Guanidines: Synthesis of Novel Histamine H3R Antagonists with Additional Breast Anticancer Activity and Cholinesterases Inhibitory Effect
Abstract
1. Introduction
2. Results and Discussion
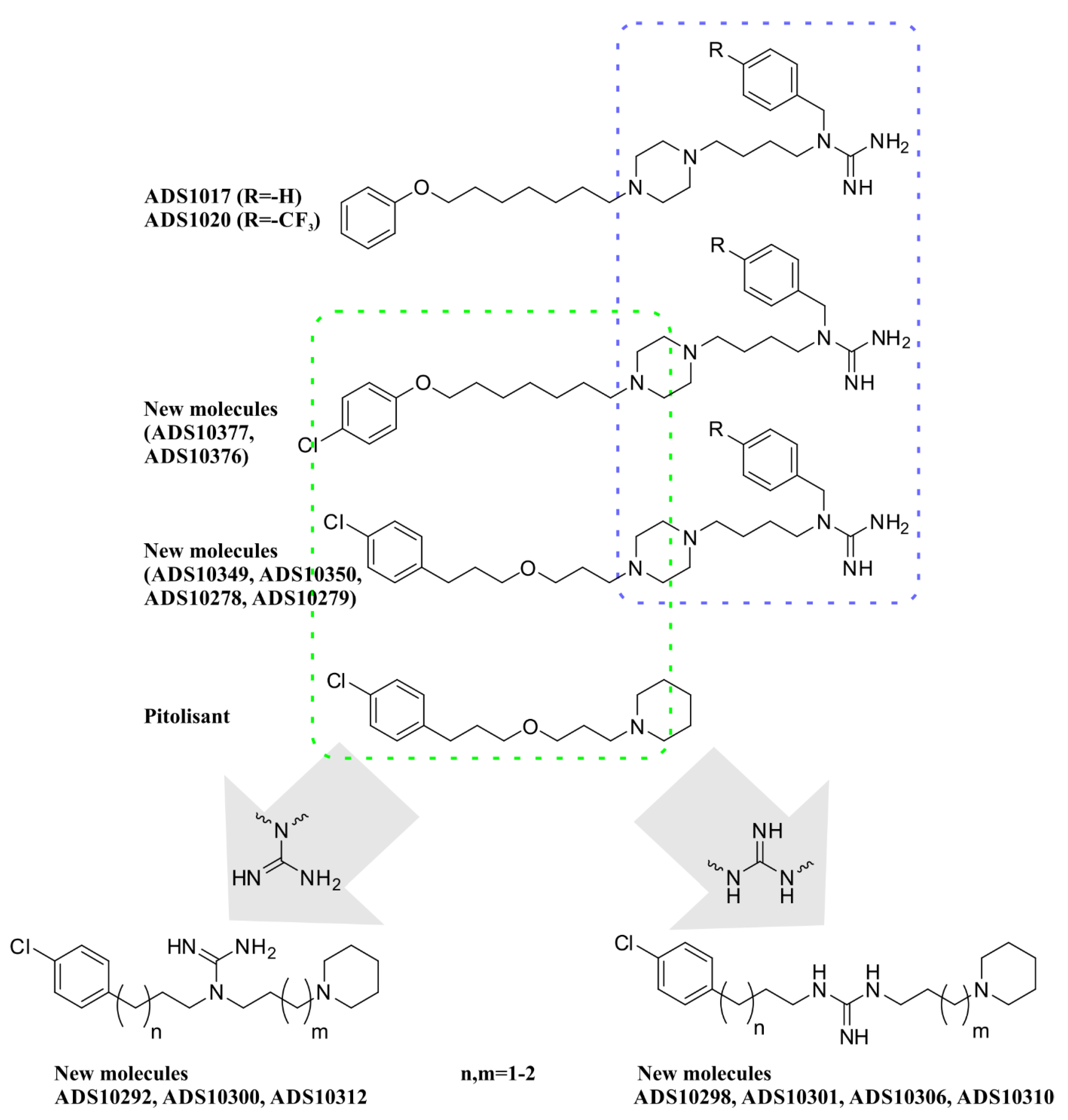
2.1. Design
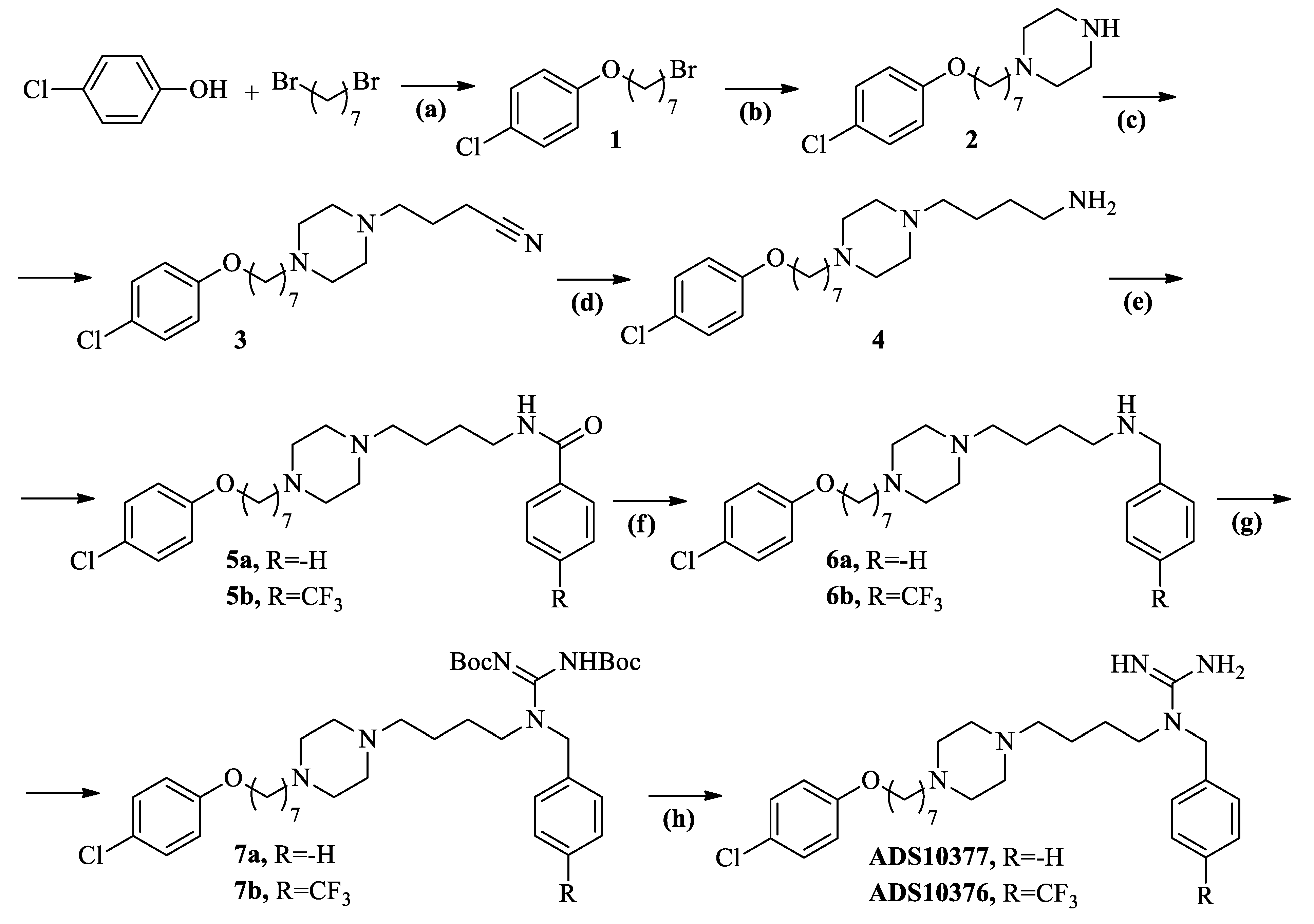
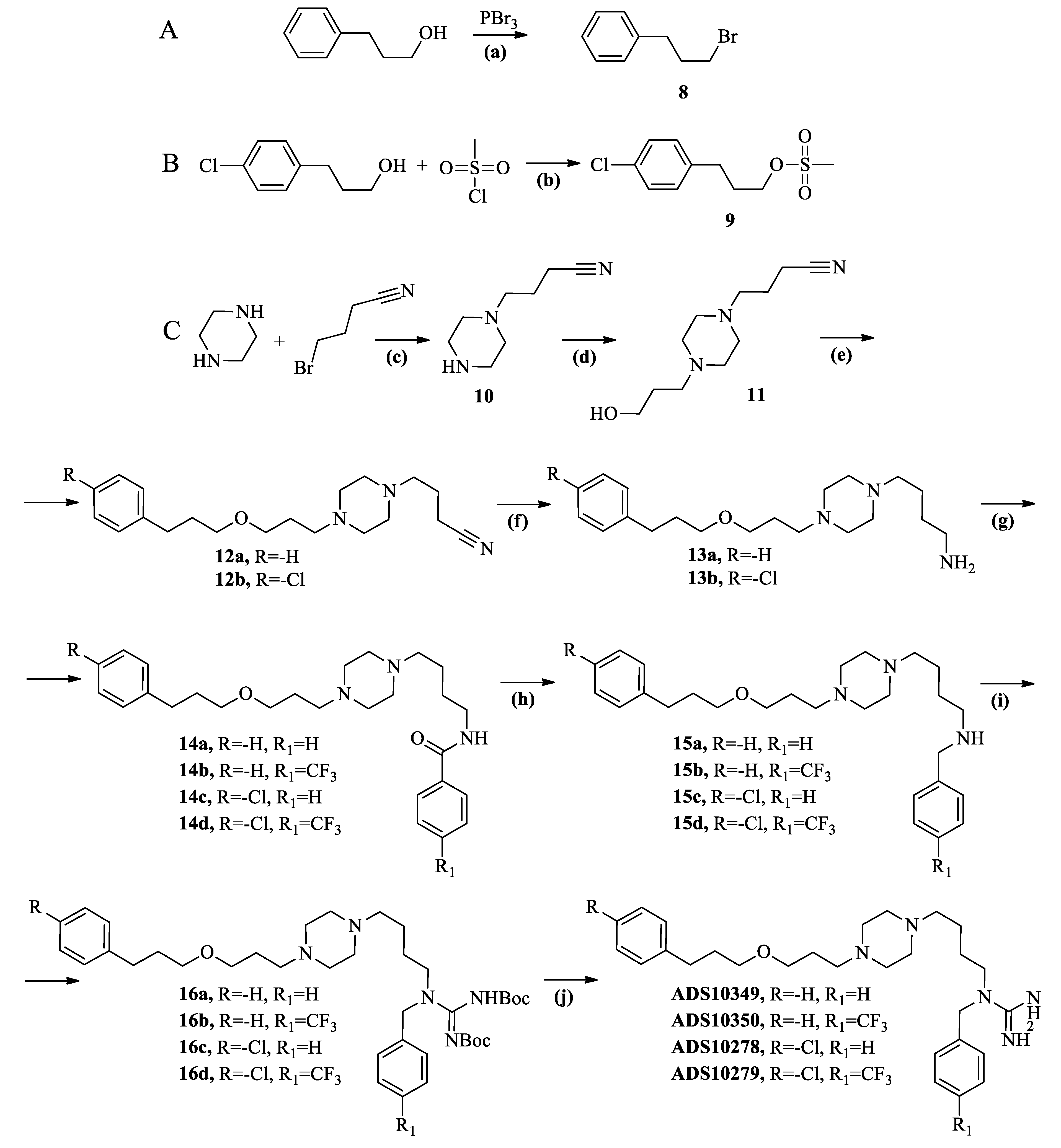
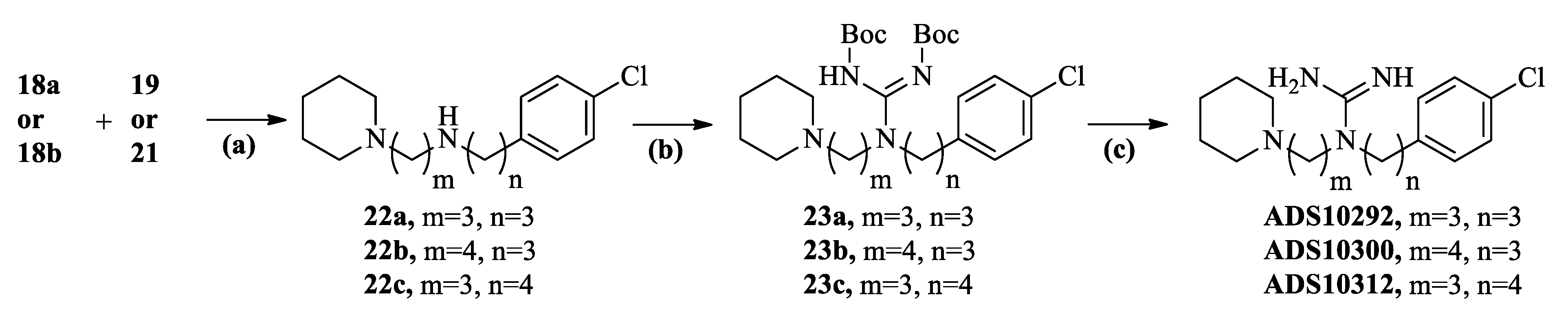
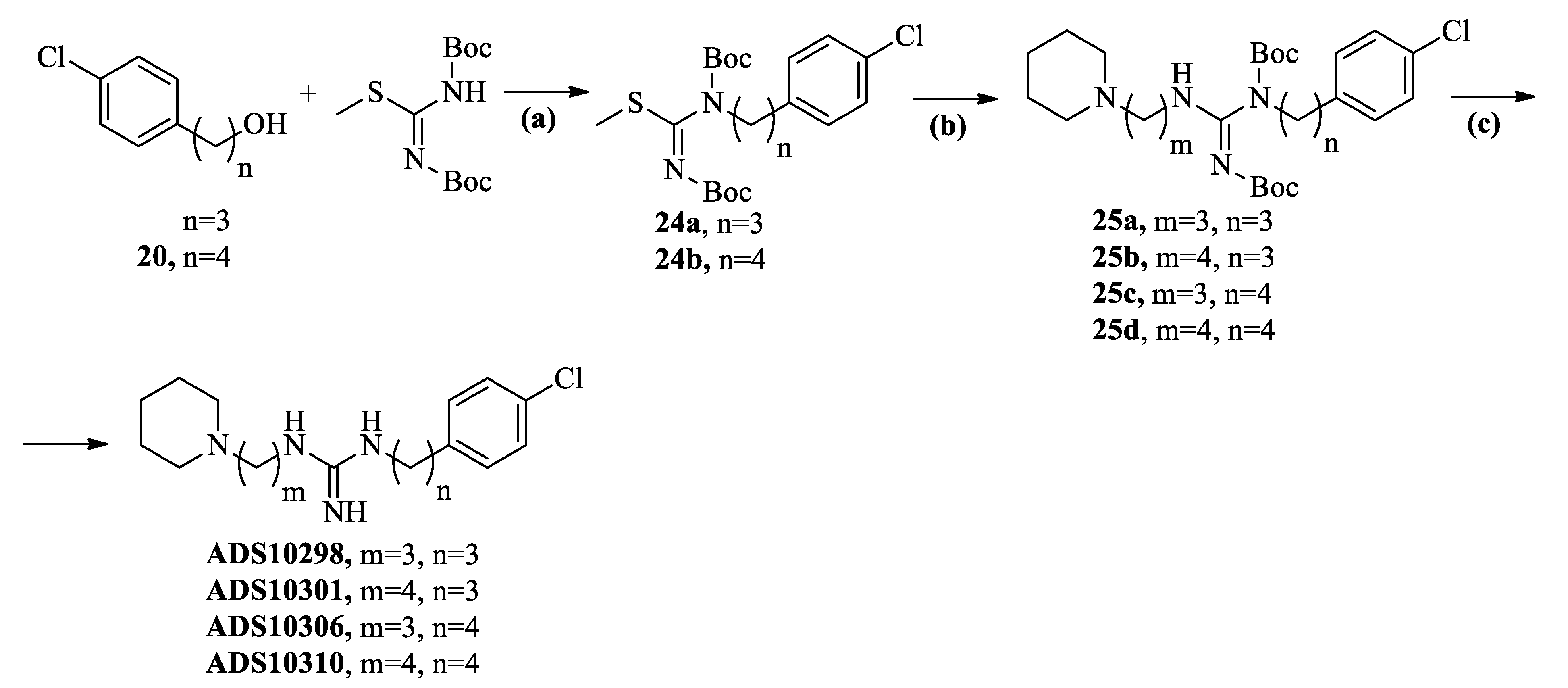
2.2. Chemistry
2.3. Pharmacology
2.3.1. Ex Vivo Screening of Histamine gpH3R/gpH1R Antagonists/Inverse Agonists on Guinea Pig Ileum and hH3R Radioligand Displacement Assay
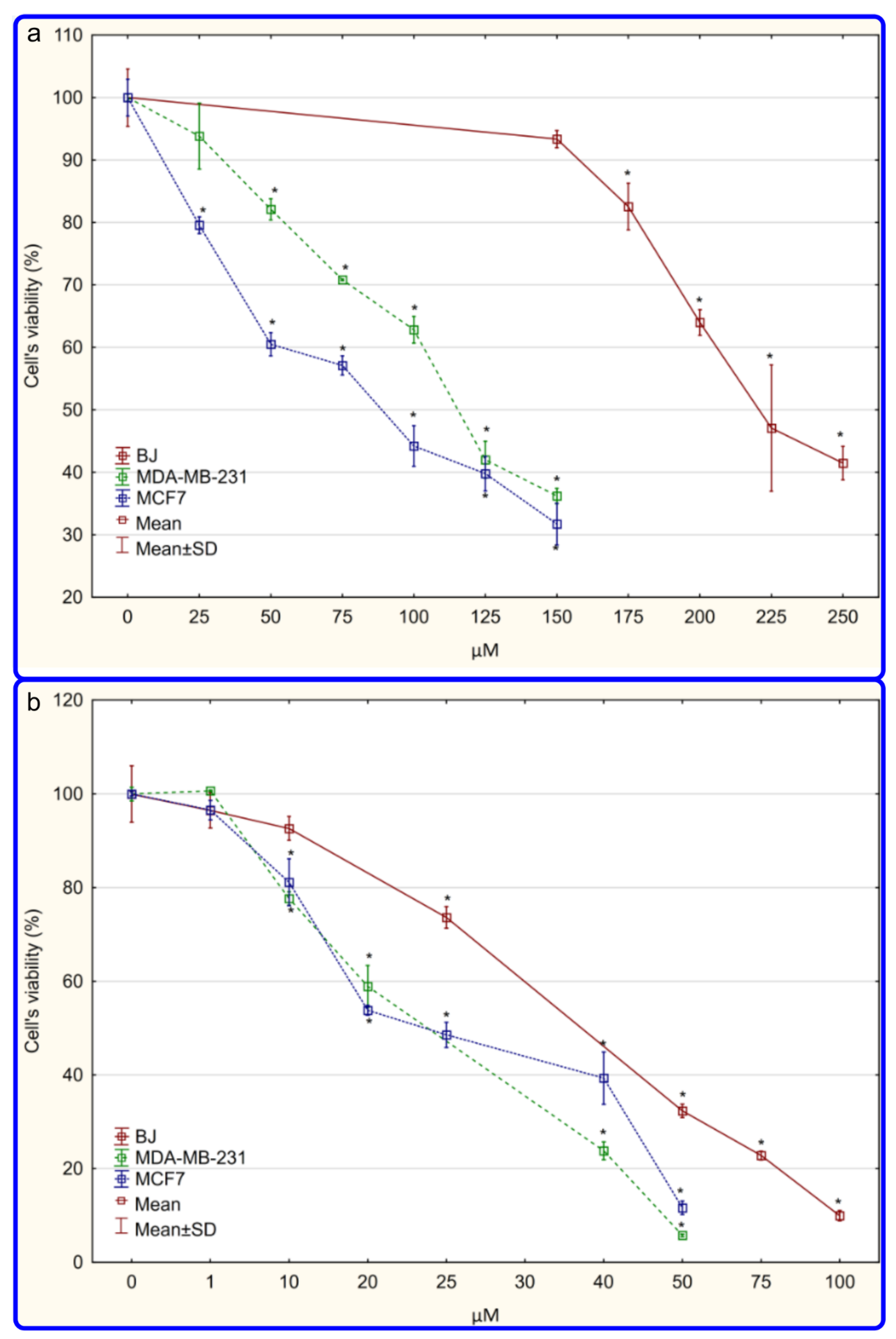
2.3.2. Cytotoxicity Analyses
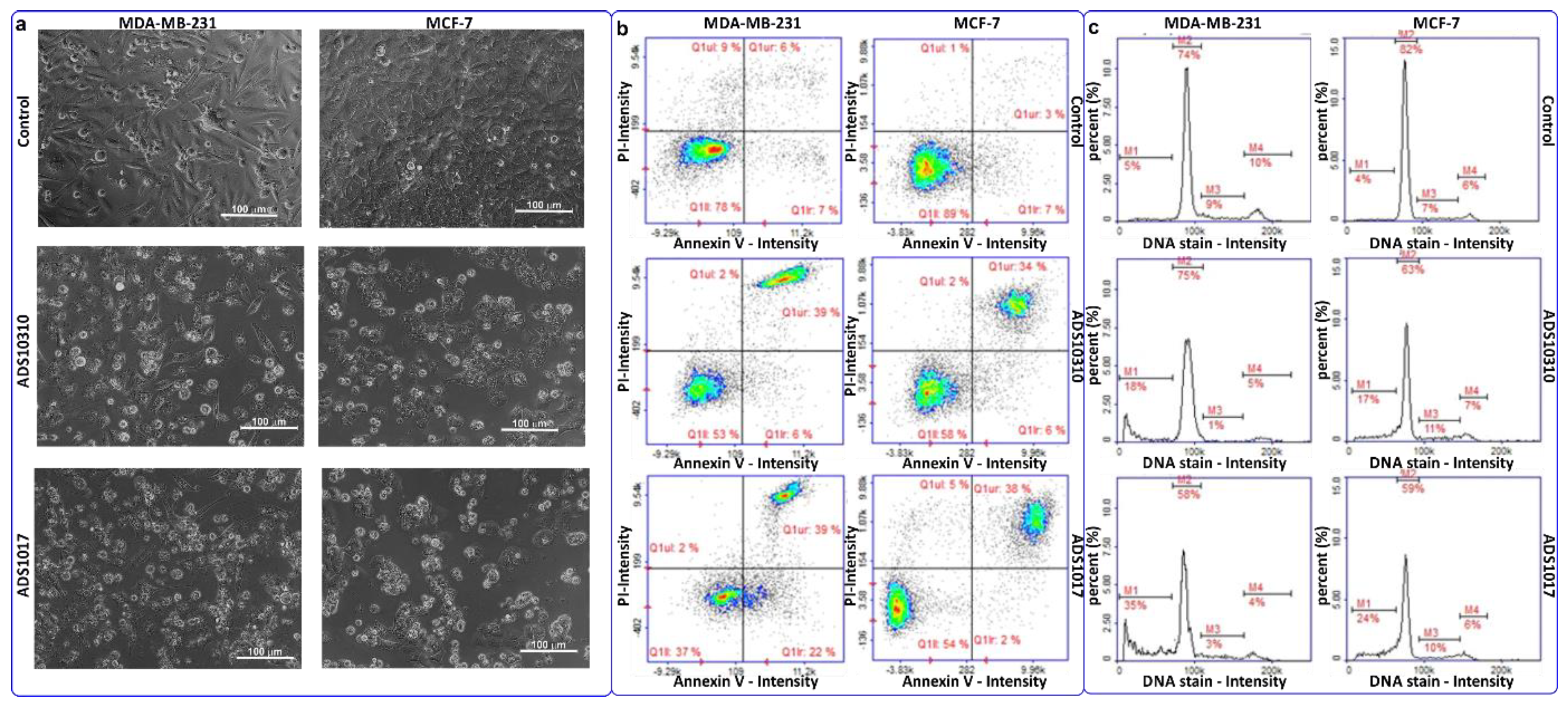
2.3.3. Apoptosis Detection
2.3.4. Cell Cycle Analysis
2.3.5. Inhibition of Electric Eel AChE and Equine Serum BuChE
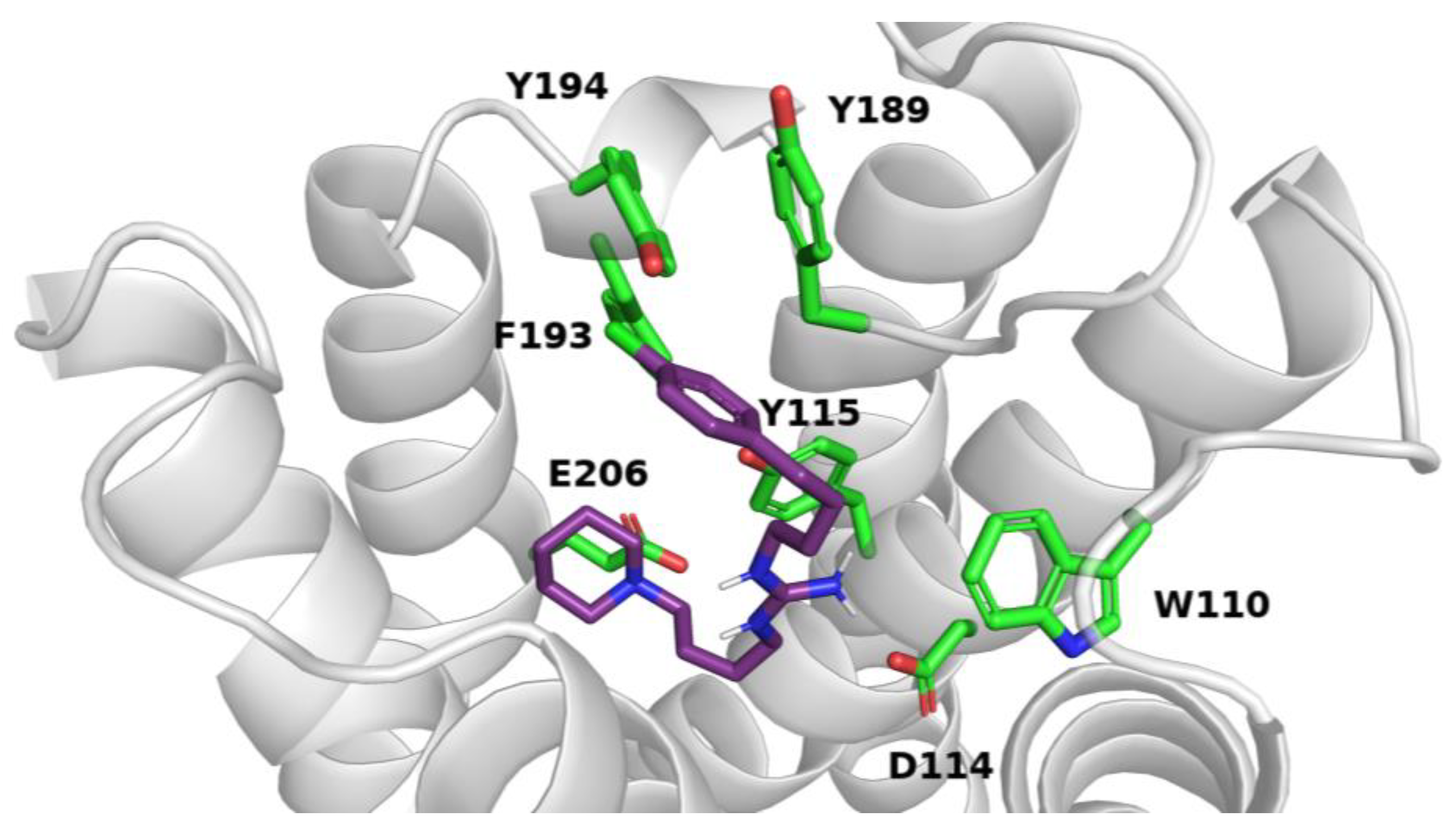
2.4. In Silico Studies
2.5. In Vitro Metabolic Stability
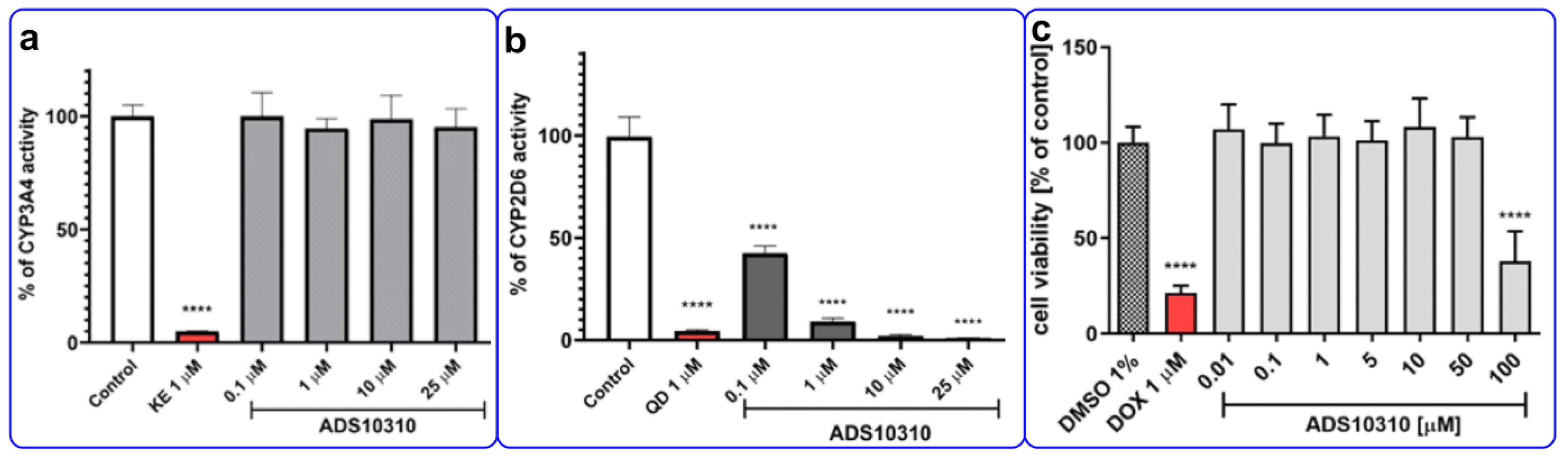
2.6. The Influence on CYP3A4 and CYP2D6 Activity
2.7. Hepatotoxicity Assay
3. Materials and Methods
3.1. Chemistry
3.2. Biological Activity
3.2.1. Cell Viability
Cell Culturing
Cytotoxicity Evaluation—MTT Test
Cell Cycle Analysis
Cell Apoptosis Assay
Statistical Analysis
3.3. In Silico Studies
3.4. In Vitro Metabolic Stability
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fang, J.; Li, Y.; Liu, R.; Pang, X.; Li, C.; Yang, R.; He, Y.; Lian, W.; Liu, A.-L.; Du, G.-H. Discovery of Multitarget-Directed Ligands against Alzheimer’s Disease through Systematic Prediction of Chemical–Protein Interactions. J. Chem. Inf. Model. 2015, 55, 149–164. [Google Scholar] [CrossRef] [PubMed]
- Berlin, M.; Boyce, C.W.; De Lera Ruiz, M. Histamine H3 Receptor as a Drug Discovery Target. J. Med. Chem. 2011, 54, 26–53. [Google Scholar] [CrossRef] [PubMed]
- Benarroch, E.E. Histamine in the CNS. Neurology 2010, 75, 1472–1479. [Google Scholar] [CrossRef] [PubMed]
- Bartholeyns, J.; Fozard, J.R. Role of histamine in tumor development. Trends Pharmacol. Sci. 1985, 6, 123–125. [Google Scholar] [CrossRef]
- Rivera, E.S.; Cricco, G.P.; Engel, N.I.; Fitzsimons, C.P.; Martín, G.A.; Bergoc, R.M. Histamine as an autocrine growth factor: An unusual role for a widespread mediator. Semin. Cancer Biol. 2000, 10, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Bartholeyns, J.; Bouclier, M. Involvement of histamine in growth of mouse and rat tumors: Antitumoral properties of monofluoromethylhistidine, an enzyme-activated irreversible inhibitor of histidine decarboxylase. Cancer Res. 1984, 44, 639–645. [Google Scholar]
- Malaviya, R.; Uckun, F.M. Histamine as an Autocrine Regulator of Leukemic Cell Proliferation. Leuk. Lymphoma 2000, 36, 367–373. [Google Scholar] [CrossRef]
- Molnar, E.L.; Cricco, G.; Martin, G.; Darvas, Z.; Hegyesi, H.; Fitzsimons, C.; Bergoc, R.; Falus, A.; Rivera, E. Histamine as a Potential Autocrine Regulator of Melanoma. Inflamm. Res. 2001, 50, 102–103. [Google Scholar] [CrossRef]
- Tomita, K.; Okabe, S. Exogenous Histamine Stimulates Colorectal Cancer Implant Growth via Immunosuppression in Mice. J. Pharmacol. Sci. 2005, 97, 116–123. [Google Scholar] [CrossRef]
- Tilly, B.C.; Tertoolen, L.G.; Remorie, R.; Ladoux, A.; Verlaan, I.; De Laat, S.W.; Moolenaar, W.H. Histamine as a growth factor and chemoattractant for human carcinoma and melanoma cells: Action through Ca2(+)-mobilizing H1 receptors. J. Cell Biol. 1990, 110, 1211–1215. [Google Scholar] [CrossRef]
- Szczepańska, K.; Kincses, A.; Vincze, K.; Szymańska, E.; Latacz, G.; Kuder, K.J.; Stark, H.; Spengler, G.; Handzlik, J.; Kieć-Kononowicz, K. N-Substituted piperazine derivatives as potential multitarget agents acting on histamine H3 receptor and cancer resistance proteins. Bioorg. Med. Chem. Lett. 2020, 30, 127522. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.-J.; Zhao, T.-Z.; Cai, W.-K.; Yang, Y.-X.; Sun, C.; Zhang, Z.; Xu, Y.-Q.; Chang, T.; Li, Z.-Y. Inhibition of histamine receptor 3 suppresses glioblastoma tumor growth, invasion, and epithelial-to-mesenchymal transition. Oncotarget 2015, 6, 17107–17120. [Google Scholar] [CrossRef] [PubMed]
- Davenas, E.; Rouleau, A.; Morisset, S.; Arrang, J.M. Autoregulation of McA-RH7777 Hepatoma Cell Proliferation by Histamine H3 Receptors. J. Pharmacol. Exp. Ther. 2008, 326, 406–413. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, S.; Sakaguchi, M.; Yoneyama, H.; Usami, Y.; Harusawa, S. Histamine H3 receptor antagonist OUP-186 attenuates the proliferation of cultured human breast cancer cell lines. Biochem. Biophys. Res. Commun. 2016, 480, 479–485. [Google Scholar] [CrossRef]
- Medina, V.; Croci, M.; Crescenti, E.; Mohamad, N.; Sanchez-Jiménez, F.; Massari, N.; Nuñez, M.; Cricco, G.; Martin, G.; Bergoc, R.; et al. The role of histamine in human mammary carcinogenesis: H3 and H4 receptors as potential therapeutic targets for breast cancer treatment. Cancer Biol. Ther. 2008, 7, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Genre, F.; Valli, E.; Medina, V.; Gutiérrez, A.; Sambuco, L.; Rivera, E.; Cricco, G.; Martín, G. Effect of histamine on the expression of metalloproteinases and cell adhesion in breast cancer cell lines. Inflamm. Res. 2009, 58, 55–56. [Google Scholar] [CrossRef] [PubMed]
- Blaya, B.; Nicolau-Galmes, F.; Jangi, S.M.; Ortega-Martinez, I.; Alonso-Tejerina, E.; Burgos-Bretones, J.; Perezyarza, G.; Asumendi, A.; Boyano, M.D. Histamine and Histamine Receptor Antagonists in Cancer Biology. Inflamm. Allergy-Drug Targets 2010, 9, 146–157. [Google Scholar] [CrossRef]
- Medina, V.; Cricco, G.; Nuñez, M.; Martín, G.; Mohamad, N.; Correa-Fiz, F.; Sanchez-Jiménez, F.; Bergoc, R.; Rivera, E.S. Histamine-mediated signaling processes in human malignant mammary cells. Cancer Biol. Ther. 2006, 5, 1462–1471. [Google Scholar] [CrossRef]
- Prast, H.; Tran, M.H.; Lamberti, C.; Fischer, H.; Kraus, M.; Grass, K.; Philippu, A. Histaminergic neurons modulate acetylcholine release in the ventral striatum: Role of H 1 and H 2 histamine receptors. Naunyn-Schmiedeberg’s Arch. Pharmacol. 1999, 360, 552–557. [Google Scholar] [CrossRef]
- Bajda, M.; Kuder, K.J.; Łażewska, D.; Kieć-Kononowicz, K.; Więckowska, A.; Ignasik, M.; Guzior, N.; Jończyk, J.; Malawska, B. Dual-Acting Diether Derivatives of Piperidine and Homopiperidine with Histamine H3 Receptor Antagonistic and Anticholinesterase Activity. Arch. Der Pharm. 2012, 345, 591–597. [Google Scholar] [CrossRef]
- Darras, F.H.; Pockes, S.; Huang, G.; Wehle, S.; Strasser, A.; Wittmann, H.J.; Nimczick, M.; Sotriffer, C.A.; Decker, M. Synthesis, Biological Evaluation, and Computational Studies of Tri- and Tetracyclic Nitrogen-Bridgehead Compounds as Potent Dual-Acting AChE Inhibitors and hH3 Receptor Antagonists. ACS Chem. Neurosci. 2014, 5, 225–242. [Google Scholar] [CrossRef] [PubMed]
- Khanfar, M.A.; Affini, A.; Lutsenko, K.; Nikolic, K.; Butini, S.; Stark, H. Multiple Targeting Approaches on Histamine H3 Receptor Antagonists. Front. Neurosci. 2016, 10, 201. [Google Scholar] [CrossRef] [PubMed]
- Guzior, N.; Wieckowska, A.; Panek, D.; Malawska, B. Recent Development of Multifunctional Agents as Potential Drug Candidates for the Treatment of Alzheimer’s Disease. Curr. Med. Chem. 2014, 22, 373–404. [Google Scholar] [CrossRef] [PubMed]
- Staszewski, M.; Nelic, D.; Jończyk, J.; Dubiel, M.; Frank, A.; Stark, H.; Bajda, M.; Jakubik, J.; Walczyński, K. Guanidine Derivatives: How Simple Structural Modification of Histamine H3R Antagonists Has Led to the Discovery of Potent Muscarinic M2R/M4R Antagonists. ACS Chem. Neurosci. 2021, 12, 2503–2519. [Google Scholar] [CrossRef] [PubMed]
- Harusawa, S.; Sawada, K.; Magata, T.; Yoneyama, H.; Araki, L.; Usami, Y.; Hatano, K.; Yamamoto, K.; Yamamoto, D.; Yamatodani, A. Synthesis and evaluation of N-alkyl-S-[3-(piperidin-1-yl)propyl]isothioureas: High affinity and human/rat species-selective histamine H3 receptor antagonists. Bioorg. Med. Chem. Lett. 2013, 23, 6415–6420. [Google Scholar] [CrossRef] [PubMed]
- Yu, K.H.; Kim, Y.S.; Kim, S.W.; Park, J.H.; Yang, S.D.; Herdering, W.; Knoechel, A. Synthesis of [18F]Fluoroclofilium as a potential cardiac imaging agent for PET studies. J. Label. Compd. Radiopharm. 2003, 46, 1151–1160. [Google Scholar] [CrossRef]
- Bihel, F.; Humbert, J.-P.; Schneider, S.; Bertin, I.; Wagner, P.; Schmitt, M.; Laboureyras, E.; Petit-Demoulière, B.; Schneider, E.; Mollereau, C.; et al. Development of a Peptidomimetic Antagonist of Neuropeptide FF Receptors for the Prevention of Opioid-Induced Hyperalgesia. ACS Chem. Neurosci. 2015, 6, 438–445. [Google Scholar] [CrossRef]
- Stark, H.; Sippl, W.; Ligneau, X.; Arrang, J.-M.; Ganellin, C.; Schwartz, J.-C.; Schunack, W. Different antagonist binding properties of human and rat histamine H3 receptors. Bioorg. Med. Chem. Lett. 2001, 11, 951–954. [Google Scholar] [CrossRef]
- Olszewska, B.; Stasiak, A.; Flores, D.M.; Fogel, W.A.; Leurs, R.; Walczyński, K. 4-Hydroxypiperidines and Their Flexible 3-(Amino)propyloxy Analogues as Non-Imidazole Histamine H3 Receptor Antagonist: Further Structure–Activity Relationship Exploration and In Vitro and In Vivo Pharmacological Evaluation. Int. J. Mol. Sci. 2018, 19, 1243. [Google Scholar] [CrossRef]
- Vollinga, R.C.; Zuiderveld, O.P.; Scheerens, H.; Bast, A.; Timmerman, H. A simple and rapid in vitro test system for the screening of histamine H3 ligands. Methods Find. Exp. Clin. Pharmacol. 1992, 14, 747–751. [Google Scholar]
- Ellman, G.L.; Courtney, K.D.; Andres, V., Jr.; Featherstone, R.M. A new and rapid colorimetric determination of acetylcholinesterase activity. Biochem. Pharmacol. 1961, 7, 88–95. [Google Scholar] [CrossRef] [PubMed]
- Jończyk, J.; Malawska, B.; Bajda, M. Hybrid approach to structure modeling of the histamine H3 receptor: Multi-level assessment as a tool for model verification. PLoS ONE 2017, 12, e0186108. [Google Scholar] [CrossRef] [PubMed]
- Więckowska, A.; Wichur, T.; Godyń, J.; Bucki, A.; Marcinkowska, M.; Siwek, A.; Więckowski, K.; Zaręba, P.; Knez, D.; Głuch-Lutwin, M.; et al. Novel Multitarget-Directed Ligands Aiming at Symptoms and Causes of Alzheimer’s Disease. ACS Chem. Neurosci. 2018, 9, 1195–1214. [Google Scholar] [CrossRef]
- Staszewski, M.; Walczyński, K. Synthesis and Preliminary Pharmacological Investigation of NewN-Substituted-N-[ω-(ω-phenoxy-alkylpiperazin-1-yl)alkyl]guanidines as Non-Imidazole Histamine H3Antagonists. Arch. Der Pharm. 2012, 345, 431–443. [Google Scholar] [CrossRef] [PubMed]
- Arunlakshana, O.; Schild, H.O. Some Quantitative Uses Of Drug Antagonists. Br. J. Pharmacol. Chemother. 1959, 14, 48–58. [Google Scholar] [CrossRef]
- Kottke, T.; Sander, K.; Weizel, L.; Schneider, E.H.; Seifert, R.; Stark, H. Receptor-Specific Functional Efficacies of Alkyl Imidazoles as Dual Histamine H3/H4 Receptor Ligands. Eur. J. Pharmacol. 2011, 654, 200–208. [Google Scholar] [CrossRef]
- Cheng, Y.-C.; Prusoff, W.H. Relationship between the inhibition constant (KI) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem. Pharmacol. 1973, 22, 3099–3108. [Google Scholar] [CrossRef]
- Peng, X.; Yang, L.; Liu, Z.; Lou, S.; Mei, S.; Li, M.; Chen, Z.; Zhang, H. Structural basis for recognition of antihistamine drug by human histamine receptor. Nat. Commun. 2022, 13, 1–9. [Google Scholar] [CrossRef]
- Lubelska, A.; Latacz, G.; Jastrzȩbska-Wiȩsek, M.; Kotańska, M.; Kurczab, R.; Partyka, A.; Maŕc, M.A.; Wilczyńska, D.; Doroz-Płonka, A.; Łazewska, D.; et al. Are the Hydantoin-1,3,5-Triazine 5-HT6R Ligands a Hope to a Find New Procognitive and Anti-Obesity Drug? Considerations Based on Primary In Vivo Assays and ADME-Tox Profile In Vitro. Molecules 2019, 24, 4472. [Google Scholar] [CrossRef]



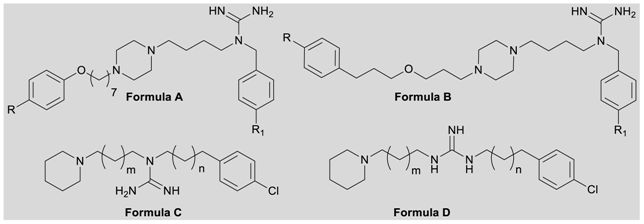 | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Cpd. | Formula/ m/n/R/R1 | hH3R -log(Ki) ± sem a | gpH3R pA2gp ± sem a | gpH1R +Atropine pA2gp ± sem a | eeAChE IC50 (μM) ± sem b | eqBuChE IC50 (μM) ± sem c | MTT IC50 [µM] | ||
| MDA-MB-231 | MCF-7 | BJ | |||||||
| ADS10377 | A/-/-/Cl/H | 6.55 ± 0.08 | 7.82 ± 0.09 | 6.91 ± 0.12 | 1.9 ± 0.1 | ||||
| ADS10376 | A/-/-/Cl/CF3 | 6.40 ± 0.16 | 8.15 ± 0.06 | 6.67 ± 0.01 | 4.9 ± 0.2 | ||||
| ADS1017 | A/-/-/H/H | 6.29 ± 0.09 | 8.21 ± 0.15 | 6.60 ± 0.14 | 5.1 ± 0.1 | 19.86 | 24.71 | 38.97 | |
| ADS1020 | A/-/-/H/CF3 | 6.36 ± 0.01 | 8.49 ± 0.05 | 6.06 ± 0.11 | |||||
| ADS10349 | B/-/-/H/H | 5.60 ± 0.03 | 7.34 ± 0.15 | 6.46 ± 0.11 | |||||
| ADS10350 | B/-/-/H/CF3 | 5.57 ± 0.07 | 7.54 ± 0.10 | 6.57 ± 0.13 | |||||
| ADS10278 | B/-/-/Cl/H | 6.49 ± 0.22 | 7.83 ± 0.07 | 6.93 ± 0.10 | 2.0 ± 0.1 | ||||
| ADS10279 | B/-/-/Cl/CF3 | 5.97 ± 0.12 | 8.28 ± 0.10 | 6.87 ± 0.16 | 4.8 ± 0.1 | ||||
| ADS10292 | C/1/1/-/- | 5.57 ± 0.05 | 7.34 ± 0.09 | 5.92 ± 0.05 | 10.9 ± 0.4 | 8.4 ± 0.2 | |||
| ADS10300 | C/2/1/-/- | 6.22 ± 0.14 | 7.32 ± 0.08 | 5.60 ± 0.01 | 2.4 ± 0.1 | ||||
| ADS10312 | C/1/2/-/- | 6.28 ± 0.05 | 7.03 ± 0.11 | 5.86 ± 0.13 | 3.5 ± 0.1 | ||||
| ADS10298 | D/1/1/-/- | 6.27 ± 0.04 | 7.38 ± 0.07 | 6.88 ± 0.07 | |||||
| ADS10301 | D/2/1/-/- | 6.72 ± 0.06 | 7.64 ± 0.15 | 6.46 ± 0.09 | 5.9 ± 0.1 | ||||
| ADS10306 | D/1/2/-/- | 6.71 ± 0.16 | 7.32 ± 0.14 | 7.17 ± 0.06 | 1.6 ± 0.0 | ||||
| ADS10310 | D/2/2/-/- | 6.90 ± 0.22 | 7.72 ± 0.09 | 6.47 ± 0.05 | 1.7 ± 0.1 | 115.16 | 82.94 | 231.47 | |
| Thioperamide | 8.81 ± 0.05 | ||||||||
| Pyrilamine | 9.13 ± 0.06 | ||||||||
| Pitolisant | 7.95 ± 0.04 | ||||||||
| Tacrine | 0.024 ± 0.0 | 0.015 ± 0.0 | |||||||
| Doxorubicin | 1.85 | 2.12 | 4.87 | ||||||
| Substrate | Molecular Mass (m/z) | Substrate Remaining | The Molecular Mass of the Metabolite (m/z) | Metabolic Pathway * |
|---|---|---|---|---|
| ADS10310 | 365.28 | 73.6% | 205.13 (M1) | decomposition and triple hydroxylation |
| 381.23 (M2) | hydroxylation | |||
| 362.08 (M3) | double-dehydrogenation | |||
| Verapamil * | 455.31 | 30.8% | 441.35 (M1) | demethylation |
| 291.328 (M2) | decomposition | |||
| 165.09 (M3) | decomposition | |||
| 441.29 (M4) | demethylation | |||
| 427.33 (M5) | double-demethylation | |||
| 277.26 (M6) | decomposition |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Staszewski, M.; Iwan, M.; Werner, T.; Bajda, M.; Godyń, J.; Latacz, G.; Korga-Plewko, A.; Kubik, J.; Szałaj, N.; Stark, H.; et al. Guanidines: Synthesis of Novel Histamine H3R Antagonists with Additional Breast Anticancer Activity and Cholinesterases Inhibitory Effect. Pharmaceuticals 2023, 16, 675. https://doi.org/10.3390/ph16050675
Staszewski M, Iwan M, Werner T, Bajda M, Godyń J, Latacz G, Korga-Plewko A, Kubik J, Szałaj N, Stark H, et al. Guanidines: Synthesis of Novel Histamine H3R Antagonists with Additional Breast Anticancer Activity and Cholinesterases Inhibitory Effect. Pharmaceuticals. 2023; 16(5):675. https://doi.org/10.3390/ph16050675
Chicago/Turabian StyleStaszewski, Marek, Magdalena Iwan, Tobias Werner, Marek Bajda, Justyna Godyń, Gniewomir Latacz, Agnieszka Korga-Plewko, Joanna Kubik, Natalia Szałaj, Holger Stark, and et al. 2023. "Guanidines: Synthesis of Novel Histamine H3R Antagonists with Additional Breast Anticancer Activity and Cholinesterases Inhibitory Effect" Pharmaceuticals 16, no. 5: 675. https://doi.org/10.3390/ph16050675
APA StyleStaszewski, M., Iwan, M., Werner, T., Bajda, M., Godyń, J., Latacz, G., Korga-Plewko, A., Kubik, J., Szałaj, N., Stark, H., Malawska, B., Więckowska, A., & Walczyński, K. (2023). Guanidines: Synthesis of Novel Histamine H3R Antagonists with Additional Breast Anticancer Activity and Cholinesterases Inhibitory Effect. Pharmaceuticals, 16(5), 675. https://doi.org/10.3390/ph16050675








