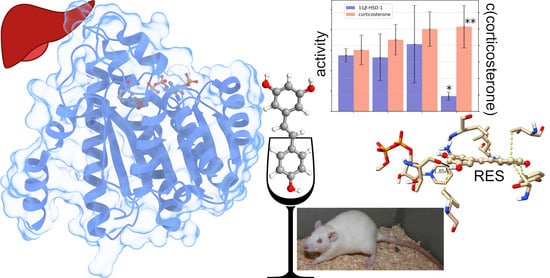
Can Resveratrol Influence the Activity of 11β-Hydroxysteroid Dehydrogenase Type 1? A Combined In Silico and In Vivo Study
Abstract
1. Introduction
2. Results
2.1. In Vivo Results
2.1.1. Effects of Trans-Resveratrol Treatment on Hepatic 11β-HSD-1 Activity and Plasma Corticosterone Levels
2.1.2. Effects of Trans-Resveratrol Treatment on the Elevated plus Maze (EPM) Test Values
2.1.3. Effects of Trans-Resveratrol Treatment on Hepatic 11β-HSD-1 Activity and Plasma Corticosterone Levels in Rats with PTSD
2.1.4. Effects of Trans-Resveratrol Treatment on EPM Test Scores in PTSD Rats
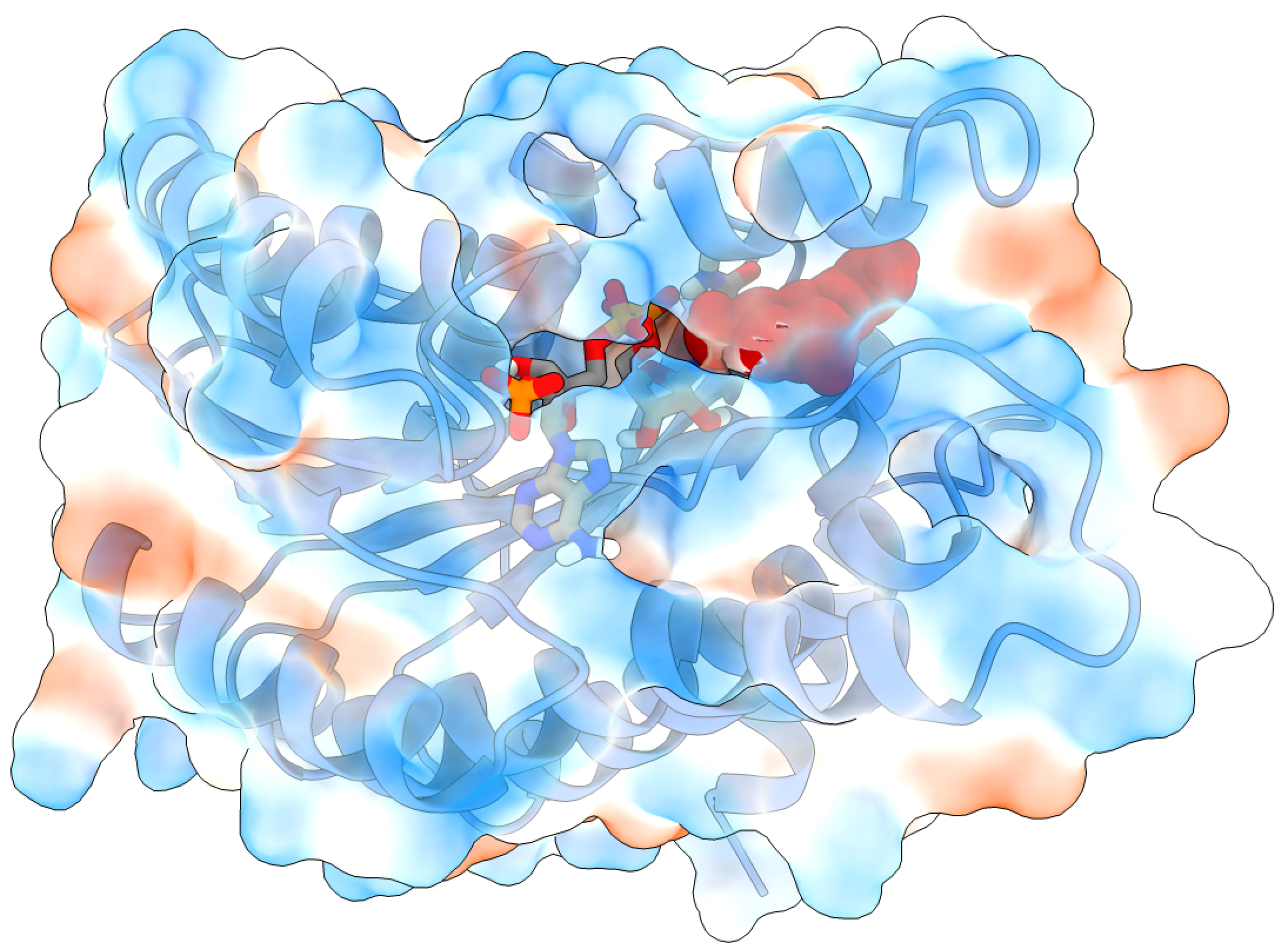
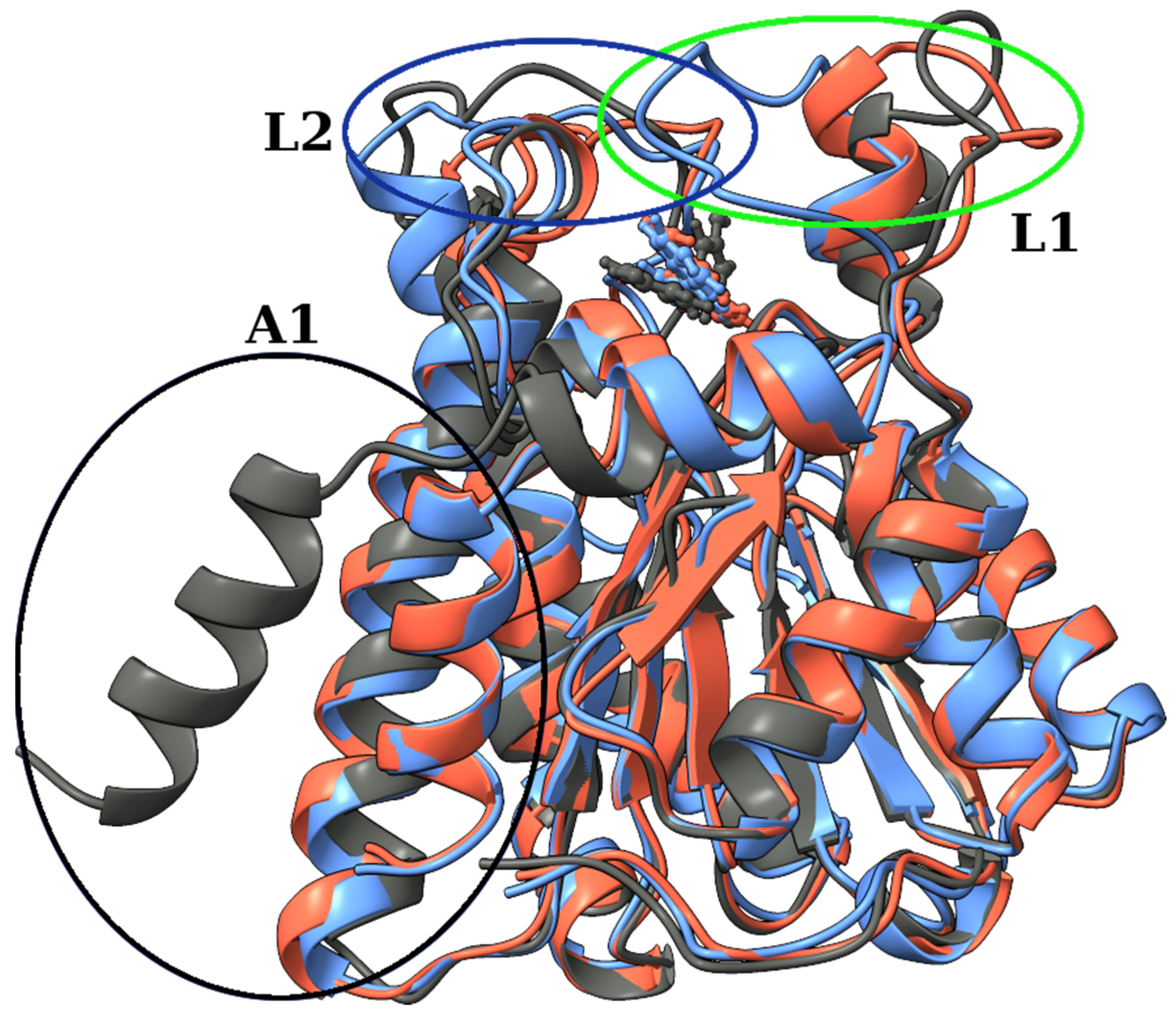
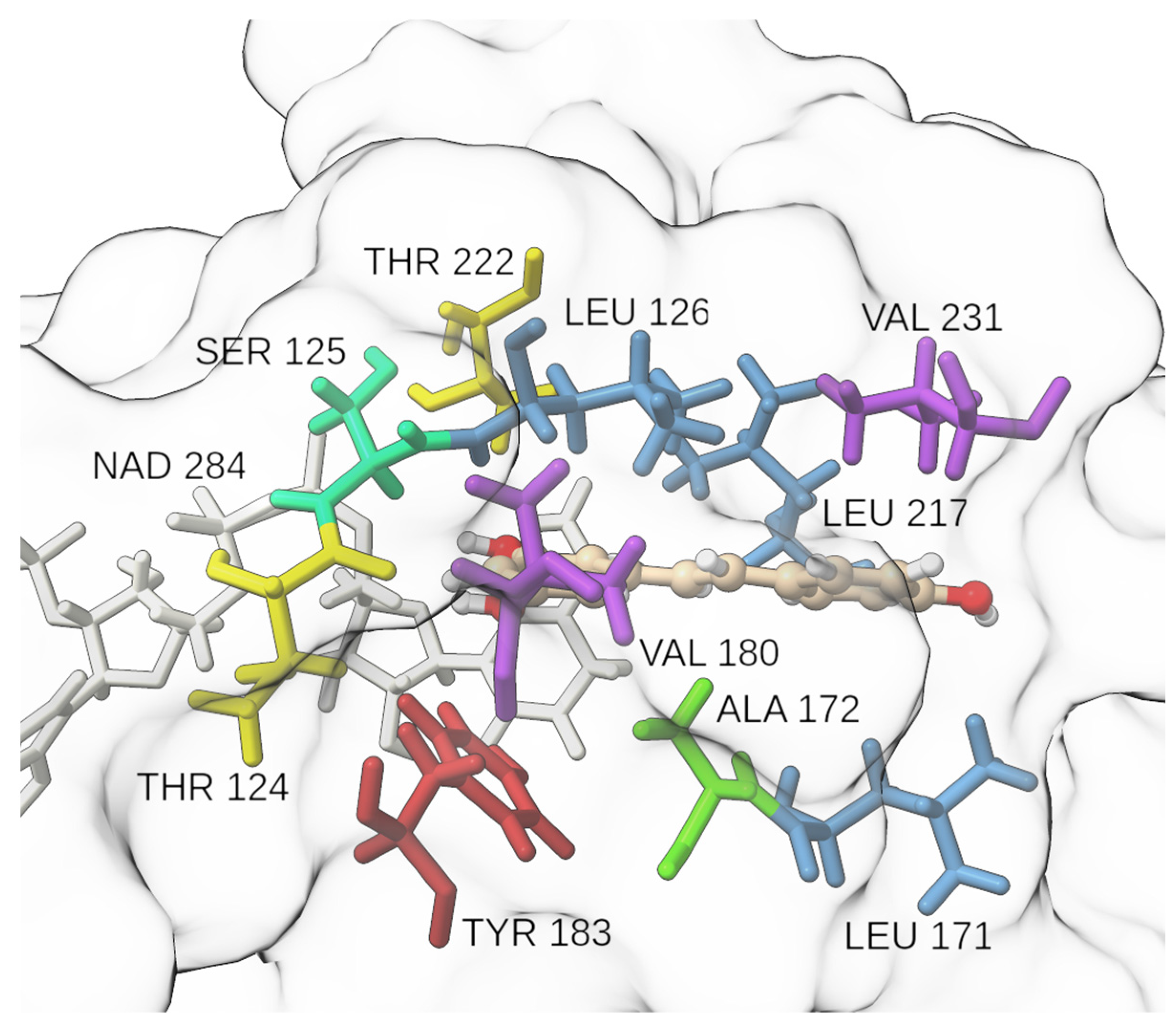
2.2. Molecular Docking and Molecular Dynamics Simulations
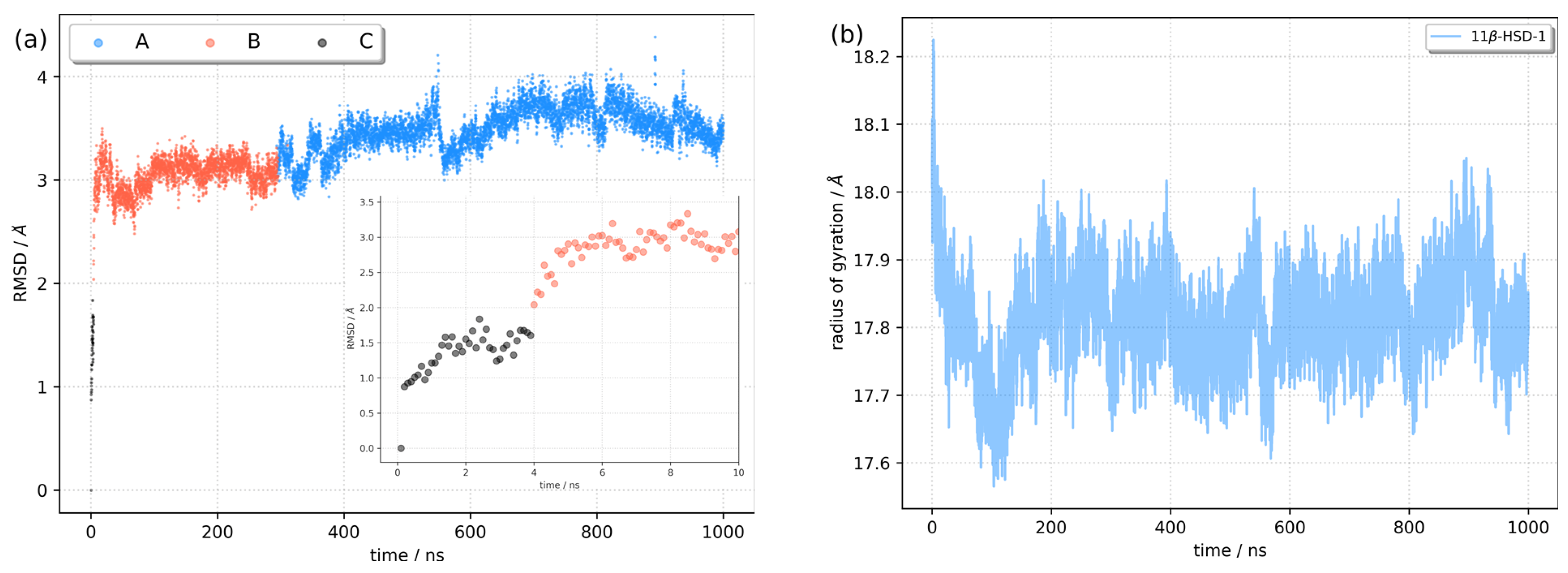
2.3. Identification of Three Conformations
2.4. Binding Free Energy Calculation
3. Discussion
4. Materials and Methods
4.1. Molecular Docking
4.2. Molecular Dynamics Simulations
4.3. Binding Free Energy Calculation
4.4. Dynamics Cross-Correlation Map (DCCM) Analysis
4.5. Cluster Analysis
4.6. Principal Component Analysis (PCA)
4.7. In Vivo Experiments
4.7.1. Protocol No. 1
- Control rats (treated with vehicle only for 10 days; n = 10);
- RES1 (treatment with a dose of 10 mg/kg for 10 days; n = 6);
- RES2 (treatment with a dose of 25 mg/kg for 10 days; n = 10);
- RES3 (treatment with a dose of 40 mg/kg for 10 days; n = 10).
4.7.2. Protocol No. 2
- Control rats (treated with vehicle only for 10 days; n = 10)
- PTSD (rats previously exposed to chronic predator stress followed by a two-week break; n = 10)
- RES + PTSD (an effective dose of resveratrol administered to rats via a tube one hour before the onset of predatory stress; n = 10). The effective dose of resveratrol was determined after protocol № 1 was performed.
4.7.3. Behavioral Assessment
4.7.4. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rana, A.; Samtiya, M.; Dhewa, T.; Mishra, V.; Aluko, R.E. Health Benefits of Polyphenols: A Concise Review. J. Food Biochem. 2022, 46, e14264. [Google Scholar] [CrossRef] [PubMed]
- Kubczak, M.; Szustka, A.; Rogalińska, M. Molecular Targets of Natural Compounds with Anti-Cancer Properties. Int. J. Mol. Sci. 2021, 22, 13659. [Google Scholar] [CrossRef] [PubMed]
- Kalliora, C.; Kyriazis, I.D.; Oka, S.; Lieu, M.J.; Yue, Y.; Area-Gomez, E.; Pol, C.J.; Tian, Y.; Mizushima, W.; Chin, A.; et al. Dual PPARα/γ Activation InhibitsSIRT1-PGC1α Axis and Causes Cardiac Dysfunction. JCI Insight 2019, 4, e129556. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.-S.; Hsu, Y.-A.; Lin, C.-H.; Wang, Y.-C.; Lin, E.-S.; Chang, C.-Y.; Chen, J.J.-Y.; Wu, M.-Y.; Lin, H.-J.; Wan, L. Fallopia Japonica and Prunella Vulgaris Inhibit Myopia Progression by Suppressing AKT and NFκB Mediated Inflammatory Reactions. BMC Complement. Med. Ther. 2022, 22, 271. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Zheng, H.; Fu, Y.; Gu, Y.; Zou, H.; Yuan, Y.; Gu, J.; Liu, Z.; Bian, J. Role of PI3K/Akt-Mediated Nrf2/HO-1 Signaling Pathway in Resveratrol Alleviation of Zearalenone-Induced Oxidative Stress and Apoptosis in TM4 Cells. Toxins 2022, 14, 733. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Ju, Y.; Yan, Z.; Ji, M.; Li, J.; Wu, Q.; Yang, M.; Sun, G. Resveratrol Attenuates Autophagy and Inflammation after Traumatic Brain Injury by Activation of PI3K/Akt/MTOR Pathway in Rats. Folia Neuropathol. 2022, 60, 153–164. [Google Scholar] [CrossRef] [PubMed]
- Jang, J.Y.; Im, E.; Kim, N.D. Mechanism of Resveratrol-Induced Programmed Cell Death and New Drug Discovery against Cancer: A Review. Int. J. Mol. Sci. 2022, 23, 13689. [Google Scholar] [CrossRef]
- Iacobini, C.; Vitale, M.; Pugliese, G.; Menini, S. Normalizing HIF-1α Signaling Improves Cellular Glucose Metabolism and Blocks the Pathological Pathways of Hyperglycemic Damage. Biomedicines 2021, 9, 1139. [Google Scholar] [CrossRef]
- Soltani, D.; Azizi, B.; Rahimi, R.; Talasaz, A.H.; Rezaeizadeh, H.; Vasheghani-Farahani, A. Mechanism-Based Targeting of Cardiac Arrhythmias by Phytochemicals and Medicinal Herbs: A Comprehensive Review of Preclinical and Clinical Evidence. Front. Cardiovasc. Med. 2022, 9, 990063. [Google Scholar] [CrossRef]
- García-Martínez, B.I.; Ruiz-Ramos, M.; Pedraza-Chaverri, J.; Santiago-Osorio, E.; Mendoza-Núñez, V.M. Influence of Age and Dose on the Effect of Resveratrol for Glycemic Control in Type 2 Diabetes Mellitus: Systematic Review and Meta-Analysis. Molecules 2022, 27, 5232. [Google Scholar] [CrossRef]
- Mongioì, L.M.; la Vignera, S.; Cannarella, R.; Cimino, L.; Compagnone, M.; Condorelli, R.A.; Calogero, A.E. The Role of Resveratrol Administration in Human Obesity. Int. J. Mol. Sci. 2021, 22, 4362. [Google Scholar] [CrossRef] [PubMed]
- Clarke, J.O.; Mullin, G.E. A Review of Complementary and Alternative Approaches to Immunomodulation. Nutr. Clin. Pract. 2008, 23, 49–62. [Google Scholar] [CrossRef] [PubMed]
- Athar, M.; Back, J.H.; Kopelovich, L.; Bickers, D.R.; Kim, A.L. Multiple Molecular Targets of Resveratrol: Anti-Carcinogenic Mechanisms. Arch. Biochem. Biophys. 2009, 486, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Islam, F.; Nafady, M.H.; Islam, M.R.; Saha, S.; Rashid, S.; Akter, A.; Or-Rashid, M.H.; Akhtar, M.F.; Perveen, A.; Ashraf, M.G.; et al. Resveratrol and Neuroprotection: An Insight into Prospective Therapeutic Approaches against Alzheimer’s Disease from Bench to Bedside. Mol. Neurobiol. 2022, 59, 4384–4404. [Google Scholar] [CrossRef] [PubMed]
- Moore, A.; Beidler, J.; Hong, M. Resveratrol and Depression in Animal Models: A Systematic Review of the Biological Mechanisms. Molecules 2018, 23, 2197. [Google Scholar] [CrossRef]
- Hsu, M.-H.; Chang, K.-A.; Chen, Y.-C.; Lin, I.-C.; Sheen, J.-M.; Huang, L.-T. Resveratrol Prevented Spatial Deficits and Rescued Disarrayed Hippocampus Asymmetric Dimethylarginine and Brain-Derived Neurotrophic Factor Levels in Young Rats with Increased Circulating Asymmetric Dimethylarginine. Neuroreport 2021, 32, 1091–1099. [Google Scholar] [CrossRef]
- Leuner, B.; Shors, T.J. Stress, Anxiety, and Dendritic Spines: What Are the Connections? Neuroscience 2013, 251, 108–119. [Google Scholar] [CrossRef]
- Domitrovic Spudic, S.; Nikolac Perkovic, M.; Uzun, S.; Nedic Erjavec, G.; Kozumplik, O.; Svob Strac, D.; Mimica, N.; Pivac, N. Reduced Plasma BDNF Concentration and Cognitive Decline in Veterans with PTSD. Psychiatry Res. 2022, 316, 114772. [Google Scholar] [CrossRef]
- Yehuda, R.; Seckl, J. Minireview: Stress-Related Psychiatric Disorders with Low Cortisol Levels: A Metabolic Hypothesis. Endocrinology 2011, 152, 4496–4503. [Google Scholar] [CrossRef]
- Chapman, K.; Holmes, M.; Seckl, J. 11β-Hydroxysteroid Dehydrogenases: Intracellular Gate-Keepers of Tissue Glucocorticoid Action. Physiol. Rev. 2013, 93, 1139–1206. [Google Scholar] [CrossRef]
- Seckl, J.R. 11β-Hydroxysteroid Dehydrogenases: Changing Glucocorticoid Action. Curr. Opin. Pharmacol. 2004, 4, 597–602. [Google Scholar] [CrossRef] [PubMed]
- Wyrwoll, C.S.; Holmes, M.C.; Seckl, J.R. 11β-Hydroxysteroid Dehydrogenases and the Brain: From Zero to Hero, a Decade of Progress. Front. Neuroendocrinol. 2011, 32, 265–286. [Google Scholar] [CrossRef]
- Bhatt, S.; Nabulsi, N.B.; Li, S.; Cai, Z.; Matuskey, D.; Bini, J.; Najafzadeh, S.; Kapinos, M.; Ropchan, J.R.; Carson, R.E.; et al. First In-Human PET Study and Kinetic Evaluation of [18F]AS2471907 for Imaging 11β-Hydroxysteroid Dehydrogenase Type 1. J. Cereb. Blood Flow Metab. 2020, 40, 695–704. [Google Scholar] [CrossRef] [PubMed]
- Tseilikman, V.; Lapshin, M.; Klebanov, I.; Chrousos, G.; Vasilieva, M.; Pashkov, A.; Fedotova, J.; Tseilikman, D.; Shatilov, V.; Manukhina, E.; et al. The Link between Activities of Hepatic 11beta-Hydroxysteroid Dehydrogenase-1 and Monoamine Oxidase-A in the Brain Following Repeated Predator Stress: Focus on Heightened Anxiety. Int. J. Mol. Sci. 2022, 23, 4881. [Google Scholar] [CrossRef]
- Wheelan, N.; Seckl, J.R.; Yau, J.L.W. 11β-Hydroxysteroid Dehydrogenase 1 Deficiency Prevents PTSD-like Memory in Young Adult Mice. Psychoneuroendocrinology 2022, 146, 105945. [Google Scholar] [CrossRef] [PubMed]
- Kabsch, W.; Sander, C. Dictionary of Protein Secondary Structure: Pattern Recognition of Hydrogen-Bonded and Geometrical Features. Biopolymers 1983, 22, 2577–2637. [Google Scholar] [CrossRef] [PubMed]
- Novak, J.; Rimac, H.; Kandagalla, S.; Grishina, M.A.; Potemkin, V.A. Can Natural Products Stop the SARS-CoV-2 Virus? A Docking and Molecular Dynamics Study of a Natural Product Database. Future Med. Chem. 2021, 13, 363–378. [Google Scholar] [CrossRef]
- Kandagalla, S.; Novak, J.; Shekarappa, S.B.; Grishina, M.A.; Potemkin, V.A.; Kumbar, B. Exploring Potential Inhibitors against Kyasanur Forest Disease by Utilizing Molecular Dynamics Simulations and Ensemble Docking. J. Biomol. Struct. Dyn. 2022, 40, 13547–13563. [Google Scholar] [CrossRef]
- Singh, A.K.; Novak, J.; Kumar, A.; Singh, H.; Thareja, S.; Pathak, P.; Grishina, M.; Verma, A.; Yadav, J.P.; Khalilullah, H.; et al. Gaussian Field-Based 3D-QSAR and Molecular Simulation Studies to Design Potent Pyrimidine–Sulfonamide Hybrids as Selective BRAFV600E Inhibitors. RSC Adv. 2022, 12, 30181–30200. [Google Scholar] [CrossRef]
- Tagawa, N.; Kubota, S.; Kato, I.; Kobayashi, Y. Resveratrol Inhibits 11β-Hydroxysteroid Dehydrogenase Type 1 Activity in Rat Adipose Microsomes. J. Endocrinol. 2013, 218, 311–320. [Google Scholar] [CrossRef]
- Yu, Y.; Wang, R.; Chen, C.; Du, X.; Ruan, L.; Sun, J.; Li, J.; Zhang, L.; O’Donnell, J.M.; Pan, J.; et al. Antidepressant-like Effect of Trans-Resveratrol in Chronic Stress Model: Behavioral and Neurochemical Evidences. J. Psychiatr. Res. 2013, 47, 315–322. [Google Scholar] [CrossRef] [PubMed]
- Hare, B.D.; Ghosal, S.; Duman, R.S. Rapid Acting Antidepressants in Chronic Stress Models: Molecular and Cellular Mechanisms. Chronic Stress 2017, 1, 247054701769731. [Google Scholar] [CrossRef]
- Tseilikman, V.; Dremencov, E.; Tseilikman, O.; Pavlovicova, M.; Lacinova, L.; Jezova, D. Role of Glucocorticoid- and Monoamine-Metabolizing Enzymes in Stress-Related Psychopathological Processes. Stress 2020, 23, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem in 2021: New Data Content and Improved Web Interfaces. Nucleic Acids Res. 2021, 49, D1388–D1395. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. J. Comput Chem 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Wang, J.; Wang, W.; Kollman, P.A.; Case, D.A. Automatic Atom Type and Bond Type Perception in Molecular Mechanical Calculations. J. Mol. Graph. Model. 2006, 25, 247–260. [Google Scholar] [CrossRef]
- Wang, J.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A.; Case, D.A. Development and Testing of a General Amber Force Field. J. Comput. Chem. 2004, 25, 1157–1174. [Google Scholar] [CrossRef]
- Tian, C.; Kasavajhala, K.; Belfon, K.A.A.; Raguette, L.; Huang, H.; Migues, A.N.; Bickel, J.; Wang, Y.; Pincay, J.; Wu, Q.; et al. Ff19SB: Amino-Acid-Specific Protein Backbone Parameters Trained against Quantum Mechanics Energy Surfaces in Solution. J. Chem. Theory Comput. 2020, 16, 528–552. [Google Scholar] [CrossRef]
- Dolinsky, T.J.; Czodrowski, P.; Li, H.; Nielsen, J.E.; Jensen, J.H.; Klebe, G.; Baker, N.A. PDB2PQR: Expanding and Upgrading Automated Preparation of Biomolecular Structures for Molecular Simulations. Nucleic Acids Res. 2007, 35, W522–W525. [Google Scholar] [CrossRef] [PubMed]
- Machado, M.R.; Pantano, S. Split the Charge Difference in Two! A Rule of Thumb for Adding Proper Amounts of Ions in MD Simulations. J. Chem. Theory Comput. 2020, 16, 1367–1372. [Google Scholar] [CrossRef] [PubMed]
- Andersen, H.C. Rattle: A “Velocity” Version of the Shake Algorithm for Molecular Dynamics Calculations. J. Comput. Phys. 1983, 52, 24–34. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle Mesh Ewald: An N⋅log(N) Method for Ewald Sums in Large Systems. J. Chem. Phys. 1993, 98, 10089. [Google Scholar] [CrossRef]
- Case, D.A.; Betz, R.M.; Cerutti, D.S.; Cheatham, T.E., III; Darden, T.A.; Duke, R.E.; Giese, T.J.; Gohlke, H.; Goetz, A.W.; Homeyer, N.; et al. Amber 2016; University of California: San Francisco, CA, USA, 2016. [Google Scholar]
- Genheden, S.; Ryde, U. The MM/PBSA and MM/GBSA Methods to Estimate Ligand-Binding Affinities. Expert Opin. Drug Discov. 2015, 10, 449–461. [Google Scholar] [CrossRef]
- Rastelli, G.; Rio, A.D.; Degliesposti, G.; Sgobba, M. Fast and Accurate Predictions of Binding Free Energies Using MM-PBSA and MM-GBSA. J. Comput. Chem. 2010, 31, 797–810. [Google Scholar] [CrossRef]
- Hünenberger, P.H.; Mark, A.E.; van Gunsteren, W.F. Fluctuation and Cross-Correlation Analysis of Protein Motions Observed in Nanosecond Molecular Dynamics Simulations. J. Mol. Biol. 1995, 252, 492–503. [Google Scholar] [CrossRef]
- McCammon, J.A. Protein Dynamics. Rep. Prog. Phys. 1984, 47, 1–46. [Google Scholar] [CrossRef]
- Roe, D.R.; Cheatham, T.E. PTRAJ and CPPTRAJ: Software for Processing and Analysis of Molecular Dynamics Trajectory Data. J. Chem. Theory Comput. 2013, 9, 3084–3095. [Google Scholar] [CrossRef]
- Pedregosa, F.; Grisel, O.; Weiss, R.; Passos, A.; Brucher, M.; Varoquax, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; et al. Scikit-Learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Li, G.; Wang, G.; Shi, J.; Xie, X.; Fei, N.; Chen, L.; Liu, N.; Yang, M.; Pan, J.; Huang, W.; et al. Trans-Resveratrol Ameliorates Anxiety-like Behaviors and Fear Memory Deficits in a Rat Model of Post-Traumatic Stress Disorder. Neuropharmacology 2018, 133, 181–188. [Google Scholar] [CrossRef] [PubMed]
- Cohen, H.; Zohar, J. An Animal Model of Posttraumatic Stress Disorder: The Use of Cut-Off Behavioral Criteria. Ann. N. Y. Acad. Sci. 2004, 1032, 167–178. [Google Scholar] [CrossRef] [PubMed]
- Lazuko, S.S.; Kuzhel, O.P.; Belyaeva, L.E.; Manukhina, E.B.; Fred Downey, H.; Tseilikman, O.B.; Komelkova, M.V.; Tseilikman, V.E. Posttraumatic Stress Disorder Disturbs Coronary Tone and Its Regulatory Mechanisms. Cell Mol. Neurobiol. 2018, 38, 209–217. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; Jabłońska, J.; Pravda, L.; Vařeková, R.S.; Thornton, J.M. PDBsum: Structural Summaries of PDB Entries. Protein Sci. 2018, 27, 129–134. [Google Scholar] [CrossRef] [PubMed]




| Variable | Control | RES1 (10 mg/kg) | RES2 (25 mg/kg) | RES3 (40 mg/kg) |
|---|---|---|---|---|
| N | 10 | 6 | 10 | 10 |
| Time spent in closed arms/s | 478.7 ± 28.37 | 471.5 ± 30.11 | 495 ± 29.59 | 342.1 ± 108.72 ** |
| Time spent in closed arms/s | 121.3 ± 28.37 | 128.5 ± 30.11 | 105 ± 29.59 | 257.9 ± 108.72 ** |
| Entries into closed arms | 9.4 ± 3.5 | 6.6 ± 1.86 | 12.5 ± 5.89 | 4.6 ± 2.5 ** |
| Entries into open arms | 5.7 ± 1.6 | 4.5 ± 1.22 | 6.6 ± 3.65 | 5.8 ± 2.9 |
| AI | 0.7 ± 0.08 | 0.71 ± 0.05 | 0.77 ± 0.09 | 0.51 ± 0.09 *** |
| Variable | Control | PTSD | PTSD + RES (40 mg/kg) |
|---|---|---|---|
| n | 10 | 10 | 10 |
| Time spent in closed arms/s | 461.4 ± 37.4 | 554 ± 25.44 *** | 451.4 ± 36.9 ## |
| Time spent in closed arms/s | 138.5 ± 37.4 | 46 ± 25.44 *** | 148.6 ± 36.9 ## |
| Entries into closed arms | 7.00 ± 4 | 5.5 ± 3.3 * | 13.3 ± 7.7 ## |
| Entries into open arms | 4.1 ± 1.5 | 2.2 ± 1.9 | 8.3 ± 6.1 ## |
| AI | 0.68 ± 0.09 | 0.83 ± 0.06 *** | 0.69 ± 0.05 ## |
| Cluster | Conformation | Cluster Population | d a | csd b | RMSD against A |
|---|---|---|---|---|---|
| 1 | A | 0.705 | 2.253 | 0.251 | 0 |
| 2 | B | 0.291 | 2.524 | 0.425 | 2.858 |
| 3 | C | 0.004 | 1.857 | 0.236 | 3.816 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Novak, J.; Tseilikman, V.E.; Tseilikman, O.B.; Lazuko, S.S.; Belyeva, L.E.; Rahmani, A.; Fedotova, J. Can Resveratrol Influence the Activity of 11β-Hydroxysteroid Dehydrogenase Type 1? A Combined In Silico and In Vivo Study. Pharmaceuticals 2023, 16, 251. https://doi.org/10.3390/ph16020251
Novak J, Tseilikman VE, Tseilikman OB, Lazuko SS, Belyeva LE, Rahmani A, Fedotova J. Can Resveratrol Influence the Activity of 11β-Hydroxysteroid Dehydrogenase Type 1? A Combined In Silico and In Vivo Study. Pharmaceuticals. 2023; 16(2):251. https://doi.org/10.3390/ph16020251
Chicago/Turabian StyleNovak, Jurica, Vadim E. Tseilikman, Olga B. Tseilikman, Svetlana S. Lazuko, Lyudmila E. Belyeva, Azam Rahmani, and Julia Fedotova. 2023. "Can Resveratrol Influence the Activity of 11β-Hydroxysteroid Dehydrogenase Type 1? A Combined In Silico and In Vivo Study" Pharmaceuticals 16, no. 2: 251. https://doi.org/10.3390/ph16020251
APA StyleNovak, J., Tseilikman, V. E., Tseilikman, O. B., Lazuko, S. S., Belyeva, L. E., Rahmani, A., & Fedotova, J. (2023). Can Resveratrol Influence the Activity of 11β-Hydroxysteroid Dehydrogenase Type 1? A Combined In Silico and In Vivo Study. Pharmaceuticals, 16(2), 251. https://doi.org/10.3390/ph16020251