Integrated Single-Cell and Transcriptome Sequencing Analyses Identify Dipeptidase 2 as an Immune-Associated Prognostic Biomarker for Lung Adenocarcinoma
Abstract
1. Introduction
2. Results
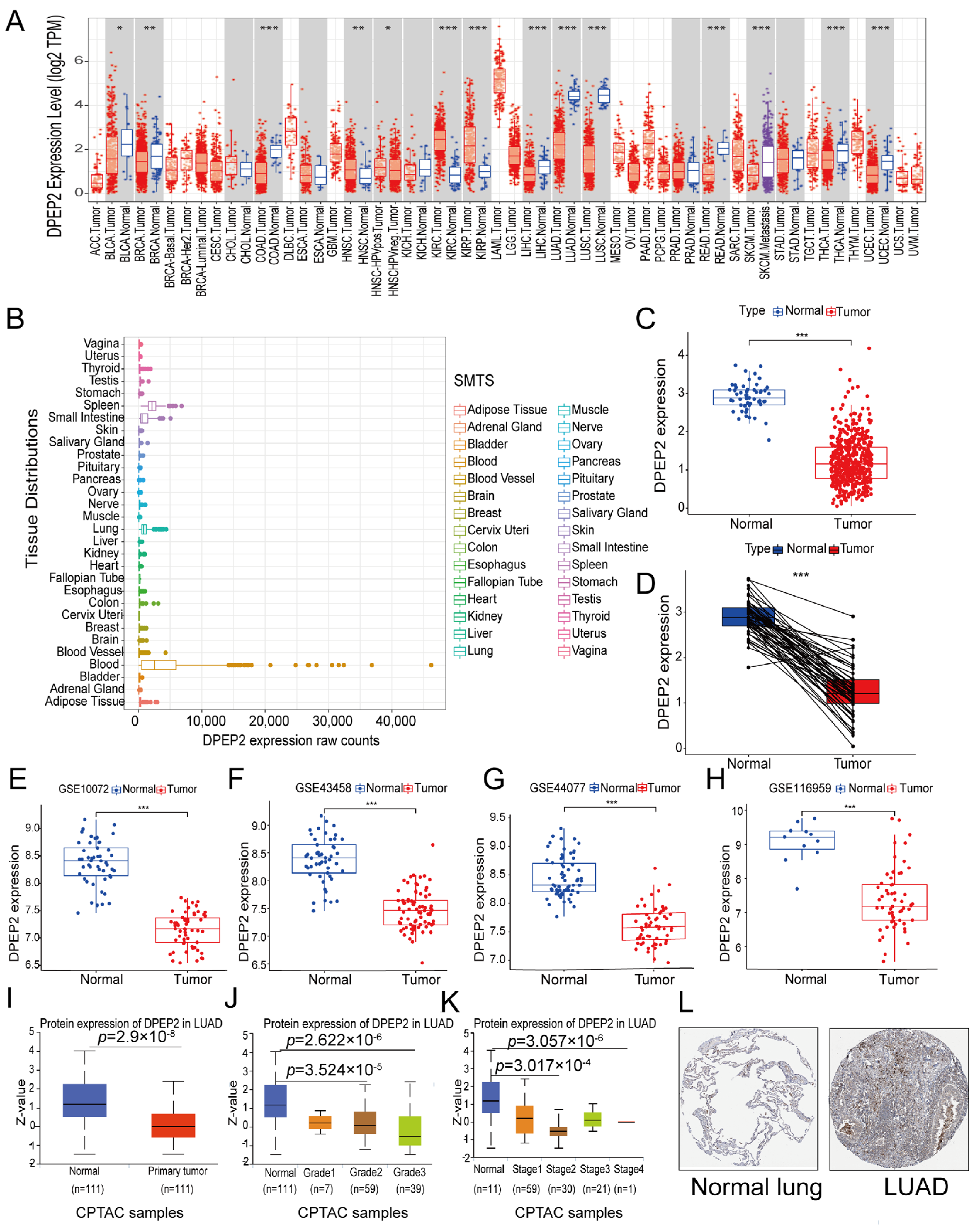
2.1. DPEP2 Expression Is Down-Regulated in LUAD
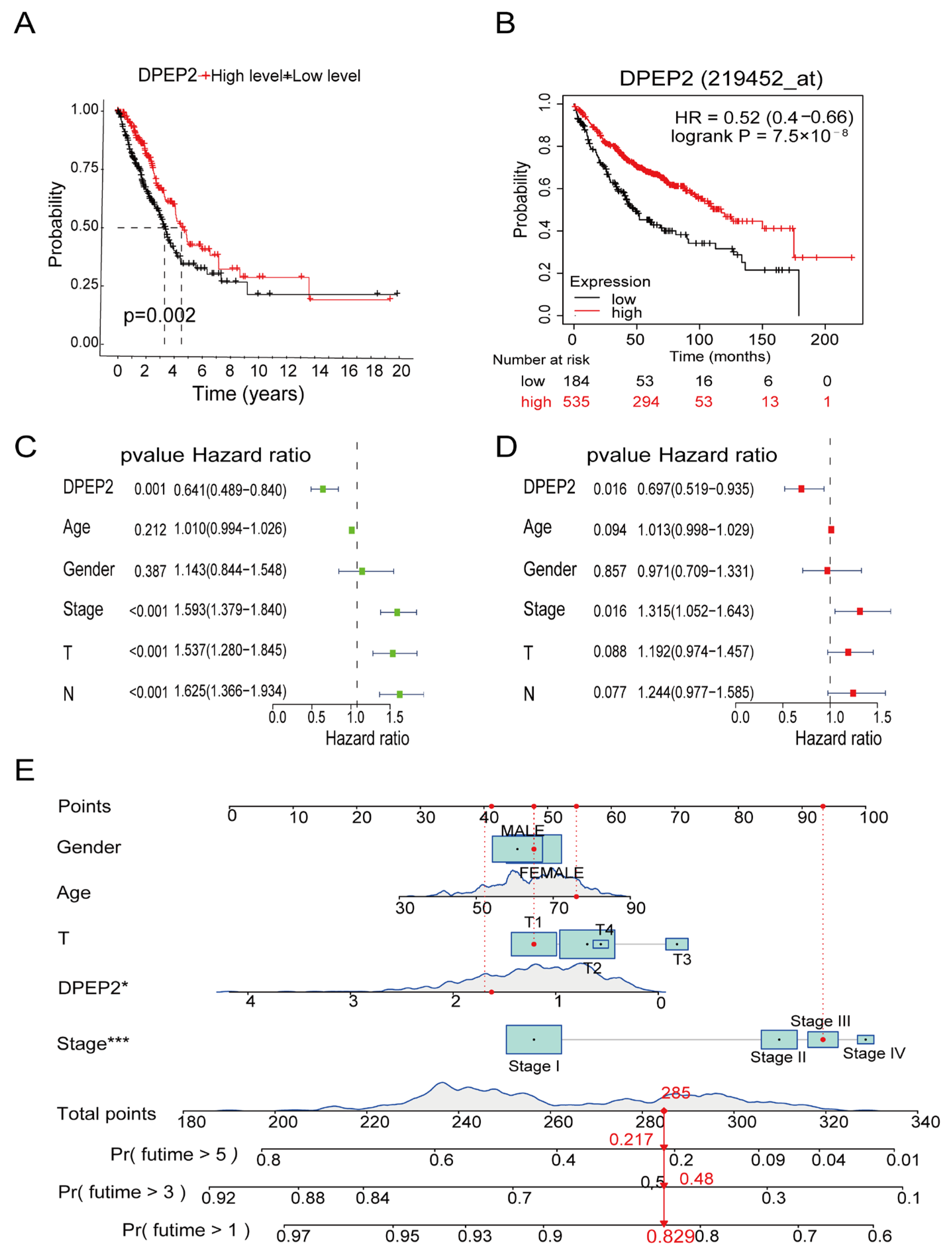
2.2. Relationships between DPEP2 and Clinicopathological Characteristics of LUAD Patients
2.3. DPEP2 Is a Favorable Independent Prognostic Factor for LUAD
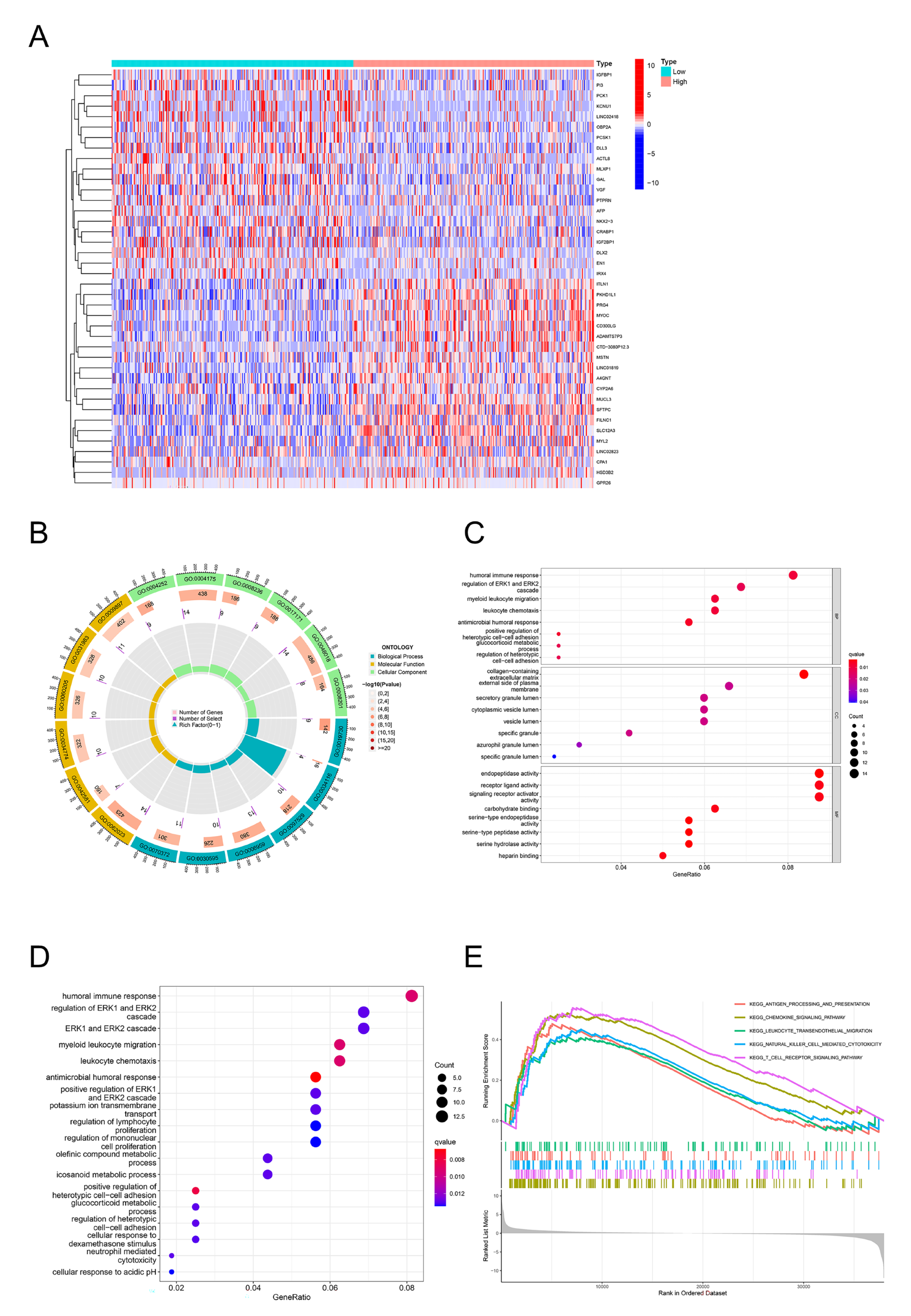
2.4. GO, KEGG, and GSEA Identify DPEP2-Related Signaling Pathways in LUAD
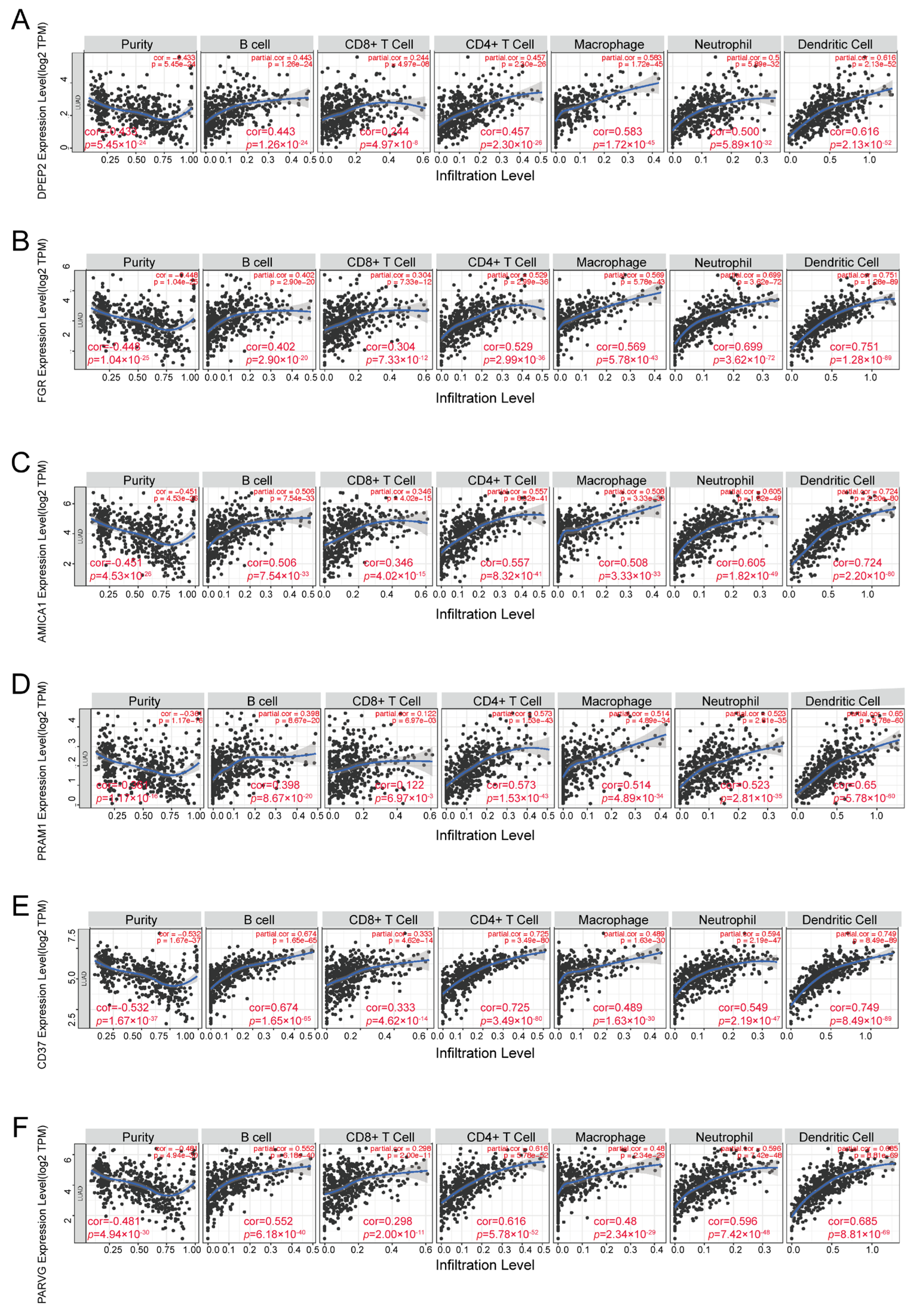
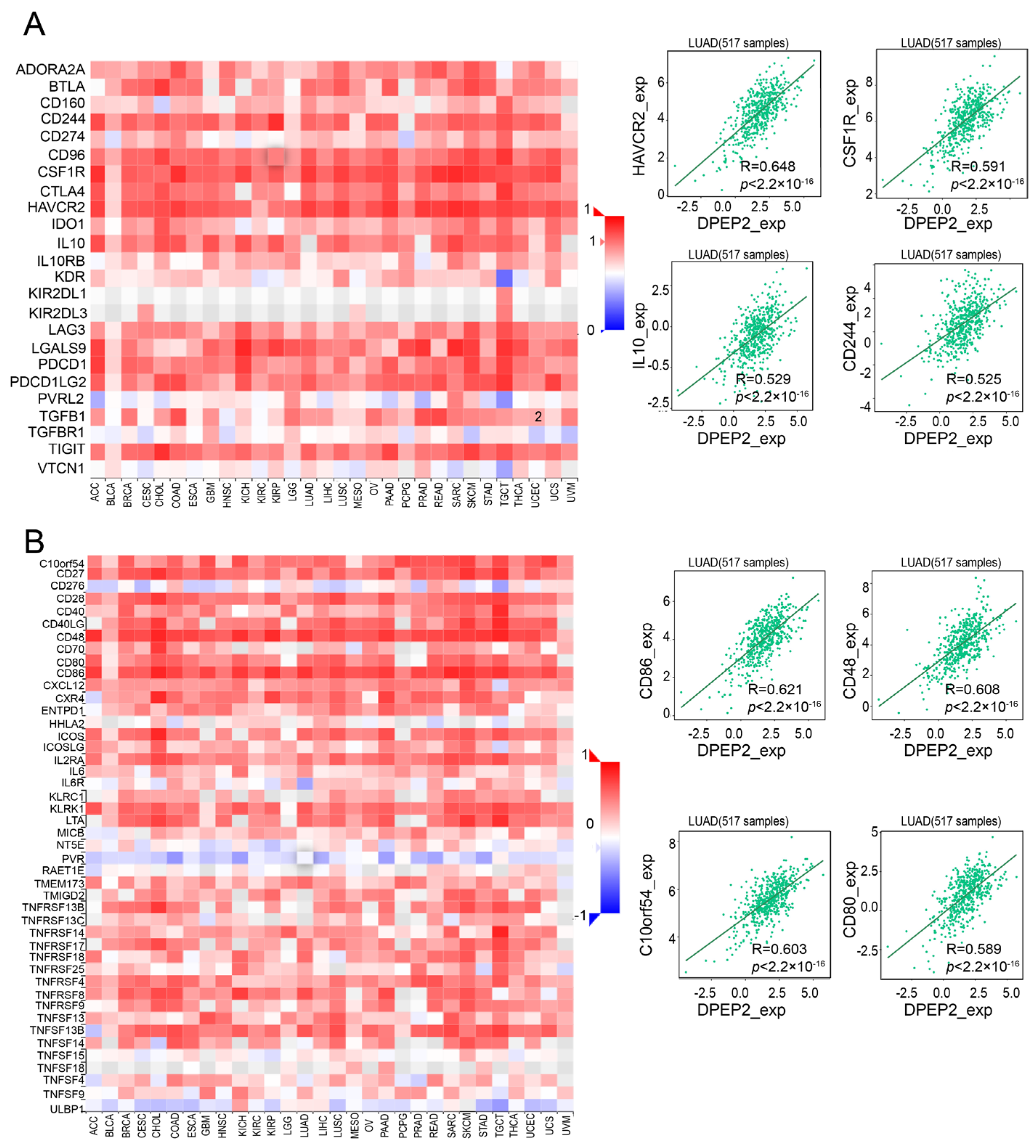
2.5. DPEP2 and Its Co-Expressed Genes Are Significantly Associated with Immunity in LUAD
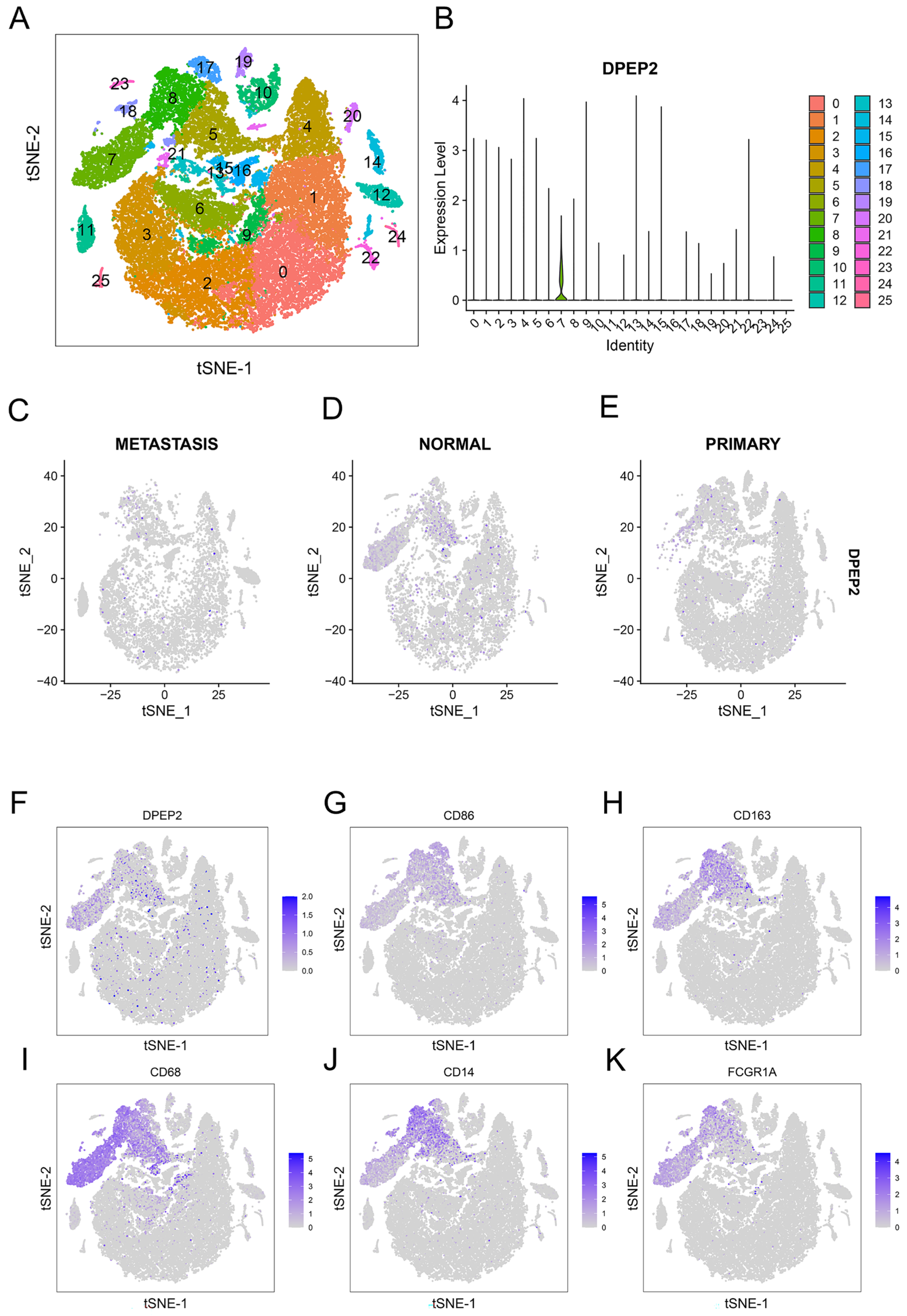
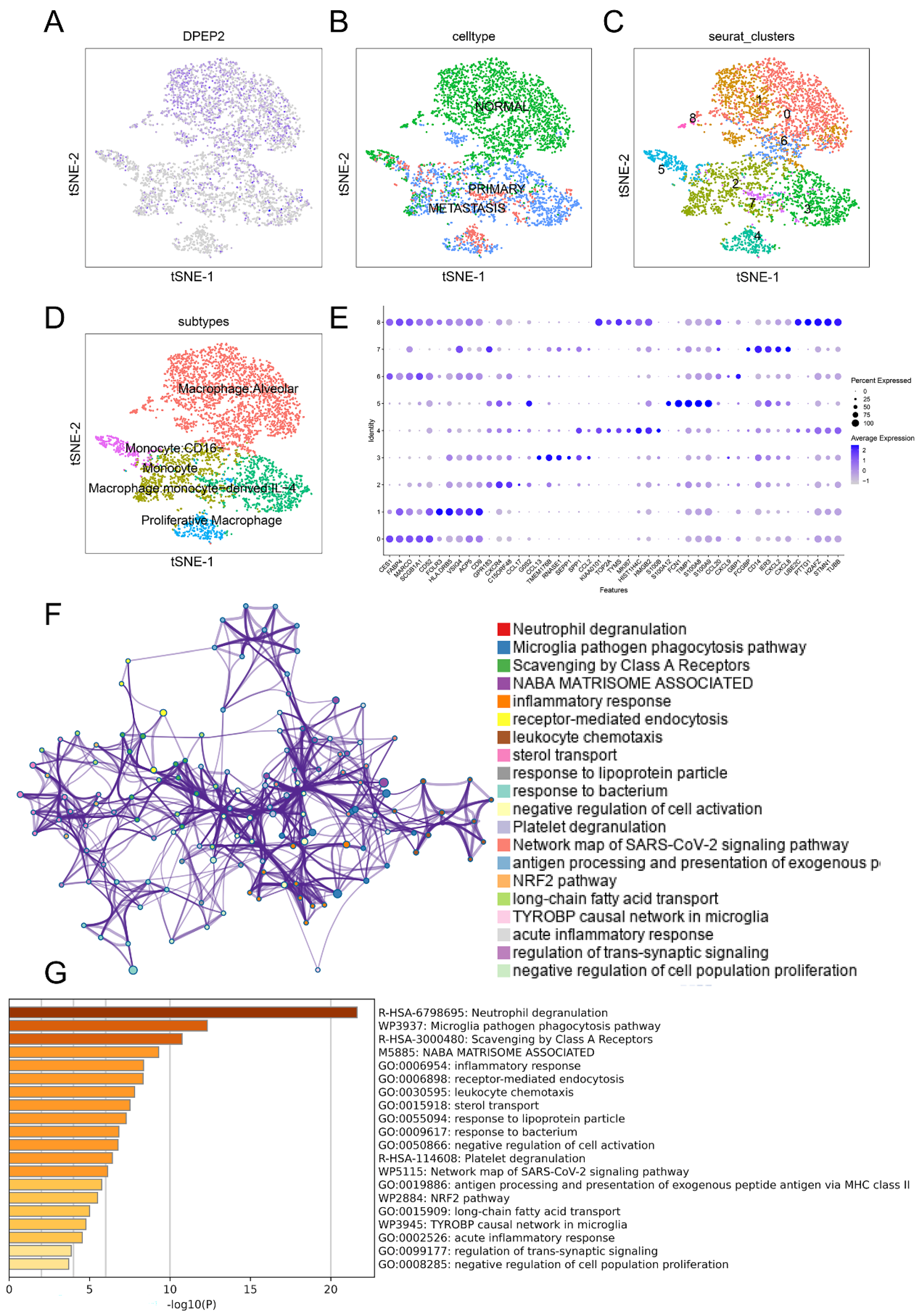
2.6. DPEP2 Is Expressed in Macrophages in the Single-Cell Transcriptome
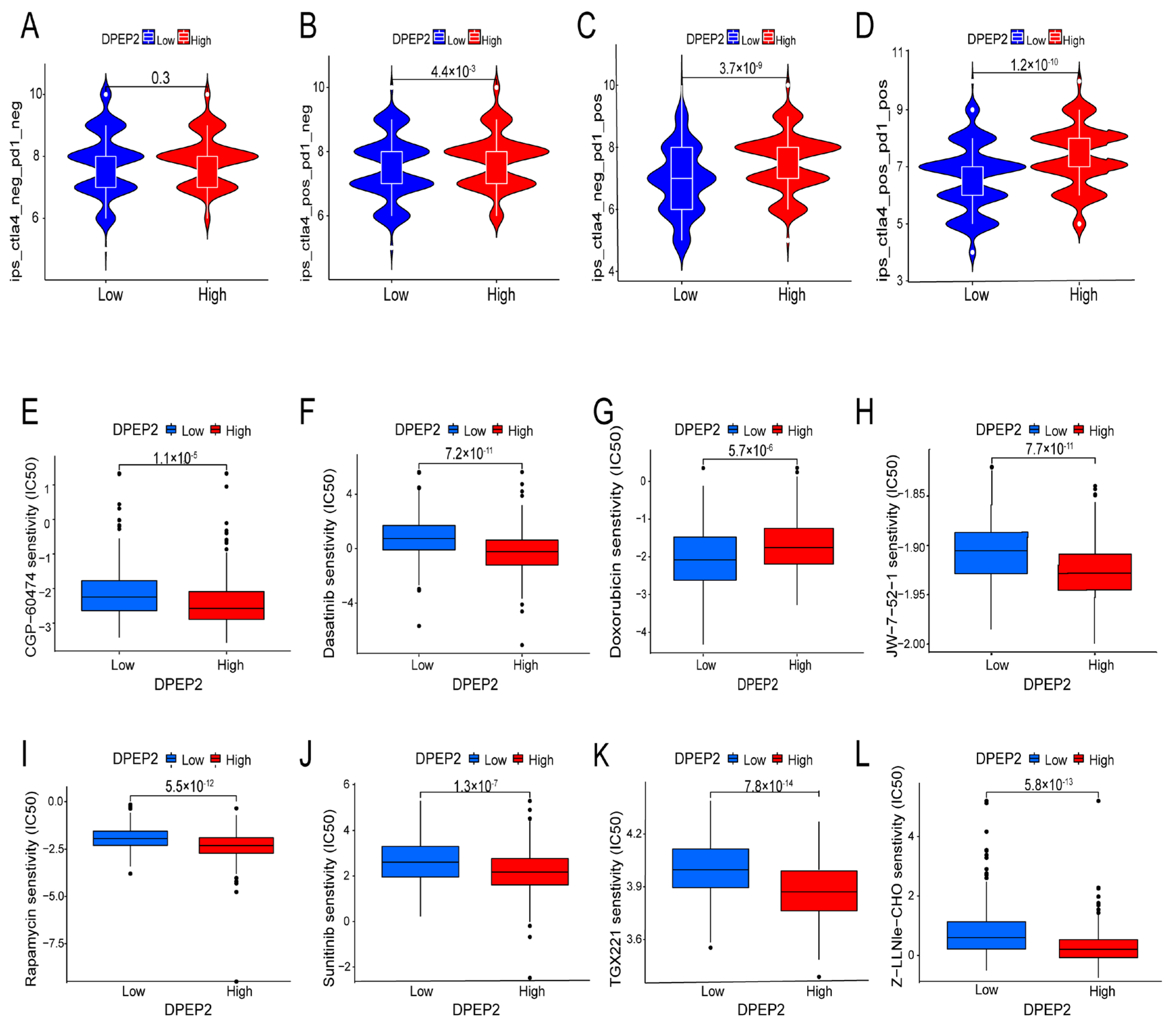
2.7. Relationship between DPEP2 and Sensitivity to Immunotherapy in LUAD
2.8. Relationship between DPEP2 and Sensitivity to Drugs in LUAD
2.9. DPEP2 Is Downregulated in Clinical LUAD Samples
3. Discussion
4. Materials and Methods
4.1. Data Collection and Integration
4.2. Demographic and Clinical Characteristics
4.3. Survival Analysis, Significant Prognostic Marker Analysis, and Nomogram
4.4. Differentially Expressed Gene Analysis
4.5. Functional Enrichment Analysis
4.6. Correlation Analysis of DPEP2 and Its Co-Expressed Genes with Immunity in LUAD
4.7. Single-Cell Transcriptome Analysis
4.8. Expression of DPEP2 and Immunotherapy and Drug Sensitivity Analysis in LUAD
4.9. Immunohistochemistry
4.10. Cell Culture and Transfection Assay for Overexpression
4.11. Wound Healing Assay
4.12. Transwell Migration and Invasion Assay
4.13. Statistical Methods
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| LUAD | Lung adenocarcinoma |
| NSCLC | Non-small cell lung cancer |
| LTD4 | Leukotriene D4 |
| DPEP2 | Dipeptidase 2 |
| LTE4 | Leukotriene E4 |
| TIMER | The Tumor Immune Estimation Resource |
| GTEx | Genotype-Tissue Expression |
| TCGA | The Cancer Genome Atlas |
| GEO | Gene Expression Omnibus |
| THPA | The Human Protein Atlas |
| AJCC | American Joint Committee on Cancer |
| OS event | Over survival event |
| DEGs | Differentially expressed genes |
| GO | Gene Ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| GSEA | Gene Set Enrichment Analysis |
| UMIs | Unique molecular identifiers |
| PCA | Principal component analysis |
| IPS | Immunophenoscores |
| TCIA | The Cancer Immunome Database |
| IC50 | The half-maximal inhibitory concentration |
| IHC | Immunohistochemistry |
| ATCC | American Type Culture Collection |
| DMEM | Dulbecco’s Modified Eagle Medium |
| FBS | Fetal bovine serum |
| PBS | Phosphate buffered saline |
| ANOVA | Analysis of variance |
References
- Carobene, L.; Spina, D.; Disanto, M.G.; Micheletto, C.; Mazzei, M.A.; Paladini, P.; Ghiribelli, C.; Bargagli, E. Lung cancer interstitial lung diseases: The lack of prognostic impact of lung cancer in, I.P.F. Intern. Emerg. Med. 2022, 17, 457–464. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Hirsch, F.R.; Scagliotti, G.V.; Mulshine, J.L.; Kwon, R.; Curran, W.J., Jr.; Wu, Y.-L.; Paz-Ares, L. Lung cancer: Current therapies and new targeted treatments. Lancet 2017, 389, 299–311. [Google Scholar] [CrossRef] [PubMed]
- CCochetti, G.; de Vermandois, J.A.R.; Maulà, V.; Cari, L.; Cagnani, R.; Suvieri, C.; Balducci, P.M.; Paladini, A.; Del Zingaro, M.; Nocentini, G.; et al. Diagnostic performance of the Bladder EpiCheck methylation test and photodynamic diagnosis-guided cystoscopy in the surveillance of high-risk non-muscle invasive bladder cancer: A single centre, prospective, blinded clinical trial. Urol. Oncol. 2022, 40, 105.e11–105.e18. [Google Scholar] [CrossRef] [PubMed]
- Brain, S.D.; Williams, T.J. Leukotrienes and inflammation. Pharmacol. Ther. 1990, 46, 57–66. [Google Scholar] [CrossRef]
- Voisin, T.; Perner, C.; Messou, M.A.; Shiers, S.; Ualiyeva, S.; Kanaoka, Y.; Price, Y.; Sokol, T.J.; Bankova, C.L.; Austen, L.G.; et al. The CysLT(2)R receptor mediates leukotriene C(4)-driven acute and chronic itch. Proc. Natl. Acad. Sci. USA 2021, 118, e2022087118. [Google Scholar] [CrossRef]
- Savari, S.; Vinnakota, K.; Zhang, Y.; Sjolander, A. Cysteinyl leukotrienes and their receptors: Bridging inflammation and colorectal cancer. World J. Gastroenterol. 2014, 20, 968–977. [Google Scholar] [CrossRef]
- van Brussel, J.P.; Oomen, M.A.; Vossebeld, P.J.; Wiemer, E.A.; Sonneveld, P.; Mickisch, G.H. Identification of multidrug resistance-associated protein 1 and glutathione as multidrug resistance mechanisms in human prostate cancer cells: Chemosensitization with leukotriene D4 antagonists and buthionine sulfoximine. BJU Int. 2004, 93, 1333–1338. [Google Scholar] [CrossRef]
- Lukic, A.; Wahlund, C.J.; Gómez, C.; Brodin, D.; Samuelsson, B.; Wheelock, C.E.; Gabrielsson, S.; Rådmark, O. Exosomes and cells from lung cancer pleural exudates transform LTC4 to LTD4, promoting cell migration and survival via CysLT1. Cancer Lett. 2019, 444, 1–8. [Google Scholar] [CrossRef]
- Magnusson, C.; Mezhybovska, M.; Lorinc, E.; Fernebro, E.; Nilbert, M.; Sjolander, A. Low expression of CysLT1R and high expression of CysLT2R mediate good prognosis in colorectal cancer. Eur. J. Cancer 2010, 46, 826–835. [Google Scholar] [CrossRef]
- Habib, G.M.; Shi, Z.Z.; Cuevas, A.A.; Lieberman, M.W. Identification of two additional members of the membrane-bound dipeptidase family. FASEB J. 2003, 17, 1313–1315. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Yue, Y.; Xiong, S. Dpep2 Emerging as a Modulator of Macrophage Inflammation Confers Protection Against CVB3-Induced Viral Myocarditis. Front. Cell. Infect. Microbiol. 2019, 9, 57. [Google Scholar] [CrossRef]
- Wu, H.; Wang, D.; Zheng, Q.; Xu, Z. Integrating SWATH-MS proteomics and transcriptome analysis to preliminarily identify three DEGs as biomarkers for proliferative diabetic retinopathy. Proteom. Clin. Appl. 2022, 16, e21000162022. [Google Scholar] [CrossRef]
- Miyata, J.; Fukunaga, K.; Kawashima, Y.; Watanabe, T.; Saitoh, A.; Hirosaki, T.; Araki, Y.; Kikawada, T.; Betsuyaku, T.; Ohara, O.; et al. Dysregulated fatty acid metabolism in nasal polyp-derived eosinophils from patients with chronic rhinosinusitis. Allergy 2019, 74, 1113–1124. [Google Scholar] [CrossRef]
- Gresham, H.D.; Dale, B.M.; Potter, J.W.; Chang, P.W.; Vines, C.M.; Lowell, C.A.; Lagenaur, C.F.; Willman, C.L. Negative regulation of phagocytosis in murine macrophages by the Src kinase family member, Fgr. J. Exp. Med. 2000, 191, 515–528. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.; Zhang, Y.; He, M.; Han, Y.; Cai, C.; Liu, S.; Liu, S.; Chen, P.; Shen, Y.; Zeng, H.; et al. AMICA1 is a diagnostic and prognostic biomarker and induces immune cells infiltration by activating cGAS-STING signaling in lung adenocarcinoma. Cancer Cell. Int. 2022, 22, 111. [Google Scholar] [CrossRef] [PubMed]
- Quail, D.F.; Joyce, J.A. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 2013, 19, 1423–1437. [Google Scholar] [CrossRef]
- Pitt, J.M.; Marabelle, A.; Eggermont, A.; Soria, J.C.; Kroemer, G.; Zitvogel, L. Targeting the tumor microenvironment: Removing obstruction to anticancer immune responses and immunotherapy. Ann. Oncol. 2016, 27, 1482–1492. [Google Scholar] [CrossRef]
- Kloosterman, D.J.; Akkari, L. Macrophages at the interface of the co-evolving cancer ecosystem. Cell 2023, 186, 1627–1651. [Google Scholar] [CrossRef]
- Dai, X.; Lu, L.; Deng, S.; Meng, J.; Wan, C.; Huang, J.; Sun, Y.; Hu, Y.; Wu, B.; Wu, G.; et al. USP7 targeting modulates anti-tumor immune response by reprogramming Tumor-associated Macrophages in Lung Cancer. Theranostics 2020, 10, 9332–9347. [Google Scholar] [CrossRef]
- Qian, B.Z.; Pollard, J.W. Macrophage diversity enhances tumor progression and metastasis. Cell 2010, 141, 39–51. [Google Scholar] [CrossRef]
- Xiang, X.; Wang, J.; Lu, D.; Xu, X. Targeting tumor-associated macrophages to synergize tumor immunotherapy. Signal Transduct. Target. Ther. 2021, 6, 75. [Google Scholar] [CrossRef] [PubMed]
- Duan, Z.; Luo, Y. Targeting macrophages in cancer immunotherapy. Signal Transduct. Target. Ther. 2021, 6, 127. [Google Scholar] [CrossRef] [PubMed]
- Pan, K.; Farrukh, H.; Chittepu, V.; Xu, H.; Pan, C.X.; Zhu, Z. CAR race to cancer immunotherapy: From CAR T, CAR NK to CAR macrophage therapy. J. Exp. Clin. Cancer Res. 2022, 41, 119. [Google Scholar] [CrossRef]
- Anderson, N.R.; Minutolo, N.G.; Gill, S.; Klichinsky, M. Macrophage-Based Approaches for Cancer Immunotherapy. Cancer Res. 2021, 81, 1201–1208. [Google Scholar] [CrossRef] [PubMed]
- Komi, D.E.A.; Redegeld, F.A. Role of Mast Cells in Shaping the Tumor Microenvironment. Clin. Rev. Allergy Immunol. 2020, 58, 313–325. [Google Scholar] [CrossRef]
- Xia, Y.; Rao, L.; Yao, H.; Wang, Z.; Ning, P.; Chen, X. Engineering Macrophages for Cancer Immunotherapy and Drug Delivery. Adv. Mater. 2020, 32, e20020542020. [Google Scholar] [CrossRef] [PubMed]
- Zheng, C.; Zheng, L.; Yoo, J.-K.; Guo, H.; Zhang, Y.; Guo, X.; Kang, B.; Hu, R.; Huang, J.Y.; Zhang, Q.; et al. Landscape of Infiltrating T Cells in Liver Cancer Revealed by Single-Cell Sequencing. Cell 2017, 169, 1342–1356.e16. [Google Scholar] [CrossRef]
- Han, J.; DePinho, R.A.; Maitra, A. Single-cell RNA sequencing in pancreatic cancer. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 451–452. [Google Scholar] [CrossRef]
- Skoulidis, F.; Goldberg, M.E.; Greenawalt, D.M.; Hellmann, M.D.; Awad, M.M.; Gainor, J.F.; Schrock, J.F.; Hartmaier, A.B.; Trabucco, R.J.; Gay, S.E.; et al. STK11/LKB1 Mutations and PD-1 Inhibitor Resistance in KRAS-Mutant Lung Adenocarcinoma. Cancer Discov. 2018, 8, 822–835. [Google Scholar] [CrossRef]
- Kennedy, L.B.; Salama, A.K.S. A review of cancer immunotherapy toxicity. CA Cancer J. Clin. 2020, 70, 86–104. [Google Scholar] [CrossRef] [PubMed]
- Anker, J.F.; Naseem, A.F.; Mok, H.; Schaeffer, A.J.; Abdulkadir, S.A.; Thumbikat, P. Multi-faceted immunomodulatory and tissue-tropic clinical bacterial isolate potentiates prostate cancer immunotherapy. Nat. Commun. 2018, 9, 1591. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, Z. The history and advances in cancer immunotherapy: Understanding the characteristics of tumor-infiltrating immune cells and their therapeutic implications. Cell. Mol. Immunol. 2020, 17, 807–821. [Google Scholar] [CrossRef] [PubMed]
- Cooper, A.J.; Sequist, L.V.; Lin, J.J. Third-generation EGFR and ALK inhibitors: Mechanisms of resistance and management. Nat. Rev. Clin. Oncol. 2022, 19, 499–514. [Google Scholar] [CrossRef]
- Solomon, B.J.; Bauer, T.M.; Mok, T.S.K.; Liu, G.; Mazieres, J.; de Marinis, F.; Goto, Y.; Kim, D.-W.; Wu, Y.-L.; Jassem, J.; et al. Efficacy and safety of first-line lorlatinib versus crizotinib in patients with advanced, ALK-positive non-small-cell lung cancer: Updated analysis of data from the phase 3, randomised, open-label CROWN study. Lancet Respir. Med. 2023, 11, 354–366. [Google Scholar] [CrossRef]
- Drilon, A.; Jenkins, C.; Iyer, S.; Schoenfeld, A.; Keddy, C.; Davare, M.A. ROS1-dependent cancers—Biology, diagnostics and therapeutics. Nat. Rev. Clin. Oncol. 2021, 18, 35–55. [Google Scholar] [CrossRef]
- Poulikakos, P.I.; Sullivan, R.J.; Yaeger, R. Molecular Pathways and Mechanisms of BRAF in Cancer Therapy. Clin. Cancer Res. 2022, 28, 4618–4628. [Google Scholar] [CrossRef]
- Remon, J.; Hendriks, L.E.; Mountzios, G.; García-Campelo, R.; Saw, S.P.; Uprety, D.; Recondo, G.; Villacampa, G.; Reck, M. MET alterations in NSCLC-Current Perspectives and Future Challenges. J. Thorac. Oncol. 2023, 18, 419–435. [Google Scholar] [CrossRef]
- Griesinger, F.; Curigliano, G.; Thomas, M.; Subbiah, V.; Baik, C.; Tan, D.; Lee, D.; Misch, D.; Garralda, E.; Kim, D.-W.; et al. Safety and efficacy of pralsetinib in RET fusion-positive non-small-cell lung cancer including as first-line therapy: Update from the ARROW trial. Ann. Oncol. 2022, 33, 1168–1178. [Google Scholar] [CrossRef]
- Holmes, E.C. Postoperative chemotherapy for non-small-cell lung cancer. Chest 1993, 103 (Suppl. 1), 30S–34S. [Google Scholar] [CrossRef]
- Sohail, M.; Sun, Z.; Li, Y.; Gu, X.; Xu, H. Research progress in strategies to improve the efficacy and safety of doxorubicin for cancer chemotherapy. Expert. Rev. Anticancer. Ther. 2021, 21, 1385–1398. [Google Scholar] [CrossRef] [PubMed]
- Socinski, M.A. The current status and evolving role of sunitinib in non-small cell lung cancer. J. Thorac. Oncol. 2008, 3 (Suppl. 2), S119–S123. [Google Scholar] [CrossRef] [PubMed]
- Johnson, F.M.; Bekele, B.N.; Feng, L.; Wistuba, I.; Tang, X.M.; Tran, H.T.; Erasmus, J.J.; Hwang, L.-L.; Takebe, N.; Blumenschein, G.R.; et al. Phase II study of dasatinib in patients with advanced non-small-cell lung cancer. J. Clin. Oncol. 2010, 28, 4609–4615. [Google Scholar] [CrossRef] [PubMed]


| Characteristic | Low Expression of DPEP2 | High Expression of DPEP2 | p |
|---|---|---|---|
| n | 267 | 268 | |
| T stage, n (%) | 0.004 | ||
| T1 | 72 (13.5%) | 103 (19.4%) | |
| T2 | 149 (28%) | 140 (26.3%) | |
| T3 | 31 (5.8%) | 18 (3.4%) | |
| T4 | 14 (2.6%) | 5 (0.9%) | |
| N stage, n (%) | 0.038 | ||
| N0 | 162 (31.2%) | 186 (35.8%) | |
| N1 | 50 (9.6%) | 45 (8.7%) | |
| N2 | 47 (9.1%) | 27 (5.2%) | |
| N3 | 1 (0.2%) | 1 (0.2%) | |
| M stage, n (%) | 0.557 | ||
| M0 | 187 (48.4%) | 174 (45.1%) | |
| M1 | 15 (3.9%) | 10 (2.6%) | |
| Pathologic stage, n (%) | 0.011 | ||
| Stage I | 129 (24.5%) | 165 (31.3%) | |
| Stage II | 65 (12.3%) | 58 (11%) | |
| Stage III | 53 (10.1%) | 31 (5.9%) | |
| Stage IV | 15 (2.8%) | 11 (2.1%) | |
| Age, n (%) | <0.001 | ||
| <=65 | 147 (28.5%) | 108 (20.9%) | |
| >65 | 110 (21.3%) | 151 (29.3%) | |
| Smoker, n (%) | 0.003 | ||
| No | 25 (4.8%) | 50 (9.6%) | |
| Yes | 236 (45.3%) | 210 (40.3%) | |
| OS event, n (%) | 0.008 | ||
| Alive | 156 (29.2%) | 187 (35%) | |
| Dead | 111 (20.7%) | 81 (15.1%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.; Zhang, Y.; Gao, X.; Zhang, Y.; Chen, Y. Integrated Single-Cell and Transcriptome Sequencing Analyses Identify Dipeptidase 2 as an Immune-Associated Prognostic Biomarker for Lung Adenocarcinoma. Pharmaceuticals 2023, 16, 871. https://doi.org/10.3390/ph16060871
Zhang X, Zhang Y, Gao X, Zhang Y, Chen Y. Integrated Single-Cell and Transcriptome Sequencing Analyses Identify Dipeptidase 2 as an Immune-Associated Prognostic Biomarker for Lung Adenocarcinoma. Pharmaceuticals. 2023; 16(6):871. https://doi.org/10.3390/ph16060871
Chicago/Turabian StyleZhang, Xiangqian, Yunfan Zhang, Xiaomei Gao, Ye Zhang, and Yongheng Chen. 2023. "Integrated Single-Cell and Transcriptome Sequencing Analyses Identify Dipeptidase 2 as an Immune-Associated Prognostic Biomarker for Lung Adenocarcinoma" Pharmaceuticals 16, no. 6: 871. https://doi.org/10.3390/ph16060871
APA StyleZhang, X., Zhang, Y., Gao, X., Zhang, Y., & Chen, Y. (2023). Integrated Single-Cell and Transcriptome Sequencing Analyses Identify Dipeptidase 2 as an Immune-Associated Prognostic Biomarker for Lung Adenocarcinoma. Pharmaceuticals, 16(6), 871. https://doi.org/10.3390/ph16060871








