. Int. J. Mol. Sci. 2021, 22, 10979. [Google Scholar] [CrossRef]
- El-Kalyoubi, S.; Agili, F. Synthesis, In Silico Prediction and In Vitro Evaluation of Antitumor Activities of Novel Pyrido [2, 3-d] pyrimidine, Xanthine and Lumazine Derivatives. Molecules 2020, 25, 5205. [Google Scholar] [CrossRef]
- El-Kalyoubi, S.; Agili, F.; Youssif, S. Novel 2-Thioxanthine and Dipyrimidopyridine Derivatives: Synthesis and Antimicrobial Activity. Molecules 2015, 20, 19263–19276. [Google Scholar] [CrossRef]
- Han, H.; Li, C.; Li, M.; Yang, L.; Zhao, S.; Wang, Z.; Liu, H.; Liu, D. Design, Synthesis, and Biological Evaluation of 8-Mercapto-3,7-Dihydro-1H-Purine-2,6-Diones as Potent Inhibitors of SIRT1, SIRT2, SIRT3, and SIRT5. Molecules 2020, 25, 2755. [Google Scholar] [CrossRef]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
- Bandyopadhyay, D.; Okan, N.A.; Bales, E.; Nascimento, L.; Cole, P.A.; Medrano, E.E. Down-regulation of p300/CBP histone acetyltransferase activates a senescence checkpoint in human melanocytes. Cancer Res. 2002, 62, 6231–6239. [Google Scholar]
- Bandyopadhyay, D.; Mishra, A.; Medrano, E.E. Overexpression of Histone Deacetylase 1 Confers Resistance to Sodium Butyrate–Mediated Apoptosis in Melanoma Cells through a p53-Mediated Pathway. Cancer Res. 2004, 64, 7706–7710. [Google Scholar] [CrossRef]
- Wang, Z.; Qin, G.; Zhao, T.C. HDAC4: Mechanism of regulation and biological functions. Epigenomics 2014, 6, 139–150. [Google Scholar] [CrossRef] [PubMed]
- Mai, A.; Perrone, A.; Nebbioso, A.; Rotili, D.; Valente, S.; Tardugno, M.; Massa, S.; De Bellis, F.; Altucci, L. Novel uracil-based 2-aminoanilide and 2-aminoanilide-like derivatives: Histone deacetylase inhibition and in-cell activities. Bioorganic Med. Chem. Lett. 2008, 18, 2530–2535. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Wang, D.; Wang, X.; Wang, Y.; Ren, F.; Chang, D.; Chang, Z.; Jia, B. Caspase 3 is Activated through Caspase 8 instead of Caspase 9 during H2O2-induced Apoptosis in HeLa Cells. Cell. Physiol. Biochem. 2011, 27, 539–546. [Google Scholar] [CrossRef]
- Denizot, F.; Lang, R. Rapid colorimetric assay for cell growth and survival: Modifications to the tetrazolium dye procedure giving improved sensitivity and reliability. J. Immunol. Methods 1986, 89, 271–277. [Google Scholar] [CrossRef]
- Thabrew, M.I.; Hughes, R.D.; Mcfarlane, I.G. Screening of Hepatoprotective Plant Components using a HepG2 Cell Cytotoxicity Assay. J. Pharm. Pharmacol. 1997, 49, 1132–1135. [Google Scholar] [CrossRef]
- Al-Rashood, S.T.; Hamed, A.R.; Hassan, G.S.; Alkahtani, H.M.; Almehizia, A.A.; Alharbi, A.; Al-Sanea, M.M.; Eldehna, W.M. Antitumor properties of certain spirooxindoles towards hepatocellular carcinoma endowed with antioxidant activity. J. Enzym. Inhib. Med. Chem. 2020, 35, 831–839. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Nagarajan, A.; Uchil, P.D. Analysis of Cell Viability by the MTT Assay. Cold Spring Harb. Protoc. 2018, 2018, pdb-prot095505. [Google Scholar] [CrossRef]
- Fournel, M.; Bonfils, C.; Hou, Y.; Yan, P.T.; Trachy-Bourget, M.-C.; Kalita, A.; Liu, J.; Lu, A.-H.; Zhou, N.Z.; Robert, M.-F. MGCD0103, a novel isotype-selective histone deacetylase inhibitor, has broad spectrum antitumor activity in vitro and in vivo. Mol. Cancer Ther. 2008, 7, 759–768. [Google Scholar] [CrossRef] [PubMed]
- Heltweg, B.; Trapp, J.; Jung, M. In vitro assays for the determination of histone deacetylase activity. Methods 2005, 36, 332–337. [Google Scholar] [CrossRef]
- Eldehna, W.M.; Hassan, G.S.; Al-Rashood, S.T.; Al-Warhi, T.; Altyar, A.E.; Alkahtani, H.M.; Almehizia, A.A.; Abdel-Aziz, H.A. Synthesis and in vitro anticancer activity of certain novel 1-(2-methyl-6-arylpyridin-3-yl)-3-phenylureas as apoptosis-inducing agents. J. Enzym. Inhib. Med. Chem. 2019, 34, 322–332. [Google Scholar] [CrossRef]
- Yousef, R.G.; Sakr, H.M.; Eissa, I.H.; Mehany, A.B.; Metwaly, A.M.; Elhendawy, M.A.; Radwan, M.M.; ElSohly, M.A.; Abulkhair, H.S.; El-Adl, K. New quinoxaline-2 (1 H)-ones as potential VEGFR-2 inhibitors: Design, synthesis, molecular docking, ADMET profile and anti-proliferative evaluations. New J. Chem. 2021, 45, 16949–16964. [Google Scholar] [CrossRef]
- Pozarowski, P.; Darzynkiewicz, Z. Analysis of Cell Cycle by Flow Cytometry. In Checkpoint Controls and Cancer: Volume 2: Activation and Regulation Protocols; Springer: Berlin, Germany, 2004; pp. 301–311. [Google Scholar]
- Lo, K.K.-W.; Lee, T.K.-M.; Lau, J.S.-Y.; Poon, W.-L.; Cheng, S.-H. Luminescent Biological Probes Derived from Ruthenium(II) Estradiol Polypyridine Complexes. Inorg. Chem. 2007, 47, 200–208. [Google Scholar] [CrossRef]
- Elkady, H.; Elwan, A.; El-Mahdy, H.A.; Doghish, A.S.; Ismail, A.; Taghour, M.S.; Elkaeed, E.B.; Eissa, I.H.; Dahab, M.A.; Mahdy, H.A.; et al. New benzoxazole derivatives as potential VEGFR-2 inhibitors and apoptosis inducers: Design, synthesis, anti-proliferative evaluation, flowcytometric analysis, and in silico studies. J. Enzym. Inhib. Med. Chem. 2022, 37, 403–416. [Google Scholar] [CrossRef]
- Eray, M.; Mättö, M.; Kaartinen, M.; Andersson, L.C.; Pelkonen, J. Flow cytometric analysis of apoptotic subpopulations with a combination of Annexin V-FITC, propidium iodide, and SYTO 17. Cytom. J. Int. Soc. Anal. Cytol. 2001, 43, 134–142. [Google Scholar] [CrossRef]
- Balah, A.; Ezzat, O.; Akool, E.-S. Vitamin E inhibits cyclosporin A-induced CTGF and TIMP-1 expression by repressing ROS-mediated activation of TGF-β/Smad signaling pathway in rat liver. Int. Immunopharmacol. 2018, 65, 493–502. [Google Scholar] [CrossRef]
- Aborehab, N.M.; Elnagar, M.R.; Waly, N.E. Gallic acid potentiates the apoptotic effect of paclitaxel and carboplatin via overexpression of Bax and P53 on the MCF-7 human breast cancer cell line. J. Biochem. Mol. Toxicol. 2020, 35, e22638. [Google Scholar] [CrossRef] [PubMed]
- Elnagar, M.R.; Walls, A.B.; Helal, G.K.; Hamada, F.M.; Thomsen, M.S.; Jensen, A.A. Functional characterization of α7 nicotinic acetylcholine and NMDA receptor signaling in SH-SY5Y neuroblastoma cells in an ERK phosphorylation assay. Eur. J. Pharmacol. 2018, 826, 106–113. [Google Scholar] [CrossRef]
- Guo, Y.; Tong, Y.; Zhu, H.; Xiao, Y.; Guo, H.; Shang, L.; Zheng, W.; Ma, S.; Liu, X.; Bai, Y. Quercetin suppresses pancreatic ductal adenocarcinoma progression via inhibition of SHH and TGF-β/Smad signaling pathways. Cell Biol. Toxicol. 2021, 37, 479–496. [Google Scholar] [CrossRef]
- Jiao, C.; Chen, W.; Tan, X.; Liang, H.; Li, J.; Yun, H.; He, C.; Chen, J.; Ma, X.; Xie, Y.; et al. Ganoderma lucidum spore oil induces apoptosis of breast cancer cells in vitro and in vivo by activating caspase-3 and caspase-9. J. Ethnopharmacol. 2020, 247, 112256. [Google Scholar] [CrossRef]
- Ma, C.; Taghour, M.S.; Belal, A.; Mehany, A.B.M.; Mostafa, N.; Nabeeh, A.; Eissa, I.H.; Al-Karmalawy, A.A. Design and Synthesis of New Quinoxaline Derivatives as Potential Histone Deacetylase Inhibitors Targeting Hepatocellular Carcinoma: In Silico, In Vitro, and SAR Studies. Front. Chem. 2021, 9, 725135. [Google Scholar] [CrossRef]
- Elkady, M.A.; Doghish, A.S.; Elshafei, A.; Elshafey, M.M. MicroRNA-567 inhibits cell proliferation and induces cell apoptosis in A549 NSCLC cells by regulating cyclin-dependent kinase 8. Saudi J. Biol. Sci. 2021, 28, 2581–2590. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).