Discovery of AI-2 Quorum Sensing Inhibitors Targeting the LsrK/HPr Protein–Protein Interaction Site by Molecular Dynamics Simulation, Virtual Screening, and Bioassay Evaluation
Abstract
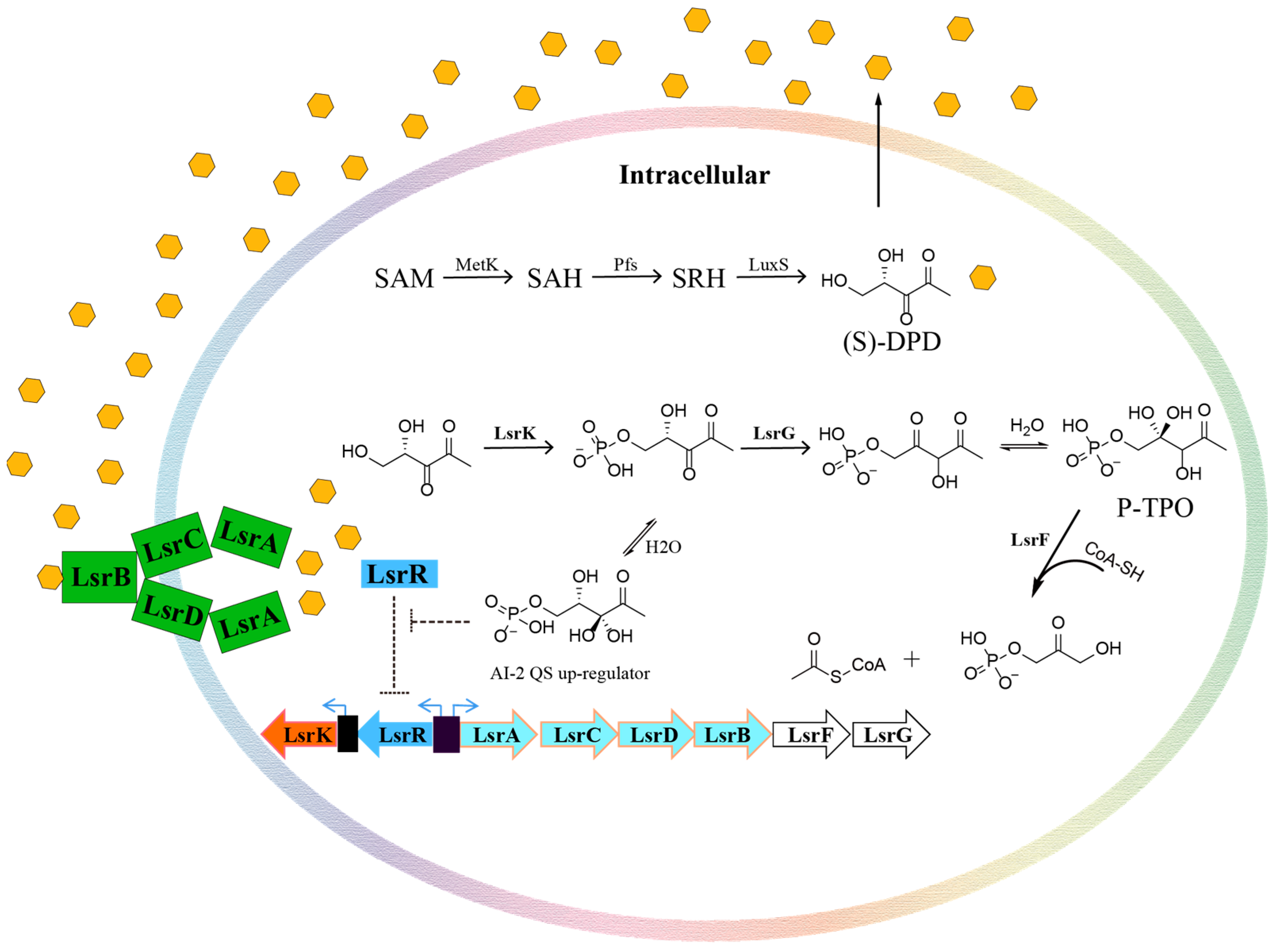
1. Introduction
2. Results and Discussion
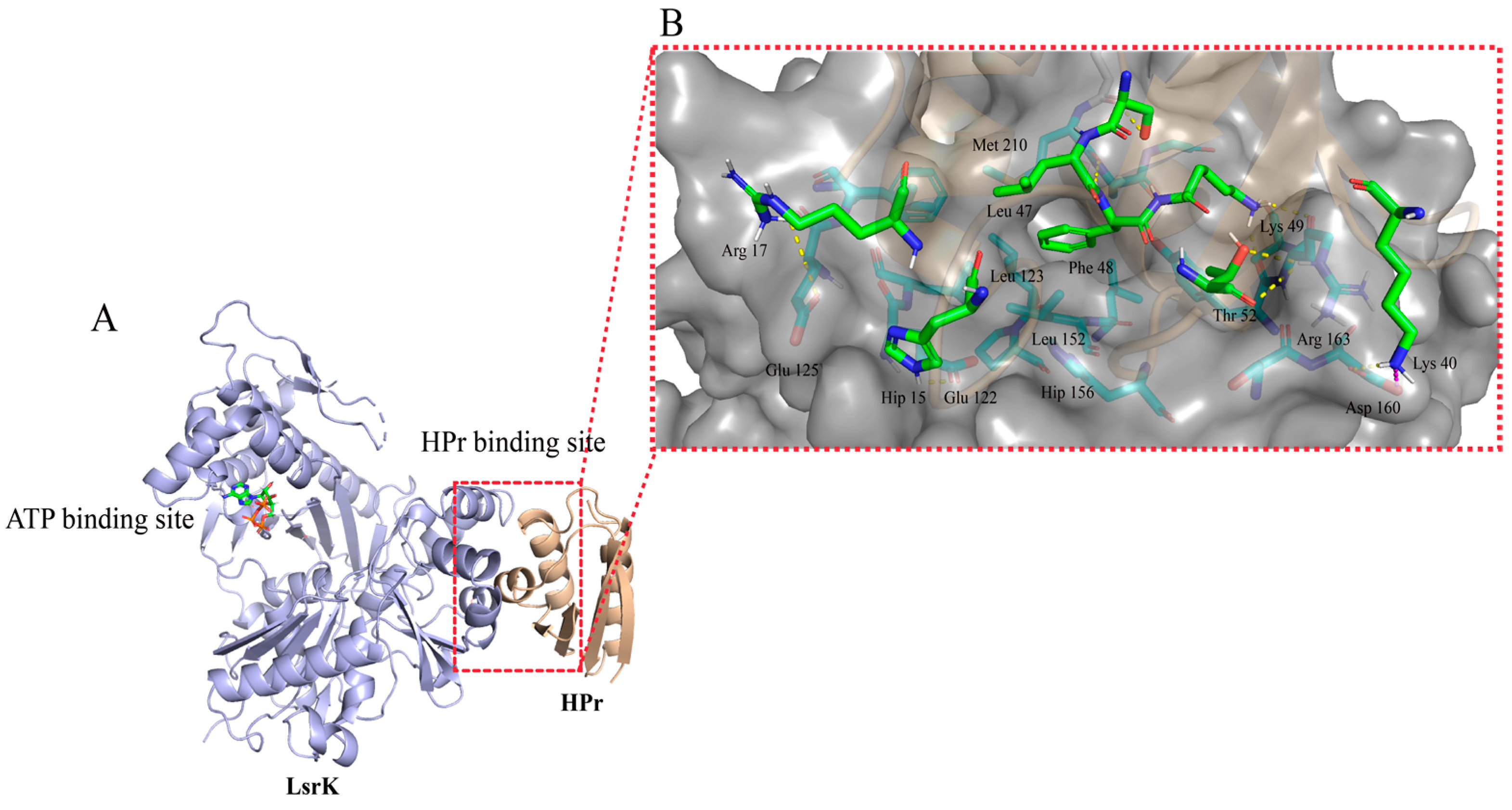
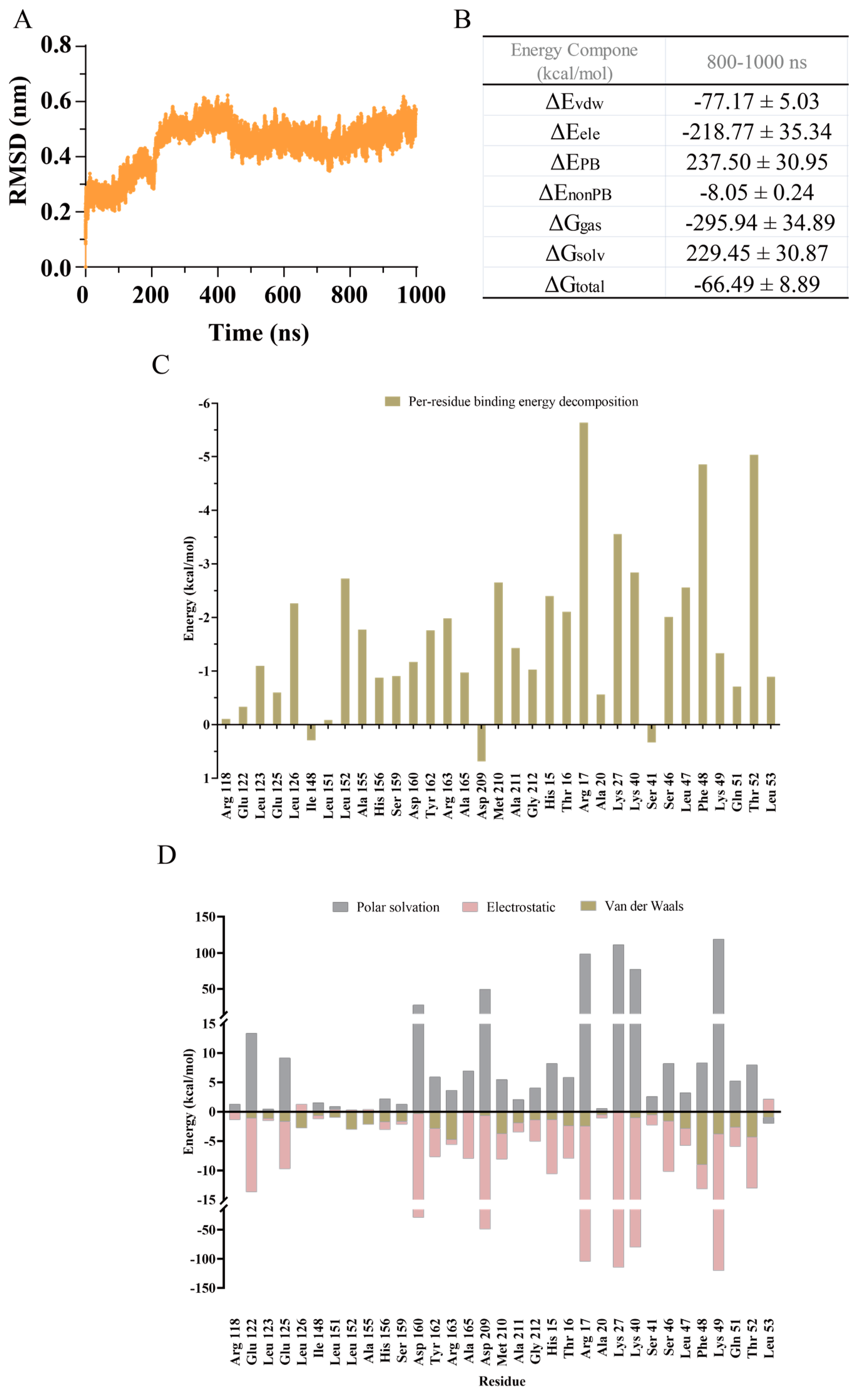
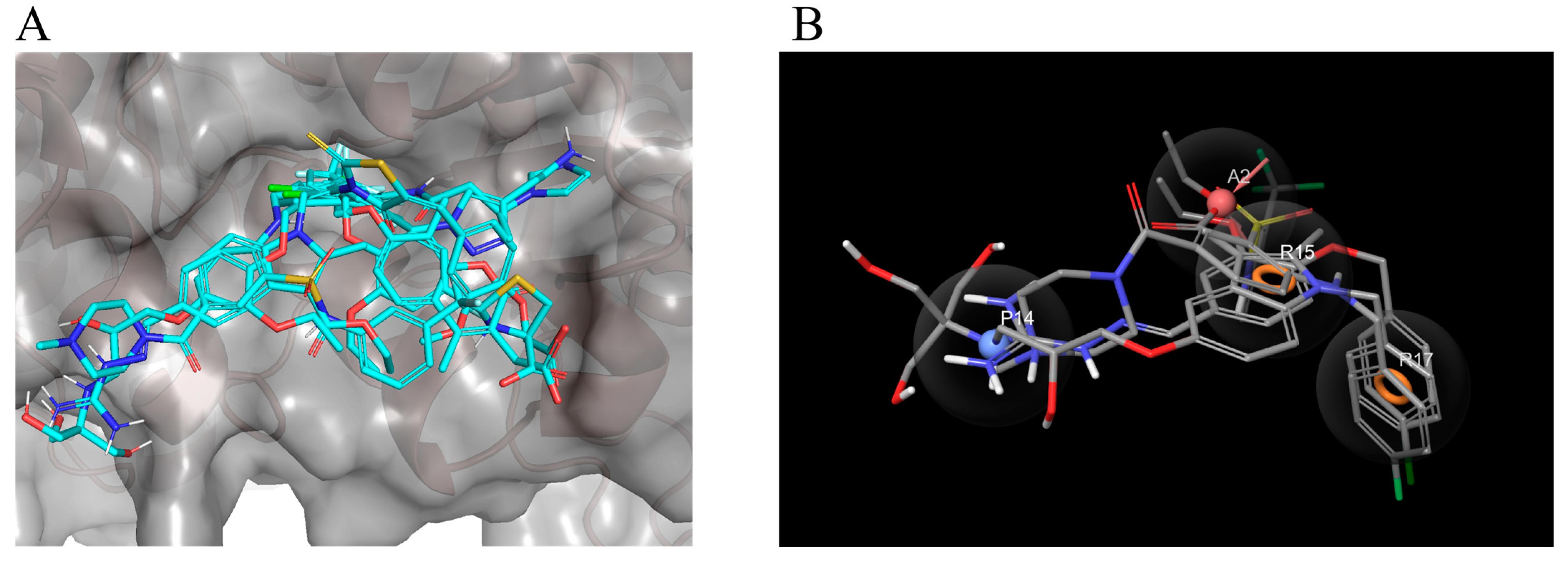
2.1. Analysis of the LsrK-HPr PPI Site
2.2. Virtual Screening Workflow (VSW)
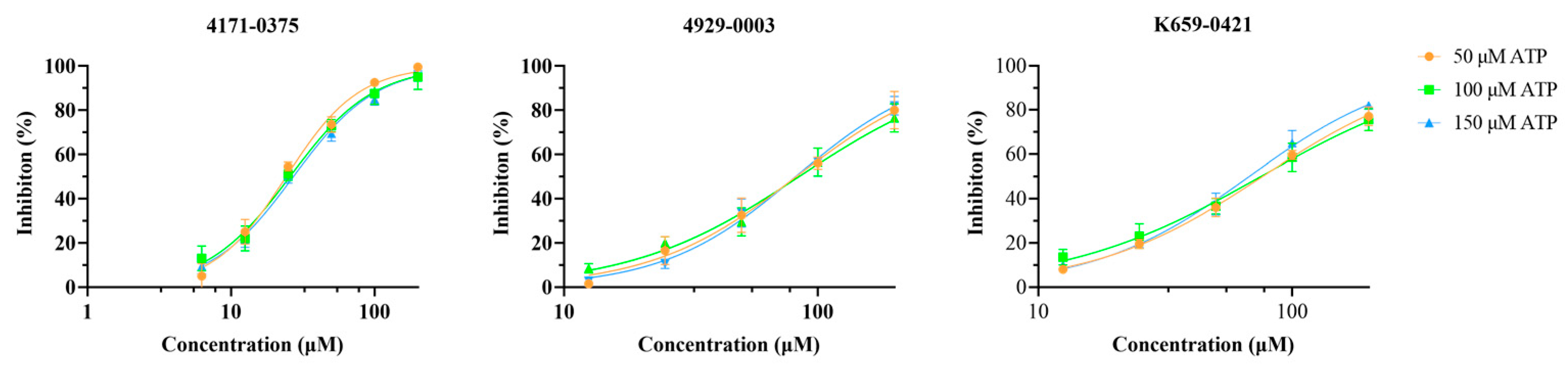
2.3. LsrK Inhibition Assay
2.4. Cell-Based AI-2-Mediated QS Interference Assay
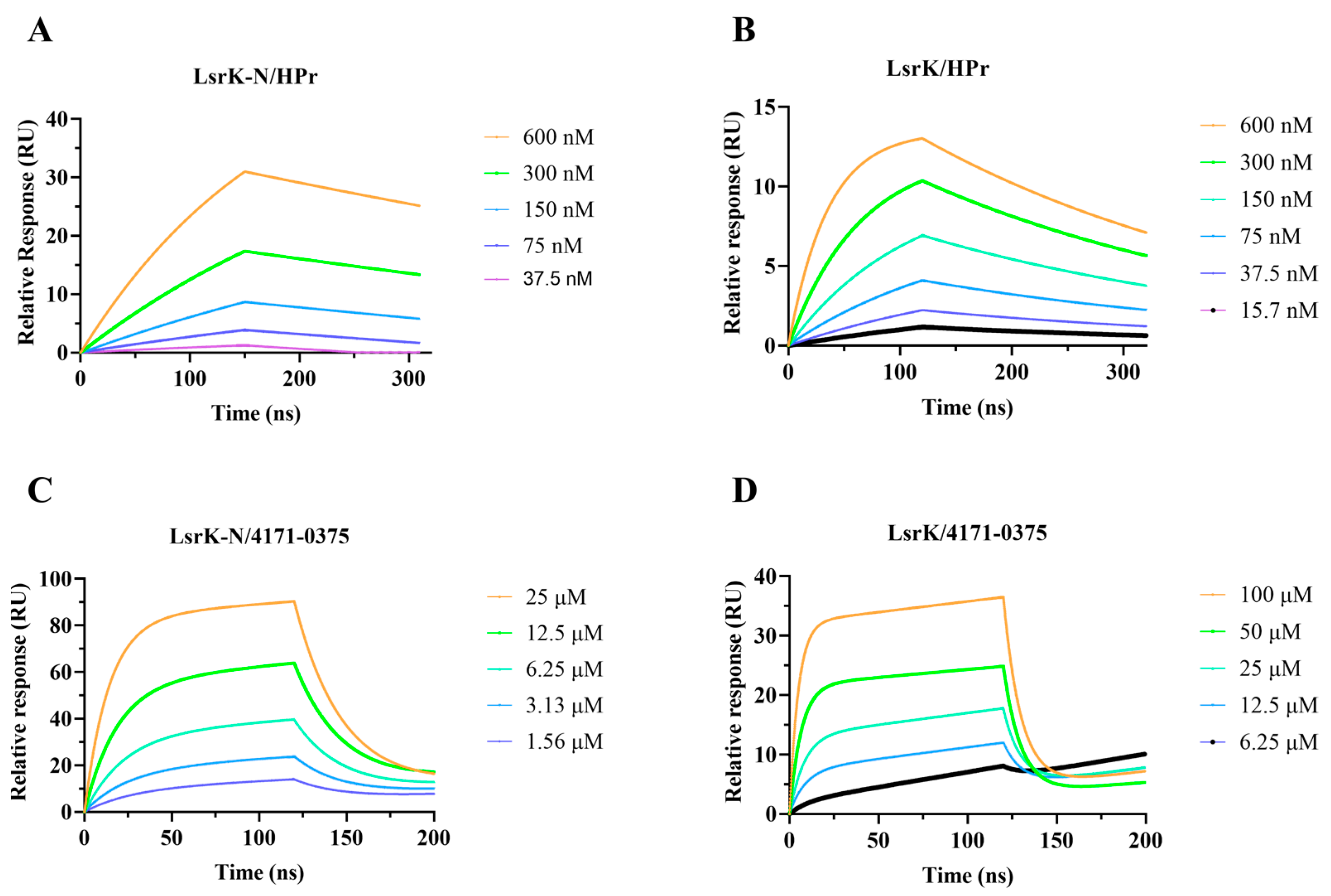
2.5. SPR Assay
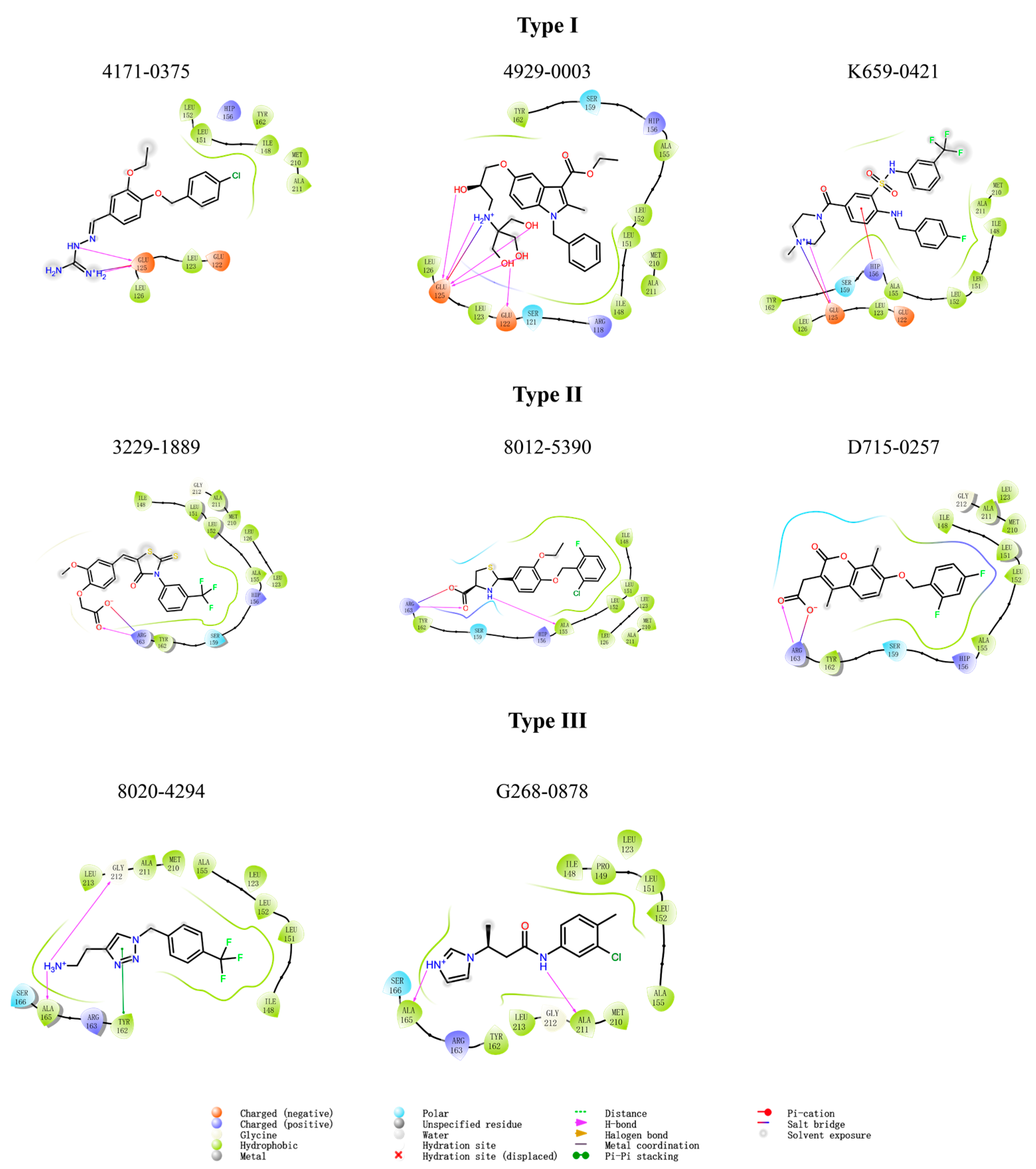
2.6. Exploration of Structure-Activity Relationships (SARs)
3. Materials and Methods
3.1. MD Simulation, Virtual Screening and Pharmacophore Hypothesis Generation
3.1.1. Protein Preparation and Binding Site Detection
3.1.2. Molecular Dynamics (MD) Simulation
3.1.3. Estimation and Decomposition of the Binding Free Energy by gmx_MMPBSA
3.1.4. Virtual Screening
3.1.5. Pharmacophore Hypothesis Generation
3.2. Bioassay Methods
3.2.1. Overexpression and Purification of LsrK, LsrK-N, and HPr
3.2.2. LsrK Inhibition Assay
3.2.3. SPR Assay
3.2.4. Cell-Based AI-2-Mediated QS Interference Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Murray, C.J.L.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Robles Aguilar, G.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global Burden of Bacterial Antimicrobial Resistance in 2019: A Systematic Analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- Bhusal, R.P.; Barr, J.J.; Subedi, D. A Metabolic Perspective into Antimicrobial Tolerance and Resistance. Lancet Microbe 2022, 3, e160–e161. [Google Scholar] [CrossRef] [PubMed]
- Pang, Z.; Raudonis, R.; Glick, B.R.; Lin, T.J.; Cheng, Z. Antibiotic Resistance in Pseudomonas Aeruginosa: Mechanisms and Alternative Therapeutic Strategies. Biotechnol. Adv. 2019, 37, 177–192. [Google Scholar] [CrossRef] [PubMed]
- Rutherford, S.T.; Bassler, B.L. Bacterial Quorum Sensing: Its Role in Virulence and Possibilities for Its Control. Cold Spring Harb. Perspect. Med. 2012, 2, a012427. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Li, M.; Yi, G.; Liao, L.; Cheng, Q.; Zhu, J.; Zhang, B.; Wang, Y.; Chen, Y.; Zeng, M. Screening Strategies for Quorum Sensing Inhibitors in Combating Bacterial Infections. J. Pharm. Anal. 2022, 12, 1–14. [Google Scholar] [CrossRef]
- Lowery, C.A.; Dickerson, T.J.; Janda, K.D. Interspecies and Interkingdom Communication Mediated by Bacterial Quorum Sensing. Chem. Soc. Rev. 2008, 37, 1337–1346. [Google Scholar] [CrossRef]
- Eickhoff, M.J.; Bassler, B.L. SnapShot: Bacterial Quorum Sensing. Cell 2018, 174, 1328-1328.e1. [Google Scholar] [CrossRef]
- Surette, M.G.; Miller, M.B.; Bassler, B.L. Quorum Sensing in Escherichia coli, Salmonella Typhimurium, and Vibrio Harveyi: A New Family of Genes Responsible for Autoinducer Production. Proc. Natl. Acad. Sci. USA 1999, 96, 1639–1644. [Google Scholar] [CrossRef]
- Waters, C.M.; Bassler, B.L. QUORUM SENSING: Cell-to-Cell Communication in Bacteria. Annu. Rev. Cell Dev. Biol. 2005, 21, 319–346. [Google Scholar] [CrossRef]
- Engebrecht, J.; Nealson, K.; Silverman, M. Bacterial Bioluminescence: Isolation and Genetic Analysis of Functions from Vibrio Fischeri. Cell 1983, 32, 773–781. [Google Scholar] [CrossRef]
- Davies, D.G.; Parsek, M.R.; Pearson, J.P.; Iglewski, B.H.; Costerton, J.W.; Greenberg, E.P. The Involvement of Cell-to-Cell Signals in the Development of a Bacterial Biofilm. Science 1998, 280, 295–298. [Google Scholar] [CrossRef]
- Ahmed, N.A.A.M.; Petersen, F.C.; Scheie, A.A. AI-2 Quorum Sensing Affects Antibiotic Susceptibility in Streptococcus Anginosus. J. Antimicrob. Chemother. 2007, 60, 49–53. [Google Scholar] [CrossRef]
- Pollitt, E.J.G.; Diggle, S.P. Defining Motility in the Staphylococci. Cell. Mol. Life Sci. 2017, 74, 2943–2958. [Google Scholar] [CrossRef]
- Val, D.L.; Cronan, J.E.J. In Vivo Evidence that S-Adenosylmethionine and Fatty Acid Synthesis Intermediates Are the Substrates for the LuxI Family of Autoinducer Synthases. J. Bacteriol. 1998, 180, 2644–2651. [Google Scholar] [CrossRef] [PubMed]
- Parsek, M.R.; Val, D.L.; Hanzelka, B.L.; Cronan, J.E.J.; Greenberg, E.P. Acyl Homoserine-Lactone Quorum-Sensing Signal Generation. Proc. Natl. Acad. Sci. USA 1999, 96, 4360–4365. [Google Scholar] [CrossRef] [PubMed]
- Latifi, A.; Foglino, M.; Tanaka, K.; Williams, P.; Lazdunski, A. A Hierarchical Quorum-Sensing Cascade in Pseudomonas Aeruginosa Links the Transcriptional Activators LasR and RhIR (VsmR) to Expression of the Stationary-Phase Sigma Factor RpoS. Mol. Microbiol. 1996, 21, 1137–1146. [Google Scholar] [CrossRef] [PubMed]
- Håvarstein, L.S.; Coomaraswamy, G.; Morrison, D.A. An Unmodified Heptadecapeptide Pheromone Induces Competence for Genetic Transformation in Streptococcus Pneumoniae. Proc. Natl. Acad. Sci. USA 1995, 92, 11140–11144. [Google Scholar] [CrossRef]
- Ji, G.; Beavis, R.C.; Novick, R.P. Cell Density Control of Staphylococcal Virulence Mediated by an Octapeptide Pheromone. Proc. Natl. Acad. Sci. USA 1995, 92, 12055–12059. [Google Scholar] [CrossRef]
- Solomon, J.M.; Lazazzera, B.A.; Grossman, A.D. Purification and Characterization of an Extracellular Peptide Factor that Affects Two Different Developmental Pathways in Bacillus Subtilis. Genes Dev. 1996, 10, 2014–2024. [Google Scholar] [CrossRef]
- Papenfort, K.; Vogel, J. Regulatory RNA in Bacterial Pathogens. Cell Host Microbe 2010, 8, 116–127. [Google Scholar] [CrossRef]
- Ledgham, F.; Ventre, I.; Soscia, C.; Foglino, M.; Sturgis, J.N.; Lazdunski, A. Interactions of the Quorum Sensing Regulator QscR: Interaction with Itself and the Other Regulators of Pseudomonas Aeruginosa LasR and RhlR. Mol. Microbiol. 2003, 48, 199–210. [Google Scholar] [CrossRef]
- Federle, M.J.; Bassler, B.L. Interspecies Communication in Bacteria. J. Clin. Investig. 2003, 112, 1291–1299. [Google Scholar] [CrossRef] [PubMed]
- Schauder, S.; Shokat, K.; Surette, M.G.; Bassler, B.L. The LuxS Family of Bacterial Autoinducers: Biosynthesis of a Novel Quorum-Sensing Signal Molecule. Mol. Microbiol. 2001, 41, 463–476. [Google Scholar] [CrossRef] [PubMed]
- Khera, R.; Mehdipour, A.R.; Bolla, J.R.; Kahnt, J.; Welsch, S.; Ermler, U.; Muenke, C.; Robinson, C.V.; Hummer, G.; Xie, H.; et al. Cryo-EM Structures of Pentameric Autoinducer-2 Exporter from Escherichia coli Reveal Its Transport Mechanism. EMBO J. 2022, 41, e109990. [Google Scholar] [CrossRef] [PubMed]
- Globisch, D.; Lowery, C.A.; McCague, K.C.; Janda, K.D. Uncharacterized 4,5-Dihydroxy-2,3-Pentanedione (DPD) Molecules Revealed through NMR Spectroscopy: Implications for a Greater Signaling Diversity in Bacterial Species. Angew. Chem. Int. Ed. Engl. 2012, 51, 4204–4208. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Schauder, S.; Potier, N.; Van Dorsselaer, A.; Pelczer, I.; Bassler, B.L.; Hughson, F.M. Structural Identification of a Bacterial Quorum-Sensing Signal Containing Boron. Nature 2002, 415, 545–549. [Google Scholar] [CrossRef] [PubMed]
- Miller, S.T.; Xavier, K.B.; Campagna, S.R.; Taga, M.E.; Semmelhack, M.F.; Bassler, B.L.; Hughson, F.M. Salmonella Typhimurium Recognizes a Chemically Distinct Form of the Bacterial Quorum-Sensing Signal AI-2. Mol. Cell 2004, 15, 677–687. [Google Scholar] [CrossRef]
- Shao, H.; James, D.; Lamont, R.J.; Demuth, D.R. Differential Interaction of Aggregatibacter (Actinobacillus) Actinomycetemcomitans LsrB and RbsB Proteins with Autoinducer 2. J. Bacteriol. 2007, 189, 5559–5565. [Google Scholar] [CrossRef]
- Taga, M.E.; Semmelhack, J.L.; Bassler, B.L. The LuxS-Dependent Autoinducer AI-2 Controls the Expression of an ABC Transporter that Functions in AI-2 Uptake in Salmonella Typhimurium. Mol. Microbiol. 2001, 42, 777–793. [Google Scholar] [CrossRef]
- Xavier, K.B.; Miller, S.T.; Lu, W.; Kim, J.H.; Rabinowitz, J.; Pelczer, I.; Semmelhack, M.F.; Bassler, B.L. Phosphorylation and Processing of the Quorum-Sensing Molecule Autoinducer-2 in Enteric Bacteria. ACS Chem. Biol. 2007, 2, 128–136. [Google Scholar] [CrossRef]
- Taga, M.E.; Miller, S.T.; Bassler, B.L. Lsr-Mediated Transport and Processing of AI-2 in Salmonella Typhimurium. Mol. Microbiol. 2003, 50, 1411–1427. [Google Scholar] [CrossRef] [PubMed]
- Xavier, K.B.; Bassler, B.L. Regulation of Uptake and Processing of the Quorum-Sensing Autoinducer AI-2 in Escherichia coli. J. Bacteriol. 2005, 187, 238–248. [Google Scholar] [CrossRef] [PubMed]
- Ha, J.-H.; Hauk, P.; Cho, K.; Eo, Y.; Ma, X.; Stephens, K.; Cha, S.; Jeong, M.; Suh, J.-Y.; Sintim, H.O.; et al. Evidence of Link between Quorum Sensing and Sugar Metabolism in Escherichia coli Revealed via Cocrystal Structures of LsrK and HPr. Sci. Adv. 2018, 4, eaar7063. [Google Scholar] [CrossRef]
- Gatta, V.; Tomasic, T.; Ilas, J.; Zidar, N.; Peterlin Masic, L.; Barancokova, M.; Frlan, R.; Anderluh, M.; Kikelj, D.; Tammela, P. A New Cell-Based AI-2-Mediated Quorum Sensing Interference Assay in Screening of LsrK-Targeted Inhibitors. Chembiochem 2020, 21, 1918–1922. [Google Scholar] [CrossRef] [PubMed]
- Medarametla, P.; Gatta, V.; Kajander, T.; Laitinen, T.; Tammela, P.; Poso, A. Structure-Based Virtual Screening of LsrK Kinase Inhibitors to Target Quorum Sensing. ChemMedChem 2018, 13, 2400–2407. [Google Scholar] [CrossRef] [PubMed]
- Stotani, S.; Gatta, V.; Medarametla, P.; Padmanaban, M.; Karawajczyk, A.; Giordanetto, F.; Tammela, P.; Laitinen, T.; Poso, A.; Tzalis, D.; et al. DPD-Inspired Discovery of Novel LsrK Kinase Inhibitors: An Opportunity to Fight Antimicrobial Resistance. J. Med. Chem. 2019, 62, 2720–2737. [Google Scholar] [CrossRef]
- Gatta, V.; Ilina, P.; Porter, A.; McElroy, S.; Tammela, P. Targeting Quorum Sensing: High-Throughput Screening to Identify Novel LsrK Inhibitors. Int. J. Mol. Sci. 2019, 20, 3112. [Google Scholar] [CrossRef]
- Scott, D.E.; Bayly, A.R.; Abell, C.; Skidmore, J. Small Molecules, Big Targets: Drug Discovery Faces the Protein-Protein Interaction Challenge. Nat. Rev. Drug Discov. 2016, 15, 533–550. [Google Scholar] [CrossRef]
- Berlinck, R.G.S.; Burtoloso, A.C.B.; Trindade-Silva, A.E.; Romminger, S.; Morais, R.P.; Bandeira, K.; Mizuno, C.M. The Chemistry and Biology of Organic Guanidine Derivatives. Nat. Prod. Rep. 2010, 27, 1871–1907. [Google Scholar] [CrossRef]
- Páll, S.; Zhmurov, A.; Bauer, P.; Abraham, M.; Lundborg, M.; Gray, A.; Hess, B.; Lindahl, E. Heterogeneous Parallelization and Acceleration of Molecular Dynamics Simulations in GROMACS. J. Chem. Phys. 2020, 153, 134110. [Google Scholar] [CrossRef]
- Lindorff-Larsen, K.; Piana, S.; Palmo, K.; Maragakis, P.; Klepeis, J.L.; Dror, R.O.; Shaw, D.E. Improved Side-Chain Torsion Potentials for the Amber Ff99SB Protein Force Field. Proteins 2010, 78, 1950–1958. [Google Scholar] [CrossRef] [PubMed]
- Berendsen, H.J.C.; Postma, J.P.M.; van Gunsteren, W.F.; DiNola, A.; Haak, J.R. Molecular Dynamics with Coupling to an External Bath. J. Chem. Phys. 1984, 81, 3684–3690. [Google Scholar] [CrossRef]
- Parrinello, M.; Rahman, A. Polymorphic Transitions in Single Crystals: A New Molecular Dynamics m Ethod. J. Appl. Phys. 1981, 52, 7182–7190. [Google Scholar] [CrossRef]
- Valdés-Tresanco, M.S.; Valdés-Tresanco, M.E.; Valiente, P.A.; Moreno, E. Gmx_MMPBSA: A New Tool to Perform End-State Free Energy Calculations with GROMACS. J. Chem. Theory Comput. 2021, 17, 6281–6291. [Google Scholar] [CrossRef]
- Rodionova, I.A.; Zhang, Z.; Mehla, J.; Goodacre, N.; Babu, M.; Emili, A.; Uetz, P.; Saier, M.H.J. The Phosphocarrier Protein HPr of the Bacterial Phosphotransferase System Globally Regulates Energy Metabolism by Directly Interacting with Multiple Enzymes in Escherichia coli. J. Biol. Chem. 2017, 292, 14250–14257. [Google Scholar] [CrossRef]
- Park, Y.-H.; Lee, C.-R.; Choe, M.; Seok, Y.-J. HPr Antagonizes the Anti-Σ70 Activity of Rsd in Escherichia coli. Proc. Natl. Acad. Sci. USA 2013, 110, 21142–21147. [Google Scholar] [CrossRef] [PubMed]
- Deutscher, J.; Aké, F.M.D.; Derkaoui, M.; Zébré, A.C.; Cao, T.N.; Bouraoui, H.; Kentache, T.; Mokhtari, A.; Milohanic, E.; Joyet, P. The Bacterial Phosphoenolpyruvate:Carbohydrate Phosphotransferase System: Regulation by Protein Phosphorylation and Phosphorylation-Dependent Protein-Protein Interactions. Microbiol. Mol. Biol. Rev. 2014, 78, 231–256. [Google Scholar] [CrossRef]
- Choe, M.; Park, Y.-H.; Lee, C.-R.; Kim, Y.-R.; Seok, Y.-J. The General PTS Component HPr Determines the Preference for Glucose over Mannitol. Sci. Rep. 2017, 7, 43431. [Google Scholar] [CrossRef]
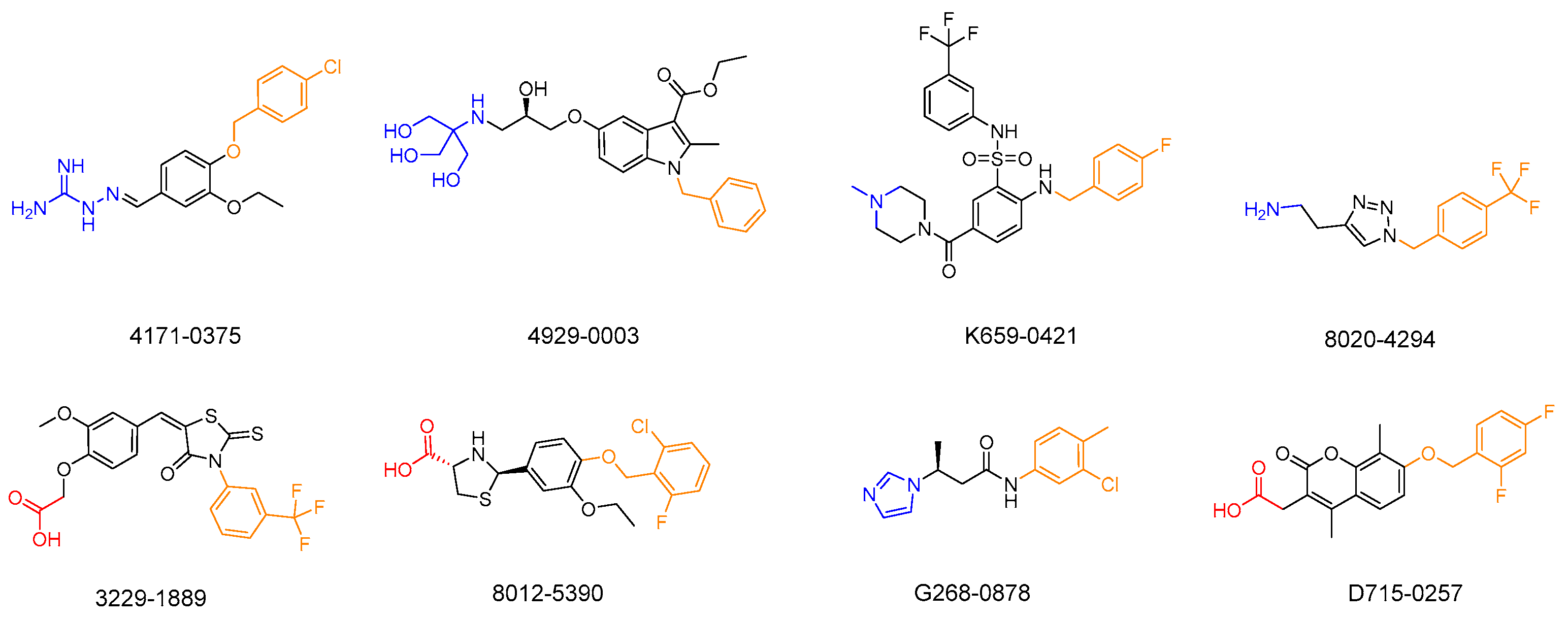
| Compound ID | LsrK Inhibition (%) | LsrK (IC50) A | WHQ02 (IC50) B | WHQ01 (IC50) C |
|---|---|---|---|---|
| 4171-0375 | 98 ± 7 | 26.13 ± 2.94 | 30.13 ± 1.62 * | 42.93 ± 1.63 * |
| 4929-0003 | 78 ± 12 | 80.75 ± 4.30 | 35.40 ± 2.29 * | 20.89 ± 1.24 * |
| K659-0421 | 75 ± 8 | 73.69 ± 5.33 | 16.98 ± 0.37 * | 7.97 ± 0.38 * |
| 8020-4294 | 65 ± 9 | 148.40 ± 6.92 | 67.25 ± 4.32 | 24.86 ± 1.10 |
| 3229-1889 | 63 ± 4 | 121.31 ± 5.37 | 14.99 ± 1.24 | 24.72 ± 1.42 |
| 8012-5390 | 60 ± 4 | 147.13 ± 5.58 | 17.68 ± 0.98 | 23.28 ± 1.05 |
| G268-0878 | 58 ± 5 | 128.73 ± 6.14 | 42.30 ± 1.42 | 30.55 ± 1.26 |
| D715-0257 | 57 ± 7 | 152.57 ± 5.43 | 22.34 ± 2.15 | 21.10 ± 1.18 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, Y.; Zeng, C.; Wen, H.; Shi, Q.; Zhao, X.; Meng, Q.; Li, X.; Xiao, J. Discovery of AI-2 Quorum Sensing Inhibitors Targeting the LsrK/HPr Protein–Protein Interaction Site by Molecular Dynamics Simulation, Virtual Screening, and Bioassay Evaluation. Pharmaceuticals 2023, 16, 737. https://doi.org/10.3390/ph16050737
Xu Y, Zeng C, Wen H, Shi Q, Zhao X, Meng Q, Li X, Xiao J. Discovery of AI-2 Quorum Sensing Inhibitors Targeting the LsrK/HPr Protein–Protein Interaction Site by Molecular Dynamics Simulation, Virtual Screening, and Bioassay Evaluation. Pharmaceuticals. 2023; 16(5):737. https://doi.org/10.3390/ph16050737
Chicago/Turabian StyleXu, Yijie, Chunlan Zeng, Huiqi Wen, Qianqian Shi, Xu Zhao, Qingbin Meng, Xingzhou Li, and Junhai Xiao. 2023. "Discovery of AI-2 Quorum Sensing Inhibitors Targeting the LsrK/HPr Protein–Protein Interaction Site by Molecular Dynamics Simulation, Virtual Screening, and Bioassay Evaluation" Pharmaceuticals 16, no. 5: 737. https://doi.org/10.3390/ph16050737
APA StyleXu, Y., Zeng, C., Wen, H., Shi, Q., Zhao, X., Meng, Q., Li, X., & Xiao, J. (2023). Discovery of AI-2 Quorum Sensing Inhibitors Targeting the LsrK/HPr Protein–Protein Interaction Site by Molecular Dynamics Simulation, Virtual Screening, and Bioassay Evaluation. Pharmaceuticals, 16(5), 737. https://doi.org/10.3390/ph16050737





