Mucosal Genes Expression in Inflammatory Bowel Disease Patients: New Insights
Abstract
1. Introduction
2. Results
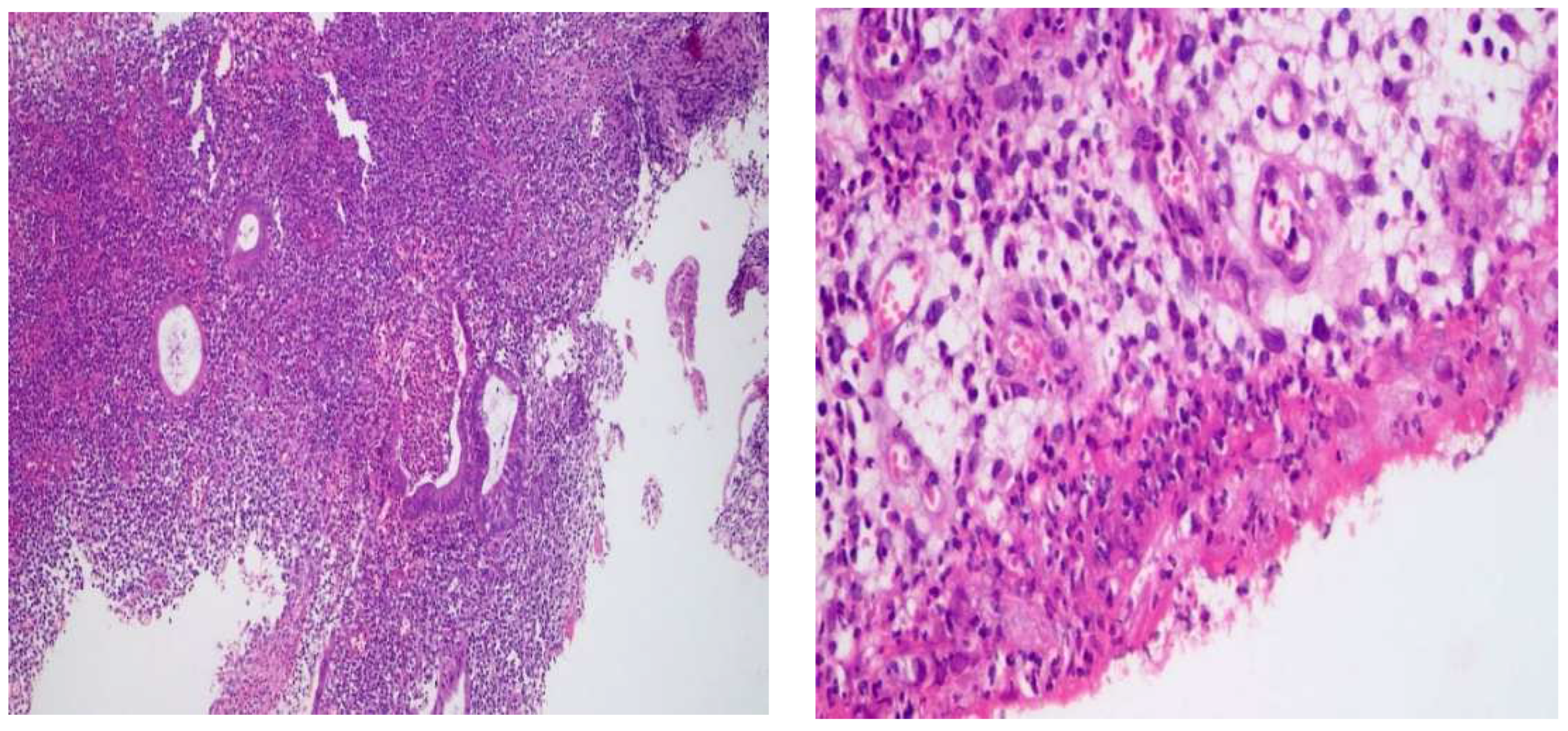
2.1. Histological Changes


2.2. Patients’ and Non-IBD Control Participants’ Characteristics
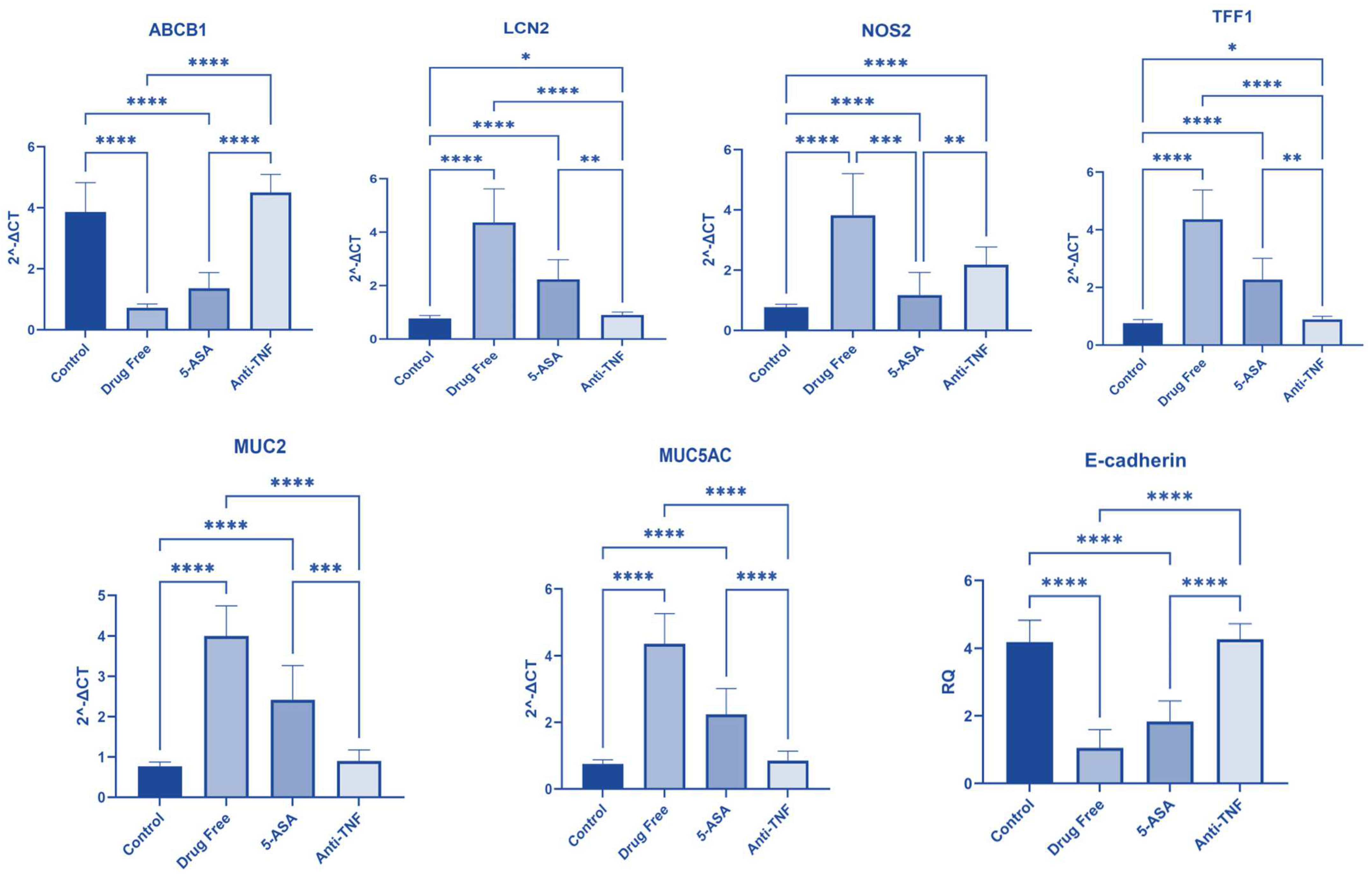
2.3. Gene Expression among Studied Groups
2.3.1. ABCB1 Gene Expression
2.3.2. LCN2 Gene Expression
2.3.3. NOS2 Gene Expression
2.3.4. TFF1 Gene Expression
2.3.5. MUC2 Gene Expression
2.3.6. MUC5AC Gene Expression
2.3.7. E-Cadherin Gene Expression
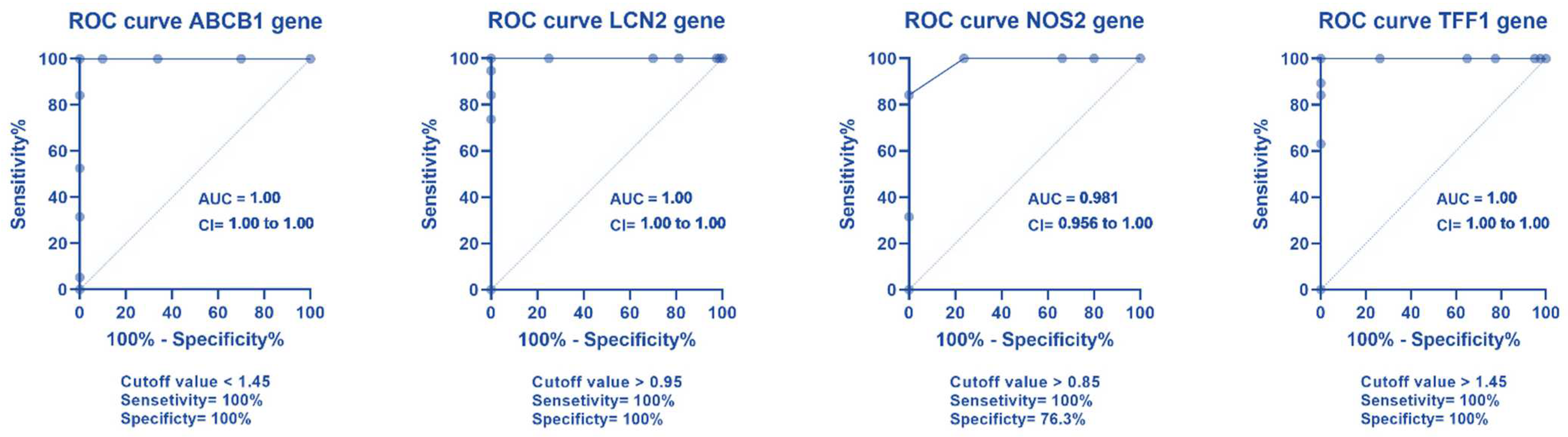
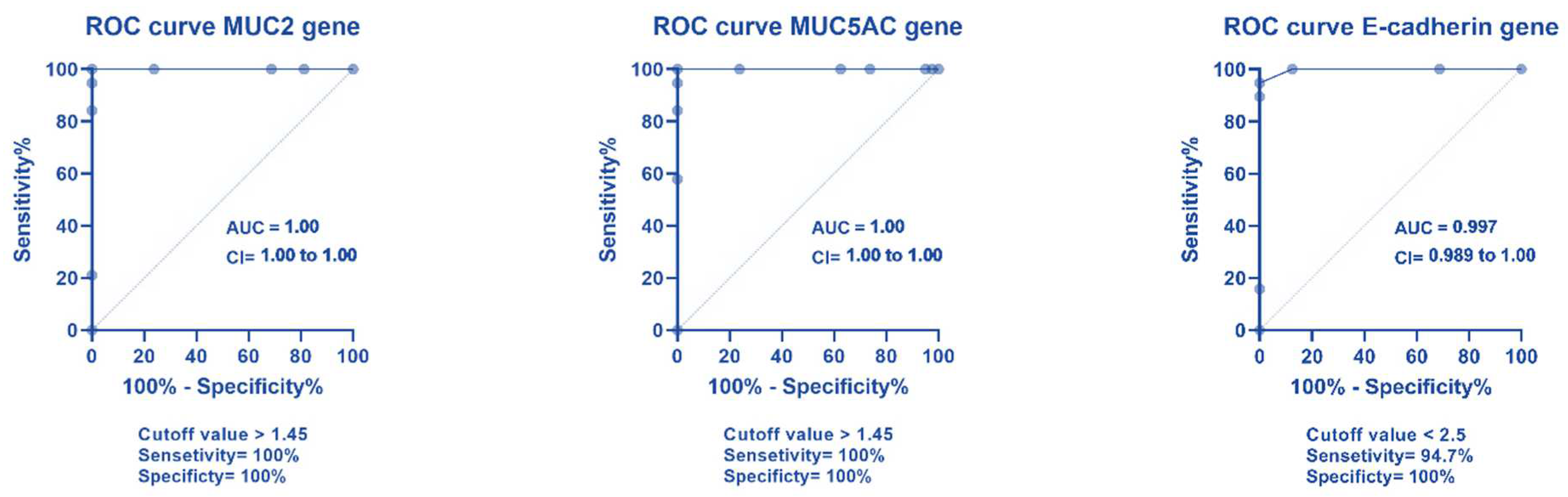
2.4. ROC Curve Analysis for Different Gene Expression
2.5. Correlation between Different Genes
3. Discussion
4. Materials and Methods
4.1. Patients
4.2. Total RNA Isolation and qPCR
4.3. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gomollón, F.; Dignass, A.; Annese, V.; Tilg, H.; Van Assche, G.; Lindsay, J.O.; Peyrin-Biroulet, L.; Cullen, G.J.; Daperno, M.; Kucharzik, T., 3rd. European evidence-based consensus on the diagnosis and management of Crohn’s disease 2016: Part 1: Diagnosis and medical management. J. Crohns Colitis 2017, 11, 3–25. [Google Scholar] [CrossRef] [PubMed]
- Thia, K.T.; Sandborn, W.J.; Harmsen, W.S.; Zinsmeister, A.R.; Loftus Jr, E.V. Risk factors associated with progression to intestinal complications of Crohn’s disease in a population-based cohort. Gastroenterology 2010, 139, 1147–1155. [Google Scholar] [CrossRef] [PubMed]
- Magro, F.; Gionchetti, P.; Eliakim, R.; Ardizzone, S.; Armuzzi, A.; Barreiro-de Acosta, M.; Burisch, J.; Gecse, K.B.; Hart, A.L.; Hindryckx, P. Third European evidence-based consensus on diagnosis and management of ulcerative colitis. Part 1: Definitions, diagnosis, extra-intestinal manifestations, pregnancy, cancer surveillance, surgery, and ileo-anal pouch disorders. J. Crohns Colitis 2017, 11, 649–670. [Google Scholar] [CrossRef] [PubMed]
- Beaugerie, L.; Seksik, P.; Nion–Larmurier, I.; Gendre, J.P.; Cosnes, J. Predictors of Crohn’s disease. Gastroenterology 2006, 130, 650–656. [Google Scholar] [CrossRef]
- Monstad, I.; Hovde, Ø.; Solberg, I.C.; Moum, B.A. Clinical course and prognosis in ulcerative colitis: Results from population-based and observational studies. Ann. Gastroenterol. Q. Publ. Hell. Soc. Gastroenterol. 2014, 27, 95. [Google Scholar]
- Alarfaj, S.J.; Mostafa, S.A.; Abdelsalam, R.A.; Negm, W.A.; El-Masry, T.A.; Hussein, I.A.; El Nakib, A.M. Helicobacter pylori Infection in Cirrhotic Patients with Portal Hypertensive Gastropathy: A New Enigma? Front. Med. 2022, 9, 902255. [Google Scholar] [CrossRef]
- Martin, J.C.; Chang, C.; Boschetti, G.; Ungaro, R.; Giri, M.; Grout, J.A.; Gettler, K.; Chuang, L.-s.; Nayar, S.; Greenstein, A.J. Single-cell analysis of Crohn’s disease lesions identifies a pathogenic cellular module associated with resistance to anti-TNF therapy. Cell 2019, 178, 1493–1508.e1420. [Google Scholar] [CrossRef]
- Marigorta, U.M.; Denson, L.A.; Hyams, J.S.; Mondal, K.; Prince, J.; Walters, T.D.; Griffiths, A.; Noe, J.D.; Crandall, W.V.; Rosh, J.R. Transcriptional risk scores link GWAS to eQTLs and predict complications in Crohn’s disease. Nat. Genet. 2017, 49, 1517–1521. [Google Scholar] [CrossRef]
- Lee, J.C.; Lyons, P.A.; McKinney, E.F.; Sowerby, J.M.; Carr, E.J.; Bredin, F.; Rickman, H.M.; Ratlamwala, H.; Hatton, A.; Rayner, T.F. Gene expression profiling of CD8+ T cells predicts prognosis in patients with Crohn disease and ulcerative colitis. J. Clin. Investig. 2011, 121, 4170–4179. [Google Scholar] [CrossRef]
- Reinisch, W.; Sandborn, W.J.; Rutgeerts, P.; Feagan, B.G.; Rachmilewitz, D.; Hanauer, S.B.; Lichtenstein, G.R.; De Villiers, W.J.; Blank, M.; Lang, Y. Long-term infliximab maintenance therapy for ulcerative colitis: The ACT-1 and-2 extension studies. Inflamm. Bowel Dis. 2012, 18, 201–211. [Google Scholar] [CrossRef]
- Reinglas, J.; Gonczi, L.; Kurt, Z.; Bessissow, T.; Lakatos, P.L. Positioning of old and new biologicals and small molecules in the treatment of inflammatory bowel diseases. World J. Gastroenterol. 2018, 24, 3567. [Google Scholar] [CrossRef]
- Atreya, R.; Neurath, M.F. Current and future targets for mucosal healing in inflammatory bowel disease. Visc. Med. 2017, 33, 82–88. [Google Scholar] [CrossRef]
- Ben-Horin, S.; Chowers, Y. loss of response to anti-TNF treatments in Crohn’s disease. Aliment. Pharmacol. Ther. 2011, 33, 987–995. [Google Scholar] [CrossRef]
- Milanesi, E.; Dobre, M.; Manuc, T.E.; Becheanu, G.; Tieranu, C.G.; Ionescu, E.M.; Manuc, M. Mucosal gene expression changes induced by anti-TNF treatment in inflammatory bowel disease patients. Drug Dev. Res. 2019, 80, 831–836. [Google Scholar] [CrossRef]
- D’Amico, F.; Netter, P.; Baumann, C.; Veltin, M.; Zallot, C.; Aimone-Gastin, I.; Danese, S.; Peyrin-Biroulet, L. Setting up a virtual calprotectin clinic in inflammatory bowel diseases: Literature review and nancy experience. J. Clin. Med. 2020, 9, 2697. [Google Scholar] [CrossRef]
- Mostafa, S.A.; Mohammad, M.H.; Negm, W.A.; Batiha, G.E.S.; Alotaibi, S.S.; Albogami, S.M.; De Waard, M.; Tawfik, N.Z.; Abdallah, H.Y. Circulating microRNA203 and its target genes’ role in psoriasis pathogenesis. Front. Med. 2022, 9, 988962. [Google Scholar] [CrossRef]
- Muise, A.M.; Walters, T.D.; Glowacka, W.K.; Griffiths, A.M.; Ngan, B.; Lan, H.; Xu, W.; Silverberg, M.; Rotin, D. Polymorphisms in E-cadherin (CDH1) result in a mis-localised cytoplasmic protein that is associated with Crohn’s disease. Gut 2009, 58, 1121–1127. [Google Scholar] [CrossRef]
- Borg-Bartolo, S.P.; Boyapati, R.K.; Satsangi, J.; Kalla, R. Precision medicine in inflammatory bowel disease: Concept, progress and challenges. F1000Research 2020, 9, 54. [Google Scholar] [CrossRef]
- Petryszyn, P.W.; Wiela-Hojeńska, A. The importance of the polymorphisms of the ABCB1 gene in disease susceptibility, behavior and response to treatment in inflammatory bowel disease: A literature review. Adv. Clin. Exp. Med. 2018, 27, 1459–1463. [Google Scholar] [CrossRef]
- Palmieri, O.; Latiano, A.; Valvano, R.; D’inca, R.; Vecchi, M.; Sturniolo, G.; Saibeni, S.; Bossa, F.; Latiano, T.; Devoto, M. Multidrug resistance 1 gene polymorphisms are not associated with inflammatory bowel disease and response to therapy in Italian patients. Aliment. Pharmacol. Ther. 2005, 22, 1129–1138. [Google Scholar] [CrossRef]
- Zintzaras, E. Is there evidence to claim or deny association between variants of the multidrug resistance gene (MDR1 or ABCB1) and inflammatory bowel disease? Inflamm. Bowel Dis. 2012, 18, 562–572. [Google Scholar] [CrossRef]
- Belliard, A.M.; Lacour, B.; Farinotti, R.; Leroy, C. Effect of tumor necrosis factor-α and interferon-γ on intestinal P-glycoprotein expression, activity, and localization in Caco-2 cells. J. Pharm. Sci. 2004, 93, 1524–1536. [Google Scholar] [CrossRef] [PubMed]
- Dobre, M.; Mănuc, T.E.; Milanesi, E.; Pleşea, I.E.; Ţieranu, E.N.; Popa, C.; Mănuc, M.; Preda, C.M.; Ţieranu, I.; Diculescu, M.M. Mucosal CCR1 gene expression as a marker of molecular activity in Crohn’s disease: Preliminary data. Rom. J. Morphol. Embryol 2017, 58, 1263–1268. [Google Scholar] [PubMed]
- Thorsvik, S.; Bakke, I.; van Beelen Granlund, A.; Røyset, E.S.; Damås, J.K.; Østvik, A.E.; Sandvik, A.K. Expression of neutrophil gelatinase-associated lipocalin (NGAL) in the gut in Crohn’s disease. Cell Tissue Res. 2018, 374, 339–348. [Google Scholar] [CrossRef] [PubMed]
- Bolignano, D.; Della Torre, A.; Lacquaniti, A.; Costantino, G.; Fries, W.; Buemi, M. Neutrophil gelatinase-associated lipocalin levels in patients with Crohn disease undergoing treatment with infliximab. J. Investig. Med. 2010, 58, 569–571. [Google Scholar] [CrossRef]
- Guihot, G.; Guimbaud, R.; Bertrand, V.; Narcy-Lambare, B.; Couturier, D.; Duée, P.-H.; Chaussade, S.; Blachier, F. Inducible nitric oxide synthase activity in colon biopsies from inflammatory areas: Correlation with inflammation intensity in patients with ulcerative colitis but not with Crohn’s disease. Amino Acids 2000, 18, 229–237. [Google Scholar] [CrossRef]
- Alotaibi, B.; Mokhtar, F.A.; El-Masry, T.A.; Elekhnawy, E.; Mostafa, S.A.; Abdelkader, D.H.; Elharty, M.E.; Saleh, A.; Negm, W.A. Antimicrobial activity of brassica rapa L. Flowers extract on gastrointestinal tract infections and antiulcer potential against Indomethacin-Induced gastric ulcer in rats supported by metabolomics profiling. J. Inflamm. Res. 2021, 14, 7411. [Google Scholar] [CrossRef]
- Senhaji, N.; Nadifi, S.; Zaid, Y.; Serrano, A.; Rodriguez, D.A.L.; Serbati, N.; Karkouri, M.; Badre, W.; Martín, J. Polymorphisms in oxidative pathway related genes and susceptibility to inflammatory bowel disease. World J. Gastroenterol. 2017, 23, 8300. [Google Scholar] [CrossRef]
- Martín, M.C.; Martinez, A.; Mendoza, J.L.; Taxonera, C.; Díaz-Rubio, M.; Fernández-Arquero, M.; de la Concha, E.G.; Urcelay, E. Influence of the inducible nitric oxide synthase gene (NOS2A) on inflammatory bowel disease susceptibility. Immunogenetics 2007, 59, 833–837. [Google Scholar] [CrossRef]
- Luther, J.; Gala, M.; Patel, S.J.; Dave, M.; Borren, N.; Xavier, R.J.; Ananthakrishnan, A.N. Loss of response to anti-tumor necrosis factor alpha therapy in Crohn’s disease is not associated with emergence of novel inflammatory pathways. Dig. Dis. Sci. 2018, 63, 738–745. [Google Scholar] [CrossRef]
- Kjellev, S. The trefoil factor family–small peptides with multiple functionalities. Cell. Mol. Life Sci. 2009, 66, 1350–1369. [Google Scholar] [CrossRef]
- Aihara, E.; Engevik, K.A.; Montrose, M.H. Trefoil factor peptides and gastrointestinal function. Annu. Rev. Physiol. 2017, 79, 357. [Google Scholar] [CrossRef]
- Longman, R.J.; Poulsom, R.; Corfield, A.P.; Warren, B.F.; Wright, N.A.; Thomas, M.G. Alterations in the composition of the supramucosal defense barrier in relation to disease severity of ulcerative colitis. J. Histochem. Cytochem. 2006, 54, 1335–1348. [Google Scholar] [CrossRef]
- Shaoul, R.; Okada, Y.; Cutz, E.; Marcon, M.A. Colonic expression of MUC2, MUC5AC, and TFF1 in inflammatory bowel disease in children. J. Pediatr. Gastroenterol. Nutr. 2004, 38, 488–493. [Google Scholar] [CrossRef]
- Vestergaard, E.; Brynskov, J.; Ejskjaer, K.; Clausen, J.; Thim, L.; Nexø, E.; Poulsen, S. Immunoassays of human trefoil factors 1 and 2: Measured on serum from patients with inflammatory bowel disease. Scand. J. Clin. Lab. Investig. 2004, 64, 146–156. [Google Scholar] [CrossRef]
- Grønbæk, H.; Vestergaard, E.M.; Hey, H.; Nielsen, J.N.; Nexø, E. Serum trefoil factors in patients with inflammatory bowel disease. Digestion 2006, 74, 33–39. [Google Scholar] [CrossRef]
- Arijs, I.; De Hertogh, G.; Machiels, K.; Van Steen, K.; Lemaire, K.; Schraenen, A.; Van Lommel, L.; Quintens, R.; Van Assche, G.; Vermeire, S. Mucosal gene expression of cell adhesion molecules, chemokines, and chemokine receptors in patients with inflammatory bowel disease before and after infliximab treatment. Off. J. Am. Coll. Gastroenterol. ACG 2011, 106, 748–761. [Google Scholar] [CrossRef]
- Clamp, J.; Fraser, G.; Read, A. Study of the carbohydrate content of mucus glycoproteins from normal and diseased colons. Clin. Sci. 1981, 61, 229–234. [Google Scholar] [CrossRef]
- Cui, M.; Zhang, M.; Liu, K. Colon-targeted drug delivery of polysaccharide-based nanocarriers for synergistic treatment of inflammatory bowel disease: A review. Carbohydr. Polym. 2021, 272, 118530. [Google Scholar] [CrossRef]
- Khulusi, S.; Hanby, A.; Marrero, J.; Patel, P.; Mendall, M.; Badve, S.; Poulsom, R.; Elia, G.; Wright, N.; Northfield, T. Expression of trefoil peptides pS2 and human spasmolytic polypeptide in gastric metaplasia at the margin of duodenal ulcers. Gut 1995, 37, 205–209. [Google Scholar] [CrossRef]



| IBD Patients | N = 80 |
| Age (mean ± SD) | 46.75 ± 6.648 |
| Sex (%M) | (N = 56) 70% |
| Sex (%F) | (N = 24) 30% |
| Smoking Behavior | |
| Non-smokers | (N = 62) 77.5% |
| Mild | (N = 12) 15% |
| Moderate | (N = 3) 3.75% |
| Severe | (N = 3) 3.75% |
| Type of Disease | |
| (%UC) | (N = 62) 77.5% |
| (%CD) | (N = 18) 22.5% |
| Disease Activity | |
| % Active | (N = 59) 73.75% |
| % Remission | (N = 21) 26.25% |
| Treatment Duration | |
| Drug-free | 23.75% (19 patients) |
| 5-ASA treatment | 48.75% (3–24 months) (31 UC patients & 8 CD patients) |
| Anti-TNF treatment | 27.5% (3–24 months) (12 UC patients & 10 CD patients) |
| Non-IBD subjects | N = 80 |
| Age (mean ± SD) | 49.78 ± 4.352 |
| Sex (%M) | (N = 55) 68.75% |
| Sex (%F) | (N = 25) 31.25% |
| Smoking Behavior | |
| Non-smokers | (N = 70) 87.5% |
| Mild | (N = 5) 6.25% |
| Moderate | (N = 2) 2.5% |
| Severe | (N = 3) 3.75% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alarfaj, S.J.; Mostafa, S.A.; Negm, W.A.; El-Masry, T.A.; Kamal, M.; Elsaeed, M.; El Nakib, A.M. Mucosal Genes Expression in Inflammatory Bowel Disease Patients: New Insights. Pharmaceuticals 2023, 16, 324. https://doi.org/10.3390/ph16020324
Alarfaj SJ, Mostafa SA, Negm WA, El-Masry TA, Kamal M, Elsaeed M, El Nakib AM. Mucosal Genes Expression in Inflammatory Bowel Disease Patients: New Insights. Pharmaceuticals. 2023; 16(2):324. https://doi.org/10.3390/ph16020324
Chicago/Turabian StyleAlarfaj, Sumaiah J., Sally Abdallah Mostafa, Walaa A. Negm, Thanaa A. El-Masry, Marwa Kamal, Mohamed Elsaeed, and Ahmed Mohamed El Nakib. 2023. "Mucosal Genes Expression in Inflammatory Bowel Disease Patients: New Insights" Pharmaceuticals 16, no. 2: 324. https://doi.org/10.3390/ph16020324
APA StyleAlarfaj, S. J., Mostafa, S. A., Negm, W. A., El-Masry, T. A., Kamal, M., Elsaeed, M., & El Nakib, A. M. (2023). Mucosal Genes Expression in Inflammatory Bowel Disease Patients: New Insights. Pharmaceuticals, 16(2), 324. https://doi.org/10.3390/ph16020324









