Overexpression of Krüppel-Like Factor 9 Enhances the Antitumor Properties of Paclitaxel in Malignant Melanoma-Derived Cell Lines
Abstract
1. Introduction
2. Results
2.1. A Rapid Proliferation of Malignant Melanoma-Derived Cell Lines Compared to Normal Melanocytes
2.2. A Dose- and Time-Dependent Effect of Paclitaxel on Cellular Viability
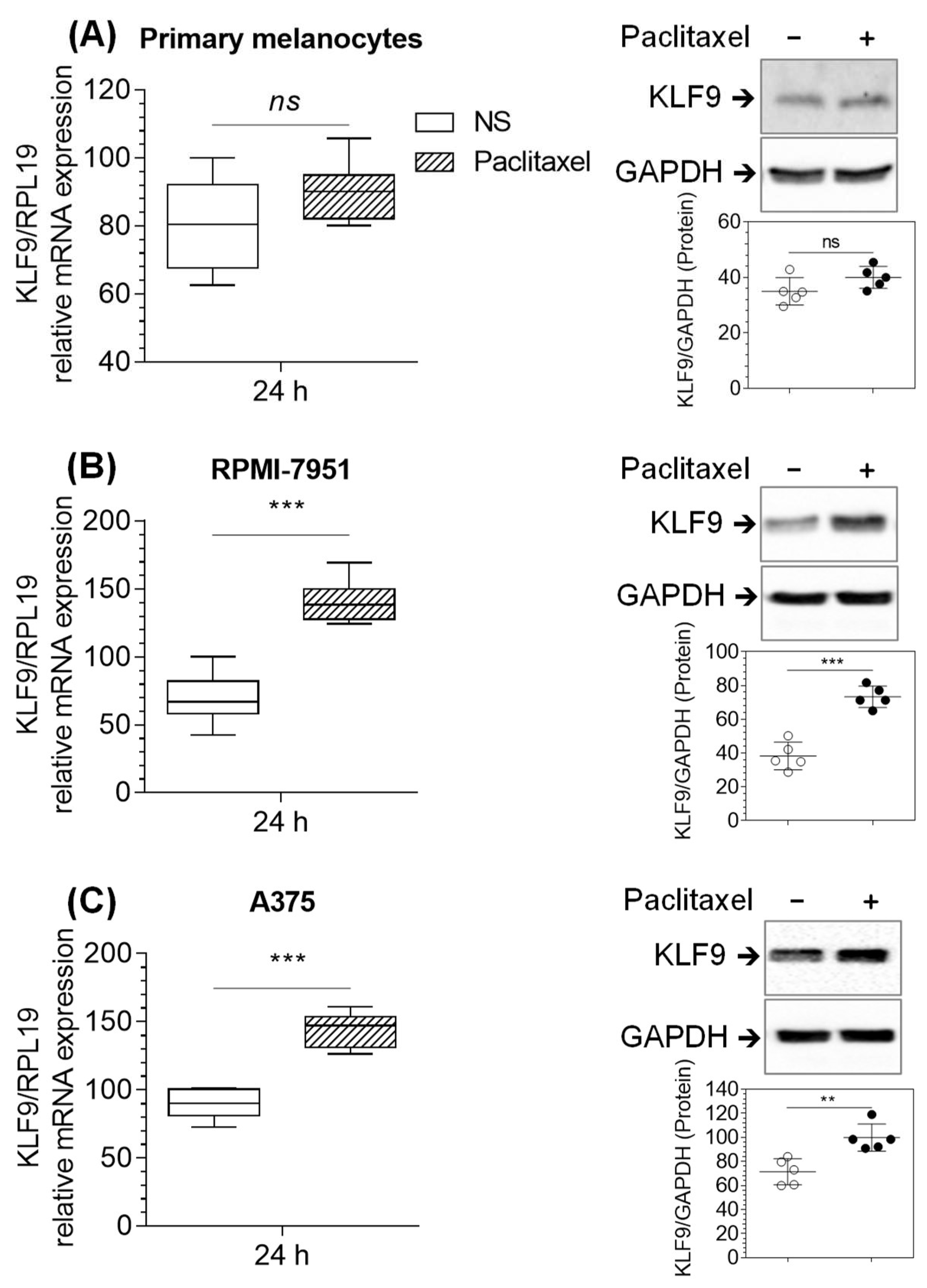
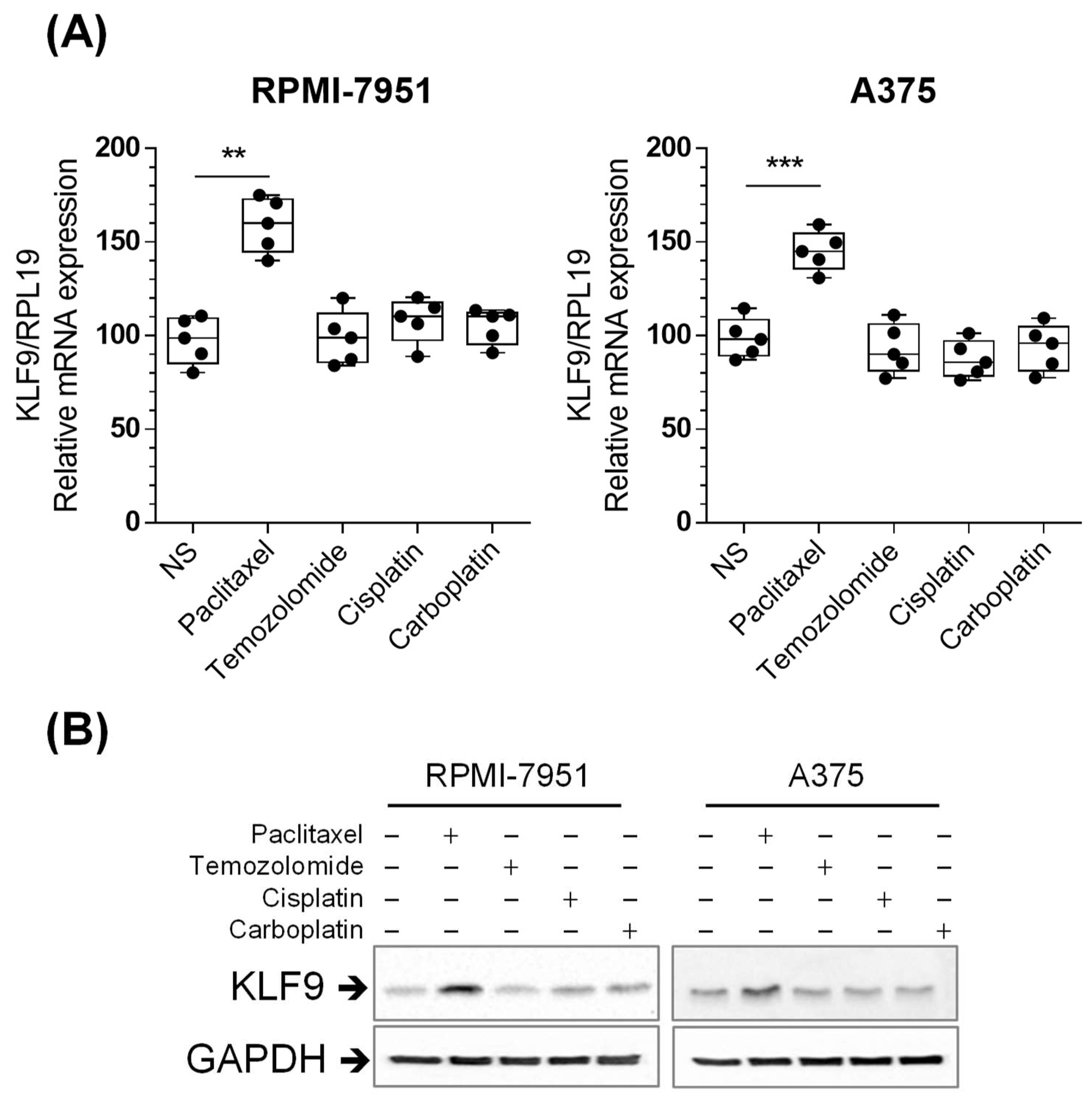
2.3. Paclitaxel Increased KLF9 mRNA and Protein Expression in RPMI-7951 and A375 Cells
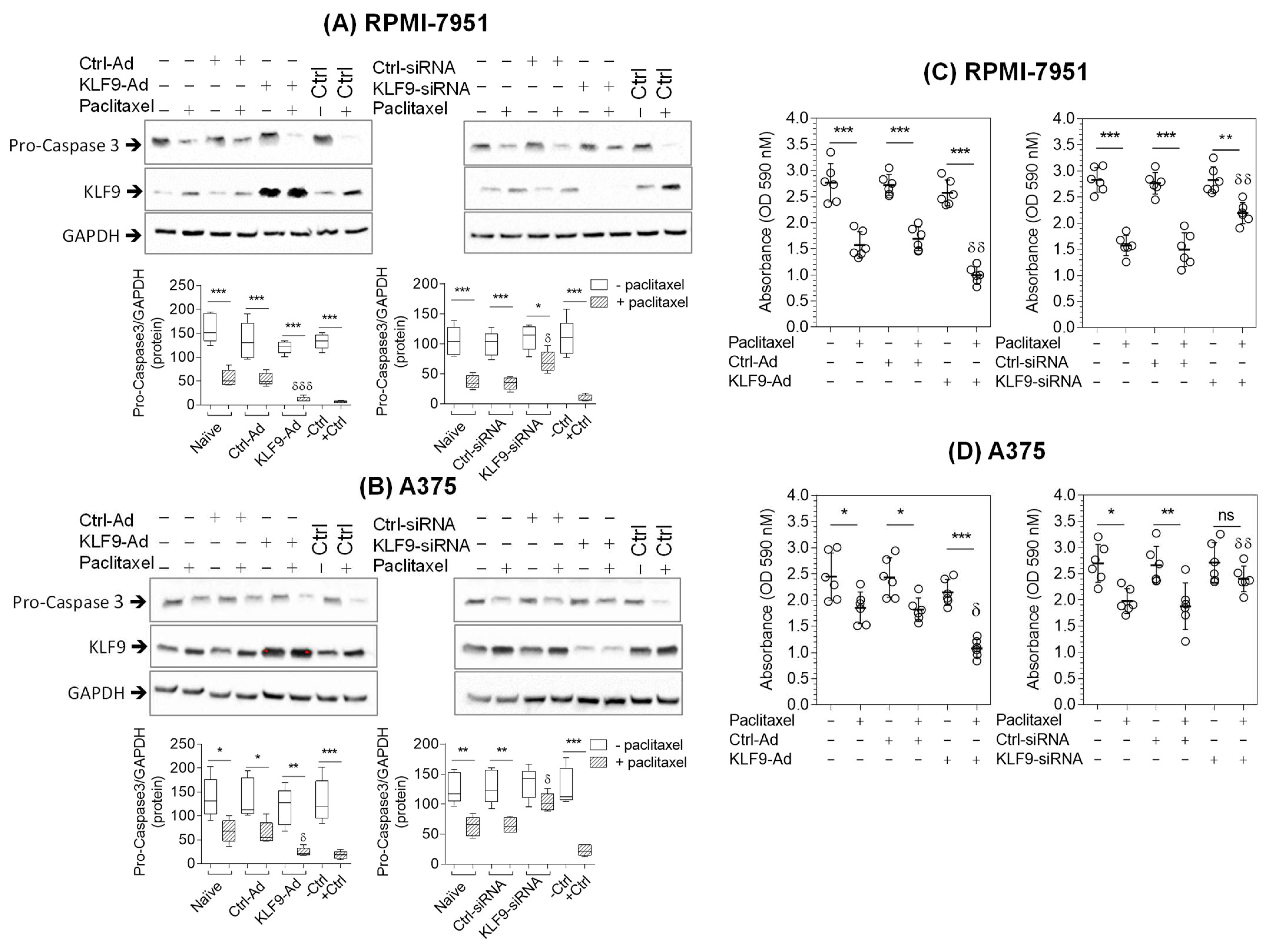
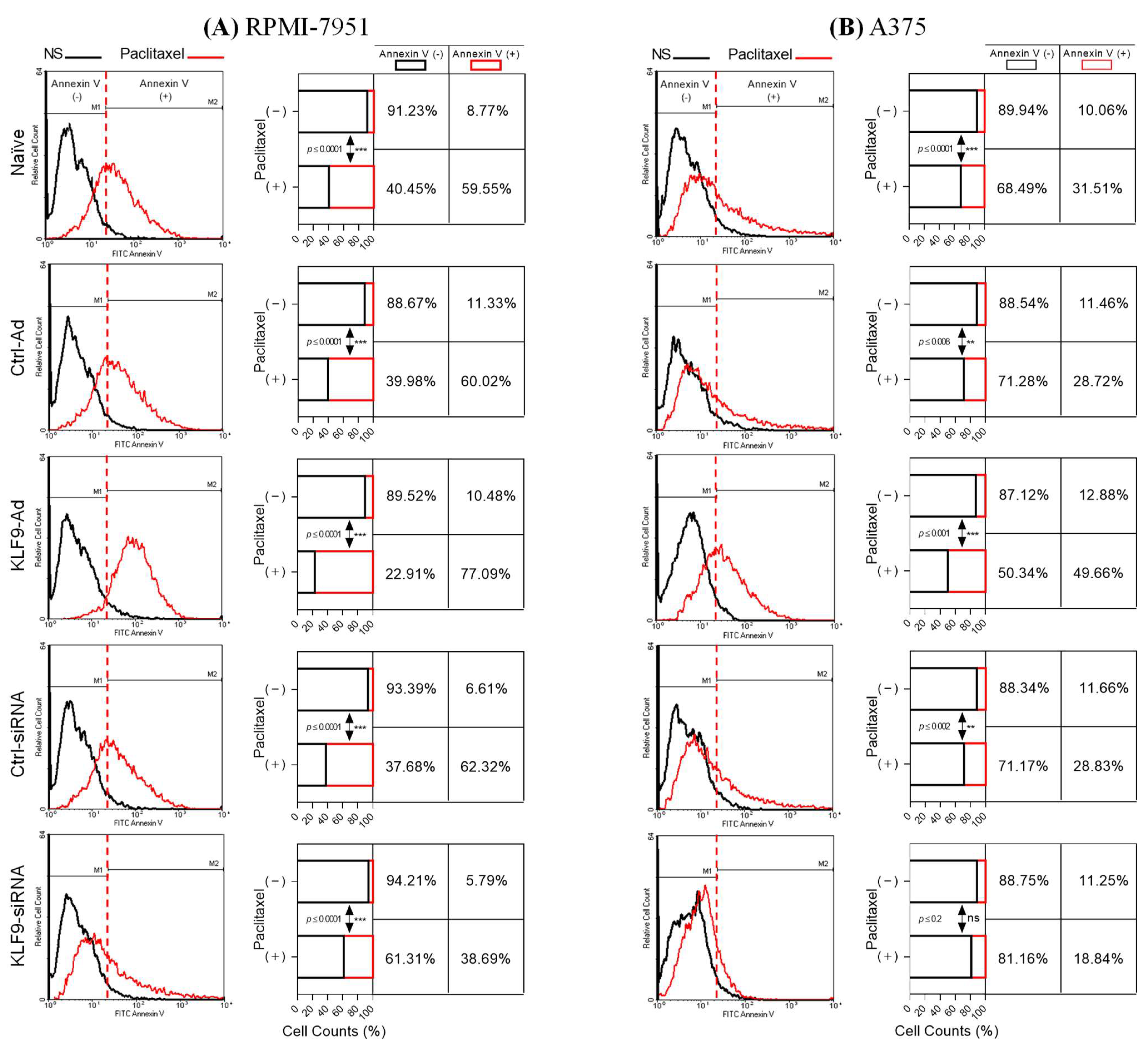
2.4. KLF9 Modulated Paclitaxel-Induced Apoptosis in RPMI-7951 and A375 Cells
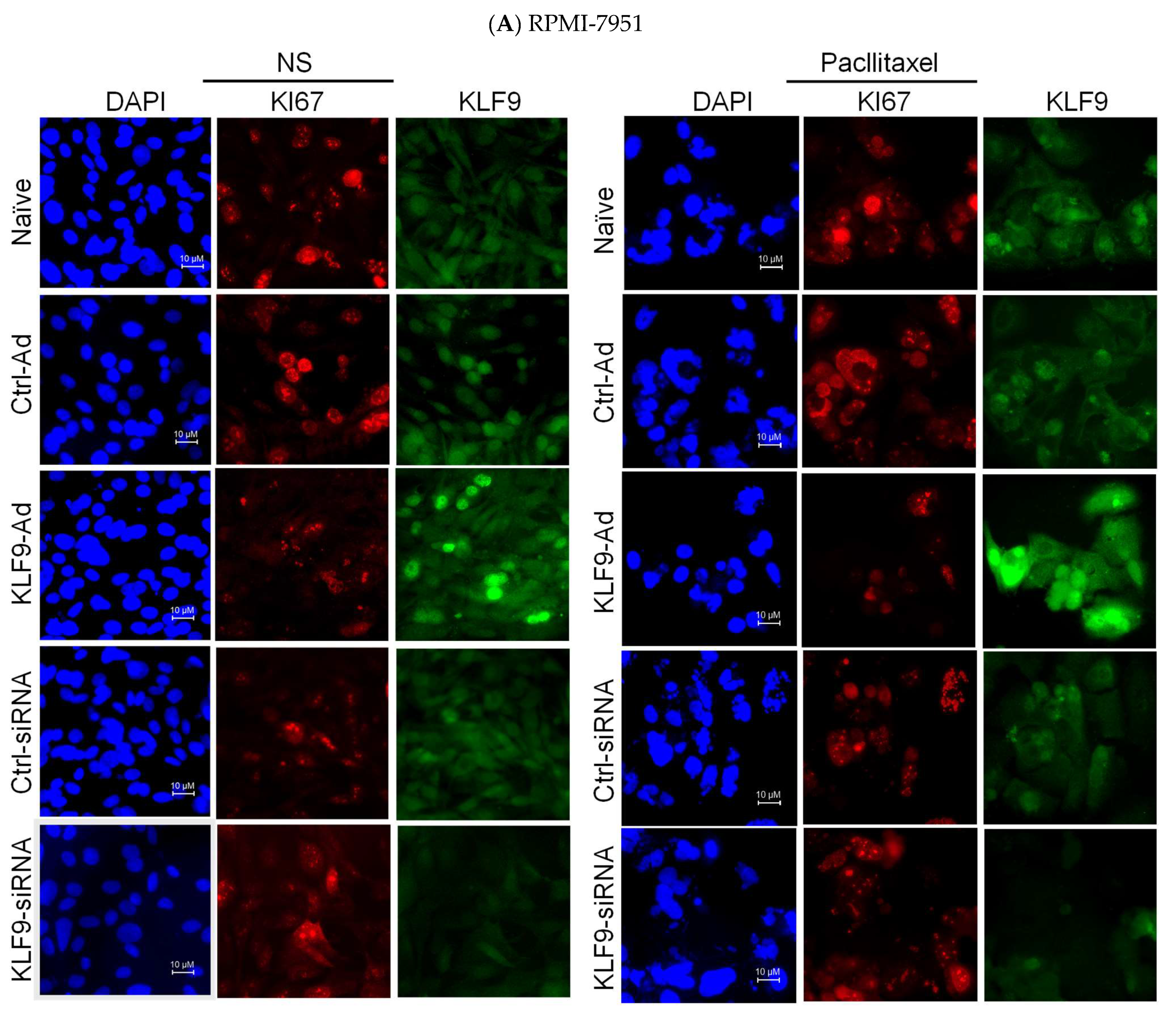
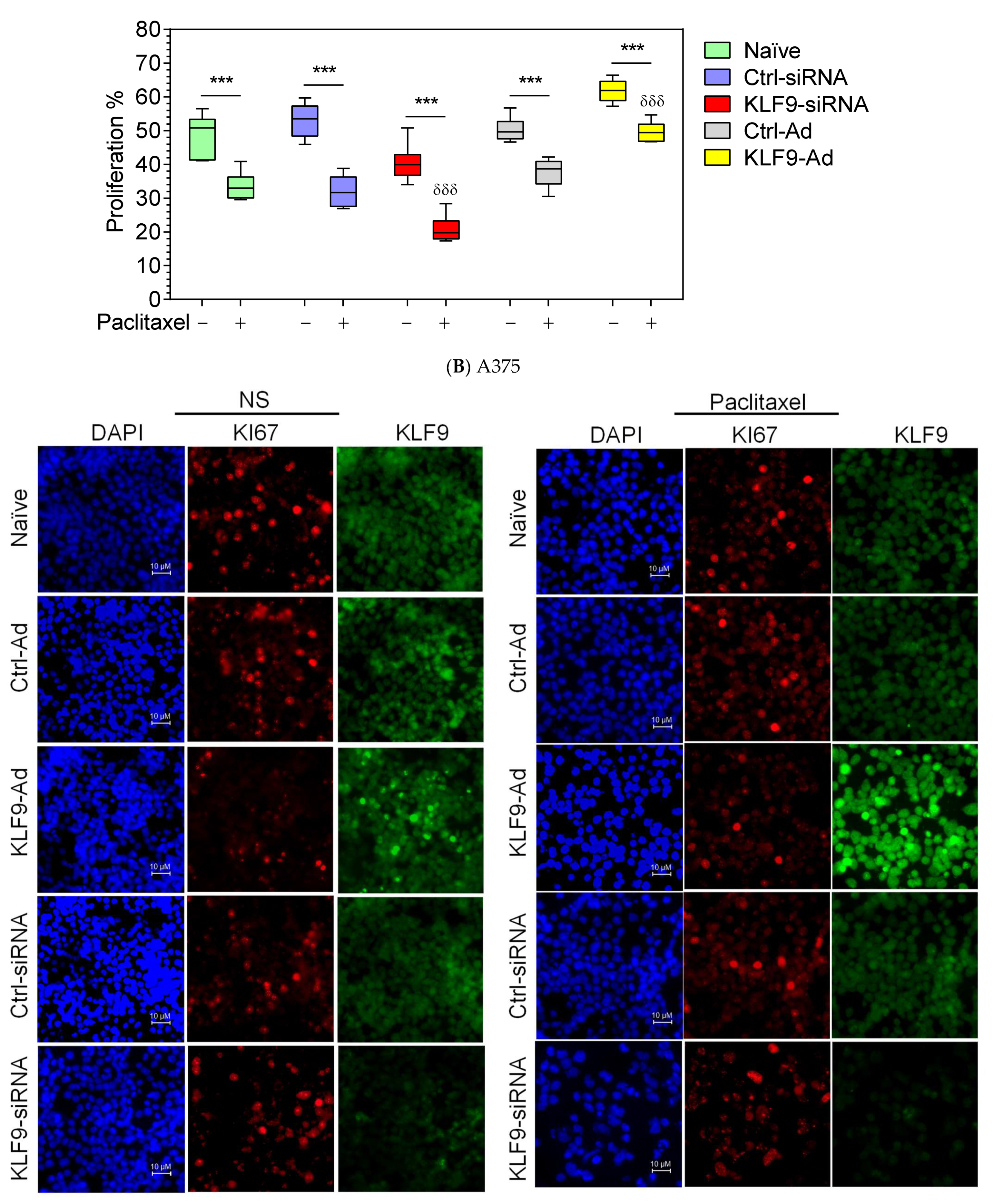
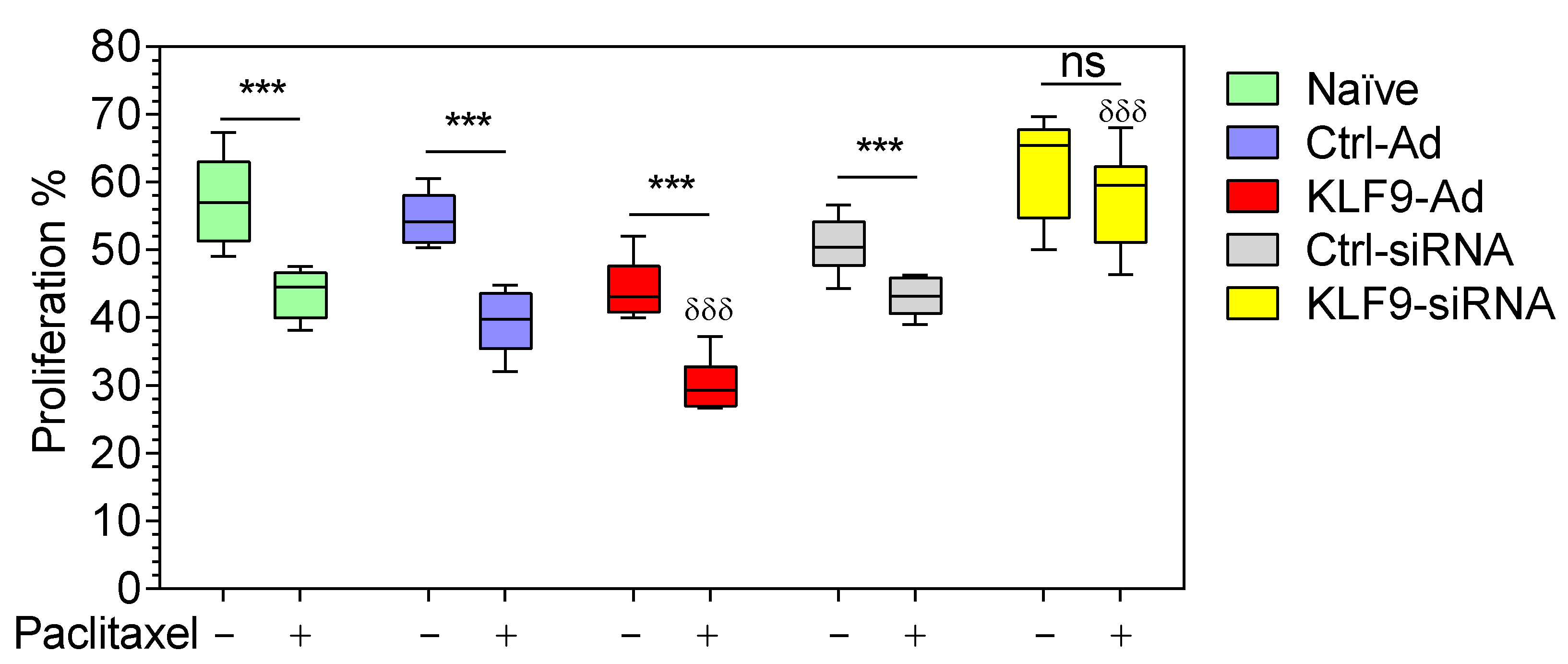
2.5. KLF9 Modulated Paclitaxel Anti-Proliferative Effect in RPMI-7951 and A375 Cells
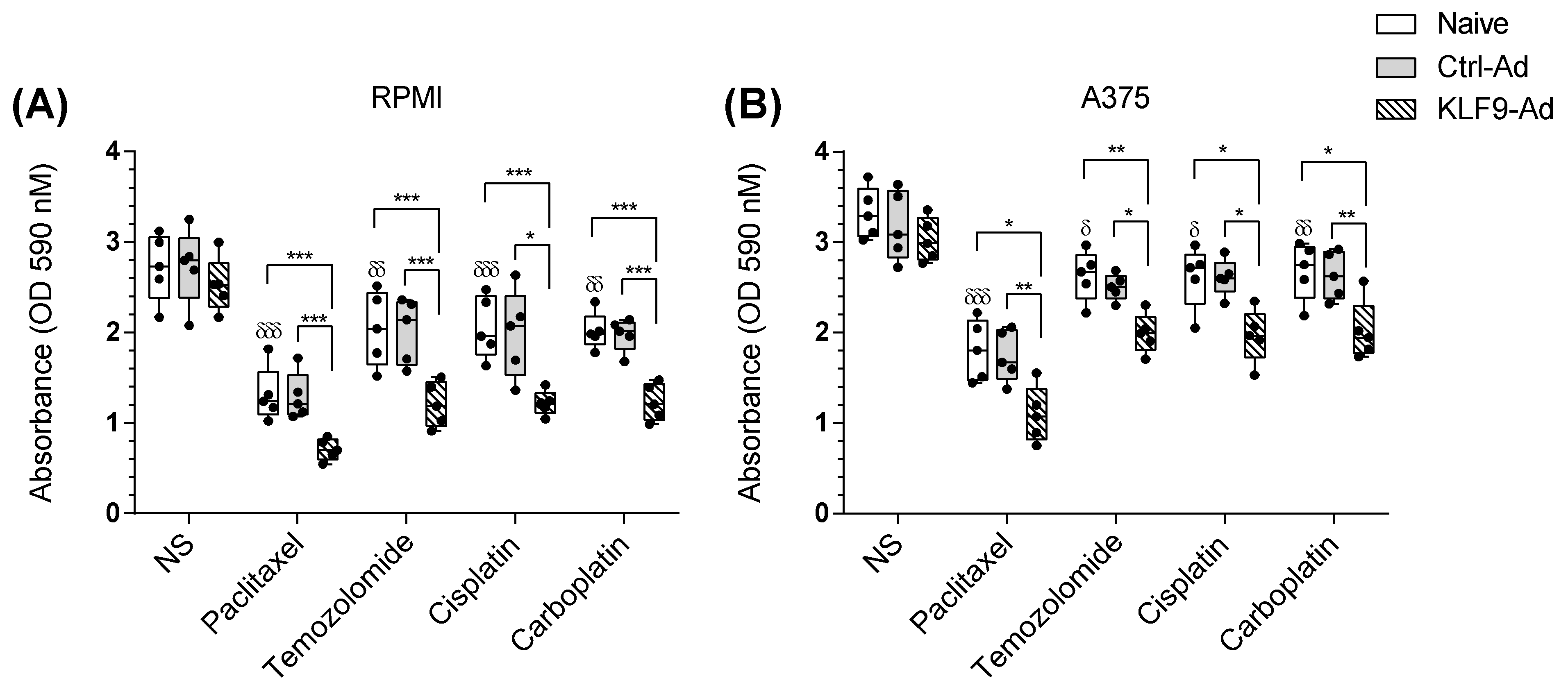
2.6. KLF9 Overexpression Potentiated Temozolomide, Cisplatin, and Carboplatin Apoptotic Effect on Malignant Melanoma Cells
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Reagents
4.2. KLF9 Adenovirus Transduction and mRNA Transfection
4.3. Cell Viability Assay
4.4. RNA Purification, cDNA Synthesis, and QRT-PCR Analysis
4.5. Cell Lysate Preparation, Total Protein Quantification, and Western Blot Analysis
4.6. Annexin V Staining and Flow Cytometry Analysis
4.7. Immunofluorescent Microscopy and Image Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Larkin, J.; Chiarion-Sileni, V.; Gonzalez, R.; Grob, J.-J.; Rutkowski, P.; Lao, C.D.; Cowey, C.L.; Schadendorf, D.; Wagstaff, J.; Dummer, R.; et al. Five-Year Survival with Combined Nivolumab and Ipilimumab in Advanced Melanoma. N. Engl. J. Med. 2019, 381, 1535–1546. [Google Scholar] [CrossRef] [PubMed]
- Lili, L.N.; Klopot, A.; Readhead, B.; Baida, G.; Dudley, J.T.; Budunova, I. Transcriptomic Network Interactions in Human Skin Treated with Topical Glucocorticoid Clobetasol Propionate. J. Investig. Dermatol. 2019, 139, 2281–2291. [Google Scholar] [CrossRef] [PubMed]
- Mostafa, M.M.; Bansal, A.; Michi, A.N.; Sasse, S.K.; Proud, D.; Gerber, A.N.; Newton, R. Genomic determinants implicated in the glucocorticoid-mediated induction of KLF9 in pulmonary epithelial cells. J. Biol. Chem. 2020, 296, 100065. [Google Scholar] [CrossRef]
- Ying, M.; Sang, Y.; Li, Y.; Guerrero-Cazares, H.; Quinones-Hinojosa, A.; Vescovi, A.L.; Eberhart, C.G.; Xia, S.; Laterra, J. KLF9, a Differentiation-Associated Transcription Factor, Suppresses Notch1 Signaling and Inhibits Glioblastoma-Initiating Stem Cells. Stem Cells 2011, 29, 20–31. [Google Scholar] [CrossRef] [PubMed]
- Imataka, H.; Sogawa, K.; Yasumoto, K.; Kikuchi, Y.; Sasano, K.; Kobayashi, A.; Hayami, M.; Fujii-Kuriyama, Y. Two regulatory proteins that bind to the basic transcription element (BTE), a GC box sequence in the promoter region of the rat P-4501A1 gene. EMBO J. 1992, 11, 3663–3671. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Li, J.; Zhang, X.; Xiong, T.; Ye, J.; Yu, J.; Gui, Y. The miR-140-5p/KLF9/KCNQ1 axis promotes the progression of renal cell carcinoma. FASEB J. 2020, 34, 10623–10639. [Google Scholar] [CrossRef]
- Spörl, F.; Korge, S.; Jürchott, K.; Wunderskirchner, M.; Schellenberg, K.; Heins, S.; Specht, A.; Stoll, C.; Klemz, R.; Maier, B.; et al. Krüppel-like factor 9 is a circadian transcription factor in human epidermis that controls proliferation of keratinocytes. Proc. Natl. Acad. Sci. USA 2012, 109, 10903–10908. [Google Scholar] [CrossRef]
- Good, K.L.; Tangye, S.G. Decreased expression of Krüppel-like factors in memory B cells induces the rapid response typical of secondary antibody responses. Proc. Natl. Acad. Sci. USA 2007, 104, 13420–13425. [Google Scholar] [CrossRef]
- Savignac, M.; Mellström, B.; Bébin, A.-G.; Oliveros, J.C.; Delpy, L.; Pinaud, E.; Naranjo, J.R. Increased B Cell Proliferation and Reduced Ig Production in DREAM Transgenic Mice. J. Immunol. 2010, 185, 7527–7536. [Google Scholar] [CrossRef]
- Tetreault, M.-P.; Yang, Y.; Katz, J.P. Krüppel-like factors in cancer. Nat. Rev. Cancer 2013, 13, 701–713. [Google Scholar] [CrossRef]
- Qiao, F.; Yao, F.; Chen, L.; Lu, C.; Ni, Y.; Fang, W.; Jin, H. Krüppel-like factor 9 was down-regulated in esophageal squamous cell carcinoma and negatively regulated beta-catenin/TCF signaling. Mol. Carcinog. 2016, 55, 280–291. [Google Scholar] [CrossRef] [PubMed]
- Bagati, A.; Moparthy, S.; Fink, E.E.; Bianchi-Smiraglia, A.; Yun, D.H.; Kolesnikova, M.; Udartseva, O.O.; Wolff, D.W.; Roll, M.V.; Lipchick, B.C.; et al. KLF9-dependent ROS regulate melanoma progression in stage-specific manner. Oncogene 2019, 38, 3585–3597. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Sun, Q.; Jiang, M.; Li, S.; Zhang, J.; Xu, Z.; Guo, D.; Gu, T.; Wang, B.; Xiao, L.; et al. KLF9 suppresses gastric cancer cell invasion and metastasis through transcriptional inhibition of MMP28. FASEB J. 2019, 33, 7915–7928. [Google Scholar] [CrossRef] [PubMed]
- DeNicola, G.M.; Karreth, F.A.; Humpton, T.J.; Gopinathan, A.; Wei, C.; Frese, K.; Mangal, D.; Yu, K.H.; Yeo, C.J.; Calhoun, E.S.; et al. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature 2011, 475, 106–109. [Google Scholar] [CrossRef] [PubMed]
- Cairns, R.A.; Harris, I.S.; Mak, T.W. Regulation of cancer cell metabolism. Nat. Rev. Cancer 2011, 11, 85–95. [Google Scholar] [CrossRef] [PubMed]
- Feitelson, M.A.; Arzumanyan, A.; Kulathinal, R.J.; Blain, S.W.; Holcombe, R.F.; Mahajna, J.; Marino, M.; Martinez-Chantar, M.L.; Nawroth, R.; Sanchez-Garcia, I.; et al. Sustained proliferation in cancer: Mechanisms and novel therapeutic targets. Semin. Cancer Biol. 2015, 35, S25–S54. [Google Scholar] [CrossRef]
- Nygren, P. What is cancer chemotherapy? Acta Oncol. 2001, 40, 166–174. [Google Scholar] [CrossRef]
- Abosedera, D.A.; Emara, S.; Tamam, O.A.; Badr, O.M.; Khalifa, S.A.; El-Seedi, H.R.; Refaey, M.S. Metabolomic profile and in vitro evaluation of the cytotoxic activity of Asphodelus microcarpus against human malignant melanoma cells A375. Arab. J. Chem. 2022, 15, 104174. [Google Scholar] [CrossRef]
- Weaver, B.A. How Taxol/paclitaxel kills cancer cells. Mol. Biol. Cell 2014, 25, 2677–2681. [Google Scholar] [CrossRef]
- McGuire, W.P.; Rowinsky, E.K.; Rosenshein, N.B.; Grumbine, F.C.; Ettinger, D.S.; Armstrong, D.K.; Donehower, R.C. Taxol: A Unique Antineoplastic Agent with Significant Activity in Advanced Ovarian Epithelial Neoplasms. Ann. Intern. Med. 1989, 111, 273–279. [Google Scholar] [CrossRef]
- Kottschade, L.A.; Suman, V.J.; Perez, D.G.; McWilliams, R.R.; Kaur, J.S.; Amatruda, T.T., III; Geoffroy, F.J.; Gross, H.M.; Cohen, P.A.; Jaslowski, A.J.; et al. A randomized phase 2 study of temozolomide and bevacizumab or nab-paclitaxel, carboplatin, and bevacizumab in patients with unresectable stage IV melanoma: A North Central Cancer Treatment Group Study, N0775. Cancer 2013, 119, 586–592. [Google Scholar] [CrossRef] [PubMed]
- González Cao, M.; Viteri, S.; Garrán, C.; Nieto, Y.; Aristu, J.; Ponz, M.; Martín Algarra, S. Response of resistant melanoma to a combination of weekly paclitaxel and bevacizumab. Clin. Transl. Oncol. 2007, 9, 119–120. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.B.; Sosman, J.A.; Fruehauf, J.P.; Linette, G.P.; Markovic, S.N.; McDermott, D.F.; Weber, J.S.; Nguyen, H.; Cheverton, P.; Chen, D.; et al. BEAM: A Randomized Phase II Study Evaluating the Activity of Bevacizumab in Combination with Carboplatin Plus Paclitaxel in Patients With Previously Untreated Advanced Melanoma. J. Clin. Oncol. 2012, 30, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Foley, E.A.; Kapoor, T.M. Microtubule attachment and spindle assembly checkpoint signalling at the kinetochore. Nat. Rev. Mol. Cell Biol. 2013, 14, 25–37. [Google Scholar] [CrossRef]
- Wang, Y.B.; Sun, B.; Han, B.; Hu, M. Retinoic acid increases the anticancer effect of paclitaxel by inducing differentiation of cancer stem cells in melanoma. Pharmazie 2018, 73, 729–732. [Google Scholar] [CrossRef]
- Arslanbaeva, L.R.; Santoro, M.M. Adaptive redox homeostasis in cutaneous melanoma. Redox Biol. 2020, 37, 101753. [Google Scholar] [CrossRef]
- Zucker, S.N.; Fink, E.E.; Bagati, A.; Mannava, S.; Bianchi-Smiraglia, A.; Bogner, P.N.; Wawrzyniak, J.A.; Foley, C.; Leonova, K.I.; Grimm, M.J.; et al. Nrf2 Amplifies Oxidative Stress via Induction of Klf9. Mol. Cell 2014, 53, 916–928. [Google Scholar] [CrossRef]
- Parga, J.; Rodriguez-Perez, A.; Garcia-Garrote, M.; Rodriguez-Pallares, J.; Labandeira-Garcia, G. Angiotensin II Induces Oxi-dative Stress and Upregulates Neuroprotective Signaling from the NRF2 and KLF9 Pathway in Dopaminergic Cells. Free Radic. Biol. Med. 2018, 129, 394–406. [Google Scholar] [CrossRef]
- Jin, L.; Chang, C.; Pawlik, K.M.; Datta, A.; Johnson, L.M.; Vu, T.; Napoli, J.L.; Datta, P.K. Serine Threonine Kinase Receptor-Associated Protein Deficiency Impairs Mouse Embryonic Stem Cells Lineage Commitment through CYP26A1-Mediated Retinoic Acid Homeostasis. Stem Cells 2018, 36, 1368–1379. [Google Scholar] [CrossRef]
- Degos, L.; Wang, Z.Y. All Trans Retinoic Acid in Acute Promyelocytic Leukemia. Oncogene 2001, 20, 7140–7145. [Google Scholar] [CrossRef]
- Xu, Y.; Shen, M.; Li, Y.; Sun, Y.; Teng, Y.; Wang, Y.; Duan, Y. The synergic antitumor effects of paclitaxel and temozolomide co-loaded in mPEG-PLGA nanoparticles on glioblastoma cells. Oncotarget 2016, 7, 20890–20901. [Google Scholar] [CrossRef] [PubMed]
- Patel, P.M.; Suciu, S.; Mortier, L.; Kruit, W.H.; Robert, C.; Schadendorf, D.; Trefzer, U.; Punt, C.J.; Dummer, R.; Davidson, N.; et al. Extended schedule, escalated dose temozolomide versus dacarbazine in stage IV melanoma: Final results of a randomised phase III study (EORTC 18032). Eur. J. Cancer 2011, 47, 1476–1483. [Google Scholar] [CrossRef] [PubMed]
- O’Day, S.J.; Eggermont, A.M.; Chiarion-Sileni, V.; Kefford, R.; Grob, J.J.; Mortier, L.; Robert, C.; Schachter, J.; Testori, A.; Mackiewicz, J.; et al. Final Results of Phase III SYMMETRY Study: Randomized, Double-Blind Trial of Elesclomol Plus Paclitaxel Versus Paclitaxel Alone as Treatment for Chemotherapy-Naive Patients With Advanced Melanoma. J. Clin. Oncol. 2016, 31, 1211–1218. [Google Scholar] [CrossRef] [PubMed]
- Flaherty, K.T.; Lee, S.J.; Zhao, F.; Schuchter, L.M.; Flaherty, L.; Kefford, R.; Atkins, M.B.; Leming, P.; Kirkwood, J.M. Phase III Trial of Carboplatin and Paclitaxel with or Without Sorafenib in Metastatic Melanoma. J. Clin. Oncol. 2012, 31, 373–379. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Gomes, F.; Lorigan, P.; Rutkowski, P.; Wernick, B.D.; Goel, N.; Zih, F.S.; Farma, J.M.; Tsai, K.K.; Algazi, A.P.; et al. The role for chemotherapy in the modern management of melanoma. Melanoma Manag. 2017, 4, 125–136. [Google Scholar] [CrossRef]
- Hallmarks of Cancer: The Next Generation|Elsevier Enhanced Reader. Available online: https://reader.elsevier.com/reader/sd/pii/S0092867411001279?token=AE490FDF0C0905CCF15B06180518CB852048F97B85C78A34BFCA84FC1F1E1917A46C407A9669638013BE5E937C389ACB&originRegion=us-east-1&originCreation=20211208182800 (accessed on 8 December 2021).
- Altonsy, M.O.; Ganguly, A.; Amrein, M.; Surmanowicz, P.; Li, S.S.; Lauzon, G.J.; Mydlarski, P.R. Beta3-Tubulin Is Critical for Microtubule Dynamics, Cell Cycle Regulation, and Spontaneous Release of Microvesicles in Human Malignant Melanoma Cells (A375). Int. J. Mol. Sci. 2020, 21, 1656. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Altonsy, M.O.; Song-Zhao, G.X.; Mostafa, M.M.; Mydlarski, P.R. Overexpression of Krüppel-Like Factor 9 Enhances the Antitumor Properties of Paclitaxel in Malignant Melanoma-Derived Cell Lines. Pharmaceuticals 2023, 16, 557. https://doi.org/10.3390/ph16040557
Altonsy MO, Song-Zhao GX, Mostafa MM, Mydlarski PR. Overexpression of Krüppel-Like Factor 9 Enhances the Antitumor Properties of Paclitaxel in Malignant Melanoma-Derived Cell Lines. Pharmaceuticals. 2023; 16(4):557. https://doi.org/10.3390/ph16040557
Chicago/Turabian StyleAltonsy, Mohammed O., George X. Song-Zhao, Mahmoud M. Mostafa, and Paule Régine Mydlarski. 2023. "Overexpression of Krüppel-Like Factor 9 Enhances the Antitumor Properties of Paclitaxel in Malignant Melanoma-Derived Cell Lines" Pharmaceuticals 16, no. 4: 557. https://doi.org/10.3390/ph16040557
APA StyleAltonsy, M. O., Song-Zhao, G. X., Mostafa, M. M., & Mydlarski, P. R. (2023). Overexpression of Krüppel-Like Factor 9 Enhances the Antitumor Properties of Paclitaxel in Malignant Melanoma-Derived Cell Lines. Pharmaceuticals, 16(4), 557. https://doi.org/10.3390/ph16040557





