Anti-Inflammatory Activity of the Constituents from the Leaves of Perilla frutescens var. acuta
Abstract
:1. Introduction
2. Results
2.1. Isolation of the Compounds
2.2. In Silico Docking Simulation
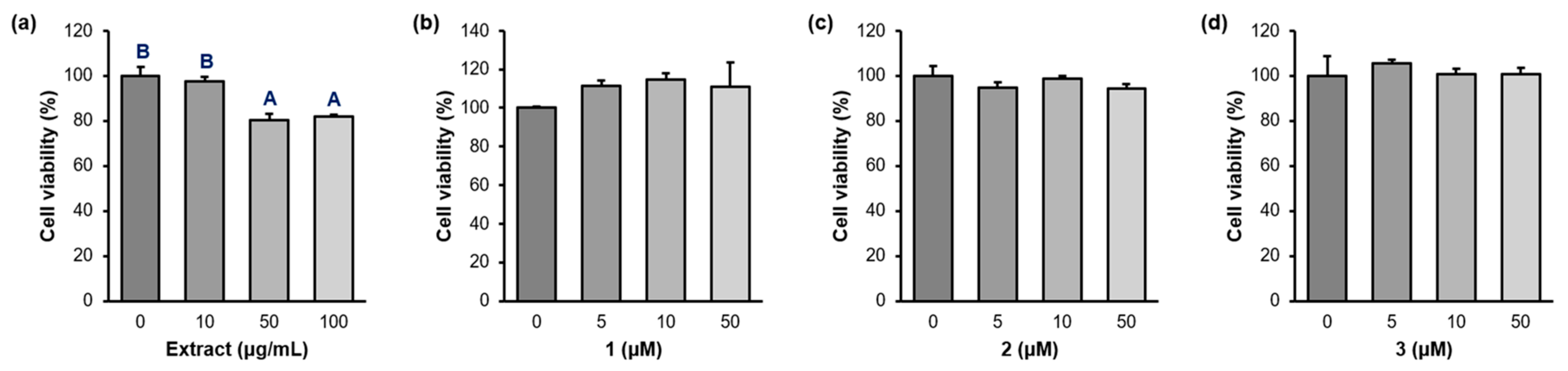
2.3. Cell Viability
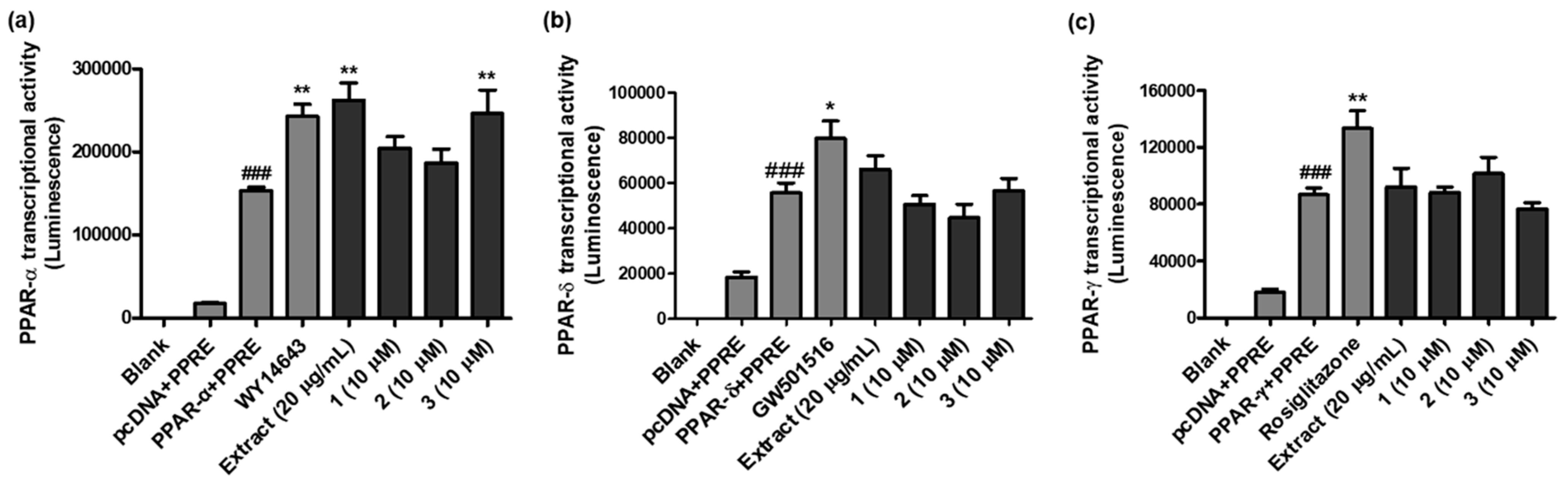
2.4. PPAR-α/δ/γ Transcriptional Activity
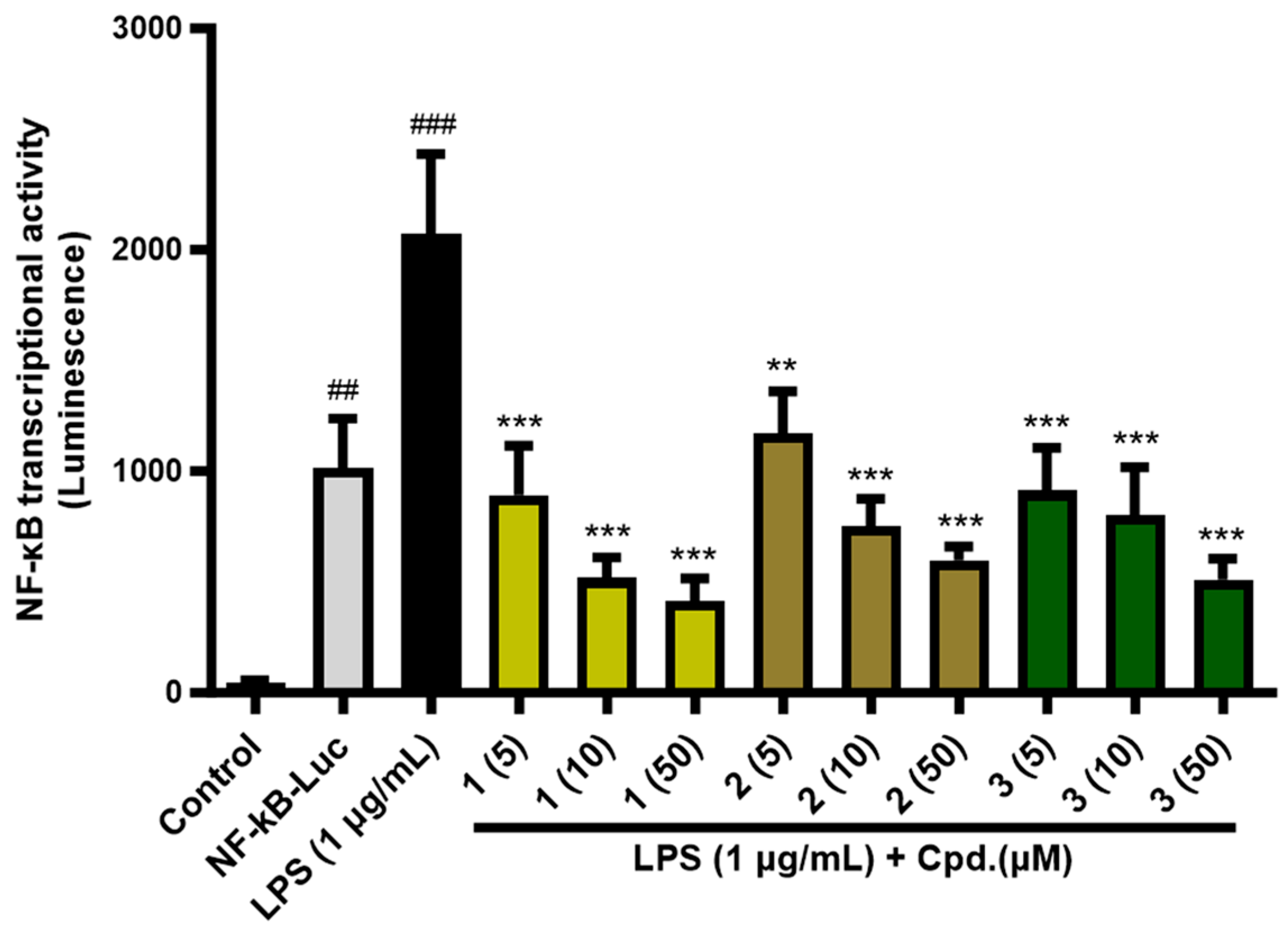
2.5. NF-κB Transcriptional Activity
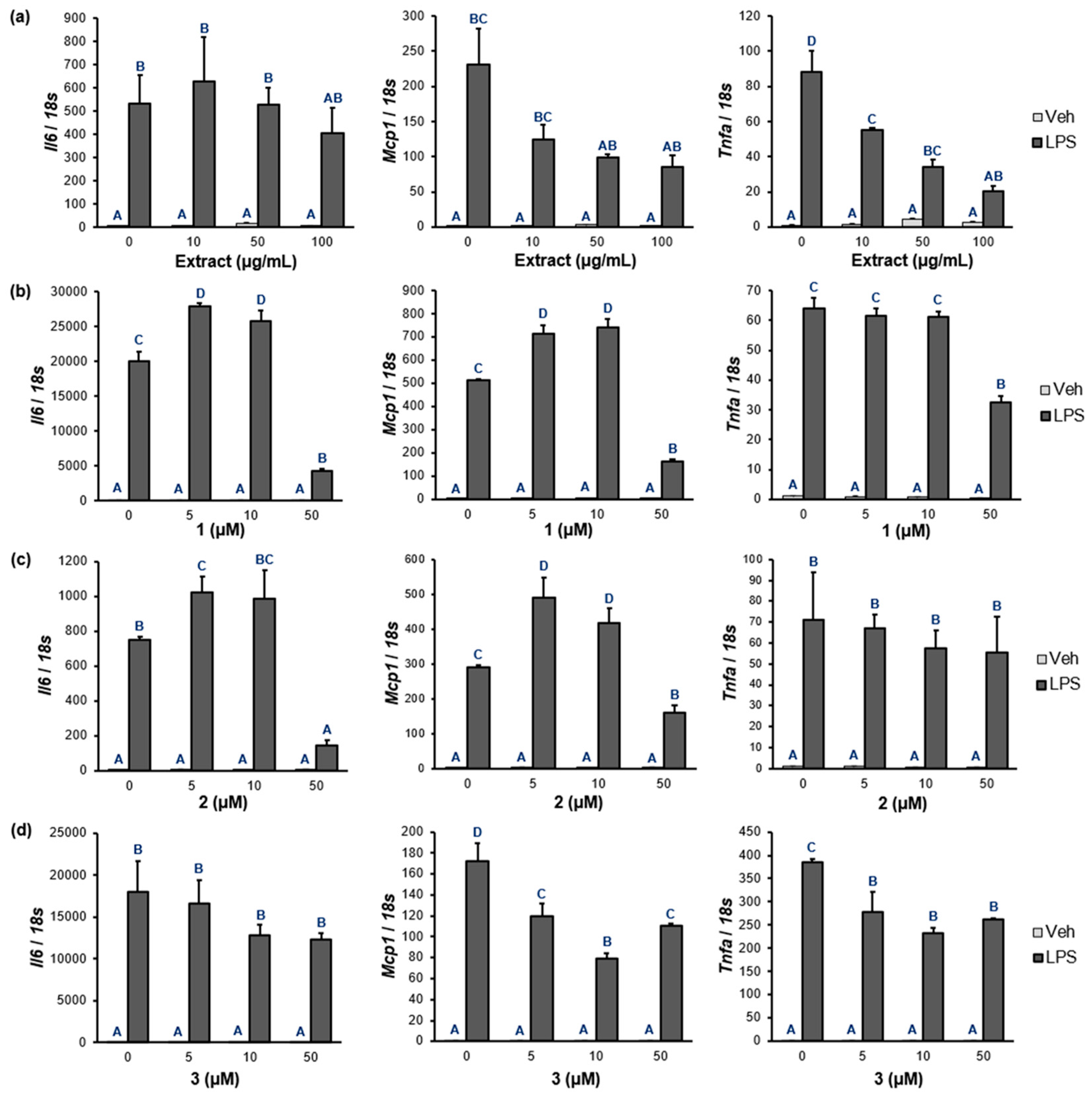
2.6. NF-κB Target Gene Expression
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. General Experimental Procedures
4.3. Extraction and Isolation
4.4. Molecular Docking
4.5. Cell Viability
4.6. PPAR and NF-κB Transcriptional Activity
4.7. NF-κB Target Gene Expression
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Plants of the World Online. Available online: https://powo.science.kew.org/taxon/urn:lsid:ipni.org:names:190343-2 (accessed on 23 August 2023).
- Ahmed, H.M. Ethnomedicinal, phytochemical, and pharmacological investigations of Perilla frutescens (L.) Britt. Molecules 2018, 24, 102. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Qiu, J.-F.; Ma, L.-J.; Hu, Y.-J.; Li, P.; Wan, J.-B. Phytochemical and phytopharmacological review of Perilla frutescens L.(Labiatae), a traditional edible-medicinal herb in China. Food Chem. Toxicol. 2017, 108, 375–391. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.P.; Lin, C.H.; Chen, Y.C.; Kao, S.H. Anti-inflammatory effects of Perilla frutescens leaf extract on lipopolysaccharide-stimulated Raw 264.7 cells. Mol. Med. Rep. 2014, 10, 1077–1083. [Google Scholar] [CrossRef] [PubMed]
- Ueda, H.; Yamazaki, M. Inhibition of tumor necrosis factor-α production by orally administering a perilla leaf extract. Biosci. Biotechnol. Biochem. 1997, 61, 1292–1295. [Google Scholar] [CrossRef]
- Chen, L.; Deng, H.; Cui, H.; Fang, J.; Zuo, Z.; Deng, J.; Li, Y.; Wang, X.; Zhao, L. Inflammatory responses and inflammation-associated diseases in organs. Oncotarget 2018, 9, 7204. [Google Scholar] [CrossRef]
- Yasin, Y.J.; Banoub, J.A.M.; Husseini, A. GBD 2017 Causes of Death Collaborators. Global, regional, and national age-sex-specific mortality for 282 causes of death in 195 countries and territories, 1980–2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet 2018, 392, 1736–1788. [Google Scholar]
- Taniguchi, K.; Karin, M. NF-κB, inflammation, immunity and cancer: Coming of age. Nat. Rev. Immunol. 2018, 18, 309–324. [Google Scholar] [CrossRef]
- Korbecki, J.; Bobiński, R.; Dutka, M. Self-regulation of the inflammatory response by peroxisome proliferator-activated receptors. Inflamm. Res. 2019, 68, 443–458. [Google Scholar] [CrossRef]
- Ricote, M.; Li, A.C.; Willson, T.M.; Kelly, C.J.; Glass, C.K. The peroxisome proliferator-activated receptor-γ is a negative regulator of macrophage activation. Nature 1998, 391, 79–82. [Google Scholar] [CrossRef]
- Arulselvan, P.; Fard, M.T.; Tan, W.S.; Gothai, S.; Fakurazi, S.; Norhaizan, M.E.; Kumar, S.S. Role of antioxidants and natural products in inflammation. Oxidative Med. Cell. Longev. 2016, 2016, 5276130. [Google Scholar] [CrossRef]
- Zuzarte, M.; Francisco, V.; Neves, B.; Liberal, J.; Cavaleiro, C.; Canhoto, J.; Salgueiro, L.; Cruz, M.T. Lavandula viridis L. Her. essential oil inhibits the inflammatory response in macrophages through blockade of NF-κB signaling cascade. Front. Pharmacol. 2022, 12, 695911. [Google Scholar] [CrossRef]
- Notarte, K.I.R.; Quimque, M.T.J.; Macaranas, I.T.; Khan, A.; Pastrana, A.M.; Villaflores, O.B.; Arturo, H.C.P.; Pilapil, D.Y.H., IV; Tan, S.M.M.; Wei, D.-Q.; et al. Attenuation of lipopolysaccharide-induced inflammatory responses through inhibition of the NF-κB pathway and the increased NRF2 level by a flavonol-enriched n-Butanol fraction from Uvaria alba. ACS Omega 2023, 8, 5377–5392. [Google Scholar] [CrossRef] [PubMed]
- Mues, R. Species specific flavone glucuronides in Elodea species. Biochem. Syst. Ecol. 1983, 11, 261–265. [Google Scholar] [CrossRef]
- Aritomi, M.; Kumori, T.; Kawasaki, T. Cyanogenic glycosides in leaves of Perilla frutescens var. acuta. Phytochemistry 1985, 24, 2438–2439. [Google Scholar] [CrossRef]
- Makino, T.; Furuta, Y.; Fujii, H.; Nakagawa, T.; Wakushima, H.; Saito, K.-i.; Kano, Y. Effect of oral treatment of Perilla frutescens and its constituents on Type-I allergy in mice. Biol. Pharm. Bull. 2001, 24, 1206–1209. [Google Scholar] [CrossRef]
- Jiang, T.; Guo, K.; Liu, L.; Tian, W.; Xie, X.; Wen, S.; Wen, C. Integrated transcriptomic and metabolomic data reveal the flavonoid biosynthesis metabolic pathway in Perilla frutescens (L.) leaves. Sci. Rep. 2020, 10, 16207. [Google Scholar] [CrossRef]
- Aritomi, M. Chemical studies on the constituents of edible plants (Part 1). Phenolic compounds in leaves of Perilla frutescens BRITTON var. acuta KUDO f. viridis MAKINO. J. Home Econ. 1982, 33, 353–359. [Google Scholar] [CrossRef]
- Petersen, M.; Simmonds, M.S. Rosmarinic acid. Phytochemistry 2003, 62, 121–125. [Google Scholar] [CrossRef]
- Ferreira, L.G.; Dos Santos, R.N.; Oliva, G.; Andricopulo, A.D. Molecular docking and structure-based drug design strategies. Molecules 2015, 20, 13384–13421. [Google Scholar] [CrossRef]
- Williams, D.H.; Stephens, E.; O’Brien, D.P.; Zhou, M. Understanding noncovalent interactions: Ligand binding energy and catalytic efficiency from ligand—Induced reductions in motion within receptors and enzymes. Angew. Chem. Int. Ed. 2004, 43, 6596–6616. [Google Scholar] [CrossRef]
- Tanaka, N.; Zhang, X.; Sugiyama, E.; Kono, H.; Horiuchi, A.; Nakajima, T.; Kanbe, H.; Tanaka, E.; Gonzalez, F.J.; Aoyama, T. Eicosapentaenoic acid improves hepatic steatosis independent of PPAR-α activation through inhibition of SREBP-1 maturation in mice. Biochem. Pharmacol. 2010, 80, 1601–1612. [Google Scholar] [CrossRef] [PubMed]
- Xiong, G.; Wu, Z.; Yi, J.; Fu, L.; Yang, Z.; Hsieh, C.; Yin, M.; Zeng, X.; Wu, C.; Lu, A. ADMETlab 2.0: An integrated online platform for accurate and comprehensive predictions of ADMET properties. Nucleic Acids Res. 2021, 49, W5–W14. [Google Scholar] [CrossRef] [PubMed]
- Delerive, P.; De Bosscher, K.; Besnard, S.; Berghe, W.V.; Peters, J.M.; Gonzalez, F.J.; Fruchart, J.-C.; Tedgui, A.; Haegeman, G.; Staels, B. Peroxisome proliferator-activated receptor α negatively regulates the vascular inflammatory gene response by negative cross-talk with transcription factors NF-κB and AP-1. J. Biol. Chem. 1999, 274, 32048–32054. [Google Scholar] [CrossRef] [PubMed]
- Planavila, A.; Iglesias, R.; Giralt, M.; Villarroya, F. Sirt1 acts in association with PPARα to protect the heart from hypertrophy, metabolic dysregulation, and inflammation. Cardiovasc. Res. 2011, 90, 276–284. [Google Scholar] [CrossRef] [PubMed]
- Grabacka, M.; Pierzchalska, M.; Płonka, P.M.; Pierzchalski, P. The role of PPAR alpha in the modulation of innate immunity. Int. J. Mol. Sci. 2021, 22, 10545. [Google Scholar] [CrossRef] [PubMed]
- Barroso, E.; Eyre, E.; Palomer, X.; Vázquez-Carrera, M. The peroxisome proliferator-activated receptor β/δ (PPAR-β/δ) agonist GW501516 prevents TNF-α-induced NF-κB activation in human HaCaT cells by reducing p65 acetylation through AMPK and SIRT1. Biochem. Pharmacol. 2011, 81, 534–543. [Google Scholar] [CrossRef]
- He, X.; Liu, W.; Shi, M.; Yang, Z.; Zhang, X.; Gong, P. Docosahexaenoic acid attenuates LPS-stimulated inflammatory response by regulating the PPAR-γ/NF-κB pathways in primary bovine mammary epithelial cells. Res. Vet. Sci. 2017, 112, 7–12. [Google Scholar] [CrossRef]
- Ramanan, S.; Kooshki, M.; Zhao, W.; Hsu, F.-C.; Robbins, M.E. PPAR-α ligands inhibit radiation-induced microglial inflammatory responses by negatively regulating NF-κB and AP-1 pathways. Free Radical Biol. Med. 2008, 45, 1695–1704. [Google Scholar] [CrossRef]
- Han, J.; Wang, D.; Ye, L.; Li, P.; Hao, W.; Chen, X.; Ma, J.; Wang, B.; Shang, J.; Li, D. Rosmarinic acid protects against inflammation and cardiomyocyte apoptosis during myocardial ischemia/reperfusion injury by activating peroxisome proliferator-activated receptor gamma. Front. Pharmacol. 2017, 8, 456. [Google Scholar] [CrossRef]
- Engels, N.S.; Waltenberger, B.; Michalak, B.; Huynh, L.; Tran, H.; Kiss, A.K.; Stuppner, H. Inhibition of pro-inflammatory functions of human neutrophils by constituents of Melodorum fruticosum leaves. Chem. Biodivers. 2018, 15, e1800269. [Google Scholar] [CrossRef]
- Jalil, J.; Sabandar, C.W.; Ahmat, N.; Jamal, J.A.; Jantan, I.; Aladdin, N.-A.; Muhammad, K.; Buang, F.; Mohamad, H.F.; Sahidin, I. Inhibitory effect of triterpenoids from Dillenia serrata (Dilleniaceae) on prostaglandin E2 production and quantitative HPLC analysis of its koetjapic acid and betulinic acid contents. Molecules 2015, 20, 3206–3220. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.; Li, X.; Fang, N.; Li, P.; Zhang, Z.; Lin, M.; Hou, Q. Perilla leaf extract (PLE) attenuates COPD airway inflammation via the TLR4/Syk/PKC/NF-κB pathway in vivo and in vitro. Front. Pharmacol. 2022, 12, 763624. [Google Scholar] [CrossRef] [PubMed]
- Oh, H.-A.; Park, C.-S.; Ahn, H.-J.; Park, Y.S.; Kim, H.-M. Effect of Perilla frutescens var. acuta Kudo and rosmarinic acid on allergic inflammatory reactions. Exp. Biol. Med. 2011, 236, 99–106. [Google Scholar] [CrossRef]
- Banno, N.; Akihisa, T.; Tokuda, H.; Yasukawa, K.; Higashihara, H.; Ukiya, M.; Watanabe, K.; Kimura, Y.; Hasegawa, J.-i.; Nishino, H. Triterpene acids from the leaves of Perilla frutescens and their anti-inflammatory and antitumor-promoting effects. Biosci. Biotechnol. Biochem. 2004, 68, 85–90. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.; Kim, S.-Y.; Kim, S.-W.; Lee, J.M.; Kim, S.-K.; Park, M.-H.; Kim, K.-H.; Oh, M.; Son, C.-G.; Jung, I.C. Efficacy of Perilla frutescens (L.) Britton var. frutescens extract on mild knee joint pain: A randomized controlled trial. Front. Pharmacol. 2023, 14, 1114410. [Google Scholar]
- Osakabe, N.; Takano, H.; Sanbongi, C.; Yasuda, A.; Yanagisawa, R.; Inoue, K.-i.; Yoshikawa, T. Anti-inflammatory and anti-allergic effect of rosmarinic acid (RA): Inhibition of seasonal allergic rhinoconjunctivitis (SAR) and its mechanism. BioFactors 2004, 21, 127–131. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Jung, E.; Koh, J.; Kim, Y.S.; Park, D. Effect of rosmarinic acid on atopic dermatitis. J. Dermatol. 2008, 35, 768–771. [Google Scholar] [CrossRef]
- Kim, J.; Kang, H.; Choi, H.; Jo, A.; Oh, D.-R.; Kim, Y.; Im, S.; Lee, S.-G.; Jeong, K.-I.; Ryu, G.-C. Aqueous extract of Perilla frutescens var. acuta relaxes the ciliary smooth muscle by increasing NO/cGMP content in vitro and in vivo. Molecules 2018, 23, 1777. [Google Scholar]
- Choi, C.Y.; Kim, J.Y.; Kang, H.W.; Jo, A.; Oh, D.-R.; Kim, Y.J.; Im, S.J.; Lee, S.G.; Lee, G.O.; Park, S.Y.; et al. Pharmaceutical Composition for Alleviating Eye Fatigue, Containing, as Active Ingredients, Luteolin-7-O-diglucuronide and Apigenin-7-O-diglucuronide Isolated from Perilla frutescens (L.) Britton var. acuta (thunb.) Kudo Leaf Extract. US Patent US-2023-0020109-A1, 19 January 2023. [Google Scholar]
- Liu, T.; Zhang, L.; Joo, D.; Sun, S.-C. NF-κB signaling in inflammation. Signal Transduct. Target. Ther. 2017, 2, 1–9. [Google Scholar] [CrossRef]
- Li, J.; Ma, J.; Wang, K.S.; Mi, C.; Wang, Z.; Piao, L.X.; Xu, G.H.; Li, X.; Lee, J.J.; Jin, X. Baicalein inhibits TNF-α-induced NF-κB activation and expression of NF-κB-regulated target gene products. Oncol. Rep. 2016, 36, 2771–2776. [Google Scholar] [CrossRef]
- Rajagopal, P.; Jayaraman, S.; Jh, S.F.; Radhakrishnan, S.; Laxman, P.A.; Muthaiah, V.P.K.; Tripathi, S.C.; Gugapriya, T.; Tarnekar, A.M.; Muthiyan, G.G. Molecular docking analysis of PPAR-γ with compounds from Ocimum tenuiflorum. Bioinformation 2021, 17, 928. [Google Scholar] [CrossRef]
- Vasileva, L.V.; Savova, M.S.; Tews, D.; Wabitsch, M.; Georgiev, M.I. Rosmarinic acid attenuates obesity and obesity-related inflammation in human adipocytes. Food Chem. Toxicol. 2021, 149, 112002. [Google Scholar] [CrossRef]
- Wang, W.; Lin, Q.; Lin, R.; Zhang, J.; Ren, F.; Zhang, J.; Ji, M.; Li, Y. PPAR-α agonist fenofibrate attenuates TNF-α-induced CD40 expression in 3T3-L1 adipocytes via the SIRT1-dependent signaling pathway. Exp. Cell Res. 2013, 319, 1523–1533. [Google Scholar] [CrossRef]
- Yeung, F.; Hoberg, J.E.; Ramsey, C.S.; Keller, M.D.; Jones, D.R.; Frye, R.A.; Mayo, M.W. Modulation of NF–κB–dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 2004, 23, 2369–2380. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, Y.; Xiao, F.; Liu, Y.; Wang, J.; Gao, H.; Rong, S.; Yao, Y.; Li, J.; Xu, G. The peroxisome proliferator-activated receptor-γ agonist pioglitazone prevents NF-κB activation in cisplatin nephrotoxicity through the reduction of p65 acetylation via the AMPK-SIRT1/p300 pathway. Biochem. Pharmacol. 2016, 101, 100–111. [Google Scholar] [CrossRef]
- Delerive, P.; Gervois, P.; Fruchart, J.-C.; Staels, B. Induction of IκBα expression as a mechanism contributing to the anti-inflammatory activities of peroxisome proliferator-activated receptor-α activators. J. Biol. Chem. 2000, 275, 36703–36707. [Google Scholar]
- Scirpo, R.; Fiorotto, R.; Villani, A.; Amenduni, M.; Spirli, C.; Strazzabosco, M. Stimulation of nuclear receptor peroxisome proliferator-activated receptor-γ limits NF-κB-dependent inflammation in mouse cystic fibrosis biliary epithelium. Hepatology 2015, 62, 1551–1562. [Google Scholar] [CrossRef]
- Stockert, J.; Wolf, A.; Kaddatz, K.; Schnitzer, E.; Finkernagel, F.; Meissner, W.; Müller-Brüsselbach, S.; Kracht, M.; Müller, R. Regulation of TAK1/TAB1-mediated IL-1β signaling by cytoplasmic PPAR-β/δ. PLoS ONE 2013, 8, e63011. [Google Scholar] [CrossRef]
- Pantsar, T.; Poso, A. Binding affinity via docking: Fact and fiction. Molecules 2018, 23, 1899. [Google Scholar] [CrossRef]
- Di Nicola, M.; Cattaneo, A.; Hepgul, N.; Di Forti, M.; Aitchison, K.J.; Janiri, L.; Murray, R.M.; Dazzan, P.; Pariante, C.M.; Mondelli, V. Serum and gene expression profile of cytokines in first-episode psychosis. Brain Behav. Immun. 2013, 31, 90–95. [Google Scholar] [CrossRef]
- Asano, O. Expression of monocyte chemoattractant protein-1 in Kawasaki disease: The anti-inflammatory effect of gamma globulin therapy. Scand. J. Immunol. 2000, 51, 98–103. [Google Scholar] [CrossRef]
- Aung, H.T.; Nikai, T.; Niwa, M.; Takaya, Y. Rosmarinic acid in Argusia argentea inhibits snake venom-induced hemorrhage. J. Nat. Med. 2010, 64, 482–486. [Google Scholar] [CrossRef]

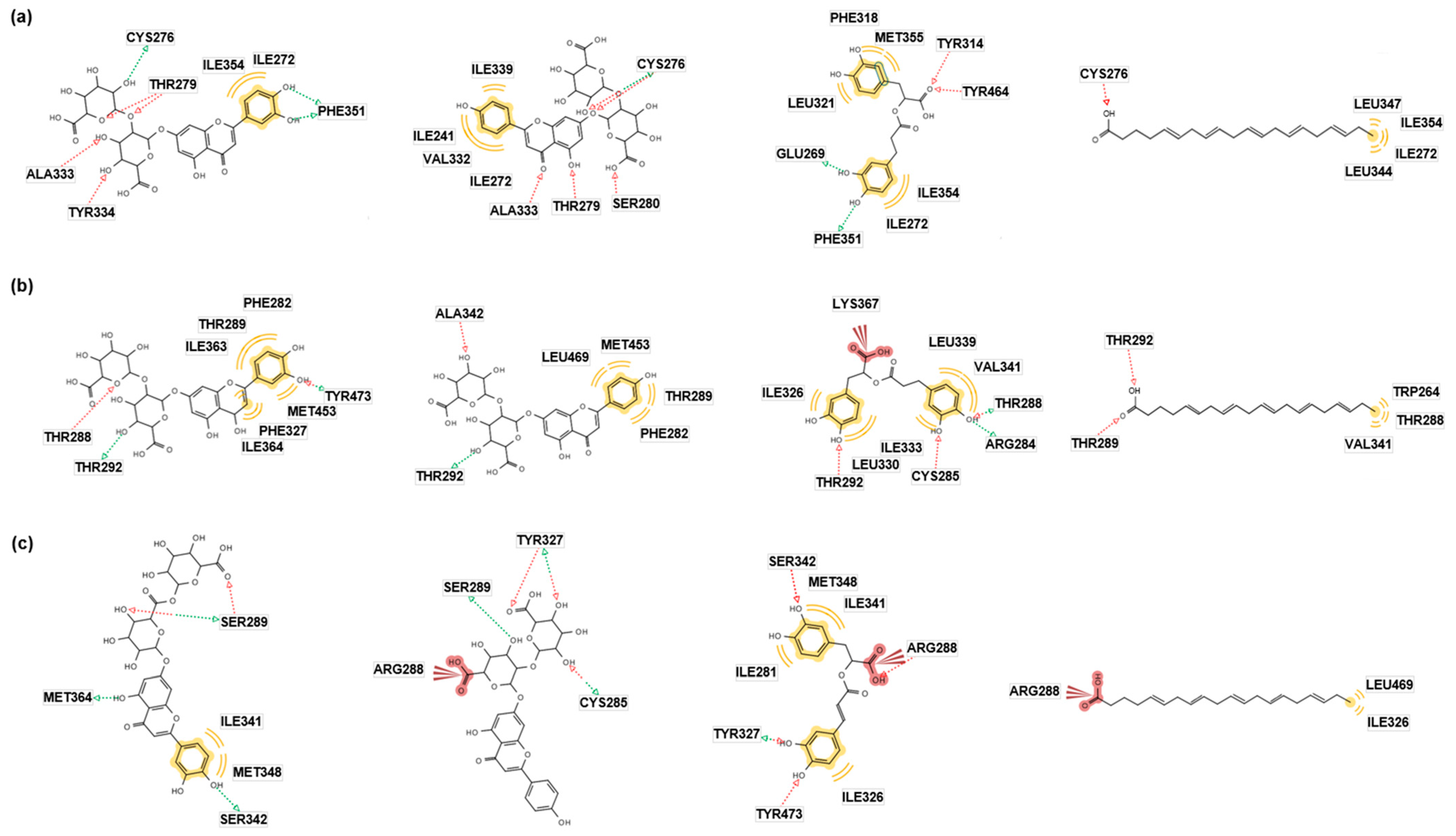
| Compound | Autodock Vina | Autodock 4 | Dock6 | |
|---|---|---|---|---|
| PPAR-α | EPA * | −6.7 | −7.8 | −35.5 |
| 1 | −7.2 | −13.2 | −41.5 | |
| 2 | −6.5 | −9.1 | −41.7 | |
| 3 | −8.6 | −9.7 | −43.0 | |
| PPAR-δ | EPA | −7.8 | −7.4 | −41.0 |
| 1 | −9.7 | −14.7 | −58.1 | |
| 2 | −9.4 | −13.2 | −43.3 | |
| 3 | −8.3 | −9.3 | −41.5 | |
| PPAR-γ | EPA | −6.8 | −8.1 | −37.2 |
| 1 | −5.8 | −9.7 | −57.1 | |
| 2 | −5.6 | −13.4 | −51.6 | |
| 3 | −7.6 | −8.9 | −40.1 |
| Category | Feature | EPA | 1 | 2 | 3 |
|---|---|---|---|---|---|
| Physicochemical properties | Molecular weight | 302.22 | 638.11 | 622.12 | 360.08 |
| Van der Waals (Volume) | 356.24 | 564.63 | 555.84 | 346.37 | |
| Density | 0.85 | 1.13 | 1.12 | 1.03 | |
| No. a of H-bond b acceptors | 2 | 18 | 17 | 8 | |
| No. of H-bond donors | 1 | 10 | 9 | 5 | |
| No. of rotatable bonds | 13 | 7 | 7 | 7 | |
| No. of rings | 0 | 5 | 5 | 2 | |
| No. of atoms in the biggest ring | 0 | 10 | 10 | 6 | |
| Pure LogS (log mol/L) | −4.42 | −4.37 | −2.94 | −2.95 | |
| LogP | 5.18 | −0.31 | 0.88 | 1.51 | |
| Medicinal chemistry | SA score c | 3.04 | 4.88 | 4.74 | 2.90 |
| MCE-18 | 0 | 127.46 | 123.92 | 30 | |
| Lipinski rule | Accepted | Rejected | Rejected | Accepted | |
| Pfizer rule | Rejected | Accepted | Accepted | Accepted | |
| GSK rule | Rejected | Rejected | Rejected | Accepted | |
| Absorption | Caco-2 cell permeability (log unit) | −5.08 | −6.90 | −6.94 | −5.80 |
| MDCK cell permeability (cm/s) | 1.7 × 10−5 | 5.96 × 10−5 | 3.30 × 10−5 | 5.00 × 10−6 | |
| F20% | 0.94 | 0.01 | 0.84 | 0.98 | |
| Distribution | Plasma protein binding (%) | 100.70 | 83.34 | 81.14 | 92.41 |
| Volume distribution (L/kg) | 0.26 | 0.61 | 0.50 | 0.36 | |
| BBB penetration probability | 0.001 | 0.054 | 0.045 | 0.021 | |
| Fu (The fraction unbound in plasma %) | 1.09 | 14.64 | 11.64 | 3.31 | |
| Metabolism | CYP1A2-inhibition probability | 0.072 | 0.116 | 0.041 | 0.251 |
| CYP1A2-substrate probability | 0.117 | 0.015 | 0.006 | 0.022 | |
| CYP2C19-inhibition probability | 0.028 | 0.048 | 0.054 | 0.064 | |
| CYP2C19-substrate probability | 0.050 | 0.034 | 0.030 | 0.034 | |
| CYP2C9-inhibition probability | 0.116 | 0.004 | 0.004 | 0.481 | |
| CYP2C9-substrate probability | 1.00 | 0.09 | 0.15 | 0.94 | |
| Excretion | Clearance (mL/min/kg) | 1.77 | 1.10 | 1.04 | 9.52 |
| Toxicity | Human hepatotoxicity probability | 0.92 | 0.14 | 0.25 | 0.59 |
| Ames toxicity probability | 0.003 | 0.305 | 0.042 | 0.235 | |
| Rat oral acute toxicity probability | 0 | 0.008 | 0.027 | 0.272 | |
| Carcinogenicity probability | 0.105 | 0.037 | 0.109 | 0.536 | |
| Respiratory toxicity probability | 0.535 | 0.014 | 0.012 | 0.034 | |
| Toxicophore rules | Acute toxicity rule (alerts) | 0 | 0 | 0 | 0 |
| Genotoxic carcinogenicity rule (alerts) | 0 | 0 | 0 | 1 | |
| Non-genotoxic carcinogenicity rule (alerts) | 0 | 0 | 0 | 1 | |
| SureChEMBL rule (alerts) | 0 | 0 | 0 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Youn, I.; Han, S.; Jung, H.J.; Noh, S.G.; Chung, H.Y.; Koo, Y.K.; Shin, S.; Seo, E.K. Anti-Inflammatory Activity of the Constituents from the Leaves of Perilla frutescens var. acuta. Pharmaceuticals 2023, 16, 1655. https://doi.org/10.3390/ph16121655
Youn I, Han S, Jung HJ, Noh SG, Chung HY, Koo YK, Shin S, Seo EK. Anti-Inflammatory Activity of the Constituents from the Leaves of Perilla frutescens var. acuta. Pharmaceuticals. 2023; 16(12):1655. https://doi.org/10.3390/ph16121655
Chicago/Turabian StyleYoun, Isoo, Sujin Han, Hee Jin Jung, Sang Gyun Noh, Hae Young Chung, Yean Kyoung Koo, Sunhye Shin, and Eun Kyoung Seo. 2023. "Anti-Inflammatory Activity of the Constituents from the Leaves of Perilla frutescens var. acuta" Pharmaceuticals 16, no. 12: 1655. https://doi.org/10.3390/ph16121655
APA StyleYoun, I., Han, S., Jung, H. J., Noh, S. G., Chung, H. Y., Koo, Y. K., Shin, S., & Seo, E. K. (2023). Anti-Inflammatory Activity of the Constituents from the Leaves of Perilla frutescens var. acuta. Pharmaceuticals, 16(12), 1655. https://doi.org/10.3390/ph16121655







