Metformin Induces Apoptosis in Human Pancreatic Cancer (PC) Cells Accompanied by Changes in the Levels of Histone Acetyltransferases (Particularly, p300/CBP-Associated Factor (PCAF) Protein Levels)
Abstract
1. Introduction
2. Results
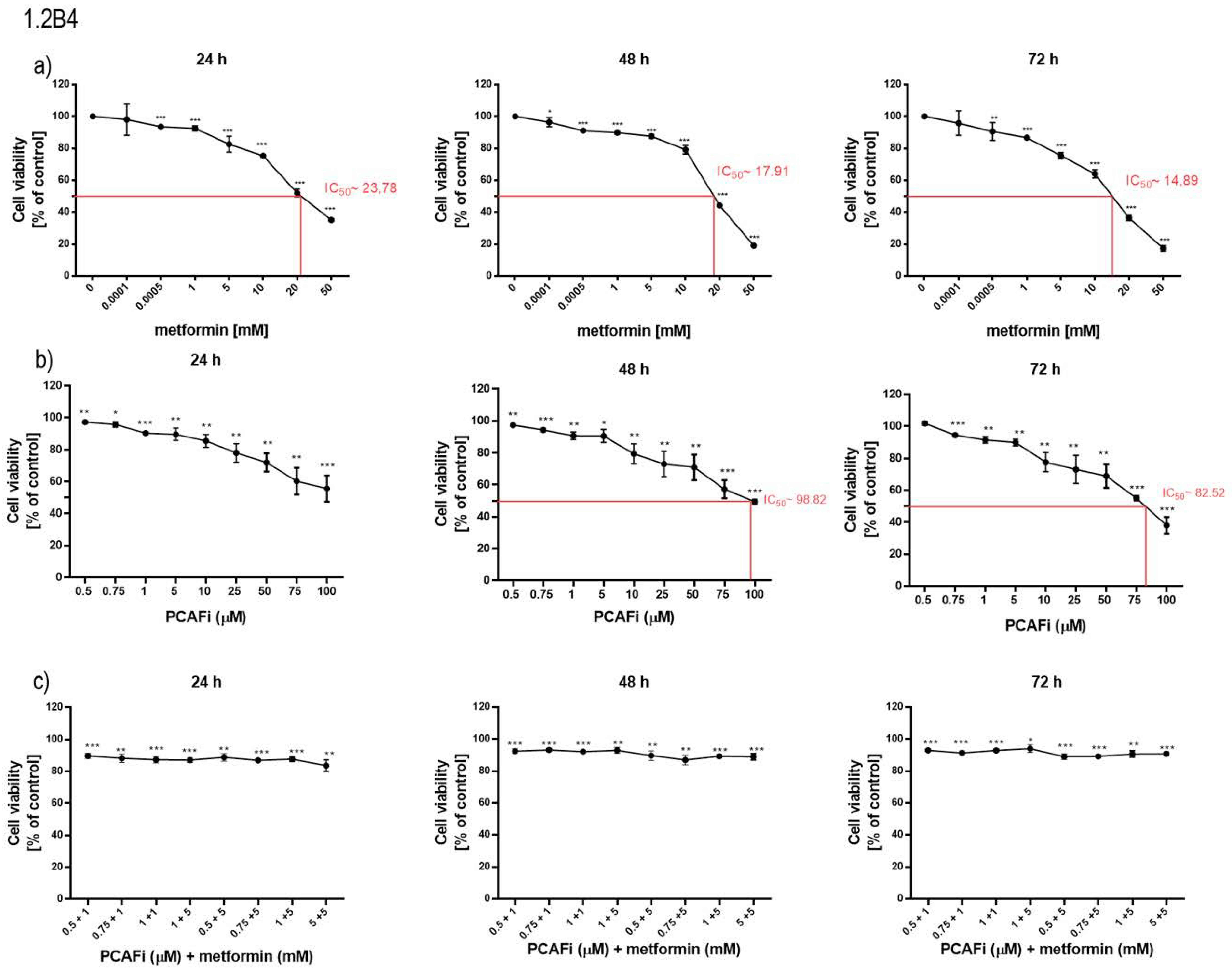
2.1. Metformin Decreased the Viability of 1.2B4 and PANC-1 Cells
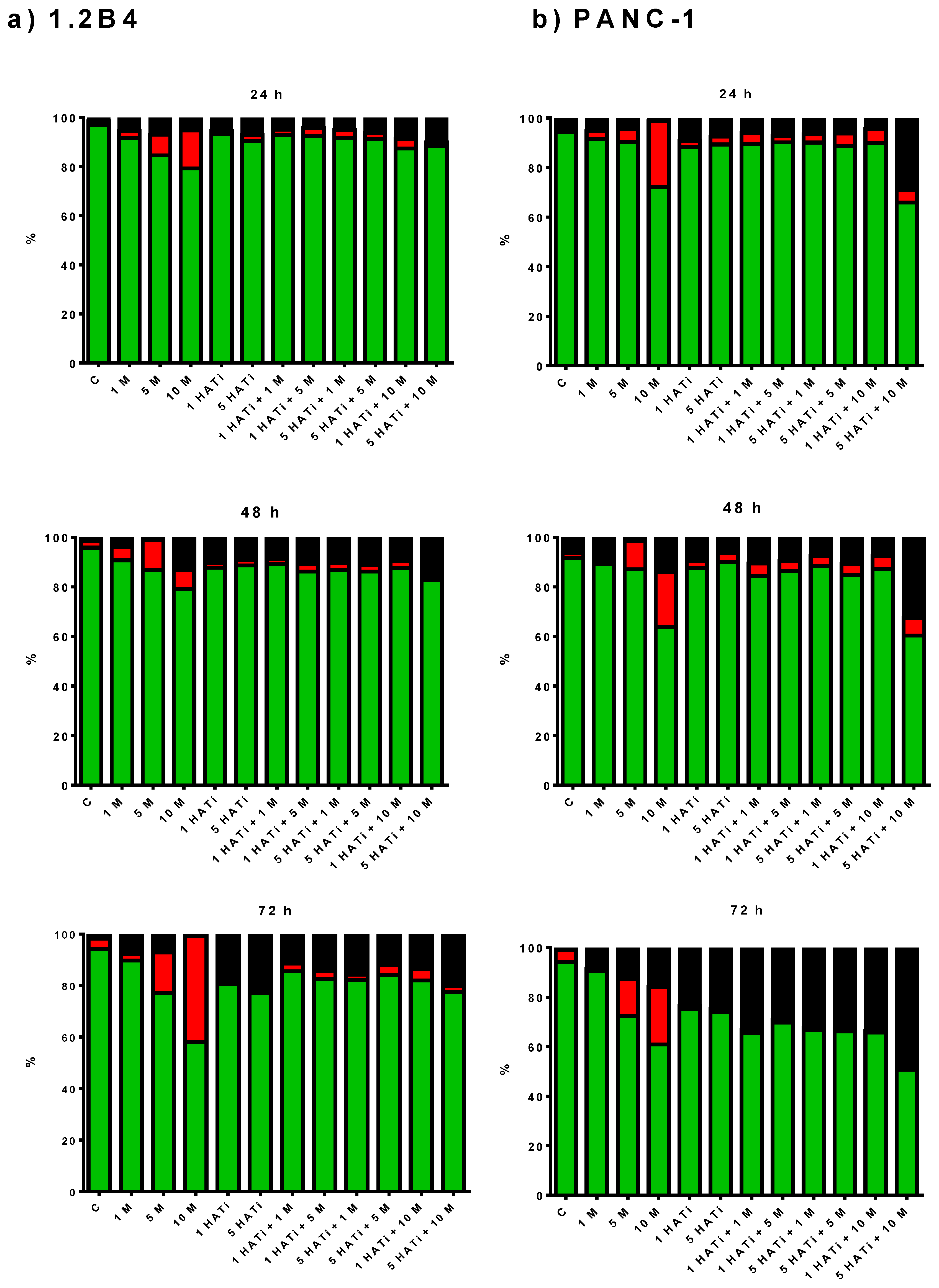
2.2. Metformin Induces Apoptosis of 1.2B4 and PANC-1 Cells
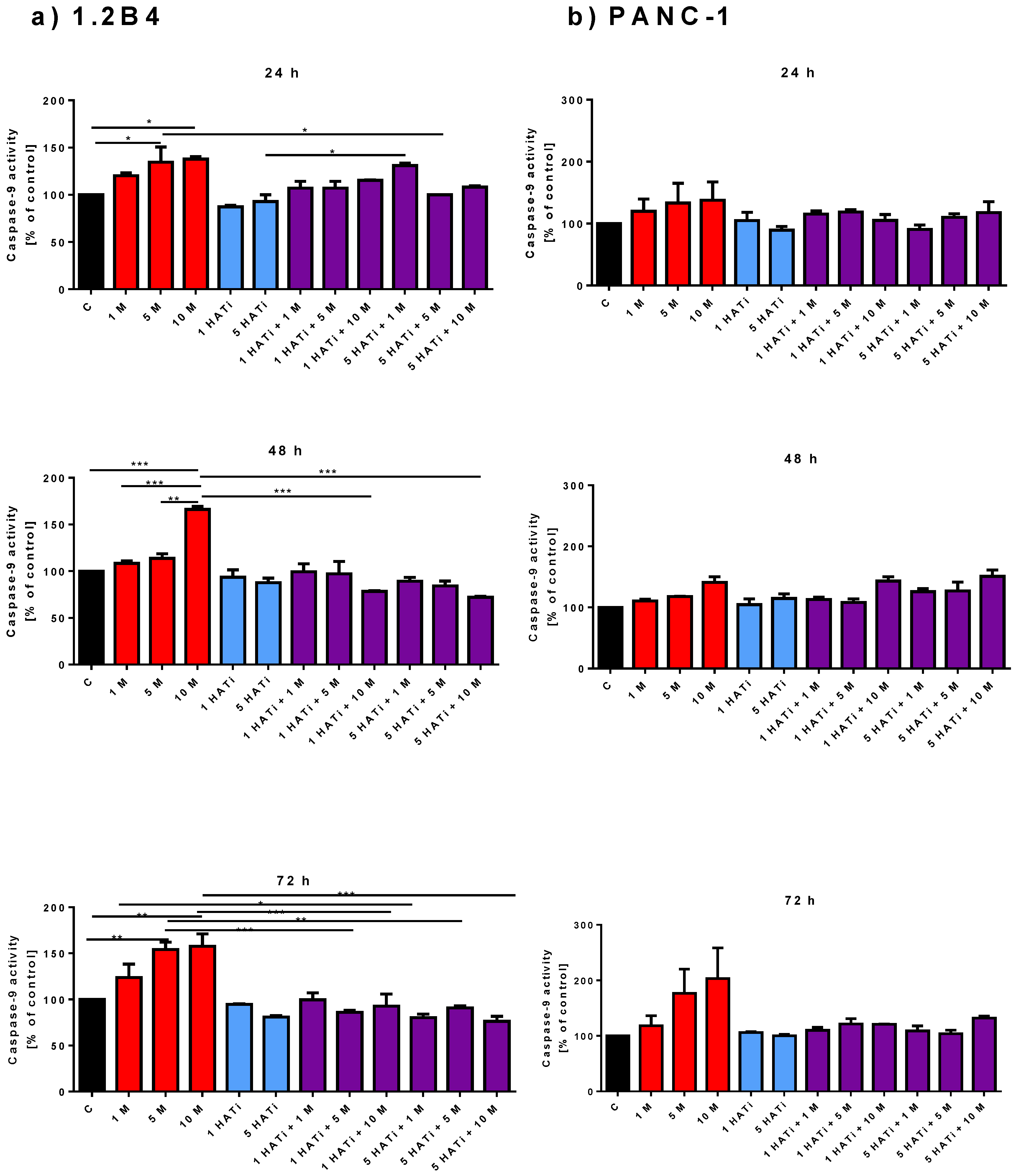
2.3. Metformin Increases Caspase-9 Activity
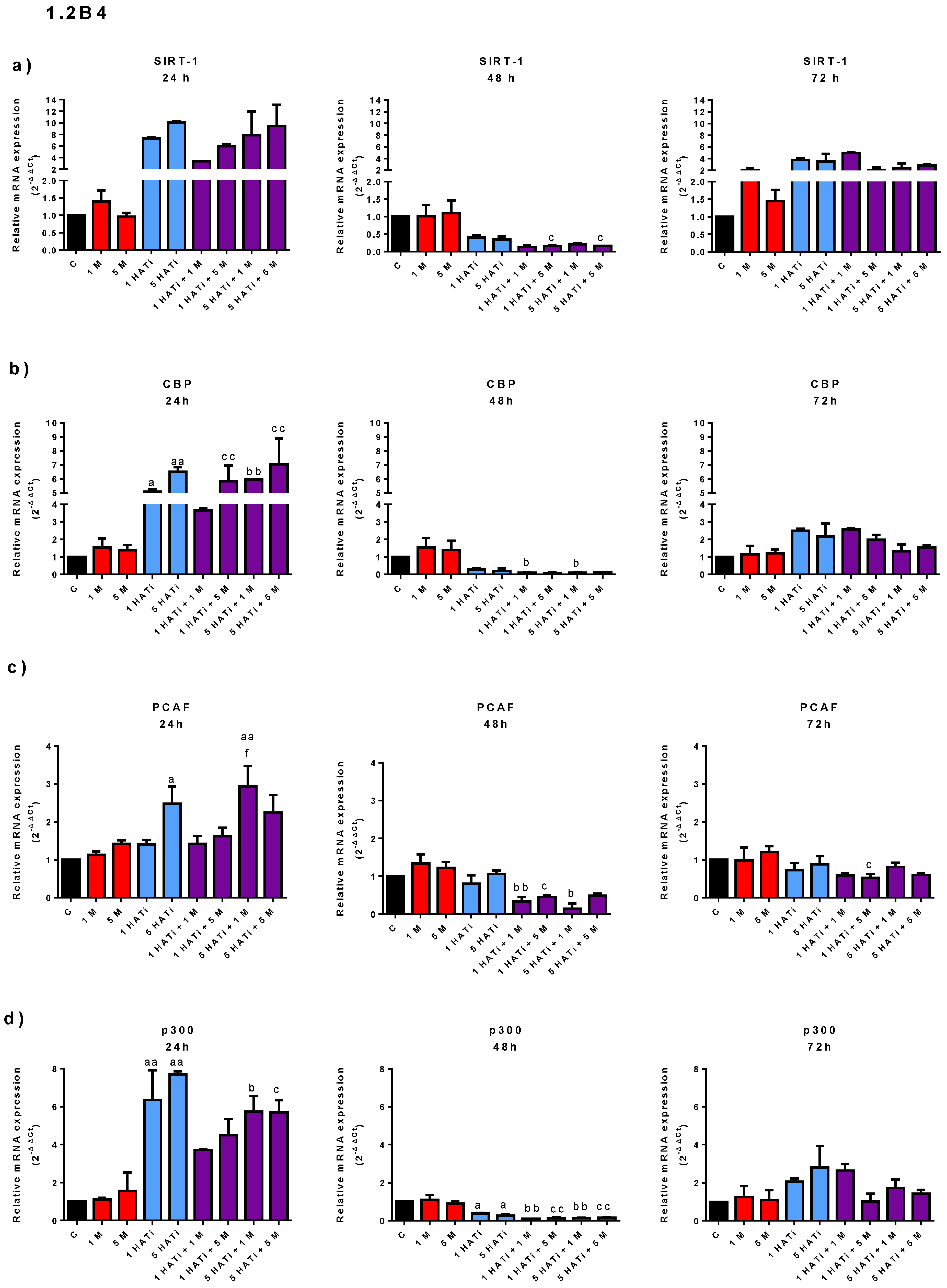
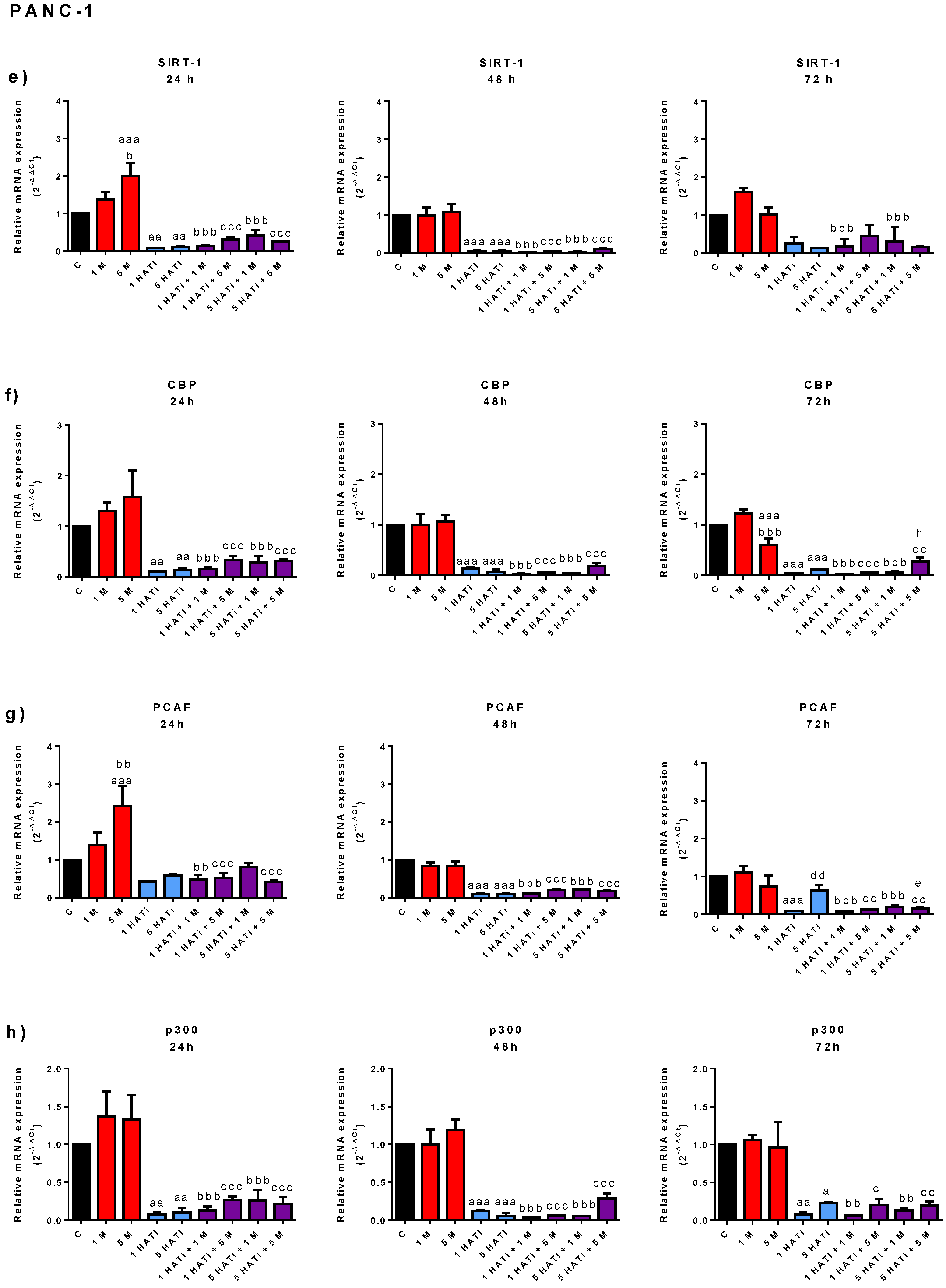
2.4. Metformin Slightly Modifies mRNA Expression of SIRT-1, PCAF, p300, and CBP in PANC-1 Cells, but Not in 1.2B4 Cells
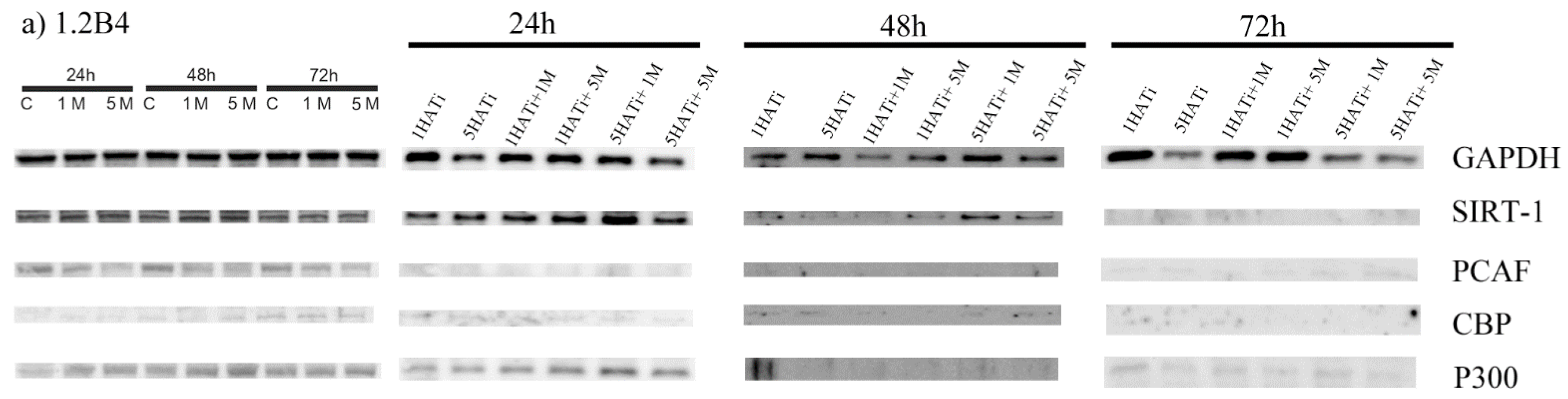
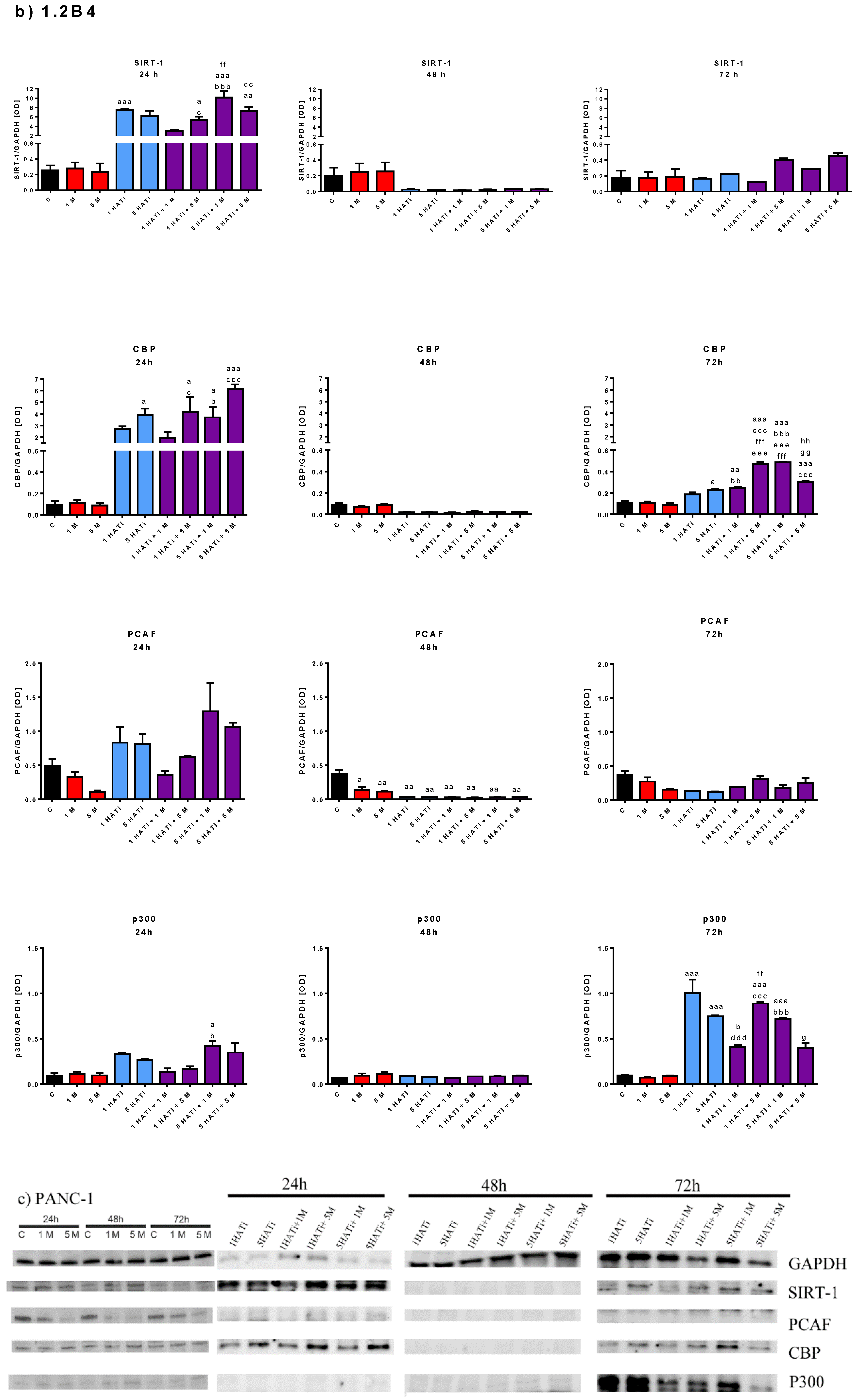
2.5. Metformin Decreases Protein Expression of PCAF in Both 1.2B4 and PANC-1 Cells
2.6. Metformin Acts via HAT to Induce Apoptosis of Human PC Cells
3. Discussion
4. Materials and Methods
4.1. Cell Lines and Treatment
4.2. The Cytotoxicity of Metformin, HATi, and Combination of HATi with Metformin—MTT Assay
4.3. Detection of Apoptosis—Flow Cytometry
4.4. Activity of Caspase-9
4.5. mRNA Expression—Total RNA Isolation and qRT-PCR
4.6. Protein Expression—Western Botting
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Leone, A.; Di Gennaro, E.; Bruzzese, F.; Avallone, A.; Budillon, A. New Perspective for an Old Antidiabetic Drug: Metformin as Anticancer Agent. Cancer Treat. Res. 2014, 159, 355–376. [Google Scholar] [CrossRef]
- Abudawood, M. Diabetes and Cancer: A Comprehensive Review. J. Res. Med. Sci. Off. J. Isfahan Univ. Med. Sci. 2019, 24, 94. [Google Scholar] [CrossRef] [PubMed]
- Evans, J.M.M.; Donnelly, L.A.; Emslie-Smith, A.M.; Alessi, D.R.; Morris, A.D. Metformin and Reduced Risk of Cancer in Diabetic Patients. BMJ 2005, 330, 1304–1305. [Google Scholar] [CrossRef] [PubMed]
- Franciosi, M.; Lucisano, G.; Lapice, E.; Strippoli, G.F.M.; Pellegrini, F.; Nicolucci, A. Metformin Therapy and Risk of Cancer in Patients with Type 2 Diabetes: Systematic Review. PLoS ONE 2013, 8, e71583. [Google Scholar] [CrossRef]
- Zhang, P.; Li, H.; Tan, X.; Chen, L.; Wang, S. Association of Metformin Use with Cancer Incidence and Mortality: A Meta-Analysis. Cancer Epidemiol. 2013, 37, 207–218. [Google Scholar] [CrossRef] [PubMed]
- Morales, D.R.; Morris, A.D. Metformin in Cancer Treatment and Prevention. Annu. Rev. Med. 2015, 66, 17–29. [Google Scholar] [CrossRef]
- Yu, H.; Zhong, X.; Gao, P.; Shi, J.; Wu, Z.; Guo, Z.; Wang, Z.; Song, Y. The Potential Effect of Metformin on Cancer: An Umbrella Review. Front. Endocrinol. 2019, 10, 617. [Google Scholar] [CrossRef] [PubMed]
- Madiraju, A.K.; Erion, D.M.; Rahimi, Y.; Zhang, X.-M.; Braddock, D.T.; Albright, R.A.; Prigaro, B.J.; Wood, J.L.; Bhanot, S.; MacDonald, M.J.; et al. Metformin Suppresses Gluconeogenesis by Inhibiting Mitochondrial Glycerophosphate Dehydrogenase. Nature 2014, 510, 542–546. [Google Scholar] [CrossRef]
- El-Mir, M.Y.; Nogueira, V.; Fontaine, E.; Avéret, N.; Rigoulet, M.; Leverve, X. Dimethylbiguanide Inhibits Cell Respiration via an Indirect Effect Targeted on the Respiratory Chain Complex I. J. Biol. Chem. 2000, 275, 223–228. [Google Scholar] [CrossRef]
- Zakikhani, M.; Dowling, R.; Fantus, I.G.; Sonenberg, N.; Pollak, M. Metformin Is an AMP Kinase-Dependent Growth Inhibitor for Breast Cancer Cells. Cancer Res. 2006, 66, 10269–10273. [Google Scholar] [CrossRef]
- Zhou, J.; Wulfkuhle, J.; Zhang, H.; Gu, P.; Yang, Y.; Deng, J.; Margolick, J.B.; Liotta, L.A.; Petricoin, E.; Zhang, Y. Activation of the PTEN/MTOR/STAT3 Pathway in Breast Cancer Stem-like Cells Is Required for Viability and Maintenance. Proc. Natl. Acad. Sci. USA 2007, 104, 16158–16163. [Google Scholar] [CrossRef] [PubMed]
- Korkaya, H.; Paulson, A.; Charafe-Jauffret, E.; Ginestier, C.; Brown, M.; Dutcher, J.; Clouthier, S.G.; Wicha, M.S. Regulation of Mammary Stem/Progenitor Cells by PTEN/Akt/Beta-Catenin Signaling. PLoS Biol. 2009, 7, e1000121. [Google Scholar] [CrossRef] [PubMed]
- Jalving, M.; Gietema, J.A.; Lefrandt, J.D.; de Jong, S.; Reyners, A.K.L.; Gans, R.O.B.; de Vries, E.G.E. Metformin: Taking Away the Candy for Cancer? Eur. J. Cancer 2010, 46, 2369–2380. [Google Scholar] [CrossRef] [PubMed]
- Gong, J.; Kelekar, G.; Shen, J.; Shen, J.; Kaur, S.; Mita, M. The Expanding Role of Metformin in Cancer: An Update on Antitumor Mechanisms and Clinical Development. Target. Oncol. 2016, 11, 447–467. [Google Scholar] [CrossRef]
- Pernicova, I.; Korbonits, M. Metformin--Mode of Action and Clinical Implications for Diabetes and Cancer. Nat. Rev. Endocrinol. 2014, 10, 143–156. [Google Scholar] [CrossRef]
- Pryor, R.; Cabreiro, F. Repurposing Metformin: An Old Drug with New Tricks in Its Binding Pockets. Biochem. J. 2015, 471, 307–322. [Google Scholar] [CrossRef]
- Safe, S.; Nair, V.; Karki, K. Metformin-Induced Anticancer Activities: Recent Insights. Biol. Chem. 2018, 399, 321–335. [Google Scholar] [CrossRef]
- Shogren-Knaak, M.; Ishii, H.; Sun, J.-M.; Pazin, M.J.; Davie, J.R.; Peterson, C.L. Histone H4-K16 Acetylation Controls Chromatin Structure and Protein Interactions. Science 2006, 311, 844–847. [Google Scholar] [CrossRef]
- Smith, B.C.; Denu, J.M. Chemical Mechanisms of Histone Lysine and Arginine Modifications. Biochim. Biophys. Acta 2009, 1789, 45–57. [Google Scholar] [CrossRef]
- Glozak, M.A.; Seto, E. Histone Deacetylases and Cancer. Oncogene 2007, 26, 5420–5432. [Google Scholar] [CrossRef]
- Bolden, J.E.; Peart, M.J.; Johnstone, R.W. Anticancer Activities of Histone Deacetylase Inhibitors. Nat. Rev. Drug Discov. 2006, 5, 769–784. [Google Scholar] [CrossRef]
- Schiltz, R.L.; Mizzen, C.A.; Vassilev, A.; Cook, R.G.; Allis, C.D.; Nakatani, Y. Overlapping but Distinct Patterns of Histone Acetylation by the Human Coactivators P300 and PCAF within Nucleosomal Substrates. J. Biol. Chem. 1999, 274, 1189–1192. [Google Scholar] [CrossRef]
- Trievel, R.C.; Li, F.Y.; Marmorstein, R. Application of a Fluorescent Histone Acetyltransferase Assay to Probe the Substrate Specificity of the Human P300/CBP-Associated Factor. Anal. Biochem. 2000, 287, 319–328. [Google Scholar] [CrossRef]
- Houtkooper, R.H.; Pirinen, E.; Auwerx, J. Sirtuins as Regulators of Metabolism and Healthspan. Nat. Rev. Mol. Cell Biol. 2012, 13, 225–238. [Google Scholar] [CrossRef] [PubMed]
- Saunders, L.R.; Verdin, E. Sirtuins: Critical Regulators at the Crossroads between Cancer and Aging. Oncogene 2007, 26, 5489–5504. [Google Scholar] [CrossRef] [PubMed]
- Rawla, P.; Sunkara, T.; Gaduputi, V. Epidemiology of Pancreatic Cancer: Global Trends, Etiology and Risk Factors. World J. Oncol. 2019, 10, 10–27. [Google Scholar] [CrossRef] [PubMed]
- Tseng, C.-H. Metformin Reduces Ovarian Cancer Risk in Taiwanese Women with Type 2 Diabetes Mellitus. Diabetes Metab. Res. Rev. 2015, 31, 619–626. [Google Scholar] [CrossRef]
- Tseng, C.-H. Metformin Significantly Reduces Incident Prostate Cancer Risk in Taiwanese Men with Type 2 Diabetes Mellitus. Eur. J. Cancer 2014, 50, 2831–2837. [Google Scholar] [CrossRef]
- Higurashi, T.; Takahashi, H.; Endo, H.; Hosono, K.; Yamada, E.; Ohkubo, H.; Sakai, E.; Uchiyama, T.; Hata, Y.; Fujisawa, N.; et al. Metformin Efficacy and Safety for Colorectal Polyps: A Double-Blind Randomized Controlled Trial. BMC Cancer 2012, 12, 118. [Google Scholar] [CrossRef]
- Sato, N.; Goggins, M. The Role of Epigenetic Alterations in Pancreatic Cancer. J. Hepato-Biliary-Pancreat. Surg. 2006, 13, 286–295. [Google Scholar] [CrossRef]
- Bridgeman, S.C.; Ellison, G.C.; Melton, P.E.; Newsholme, P.; Mamotte, C.D.S. Epigenetic Effects of Metformin: From Molecular Mechanisms to Clinical Implications. Diabetes Obes. Metab. 2018, 20, 1553–1562. [Google Scholar] [CrossRef] [PubMed]
- Libby, G.; Donnelly, L.A.; Donnan, P.T.; Alessi, D.R.; Morris, A.D.; Evans, J.M.M. New Users of Metformin Are at Low Risk of Incident Cancer. Diabetes Care 2009, 32, 1620–1625. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.Y.; Kim, Y.S.; Kim, M.; Choi, M.Y.; Roh, G.S.; Lee, D.H.; Kim, H.J.; Kang, S.S.; Cho, G.J.; Shin, J.K.; et al. Metformin Inhibits Cervical Cancer Cell Proliferation via Decreased AMPK O-GlcNAcylation. Anim. Cells Syst. 2019, 23, 302–309. [Google Scholar] [CrossRef] [PubMed]
- Erices, R.; Bravo, M.L.; Gonzalez, P.; Oliva, B.; Racordon, D.; Garrido, M.; Ibañez, C.; Kato, S.; Brañes, J.; Pizarro, J.; et al. Metformin, at Concentrations Corresponding to the Treatment of Diabetes, Potentiates the Cytotoxic Effects of Carboplatin in Cultures of Ovarian Cancer Cells. Reprod. Sci. 2013, 20, 1433–1446. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Wei, J.; Wan, J.; Wang, W.; Wang, L.; Yuan, Y.; Yang, Z.; Liu, X.; Ming, L. Low Glucose and Metformin-Induced Apoptosis of Human Ovarian Cancer Cells Is Connected to ASK1 via Mitochondrial and Endoplasmic Reticulum Stress-Associated Pathways. J. Exp. Clin. Cancer Res. CR 2019, 38, 77. [Google Scholar] [CrossRef] [PubMed]
- Kato, K.; Gong, J.; Iwama, H.; Kitanaka, A.; Tani, J.; Miyoshi, H.; Nomura, K.; Mimura, S.; Kobayashi, M.; Aritomo, Y.; et al. The Antidiabetic Drug Metformin Inhibits Gastric Cancer Cell Proliferation In Vitro and In Vivo. Mol. Cancer Ther. 2012, 11, 549–560. [Google Scholar] [CrossRef]
- Mogavero, A.; Maiorana, M.V.; Zanutto, S.; Varinelli, L.; Bozzi, F.; Belfiore, A.; Volpi, C.C.; Gloghini, A.; Pierotti, M.A.; Gariboldi, M. Metformin Transiently Inhibits Colorectal Cancer Cell Proliferation as a Result of Either AMPK Activation or Increased ROS Production. Sci. Rep. 2017, 7, 15992. [Google Scholar] [CrossRef]
- Deng, X.-S.; Wang, S.; Deng, A.; Liu, B.; Edgerton, S.M.; Lind, S.E.; Wahdan-Alaswad, R.; Thor, A.D. Metformin Targets Stat3 to Inhibit Cell Growth and Induce Apoptosis in Triple-Negative Breast Cancers. Cell Cycle Georget. Tex 2012, 11, 367–376. [Google Scholar] [CrossRef]
- Zordoky, B.N.M.; Bark, D.; Soltys, C.L.; Sung, M.M.; Dyck, J.R.B. The Anti-Proliferative Effect of Metformin in Triple-Negative MDA-MB-231 Breast Cancer Cells Is Highly Dependent on Glucose Concentration: Implications for Cancer Therapy and Prevention. Biochim. Biophys. Acta 2014, 1840, 1943–1957. [Google Scholar] [CrossRef]
- Tseng, H.-W.; Li, S.-C.; Tsai, K.-W. Metformin Treatment Suppresses Melanoma Cell Growth and Motility Through Modulation of MicroRNA Expression. Cancers 2019, 11, 209. [Google Scholar] [CrossRef]
- Wang, L.-W.; Li, Z.-S.; Zou, D.-W.; Jin, Z.-D.; Gao, J.; Xu, G.-M. Metformin Induces Apoptosis of Pancreatic Cancer Cells. World J. Gastroenterol. 2008, 14, 7192–7198. [Google Scholar] [CrossRef] [PubMed]
- Nair, V.; Pathi, S.; Jutooru, I.; Sreevalsan, S.; Basha, R.; Abdelrahim, M.; Samudio, I.; Safe, S. Metformin Inhibits Pancreatic Cancer Cell and Tumor Growth and Downregulates Sp Transcription Factors. Carcinogenesis 2013, 34, 2870–2879. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, R.; Tomosugi, M.; Horinaka, M.; Sowa, Y.; Sakai, T. Metformin Causes G1-Phase Arrest via Down-Regulation of MiR-221 and Enhances TRAIL Sensitivity through DR5 Up-Regulation in Pancreatic Cancer Cells. PLoS ONE 2015, 10, e0125779. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.-W.; Zhou, N.; Jin, F.; Wang, R.; Zhao, J.-Q. Metformin Reduces Pancreatic Cancer Cell Proliferation and Increases Apoptosis through MTOR Signaling Pathway and Its Dose-Effect Relationship. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 5336–5344. [Google Scholar] [CrossRef]
- Chen, K.; Qian, W.; Jiang, Z.; Cheng, L.; Li, J.; Sun, L.; Zhou, C.; Gao, L.; Lei, M.; Yan, B.; et al. Metformin Suppresses Cancer Initiation and Progression in Genetic Mouse Models of Pancreatic Cancer. Mol. Cancer 2017, 16, 131. [Google Scholar] [CrossRef]
- Lomberk, G.; Dusetti, N.; Iovanna, J.; Urrutia, R. Emerging Epigenomic Landscapes of Pancreatic Cancer in the Era of Precision Medicine. Nat. Commun. 2019, 10, 3875. [Google Scholar] [CrossRef]
- Paradise, B.D.; Barham, W.; Fernandez-Zapico, M.E. Targeting Epigenetic Aberrations in Pancreatic Cancer, a New Path to Improve Patient Outcomes? Cancers 2018, 10, 128. [Google Scholar] [CrossRef]
- Jin, J.; Chu, Z.; Ma, P.; Meng, Y.; Yang, Y. SIRT1 Promotes the Proliferation and Metastasis of Human Pancreatic Cancer Cells. Tumour Biol. J. Int. Soc. Oncodevelopmental Biol. Med. 2017, 39, 1010428317691180. [Google Scholar] [CrossRef]
- Li, Y.-H.; Li, Y.-X.; Li, M.; Song, S.; Ge, Y.; Jin, J.; Li, X.; Tan, X.; Ye, J. The Ras-ERK1/2 Signaling Pathway Regulates H3K9ac through PCAF to Promote the Development of Pancreatic Cancer. Life Sci. 2020, 256, 117936. [Google Scholar] [CrossRef]
- Yu, X.; Mao, W.; Zhai, Y.; Tong, C.; Liu, M.; Ma, L.; Yu, X.; Li, S. Anti-Tumor Activity of Metformin: From Metabolic and Epigenetic Perspectives. Oncotarget 2017, 8, 5619–5628. [Google Scholar] [CrossRef]
- Imai, S.-I.; Kiess, W. Therapeutic Potential of SIRT1 and NAMPT-Mediated NAD Biosynthesis in Type 2 Diabetes. Front. Biosci. Landmark Ed. 2009, 14, 2983–2995. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.; Cui, J.; Zhang, J.-G.; Qin, Q.; Chen, Q.; Yin, T.; Deng, S.-C.; Liu, Y.; Liu, L.; Wang, B.; et al. SIRT1 RNAi Knockdown Induces Apoptosis and Senescence, Inhibits Invasion and Enhances Chemosensitivity in Pancreatic Cancer Cells. Gene Ther. 2011, 18, 920–928. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhao, G.; Qin, Q.; Wang, B.; Liu, L.; Liu, Y.; Deng, S.; Tian, K.; Wang, C. Nicotinamide Prohibits Proliferation and Enhances Chemosensitivity of Pancreatic Cancer Cells through Deregulating SIRT1 and Ras/Akt Pathways. Pancreatol. Off. J. Int. Assoc. Pancreatol. IAP Al 2013, 13, 140–146. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Cheng, X.; Wang, D.; Peng, M.; Xue, Z.; Da, Y.; Zhang, N.; Yao, Z.; Li, M.; Xu, A.; et al. Adiponectin Promotes Pancreatic Cancer Progression by Inhibiting Apoptosis via the Activation of AMPK/Sirt1/PGC-1α Signaling. Oncotarget 2014, 5, 4732–4745. [Google Scholar] [CrossRef]
- Liu, Y.; Li, X.; Zhu, S.; Zhang, J.-g.; Yang, M.; Qin, Q.; Deng, S.-c.; Wang, B.; Tian, K.; Liu, L.; et al. Ectopic Expression of MiR-494 Inhibited the Proliferation, Invasion and Chemoresistance of Pancreatic Cancer by Regulating SIRT1 and c-Myc. Gene Ther. 2015, 22, 729–738. [Google Scholar] [CrossRef]
- Wang, F.; Li, H.; Yan, X.-G.; Zhou, Z.-W.; Yi, Z.-G.; He, Z.-X.; Pan, S.-T.; Yang, Y.-X.; Wang, Z.-Z.; Zhang, X.; et al. Alisertib Induces Cell Cycle Arrest and Autophagy and Suppresses Epithelial-to-Mesenchymal Transition Involving PI3K/Akt/MTOR and Sirtuin 1-Mediated Signaling Pathways in Human Pancreatic Cancer Cells. Drug Des. Dev. Ther. 2015, 9, 575–601. [Google Scholar] [CrossRef]
- Zhang, E.; Guo, Q.; Gao, H.; Xu, R.; Teng, S.; Wu, Y. Metformin and Resveratrol Inhibited High Glucose-Induced Metabolic Memory of Endothelial Senescence through SIRT1/P300/P53/P21 Pathway. PLoS ONE 2015, 10, e0143814. [Google Scholar] [CrossRef]
- Lin, Z.; Fang, D. The Roles of SIRT1 in Cancer. Genes Cancer 2013, 4, 97–104. [Google Scholar] [CrossRef]
- Yi, G.; He, Z.; Zhou, X.; Xian, L.; Yuan, T.; Jia, X.; Hong, J.; He, L.; Liu, J. Low Concentration of Metformin Induces a P53-Dependent Senescence in Hepatoma Cells via Activation of the AMPK Pathway. Int. J. Oncol. 2013, 43, 1503–1510. [Google Scholar] [CrossRef]
- He, L.; Sabet, A.; Djedjos, S.; Miller, R.; Sun, X.; Hussain, M.A.; Radovick, S.; Wondisford, F.E. Metformin and Insulin Suppress Hepatic Gluconeogenesis through Phosphorylation of CREB Binding Protein. Cell 2009, 137, 635–646. [Google Scholar] [CrossRef]
- Zhang, Y.; Qiu, J.; Wang, X.; Zhang, Y.; Xia, M. AMP-Activated Protein Kinase Suppresses Endothelial Cell Inflammation through Phosphorylation of Transcriptional Coactivator P300. Arterioscler. Thromb. Vasc. Biol. 2011, 31, 2897–2908. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Hong, Y.H.; Shen, X.Q.; Frankowski, C.; Camp, H.S.; Leff, T. Regulation of Transcription by AMP-Activated Protein Kinase: Phosphorylation of P300 Blocks Its Interaction with Nuclear Receptors. J. Biol. Chem. 2001, 276, 38341–38344. [Google Scholar] [CrossRef] [PubMed]
- Tachibana, K.; Yamasaki, D.; Ishimoto, K.; Doi, T. The Role of PPARs in Cancer. PPAR Res. 2008, 2008, 102737. [Google Scholar] [CrossRef] [PubMed]
- Eckner, R. P53-Dependent Growth Arrest and Induction of P21: A Critical Role for PCAF-Mediated Histone Acetylation. Cell Cycle Georget. Tex 2012, 11, 2591–2592. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Gai, X.; Ding, F.; Lu, Z.; Tu, K.; Yao, Y.; Liu, Q. Histone Acetyltransferase PCAF Up-Regulated Cell Apoptosis in Hepatocellular Carcinoma via Acetylating Histone H4 and Inactivating AKT Signaling. Mol. Cancer 2013, 12, 96. [Google Scholar] [CrossRef]
- Gai, X.; Tu, K.; Li, C.; Lu, Z.; Roberts, L.R.; Zheng, X. Histone Acetyltransferase PCAF Accelerates Apoptosis by Repressing a GLI1/BCL2/BAX Axis in Hepatocellular Carcinoma. Cell Death Dis. 2015, 6, e1712. [Google Scholar] [CrossRef]
- Fei, H.-J.; Zu, L.-D.; Wu, J.; Jiang, X.-S.; Wang, J.-L.; Chin, Y.E.; Fu, G.-H. PCAF Acts as a Gastric Cancer Suppressor through a Novel PCAF-P16-CDK4 Axis. Am. J. Cancer Res. 2016, 6, 2772–2786. [Google Scholar]
- Wan, J.; Xu, W.; Zhan, J.; Ma, J.; Li, X.; Xie, Y.; Wang, J.; Zhu, W.-G.; Luo, J.; Zhang, H. PCAF-Mediated Acetylation of Transcriptional Factor HOXB9 Suppresses Lung Adenocarcinoma Progression by Targeting Oncogenic Protein JMJD6. Nucleic Acids Res. 2016, 44, 10662–10675. [Google Scholar] [CrossRef]
- Malatesta, M.; Steinhauer, C.; Mohammad, F.; Pandey, D.P.; Squatrito, M.; Helin, K. Histone Acetyltransferase PCAF Is Required for Hedgehog-Gli-Dependent Transcription and Cancer Cell Proliferation. Cancer Res. 2013, 73, 6323–6333. [Google Scholar] [CrossRef]
- Zhang, S.; Sun, G.; Wang, Z.; Wan, Y.; Guo, J.; Shi, L. PCAF-Mediated Akt1 Acetylation Enhances the Proliferation of Human Glioblastoma Cells. Tumour Biol. J. Int. Soc. Oncodevelopmental Biol. Med. 2015, 36, 1455–1462. [Google Scholar] [CrossRef]
- Patel, J.H.; Du, Y.; Ard, P.G.; Phillips, C.; Carella, B.; Chen, C.-J.; Rakowski, C.; Chatterjee, C.; Lieberman, P.M.; Lane, W.S.; et al. The C-MYC Oncoprotein Is a Substrate of the Acetyltransferases HGCN5/PCAF and TIP60. Mol. Cell. Biol. 2004, 24, 10826–10834. [Google Scholar] [CrossRef] [PubMed]
- Xenaki, G.; Ontikatze, T.; Rajendran, R.; Stratford, I.J.; Dive, C.; Krstic-Demonacos, M.; Demonacos, C. PCAF Is an HIF-1alpha Cofactor That Regulates P53 Transcriptional Activity in Hypoxia. Oncogene 2008, 27, 5785–5796. [Google Scholar] [CrossRef] [PubMed]
- Love, I.M.; Sekaric, P.; Shi, D.; Grossman, S.R.; Androphy, E.J. The Histone Acetyltransferase PCAF Regulates P21 Transcription through Stress-Induced Acetylation of Histone H3. Cell Cycle Georget. Tex 2012, 11, 2458–2466. [Google Scholar] [CrossRef] [PubMed]
- Poux, A.N.; Marmorstein, R. Molecular Basis for Gcn5/PCAF Histone Acetyltransferase Selectivity for Histone and Nonhistone Substrates. Biochemistry 2003, 42, 14366–14374. [Google Scholar] [CrossRef]
- Lim, J.-H.; West, K.L.; Rubinstein, Y.; Bergel, M.; Postnikov, Y.V.; Bustin, M. Chromosomal Protein HMGN1 Enhances the Acetylation of Lysine 14 in Histone H3. EMBO J. 2005, 24, 3038–3048. [Google Scholar] [CrossRef]
- Zhao, J.; Gong, A.-Y.; Zhou, R.; Liu, J.; Eischeid, A.N.; Chen, X.-M. Downregulation of PCAF by MiR-181a/b Provides Feedback Regulation to TNF-α-Induced Transcription of Proinflammatory Genes in Liver Epithelial Cells. J. Immunol. 2012, 188, 1266–1274. [Google Scholar] [CrossRef]
- Gong, A.-Y.; Eischeid, A.N.; Xiao, J.; Zhao, J.; Chen, D.; Wang, Z.-Y.; Young, C.Y.; Chen, X.-M. MiR-17-5p Targets the P300/CBP-Associated Factor and Modulates Androgen Receptor Transcriptional Activity in Cultured Prostate Cancer Cells. BMC Cancer 2012, 12, 492. [Google Scholar] [CrossRef]
- Pichiorri, F.; Suh, S.-S.; Ladetto, M.; Kuehl, M.; Palumbo, T.; Drandi, D.; Taccioli, C.; Zanesi, N.; Alder, H.; Hagan, J.P.; et al. MicroRNAs Regulate Critical Genes Associated with Multiple Myeloma Pathogenesis. Proc. Natl. Acad. Sci. USA 2008, 105, 12885–12890. [Google Scholar] [CrossRef]
- Li, J.; Chen, C.; Ma, X.; Geng, G.; Liu, B.; Zhang, Y.; Zhang, S.; Zhong, F.; Liu, C.; Yin, Y.; et al. Long Noncoding RNA NRON Contributes to HIV-1 Latency by Specifically Inducing Tat Protein Degradation. Nat. Commun. 2016, 7, 11730. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, W.; Zhu, W.; Dong, J.; Cheng, Y.; Yin, Z.; Shen, F. Mechanisms and Functions of Long Non-Coding RNAs at Multiple Regulatory Levels. Int. J. Mol. Sci. 2019, 20, 5573. [Google Scholar] [CrossRef]
- Zhao, B.S.; Roundtree, I.A.; He, C. Post-Transcriptional Gene Regulation by MRNA Modifications. Nat. Rev. Mol. Cell Biol. 2017, 18, 31–42. [Google Scholar] [CrossRef] [PubMed]
- Santos-Rosa, H.; Valls, E.; Kouzarides, T.; Martínez-Balbás, M. Mechanisms of P/CAF Auto-Acetylation. Nucleic Acids Res. 2003, 31, 4285–4292. [Google Scholar] [CrossRef] [PubMed]
- Dasuri, K.; Zhang, L.; Keller, J.N. Oxidative Stress, Neurodegeneration, and the Balance of Protein Degradation and Protein Synthesis. Free Radic. Biol. Med. 2013, 62, 170–185. [Google Scholar] [CrossRef] [PubMed]
- Yi, Y.; Chen, D.; Ao, J.; Sun, S.; Wu, M.; Li, X.; Bergholz, J.; Zhang, Y.; Xiao, Z.-X. Metformin Promotes AMP-Activated Protein Kinase-Independent Suppression of ΔNp63α Protein Expression and Inhibits Cancer Cell Viability. J. Biol. Chem. 2017, 292, 5253–5261. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Zeng, S.X.; Lee, H.; Lu, H. MDM2 Mediates P300/CREB-Binding Protein-Associated Factor Ubiquitination and Degradation. J. Biol. Chem. 2004, 279, 20035–20043. [Google Scholar] [CrossRef]
- Skelding, K.A.; Rostas, J.A.P.; Verrills, N.M. Controlling the Cell Cycle: The Role of Calcium/Calmodulin-Stimulated Protein Kinases I and II. Cell Cycle Georget. Tex 2011, 10, 631–639. [Google Scholar] [CrossRef]
- Mazumder, S.; DuPree, E.L.; Almasan, A. A Dual Role of Cyclin E in Cell Proliferation and Apotosis May Provide a Target for Cancer Therapy. Curr. Cancer Drug Targets 2004, 4, 65–75. [Google Scholar] [CrossRef]
- Arif, M.; Vedamurthy, B.M.; Choudhari, R.; Ostwal, Y.B.; Mantelingu, K.; Kodaganur, G.S.; Kundu, T.K. Nitric Oxide-Mediated Histone Hyperacetylation in Oral Cancer: Target for a Water-Soluble HAT Inhibitor, CTK7A. Chem. Biol. 2010, 17, 903–913. [Google Scholar] [CrossRef] [PubMed]
- Gradiz, R.; Silva, H.C.; Carvalho, L.; Botelho, M.F.; Mota-Pinto, A. MIA PaCa-2 and PANC-1 - Pancreas Ductal Adenocarcinoma Cell Lines with Neuroendocrine Differentiation and Somatostatin Receptors. Sci. Rep. 2016, 6, 21648. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods San Diego Calif 2001, 25, 402–408. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Szymczak-Pajor, I.; Drzewoski, J.; Świderska, E.; Strycharz, J.; Gabryanczyk, A.; Kasznicki, J.; Bogdańska, M.; Śliwińska, A. Metformin Induces Apoptosis in Human Pancreatic Cancer (PC) Cells Accompanied by Changes in the Levels of Histone Acetyltransferases (Particularly, p300/CBP-Associated Factor (PCAF) Protein Levels). Pharmaceuticals 2023, 16, 115. https://doi.org/10.3390/ph16010115
Szymczak-Pajor I, Drzewoski J, Świderska E, Strycharz J, Gabryanczyk A, Kasznicki J, Bogdańska M, Śliwińska A. Metformin Induces Apoptosis in Human Pancreatic Cancer (PC) Cells Accompanied by Changes in the Levels of Histone Acetyltransferases (Particularly, p300/CBP-Associated Factor (PCAF) Protein Levels). Pharmaceuticals. 2023; 16(1):115. https://doi.org/10.3390/ph16010115
Chicago/Turabian StyleSzymczak-Pajor, Izabela, Józef Drzewoski, Ewa Świderska, Justyna Strycharz, Anna Gabryanczyk, Jacek Kasznicki, Marta Bogdańska, and Agnieszka Śliwińska. 2023. "Metformin Induces Apoptosis in Human Pancreatic Cancer (PC) Cells Accompanied by Changes in the Levels of Histone Acetyltransferases (Particularly, p300/CBP-Associated Factor (PCAF) Protein Levels)" Pharmaceuticals 16, no. 1: 115. https://doi.org/10.3390/ph16010115
APA StyleSzymczak-Pajor, I., Drzewoski, J., Świderska, E., Strycharz, J., Gabryanczyk, A., Kasznicki, J., Bogdańska, M., & Śliwińska, A. (2023). Metformin Induces Apoptosis in Human Pancreatic Cancer (PC) Cells Accompanied by Changes in the Levels of Histone Acetyltransferases (Particularly, p300/CBP-Associated Factor (PCAF) Protein Levels). Pharmaceuticals, 16(1), 115. https://doi.org/10.3390/ph16010115





