Discovery and Characterization of a Dual-Function Peptide Derived from Bitter Gourd Seed Protein Using Two Orthogonal Bioassay-Guided Fractionations Coupled with In Silico Analysis
Abstract
:1. Introduction
2. Results and Discussion
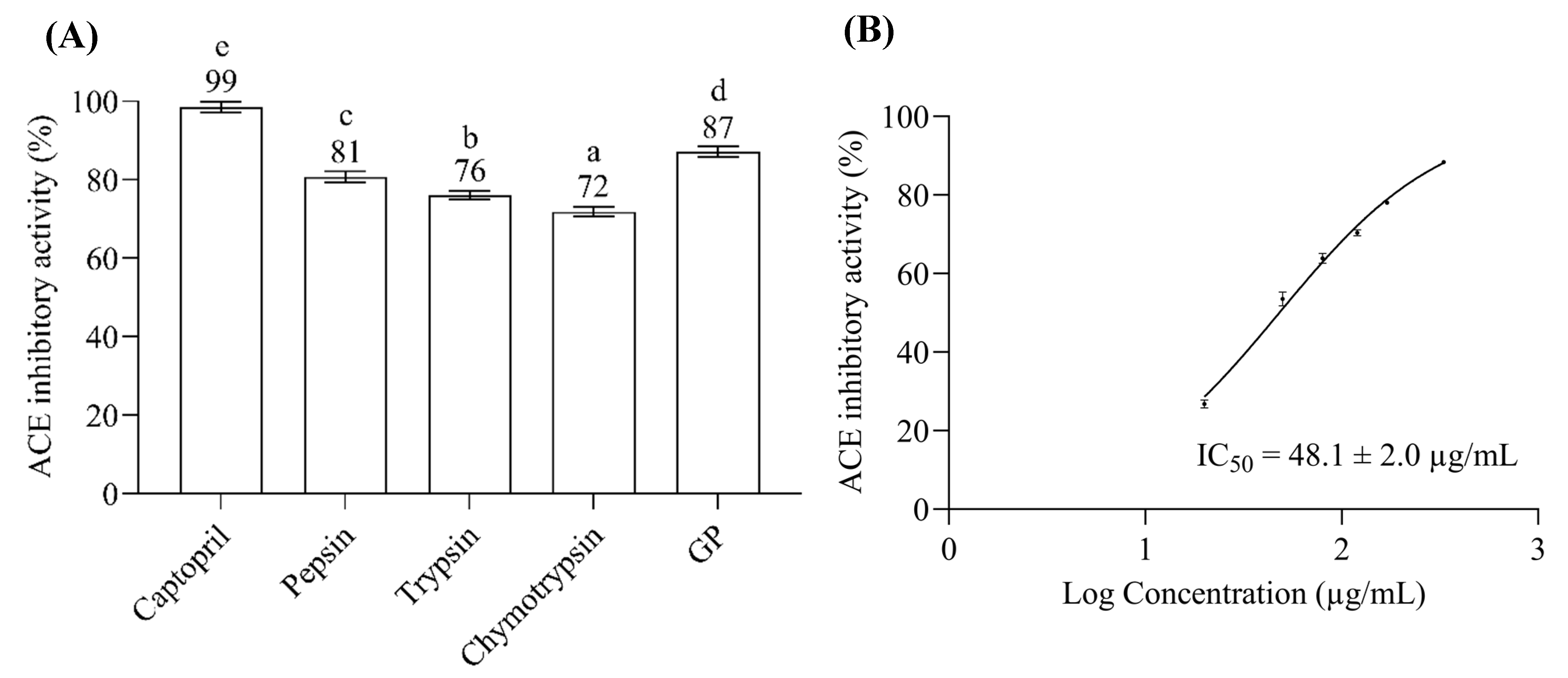
2.1. ACE Inhibitory Assay of Bitter Gourd Seed Protein (BGSP) Hydrolysate
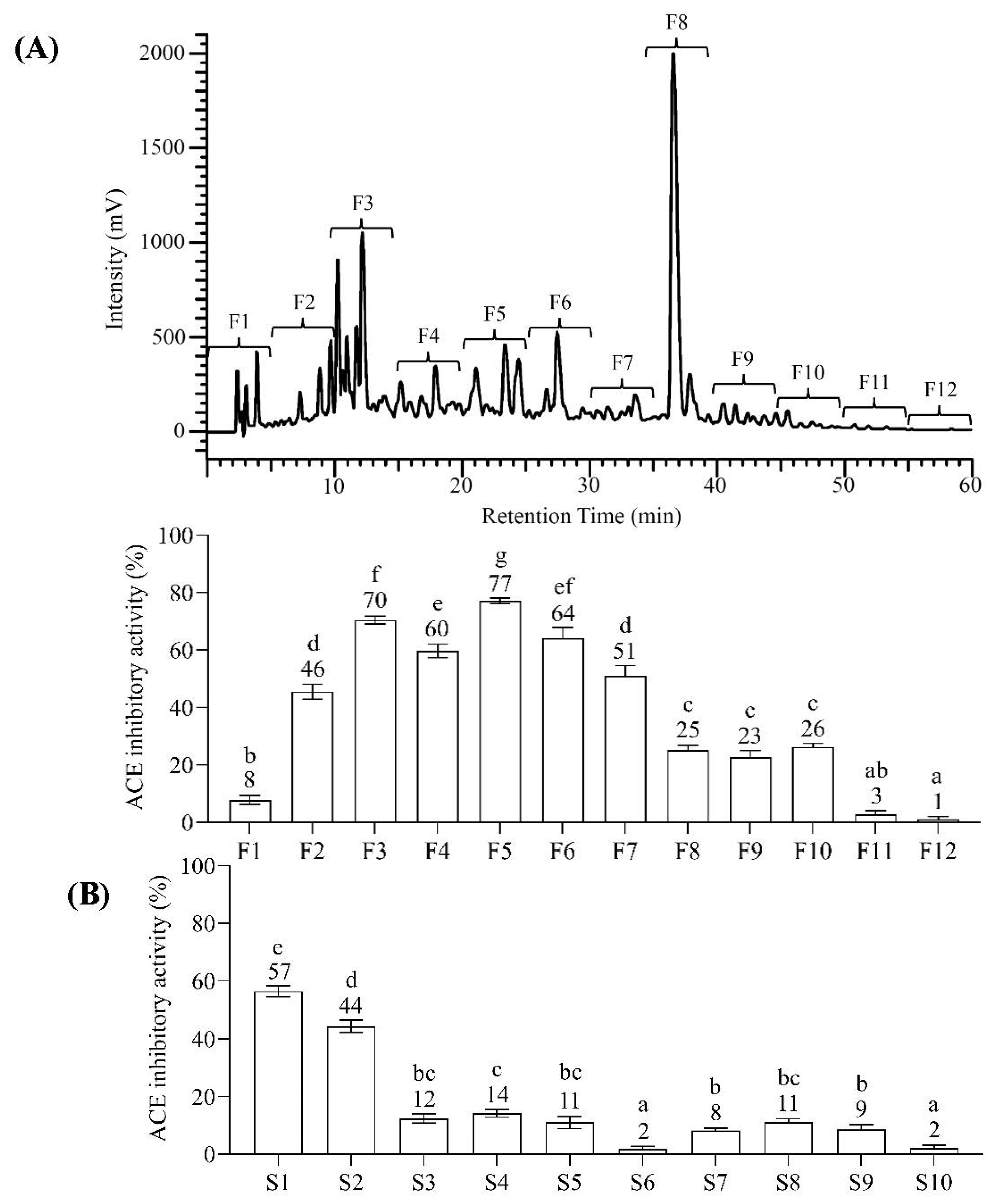
2.2. Bioassay-Guided Fractionation of BGSP-GP Hydrolysate
2.3. Peptide Identification and In Silico Analysis of ACEI Peptide Candidates
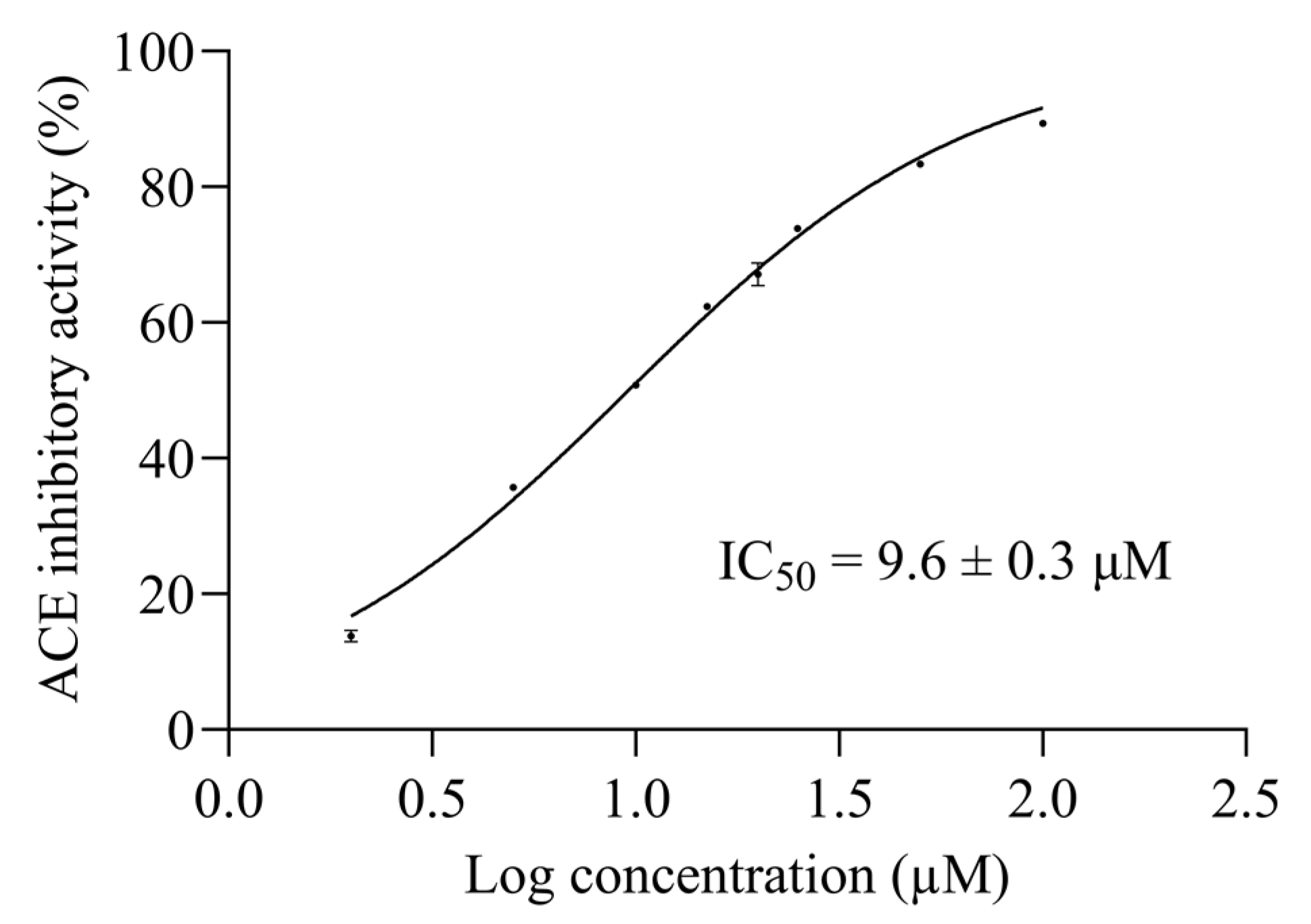
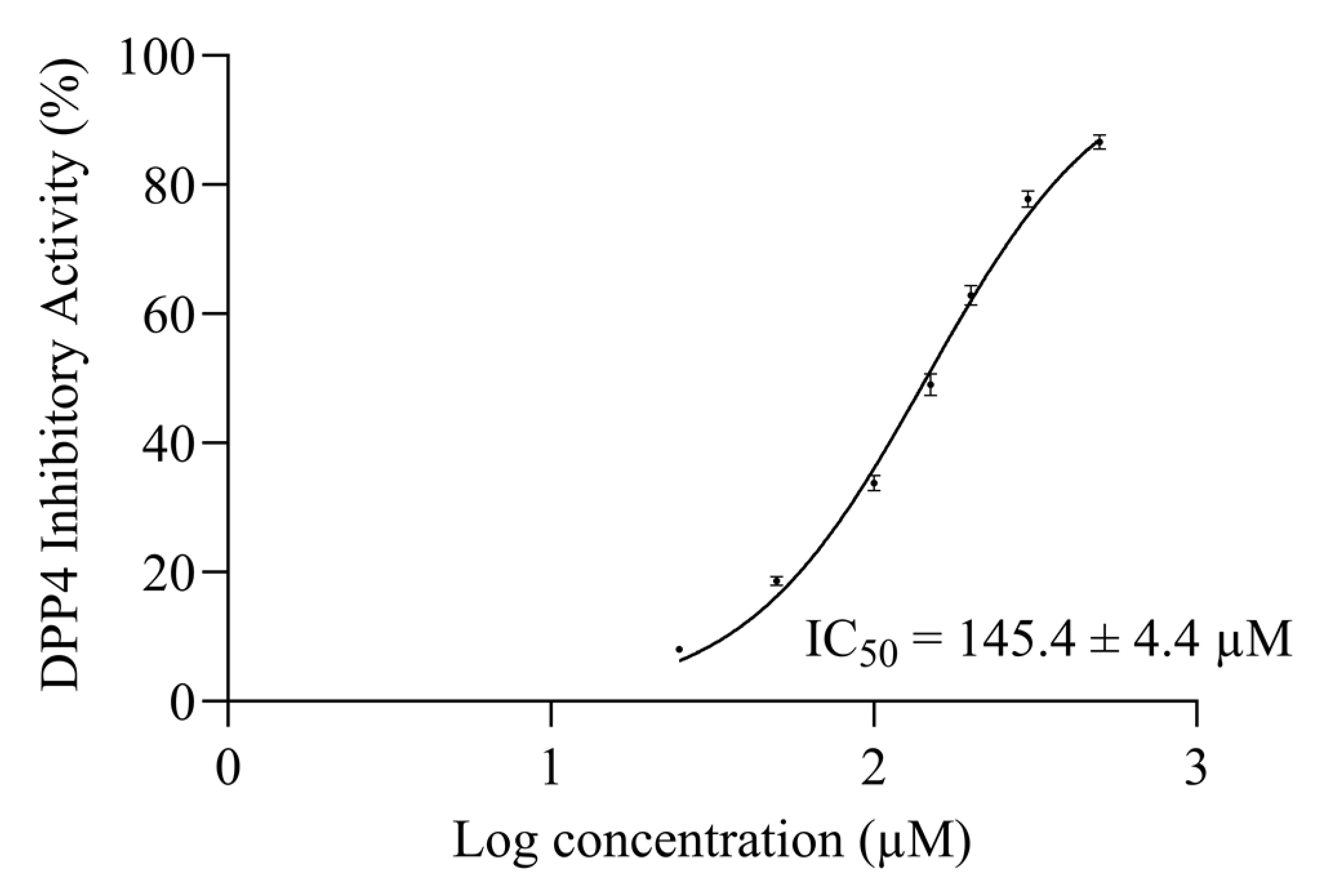
2.4. AW6 Half-Maximal ACE and DPP4 Inhibitory Concentration
2.5. Inhibitory Mechanism of AW6 toward ACE and DPP4
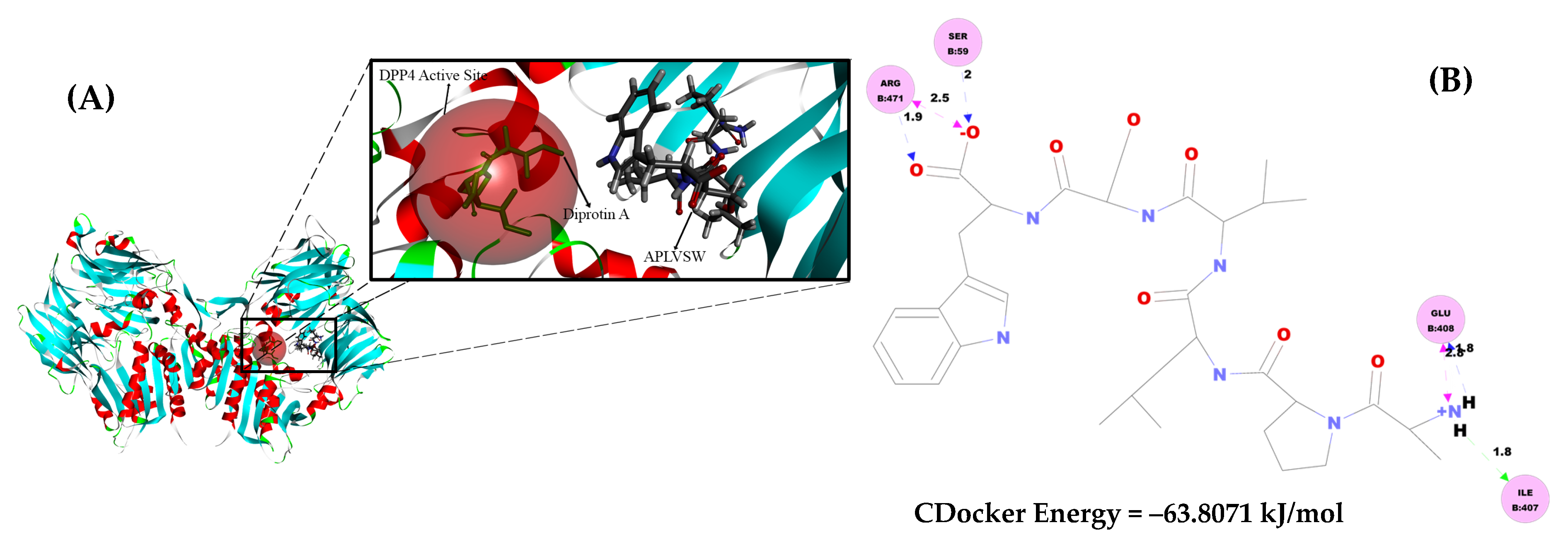
2.6. Intermolecular Interaction Study of AW6 toward ACE and DPP4 Using Molecular Docking Simulation
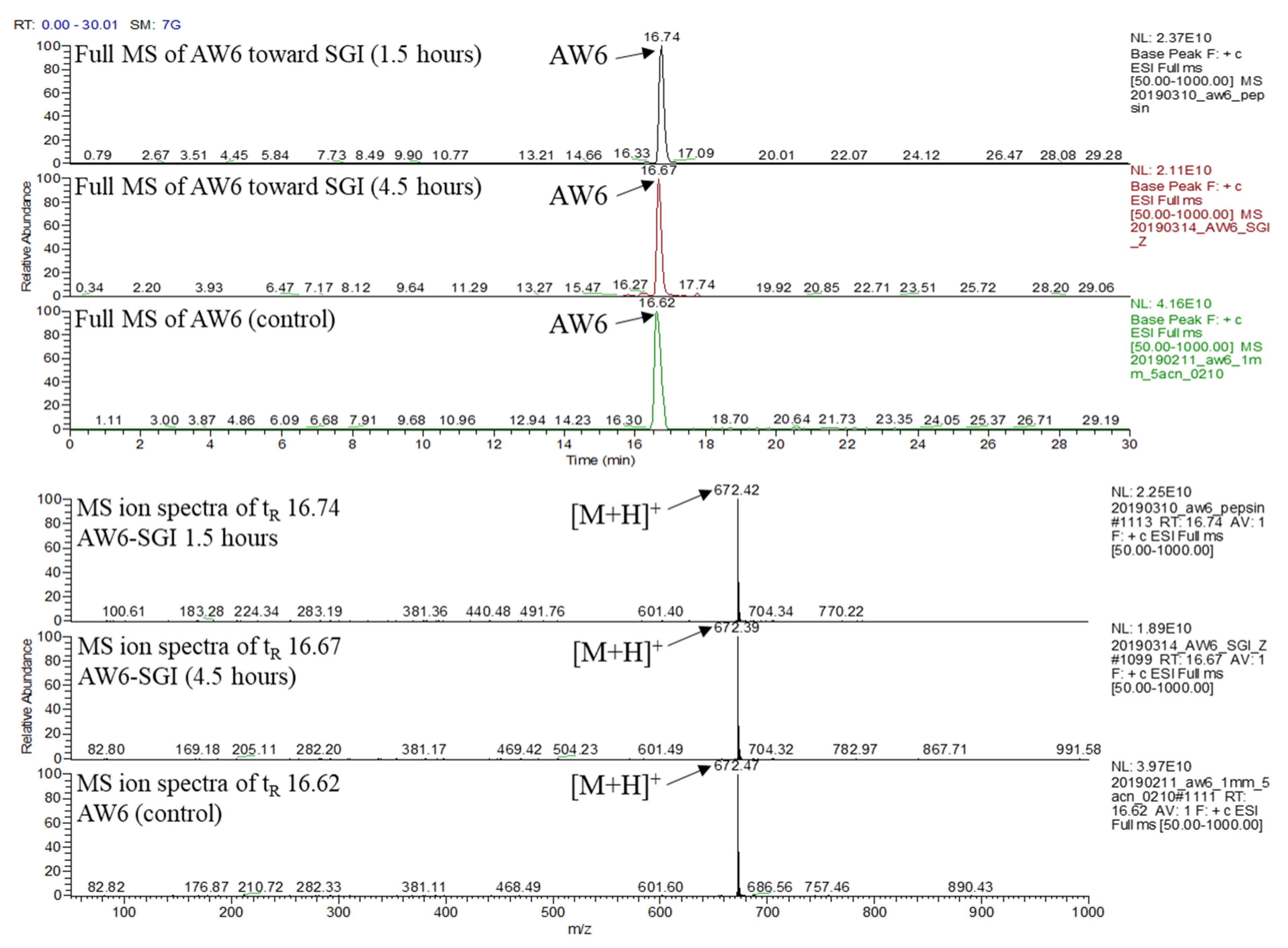
2.7. In Vitro Simulated Gastrointestinal (SGI) Digestion Stability of AW6
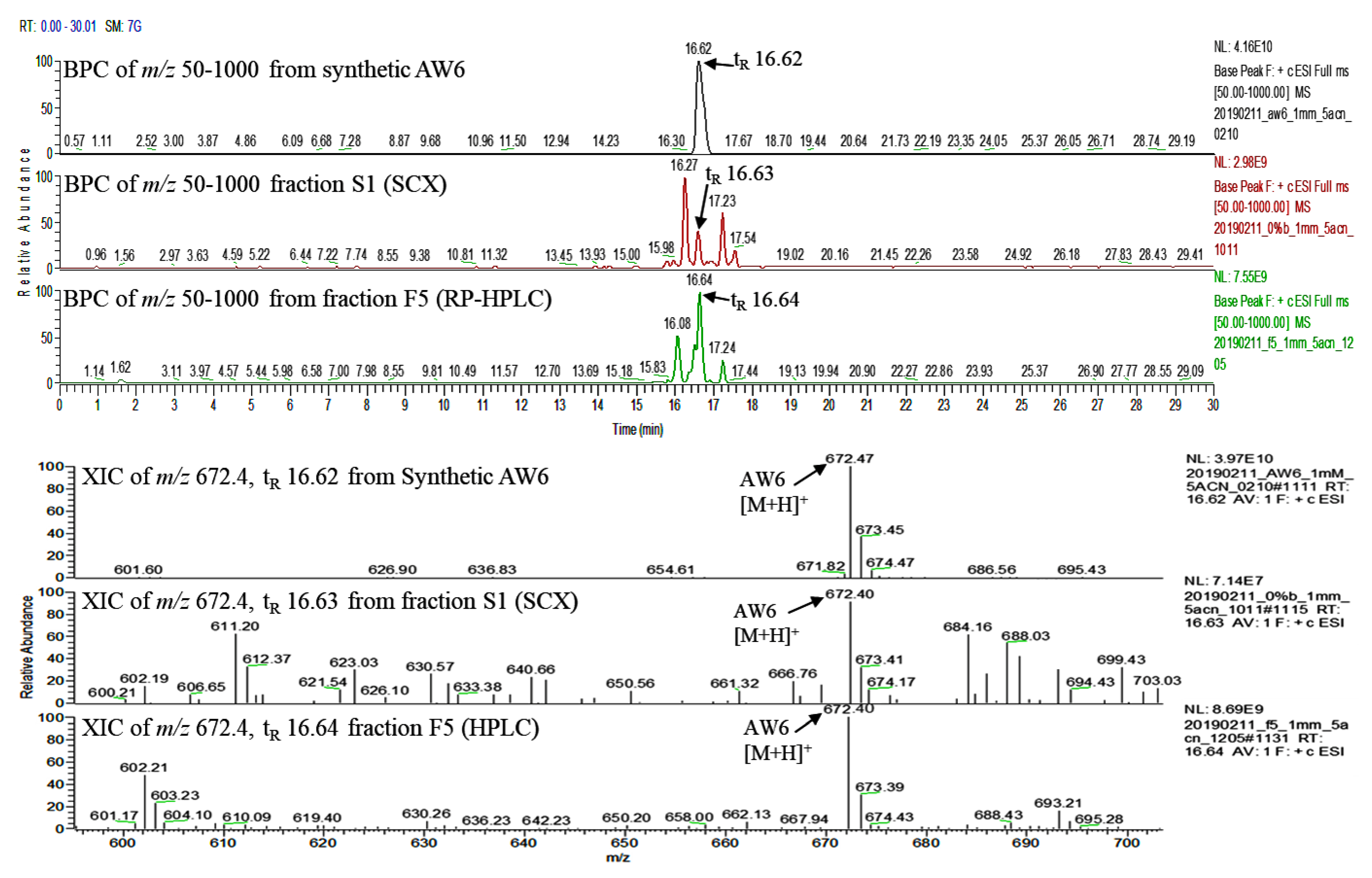
2.8. Quantification of AW6 in BGSP-GP Hydrolysate, Fraction F5 (RP-HPLC), and Fraction S1 (SCX)
3. Materials and Methods
3.1. Materials
3.2. Bitter Gourd Seed Protein (BGSP) Extraction
3.3. Bitter Gourd Seed Protein Hydrolysate Preparation
3.4. Hydrolysis Degree Analysis of BGSP Hydrolysate
3.5. Determination of Angiotensin-I-Converting Enzyme Inhibitory (ACEI) Activity and Its Half-Maximal Inhibitory Concentration
3.6. Determination of Dipeptidyl Peptidase-IV Inhibitory (DPP4I) Activity and Its Half-Maximal Inhibitory Concentration
3.7. Two Orthogonal Bioassay-Guided Fractionations of BGSP-GP
3.8. Identification of Peptide Using LC-MS/MS Analysis Coupled with De Novo Peptide Sequencing and Database-Assisted Matching
3.9. In Silico Analysis Prediction of Peptide Toxicity and Biological Activity
3.10. Synthetic APLVSW (AW6) Preparation and Purification
3.11. Investigation of APLVSW (AW6) Inhibition Mechanism toward ACE and DPP4
3.12. Molecular Level Interaction Study of AW6 toward ACE and DPP4 through Simulation of Molecular Docking
3.13. AW6 Stability toward Gastrointestinal Protease Digestion
3.14. Multiple-Reaction Monitoring (MRM) Analysis of AW6 in Hydrolysate and Fractions
3.15. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chappell, M.C. Biochemical evaluation of the renin-angiotensin system: The good, bad, and absolute? Am. J. Physiol. Heart Circ. Physiol. 2016, 310, H137–H152. [Google Scholar] [CrossRef]
- Jiang, F.; Yang, J.; Zhang, Y.; Dong, M.; Wang, S.; Zhang, Q.; Liu, F.F.; Zhang, K.; Zhang, C. Angiotensin-converting enzyme 2 and angiotensin 1–7: Novel therapeutic targets. Nat. Rev. Cardiol. 2014, 11, 413–426. [Google Scholar] [CrossRef]
- Smith, C.G.; Vane, J.R. The Discovery of Captopril. FASEB J. 2003, 17, 788–789. [Google Scholar] [CrossRef]
- Fitzgerald, R.J.; Murray, B.A. Bioactive peptides and lactic fermentations. Int. J. Dairy Technol. 2006, 59, 118–125. [Google Scholar] [CrossRef]
- Setayesh-Mehr, Z.; Asoodeh, A. The inhibitory activity of HL-7 and HL-10 peptide from scorpion venom (Hemiscorpius lepturus) on angiotensin converting enzyme: Kinetic and docking study. Bioorg. Chem. 2017, 75, 30–37. [Google Scholar] [CrossRef] [PubMed]
- Kharazmi-Khorassani, J.; Asoodeh, A.; Tanzadehpanah, H. Antioxidant and angiotensin-converting enzyme (ACE) inhibitory activity of thymosin alpha-1 (Thα1) peptide. Bioorg. Chem. 2019, 87, 743–752. [Google Scholar] [CrossRef] [PubMed]
- Mirzaei, M.; Mirdamadi, S.; Safavi, M.; Hadizadeh, M. In vitro and in silico studies of novel synthetic ACE-inhibitory peptides derived from Saccharomyces cerevisiae protein hydrolysate. Bioorg. Chem. 2019, 87, 647–654. [Google Scholar] [CrossRef]
- Bhat, Z.F.; Kumar, S.; Bhat, H.F. Antihypertensive peptides of animal origin: A review. Crit. Rev. Food Sci. Nutr. 2017, 57, 566–578. [Google Scholar] [CrossRef]
- Daskaya-Dikmen, C.; Yucetepe, A.; Karbancioglu-Guler, F.; Daskaya, H.; Ozcelik, B. Angiotensin-I-Converting Enzyme (ACE)-Inhibitory Peptides from Plants. Nutrients 2017, 9, 316. [Google Scholar] [CrossRef] [PubMed]
- Gress, T.W.; Nieto, F.J.; Shahar, E.; Wofford, M.R.; Brancati, F.L. Hypertension and Antihypertensive Therapy as Risk Factors for Type 2 Diabetes Mellitus. N. Engl. J. Med. 2000, 342, 905–912. [Google Scholar] [CrossRef]
- Tönnies, T.; Brinks, R.; Isom, S.; Dabelea, D.; Divers, J.; Mayer-Davis, E.J.; Lawrence, J.M.; Pihoker, C.; Dolan, L.; Liese, A.D.; et al. Projections of Type 1 and Type 2 Diabetes Burden in the U.S. Population Aged < 20 Years Through 2060: The SEARCH for Diabetes in Youth Study. Diabetes Care 2022, 46, 313–320. [Google Scholar]
- Deacon, C.F. Peptide degradation and the role of DPP-4 inhibitors in the treatment of type 2 diabetes. Peptides 2018, 100, 150–157. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Su, K.; Mao, X.; Zhang, X. Separation and identification of enzyme inhibition peptides from dark tea protein. Bioorg. Chem. 2020, 99, 103772. [Google Scholar] [CrossRef]
- Wang, K.; Yang, X.; Lou, W.; Zhang, X. Discovery of dipeptidyl peptidase 4 inhibitory peptides from Largemouth bass (Micropterus salmoides) by a comprehensive approach. Bioorg. Chem. 2020, 105, 104432. [Google Scholar] [CrossRef] [PubMed]
- Rivero-Pino, F.; Espejo-Carpio, F.J.; Guadix, E.M. Identification of dipeptidyl peptidase IV (DPP-IV) inhibitory peptides from vegetable protein sources. Food Chem. 2021, 354, 129473. [Google Scholar] [CrossRef]
- Lee, S.W.; Kim, H.C.; Lee, J.-M.; Yun, Y.M.; Lee, J.Y.; Suh, I. Association between changes in systolic blood pressure and incident diabetes in a community-based cohort study in Korea. Hypertens. Res. 2017, 40, 710–716. [Google Scholar] [CrossRef]
- Naik, M.; Natarajan, V.; Modupalli, N.; Thangaraj, S.; Rawson, A. Pulsed ultrasound assisted extraction of protein from defatted Bitter melon seeds (Momardica charantia L.) meal: Kinetics and quality measurements. LWT 2022, 155, 112997. [Google Scholar] [CrossRef]
- Saeed, F.; Afzaal, M.; Niaz, B.; Arshad, M.U.; Tufail, T.; Hussain, M.B.; Javed, A. Bitter melon (Momordica charantia): A natural healthy vegetable. Int. J. Food Prop. 2018, 21, 1270–1290. [Google Scholar] [CrossRef]
- Fuangchan, A.; Sonthisombat, P.; Seubnukarn, T.; Chanouan, R.; Chotchaisuwat, P.; Sirigulsatien, V.; Ingkaninan, K.; Plianbangchang, P.; Haines, S.T. Hypoglycemic effect of bitter melon compared with metformin in newly diagnosed type 2 diabetes patients. J. Ethnopharmacol. 2011, 134, 422–428. [Google Scholar] [CrossRef]
- Priyanto, A.D.; Doerksen, R.J.; Chang, C.-I.; Sung, W.-C.; Widjanarko, S.B.; Kusnadi, J.; Lin, Y.-C.; Wang, T.-C.; Hsu, J.-L. Screening, discovery, and characterization of angiotensin-I converting enzyme inhibitory peptides derived from proteolytic hydrolysate of bitter melon seed proteins. J. Proteom. 2015, 128, 424–435. [Google Scholar] [CrossRef]
- Nielsen, P.M. Functionality of protein hydrolysates. In Food Proteins and Their Applications, 1st ed.; Damodaran, S., Ed.; CRC Press: Boca Raton, FL, USA, 2017; pp. 443–472. [Google Scholar]
- Ngamsuk, S.; Huang, T.-C.; Hsu, J.-L. ACE Inhibitory Activity and Molecular Docking of Gac Seed Protein Hydrolysate Purified by HILIC and RP-HPLC. Molecules 2020, 25, 4635. [Google Scholar] [CrossRef] [PubMed]
- Herraiz, T. Sample preparation and reversed phase-high performance liquid chromatography analysis of food-derived peptides. Anal. Chim. Acta 1997, 352, 119–139. [Google Scholar] [CrossRef]
- Kim, S.; Lim, S.-D. Separation and purification of lipase inhibitory peptide from fermented milk by Lactobacillus plantarum Q180. Food Sci. Anim. Resour. 2020, 40, 87. [Google Scholar] [CrossRef] [PubMed]
- Krippendorff, B.-F.; Neuhaus, R.; Lienau, P.; Reichel, A.; Huisinga, W. Mechanism-Based Inhibition: Deriving KI and kinact Directly from Time-Dependent IC50 Values. SLAS Discov. 2009, 14, 913–923. [Google Scholar] [CrossRef]
- Cortés, A.; Cascante, M.; Cárdenas, M.L.; Cornish-Bowden, A. Relationships between inhibition constants, inhibitor concentrations for 50% inhibition and types of inhibition: New ways of analysing data. Biochem. J. 2001, 357, 263–268. [Google Scholar] [CrossRef]
- Shih, Y.-H.; Chen, F.-A.; Wang, L.-F.; Hsu, J.-L. Discovery and Study of Novel Antihypertensive Peptides Derived from Cassia obtusifolia Seeds. J. Agric. Food Chem. 2019, 67, 7810–7820. [Google Scholar] [CrossRef] [PubMed]
- Gangopadhyay, N.; Wynne, K.; O’Connor, P.; Gallagher, E.; Brunton, N.P.; Rai, D.K.; Hayes, M. In silico and in vitro analyses of the angiotensin-I converting enzyme inhibitory activity of hydrolysates generated from crude barley (Hordeum vulgare) protein concentrates. Food Chem. 2016, 203, 367–374. [Google Scholar] [CrossRef]
- Wang, X.; Chen, H.; Fu, X.; Li, S.; Wei, J. A novel antioxidant and ACE inhibitory peptide from rice bran protein: Biochemical characterization and molecular docking study. LWT 2017, 75, 93–99. [Google Scholar] [CrossRef]
- Xu, F.; Yao, Y.; Xu, X.; Wang, M.; Pan, M.; Ji, S.; Wu, J.; Jiang, D.; Ju, X.; Wang, L. Identification and Quantification of DPP-IV-Inhibitory Peptides from Hydrolyzed-Rapeseed-Protein-Derived Napin with Analysis of the Interactions between Key Residues and Protein Domains. J. Agric. Food Chem. 2019, 67, 3679–3690. [Google Scholar] [CrossRef]
- Lammi, C.; Zanoni, C.; Arnoldi, A.; Vistoli, G. Peptides Derived from Soy and Lupin Protein as Dipeptidyl-Peptidase IV Inhibitors: In Vitro Biochemical Screening and in Silico Molecular Modeling Study. J. Agric. Food Chem. 2016, 64, 9601–9606. [Google Scholar] [CrossRef]
- Nong, N.T.P.; Hsu, J.-L. Characteristics of Food Protein-Derived Antidiabetic Bioactive Peptides: A Literature Update. Int. J. Mol. Sci. 2021, 22, 9508. [Google Scholar] [CrossRef]
- Natesh, R.; Schwager, S.L.; Evans, H.R.; Sturrock, E.D.; Acharya, K.R. Structural details on the binding of antihypertensive drugs captopril and enalaprilat to human testicular angiotensin I-converting enzyme. Biochemistry 2004, 43, 8718–8724. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Zhou, L.; Zhang, Y.; Sheng, N.-J.; Wang, Z.-K.; Wu, T.-Z.; Wang, X.-Z.; Wu, H. Rapid Identification of Dipeptidyl Peptidase-IV (DPP-IV) Inhibitory Peptides from Ruditapes philippinarum Hydrolysate. Molecules 2017, 22, 1714. [Google Scholar] [CrossRef]
- Hiramatsu, H.; Yamamoto, A.; Kyono, K.; Higashiyama, Y.; Fukushima, C.; Shima, H.; Sugiyama, S.; Inaka, K.; Shimizu, R. The crystal structure of human dipeptidyl peptidase IV (DPPIV) complex with diprotin A. J. Biol. Chem. 2004, 385, 561–564. [Google Scholar] [CrossRef]
- Jao, C.-L.; Huang, S.-L.; Hsu, K.-C. Angiotensin I-converting enzyme inhibitory peptides: Inhibition mode, bioavailability, and antihypertensive effects. BioMedicine 2012, 2, 130–136. [Google Scholar] [CrossRef]
- Vermeirssen, V.; Camp, J.V.; Verstraete, W. Bioavailability of angiotensin I converting enzyme inhibitory peptides. Br. J. Nutr. 2004, 92, 357–366. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Li, B. The effect of molecular weights on the survivability of casein-derived antioxidant peptides after the simulated gastrointestinal digestion. Innov. Food Sci. Emerg. Technol. 2012, 16, 341–348. [Google Scholar] [CrossRef]
- Ningrum, S.; Sutrisno, A.; Hsu, J.-L. An exploration of angiotensin-converting enzyme (ACE) inhibitory peptides derived from gastrointestinal protease hydrolysate of milk using a modified bioassay-guided fractionation approach coupled with in silico analysis. J. Dairy Sci. 2022, 105, 1913–1928. [Google Scholar] [CrossRef]
- Sutopo, C.C.Y.; Aznam, N.; Arianingrum, R.; Hsu, J.-L. Screening potential hypertensive peptides using two consecutive bioassay-guided SPE fractionations and identification of an ACE inhibitory peptide, DHSTAVW (DW7), derived from pearl garlic protein hydrolysate. Peptides 2023, 167, 171046. [Google Scholar] [CrossRef]
- Hatanaka, T.; Inoue, Y.; Arima, J.; Kumagai, Y.; Usuki, H.; Kawakami, K.; Kimura, M.; Mukaihara, T. Production of dipeptidyl peptidase IV inhibitory peptides from defatted rice bran. Food Chem. 2012, 134, 797–802. [Google Scholar] [CrossRef]
- Sutopo, C.C.Y.; Sutrisno, A.; Wang, L.-F.; Hsu, J.-L. Identification of a potent Angiotensin-I converting enzyme inhibitory peptide from Black cumin seed hydrolysate using orthogonal bioassay-guided fractionations coupled with in silico screening. Process Biochem. 2020, 95, 204–213. [Google Scholar] [CrossRef]
- Cushman, D.W.; Cheung, H.S. Spectrophotometric assay and properties of the angiotensin-converting enzyme of rabbit lung. Biochem. Pharmacol. 1971, 20, 1637–1648. [Google Scholar] [CrossRef] [PubMed]
- Nong, N.T.P.; Chen, Y.-K.; Shih, W.-L.; Hsu, J.-L. Characterization of Novel Dipeptidyl Peptidase-IV Inhibitory Peptides from Soft-Shelled Turtle Yolk Hydrolysate Using Orthogonal Bioassay-Guided Fractionations Coupled with In Vitro and In Silico Study. Pharmaceuticals 2020, 13, 308. [Google Scholar] [CrossRef]
- Pujiastuti, D.Y.; Shih, Y.-H.; Chen, W.-L.; Sukoso; Hsu, J.-L. Screening of angiotensin-I converting enzyme inhibitory peptides derived from soft-shelled turtle yolk using two orthogonal bioassay-guided fractionations. J. Funct. Foods 2017, 28, 36–47. [Google Scholar] [CrossRef]
- Mooney, C.; Haslam, N.J.; Pollastri, G.; Shields, D.C. Towards the Improved Discovery and Design of Functional Peptides: Common Features of Diverse Classes Permit Generalized Prediction of Bioactivity. PLoS ONE 2012, 7, e45012. [Google Scholar] [CrossRef] [PubMed]
- Minkiewicz, P.; Iwaniak, A.; Darewicz, M. BIOPEP-UWM Database of Bioactive Peptides: Current Opportunities. Int. J. Mol. Sci. 2019, 20, 5978. [Google Scholar] [CrossRef]
- Gupta, S.; Kapoor, P.; Chaudhary, K.; Gautam, A.; Kumar, R.; Open Source Drug Discovery Consortium; Raghava, G.P.S. In Silico Approach for Predicting Toxicity of Peptides and Proteins. PLoS ONE 2013, 8, e73957. [Google Scholar] [CrossRef]



| Peptide Sequence | Peptide Length | ToxinPred | * BIOPEP (B) | PeptideRanker |
|---|---|---|---|---|
| APLVSW | 6 | Non-Toxin | 0.00122 | 0.516023 |
| EPTTSDVVVAGEFDQGSGSMR | 21 | Non-Toxin | 0.00006 | 0.172646 |
| ATISLENSW | 9 | Non-Toxin | - | 0.191382 |
| AW6 (µg) | Purification Fold | |
|---|---|---|
| GP-BGSP hydrolysate | 1.8 ± 0.2 a | 1.0 |
| RP-HPLC fraction F5 | 9.8 ± 0.3 c | 5.4 |
| SCX fraction S1 | 2.5 ± 0.2 b | 1.4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hung, W.-T.; Sutopo, C.C.Y.; Wu, M.-L.; Hsu, J.-L. Discovery and Characterization of a Dual-Function Peptide Derived from Bitter Gourd Seed Protein Using Two Orthogonal Bioassay-Guided Fractionations Coupled with In Silico Analysis. Pharmaceuticals 2023, 16, 1629. https://doi.org/10.3390/ph16111629
Hung W-T, Sutopo CCY, Wu M-L, Hsu J-L. Discovery and Characterization of a Dual-Function Peptide Derived from Bitter Gourd Seed Protein Using Two Orthogonal Bioassay-Guided Fractionations Coupled with In Silico Analysis. Pharmaceuticals. 2023; 16(11):1629. https://doi.org/10.3390/ph16111629
Chicago/Turabian StyleHung, Wei-Ting, Christoper Caesar Yudho Sutopo, Mei-Li Wu, and Jue-Liang Hsu. 2023. "Discovery and Characterization of a Dual-Function Peptide Derived from Bitter Gourd Seed Protein Using Two Orthogonal Bioassay-Guided Fractionations Coupled with In Silico Analysis" Pharmaceuticals 16, no. 11: 1629. https://doi.org/10.3390/ph16111629
APA StyleHung, W.-T., Sutopo, C. C. Y., Wu, M.-L., & Hsu, J.-L. (2023). Discovery and Characterization of a Dual-Function Peptide Derived from Bitter Gourd Seed Protein Using Two Orthogonal Bioassay-Guided Fractionations Coupled with In Silico Analysis. Pharmaceuticals, 16(11), 1629. https://doi.org/10.3390/ph16111629









