Pro-Apoptotic Activity of Epi-Obtusane against Cervical Cancer: Nano Formulation, In Silico Molecular Docking, and Pharmacological Network Analysis
Abstract
:1. Introduction
2. Results and Discussion
2.1. In Silico-Based Determination of the Bioactivity
2.2. Epi-Obtusane-Containing Liposomes
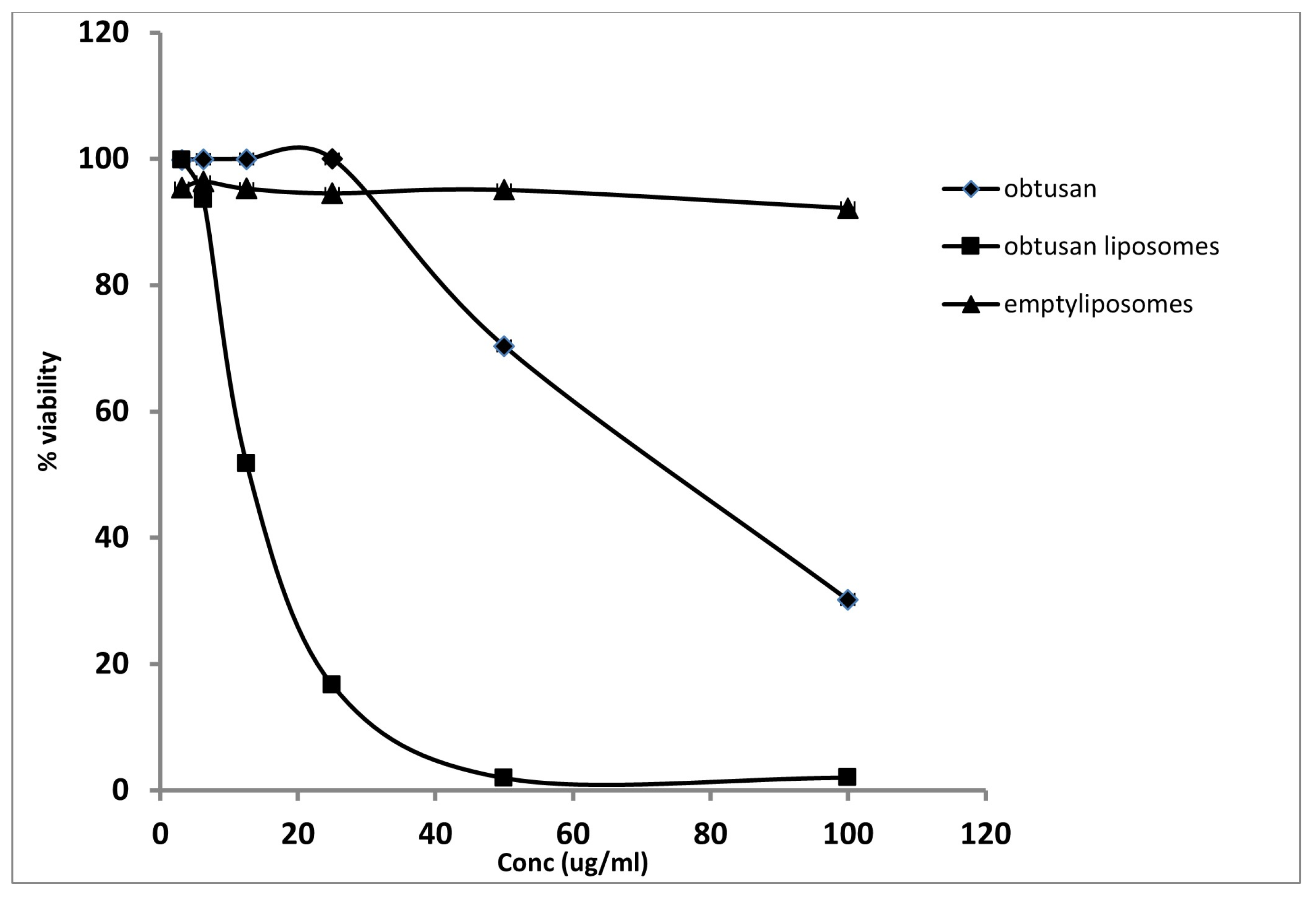
2.3. Antiproliferative Activity of Epi-Obtusane and Its Liposomal Formulation
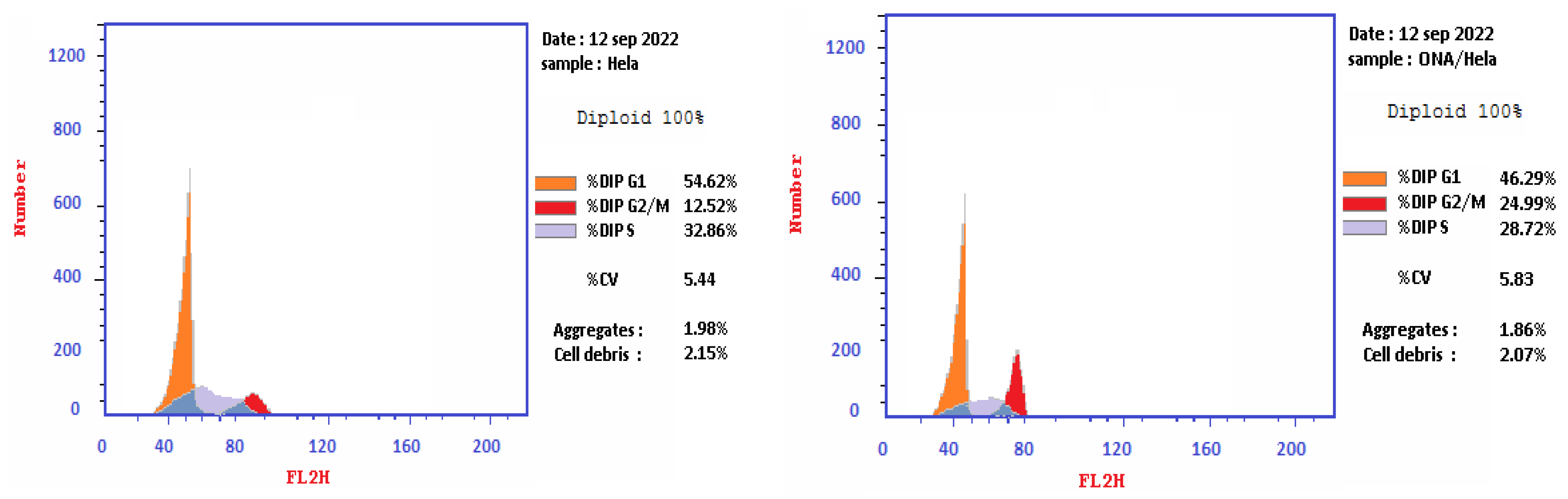
2.4. Cell Cycle Analysis on HeLa Cell Line Treated with Epi-Obtusane
2.5. Apoptosis Determination by Annexin-V Assay
2.6. Pharmacological Network
2.6.1. Collection of Potential Targets for Antitumor Activity
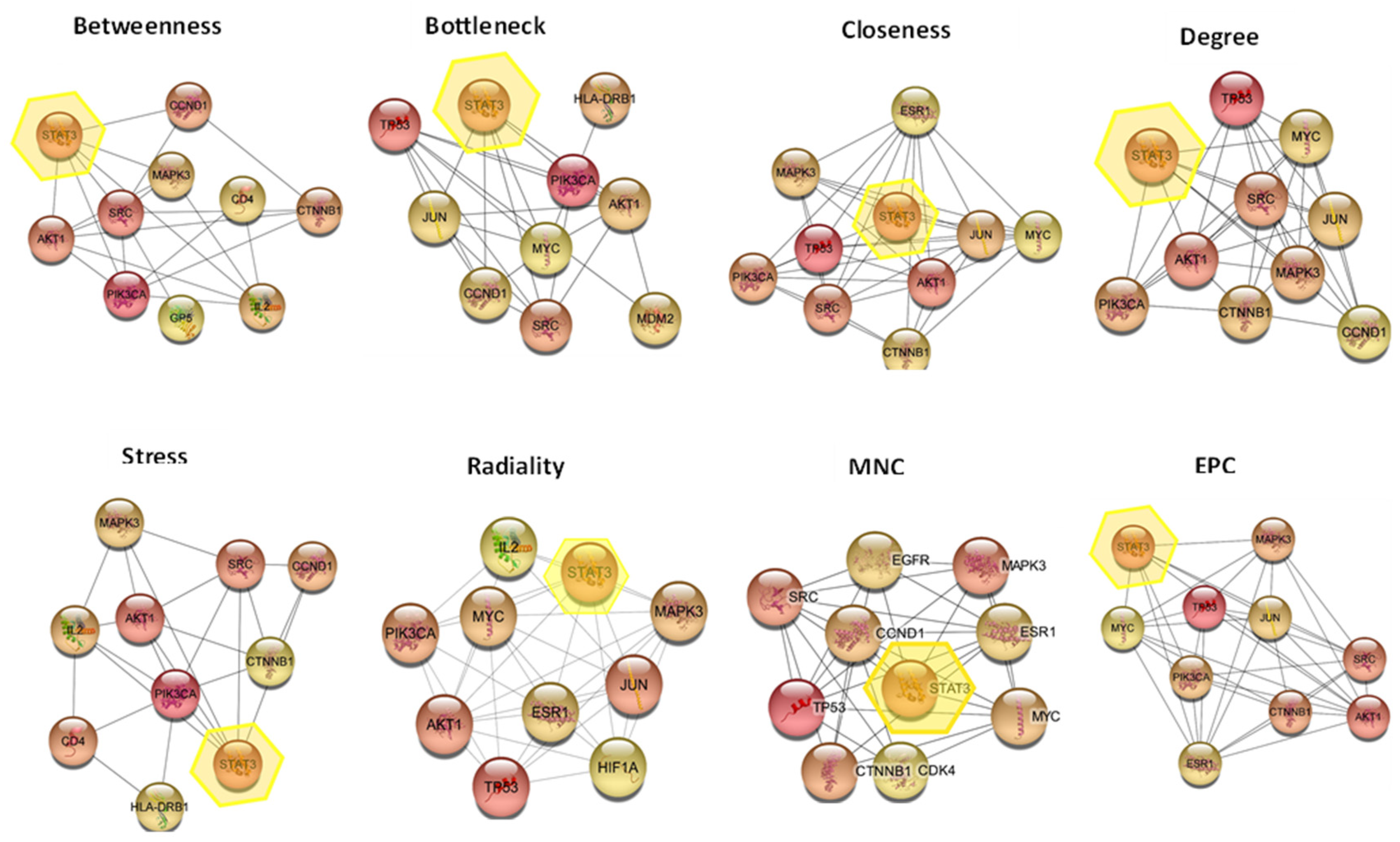
2.6.2. Construction of Protein-Protein Interaction (PPI) Network
2.6.3. Hub Gene Expression Analysis
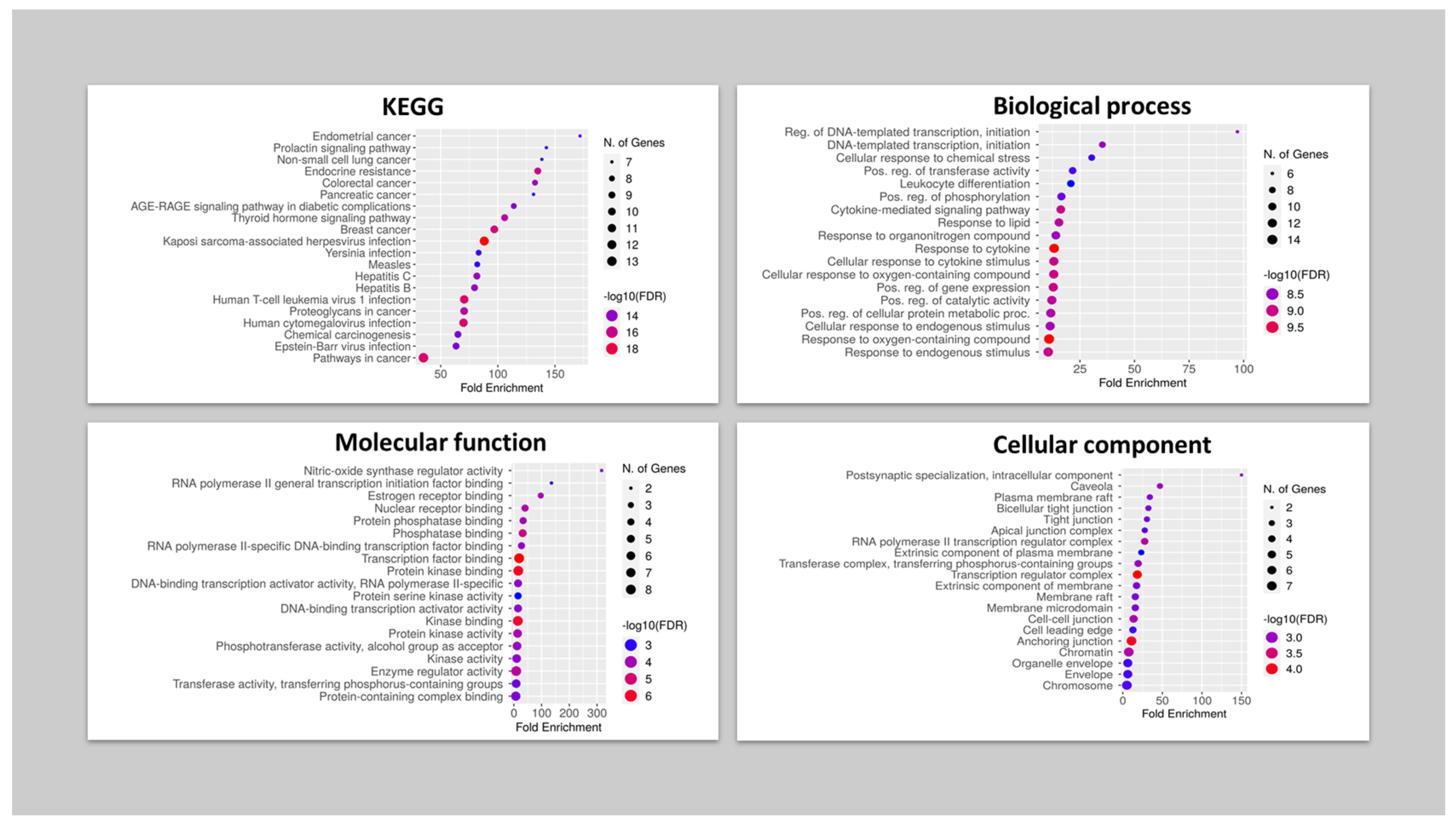
2.6.4. Gene Ontology and Enrichment Analysis
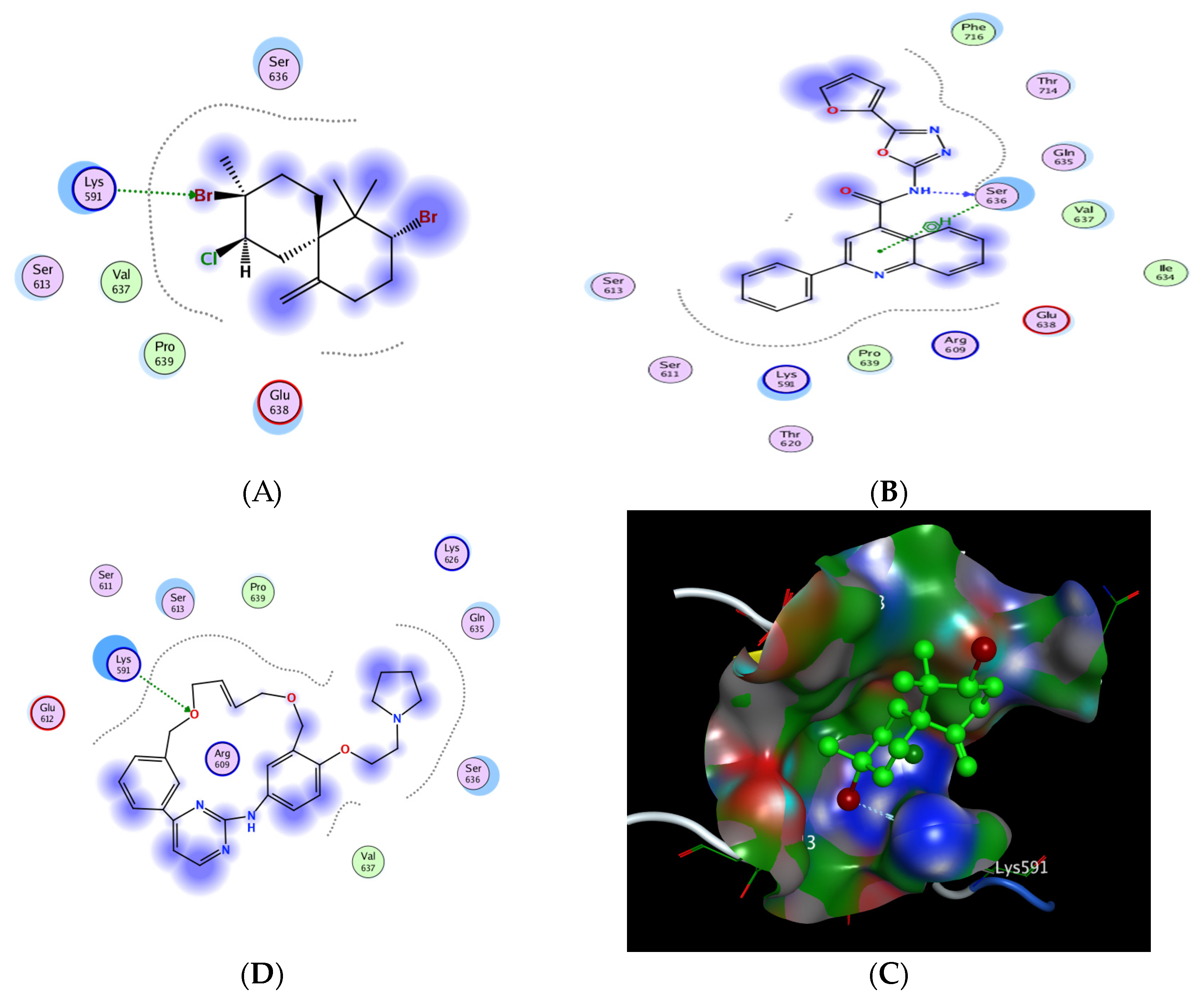
2.6.5. In Silico Molecular Docking of Epi-Obtusane and STAT 3 Receptor
2.6.6. Effect of Epi-Obtusane on STAT3 Total in HeLa Cells
3. Materials and Methods
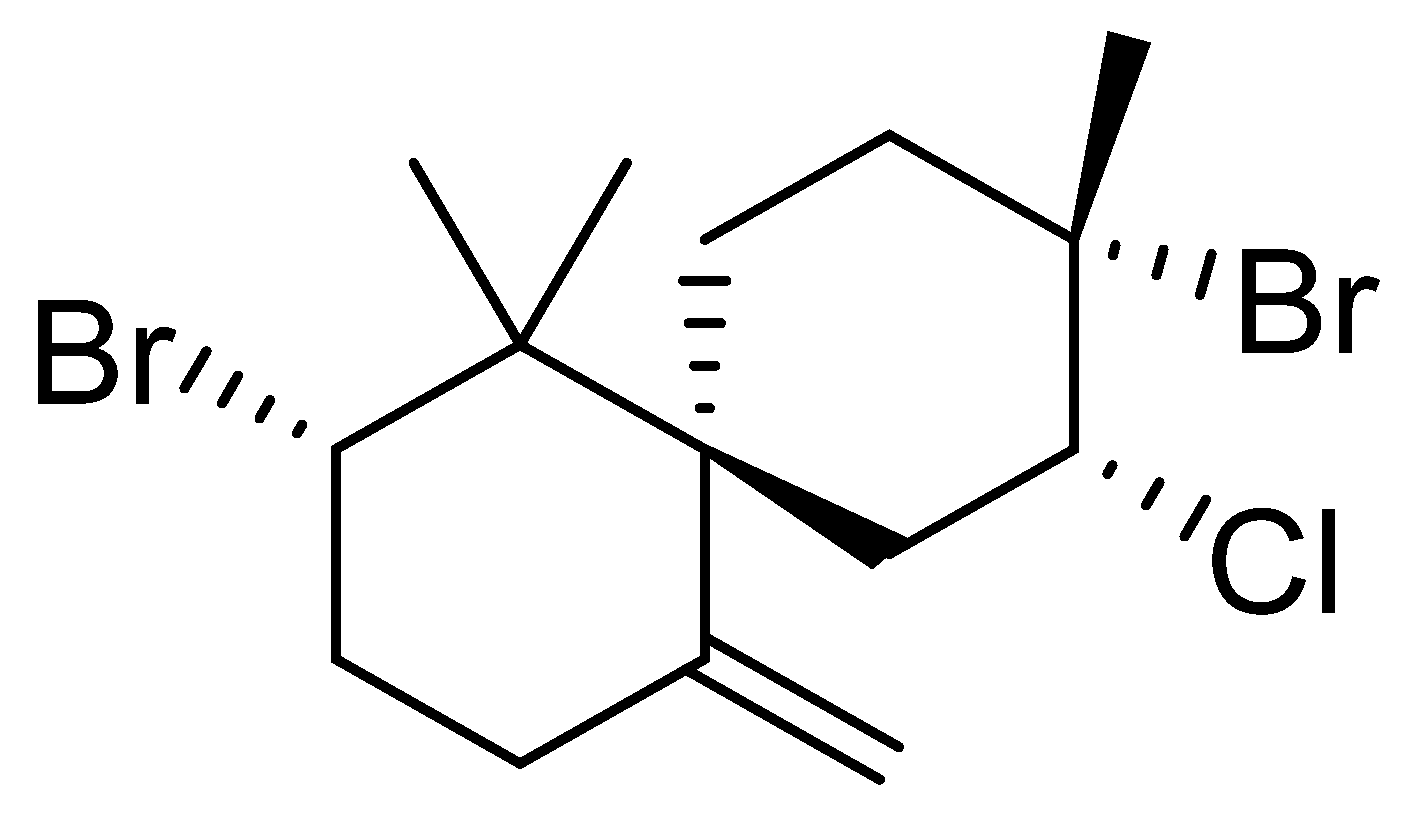
3.1. Epi-Obtusane Material
3.2. Preparation of Epi-Obtusane-Containing Liposomes
3.3. Characterization of Epi-Obtusane- Containing Liposomes
3.4. Antiproliferative Assay
3.5. Cell Cycle Analysis on HeLa Cell Line Treated with Epi-Obtusane
3.6. Apoptosis Determination by Annexin-V Assay
3.7. In Silico Molecular Docking of Epi-Obtusane and STAT 3 Protein
3.8. Effect of Epi-Obtusane on STAT3 and p-STAT3 Total in HeLa Cells
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hanahan, D. Hallmarks of cancer: New dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2021. Ca Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Subavathy, P.; Shibana, C. Anticancer Activity of Turbo brunneus, Cypraea annulus and Babylonia spirata on MCF-7 Cell Line. Asian J. Biol. Life Sci. 2021, 10, 118–122. [Google Scholar] [CrossRef]
- Ameen, F.; AlNAdhari, S.; Al-Homaidan, A.A. Marine fungi showing multifunctional activity against human pathogenic microbes and cancer. PLoS ONE 2022, 17, e0276926. [Google Scholar] [CrossRef] [PubMed]
- Kachhwaha, N.; Sharma, M.K.; Khandelwal, R.; Kaushik, P. Marine algal bioactive metabolites and their pharmacological applications. Ther. Implic. Nat. Bioact. Compd. 2022, 3, 118–134. [Google Scholar]
- Soerjomataram, I.; Bray, F. Planning for tomorrow: Global cancer incidence and the role of prevention 2020–2070. Nat. Rev. Clin. Oncol. 2021, 18, 663–672. [Google Scholar] [CrossRef]
- Balwan, W.K.; Kour, S. Lifestyle Diseases: The Link between Modern Lifestyle and Threat to Public Health. Saudi J. Med. Pharm. Sci. 2021, 7, 179–184. [Google Scholar] [CrossRef]
- Abou-Taleb, H.A.; Sayed, A.M.; Refaat, H.; Alsenani, F.; Alaaeldin, E.; Mokhtar, F.A.; Abdelmohsen, U.R.; Shady, N.H. Network Pharmacological Analysis of the Red Sea Sponge Hyrtios erectus Extract to Reveal Anticancer Efficacy of Corresponding Loaded Niosomes. Mar. Drugs 2022, 20, 628. [Google Scholar] [CrossRef]
- Ferrall, L.; Lin, K.Y.; Roden, R.; Hung, C.-F.; Wu, T.-C. Cervical Cancer Immunotherapy: Facts and Hopes Immunotherapy for Cervical Cancer. Clin. Cancer Res. 2021, 27, 4953–4973. [Google Scholar] [CrossRef]
- Thomas, A.; Takahashi, N.; Rajapakse, V.N.; Zhang, X.; Sun, Y.; Ceribelli, M.; Wilson, K.M.; Zhang, Y.; Beck, E.; Sciuto, L.; et al. Therapeutic targeting of ATR yields durable regressions in small cell lung cancers with high replication stress. Cancer Cell 2021, 39, 566–579.e7. [Google Scholar] [CrossRef]
- Ma, Y.-L.; Zhang, Y.-S.; Zhang, F.; Zhang, Y.-Y.; Thakur, K.; Zhang, J.-G.; Wei, Z.-J. Methyl protodioscin from Polygonatum sibiricum inhibits cervical cancer through cell cycle arrest and apoptosis induction. Food Chem. Toxicol. 2019, 132, 110655. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.-Y.; Wang, F.-X.; Jia, K.-K.; Kong, L.-D. Natural Product Interventions for Chemotherapy and Radiotherapy-Induced Side Effects. Front. Pharmacol. 2018, 9, 1253. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Shahrajabian, M.H. Therapeutic Potential of Phenolic Compounds in Medicinal Plants—Natural Health Products for Human Health. Molecules 2023, 28, 1845. [Google Scholar] [CrossRef]
- Quazi, A.; Patwekar, M.; Patwekar, F.; Alghamdi, S.; Rajab, B.S.; Babalghith, A.O.; Islam, F.J. In vitro alpha-amylase enzyme assay of hydroalcoholic polyherbal extract: Proof of concept for the development of polyherbal teabag formulation for the treatment of diabetes. Evid. Based Complement. Altern. Med. 2022, 2022, 1577957. [Google Scholar] [CrossRef] [PubMed]
- Abdou, R.; Alqahtani, A.M.; Attia, G.H. Anticancer natural products from Aspergillus neoniger, an endophyte of Ficus carica. Bull. Natl. Res. Cent. 2021, 45, 74. [Google Scholar] [CrossRef]
- Banerjee, P.; Mandhare, A.; Bagalkote, V. Marine natural products as source of new drugs: An updated patent review (July 2018–July 2021). Expert Opin. Ther. Patents 2022, 32, 317–363. [Google Scholar] [CrossRef]
- Chen, S.; Cai, R.; Liu, Z.; Cui, H.; She, Z. Secondary metabolites from mangrove-associated fungi: Source, chemistry and bioactivities. Nat. Prod. Rep. 2022, 39, 560–595. [Google Scholar] [CrossRef]
- Pinedo-Rivilla, C.; Aleu, J.; Durán-Patrón, R. Cryptic Metabolites from Marine-Derived Microorganisms Using OSMAC and Epigenetic Approaches. Mar. Drugs 2022, 20, 84. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P. The STRING database in 2017: Quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res. 2016, 2016, gkw937. [Google Scholar] [CrossRef]
- Šimat, V.; Rathod, N.B.; Čagalj, M.; Hamed, I.; Mekinić, I.G. Astaxanthin from Crustaceans and Their Byproducts: A Bioactive Metabolite Candidate for Therapeutic Application. Mar. Drugs 2022, 20, 206. [Google Scholar] [CrossRef]
- Yang, W.; Zhao, P.; Li, X.; Guo, L.; Gao, W. The potential roles of natural plant polysaccharides in inflammatory bowel disease: A review. Carbohydr. Polym. 2022, 277, 118821. [Google Scholar] [CrossRef] [PubMed]
- Najmi, A.; Javed, S.A.; Al Bratty, M.; Alhazmi, H.A. Modern Approaches in the Discovery and Development of Plant-Based Natural Products and Their Analogues as Potential Therapeutic Agents. Molecules 2022, 27, 349. [Google Scholar] [CrossRef] [PubMed]
- Twaij, B.M.; Hasan, M. Bioactive secondary metabolites from plant sources: Types, synthesis, and their therapeutic uses. Int. J. Plant Biol. 2022, 13, 4–14. [Google Scholar] [CrossRef]
- Sedeek, A.; Ismail, M.; Elsayed, T.; Ramadan, M. Recent methods for discovering novel bioactive metabolites, specifically antimicrobial agents, from marine-associated micro-organisms. Lett. Appl. Microbiol. 2022, 75, 511–525. [Google Scholar] [CrossRef]
- Hannan, M.A.; Dash, R.; Haque, M.N.; Mohibbullah, M.; Sohag, A.A.M.; Rahman, M.A.; Uddin, M.J.; Alam, M.; Moon, I. Neuroprotective potentials of marine algae and their bioactive metabolites: Pharmacological insights and therapeutic advances. Mar. Drugs 2020, 18, 347. [Google Scholar] [CrossRef]
- Pradhan, B.; Nayak, R.; Patra, S.; Jit, B.P.; Ragusa, A.; Jena, M. Bioactive Metabolites from Marine Algae as Potent Pharmacophores against Oxidative Stress-Associated Human Diseases: A Comprehensive Review. Molecules 2020, 26, 37. [Google Scholar] [CrossRef]
- Karthikeyan, A.; Joseph, A.; Nair, B.G. Promising bioactive compounds from the marine environment and their potential effects on various diseases. J. Genet. Eng. Biotechnol. 2022, 20, 1–14. [Google Scholar] [CrossRef]
- Li, T.; Ding, T.; Li, J. Medicinal Purposes: Bioactive Metabolites from Marine-derived Organisms. Mini-Rev. Med. Chem. 2019, 19, 138–164. [Google Scholar] [CrossRef]
- Gunathilaka, T.L.; Samarakoon, K.; Ranasinghe, P.; Peiris, L.D.C. Antidiabetic Potential of Marine Brown Algae—A Mini Review. J. Diabetes Res. 2020, 2020, 1230218. [Google Scholar] [CrossRef]
- Abdelhafez, O.H.; Fahim, J.R.; El Masri, R.R.; Salem, M.A.; Desoukey, S.Y.; Ahmed, S.; Kamel, M.S.; Pimentel-Elardo, S.M.; Nodwell, J.R.; Abdelmohsen, U.R. Chemical and biological studies on the soft coral Nephthea sp. RSC Adv. 2021, 11, 23654–23663. [Google Scholar] [CrossRef]
- Sameeh, M.Y.; Mohamed, A.A.; Elazzazy, A.M.J. Polyphenolic contents and antimicrobial activity of different extracts of Padina boryana Thivy and Enteromorpha sp marine algae. J. App. Pharm. Sci. 2016, 6, 087–092. [Google Scholar] [CrossRef]
- Alghamdi, A.A.; Al-Hazmi, A.; Almalki, A.A.; Alsubaihi, A.A.; Anagreyyah, S.A.; Qasem, A.H.; Anajirih, N.A.; Allahyani, M.; Alyamani, R.A.; Albanghali, M.A.J. Antioxidant activity derived from marine green-lipped mussel Perna canaliculus extracts in mice. BioMed Res. Int. 2021, 2021, 1622270. [Google Scholar] [CrossRef] [PubMed]
- El-Beltagi, H.S.; Mohamed, A.A.; Mohamed, H.I.; Ramadan, K.M.; Barqawi, A.A.; Mansour, A.T.J. Phytochemical and potential properties of seaweeds and their recent applications: A review. Mar. Drugs 2022, 20, 342. [Google Scholar] [CrossRef] [PubMed]
- Hegazy, M.-E.F.; Moustfa, A.Y.; Mohamed, A.E.-H.H.; Alhammady, M.A.; Elbehairi, S.E.I.; Ohta, S.; Paré, P.W. New cytotoxic halogenated sesquiterpenes from the Egyptian sea hare, Aplysia oculifera. Tetrahedron Lett. 2014, 55, 1711–1714. [Google Scholar] [CrossRef]
- Liang, X.; Luo, D.; Luesch, H. Advances in exploring the therapeutic potential of marine natural products. Pharmacol. Res. 2019, 147, 104373. [Google Scholar] [CrossRef]
- Abdel-Wahab, N.M.; Gomaa, A.A.-R.; Mostafa, Y.A.; Hajjar, D.; Makki, A.A.; Alaaeldin, E.; Refaat, H.; Bringmann, G.; Zayed, A.; Abdelmohsen, U.R.; et al. Diterpenoids profile of the marine sponge Chelonaplysilla erecta and candidacy as potential antitumor drugs investigated by molecular docking and pharmacokinetic studies. Nat. Prod. Res. 2022, 37, 598–602. [Google Scholar] [CrossRef]
- Iqbal, K.; Khalid, S.; A McElroy, C.; Adnan, M.; Khan, G.M.; Dar, M.J. Triple-combination therapy for cutaneous leishmaniasis using detergent-free, hyaluronate-coated elastic nanovesicles. Nanomedicine 2022, 17, 1429–1447. [Google Scholar] [CrossRef]
- Khalid, S.; Salman, S.; Iqbal, K.; Rehman, F.U.; Ullah, I.; Satoskar, A.R.; Khan, G.M.; Dar, M.J. Surfactant free synthesis of cationic nano-vesicles: A safe triple drug loaded vehicle for the topical treatment of cutaneous leishmaniasis. Nanomed. Nanotechnol. Biol. Med. 2022, 40, 102490. [Google Scholar] [CrossRef]
- Dar, M.J.; McElroy, C.A.; Khan, M.I.; Satoskar, A.R.; Khan, G.M. Development and evaluation of novel miltefosine-polyphenol co-loaded second generation nano-transfersomes for the topical treatment of cutaneous leishmaniasis. Expert Opin. Drug Deliv. 2019, 17, 97–110. [Google Scholar] [CrossRef]
- Chen, M.; Lan, H.; Jin, K.; Chen, Y. Responsive nanosystems for targeted therapy of ulcerative colitis: Current practices and future perspectives. Drug Deliv. 2023, 30, 2219427. [Google Scholar] [CrossRef]
- Cheng, X.; Yan, H.; Pang, S.; Ya, M.; Qiu, F.; Qin, P.; Zeng, C.; Lu, Y. Liposomes as Multifunctional Nano-Carriers for Medicinal Natural Products. Front. Chem. 2022, 10, 963004. [Google Scholar] [CrossRef] [PubMed]
- Rezaul, M.; Shishir, M.R.I.; Karim, N.; Gowd, V.; Zheng, X.; Chen, W. Liposomal delivery of natural product: A promising approach in health research. Trends Food Sci. Technol. 2019, 85, 177–200. [Google Scholar] [CrossRef]
- Allam, A.E.; Amen, Y.; Ashour, A.; Assaf, H.K.; Hassan, H.A.; Abdel-Rahman, I.M.; Sayed, A.M.; Shimizu, K. In silico study of natural compounds from sesame against COVID-19 by targeting M pro, PL pro and RdRp. RSC Adv. 2021, 11, 22398–22408. [Google Scholar] [CrossRef] [PubMed]
- Mekhilef, S.; Saidur, R.; Kamalisarvestani, M. Effect of dust, humidity and air velocity on efficiency of photovoltaic cells. Renew. Sustain. Energy Rev. 2012, 16, 2920–2925. [Google Scholar] [CrossRef]
- Alnusaire, T.S.; Sayed, A.M.; Elmaidomy, A.H.; Al-Sanea, M.M.; Albogami, S.; Albqmi, M.; Alowaiesh, B.F.; Mostafa, E.M.; Musa, A.; Youssif, K. An in vitro and in silico study of the enhanced antiproliferative and pro-oxidant potential of Olea europaea L. cv. Arbosana leaf extract via elastic nanovesicles (spanlastics). Antioxidants 2021, 10, 1860. [Google Scholar] [CrossRef]
- Alaaeldin, E.; Mostafa, M.; Mansour, H.F.; Soliman, G.M. Spanlastics as an efficient delivery system for the enhancement of thymoquinone anticancer efficacy: Fabrication and cytotoxic studies against breast cancer cell lines. J. Drug Deliv. Sci. Technol. 2021, 65, 102725. [Google Scholar] [CrossRef]
- Immordino, M.L.; Dosio, F.; Cattel, L. Stealth liposomes: Review of the basic science, rationale, and clinical applications, existing and potential. Int. J. Nanomed. 2006, 1, 297–315. [Google Scholar]
- Vemuri, S.; Rhodes, C. Preparation and characterization of liposomes as therapeutic delivery systems: A review. Pharm. Acta Helvetiae 1995, 70, 95–111. [Google Scholar] [CrossRef]
- Salatin, S.; Maleki Dizaj, S.; Yari Khosroushahi, A. Effect of the surface modification, size, and shape on cellular uptake of nanoparticles. Cell Biol. Int. 2015, 39, 881–890. [Google Scholar] [CrossRef]
- Chang, N.; Ahn, S.H.; Kong, D.-S.; Lee, H.W.; Nam, D.-H. The role of STAT3 in glioblastoma progression through dual influences on tumor cells and the immune microenvironment. Mol. Cell. Endocrinol. 2017, 451, 53–65. [Google Scholar] [CrossRef]
- Mostofa, A.G.; Punganuru, S.R.; Madala, H.R.; Al-Obaide, M.; Srivenugopal, K.S. The Process and Regulatory Components of Inflammation in Brain Oncogenesis. Biomolecules 2017, 7, 34. [Google Scholar] [CrossRef] [PubMed]
- Rebhan, M.; Chalifa-Caspi, V.; Prilusky, J.; Lancet, D.J.B.U.; Center, G. GeneCards: Encyclopedia for Genes, Proteins and Diseases; Weizmann Institute of Science: London, UK, 1997. [Google Scholar]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Shady, N.H.; Mostafa, N.M.; Fayez, S.; Abdel-Rahman, I.M.; Maher, S.A.; Zayed, A.; Saber, E.A.; Khowdiary, M.M.; Elrehany, M.A.; Alzubaidi, M. Mechanistic Wound Healing and Antioxidant Potential of Moringa oleifera Seeds Extract Supported by Metabolic Profiling, In Silico Network Design, Molecular Docking, and In Vivo Studies. Antioxidants 2022, 11, 1743. [Google Scholar] [CrossRef] [PubMed]
- Chin, C.-H.; Chen, S.-H.; Wu, H.-H.; Ho, C.-W.; Ko, M.-T.; Lin, C.-Y. cytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 2014, 8 (Suppl. 4), S11. [Google Scholar] [CrossRef] [PubMed]
- Vaghasia, H.; Sakaria, S.; Prajapati, J.; Saraf, M.; Rawal, R.M. Interactive bioinformatics analysis for the screening of hub genes and molecular docking of phytochemicals present in kitchen spices to inhibit CDK1 in cervical cancer. Comput. Biol. Med. 2022, 149, 105994. [Google Scholar] [CrossRef]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef]
- Shi, S.; Ma, H.-Y.; Zhang, Z.-G. Clinicopathological and prognostic value of STAT3/p-STAT3 in cervical cancer: A meta and bioinformatics analysis. Pathol.-Res. Pract. 2021, 227, 153624. [Google Scholar] [CrossRef]
- Chiao, J.; Melikian, M.; Han, L.; Xue, C.; Tsao, A.; Wang, L.; Mencher, S.K.; Fallon, J.; Solangi, K.; Bertho, G.; et al. Interaction of a small molecule Natura-α and STAT3-SH2 domain to block Y705 phosphorylation and inhibit lupus nephritis. Biochem. Pharmacol. 2015, 99, 123–131. [Google Scholar] [CrossRef]
- Fu, X.; Sun, Z.; Long, Q.; Tan, W.; Ding, H.; Liu, X.; Wu, L.; Wang, Y.; Zhang, W. Glycosides from Buyang Huanwu Decoction inhibit atherosclerotic inflammation via JAK/STAT signaling pathway. Phytomedicine 2022, 105, 154385. [Google Scholar] [CrossRef]
- Aziz, A.; Sarwar, S.; Akter, T.; Uddin, S.; Xun, S.; Zhu, Y.; Islam, M.S.; Hongjie, Z. Polyphenolic molecules targeting STAT3 pathway for the treatment of cancer. Life Sci. 2021, 268, 118999. [Google Scholar] [CrossRef]
- Bharadwaj, U.; Kasembeli, M.M.; Tweardy, D.J. STAT3 Inhibitors in Cancer: A Comprehensive Update. In STAT Inhibitors in Cancer; Springer: New York, NY, USA, 2016; pp. 95–161. [Google Scholar] [CrossRef]
- Matsuno, K.; Masuda, Y.; Uehara, Y.; Sato, H.; Muroya, A.; Takahashi, O.; Yokotagawa, T.; Furuya, T.; Okawara, T.; Otsuka, M.; et al. Identification of a New Series of STAT3 Inhibitors by Virtual Screening. ACS Med. Chem. Lett. 2010, 1, 371–375. [Google Scholar] [CrossRef] [PubMed]
- Thilakasiri, P.S.; Dmello, R.S.; Nero, T.L.; Parker, M.W.; Ernst, M.; Chand, A.L. Repurposing of drugs as STAT3 inhibitors for cancer therapy. Semin. Cancer Biol. 2019, 68, 31–46. [Google Scholar] [CrossRef] [PubMed]
- Khanam, R.; Hejazi, I.I.; Shahabuddin, S.; Bhat, A.R.; Athar, F. Pharmacokinetic evaluation, molecular docking and in vitro biological evaluation of 1,3,4-oxadiazole derivatives as potent antioxidants and STAT3 inhibitors. J. Pharm. Anal. 2018, 9, 133–141. [Google Scholar] [CrossRef]
- Yu, H.; Lee, H.; Herrmann, A.; Buettner, R.; Jove, R. Revisiting STAT3 signalling in cancer: New and unexpected biological functions. Nat. Rev. Cancer 2014, 14, 736–746. [Google Scholar] [CrossRef] [PubMed]
- Xiong, H.; Zhang, Z.-G.; Tian, X.-Q.; Sun, D.-F.; Liang, Q.-C.; Zhang, Y.-J.; Lu, R.; Chen, Y.-X.; Fang, J.-Y. Inhibition of JAK1, 2/STAT3 Signaling Induces Apoptosis, Cell Cycle Arrest, and Reduces Tumor Cell Invasion in Colorectal Cancer Cells. Neoplasia 2008, 10, 287–297. [Google Scholar] [CrossRef]
- Refaat, H.; Naguib, Y.W.; Elsayed, M.M.A.; Sarhan, H.A.A.; Alaaeldin, E. Modified Spraying Technique and Response Surface Methodology for the Preparation and Optimization of Propolis Liposomes of Enhanced Anti-Proliferative Activity against Human Melanoma Cell Line A375. Pharmaceutics 2019, 11, 558. [Google Scholar] [CrossRef]
- Bozzuto, G.; Molinari, A. Liposomes as nanomedical devices. Int. J. Nanomed. 2015, 10, 975–999. [Google Scholar] [CrossRef]
- Mostafa, M.; Alaaeldin, E.; Aly, U.F.; Sarhan, H.A. Optimization and Characterization of Thymoquinone-Loaded Liposomes with Enhanced Topical Anti-inflammatory Activity. Aaps Pharmscitech 2018, 19, 3490–3500. [Google Scholar] [CrossRef]
- Skehan, P.; Storeng, R.; Scudiero, D.; Monks, A.; McMahon, J.; Vistica, D.; Warren, J.T.; Bokesch, H.; Kenney, S.; Boyd, M.R. New Colorimetric Cytotoxicity Assay for Anticancer-Drug Screening. J. Natl. Cancer Inst. 1990, 82, 1107–1112. [Google Scholar] [CrossRef]



| No. | Sample Data | DNA Content | |||
|---|---|---|---|---|---|
| Code | %G0-G1 | %S | %G2/M | Comment | |
| 1 | Epi-obtusane/HeLa | 46.29 | 28.72 | 24.99 | cell growth arrest @ G2/M phase |
| 2 | Cont. HeLa | 54.62 | 32.86 | 12.52 | |
| Compound | Apoptosis | Necrosis | |||
|---|---|---|---|---|---|
| Total | Early | Late | |||
| 1 | Epi-obtusane/HeLa | 39.61 | 22.24 | 12.51 | 4.86 |
| 2 | Control | 1.84 | 0.37 | 0.18 | 1.29 |
| Name | Occurrence |
|---|---|
| STAT3 | 8 |
| SRC | 7 |
| PIK3CA | 6 |
| AKT1 | 6 |
| JUN | 6 |
| CTNNB1 | 6 |
| MAPK3 | 6 |
| MYC | 6 |
| TP53 | 6 |
| CCND1 | 6 |
| ESR1 | 4 |
| il2 | 3 |
| HLA-DRB1 | 2 |
| CXCL8 | 2 |
| CD4 | 2 |
| CDK4 | 2 |
| No. | Compound | S a kcal/mole | Amino Acid Bond | Distance Å | E (Kcal Mol) |
|---|---|---|---|---|---|
| 1 | Epi-obtusane | −3.606 | Lys 591/H-acceptor | 3.1 | −1.3 |
| 2 | STX-0119 | −5.026 | Ser 636/H-donor | 2.9 | −6.1 |
| Ser 636/pi H | 4.43 | −0.8 | |||
| 3 | Pacritinib | −4.991 | Lys 591/H-acceptor | 3.3 | −1.7 |
| No. | Code | STAT3 ng/mL | nM | Folds | p-STAT3 ng/mL | nM | Folds |
|---|---|---|---|---|---|---|---|
| 1 | Epi-Obtusane/HeLa | 6.168 ± 0.31 | 15.4 | 0.45 | 19.94 ± 0.37 | 50.35 | 0.44 |
| 2 | Pacritinib/HeLa | 2.253 ± 0.04 | 4.7 | 0.16 | 13.09 ± 0.67 | 27.69 | 0.28 |
| 3 | Control | 13.67 ± 1.01 | 1 | 45.24 ± 2.03 | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abdelhafez, O.H.; Abdel-Rahman, I.M.; Alaaeldin, E.; Refaat, H.; El-Sayed, R.; Al-Harbi, S.A.; Shawky, A.M.; Hegazy, M.-E.F.; Moustafa, A.Y.; Shady, N.H. Pro-Apoptotic Activity of Epi-Obtusane against Cervical Cancer: Nano Formulation, In Silico Molecular Docking, and Pharmacological Network Analysis. Pharmaceuticals 2023, 16, 1578. https://doi.org/10.3390/ph16111578
Abdelhafez OH, Abdel-Rahman IM, Alaaeldin E, Refaat H, El-Sayed R, Al-Harbi SA, Shawky AM, Hegazy M-EF, Moustafa AY, Shady NH. Pro-Apoptotic Activity of Epi-Obtusane against Cervical Cancer: Nano Formulation, In Silico Molecular Docking, and Pharmacological Network Analysis. Pharmaceuticals. 2023; 16(11):1578. https://doi.org/10.3390/ph16111578
Chicago/Turabian StyleAbdelhafez, Omnia Hesham, Islam M. Abdel-Rahman, Eman Alaaeldin, Hesham Refaat, Refat El-Sayed, Sami A. Al-Harbi, Ahmed M. Shawky, Mohamed-Elamir F. Hegazy, Alaa Y. Moustafa, and Nourhan Hisham Shady. 2023. "Pro-Apoptotic Activity of Epi-Obtusane against Cervical Cancer: Nano Formulation, In Silico Molecular Docking, and Pharmacological Network Analysis" Pharmaceuticals 16, no. 11: 1578. https://doi.org/10.3390/ph16111578
APA StyleAbdelhafez, O. H., Abdel-Rahman, I. M., Alaaeldin, E., Refaat, H., El-Sayed, R., Al-Harbi, S. A., Shawky, A. M., Hegazy, M.-E. F., Moustafa, A. Y., & Shady, N. H. (2023). Pro-Apoptotic Activity of Epi-Obtusane against Cervical Cancer: Nano Formulation, In Silico Molecular Docking, and Pharmacological Network Analysis. Pharmaceuticals, 16(11), 1578. https://doi.org/10.3390/ph16111578







