LC–MS/MS Phytochemical Profiling, Antioxidant Activity, and Cytotoxicity of the Ethanolic Extract of Atriplex halimus L. against Breast Cancer Cell Lines: Computational Studies and Experimental Validation
Abstract
1. Introduction
2. Results and Discussion
2.1. LC–MS/MS Phytochemical Profiling Results
2.2. Pharmacokinetic Properties (ADME) of AHEE
2.3. In Silico Toxicity Prediction (Using Pro-Tox II)
2.4. In Silico Prediction of a Protein-Target-Based Antioxidant and Cytotoxic Mechanisms by Molecular Docking Analysis
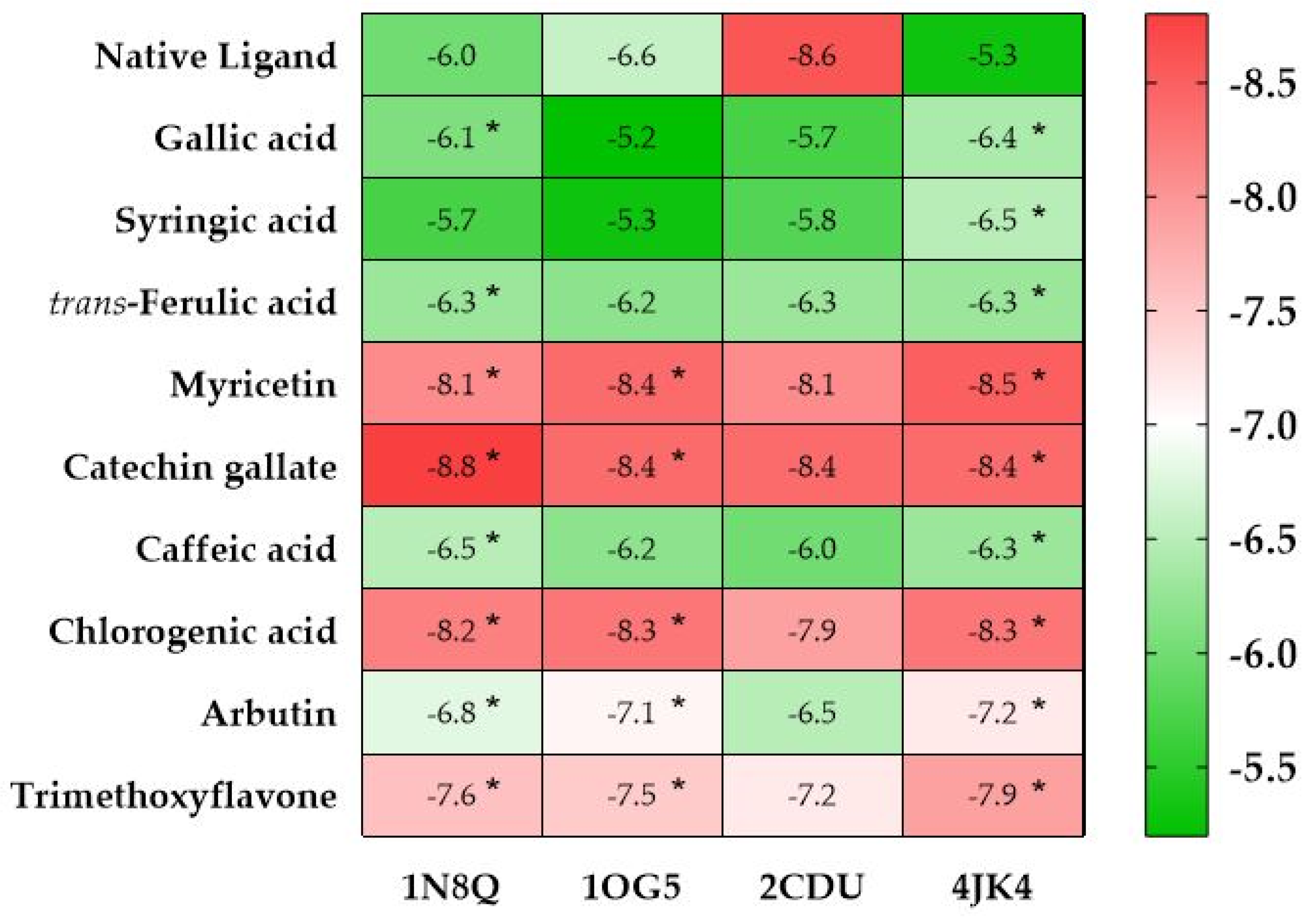
2.4.1. In Silico Prediction of the Antioxidant Activity of AHEE
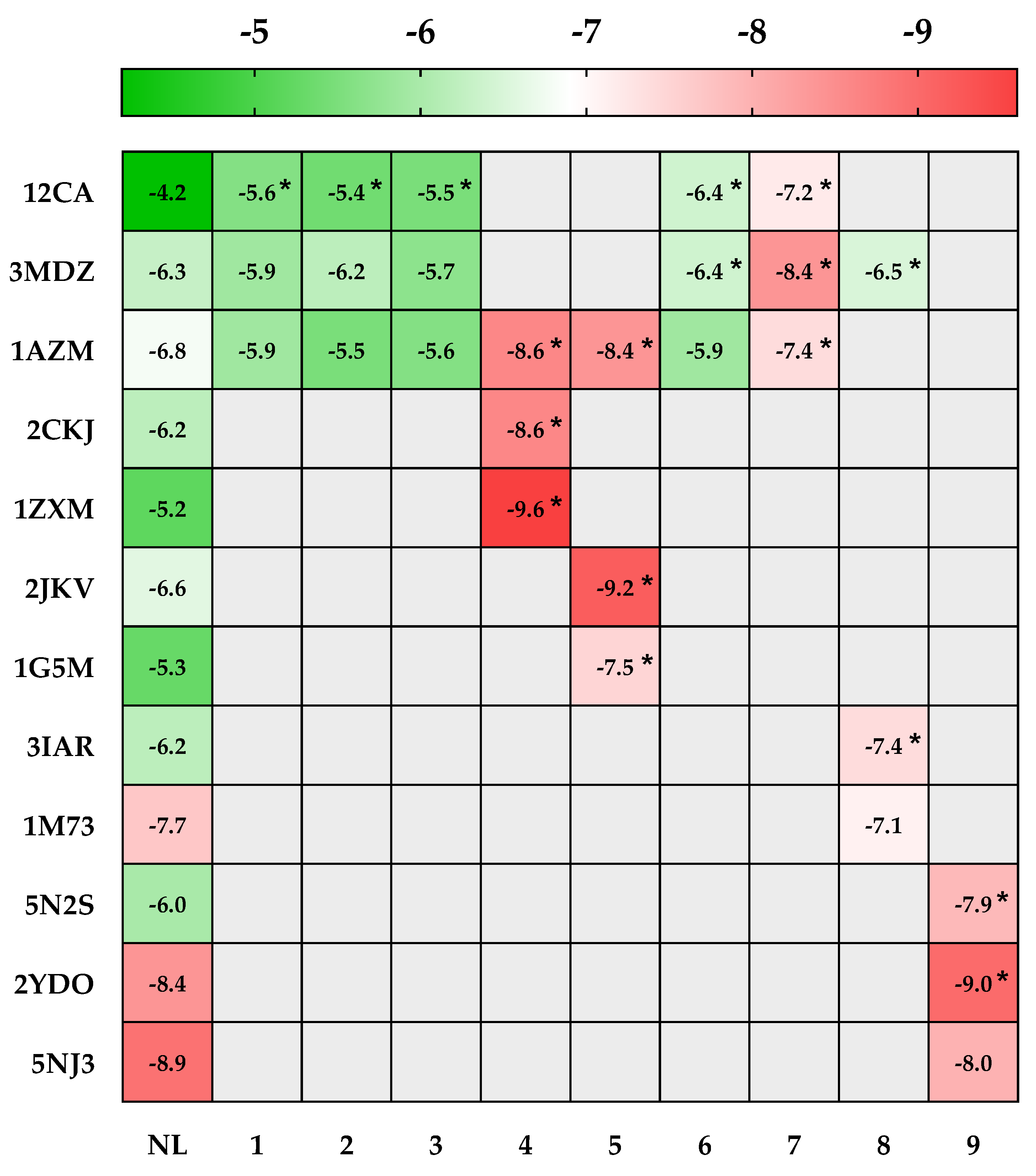
2.4.2. In Silico Prediction of the Cytotoxic Potential of AHEE
2.5. Experimental Validation of the Antioxidant and the Cytotoxic Activity of AHEE
2.5.1. Antioxidant Activity
2.5.2. Cytotoxicity of AHEE against Breast Cancer Cell Lines (MCF-7 and MDA-MB-231)
3. Materials and Methods
3.1. Plant Material and Extraction
3.2. LC–MS/MS Profiling of AHEE
3.3. Prediction of the Pharmacokinetic Properties and Toxicity of AHEE’s Bioactive Compounds
3.4. Molecular Docking Prediction of the Antioxidant and Cytotoxic Activity of AHEE
3.4.1. Molecular Docking General Procedure
3.4.2. Ligand-Based Target Prediction with SwissTarget Prediction
3.5. Anticancer Activity
3.5.1. Cell Culture
3.5.2. Cell Viability by MTT Assay
3.6. Antioxidant Activity
3.6.1. 2,2-Diphenyl-1-Picrylhydrazil Free Radical Scavenging Assay
3.6.2. β-Carotene Bleaching Assay
3.6.3. ABTS Scavenging Activity Assay
3.6.4. Iron Chelation
3.6.5. Total Antioxidant Capacity
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Samadi, P.; Saki, S.; Dermani, F.K.; Pourjafar, M.; Saidijam, M. Emerging Ways to Treat Breast Cancer: Will Promises Be Met? Cell. Oncol. 2018, 41, 605–621. [Google Scholar] [CrossRef]
- You, J.S.; Jones, P.A. Cancer Genetics and Epigenetics: Two Sides of the Same Coin? Cancer Cell 2012, 22, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Ferlay, J.; Shin, H.R.; Bray, F.; Forman, D.; Mathers, C.; Parkin, D.M. GLOBOCAN 2008, Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 10. Int. J. Cancer 2008, 127, 2893–2917. [Google Scholar] [CrossRef]
- Castagné, R.; Kelly-Irving, M.; Campanella, G.; Guida, F.; Krogh, V.; Palli, D.; Panico, S.; Sacerdote, C.; Tumino, R.; Kleinjans, J. Biological Marks of Early-Life Socioeconomic Experience Is Detected in the Adult Inflammatory Transcriptome. Sci. Rep. 2016, 6, 38705. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Wang, Y.; Kiani, M.F.; Wang, B. Classification, Treatment Strategy, and Associated Drug Resistance in Breast Cancer. Clin. Breast Cancer 2016, 16, 335–343. [Google Scholar] [CrossRef]
- Siegal, R.; Miller, K.D.; Jemal, A. Cancer Statistics, 2012. CA Cancer J Clin 2014, 64, 9–29. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA. Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Viale, P.H. The American Cancer Society’s Facts & Figures: 2020 Edition. J. Adv. Pract. Oncol. 2020, 11, 135. [Google Scholar]
- Turashvili, G.; Brogi, E. Tumor Heterogeneity in Breast Cancer. Front. Med. 2017, 4, 227. [Google Scholar] [CrossRef]
- Maughan, K.L.; Lutterbie, M.A.; Ham, P.S. Breast Cancer Treatment. Am. Fam. Physician 2010, 81, 1339–1346. [Google Scholar]
- Chakraborty, S.; Rahman, T. The Difficulties in Cancer Treatment. Ecancermedicalscience 2012, 6, ed16. [Google Scholar] [PubMed]
- Rosenberg, B.; Vancamp, L.; Trosko, J.E.; Mansour, V.H. Platinum Compounds: A New Class of Potent Antitumour Agents. Nature 1969, 222, 385–386. [Google Scholar] [CrossRef]
- Conte, E.; Bresciani, E.; Rizzi, L.; Cappellari, O.; De Luca, A.; Torsello, A.; Liantonio, A. Cisplatin-Induced Skeletal Muscle Dysfunction: Mechanisms and Counteracting Therapeutic Strategies. Int. J. Mol. Sci. 2020, 21, 1242. [Google Scholar] [CrossRef]
- Giacomini, I.; Ragazzi, E.; Pasut, G.; Montopoli, M. The Pentose Phosphate Pathway and Its Involvement in Cisplatin Resistance. Int. J. Mol. Sci. 2020, 21, 937. [Google Scholar] [CrossRef] [PubMed]
- Zeidan, Y.H.; Jenkins, R.W.; Hannun, Y.A. Remodeling of Cellular Cytoskeleton by the Acid Sphingomyelinase/Ceramide Pathway. J. Cell Biol. 2008, 181, 335–350. [Google Scholar] [CrossRef]
- Wang, Q.-E.; Milum, K.; Han, C.; Huang, Y.-W.; Wani, G.; Thomale, J.; Wani, A.A. Differential Contributory Roles of Nucleotide Excision and Homologous Recombination Repair for Enhancing Cisplatin Sensitivity in Human Ovarian Cancer Cells. Mol. Cancer 2011, 10, 24. [Google Scholar] [CrossRef]
- Florea, A.-M.; Büsselberg, D. Cisplatin as an Anti-Tumor Drug: Cellular Mechanisms of Activity, Drug Resistance and Induced Side Effects. Cancers 2011, 3, 1351–1371. [Google Scholar] [CrossRef]
- Zhan, Y.; Wang, H.; Ning, Y.; Zheng, H.; Liu, S.; Yang, Y.; Zhou, M.; Fan, S. Understanding the Roles of Stress Granule during Chemotherapy for Patients with Malignant Tumors. Am. J. Cancer Res. 2020, 10, 2226. [Google Scholar]
- Chavrier, P.; Mamessier, É.; Aulas, A. Stress Granules, Emerging Players in Cancer Research. Med. Sci. M/S 2021, 37, 735–741. [Google Scholar]
- Hu, T.; Hou, W.; Xiao, E.; Long, M. Mechanism and Effect of Stress Granule Formation in Cancer and Its Potential Roles in Breast Cancer Therapy. Genes Dis. 2021, 9, 659–667. [Google Scholar] [CrossRef]
- Aulas, A.; Finetti, P.; Lyons, S.M.; Bertucci, F.; Birnbaum, D.; Acquaviva, C.; Mamessier, E. Revisiting the Concept of Stress in the Prognosis of Solid Tumors: A Role for Stress Granules Proteins? Cancers 2020, 12, 2470. [Google Scholar] [CrossRef] [PubMed]
- Brown, N.S.; Bicknell, R. Hypoxia and Oxidative Stress in Breast Cancer Oxidative Stress-Its Effects on the Growth, Metastatic Potential and Response to Therapy of Breast Cancer. Breast Cancer Res. 2001, 3, 323. [Google Scholar] [CrossRef]
- Klaunig, J.E. Oxidative Stress and Cancer. Curr. Pharm. Des. 2018, 24, 4771–4778. [Google Scholar] [CrossRef] [PubMed]
- Visconti, R.; Grieco, D. New Insights on Oxidative Stress in Cancer. Curr. Opin. Drug Discov. Devel. 2009, 12, 240–245. [Google Scholar] [PubMed]
- Prakash, O.M.; Kumar, A.; Kumar, P. Anticancer Potential of Plants and Natural Products. Am. J. Pharmacol. Sci. 2013, 1, 104–115. [Google Scholar] [CrossRef]
- Zakaria, Z.; Gan, S.H.; Mohamed, M. In Vitro Studies of Asian Medicinal Plants with Potential Activity against Breast Cancer. J. Appl. Biol. Biotechnol. 2018, 6, 4–5. [Google Scholar]
- Lee, K.W.; Ching, S.M.; Hoo, F.K.; Ramachandran, V.; Swamy, M.K. Traditional Medicinal Plants and Their Therapeutic Potential against Major Cancer Types. In Anticancer Plants: Natural Products and Biotechnological Implements; Springer: Singapore, 2018; pp. 383–410. [Google Scholar]
- Hofer, D.; Schwach, G.; Ghaffari Tabrizi-Wizsy, N.; Sadjak, A.; Sturm, S.; Stuppner, H.; Pfragner, R. Christia Vespertilionis Plant Extracts as Novel Antiproliferative Agent against Human Neuroendocrine Tumor Cells. Oncol. Rep. 2013, 29, 2219–2226. [Google Scholar] [CrossRef]
- Roy, A.; Jauhari, N.; Bharadvaja, N. Medicinal Plants as a Potential Source of Chemopreventive Agents. In Anticancer Plants: Natural Products and Biotechnological Implements; Springer: Singapore, 2018; pp. 109–139. [Google Scholar]
- Arora, D.S.; Chandra, P. Antioxidant Activity of Aspergillus fumigatus. Int. Sch. Res. Not. 2011, 2011, 619395. [Google Scholar] [CrossRef]
- Kiokias, S.; Proestos, C.; Oreopoulou, V. Phenolic Acids of Plant Origin—A Review on Their Antioxidant Activity in Vitro (O/W Emulsion Systems) along with Their In Vivo Health Biochemical Properties. Foods 2020, 9, 534. [Google Scholar] [CrossRef]
- Bencheikh, N.; Elbouzidi, A.; Kharchoufa, L.; Ouassou, H.; Alami Merrouni, I.; Mechchate, H.; Es-Safi, I.; Hano, C.; Addi, M.; Bouhrim, M. Inventory of Medicinal Plants Used Traditionally to Manage Kidney Diseases in North-Eastern Morocco: Ethnobotanical Fieldwork and Pharmacological Evidence. Plants 2021, 10, 1966. [Google Scholar] [CrossRef]
- Elbouzidi, A.; Bencheikh, N.; Seddoqi, S.; Bouhrim, M.; Bouramdane, Y.; Addi, M. Investigation of the Allelopathic Effect of Matricaria Chamomilla L. Parts’ Aqueous Extracts on Germination and Seedling Growth of Two Moroccan Varieties of Durum Wheat. Int. J. Agron. 2021, 2021, 4451181. [Google Scholar] [CrossRef]
- Bouchikh-Boucif, Y.; Labani, A.; Benabdeli, K.; Boidielouane, S. Allelopathic Effects of Shoot and Root Extracts From Three Alien and Native Chenopodiaceae Species on Lettuce Seed Germination. Ecol. Balk. 2014, 6, 51–55. [Google Scholar]
- Slama, K.; Boumendjel, M.; Taibi, F.; Boumendjel, A.; Messarah, M. Atriplex halimus Aqueous Extract Abrogates Carbon Tetrachloride-Induced Hepatotoxicity by Modulating Biochemical and Histological Changes in Rats. Arch. Physiol. Biochem. 2020, 126, 49–60. [Google Scholar] [CrossRef] [PubMed]
- Benhammou, N.; Bekkara, F.A.; Panovska, T.K. Antioxidant Activity of Methanolic Extracts and Some Bioactive Compounds of Atriplex halimus. Comptes Rendus Chim. 2009, 12, 1259–1266. [Google Scholar] [CrossRef]
- Mechaala, S.; Bouatrous, Y.; Adouane, S. Traditional Knowledge and Diversity of Wild Medicinal Plants in El Kantara’s Area (Algerian Sahara Gate): An Ethnobotany Survey. Acta Ecol. Sin. 2021, 42, 33–45. [Google Scholar] [CrossRef]
- Walker, D.J.; Lutts, S.; Sánchez-García, M.; Correal, E. Atriplex halimus L.: Its Biology and Uses. J. Arid Environ. 2014, 100, 111–121. [Google Scholar] [CrossRef]
- Al-Senosy, N.K.; Abou-Eisha, A.; Ahmad, E.S. In Vitro Antiproliferation Effect of Atriplex halimus L. Crude Extract on Human Cell Lines by Induction of Apoptosis and G2/M Phase Arrest. Egypt. Acad. J. Biol. Sci. C Physiol. Mol. Biol. 2018, 10, 115–126. [Google Scholar] [CrossRef]
- Capua, C.J.; Hopson, N.P.; Stewart, C.M.M.; Johnston, G.R.; O’Neill, K.L.; Schaalje, G.B.; Lee, C.M.; Booth, G.M. Cytotoxicity of Atriplex confertifolia. J. Toxicol. 2010, 2010, 976548. [Google Scholar] [CrossRef]
- Ferruzzi, M.G.; Lobo, J.K.; Janle, E.M.; Cooper, B.; Simon, J.E.; Wu, Q.-L.; Welch, C.; Ho, L.; Weaver, C.; Pasinetti, G.M. Bioavailability of Gallic Acid and Catechins from Grape Seed Polyphenol Extract Is Improved by Repeated Dosing in Rats: Implications for Treatment in Alzheimer’s Disease. J. Alzheimers. Dis. 2009, 18, 113–124. [Google Scholar] [CrossRef]
- Kivilompolo, M.; Obůrka, V.; Hyötyläinen, T. Comparison of GC–MS and LC–MS Methods for the Analysis of Antioxidant Phenolic Acids in Herbs. Anal. Bioanal. Chem. 2007, 388, 881–887. [Google Scholar] [CrossRef]
- Wen, C.; Wang, D.; Li, X.; Huang, T.; Huang, C.; Hu, K. Targeted Isolation and Identification of Bioactive Compounds Lowering Cholesterol in the Crude Extracts of Crabapples Using UPLC-DAD-MS-SPE/NMR Based on Pharmacology-Guided PLS-DA. J. Pharm. Biomed. Anal. 2018, 150, 144–151. [Google Scholar] [CrossRef] [PubMed]
- Ousji, O.; Sleno, L. Structural Elucidation of Novel Stable and Reactive Metabolites of Green Tea Catechins and Alkyl Gallates by LC-MS/MS. Antioxidants 2022, 11, 1635. [Google Scholar] [CrossRef]
- Zhang, D.-Y.; Yao, X.-H.; Duan, M.-H.; Luo, M.; Zhao, C.-J.; Zu, Y.-G.; Fu, Y.-J. An Effective Homogenate-Assisted Negative Pressure Cavitation Extraction for the Determination of Phenolic Compounds in Pyrola by LC-MS/MS and the Evaluation of Its Antioxidant Activity. Food Funct. 2015, 6, 3323–3333. [Google Scholar] [CrossRef]
- Sharaf, M.; Mansour, R.M.A.; Saleh, N.A.M. Exudate Flavonoids from Aerial Parts of Four Cleome Species. Biochem. Syst. Ecol. 1992, 20, 443–448. [Google Scholar] [CrossRef]
- Han, C.; Wang, B. Factors That Impact the Developability of Drug Candidates: An Overview. Drug Deliv. Princ. Appl. 2005, 1, 1–15. [Google Scholar]
- Waring, M.J.; Arrowsmith, J.; Leach, A.R.; Leeson, P.D.; Mandrell, S.; Owen, R.M.; Pairaudeau, G.; Pennie, W.D.; Pickett, S.D.; Wang, J. An Analysis of the Attrition of Drug Candidates from Four Major Pharmaceutical Companies. Nat. Rev. Drug Discov. 2015, 14, 475–486. [Google Scholar] [CrossRef]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and Computational Approaches to Estimate Solubility and Permeability in Drug Discovery and Development Settings. Adv. Drug Deliv. Rev. 1997, 23, 3–25. [Google Scholar] [CrossRef]
- Ghose, A.K.; Viswanadhan, V.N.; Wendoloski, J.J. A Knowledge-Based Approach in Designing Combinatorial or Medicinal Chemistry Libraries for Drug Discovery. 1. A Qualitative and Quantitative Characterization of Known Drug Databases. J. Comb. Chem. 1999, 1, 55–68. [Google Scholar] [CrossRef]
- Veber, D.F.; Johnson, S.R.; Cheng, H.-Y.; Smith, B.R.; Ward, K.W.; Kopple, K.D. Molecular Properties That Influence the Oral Bioavailability of Drug Candidates. J. Med. Chem. 2002, 45, 2615–2623. [Google Scholar] [CrossRef]
- Egan, W.J.; Merz, K.M.; Baldwin, J.J. Prediction of Drug Absorption Using Multivariate Statistics. J. Med. Chem. 2000, 43, 3867–3877. [Google Scholar] [CrossRef]
- Martin, Y.C. A Bioavailability Score. J. Med. Chem. 2005, 48, 3164–3170. [Google Scholar] [CrossRef] [PubMed]
- Hubatsch, I.; Ragnarsson, E.G.E.; Artursson, P. Determination of Drug Permeability and Prediction of Drug Absorption in Caco-2 Monolayers. Nat. Protoc. 2007, 2, 2111–2119. [Google Scholar] [CrossRef] [PubMed]
- Urzì Brancati, V.; Scarpignato, C.; Minutoli, L.; Pallio, G. Use of Pharmacogenetics to Optimize Immunosuppressant Therapy in Kidney-Transplanted Patients. Biomedicines 2022, 10, 1798. [Google Scholar] [CrossRef]
- Yin, J.; Wang, J. Renal Drug Transporters and Their Significance in Drug–Drug Interactions. Acta Pharm. Sin. B 2016, 6, 363–373. [Google Scholar] [CrossRef]
- Pires, D.E.V.; Blundell, T.L.; Ascher, D.B. PkCSM: Predicting Small-Molecule Pharmacokinetic and Toxicity Properties Using Graph-Based Signatures. J. Med. Chem. 2015, 58, 4066–4072. [Google Scholar] [CrossRef]
- Sahota, T.; Danhof, M.; Della Pasqua, O. Pharmacology-Based Toxicity Assessment: Towards Quantitative Risk Prediction in Humans. Mutagenesis 2016, 31, 359–374. [Google Scholar] [CrossRef]
- Khaoula, Z.; Ali, B.D. Preventive and Curative Effects of Atriplex halimus L. Aqueous Extract on Benzene Provoked Hepatic Injury in Rats. J. Drug Deliv. Ther. 2020, 10, 217–222. [Google Scholar] [CrossRef]
- Senapathy, J.G.; Jayanthi, S.; Viswanathan, P.; Umadevi, P.; Nalini, N. Effect of Gallic Acid on Xenobiotic Metabolizing Enzymes in 1, 2-Dimethyl Hydrazine Induced Colon Carcinogenesis in Wistar Rats—A Chemopreventive Approach. Food Chem. Toxicol. 2011, 49, 887–892. [Google Scholar] [CrossRef]
- Zeng, X.; Liu, H.; Huang, Z.; Dong, P.; Chen, X. Anticancer Effect of Arbutin on Diethylnitrosamine-Induced Liver Carcinoma in Rats via the GRP and GADD Pathway. J. Environ. Pathol. Toxicol. Oncol. 2022, 41, 15–26. [Google Scholar] [CrossRef]
- Verma, S.; Singh, A.; Mishra, A. Gallic Acid: Molecular Rival of Cancer. Environ. Toxicol. Pharmacol. 2013, 35, 473–485. [Google Scholar] [CrossRef]
- Rodgers, E.H.; Grant, M.H. The Effect of the Flavonoids, Quercetin, Myricetin and Epicatechin on the Growth and Enzyme Activities of MCF7 Human Breast Cancer Cells. Chem. Biol. Interact. 1998, 116, 213–228. [Google Scholar] [CrossRef]
- Xie, Y.; Wang, Y.; Xiang, W.; Wang, Q.; Cao, Y. Molecular Mechanisms of the Action of Myricetin in Cancer. Mini Rev. Med. Chem. 2020, 20, 123–133. [Google Scholar] [CrossRef] [PubMed]
- Czeczot, H.; Tudek, B.; Kusztelak, J.; Szymczyk, T.; Dobrowolska, B.; Glinkowska, G.; Malinowski, J.; Strzelecka, H. Isolation and Studies of the Mutagenic Activity in the Ames Test of Flavonoids Naturally Occurring in Medical Herbs. Mutat. Res. Toxicol. 1990, 240, 209–216. [Google Scholar] [CrossRef]
- Resende, F.A.; Vilegas, W.; Dos Santos, L.C.; Varanda, E.A. Mutagenicity of Flavonoids Assayed by Bacterial Reverse Mutation (Ames) Test. Molecules 2012, 17, 5255–5268. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, L.G.; Dos Santos, R.N.; Oliva, G.; Andricopulo, A.D. Molecular Docking and Structure-Based Drug Design Strategies. Molecules 2015, 20, 13384–13421. [Google Scholar] [CrossRef] [PubMed]
- López-Vallejo, F.; Caulfield, T.; Martínez-Mayorga, K.; Giulianotti, M.A.; Nefzi, A.; Houghten, R.A.; Medina-Franco, J.L. Integrating Virtual Screening and Combinatorial Chemistry for Accelerated Drug Discovery. Comb. Chem. High Throughput Screen. 2011, 14, 475–487. [Google Scholar] [CrossRef]
- Huang, S.-Y.; Zou, X. Advances and Challenges in Protein-Ligand Docking. Int. J. Mol. Sci. 2010, 11, 3016–3034. [Google Scholar] [CrossRef]
- Semidalas, C.; Semidalas, E.; Matsoukas, M.T.; Nixarlidis, C.; Zoumpoulakis, P. In Silico Studies Reveal the Mechanisms behind the Antioxidant and Anti-Inflammatory Activities of Hydroxytyrosol. Med. Chem. Res. 2016, 25, 2498–2511. [Google Scholar] [CrossRef]
- Feng, R.; Zhou, X.; Or, P.M.Y.; Ma, J.-Y.; Tan, X.-S.; Fu, J.; Ma, C.; Shi, J.-G.; Che, C.-T.; Wang, Y. Enzyme Kinetic and Molecular Docking Studies on the Metabolic Interactions of 1-Hydroxy-2, 3, 5-Trimethoxy-Xanthone, Isolated from Halenia elliptica D. Don, with Model Probe Substrates of Human Cytochrome P450 Enzymes. Phytomedicine 2012, 19, 1125–1133. [Google Scholar] [CrossRef]
- Gao, H.; Li, J.; Sivakumar, D.; Kim, T.-S.; Patel, S.K.S.; Kalia, V.C.; Kim, I.-W.; Zhang, Y.-W.; Lee, J.-K. NADH Oxidase from Lactobacillus Reuteri: A Versatile Enzyme for Oxidized Cofactor Regeneration. Int. J. Biol. Macromol. 2019, 123, 629–636. [Google Scholar] [CrossRef]
- Sutomo, S.; Pratama, M.R.F. Measuring the Potential Antioxidant Activity of Methyl Gallate: Molecular Docking Study. Thai J. Pharm. Sci. 2020, 44, 14–22. [Google Scholar]
- Spiteller, G. On the Chemistry of Oxidative Stress. J. Lipid Mediat. 1993, 7, 199–221. [Google Scholar] [PubMed]
- Kandsi, F.; Elbouzidi, A.; Lafdil, F.Z.; Meskali, N.; Azghar, A.; Addi, M.; Hano, C.; Maleb, A.; Gseyra, N. Antibacterial and Antioxidant Activity of Dysphania ambrosioides (L.) Mosyakin and Clemants Essential Oils: Experimental and Computational Approaches. Antibiotics 2022, 11, 482. [Google Scholar] [CrossRef] [PubMed]
- da Silva Costa, J.; da Silva Ramos, R.; da Silva Lopes Costa, K.; do Socorro Barros Brasil, D.; de Paula da Silva, C.H.T.; Ferreira, E.F.B.; dos Santos Borges, R.; Campos, J.M.; da Cruz Macêdo, W.J.; dos Santos, C.B.R. An in Silico Study of the Antioxidant Ability for Two Caffeine Analogs Using Molecular Docking and Quantum Chemical Methods. Molecules 2018, 23, 2801. [Google Scholar]
- Daina, A.; Michielin, O.; Zoete, V. SwissTargetPrediction: Updated Data and New Features for Efficient Prediction of Protein Targets of Small Molecules. Nucleic Acids Res. 2019, 47, W357–W364. [Google Scholar] [CrossRef]
- Zhang, J.L.; Zheng, Q.C.; Zhang, H.X. Theoretical Improvement of the Specific Inhibitor of Human Carbonic Anhydrase VII. Comput. Biol. Chem. 2011, 35, 50–56. [Google Scholar] [CrossRef]
- Supuran, C.T. Carbonic Anhydrase Inhibition and the Management of Hypoxic Tumors. Metabolites 2017, 7, 48. [Google Scholar] [CrossRef]
- Lee, S.-H.; Griffiths, J.R. How and Why Are Cancers Acidic? Carbonic Anhydrase IX and the Homeostatic Control of Tumour Extracellular PH. Cancers 2020, 12, 1616. [Google Scholar] [CrossRef]
- Del Giudice, R.; Monti, D.M.; Truppo, E.; Arciello, A.; Supuran, C.T.; De Simone, G.; Monti, S.M. Human Carbonic Anhydrase VII Protects Cells from Oxidative Damage. Biol. Chem. 2013, 394, 1343–1348. [Google Scholar] [CrossRef]
- Ruusuvuori, E.; Li, H.; Huttu, K.; Palva, J.M.; Smirnov, S.; Rivera, C.; Kaila, K.; Voipio, J. Carbonic Anhydrase Isoform VII Acts as a Molecular Switch in the Development of Synchronous Gamma-Frequency Firing of Hippocampal CA1 Pyramidal Cells. J. Neurosci. 2004, 24, 2699–2707. [Google Scholar] [CrossRef]
- Bortolotti, M.; Polito, L.; Battelli, M.G.; Bolognesi, A. Xanthine Oxidoreductase: One Enzyme for Multiple Physiological Tasks. Redox Biol. 2021, 41, 101882. [Google Scholar] [CrossRef] [PubMed]
- Battelli, M.G.; Polito, L.; Bortolotti, M.; Bolognesi, A. Xanthine Oxidoreductase in Cancer: More than a Differentiation Marker. Cancer Med. 2016, 5, 546–557. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Zhang, G.; Liao, Y.; Gong, D. Myricetin Inhibits the Generation of Superoxide Anion by Reduced Form of Xanthine Oxidase. Food Chem. 2017, 221, 1569–1577. [Google Scholar] [CrossRef] [PubMed]
- Delgado, J.L.; Hsieh, C.-M.; Chan, N.-L.; Hiasa, H. Topoisomerases as Anticancer Targets. Biochem. J. 2018, 475, 373–398. [Google Scholar] [CrossRef]
- Skok, Z.; Zidar, N.; Kikelj, D.; Ilas, J. Dual Inhibitors of Human DNA Topoisomerase II and Other Cancer-Related Targets. J. Med. Chem. 2019, 63, 884–904. [Google Scholar] [CrossRef]
- Hanau, S.; Montin, K.; Cervellati, C.; Magnani, M.; Dallocchio, F. 6-Phosphogluconate Dehydrogenase Mechanism: Evidence for Allosteric Modulation by Substrate. J. Biol. Chem. 2010, 285, 21366–21371. [Google Scholar] [CrossRef]
- Lin, R.; Elf, S.; Shan, C.; Kang, H.-B.; Ji, Q.; Zhou, L.; Hitosugi, T.; Zhang, L.; Zhang, S.; Seo, J.H. 6-Phosphogluconate Dehydrogenase Links Oxidative PPP, Lipogenesis and Tumour Growth by Inhibiting LKB1–AMPK Signalling. Nat. Cell Biol. 2015, 17, 1484–1496. [Google Scholar] [CrossRef]
- O’Neill, J.; Manion, M.; Schwartz, P.; Hockenbery, D.M. Promises and Challenges of Targeting Bcl-2 Anti-Apoptotic Proteins for Cancer Therapy. Biochim. Biophys. Acta (BBA)-Rev. Cancer 2004, 1705, 43–51. [Google Scholar] [CrossRef]
- Jagani, H.; Kasinathan, N.; Meka, S.R.; Venkata Rao, J. Antiapoptotic Bcl-2 Protein as a Potential Target for Cancer Therapy: A Mini Review. Artif. Cells Nanomed. Biotechnol. 2016, 44, 1212–1221. [Google Scholar] [CrossRef]
- Kutryb-Zajac, B.; Koszalka, P.; Mierzejewska, P.; Bulinska, A.; Zabielska, M.A.; Brodzik, K.; Skrzypkowska, A.; Zelazek, L.; Pelikant-Malecka, I.; Slominska, E.M. Adenosine Deaminase Inhibition Suppresses Progression of 4T1 Murine Breast Cancer by Adenosine Receptor-dependent Mechanisms. J. Cell. Mol. Med. 2018, 22, 5939–5954. [Google Scholar] [CrossRef]
- Zhulai, G.; Oleinik, E.; Shibaev, M.; Ignatev, K. Adenosine-Metabolizing Enzymes, Adenosine Kinase and Adenosine Deaminase, in Cancer. Biomolecules 2022, 12, 418. [Google Scholar] [CrossRef]
- Durak, I.; Biri, H.; Devrim, E.; Sözen, S.; Avci, A. Aqueous Extract of Urtica Dioica Makes Significant Inhibition on Adenosine Deaminase Activity in Prostate Tissue from Patients with Prostate Cancer. Cancer Biol. Ther. 2004, 3, 855–857. [Google Scholar] [CrossRef] [PubMed]
- Afshar, A.S.; Nematpour, F.S.; Meshkani, M.; Khafi, A. Growth Inhibition of Human Breast Cancer Cells and Down-Regulation of ODC1 and ADA Genes by Nepeta Binaloudensis. Rev. Bras. Farmacogn. 2017, 27, 84–90. [Google Scholar] [CrossRef]
- Shanta, B. Purine Nucleoside Phosphorylase Inhibitors as Novel Immuno-Oncology Agent and Vaccine Adjuvant. Int. J. Immunol. Immunother. 2020, 7, 043. [Google Scholar] [CrossRef]
- Gessi, S.; Merighi, S.; Sacchetto, V.; Simioni, C.; Borea, P.A. Adenosine Receptors and Cancer. Biochim. Biophys. Acta-Biomembr. 2011, 1808, 1400–1412. [Google Scholar] [CrossRef] [PubMed]
- Gillet, J.-P.; Gottesman, M.M. Mechanisms of Multidrug Resistance in Cancer. In Multi-Drug Resistance in Cancer; Springer: Berlin/Heidelberg, Germany, 2010; pp. 47–76. [Google Scholar]
- Housman, G.; Byler, S.; Heerboth, S.; Lapinska, K.; Longacre, M.; Snyder, N.; Sarkar, S. Drug Resistance in Cancer: An Overview. Cancers 2014, 6, 1769–1792. [Google Scholar] [CrossRef] [PubMed]
- De Luca, L.; Mancuso, F.; Ferro, S.; Buemi, M.R.; Angeli, A.; Del Prete, S.; Capasso, C.; Supuran, C.T.; Gitto, R. Inhibitory Effects and Structural Insights for a Novel Series of Coumarin-Based Compounds That Selectively Target Human CA IX and CA XII Carbonic Anhydrases. Eur. J. Med. Chem. 2018, 143, 276–282. [Google Scholar] [CrossRef] [PubMed]
- Abotaleb, M.; Liskova, A.; Kubatka, P.; Büsselberg, D. Therapeutic Potential of Plant Phenolic Acids in the Treatment of Cancer. Biomolecules 2020, 10, 221. [Google Scholar] [CrossRef] [PubMed]
- Houglum, K.; Filip, M.; Witztum, J.L.; Chojkier, M. Malondialdehyde and 4-Hydroxynonenal Protein Adducts in Plasma and Liver of Rats with Iron Overload. J. Clin. Investig. 1990, 86, 1991–1998. [Google Scholar] [CrossRef]
- Sabir, S.M.; Ahmad, S.D.; Hamid, A.; Khan, M.Q.; Athayde, M.L.; Santos, D.B.; Boligon, A.A.; Rocha, J.B.T. Antioxidant and Hepatoprotective Activity of Ethanolic Extract of Leaves of Solidago Microglossa Containing Polyphenolic Compounds. Food Chem. 2012, 131, 741–747. [Google Scholar] [CrossRef]
- Oyaizu, M. Studies on Products of Browning Reaction Antioxidative Activities of Products of Browning Reaction Prepared from Glucosamine. Jpn. J. Nutr. Diet. 1986, 44, 307–315. [Google Scholar] [CrossRef]
- Bouaziz, S.; Amri, M.; Taibi, N.; Zeghir-Bouteldja, R.; Benkhaled, A.; Mezioug, D.; Touil-Boukoffa, C. Protoscolicidal Activity of Atriplex halimus Leaves Extract against Echinococcus Granulosus Protoscoleces. Exp. Parasitol. 2021, 229, 108155. [Google Scholar] [CrossRef] [PubMed]
- Cao, G.; Sofic, E.; Prior, R.L. Antioxidant and Prooxidant Behavior of Flavonoids: Structure-Activity Relationships. Free Radic. Biol. Med. 1997, 22, 749–760. [Google Scholar] [CrossRef]
- Dorman, H.J.D.; Koşar, M.; Kahlos, K.; Holm, Y.; Hiltunen, R. Antioxidant Properties and Composition of Aqueous Extracts from Mentha Species, Hybrids, Varieties, and Cultivars. J. Agric. Food Chem. 2003, 51, 4563–4569. [Google Scholar] [CrossRef]
- Emam, S.S. Bioactive Constituents of Atriplex halimus Plant. J. Nat. Prod. 2011, 4, 25–41. [Google Scholar]
- Clauser, M.; Dall’Acqua, S.; Loi, M.C.; Innocenti, G. Phytochemical Investigation on Atriplex halimus L. from Sardinia. Nat. Prod. Res. 2013, 27, 1940–1944. [Google Scholar] [CrossRef]
- Benarba, B. Use of Medicinal Plants by Breast Cancer Patients in Algeria. EXCLI J. 2015, 14, 1164–1166. [Google Scholar] [CrossRef]
- Benarba, B.; Belabid, L.; Righi, K.; Bekkar, A.A.; Elouissi, M.; Khaldi, A.; Hamimed, A. Ethnobotanical Study of Medicinal Plants Used by Traditional Healers in Mascara (North West of Algeria). J. Ethnopharmacol. 2015, 175, 626–637. [Google Scholar] [CrossRef]
- Zennaf, I.; Touil, A.T.; Meddah, B.; Mokhtar, M. Ethnobotanical and Phytochemical Study of the Medicinal Plant Atriplex halimus and Its Importance in the Traditional Algerian Pharmacopoeia. Fr.-Ukr. J. Chem. 2022, 10, 60–69. [Google Scholar] [CrossRef]
- Hosny, M.; Fawzy, M.; Abdelfatah, A.M.; Fawzy, E.E.; Eltaweil, A.S. Comparative Study on the Potentialities of Two Halophytic Species in the Green Synthesis of Gold Nanoparticles and Their Anticancer, Antioxidant and Catalytic Efficiencies. Adv. Powder Technol. 2021, 32, 3220–3233. [Google Scholar] [CrossRef]
- Mechchate, H.; Es-Safi, I.; Conte, R.; Hano, C.; Amaghnouje, A.; Jawhari, F.Z.; Radouane, N.; Bencheikh, N.; Grafov, A.; Bousta, D. In Vivo and In Vitro Antidiabetic and Anti-Inflammatory Properties of Flax (Linum usitatissimum L.) Seed Polyphenols. Nutrients 2021, 13, 2759. [Google Scholar] [CrossRef] [PubMed]
- Amaghnouje, A.; Mechchate, H.; Es-Safi, I.; Boukhira, S.; Aliqahtani, A.S.; Noman, O.M.; Nasr, F.A.; Conte, R.; Calarco, A.; Bousta, D. Subacute Assessment of the Toxicity and Antidepressant-like Effects of Origanum majorana L. Polyphenols in Swiss Albino Mice. Molecules 2020, 25, 5653. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A Free Web Tool to Evaluate Pharmacokinetics, Drug-Likeness and Medicinal Chemistry Friendliness of Small Molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef] [PubMed]
- Kandsi, F.; Lafdil, F.Z.; Elbouzidi, A.; Bouknana, S.; Miry, A.; Addi, M.; Conte, R.; Hano, C.; Gseyra, N. Evaluation of Acute and Subacute Toxicity and LC-MS/MS Compositional Alkaloid Determination of the Hydroethanolic Extract of Dysphania ambrosioides (L.) Mosyakin and Clemants Flowers. Toxins 2022, 14, 475. [Google Scholar] [CrossRef]
- Banerjee, P.; Eckert, A.O.; Schrey, A.K.; Preissner, R. ProTox-II: A Webserver for the Prediction of Toxicity of Chemicals. Nucleic Acids Res. 2018, 46, W257–W263. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Cosconati, S.; Forli, S.; Perryman, A.L.; Harris, R.; Goodsell, D.S.; Olson, A.J. Virtual Screening with AutoDock: Theory and Practice. Expert Opin. Drug Discov. 2010, 5, 597–607. [Google Scholar] [CrossRef] [PubMed]
- Cereto-Massagué, A.; Ojeda, M.J.; Valls, C.; Mulero, M.; Pujadas, G.; Garcia-Vallve, S. Tools for in Silico Target Fishing. Methods 2015, 71, 98–103. [Google Scholar] [CrossRef]
- Willett, P.; Winterman, V. A Comparison of Some Measures for the Determination of Inter-Molecular Structural Similarity Measures of Inter-Molecular Structural Similarity. Quant. Struct. Relatsh. 1986, 5, 18–25. [Google Scholar] [CrossRef]
- Tibell, L.; Forsman, C.; Simonsson, I.; Lindskog, S. The Inhibition of Human Carbonic Anhydrase II by Some Organic Compounds. Biochim. Biophys. Acta (BBA)/Protein Struct. Mol. 1985, 829, 202–208. [Google Scholar] [CrossRef]
- Vullo, D.; Voipio, J.; Innocenti, A.; Rivera, C.; Ranki, H.; Scozzafava, A.; Kaila, K.; Supuran, C. Carbonic Anhydrase Inhibitors. Inhibition of the Human Cytosolic Isozyme VII with Aromatic and Heterocyclic Sulfonamides. Bioorg. Med. Chem. Lett. 2005, 15, 971–976. [Google Scholar] [CrossRef] [PubMed]
- Temperini, C.; Cecchi, A.; Scozzafava, A.; Supuran, C.T. Carbonic Anhydrase Inhibitors. Sulfonamide Diuretics Revisited—Old Leads for New Applications? Org. Biomol. Chem. 2008, 6, 2499–2506. [Google Scholar] [CrossRef]
- Stevens, C.R.; Millar, T.M.; Clinch, J.G.; Kanczler, J.M.; Bodamyali, T.; Blake, D.R. Antibacterial Properties of Xanthine Oxidase in Human Milk. Lancet 2000, 356, 829–830. [Google Scholar] [CrossRef]
- Selvakumar, J.N.; Chandrasekaran, S.D.; Doss, G.P.C.; Kumar, T.D. Inhibition of the ATPase Domain of Human Topoisomerase IIa on HepG2 Cells by 1, 2-Benzenedicarboxylic Acid, Mono (2-Ethylhexyl) Ester: Molecular Docking and Dynamics Simulations. Curr. Cancer Drug Targets 2019, 19, 495–503. [Google Scholar] [CrossRef] [PubMed]
- Lebda, M.; Taha, N.; Korshom, M.; Mandour, A.E.-W.; El-Tabbakh, M. Biochemical Studies on Olive Leaves and Oil against High Sucrose Diet in Rabbits. Biochem. Lett. 2013, 9, 53–66. [Google Scholar] [CrossRef]
- Mullard, A. Pioneering Apoptosis-Targeted Cancer Drug Poised for FDA Approval: AbbVie’s BCL-2 Inhibitor Venetoclax--the Leading Small-Molecule Protein-Protein Interaction Inhibitor--Could Soon Become the First Marketed Drug to Directly Target the Ability of Cancer Ce. Nat. Rev. Drug Discov. 2016, 15, 147–150. [Google Scholar] [CrossRef]
- Jackson, R.C.; Leopold, W.R.; Ross, D.A. The Biochemical Pharmacology of (2′-R)-Chloropentostatin, a Novel Inhibitor of Adenosine Deaminase. Adv. Enzyme Regul. 1986, 25, 125–139. [Google Scholar] [CrossRef]
- Alonso, R.; Villamor, N.; Upshaw, R.; Bantia, S.; Mehrling, T.; Campo, E.; Montserrat, E.; Colomer, D. The Purine Nucleoside Phosphorylase Inhibitor Forodesine Induces P53-Independent Apoptosis in Chronic Lymphocytic Leukemia through Caspase-8 Activation, Mcl-1 Downregulation, Conformational Changes of Bax and Bak and ROS Generation. Mol. Cancer Ther. 2007, 6, A41. [Google Scholar]
- Cheng, R.K.Y.; Segala, E.; Robertson, N.; Deflorian, F.; Doré, A.S.; Errey, J.C.; Fiez-Vandal, C.; Marshall, F.H.; Cooke, R.M. Structures of Human A1 and A2A Adenosine Receptors with Xanthines Reveal Determinants of Selectivity. Structure 2017, 25, 1275–1285.e4. [Google Scholar] [CrossRef]
- Marchetti, S.; de Vries, N.A.; Buckle, T.; Bolijn, M.J.; van Eijndhoven, M.A.J.; Beijnen, J.H.; Mazzanti, R.; van Tellingen, O.; Schellens, J.H.M. Effect of the ATP-Binding Cassette Drug Transporters ABCB1, ABCG2, and ABCC2 on Erlotinib Hydrochloride (Tarceva) Disposition in in Vitro and in Vivo Pharmacokinetic Studies Employing Bcrp1−/−/Mdr1a/1b−/− (Triple-Knockout) and Wild-Type Mice. Mol. Cancer Ther. 2008, 7, 2280–2287. [Google Scholar] [CrossRef]
- Chaudhary, S.; Chandrashekar, K.S.; Pai, K.S.R.; Setty, M.M.; Devkar, R.A.; Reddy, N.D.; Shoja, M.H. Evaluation of Antioxidant and Anticancer Activity of Extract and Fractions of Nardostachys jatamansi DC in Breast Carcinoma. BMC Complement. Altern. Med. 2015, 15, 50. [Google Scholar] [CrossRef] [PubMed]
- Mosmann, T. Rapid Colorimetric Assay for Cellular Growth and Survival: Application to Proliferation and Cytotoxicity Assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
- Zrouri, H.; Elbouzidi, A.; Bouhrim, M.; Bencheikh, N.; Kharchoufa, L.; Ouahhoud, S.; Ouassou, H.; El Assri, S.; Choukri, M. Phytochemical Analysis, Antioxidant Activity, and Nephroprotective Effect of the Raphanus Sativus Aqueous Extract. Mediterr. J. Chem. 2021, 11, 84. [Google Scholar] [CrossRef]
- Rădulescu, M.; Jianu, C.; Lukinich-Gruia, A.T.; Mioc, M.; Mioc, A.; Șoica, C.; Stana, L.G. Chemical Composition, in Vitro and in Silico Antioxidant Potential of Melissa Officinalis Subsp. Officinalis Essential Oil. Antioxidants 2021, 10, 1081. [Google Scholar] [CrossRef]
- Nakyai, W.; Pabuprapap, W.; Sroimee, W.; Ajavakom, V.; Yingyongnarongkul, B.E.; Suksamrarn, A. Anti-Acne Vulgaris Potential of the Ethanolic Extract of Mesua ferrea L. Flowers. Cosmetics 2021, 8, 107. [Google Scholar] [CrossRef]
- Prieto, P.; Pineda, M.; Aguilar, M. Spectrophotometric Quantitation of Antioxidant Capacity through the Formation of a Phosphomolybdenum Complex: Specific Application to the Determination of Vitamin E. Anal. Biochem. 1999, 269, 337–341. [Google Scholar] [CrossRef]
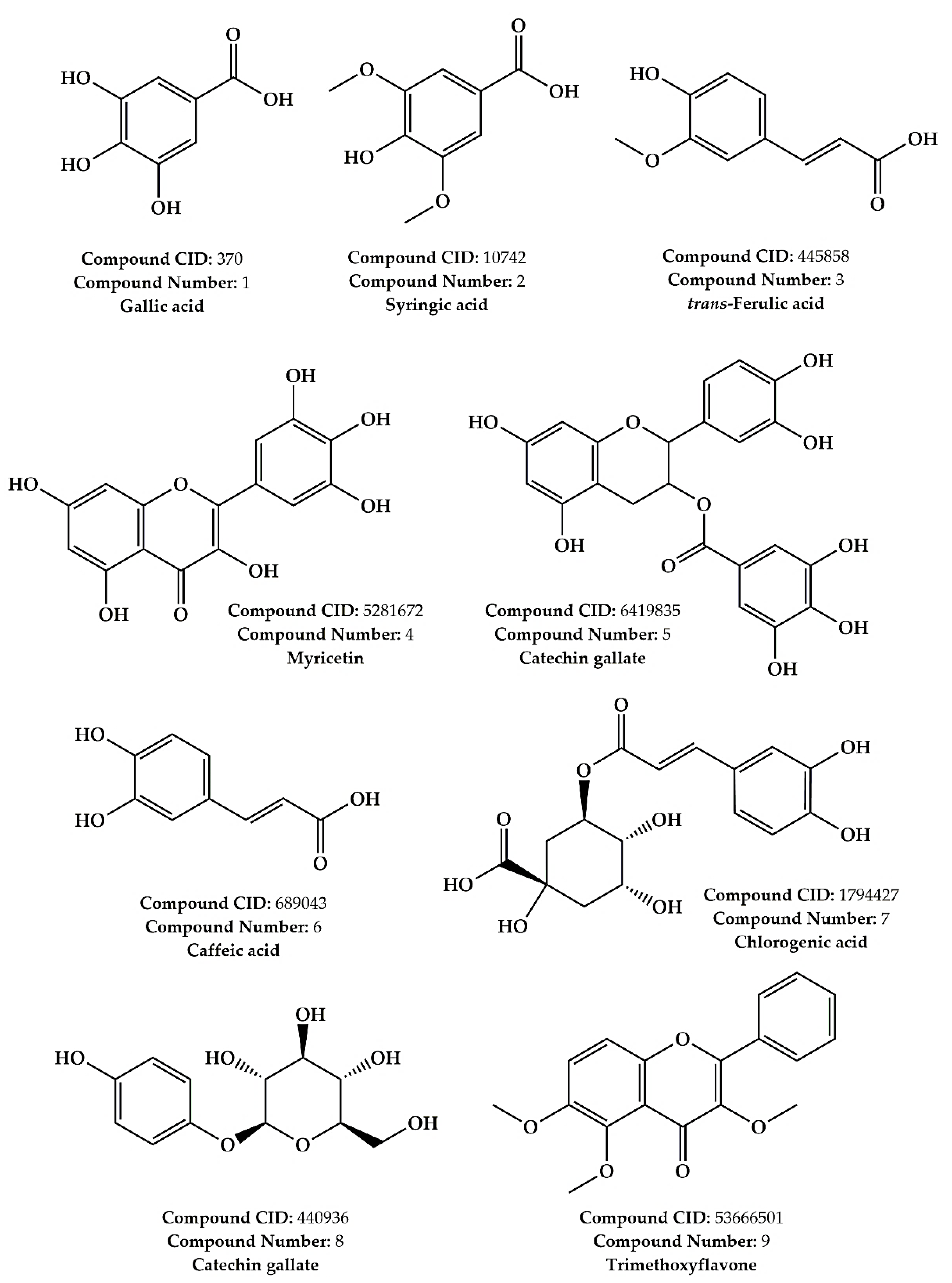


| N° | Molecule | Molecular Formula | Selected [M−H]− | Literature [M−H]− | RT (min) | Abundance |
|---|---|---|---|---|---|---|
| 1 | Gallic acid | C7H6O5 | 168.90 | 169.00 [41] | 1.586 | +++ |
| 2 | Syringic acid | C9H10O5 | 198.90 | 197.05 [42] | 1.478 | ++ |
| 3 | trans-Ferulic acid | C10H10O4 | 193.00 | 193.05 [42] | 1.191 | ++ |
| 4 | Myricetin | C15H10O8 | 317.00 | 317.01 [43] | 0.330 | + |
| 5 | Catechin gallate | C22H18O10 | 441.00 | 441.08 [44] | 1.208 | + |
| 6 | Caffeic acid | C9H8O4 | 179.00 | 179.03 [42] | 1.378 | + |
| 7 | Chlorogenic acid | C16H18O9 | 353.00 | 353.09 [42] | 1.117 | + |
| 8 | Arbutin | C12H16O7 | 271.20 | 271.20 [45] | 1.304 | + |
| 9 | Trimethoxyflavone | C18H16O5 | 312.00 | 315.00 [46] | 1.380 | + |
| Lipinski | Ghose | Veber | Egan | Bioavailability Score | |
|---|---|---|---|---|---|
| Gallic acid | Yes | No (2 violations) | Yes | Yes | 0.56 |
| Syringic acid | Yes | Yes | Yes | Yes | 0.56 |
| trans-Ferulic acid | Yes | Yes | Yes | Yes | 0.85 |
| Myricetin | Yes (1 violation) | Yes | No (1 violation) | No (1 violation) | 0.55 |
| Catechin gallate | Yes (1 violation) | Yes | No (1 violation) | No (1 violation) | 0.55 |
| Caffeic acid | Yes | Yes | Yes | Yes | 0.56 |
| Chlorogenic acid | Yes | No (1 violation) | No (1 violation) | No (1 violation) | 0.11 |
| Arbutin | Yes | No (1 violation) | Yes | Yes | 0.55 |
| Trimethoxyflavone | Yes | Yes | Yes | Yes | 0.55 |
| Water Solubility | Caco-2 Permeability | Intestinal Absorption | Skin Permeability | P-gp Substrate | P-gp I Inhibitor | P-gp II Inhibitor | |
|---|---|---|---|---|---|---|---|
| Units | Log mol/L | Log Papp in 10−6 cm/s | % | cm/s | Categorical (Yes/No) | ||
| Gallic acid | −1.64 | −0.08 | 43.37 | −2.73 | No | No | No |
| Syringic acid | −1.84 | 0.49 | 73.07 | −2.73 | No | No | No |
| Trans-ferulic acid | −2.11 | 0.17 | 93.68 | −2.72 | No | No | No |
| Myricetin | −3.01 | 0.09 | 65.93 | −2.73 | No | No | No |
| Catechin gallate | −3.70 | −1.26 | 62.09 | −2.73 | No | No | Yes |
| Caffeic acid | −1.89 | 0.63 | 69.40 | −2.72 | No | No | No |
| Chlorogenic acid | −1.62 | −0.84 | 36.37 | −2.73 | No | No | No |
| Arbutin | −0.71 | 0.00 | 38.02 | −2.80 | No | No | No |
| Trimethoxyflavone | −4.11 | 1.40 | 98.07 | −2.57 | No | Yes | Yes |
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | |
|---|---|---|---|---|---|---|---|---|---|
| VDss (human) | −1.85 | −1.44 | −1.36 | 1.31 | 0.66 | −1.09 | 0.58 | 0.02 | −0.29 |
| BBB permeability (Log BB) | −1.10 | −0.19 | −0.23 | −1.49 | −1.84 | −0.64 | −1.40 | −0.96 | −0.20 |
| CNS permeability (Log PS) | −3.74 | −2.70 | −2.61 | −3.70 | −3.74 | −2.60 | −3.85 | −3.55 | −2.14 |
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | |
|---|---|---|---|---|---|---|---|---|---|
| CYP2D6 substrate | No | No | No | No | No | No | No | No | No |
| CYP3A4 substrate | No | No | No | No | No | No | No | No | Yes |
| CYP2D6 inhibitor | No | No | No | No | No | No | No | No | Yes |
| CYP3A4 inhibitor | No | No | No | Yes | No | No | No | No | Yes |
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | |
|---|---|---|---|---|---|---|---|---|---|
| Total clearence (Log mL/min/Kg) | 0.51 | 0.64 | 0.62 | 0.42 | −0.16 | 0.50 | 0.30 | 0.52 | 0.28 |
| Renal OCT2 substrate | No | No | No | No | No | No | No | No | Yes |
| Molecules | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 |
|---|---|---|---|---|---|---|---|---|---|
| LD50 (mg/Kg) | 2000 | 1700 | 1772 | 159 | 1000 | 2980 | 5000 | 2500 | 5000 |
| Class | 4 | 4 | 4 | 3 | 4 | 5 | 5 | 5 | 5 |
| Hepatotoxicity | Inactive | Inactive | Inactive | Inactive | Inactive | Inactive | Inactive | Inactive | Inactive |
| Carcinogenicity | Active | Inactive | Inactive | Active | Inactive | Active | Inactive | Inactive | Inactive |
| Immunotoxicity | Inactive | Inactive | Active | Inactive | Inactive | Inactive | Active | Inactive | Inactive |
| Mutagenicity | Inactive | Inactive | Inactive | Active | Inactive | Inactive | Inactive | Inactive | Inactive |
| Cytotoxicity | Inactive | Inactive | Inactive | Inactive | Inactive | Inactive | Inactive | Inactive | Inactive |
| Extract/Reference | DPPH Scavenging Capacity IC50 (mg/mL) | β-Carotene Bleaching Assay (mg/mL) | ABTS Scavenging (TE µmol/mL) | Iron Chelation | Total Antioxidant Capacity * |
|---|---|---|---|---|---|
| AHEE | 0.36 ± 0.05 | 2.91 ± 0.14 | 44.10 ± 2.92 | 27.40 ± 1.46 | 124 ± 1.27 |
| Ascorbic acid (AA) | 0.19 ± 0.02 | - | 5.04 ± 0.78 | 0.94 ± 0.02 | - |
| Butylated hydroxyanisole (BHA) | - | 0.095 ± 0.00 | - | - | - |
| Treatments | IC50 Value ± SD (µg/mL) * | Selectivity Index ** | |||
|---|---|---|---|---|---|
| MCF-7 | MDA-MB-231 | PBMC | MCF-7 | MDA-MB-231 | |
| AHEE | 27.85 ± 3.14 | 51.95 ± 7.03 | 743.6 ± 9.55 | 26.7 | 14.31 |
| Cisplatin | 3.66 ± 1.05 | 1.60 ± 1.19 | 29.83 ± 1.19 | 8.15 | 18.64 |
| PDB IDs | Protein Target Name | Resolution | Native Ligands | Ligands | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | ||||
| 12CA | Human carbonic anhydrase II | 2.40 Å | Tetrazole [123] | X | X | X | X | X | ||||
| 3MDZ | Human carbonic anhydrase VII | 2.32 Å | Acetazolamide [124] | X | X | X | X | X | X | |||
| 1AZM | Human carbonic anhydrase I | 2.00 Å | Furosemide [125] | X | X | X | X | X | X | X | ||
| 2CKJ | Human milk xanthine oxidoreductase | 3.59 Å | Oxypurinol [126] | X | ||||||||
| 1ZXM | Human Topo IIa ATPase/AMP-PNP | 1.87 Å | 1,2-Benzenedicarboxylic acid, mono(2-ethylhexyl) ester [127] | X | ||||||||
| 2JKV | Human phosphogluconate dehydrogenase | 2.53 Å | NADPH [128] | X | ||||||||
| 1G5M | Human antiapoptotic protein BCL-2 | 1.80 Å | ABT-737 [129] | X | ||||||||
| 3IAR | Human adenosine deaminase | 1.52 Å | 2′-Deoxyadenosine [130] | X | ||||||||
| 1M73 | Human purine nucleoside phosphorylase (PNP) | 2.30 Å | Forodesine [131] | X | ||||||||
| 5N2S | Human A1 adenosine receptor | 3.30 Å | PSB36 [132] | X | ||||||||
| 2YDO | Human A2A adenosine receptor | 3.00 Å | Istradefylline [132] | X | ||||||||
| 5NJ3 | ABC transporter | 3.78 Å | Gefitinib [133] | X | ||||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Elbouzidi, A.; Ouassou, H.; Aherkou, M.; Kharchoufa, L.; Meskali, N.; Baraich, A.; Mechchate, H.; Bouhrim, M.; Idir, A.; Hano, C.; et al. LC–MS/MS Phytochemical Profiling, Antioxidant Activity, and Cytotoxicity of the Ethanolic Extract of Atriplex halimus L. against Breast Cancer Cell Lines: Computational Studies and Experimental Validation. Pharmaceuticals 2022, 15, 1156. https://doi.org/10.3390/ph15091156
Elbouzidi A, Ouassou H, Aherkou M, Kharchoufa L, Meskali N, Baraich A, Mechchate H, Bouhrim M, Idir A, Hano C, et al. LC–MS/MS Phytochemical Profiling, Antioxidant Activity, and Cytotoxicity of the Ethanolic Extract of Atriplex halimus L. against Breast Cancer Cell Lines: Computational Studies and Experimental Validation. Pharmaceuticals. 2022; 15(9):1156. https://doi.org/10.3390/ph15091156
Chicago/Turabian StyleElbouzidi, Amine, Hayat Ouassou, Marouane Aherkou, Loubna Kharchoufa, Nada Meskali, Abdellah Baraich, Hamza Mechchate, Mohamed Bouhrim, Abderrazak Idir, Christophe Hano, and et al. 2022. "LC–MS/MS Phytochemical Profiling, Antioxidant Activity, and Cytotoxicity of the Ethanolic Extract of Atriplex halimus L. against Breast Cancer Cell Lines: Computational Studies and Experimental Validation" Pharmaceuticals 15, no. 9: 1156. https://doi.org/10.3390/ph15091156
APA StyleElbouzidi, A., Ouassou, H., Aherkou, M., Kharchoufa, L., Meskali, N., Baraich, A., Mechchate, H., Bouhrim, M., Idir, A., Hano, C., Zrouri, H., & Addi, M. (2022). LC–MS/MS Phytochemical Profiling, Antioxidant Activity, and Cytotoxicity of the Ethanolic Extract of Atriplex halimus L. against Breast Cancer Cell Lines: Computational Studies and Experimental Validation. Pharmaceuticals, 15(9), 1156. https://doi.org/10.3390/ph15091156








