In Silico Prediction of Anti-Infective and Cell-Penetrating Peptides from Thalassophryne nattereri Natterin Toxins
Abstract
:1. Introduction
2. Results and Discussion
2.1. Identification of Potential Natterin-Derived AMPs and CPPs
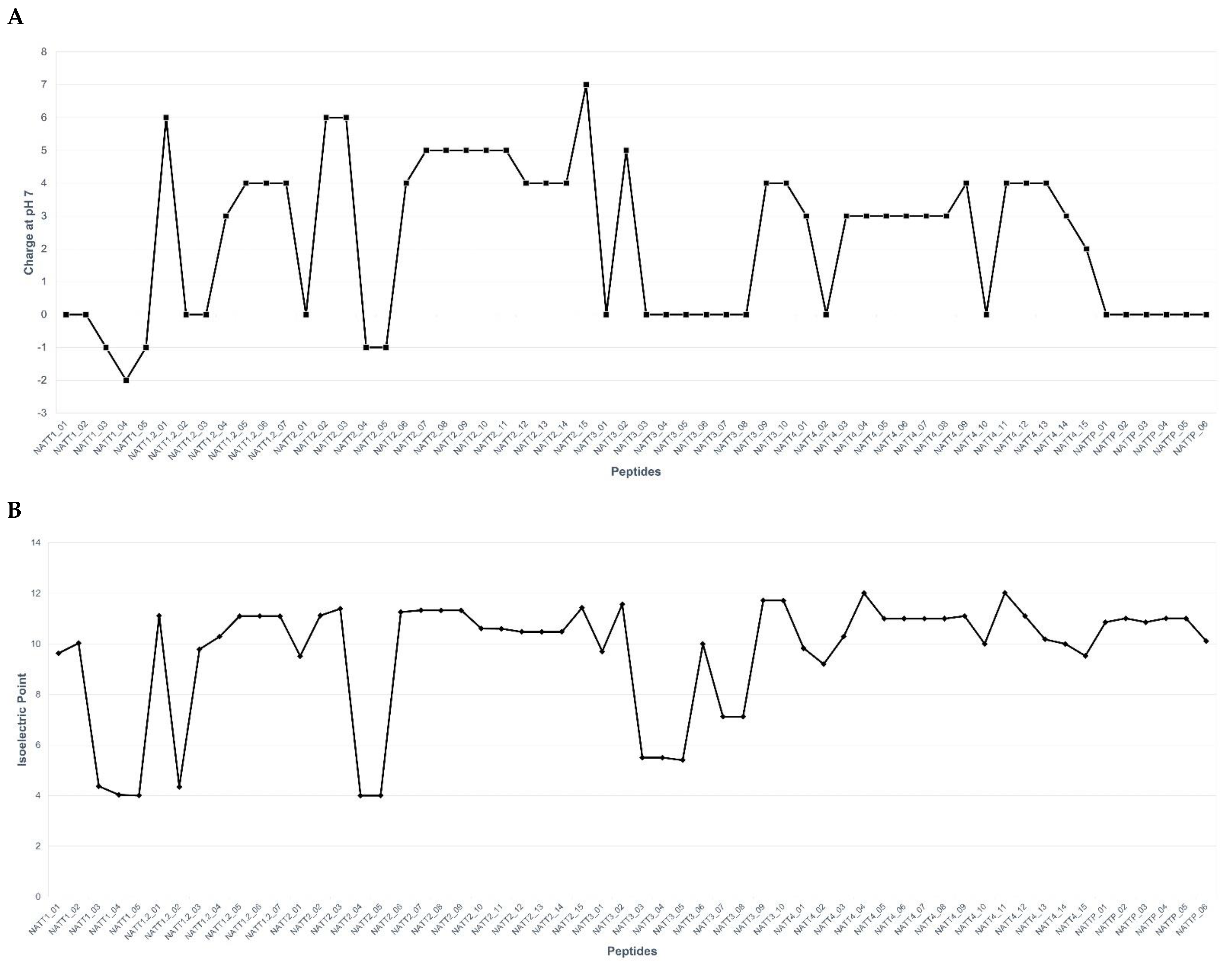
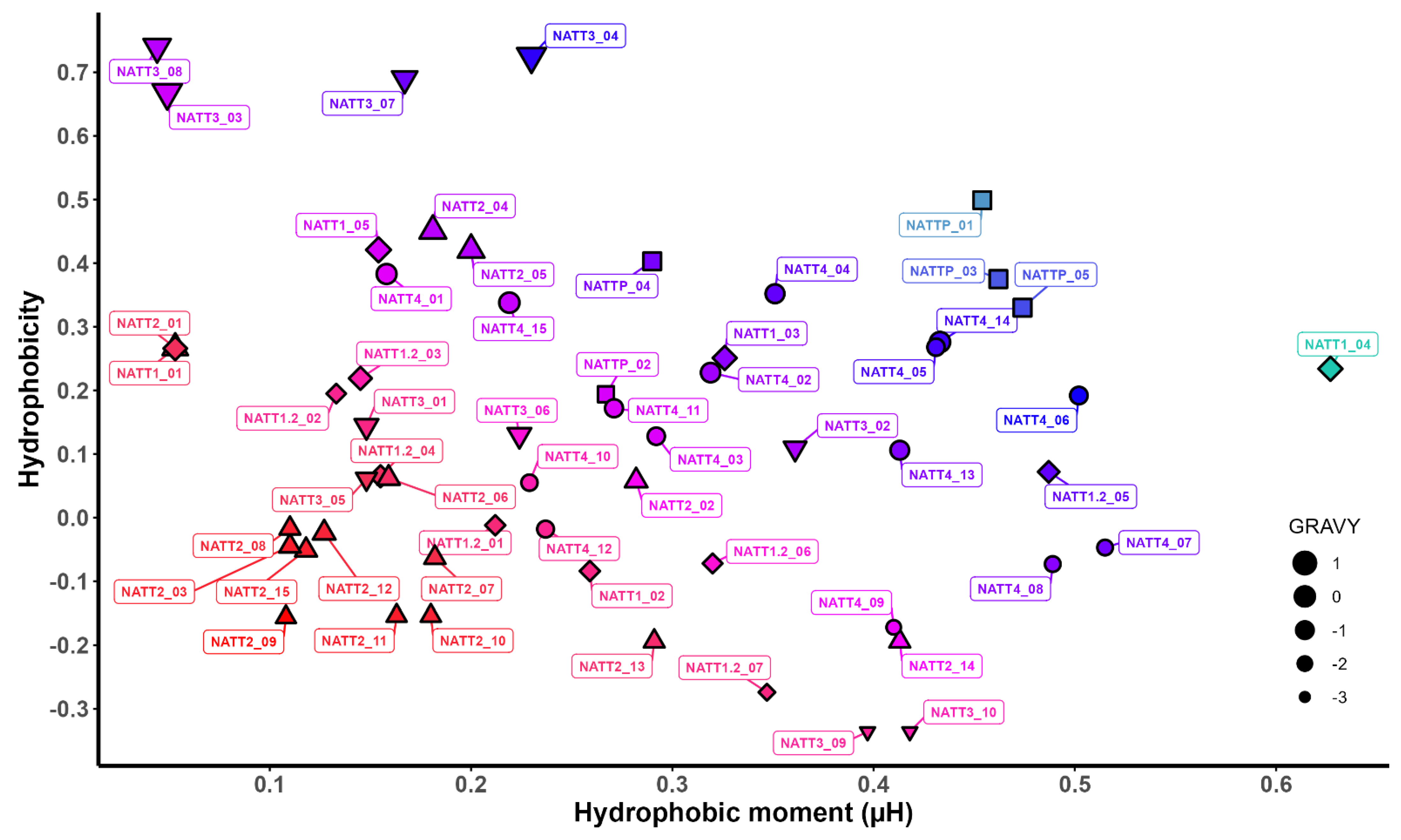
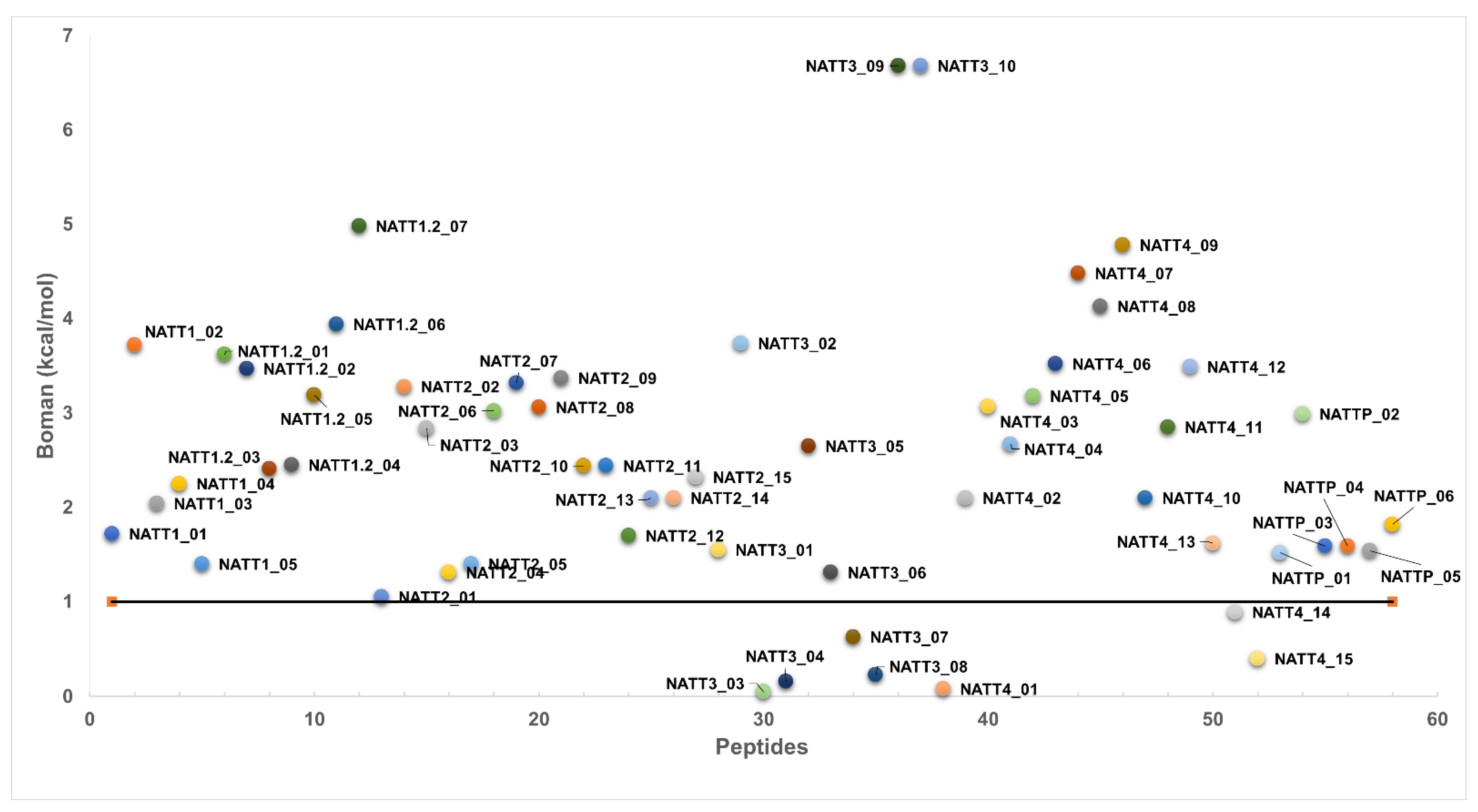
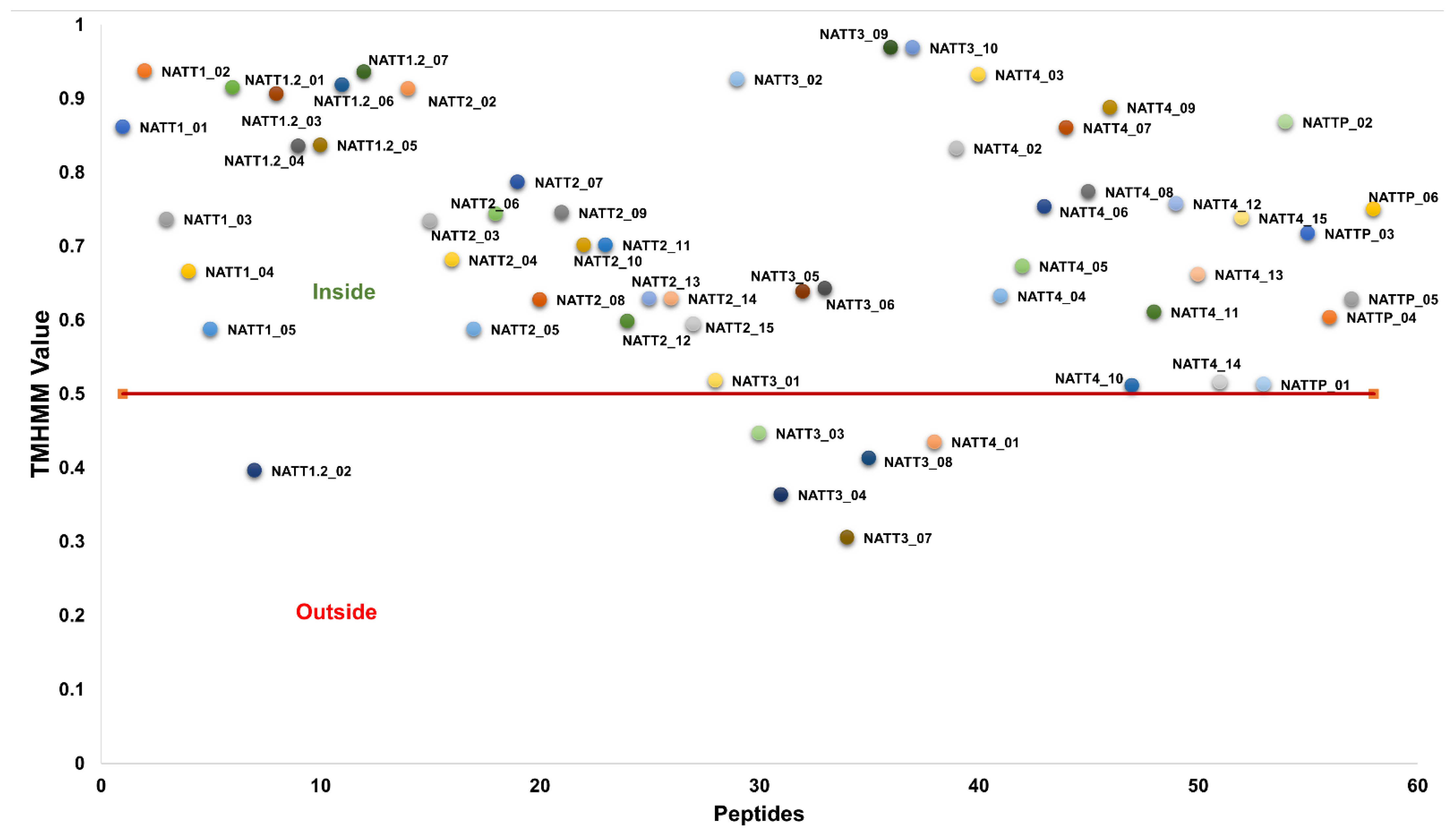
2.2. Physicochemical Properties and Membrane-Binding Potential
2.3. Prediction of Biological Activities
2.3.1. Immunogenicity, Allergenicity, and Toxicity
2.3.2. Antiviral and Anticancer Potential
2.3.3. Prediction of ADMET Properties
2.4. Medicinal Chemistry Studies
2.5. Prediction of Peptide Structures
3. Materials and Methods
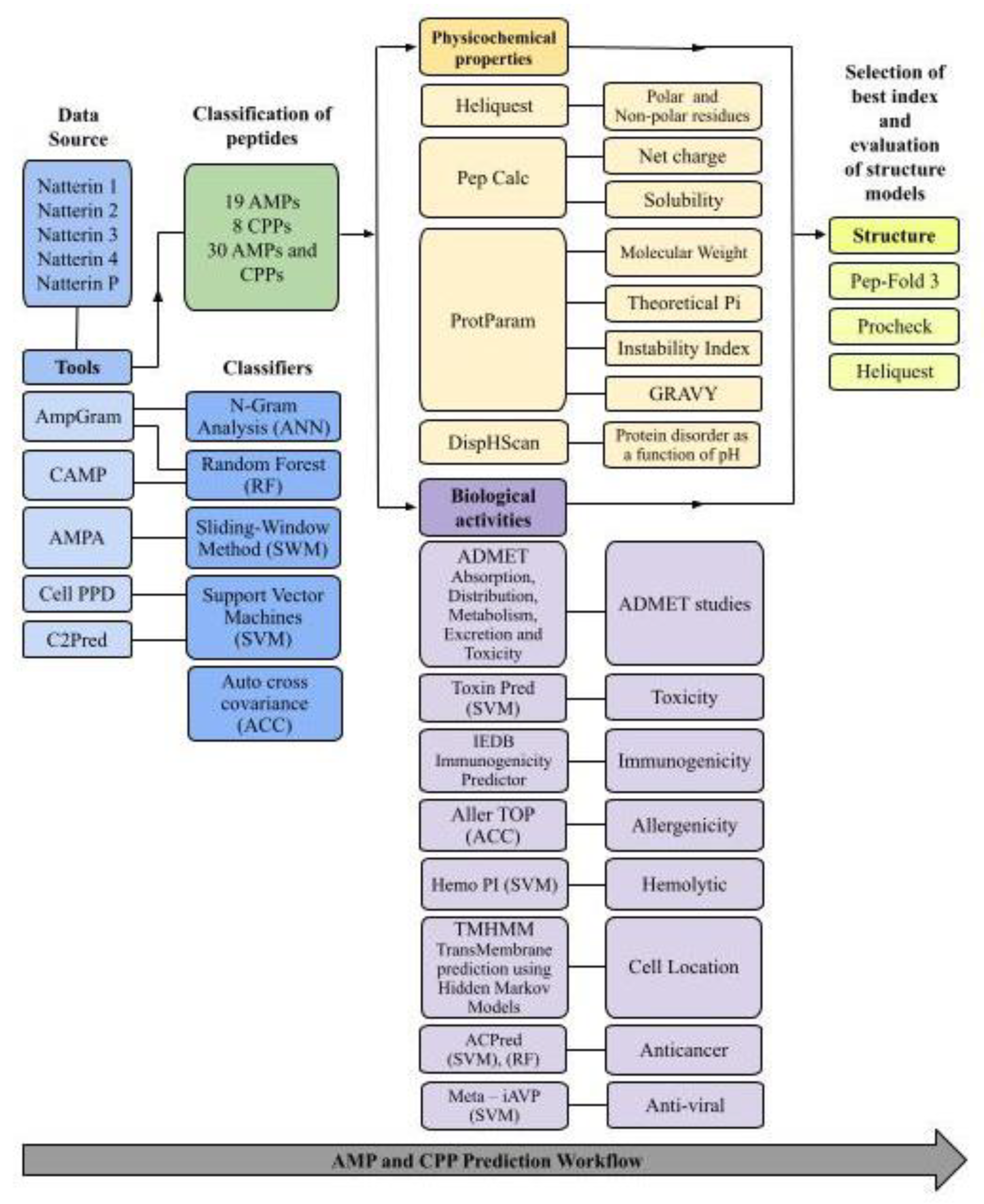
3.1. Study Design
3.2. Prediction of BAPs
3.3. Physicochemical Properties
3.4. Evaluation of the Membrane-Binding Ability of BAPs
3.5. Assessment of Immunogenicity, Toxicity, Allergenicity, and Anticancer and Antiviral Properties
3.6. Hemolytic Activity and Half-Life
3.7. Prediction of ADMET and Medicinal Chemistry Parameters
3.8. Prediction of Peptide Structure
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pennington, M.W.; Czerwinski, A.; Norton, R.S. Peptide Therapeutics from Venom: Current Status and Potential. Bioorg. Med. Chem. 2018, 26, 2738–2758. [Google Scholar] [CrossRef] [PubMed]
- Usmani, S.S.; Bedi, G.; Samuel, J.S.; Singh, S.; Kalra, S.; Kumar, P.; Ahuja, A.A.; Sharma, M.; Gautam, A.; Raghava, G.P.S. THPdb: Database of FDA-Approved Peptide and Protein Therapeutics. PLoS ONE 2017, 12, e0181748. [Google Scholar] [CrossRef] [PubMed]
- Herzig, V.; Cristofori-Armstrong, B.; Israel, M.R.; Nixon, S.A.; Vetter, I.; King, G.F. Animal Toxins—Nature’s Evolutionary-Refined Toolkit for Basic Research and Drug Discovery. Biochem. Pharmacol. 2020, 181, 114096. [Google Scholar] [CrossRef] [PubMed]
- Slagboom, J.; Kaal, C.; Arrahman, A.; Vonk, F.J.; Somsen, G.W.; Calvete, J.J.; Wüster, W.; Kool, J. Analytical Strategies in Venomics. Microchem. J. 2022, 175, 107187. [Google Scholar] [CrossRef]
- Padhi, A.; Sengupta, M.; Sengupta, S.; Roehm, K.H.; Sonawane, A. Antimicrobial Peptides and Proteins in Mycobacterial Therapy: Current Status and Future Prospects. Tuberculosis 2014, 94, 363–373. [Google Scholar] [CrossRef]
- Buchwald, H.; Dorman, R.B.; Rasmus, N.F.; Michalek, V.N.; Landvik, N.M.; Ikramuddin, S. Effects on GLP-1, PYY, and Leptin by Direct Stimulation of Terminal Ileum and Cecum in Humans: Implications for Ileal Transposition. Surg. Obes. Relat. Dis. 2014, 10, 780–786. [Google Scholar] [CrossRef]
- Giordano, C.; Marchiò, M.; Timofeeva, E.; Biagini, G. Neuroactive Peptides as Putative Mediators of Antiepileptic Ketogenic Diets. Front. Neurol. 2014, 5, 63. [Google Scholar] [CrossRef]
- Fosgerau, K.; Hoffmann, T. Peptide Therapeutics: Current Status and Future Directions. Drug Discov. Today 2015, 20, 122–128. [Google Scholar] [CrossRef] [PubMed]
- Fjell, C.D.; Hiss, J.A.; Hancock, R.E.W.; Schneider, G. Designing Antimicrobial Peptides: Form Follows Function. Nat. Rev. Drug Discov. 2012, 11, 37–51. [Google Scholar] [CrossRef]
- Diochot, S.; Baron, A.; Salinas, M.; Douguet, D.; Scarzello, S.; Dabert-Gay, A.S.; Debayle, D.; Friend, V.; Alloui, A.; Lazdunski, M.; et al. Black Mamba Venom Peptides Target Acid-Sensing Ion Channels to Abolish Pain. Nature 2012, 490, 552–555. [Google Scholar] [CrossRef]
- Hancock, R.E.W.; Sahl, H.G. Antimicrobial and Host-Defense Peptides as New Anti-Infective Therapeutic Strategies. Nat. Biotechnol. 2006, 24, 1551–1557. [Google Scholar] [CrossRef] [PubMed]
- Holford, N.; Channon, S.; Heron, J.; Jones, I. The Impact of Postpartum Psychosis on Partners 11 Medical and Health Sciences 1117 Public Health and Health Services. BMC Pregnancy Childbirth 2018, 18, 414. [Google Scholar] [CrossRef]
- Ziegman, R.; Alewood, P. Bioactive Components in Fish Venoms. Toxins 2015, 7, 1497–1531. [Google Scholar] [CrossRef] [PubMed]
- Facó, P.E.; Bezerra, G.P.; Ferreira Barbosa, P.S.; Maria Costa Martins, A.; Guimarães, J.A.; Ferreira, M.L.; Serra, H.; Monteiro, A. Epidemiology of the injuries caused by Thalassophryne nattereri (niquim) in Ceara State (1992–2002). Rev. Soc. Bras. Med. Trop. 2005, 38, 479–482. [Google Scholar] [CrossRef]
- Haddad, V., Jr.; Pardal, P.P.O.; Cardoso, J.L.C.; Martins, I.A. Case Report the Venomous Toadfish Thalassophryne Nattereri (Niquim or Miquim): Report of 43 Injuries Provoked in Fishermen of Salinópolis (Pará State) and Aracaju (Sergipe State), Brazil. Rev. Do Inst. De Med. Trop. De São Paulo 2003, 45, 221–223. [Google Scholar]
- Lopes-Ferreira, M.; Grund, L.Z.; Lima, C. Thalassophryne Nattereri fish Venom: From the Envenoming to the Understanding of the Immune System. J. Venom. Anim. Toxins Incl. Trop. Dis. 2014, 20, 1–12. [Google Scholar] [CrossRef]
- Magalhães, G.S.; Junqueira-de-Azevedo, I.L.M.; Lopes-Ferreira, M.; Lorenzini, D.M.; Ho, P.L.; Moura-da-Silva, A.M. Transcriptome Analysis of Expressed Sequence Tags from the Venom Glands of the Fish Thalassophryne Nattereri. Biochimie 2006, 88, 693–699. [Google Scholar] [CrossRef]
- Lopes-Ferreira, M.; Emim, J.A.D.S.; Oliveira, V.; Puzer, L.; Cezari, M.H.; Araújo, M.D.S.; Juliano, L.; Lapa, A.J.; Souccar, C.; Moura-Da-Silva, A.M. Kininogenase Activity of Thalassophryne Nattereri Fish Venom. Biochem. Pharmacol. 2004, 68, 2151–2157. [Google Scholar] [CrossRef]
- Magalhães, G.S.; Lopes-Ferreira, M.; Junqueira-De-Azevedo, I.L.M.; Spencer, P.J.; Araújo, M.S.; Portaro, F.C.V.; Ma, L.; Valente, R.H.; Juliano, L.; Fox, J.W.; et al. Natterins, a New Class of Proteins with Kininogenase Activity Characterized from Thalassophryne Nattereri Fish Venom. Biochimie 2005, 87, 687–699. [Google Scholar] [CrossRef]
- Lima, C.; Falcao, M.A.P.; Andrade-Barros, A.I.; Seni-Silva, A.C.; Grund, L.Z.; Balogh, E.; Conceiçao, K.; Queniaux, V.F.; Ryffel, B.; Lopes-Ferreira, M. Natterin an Aerolysin-like Fish Toxin Drives IL-1β-Dependent Neutrophilic Inflammation Mediated by Caspase-1 and Caspase-11 Activated by the Inflammasome Sensor NLRP6. Int. Immunopharmacol. 2021, 91, 107287. [Google Scholar] [CrossRef]
- Lewis, R.J.; Garcia, M.L. Therapeutic Potential of Venom Peptides. Nat. Rev. Drug Discov. 2003, 2, 790–802. [Google Scholar] [CrossRef] [PubMed]
- Heitz, F.; Morris, M.C.; Divita, G. Themed Section: Vector Design and Drug Delivery Review Twenty Years of Cell-Penetrating Peptides: From Molecular Mechanisms to Therapeutics. Br. J. Pharmacol. 2009, 157, 195–206. [Google Scholar] [CrossRef] [PubMed]
- Säälik, P.; Elmquist, A.; Hansen, M.; Padari, K.; Saar, K.; Viht, K.; Langel, Ü.; Pooga, M. Protein Cargo Delivery Properties of Cell-Penetrating Peptides. A Comparative Study. Bioconjugate Chem. 2004, 15, 1246–1253. [Google Scholar] [CrossRef]
- Pooga, M.; Soomets, U.; Hällbrink, M.; Valkna, A.; Saar, K.; Rezaei, K.; Kahl, U.; Hao, J.X.; Xu, X.J.; Wiesenfeld-Hallin, Z.; et al. Cell penetrating PNA constructs regulate galanin receptor levels and modify pain transmission in vivo. Nat. Biotechnol. 1998, 16, 857–861. [Google Scholar] [CrossRef] [PubMed]
- Vives, E. Present and Future of Cell-Penetrating Peptide Mediated Delivery Systems: “Is the Trojan Horse Too Wild to Go Only to Troy?”. J. Control. Release 2005, 109, 77–85. [Google Scholar] [CrossRef]
- Reddy, K.V.R.; Yedery, R.D.; Aranha, C. Antimicrobial Peptides: Premises and Promises. Int. J. Antimicrob. Agents 2004, 24, 536–547. [Google Scholar] [CrossRef]
- Zasloff, M. Antimicrobial Peptides of Multicellular Organisms: My Perspective. In Advances in Experimental Medicine and Biology; Springer: New York, NY, USA, 2019; Volume 1117, pp. 3–6. [Google Scholar]
- Ganz, T. Defensins: Antimicrobial Peptides of Innate Immunity. Nat. Rev. Immunol. 2003, 3, 710–720. [Google Scholar] [CrossRef]
- Ellerby, H.M.; Bredesen, D.E.; Fujimura, S.; John, V. Hunter-Killer Peptide (HKP) for Targeted Therapy. J. Med. Chem. 2008, 51, 5887–5892. [Google Scholar] [CrossRef]
- Kolonin, M.G.; Saha, P.K.; Chan, L.; Pasqualini, R.; Arap, W. Reversal of Obesity by Targeted Ablation of Adipose Tissue. Nat. Med. 2004, 10, 625–632. [Google Scholar] [CrossRef]
- Barnhart, K.F.; Christianson, D.R.; Hanley, P.W.; Driessen, W.H.P.; Bernacky, B.J.; Baze, W.B.; Wen, S.; Tian, M.; Ma, J.; Kolonin, M.G.; et al. A Peptidomimetic Targeting White Fat Causes Weight Loss and Improved Insulin Resistance in Obese Monkeys. Sci. Transl. Med. 2011, 3, 108ra112. [Google Scholar] [CrossRef]
- Ellerby, H.; Arap, W.; Ellerby, L.M.; Kain, R.; Andrusiak, R.; Rio, G.D.; Krajewski, S.; Lombardo, C.R.; Rao, R.; Ruoslahti, E.; et al. Anti-cancer activity of targeted pro-apoptotic peptides. Nat. Med. 1999, 5, 1032–1038. [Google Scholar] [CrossRef] [PubMed]
- Morris, M.C.; Depollier, J.; Mery, J.; Heitz, F.; Divita, G. A Peptide Carrier for the Delivery of Biologically Active Proteins into Mammalian Cells. Nat. Biotechnol. 2001, 19, 1173–1176. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Wang, Y.; Zhang, X.; Zhang, W.; Guo, S.; Jin, F. Recent Progress of Cell-Penetrating Peptides as New Carriers for Intracellular Cargo Delivery. J. Control. Release 2014, 174, 126–136. [Google Scholar] [CrossRef] [PubMed]
- Gautam, A.; Singh, H.; Tyagi, A.; Chaudhary, K.; Kumar, R.; Kapoor, P.; Raghava, G.P.S. CPPsite: A Curated Database of Cell Penetrating Peptides. Database 2012, 2012, bas015. [Google Scholar] [CrossRef] [PubMed]
- Farnaud, S.; Evans, R.W. Lactoferrin—A Multifunctional Protein with Antimicrobial Properties. Mol. Immunol. 2003, 40, 395–405. [Google Scholar] [CrossRef]
- Loose, C.; Jensen, K.; Rigoutsos, I.; Stephanopoulos, G. A Linguistic Model for the Rational Design of Antimicrobial Peptides. Nature 2006, 443, 867–869. [Google Scholar] [CrossRef]
- Brand, G.D.; Ramada, M.H.S.; Genaro-Mattos, T.C.; Bloch, C. Towards an Experimental Classification System for Membrane Active Peptides. Sci. Rep. 2018, 8, 1194. [Google Scholar] [CrossRef]
- Huang, Y.W.; Lee, H.J. Cell-Penetrating Peptides for Medical Theranostics and Targeted Drug Delivery. In Peptide Applications in Biomedicine, Biotechnology and Bioengineering; Elsevier Inc.: Amsterdam, The Netherlands, 2018; pp. 359–370. ISBN 9780081007426. [Google Scholar]
- Porosk, L.; Gaidutšik, I.; Langel, Ü. Approaches for the Discovery of New Cell-Penetrating Peptides. Expert Opin. Drug Discov. 2021, 16, 553–565. [Google Scholar] [CrossRef]
- Cardoso, M.H.; Orozco, R.Q.; Rezende, S.B.; Rodrigues, G.; Oshiro, K.G.N.; Cândido, E.S.; Franco, O.L. Computer-Aided Design of Antimicrobial Peptides: Are We Generating Effective Drug Candidates? Front. Microbiol. 2020, 10, 3097. [Google Scholar] [CrossRef]
- Lertampaiporn, S.; Vorapreeda, T.; Hongsthong, A.; Thammarongtham, C. Ensemble-Amppred: Robust Amp Prediction and Recognition Using the Ensemble Learning Method with a New Hybrid Feature for Differentiating Amps. Genes 2021, 12, 137. [Google Scholar] [CrossRef]
- Tyagi, A.; Kapoor, P.; Kumar, R.; Chaudhary, K.; Gautam, A.; Raghava, G.P.S. In Silico Models for Designing and Discovering Novel Anticancer Peptides. Sci. Rep. 2013, 3, 2984. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Oliveira, E.C.L.; Santana, K.; Josino, L.; Lima e Lima, A.H.; de Souza de Sales, C., Jr. Predicting Cell-Penetrating Peptides Using Machine Learning Algorithms and Navigating in Their Chemical Space. Sci. Rep. 2021, 11, 7628. [Google Scholar] [CrossRef]
- Robles-Loaiza, A.A.; Pinos-Tamayo, E.A.; Mendes, B.; Ortega-Pila, J.A.; Proaño-Bolaños, C.; Plisson, F.; Teixeira, C.; Gomes, P.; Almeida, J.R. Traditional and Computational Screening of Non-Toxic Peptides and Approaches to Improving Selectivity. Pharmaceuticals 2022, 15, 323. [Google Scholar] [CrossRef] [PubMed]
- Gautam, A.; Chaudhary, K.; Kumar, R.; Sharma, A.; Kapoor, P.; Tyagi, A.; Raghava, G.P.S. In Silico Approaches for Designing Highly Effective Cell Penetrating Peptides. J. Transl. Med. 2013, 11, 74. [Google Scholar] [CrossRef] [PubMed]
- Gaspar, D.; Salomé Veiga, A.; Castanho, M.A.R.B. From Antimicrobial to Anticancer Peptides. A Review. Front. Microbiol. 2013, 4, 294. [Google Scholar] [CrossRef] [PubMed]
- Maccari, G.; di Luca, M.; Nifosì, R. In Silico Design of Antimicrobial Peptides. Methods Mol. Biol. 2015, 1268, 195–219. [Google Scholar] [CrossRef]
- Wishart, D.S.; Feunang, Y.D.; Guo, A.C.; Lo, E.J.; Marcu, A.; Grant, J.R.; Sajed, T.; Johnson, D.; Li, C.; Sayeeda, Z.; et al. DrugBank 5.0: A Major Update to the DrugBank Database for 2018. Nucleic Acids Res. 2018, 46, D1074–D1082. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhang, M.; Qiu, S.; Wang, J.; Peng, J.; Zhao, P.; Zhu, R.; Wang, H.; Li, Y.; Wang, K.; et al. Antimicrobial Activity and Stability of the D-Amino Acid Substituted Derivatives of Antimicrobial Peptide Polybia-MPI. AMB Express 2016, 6, 122. [Google Scholar] [CrossRef]
- Pachón-Ibáñez, M.E.; Smani, Y.; Pachón, J.; Sánchez-Céspedes, J. Perspectives for Clinical Use of Engineered Human Host Defense Antimicrobial Peptides. FEMS Microbiol. Rev. 2017, 41, 323–342. [Google Scholar] [CrossRef]
- Lombardi, L.; Maisetta, G.; Batoni, G.; Tavanti, A. Insights into the Antimicrobial Properties of Hepcidins: Advantages and Drawbacks as Potential Therapeutic Agents. Molecules 2015, 20, 6319–6341. [Google Scholar] [CrossRef]
- Oshiro, K.G.N.; Cândido, E.S.; Chan, L.Y.; Torres, M.D.T.; Monges, B.E.D.; Rodrigues, S.G.; Porto, W.F.; Ribeiro, S.M.; Henriques, S.T.; Lu, T.K.; et al. Computer-Aided Design of Mastoparan-like Peptides Enables the Generation of Nontoxic Variants with Extended Antibacterial Properties. J. Med. Chem. 2019, 62, 8140–8151. [Google Scholar] [CrossRef] [PubMed]
- Vlieghe, P.; Lisowski, V.; Martinez, J.; Khrestchatisky, M. Synthetic Therapeutic Peptides: Science and Market. Drug Discov. Today 2010, 15, 40–56. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.J.; Cho, C.H. Peptides as Targeting Probes against Tumor Vasculature for Diagnosis and Drug Delivery. J. Transl. Med. 2012, 10 (Suppl S1), S1. [Google Scholar] [CrossRef] [PubMed]
- Milletti, F. Cell-Penetrating Peptides: Classes, Origin, and Current Landscape. Drug Discov. Today 2012, 17, 850–860. [Google Scholar] [CrossRef]
- Calis, J.J.A.; Maybeno, M.; Greenbaum, J.A.; Weiskopf, D.; de Silva, A.D.; Sette, A.; Keşmir, C.; Peters, B. Properties of MHC Class I Presented Peptides That Enhance Immunogenicity. PLoS Comput. Biol. 2013, 9, e1003266. [Google Scholar] [CrossRef] [PubMed]
- Thundimadathil, J. Cancer Treatment Using Peptides: Current Therapies and Future Prospects. J. Amino Acids 2012, 2012, 967347. [Google Scholar] [CrossRef]
- Smolarczyk, R.; Cichoń, T.; Szala, S. Peptydy-Nowa Klasa Leków Przeciwnowotworowych* Peptides: A New Class of Anticancer Drugs. Postepy Hig. Med. Dosw. 2009, 63, 360–368. [Google Scholar]
- Li, Z.J.; Cho, C.H. Development of Peptides as Potential Drugs for Cancer Therapy. Curr. Pharm. Des. 2010, 16, 1180–1189. [Google Scholar] [CrossRef]
- Thayer, A.M. Making peptides at large scale. Chem. Eng. News 2011, 89, 81–85. [Google Scholar] [CrossRef]
- Lau, J.L.; Dunn, M.K. Therapeutic Peptides: Historical Perspectives, Current Development Trends, and Future Directions. Bioorg. Med. Chem. 2018, 26, 2700–2707. [Google Scholar] [CrossRef]
- Pirtskhalava, M.; Vishnepolsky, B.; Grigolava, M.; Managadze, G. Physicochemical Features and Peculiarities of Interaction of Amp with the Membrane. Pharmaceuticals 2021, 14, 471. [Google Scholar] [CrossRef] [PubMed]
- Tan, T.; Wu, D.; Li, W.; Zheng, X.; Li, W.; Shan, A. High Specific Selectivity and Membrane-Active Mechanism of Synthetic Cationic Hybrid Antimicrobial Peptides Based on the Peptide FV7. Int. J. Mol. Sci. 2017, 18, 339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsuzaki, K. Control of Cell Selectivity of Antimicrobial Peptides. Biochim. Et Biophys. Acta Biomembr. 2009, 1788, 1687–1692. [Google Scholar] [CrossRef] [PubMed]
- Matsuzaki, K.; Sugishita, K.-I.; Fujii, N.; Miyajima, K. Molecular Basis for Membrane Selectivity of an Antimicrobial Peptide, Magainin 2. Biochemistry 1995, 34, 3423–3429. [Google Scholar] [CrossRef]
- Yeaman, M.R.; Yount, N.Y. Mechanisms of Antimicrobial Peptide Action and Resistance. Pharmacol. Rev. 2003, 55, 27–55. [Google Scholar] [CrossRef]
- Jiménez-Rodríguez, A.; Guardado-Félix, D.; Antunes-Ricardo, M. Challenges and Strategies for Topical and Transdermal Delivery of Bioactive Peptides. Crit. Rev. Ther. Drug Carr. Syst. 2022, 39, 1–31. [Google Scholar] [CrossRef]
- Kumar, P.; Kizhakkedathu, J.N.; Straus, S.K. Antimicrobial Peptides: Diversity, Mechanism of Action and Strategies to Improve the Activity and Biocompatibility in Vivo. Biomolecules 2018, 8, 4. [Google Scholar] [CrossRef]
- Klepach, A.; Tran, H.; Ahmad Mohammed, F.; ElSayed, M.E.H. Characterization and Impact of Peptide Physicochemical Properties on Oral and Subcutaneous Delivery. Adv. Drug Deliv. Rev. 2022, 186, 114322. [Google Scholar] [CrossRef]
- Jobin, M.L.; Blanchet, M.; Henry, S.; Chaignepain, S.; Manigand, C.; Castano, S.; Lecomte, S.; Burlina, F.; Sagan, S.; Alves, I.D. The Role of Tryptophans on the Cellular Uptake and Membrane Interaction of Arginine-Rich Cell Penetrating Peptides. Biochim. Et Biophys. Acta Biomembr. 2015, 1848, 593–602. [Google Scholar] [CrossRef]
- Boman, H.G. Antibacterial Peptides: Basic Facts and Emerging Concepts. J. Intern. Med. 2003, 254, 197–215. [Google Scholar] [CrossRef]
- Nasiri, F.; Atanaki, F.F.; Behrouzi, S.; Kavousi, K.; Bagheri, M. CpACpP: In Silico Cell-Penetrating Anticancer Peptide Prediction Using a Novel Bioinformatics Framework. ACS Omega 2021, 6, 19846–19859. [Google Scholar] [CrossRef] [PubMed]
- Behzadi, M.; Arasteh, S.; Bagheri, M. Palmitoylation of Membrane-Penetrating Magainin Derivatives Reinforces Necroptosis in A549 Cells Dependent on Peptide Conformational Propensities. ACS Appl. Mater. Interfaces 2020, 12, 56815–56829. [Google Scholar] [CrossRef]
- Doneva, N.; Doytchinova, I.; Dimitrov, I. Predicting Immunogenicity Risk in Biopharmaceuticals. Symmetry 2021, 13, 388. [Google Scholar] [CrossRef]
- Dhanda, S.K.; Mahajan, S.; Paul, S.; Yan, Z.; Kim, H.; Jespersen, M.C.; Jurtz, V.; Andreatta, M.; Greenbaum, J.A.; Marcatili, P.; et al. IEDB-AR: Immune Epitope Database—Analysis Resource in 2019. Nucleic Acids Res. 2019, 47, W502–W506. [Google Scholar] [CrossRef] [PubMed]
- Kuriakose, A.; Chirmule, N.; Nair, P. Immunogenicity of Biotherapeutics: Causes and Association with Posttranslational Modifications. J. Immunol. Res. 2016, 2016, 1298473. [Google Scholar] [CrossRef] [PubMed]
- Shankar, G.; Arkin, S.; Cocea, L.; Devanarayan, V.; Kirshner, S.; Kromminga, A.; Quarmby, V.; Richards, S.; Schneider, C.K.; Subramanyam, M.; et al. Assessment and Reporting of the Clinical Immunogenicity of Therapeutic Proteins and Peptides—Harmonized Terminology and Tactical Recommendations. AAPS J. 2014, 16, 658–673. [Google Scholar] [CrossRef]
- Dimitrov, I.; Bangov, I.; Flower, D.R.; Doytchinova, I. AllerTOP v.2—A Server for in Silico Prediction of Allergens. J. Mol. Modeling 2014, 20, 2278. [Google Scholar] [CrossRef]
- Gupta, S.; Kapoor, P.; Chaudhary, K.; Gautam, A.; Kumar, R.; Raghava, G.P.S. In Silico Approach for Predicting Toxicity of Peptides and Proteins. PLoS ONE 2013, 8, e73957. [Google Scholar] [CrossRef]
- Gupta, S.; Kapoor, P.; Chaudhary, K.; Gautam, A.; Kumar, R.; Raghava, G.P.S. Peptide Toxicity Prediction. Methods Mol. Biol. 2015, 1268, 143–157. [Google Scholar] [CrossRef]
- Chowdhury, A.S.; Reehl, S.M.; Kehn-Hall, K.; Bishop, B.; Webb-Robertson, B.J.M. Better Understanding and Prediction of Antiviral Peptides through Primary and Secondary Structure Feature Importance. Sci. Rep. 2020, 10, 19260. [Google Scholar] [CrossRef]
- Gleenberg, I.O.; Herschhorn, A.; Hizi, A. Inhibition of the Activities of Reverse Transcriptase and Integrase of Human Immunodeficiency Virus Type-1 by Peptides Derived from the Homologous Viral Protein R (Vpr). J. Mol. Biol. 2007, 369, 1230–1243. [Google Scholar] [CrossRef]
- Qureshi, A.; Kaur, G.; Kumar, M. AVCpred: An Integrated Web Server for Prediction and Design of Antiviral Compounds. Chem. Biol. Drug Des. 2017, 89, 74–83. [Google Scholar] [CrossRef]
- Littler, E.; Oberg, B. Achievements and challenges in antiviral drug discovery. Antiviral Chem. Chemother. 2005, 16, 155–168. [Google Scholar] [CrossRef]
- Chiangjong, W.; Chutipongtanate, S.; Hongeng, S. Anticancer Peptide: Physicochemical Property, Functional Aspect and Trend in Clinical Application (Review). Int. J. Oncol. 2020, 57, 678–696. [Google Scholar] [CrossRef]
- Sok, M.; Šentjurc, M.; Schara, M. Membrane fluidity Characteristics of Human Lung Cancer. Cancer Lett. 1999, 139, 215–220. [Google Scholar] [CrossRef]
- Gaspar, D.; Freire, J.M.; Pacheco, T.R.; Barata, J.T.; Castanho, M.A.R.B. Apoptotic Human Neutrophil Peptide-1 Anti-Tumor Activity Revealed by Cellular Biomechanics. Biochim. Et Biophys. Acta Mol. Cell Res. 2015, 1853, 308–316. [Google Scholar] [CrossRef]
- Deslouches, B.; Di, Y.P. Antimicrobial Peptides with Selective Antitumor Mechanisms: Prospect for Anticancer Applications. Oncotarget 2017, 8, 46635. [Google Scholar] [CrossRef]
- Dobrzy´nskadobrzy´nska, I.; Szachowicz-Petelska, B.; Law Sulkowski, S.; Figaszewski, Z. Changes in Electric Charge and Phospholipids Composition in Human Colorectal Cancer Cells; Springer: Berlin/Heidelberg, Germany, 2005; Volume 276. [Google Scholar]
- Figueiredo, C.R.; Matsuo, A.L.; Massaoka, M.H.; Polonelli, L.; Travassos, L.R. Anti-Tumor Activities of Peptides Corresponding to Conserved Complementary Determining Regions from Different Immunoglobulins. Peptides 2014, 59, 14–19. [Google Scholar] [CrossRef]
- Cooper, B.M.; Iegre, J.; O’Donovan, D.H.; Ölwegård Halvarsson, M.; Spring, D.R. Peptides as a Platform for Targeted Therapeutics for Cancer: Peptide-Drug Conjugates (PDCs). Chem. Soc. Rev. 2021, 50, 1480–1494. [Google Scholar] [CrossRef] [PubMed]
- Xiong, G.; Wu, Z.; Yi, J.; Fu, L.; Yang, Z.; Hsieh, C.; Yin, M.; Zeng, X.; Wu, C.; Lu, A.; et al. ADMETlab 2.0: An Integrated Online Platform for Accurate and Comprehensive Predictions of ADMET Properties. Nucleic Acids Res. 2021, 49, W5–W14. [Google Scholar] [CrossRef] [PubMed]
- Bockus, A.T.; Mcewen, C.M.; Lokey, R.S. Form and Function in Cyclic Peptide Natural Products: A Pharmacoki-Netic Perspective. Curr. Top. Med. Chem. 2013, 13, 821–836. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Eufracio, B.I.; Palomino-Hernández, O.; Houghten, R.A.; Medina-Franco, J.L. Exploring the Chemical Space of Peptides for Drug Discovery: A Focus on Linear and Cyclic Penta-Peptides. Mol. Divers. 2018, 22, 259–267. [Google Scholar] [CrossRef] [PubMed]
- Santos, G.B.; Ganesan, A.; Emery, F.S. Oral Administration of Peptide-Based Drugs: Beyond Lipinski’s Rule. ChemMedChem 2016, 11, 2245–2251. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Zoete, V. A Boiled-Egg to Predict Gastrointestinal Absorption and Brain Penetration of Small Molecules. ChemMedChem 2016, 11, 1117–1121. [Google Scholar] [CrossRef] [PubMed]
- Lipinski, C.A.; Dominy, B.W.; Feeney, P.J. Drug Delivery Reviews Experimental and Computational Approaches to Estimate Solubility and Permeability in Drug Discovery and Development Settings. Adv. Drug Deliv. Rev. 1997, 23, 3–25. [Google Scholar] [CrossRef]
- Veber, D.F.; Johnson, S.R.; Cheng, H.Y.; Smith, B.R.; Ward, K.W.; Kopple, K.D. Molecular Properties That Influence the Oral Bioavailability of Drug Candidates. J. Med. Chem. 2002, 45, 2615–2623. [Google Scholar] [CrossRef]
- Doak, B.C.; Over, B.; Giordanetto, F.; Kihlberg, J. Oral Druggable Space beyond the Rule of 5: Insights from Drugs and Clinical Candidates. Chem. Biol. 2014, 21, 1115–1142. [Google Scholar] [CrossRef]
- Lovering, F.; Bikker, J.; Humblet, C. Escape from Flatland: Increasing Saturation as an Approach to Improving Clinical Success. J. Med. Chem. 2009, 52, 6752–6756. [Google Scholar] [CrossRef]
- Ritchie, T.J.; MacDonald, S.J.F.; Young, R.J.; Pickett, S.D. The Impact of Aromatic Ring Count on Compound Developability: Further Insights by Examining Carbo- and Hetero-Aromatic and -Aliphatic Ring Types. Drug Discov. Today 2011, 16, 164–171. [Google Scholar] [CrossRef]
- Gautier, R.; Douguet, D.; Antonny, B.; Drin, G. HELIQUEST: A Web Server to Screen Sequences with Specific α-Helical Properties. Bioinformatics 2008, 24, 2101–2102. [Google Scholar] [CrossRef]
- Santos, J.; Iglesias, V.; Pintado, C.; Santos-Suárez, J.; Ventura, S. Disphred: A Server to Predict Ph-Dependent Order–Disorder Transitions in Intrinsically Disordered Proteins. Int. J. Mol. Sci. 2020, 21, 5814. [Google Scholar] [CrossRef] [PubMed]
- Khandogin, J.; Chen, J.; Brooks III, C.L. Exploring Atomistic Details of PH-Dependent Peptide Folding. Proc. Natl. Acad. Sci. USA 2006, 103, 18546–18550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Burdukiewicz, M.; Sidorczuk, K.; Rafacz, D.; Pietluch, F.; Chilimoniuk, J.; Rödiger, S.; Gagat, P. Proteomic Screening for Prediction and Design of Antimicrobial Peptides with Ampgram. Int. J. Mol. Sci. 2020, 21, 4310. [Google Scholar] [CrossRef] [PubMed]
- Thomas, S.; Karnik, S.; Barai, R.S.; Jayaraman, V.K.; Idicula-Thomas, S. CAMP: A Useful Resource for Research on Antimicrobial Peptides. Nucleic Acids Res. 2009, 38, D774–D780. [Google Scholar] [CrossRef]
- Torrent, M.; Pulido, D.; de La Torre, B.G.; García-Mayoral, M.F.; Nogués, M.V.; Bruix, M.; Andreu, D.; Boix, E. Refining the Eosinophil Cationic Protein Antibacterial Pharmacophore by Rational Structure Minimization. J. Med. Chem. 2011, 54, 5237–5244. [Google Scholar] [CrossRef]
- Tang, H.; Su, Z.D.; Wei, H.H.; Chen, W.; Lin, H. Prediction of Cell-Penetrating Peptides with Feature Selection Techniques. Biochem. Biophys. Res. Commun. 2016, 477, 150–154. [Google Scholar] [CrossRef]
- Schaduangrat, N.; Nantasenamat, C.; Prachayasittikul, V.; Shoombuatong, W. ACPred: A Computational Tool for the Prediction and Analysis of Anticancer Peptides. Molecules 2019, 24, 1973. [Google Scholar] [CrossRef]
- Schaduangrat, N.; Nantasenamat, C.; Prachayasittikul, V.; Shoombuatong, W. Meta-Iavp: A Sequence-Based Meta-Predictor for Improving the Prediction of Antiviral Peptides Using Effective Feature Representation. Int. J. Mol. Sci. 2019, 20, 5743. [Google Scholar] [CrossRef]
- Baell, J.B.; Holloway, G.A. New Substructure Filters for Removal of Pan Assay Interference Compounds (PAINS) from Screening Libraries and for Their Exclusion in Bioassays. J. Med. Chem. 2010, 53, 2719–2740. [Google Scholar] [CrossRef]
- Lamiable, A.; Thevenet, P.; Rey, J.; Vavrusa, M.; Derreumaux, P.; Tuffery, P. PEP-FOLD3: Faster Denovo Structure Prediction for Linear Peptides in Solution and in Complex. Nucleic Acids Res. 2016, 44, W449–W454. [Google Scholar] [CrossRef] [PubMed]
- Roman Laskowski, B.A.; Macarthur, M.W.; Thornton, J.M. Computer Programs PROCHECK: A Program to Check the Stereochemicai Quality of Protein Structures. J. Appl. Crystallogr. 1983, 13, 283–291. [Google Scholar]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef] [PubMed]
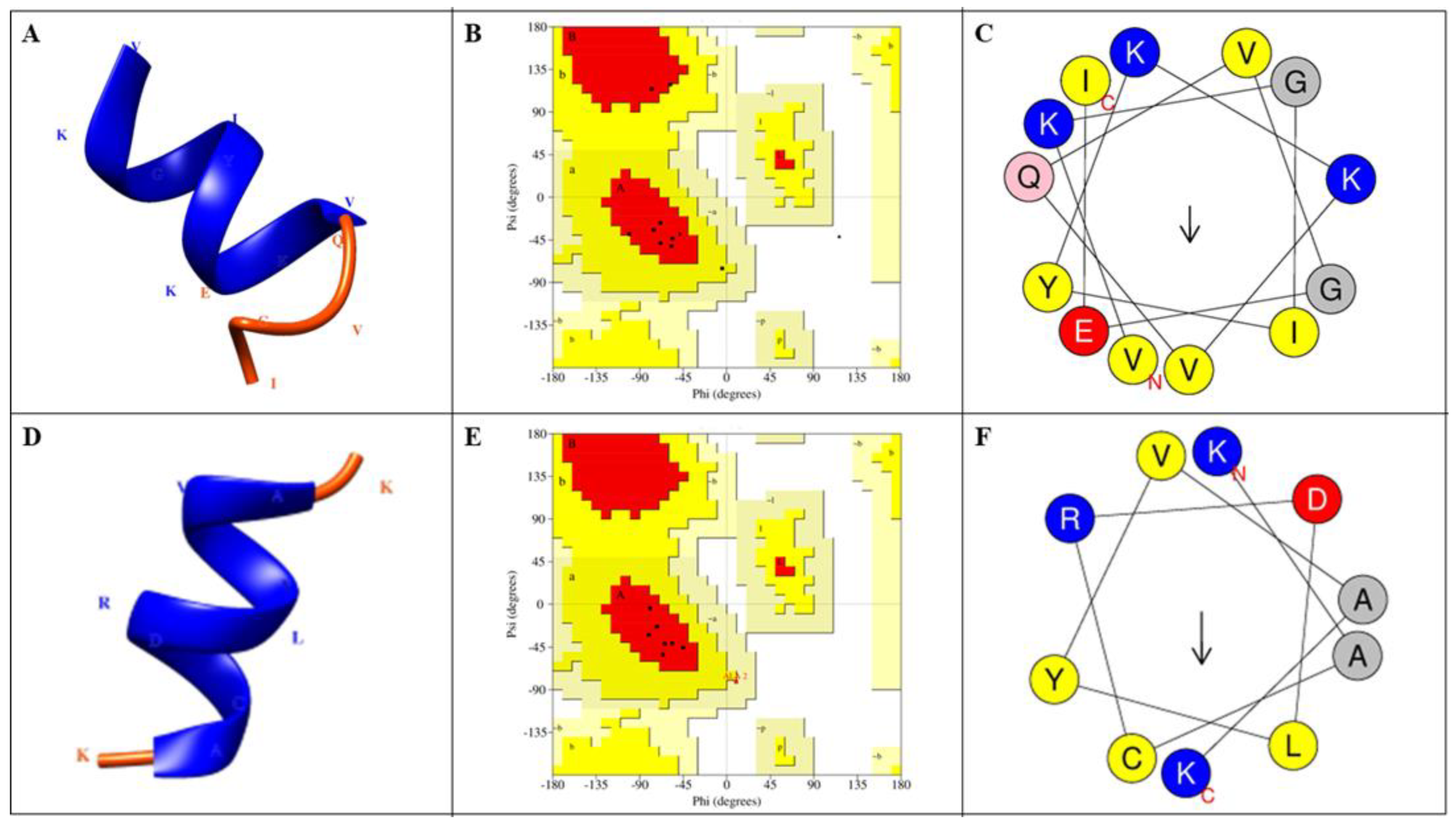
| Peptides | AMP Prediction | CPP Prediction | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Name | Sequence | AA (n) | CAMP | AMPA | AmpGram | C2Pred | CELL PPD | ||
| Prediction | Probability | Prediction | SVM Score | ||||||
| NATT1_01 | TCKTNRIYVGKGAY | 14 | 0 | AMP | 0.750 | Non-CPP | 0.836 | Non-CPP | −0.380 |
| NATT1_02 | MRKSTVNNKQCKEVTK | 16 | 0 | AMP | 0.492 | CPP | 0.530 | Non-CPP | −0.250 |
| NATT1_03 | VNKDVIEQTM | 10 | 0.501 | - | 0.047 | Non-CPP | 0.942 | Non-CPP | −0.780 |
| NATT1_04 | DVIEQTMKDV | 10 | 0.549 | - | 0.005 | Non-CPP | 0.912 | Non-CPP | −0.640 |
| NATT1_05 | TESQSYMVTV | 10 | 0.547 | - | 0.000 | CPP | 0.756 | Non-CPP | −0.820 |
| NATT1.2_01 | RTYRGGKKTQTTTKGVYRTTQV | 22 | 0 | AMP | 0.531 | Non-CPP | 0.524 | Non-CPP | −0.350 |
| NATT1.2_02 | STNDETNLHW | 10 | 0.524 | - | 0.000 | Non-CPP | 0.732 | Non-CPP | −0.780 |
| NATT1.2_03 | CKTNRIYVGK | 10 | 0.603 | - | 0.921 | Non-CPP | 0.657 | Non-CPP | −0.100 |
| NATT1.2_04 | KTNRIYVGKG | 10 | 0.544 | - | 0.561 | Non-CPP | 0.784 | Non-CPP | −0.120 |
| NATT1.2_05 | LIRTYRGGKK | 10 | 0.699 | - | 0.544 | CPP | 0.882 | CPP | 0.300 |
| NATT1.2_06 | IRTYRGGKKT | 10 | 0.613 | - | 0.541 | CPP | 0.864 | CPP | 0.010 |
| NATT1.2_07 | RTYRGGKKTQ | 10 | 0.526 | - | 0.413 | CPP | 0.537 | CPP | 0.000 |
| NATT2_01 | TCKTNKIYVGKGAY | 14 | 0 | AMP | 0.996 | Non-CPP | 0.835 | Non-CPP | −0.460 |
| NATT2_02 | RTYRGGKKTQTTTKGVYRTIQV | 22 | 0 | AMP | 0.530 | CPP | 0.655 | Non-CPP | −0.340 |
| NATT2_03 | TLRPKLKSKKPAK | 13 | 0 | AMP | 1000 | CPP | 0.947 | CPP | 0.630 |
| NATT2_04 | TETQSYMVTV | 10 | 0.684 | - | 0.000 | Non-CPP | 0.238 | Non-CPP | −0.810 |
| NATT2_05 | ETQSYMVTVS | 10 | 0.542 | - | 0.000 | Non-CPP | 0.238 | Non-CPP | −0.710 |
| NATT2_06 | TTLRPKLKSK | 10 | 0.505 | - | 0.945 | CPP | 0.978 | CPP | 0.300 |
| NATT2_07 | TLRPKLKSKK | 10 | 0.602 | - | 0.991 | CPP | 0.952 | CPP | 0.540 |
| NATT2_08 | LRPKLKSKKP | 10 | 0.565 | - | 0.987 | CPP | 0.952 | CPP | 0.460 |
| NATT2_09 | RPKLKSKKPA | 10 | 0.533 | - | 0.975 | CPP | 0.929 | CPP | 0.290 |
| NATT2_10 | PKLKSKKPAK | 10 | 0.638 | - | 1000 | CPP | 0.929 | CPP | 0.510 |
| NATT2_11 | KLKSKKPAKP | 10 | 0.627 | - | 1000 | CPP | 0.929 | CPP | 0.510 |
| NATT2_12 | LKSKKPAKPA | 10 | 0.573 | - | 1000 | CPP | 0.908 | CPP | 0.100 |
| NATT2_13 | KSKKPAKPAG | 10 | 0.529 | - | 1000 | CPP | 0.682 | CPP | 0.200 |
| NATT2_14 | SKKPAKPAGK | 10 | 0.556 | - | 1000 | CPP | 0.682 | CPP | 0.150 |
| NATT2_15 | LRPKLKSKKPAKPAGK | 16 | 0 | - | 1000 | CPP | 0.878 | CPP | 0.180 |
| NATT3_01 | VYVGKNKYGLGKVHTKHE | 18 | 0 | AMP | 0.996 | Non-CPP | 0.186 | Non-CPP | −0.520 |
| NATT3_02 | MTRTYRNGQKRTTSITGTYRAIQ | 23 | 0 | AMP | 0.015 | CPP | 0.838 | Non-CPP | −0.220 |
| NATT3_03 | YVCSCGCSSG | 10 | 0.574 | - | 0.577 | Non-CPP | 0.184 | Non-CPP | −0.680 |
| NATT3_04 | CSCGCSSGFY | 10 | 0.548 | - | 0.406 | Non-CPP | 0.204 | Non-CPP | −0.650 |
| NATT3_05 | HYAYGETEKT | 10 | 0.501 | - | 0.001 | CPP | 0.508 | Non-CPP | −0.510 |
| NATT3_06 | KYGLGKVHTK | 10 | 0.546 | - | 0.993 | Non-CPP | 0.294 | Non-CPP | −0.120 |
| NATT3_07 | PPNHYCPVTM | 10 | 0.582 | - | 0.949 | Non-CPP | 0.198 | Non-CPP | −0.550 |
| NATT3_08 | PNHYCPVTMV | 10 | 0.538 | - | 0.885 | Non-CPP | 0.246 | Non-CPP | −0.410 |
| NATT3_09 | TRTYRNGQKR | 10 | 0.531 | - | 0.168 | CPP | 0.843 | CPP | 0.190 |
| NATT3_10 | RTYRNGQKRT | 10 | 0.528 | - | 0.166 | Non-CPP | 0.486 | CPP | 0.100 |
| NATT4_01 | LYVAKNKYGLGKL | 13 | 0.772 | - | 0.989 | Non-CPP | 0.089 | Non-CPP | −0.270 |
| NATT4_02 | KACRDLYVAK | 10 | 0 | - | 0.443 | Non-CPP | 0.144 | CPP | 0.030 |
| NATT4_03 | KITNVRYNMK | 10 | 0 | - | 0.045 | Non-CPP | 0.406 | CPP | 0.070 |
| NATT4_04 | IPFTGRLTRK | 10 | 0 | - | 0.418 | Non-CPP | 0.494 | CPP | 0.140 |
| NATT4_05 | PFTGRLTRKY | 10 | 0 | - | 0.442 | CPP | 0.751 | CPP | 0.750 |
| NATT4_06 | FTGRLTRKYS | 10 | 0 | - | 0.358 | CPP | 0.751 | CPP | 0.010 |
| NATT4_07 | TGRLTRKYSN | 10 | 0 | - | 0.361 | CPP | 0.524 | CPP | 0.030 |
| NATT4_08 | GRLTRKYSNG | 10 | 0.519 | - | 0.406 | CPP | 0.746 | CPP | 0.040 |
| NATT4_09 | RLTRKYSNGK | 10 | 0 | - | 0.412 | CPP | 0.830 | CPP | 0.160 |
| NATT4_10 | KNKYGLGKLHQS | 12 | 0 | AMP | 0.989 | CPP | 0.604 | Non-CPP | −0.160 |
| NATT4_11 | KANIPFTGRLTRK | 13 | 0.516 | - | 0.449 | CPP | 0.702 | CPP | 0.050 |
| NATT4_12 | GRLTRKYSNGKVT | 13 | 0.519 | - | 0.432 | CPP | 0.804 | Non-CPP | −0.110 |
| NATT4_13 | KVTSSSVKGIYKK | 13 | 0.601 | - | 0.908 | Non-CPP | 0.231 | Non-CPP | −0.050 |
| NATT4_14 | VTSSSVKGIYKKV | 13 | 0.508 | - | 0.971 | Non-CPP | 0.231 | Non-CPP | −0.430 |
| NATT4_15 | VKGIYKKVQVGEI | 13 | 0.746 | - | 0.919 | Non-CPP | 0.186 | Non-CPP | −0.620 |
| NATTP_01 | LGQALIPRCRKMP | 13 | 0.609 | - | 0.986 | Non-CPP | 0.468 | CPP | 0.150 |
| NATTP_02 | RCRKMPGVKM | 10 | 0 | - | 0.634 | CPP | 0.767 | CPP | 0.010 |
| NATTP_03 | QALIPRCRKMPGV | 13 | 0.526 | - | 0.990 | CPP | 0.547 | Non-CPP | −0.090 |
| NATTP_04 | ALIPRCRKMPGVK | 13 | 0.771 | - | 0.990 | CPP | 0.547 | CPP | 0.280 |
| NATTP_05 | LIPRCRKMPGVKM | 13 | 0.645 | AMP | 0.893 | CPP | 0.563 | CPP | 0.050 |
| Inference/Reference range | >0.5: AMP | - | >0.5: AMP | >0.5: CPP | SVM score >0: CPP | ||||
| <0.5: non-AMP | <0.5: non-AMP | <0.5: non-CPP | SVM score <0: non-CPP | ||||||
| Peptides | MW (g/mol) | Polar Residues + GLY (n/%) | Uncharged Residues + GLY | Charged Residues | Non-Polar Residues (n/%) | |
|---|---|---|---|---|---|---|
| Name | Sequence | |||||
| NATT1_01 | TCKTNRIYVGKGAY | 1573.83 | 8/57.14 | THR 2, ASN 1, GLY 2 | LYS 2, ARG 1, | 6/42.86 |
| NATT1_02 | MRKSTVNNKQCKEVTK | 1894.24 | 12/75.00 | GLN 1, SER 1, THR 2, ASN 2, GLY 0 | LYS 4, ARG 1, GLU 1 | 4/25.00 |
| NATT1_03 | VNKDVIEQTM | 1176.35 | 6/60.00 | GLN 1, THR 1, ASN 1, GLY 0 | LYS 1, GLU 1, ASP 1, | 4/40.00 |
| NATT1_04 | DVIEQTMKDV | 1177.34 | 6/60.00 | GLN 1, THR 1, GLY 0 | LYS 1, GLU 1, ASP 2, | 4/40.00 |
| NATT1_05 | TESQSYMVTV | 1144.26 | 6/60.00 | GLN 1, SER 2, THR 2, GLY 0 | GLU 1, | 4/40.00 |
| NATT1.2_01 | RTYRGGKKTQTTTKGVYRTTQV | 2530.87 | 18/81.82 | GLN 2, THR 7, GLY 3 | LYS 3, ARG 3, | 4/18.18 |
| NATT1.2_02 | STNDETNLHW | 1216.23 | 8/80.00 | HIS 1, SER 1, THR 2, ASN 2, GLY 0 | GLU 1, ASP 1, | 2/20.00 |
| NATT1.2_03 | CKTNRIYVGK | 1181.42 | 6/60.00 | THR 1, ASN 1, GLY 1 | LYS 2, ARG 1, | 4/40.00 |
| NATT1.2_04 | KTNRIYVGKG | 1135.33 | 7/70.00 | THR 1, ASN 1, GLY 2 | LYS 2, ARG 1, | 3/30.00 |
| NATT1.2_05 | LIRTYRGGKK | 1191.44 | 7/70.00 | THR 1, GLY 2 | LYS 2, ARG 2, | 3/30.00 |
| NATT1.2_06 | IRTYRGGKKT | 1179.39 | 8/80.00 | THR 2, GLY 2 | LYS 2, ARG 2, | 2/20.00 |
| NATT1.2_07 | RTYRGGKKTQ | 1194.36 | 9/90.00 | GLN 1, THR 2, GLY 2 | LYS 2, ARG 2, | 1/10.00 |
| NATT2_01 | TCKTNKIYVGKGAY | 1545.82 | 8/57.14 | THR 2, ASN 1, GLY 2 | LYS 3, | 6/42.86 |
| NATT2_02 | RTYRGGKKTQTTTKGVYRTIQV | 2542.92 | 17/77.27 | GLN 2, THR 6, GLY 3 | LYS 3, ARG 3, | 5/22.73 |
| NATT2_03 | TLRPKLKSKKPAK | 1494.89 | 8/61.54 | SER 1, THR 1, GLY 0 | LYS 5, ARG 1, | 5/38.46 |
| NATT2_04 | TETQSYMVTV | 1158.29 | 6/60.00 | GLN 1, SER 1, THR 3, GLY 0 | GLU 1, | 4/40.00 |
| NATT2_05 | ETQSYMVTVS | 1144.26 | 6/60.00 | GLN 1, SER 2, THR 2, GLY 0 | GLU 1, | 4/40.00 |
| NATT2_06 | TTLRPKLKSK | 1171.45 | 7/70.00 | SER 1, THR 2, GLY 0 | LYS 3, ARG 1, | 3/30.00 |
| NATT2_07 | TLRPKLKSKK | 1198.52 | 7/70.00 | SER 1, THR 1, GLY 0 | LYS 4, ARG 1, | 3/30.00 |
| NATT2_08 | LRPKLKSKKP | 1194.53 | 6/60.00 | SER 1, GLY 0 | LYS 4, ARG 1, | 4/40.00 |
| NATT2_09 | RPKLKSKKPA | 1152.45 | 6/60.00 | SER 1, GLY 0 | LYS 4, ARG 1, | 4/40.00 |
| NATT2_10 | PKLKSKKPAK | 1124.44 | 6/60.00 | SER 1, GLY 0 | LYS 5, | 4/40.00 |
| NATT2_11 | KLKSKKPAKP | 1124.44 | 6/60.00 | SER 1, GLY 0 | LYS 5, | 4/40.00 |
| NATT2_12 | LKSKKPAKPA | 1067.34 | 5/50.00 | SER 1, GLY 0 | LYS 4, | 5/50.00 |
| NATT2_13 | KSKKPAKPAG | 1011.23 | 6/60.00 | SER 1, GLY 1 | LYS 4, | 4/40.00 |
| NATT2_14 | SKKPAKPAGK | 1011.23 | 6/60.00 | SER 1, GLY 1 | LYS 4, | 4/40.00 |
| NATT2_15 | LRPKLKSKKPAKPAGK | 1747.20 | 9/56.25 | SER 1, GLY 1 | LYS 6, ARG 1, | 7/43.75 |
| NATT3_01 | VYVGKNKYGLGKVHTKHE | 2057.38 | 12/66.67 | HIS 2, THR 1, ASN 1, GLY 3 | LYS 4, GLU 1, | 6/33.33 |
| NATT3_02 | MTRTYRNGQKRTTSITGTYRAIQ | 2704.06 | 17/73.91 | GLN 2, SER 1, THR 6, ASN 1, GLY 2 | LYS 1, ARG 4, | 6/26.09 |
| NATT3_03 | YVCSCGCSSG | 965.08 | 5/50.00 | SER 3, GLY 2 | - | 5/50.00 |
| NATT3_04 | CSCGCSSGFY | 1013.12 | 5/50.00 | SER 3, GLY 2 | - | 5/50.00 |
| NATT3_05 | HYAYGETEKT | 1198.25 | 7/70.00 | HIS 1, THR 2, GLY 1 | LYS 1, GLU 2, | 3/30.00 |
| NATT3_06 | KYGLGKVHTK | 1130.36 | 7/70.00 | HIS 1, THR 1, GLY 2 | LYS 3, | 3/30.00 |
| NATT3_07 | PPNHYCPVTM | 1158.36 | 3/30.00 | HIS 1, THR 1, ASN 1, GLY 0 | - | 7/70.00 |
| NATT3_08 | PNHYCPVTMV | 1160.37 | 3/30.00 | HIS 1, THR 1, ASN 1, GLY 0 | - | 7/70.00 |
| NATT3_09 | TRTYRNGQKR | 1279.42 | 9/90.00 | GLN 1, THR 2, ASN 1, GLY 1 | LYS 1, ARG 3, | 1/10.00 |
| NATT3_10 | RTYRNGQKRT | 1279.42 | 9/90.00 | GLN 1, THR 2, ASN 1, GLY 1 | LYS 1, ARG 3, | 1/10.00 |
| NATT4_01 | LYVAKNKYGLGKL | 1466.79 | 6/46.15 | ASN 1, GLY 2 | LYS 3, | 7/53.85 |
| NATT4_02 | KACRDLYVAK | 1166.4 | 4/40.00 | GLY 0 | LYS 2, ARG 1, ASP 1, | 6/60.00 |
| NATT4_03 | KITNVRYNMK | 1233.25 | 6/60.00 | THR 1, ASN 2, GLY 0 | LYS 2, ARG 1, | 4/40.00 |
| NATT4_04 | IPFTGRLTRK | 1188.44 | 6/60.00 | THR 2, GLY 1 | LYS 1, ARG 2, | 4/40.00 |
| NATT4_05 | PFTGRLTRKY | 1238.46 | 6/60.00 | THR 2, GLY 1 | LYS 1, ARG 2, | 4/40.00 |
| NATT4_06 | FTGRLTRKYS | 1228.42 | 7/70.00 | SER 1, THR 2, GLY 1 | LYS 1, ARG 2, | 3/30.00 |
| NATT4_07 | TGRLTRKYSN | 1195.34 | 8/80.00 | SER 1, THR 2, ASN 1, GLY 1 | LYS 1, ARG 2, | 2/20.00 |
| NATT4_08 | GRLTRKYSNG | 1151.29 | 8/80.00 | SER 1, THR 1, ASN 1, GLY 2 | LYS 1, ARG 2, | 2/20.00 |
| NATT4_09 | RLTRKYSNGK | 1222.41 | 8/80.00 | SER 1, THR 1, ASN 1, GLY 1 | LYS 2, ARG 2, | 2/20.00 |
| NATT4_10 | KNKYGLGKLHQS | 1372.59 | 9/75.00 | GLN 1, HIS 1, SER 1, ASN 1, GLY 2 | LYS 3, | 3/25.00 |
| NATT4_11 | KANIPFTGRLTRK | 1501.8 | 8/61.54 | THR 2, ASN 1, GLY 1 | LYS 2, ARG 2, | 5/38.46 |
| NATT4_12 | GRLTRKYSNGKVT | 1479.7 | 10/76.92 | SER 1, THR 2, ASN 1, GLY 2 | LYS 2, ARG 2, | 3/23.08 |
| NATT4_13 | KVTSSSVKGIYKK | 1424.7 | 9/69.23 | SER 3, THR 1, GLY 1 | LYS 4, | 4/30.77 |
| NATT4_14 | VTSSSVKGIYKKV | 1395.66 | 8/61.54 | SER 3, THR 1, GLY 1 | LYS 3, | 5/38.46 |
| NATT4_15 | VKGIYKKVQVGEI | 1460.78 | 7/53.85 | GLN 1, GLY 2 | LYS 3, GLU 1, | 6/46.15 |
| NATTP_01 | LGQALIPRCRKMP | 1482.87 | 5/38.46 | GLN 1, GLY 1 | LYS 1, ARG 2, | 8/61.54 |
| NATTP_02 | RCRKMPGVKM | 1205.56 | 5/50.00 | GLY 1 | LYS 2, ARG 2, | 5/50.00 |
| NATTP_03 | QALIPRCRKMPGV | 1468.84 | 5/38.46 | GLN 1, GLY 1 | LYS 1, ARG 2, | 8/61.54 |
| NATTP_04 | ALIPRCRKMPGVK | 1468.89 | 5/38.46 | GLY 1 | LYS 2, ARG 2, | 8/61.54 |
| NATTP_05 | LIPRCRKMPGVKM | 1259.0 | 5/38.46 | GLY 1 | LYS 2, ARG 2, | 8/61.54 |
| Peptides | Immunogenicity | Allergenicity | Hemolysis (%) | T1/2 Escherichia coli | T1/2 in Mammalian (in hours) | Antiviral | Anticancer | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Name | Sequence | Prediction | Probability | Prediction | Probability | |||||
| NATT1_01 | TCKTNRIYVGKGAY | 6082 | Non-allergen | 0.48 | >10 h | 7.2 | Non-AVP | 0.344 | ACP | 0.982 |
| NATT1_02 | MRKSTVNNKQCKEVTK | −6046 | Allergen | 0.49 | >10 h | 30 | Non-AVP | 0 | Non-ACP | 0.414 |
| NATT1_03 | VNKDVIEQTM | 14,129 | Non-allergen | 0.49 | >10 h | 100 | Non-AVP | 0.31 | ACP | 0.692 |
| NATT1_04 | DVIEQTMKDV | −26,539 | Allergen | 0.49 | >10 h | 1.1 | Non-AVP | 0.282 | ACP | 0.695 |
| NATT1_05 | TESQSYMVTV | −44,274 | Allergen | 0.49 | >10 h | 7.2 | Non-AVP | 0.112 | Non-ACP | 0.639 |
| NATT1.2_01 | RTYRGGKKTQTTTKGVYRTTQV | −3531 | Allergen | 0.49 | 2 min | 1 | Non-AVP | 0.004 | ACP | 0.933 |
| NATT1.2_02 | STNDETNLHW | 15,897 | Non-allergen | 0.49 | >10 h | 1.9 | Non-AVP | 0.068 | Non-ACP | 0.837 |
| NATT1.2_03 | CKTNRIYVGK | 23,725 | Non-allergen | 0.48 | >10 h | 1.2 | Non-AVP | 0.008 | ACP | 0.983 |
| NATT1.2_04 | KTNRIYVGKG | 11,744 | Non-allergen | 0.48 | 3 min | 1.3 | Non-AVP | 0 | ACP | 0.947 |
| NATT1.2_05 | LIRTYRGGKK | 3716 | Non-allergen | 0.46 | 2 min | 5.5 | AVP | 0.964 | ACP | 0.911 |
| NATT1.2_06 | IRTYRGGKKT | −18,382 | Non-allergen | 0.49 | >10 h | 20 | AVP | 0.962 | ACP | 0.906 |
| NATT1.2_07 | RTYRGGKKTQ | −24,544 | Non-allergen | 0.49 | 2 min | 1 | AVP | 0.668 | ACP | 0.686 |
| NATT2_01 | TCKTNKIYVGKGAY | −19,958 | Non-allergen | 0.49 | >10 h | 7.2 | Non-AVP | 0 | ACP | 0.994 |
| NATT2_02 | RTYRGGKKTQTTTKGVYRTIQV | −27,354 | Allergen | 0.49 | 2 min | 1 | Non-AVP | 0.068 | ACP | 0.944 |
| NATT2_03 | TLRPKLKSKKPAK | −98,576 | Non-allergen | 0.48 | >10 h | 7.2 | Non-AVP | 0.008 | ACP | 0.67 |
| NATT2_04 | TETQSYMVTV | −37,644 | Allergen | 0.49 | >10 h | 7.2 | Non-AVP | 0.068 | Non-ACP | 0.5 |
| NATT2_05 | ETQSYMVTVS | −28,293 | Allergen | 0.49 | >10 h | 1 | Non-AVP | 0.112 | Non-ACP | 0.653 |
| NATT2_06 | TTLRPKLKSK | −46,142 | Non-allergen | 0.49 | >10 h | 7.2 | AVP | 0.524 | ACP | 0.836 |
| NATT2_07 | TLRPKLKSKK | −68,378 | Non-allergen | 0.48 | >10 h | 7.2 | AVP | 0.616 | ACP | 0.703 |
| NATT2_08 | LRPKLKSKKP | −90,513 | Non-allergen | 0.49 | 2 min | 5.5 | AVP | 0.696 | Non-ACP | 0.345 |
| NATT2_09 | RPKLKSKKPA | −84,374 | Non-allergen | 0.49 | 2 min | 1 | Non-AVP | 0 | Non-ACP | 0.445 |
| NATT2_10 | PKLKSKKPAK | −7812 | Non-allergen | 0.49 | ND | >20 | Non-AVP | 0 | ACP | 0.895 |
| NATT2_11 | KLKSKKPAKP | −75,989 | Non-allergen | 0.49 | 3 min | 1.3 | Non-AVP | 0 | ACP | 0.894 |
| NATT2_12 | LKSKKPAKPA | −64,315 | Allergen | 0.49 | 2 min | 5.5 | Non-AVP | 0.318 | ACP | 0.757 |
| NATT2_13 | KSKKPAKPAG | −4492 | Allergen | 0.49 | 3 min | 1.3 | AVP | 0.654 | ACP | 0.919 |
| NATT2_14 | SKKPAKPAGK | −21,068 | Allergen | 0.49 | 10 h | 1.9 | AVP | 0.654 | ACP | 0.992 |
| NATT2_15 | LRPKLKSKKPAKPAGK | −0.91 | Non-allergen | 0.48 | 2 min | 5.5 | Non-AVP | 0 | ACP | 0.848 |
| NATT3_01 | VYVGKNKYGLGKVHTKHE | −59,206 | Allergen | 0.48 | >10 h | 100 | Non-AVP | 0.332 | ACP | 0.957 |
| NATT3_02 | MTRTYRNGQKRTTSITGTYRAIQ | 16,556 | Non-allergen | 0.49 | >10 h | 30 | Non-AVP | 0.44 | Non-ACP | 0.444 |
| NATT3_03 | YVCSCGCSSG | −4905 | Allergen | 0.49 | 2 min | 2.8 | Non-AVP | 0.398 | ACP | 0.996 |
| NATT3_04 | CSCGCSSGFY | −25,573 | Non-allergen | 0.49 | >10 h | 1.2 | Non-AVP | 0.104 | ACP | 0.998 |
| NATT3_05 | HYAYGETEKT | 13,452 | Allergen | 0.49 | >10 h | 3.5 | Non-AVP | 0.006 | ACP | 0.918 |
| NATT3_06 | KYGLGKVHTK | −8.832 | Allergen | 0.48 | 3 min | 1.3 | Non-AVP | 0.218 | ACP | 0.99 |
| NATT3_07 | PPNHYCPVTM | 2.143 | Non-allergen | 0.49 | ND | >20 | Non-AVP | 0 | Non-ACP | 0.873 |
| NATT3_08 | PNHYCPVTMV | 875.0 | Allergen | 0.49 | ND | >20 | Non-AVP | 0.126 | Non-ACP | 0.757 |
| NATT3_09 | TRTYRNGQKR | −13.888 | Non-allergen | 0.48 | >10 h | 7.2 | Non-AVP | 0.46 | Non-ACP | 0.672 |
| NATT3_10 | RTYRNGQKRT | −18.322 | Non-allergen | 0.48 | 2 min | 1 | Non-AVP | 0.46 | Non-ACP | 0.666 |
| NATT4_01 | LYVAKNKYGLGKL | −0.45197 | Allergen | 0.47 | 2 min | 5.5 | AVP | 0.998 | ACP | 0.838 |
| NATT4_02 | KACRDLYVAK | 996.0 | Non-allergen | 0.49 | 3 min | 1.3 | AVP | 0.678 | ACP | 0.725 |
| NATT4_03 | KITNVRYNMK | −1.485 | Non-allergen | 0.49 | 3 min | 1.3 | Non-AVP | 0.154 | ACP | 0.674 |
| NATT4_04 | IPFTGRLTRK | 21.302 | Allergen | 0.49 | >10 h | 20 | Non-AVP | 0.044 | ACP | 0.787 |
| NATT4_05 | PFTGRLTRKY | 0.4052 | Non-allergen | 0.50 | ND | >20 | Non-AVP | 0.004 | ACP | 0.787 |
| NATT4_06 | FTGRLTRKYS | −4.536 | Non-allergen | 0.49 | 2 min | 1.1 | AVP | 0.542 | ACP | 0.9 |
| NATT4_07 | TGRLTRKYSN | −0.20894 | Non-allergen | 0.49 | >10 h | 7.2 | Non-AVP | 0.028 | Non-ACP | 0.375 |
| NATT4_08 | GRLTRKYSNG | −0.27102 | Allergen | 0.49 | >10 h | 30 | AVP | 0.876 | ACP | 0.702 |
| NATT4_09 | RLTRKYSNGK | −0.29031 | Non-allergen | 0.49 | 2 min | 1 | Non-AVP | 0.412 | ACP | 0.747 |
| NATT4_10 | KNKYGLGKLHQS | −0.27934 | Non-allergen | 0.49 | 3 min | 1.3 | AVP | 0.998 | ACP | 0.703 |
| NATT4_11 | KANIPFTGRLTRK | 0.40878 | Non-allergen | 0.48 | 3 min | 1.3 | AVP | 0.696 | Non-ACP | 0.493 |
| NATT4_12 | GRLTRKYSNGKVT | −0.42112 | Non-allergen | 0.49 | >10 h | 30 | Non-AVP | 0.126 | Non-ACP | 0.387 |
| NATT4_13 | KVTSSSVKGIYKK | −0.61671 | Allergen | 0.51 | 3 min | 1.3 | Non-AVP | 0 | ACP | 0.999 |
| NATT4_14 | VTSSSVKGIYKKV | −0.69995 | Allergen | 0.49 | >10 h | 100 | Non-AVP | 0 | ACP | 0.999 |
| NATT4_15 | VKGIYKKVQVGEI | −0.22532 | Allergen | 0.49 | >10 h | 100 | AVP | 1 | ACP | 0.997 |
| NATTP_01 | LGQALIPRCRKMP | −0.012821 | Allergen | 0.48 | 2 min | 5.5 | Non-AVP | 0 | Non-ACP | 0.005 |
| NATTP_02 | RCRKMPGVKM | −0.44126 | Non-allergen | 0.49 | 2 min | 1 | Non-AVP | 0.07 | Non-ACP | 0.434 |
| NATTP_03 | QALIPRCRKMPGV | −0.19704 | Non-allergen | 0.46 | 10 h | 0.8 | Non-AVP | 0.268 | Non-ACP | 0.015 |
| NATTP_04 | ALIPRCRKMPGVK | −0.25838 | Non-allergen | 0.46 | >10 h | 4.4 | Non-AVP | 0.282 | Non-ACP | 0.098 |
| NATTP_05 | LIPRCRKMPGVKM | −0.40468 | Non-allergen | 0.47 | 2 min | 5.5 | AVP | 0.506 | Non-ACP | 0.241 |
| Inference/Reference Range | - | SVM method | >0.5: likely hemolytic <0.5: unlikely hemolytic | ND: not determined | <0.5: low probability >0.5: high probability | <0.5: low probability >0.5: high probability | ||||
| Peptides | Absorption | Distribution | Metabolism | Excretion | Toxicity | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Name | Sequence | HIA (%) | Caco-2 Permeability (cm/s) | VD (L/Kg) | BBB Penetration (%) | PPB(%) | CYP1A2-(I) | CYP1A2-(S) | CYP3A4-(I) | CYP3A4-(S) | CL (mL/min/Kg) | Half-Life | hERG Blockers | DILI Liver Injury | AMES | Carcinogenicity | Skin Sensitization |
| NATT1_01 | TCKTNRIYVGKGAY | 0.996 | −7.19 | 0.46 | 0.038 | 22.19 | 0 | 0 | 0.004 | 0.007 | 0.645 | 0.718 | 0.012 | 0.001 | 0.007 | 0.029 | 0.058 |
| NATT1_02 | MRKSTVNNKQCKEVTK | 0.997 | −7.377 | 0.106 | 0.025 | 17.92 | 0 | 0 | 0 | 0.002 | −0.487 | 0.799 | 0.001 | 0 | 0.043 | 0.041 | 0.06 |
| NATT1_03 | VNKDVIEQTM | 0.986 | −7.908 | 0.601 | 0.029 | 10.42 | 0 | 0 | 0.006 | 0.007 | 0.976 | 0.833 | 0 | 0.004 | 0.008 | 0.43 | 0.07 |
| NATT1_04 | DVIEQTMKDV | 0.998 | −8.074 | 0.657 | 0.018 | 9.97 | 0 | 0 | 0.006 | 0.007 | 1.171 | 0.914 | 0 | 0.005 | 0.006 | 0.406 | 0.092 |
| NATT1_05 | TESQSYMVTV | 0.979 | −8.024 | 0.428 | 0.042 | 18.47 | 0 | 0 | 0.008 | 0.009 | 0.919 | 0.88 | 0.001 | 0.02 | 0.005 | 0.074 | 0.035 |
| NATT1.2_01 | RTYRGGKKTQTTTKGVYRTTQV | 1 | −7.369 | 0.058 | 0.013 | 28.70 | 0 | 0 | 0.001 | 0.001 | −1.957 | 0.892 | 0 | 0 | 0.001 | 0.005 | 0.005 |
| NATT1.2_02 | STNDETNLHW | 0.486 | −7.794 | 0.499 | 0.068 | 16 | 0 | 0 | 0.011 | 0.006 | 0.821 | 0.904 | 0.001 | 0.012 | 0.011 | 0.095 | 0.06 |
| NATT1.2_03 | CKTNRIYVGK | 0.976 | −7.067 | 0.557 | 0.035 | 9.146 | 0 | 0 | 0.006 | 0.012 | 1.101 | 0.694 | 0.033 | 0.001 | 0.008 | 0.106 | 0.085 |
| NATT1.2_04 | KTNRIYVGKG | 0.977 | −6.821 | 0.538 | 0.058 | 11.64 | 0 | 0 | 0.007 | 0.013 | 1.012 | 0.757 | 0.033 | 0.001 | 0.008 | 0.072 | 0.087 |
| NATT1.2_05 | LIRTYRGGKK | 0.991 | −6.768 | 0.569 | 0.048 | 14.74 | 0 | 0 | 0.012 | 0.013 | 1.067 | 0.782 | 0.036 | 0.001 | 0.007 | 0.079 | 0.116 |
| NATT1.2_06 | IRTYRGGKKT | 0.997 | −7.141 | 0.526 | 0.056 | 19.12 | 0 | 0 | 0.009 | 0.012 | 0.749 | 0.807 | 0.031 | 0.001 | 0.005 | 0.04 | 0.109 |
| NATT1.2_07 | RTYRGGKKTQ | 0.987 | −6.754 | 0.501 | 0.071 | 22.28 | 0 | 0 | 0.005 | 0.008 | 0.387 | 0.752 | 0.02 | 0 | 0.01 | 0.065 | 0.098 |
| NATT2_01 | TCKTNKIYVGKGAY | 0.999 | −7.282 | 0.475 | 0.027 | 22.20 | 0 | 0 | 0.006 | 0.007 | 0.745 | 0.812 | 0.004 | 0 | 0.01 | 0.039 | 0.069 |
| NATT2_02 | RTYRGGKKTQTTTKGVYRTIQV | 1 | −7.271 | 0.085 | 0.011 | 31.57 | 0 | 0 | 0.001 | 0.001 | −1.65 | 0.887 | 0 | 0 | 0.001 | 0.008 | 0.006 |
| NATT2_03 | TLRPKLKSKKPAK | 1 | −7.301 | 0.232 | 0.014 | 24.84 | 0 | 0 | 0.001 | 0.005 | −0.097 | 0.857 | 0.006 | 0 | 0.008 | 0.009 | 0.211 |
| NATT2_04 | TETQSYMVTV | 0.99 | −7.989 | 0.41 | 0.041 | 20.64 | 0 | 0 | 0.01 | 0.01 | 0.971 | 0.89 | 0 | 0.018 | 0.003 | 0.049 | 0.021 |
| NATT2_05 | ETQSYMVTVS | 0.967 | −7.952 | 0.442 | 0.043 | 18.39 | 0 | 0 | 0.008 | 0.01 | 0.83 | 0.906 | 0.001 | 0.016 | 0.004 | 0.069 | 0.028 |
| NATT2_06 | TTLRPKLKSK | 0.991 | −7.053 | 0.416 | 0.026 | 18.04 | 0 | 0 | 0.006 | 0.015 | 0.834 | 0.757 | 0.023 | 0.002 | 0.009 | 0.033 | 0.165 |
| NATT2_07 | TLRPKLKSKK | 0.987 | −7.063 | 0.349 | 0.043 | 18.41 | 0 | 0 | 0.005 | 0.013 | 0.956 | 0.742 | 0.029 | 0.002 | 0.047 | 0.031 | 0.244 |
| NATT2_08 | LRPKLKSKKP | 0.988 | −7.162 | 0.397 | 0.078 | 17.95 | 0 | 0.001 | 0.005 | 0.014 | 1.05 | 0.705 | 0.038 | 0.003 | 0.014 | 0.038 | 0.295 |
| NATT2_09 | RPKLKSKKPA | 0.995 | −6.974 | 0.403 | 0.069 | 19.56 | 0 | 0.001 | 0.004 | 0.013 | 0.871 | 0.704 | 0.031 | 0.004 | 0.016 | 0.042 | 0.274 |
| NATT2_10 | PKLKSKKPAK | 0.999 | −7.045 | 0.352 | 0.037 | 25.27 | 0 | 0.001 | 0.005 | 0.015 | 1.085 | 0.85 | 0.006 | 0.002 | 0.765 | 0.032 | 0.281 |
| NATT2_11 | KLKSKKPAKP | 0.998 | −7.195 | 0.382 | 0.054 | 20.29 | 0 | 0.001 | 0.005 | 0.016 | 0.761 | 0.833 | 0.008 | 0.003 | 0.214 | 0.054 | 0.341 |
| NATT2_12 | LKSKKPAKPA | 0.995 | −7.388 | 0.455 | 0.038 | 17.91 | 0 | 0.001 | 0.006 | 0.017 | 0.951 | 0.857 | 0.003 | 0.003 | 0.065 | 0.05 | 0.304 |
| NATT2_13 | KSKKPAKPAG | 0.998 | −7.252 | 0.480 | 0.038 | 22.41 | 0 | 0.001 | 0.005 | 0.016 | 0.658 | 0.864 | 0.005 | 0.003 | 0.053 | 0.047 | 0.308 |
| NATT2_14 | SKKPAKPAGK | 0.998 | −7.272 | 0.504 | 0.04 | 22.67 | 0 | 0.001 | 0.005 | 0.016 | 0.797 | 0.835 | 0.003 | 0.002 | 0.031 | 0.085 | 0.333 |
| NATT2_15 | LRPKLKSKKPAKPAGK | 1 | −7.451 | 0.168 | 0.024 | 23.02 | 0 | 0 | 0.005 | 0 | 0.248 | 0.821 | 0.006 | 0 | 0.015 | 0.015 | 0.305 |
| NATT3_01 | VYVGKNKYGLGKVHTKHE | 0.999 | −7.324 | 0.516 | 0.021 | 28.76 | 0 | 0 | 0.004 | 0.003 | 0.237 | 0.954 | 0.006 | 0.001 | 0.004 | 0.003 | 0.072 |
| NATT3_02 | MTRTYRNGQKRTTSITGTYRAIQ | 1 | −7.698 | 0.009 | 0.009 | 31.78 | 0 | 0 | 0 | 0.001 | −2.266 | 0.871 | 0 | 0 | 0.001 | 0.017 | 0.004 |
| NATT3_03 | YVCSCGCSSG | 0.727 | −7.33 | 0.308 | 0.006 | 21.88 | 0 | 0 | 0.008 | 0.012 | 0.939 | 0.825 | 0.005 | 0.006 | 0.941 | 0.008 | 0.236 |
| NATT3_04 | CSCGCSSGFY | 0.8 | −8.059 | 0.391 | 0.009 | 21.41 | 0 | 0 | 0.01 | 0.011 | 1.039 | 0.821 | 0.011 | 0.007 | 0.84 | 0.03 | 0.289 |
| NATT3_05 | HYAYGETEKT | 0.993 | −7.512 | 0.589 | 0.031 | 36.88 | 0 | 0 | 0.015 | 0.008 | 1.16 | 0.956 | 0.006 | 0.007 | 0.002 | 0.038 | 0.053 |
| NATT3_06 | KYGLGKVHTK | 0.994 | −6.868 | 0.571 | 0.062 | 17.77 | 0 | 0.003 | 0.015 | 0.016 | 1.256 | 0.928 | 0.044 | 0.002 | 0.008 | 0.007 | 0.136 |
| NATT3_07 | PPNHYCPVTM | 0.995 | −6.867 | 0.475 | 0.029 | 30.59 | 0 | 0.006 | 0.009 | 0.018 | 1.412 | 0.853 | 0.006 | 0.833 | 0.006 | 0.033 | 0.077 |
| NATT3_08 | PNHYCPVTMV | 0.983 | −6.877 | 0.458 | 0.033 | 29.24 | 0 | 0.005 | 0.01 | 0.017 | 1.429 | 0.844 | 0.006 | 0.899 | 0.763 | 0.025 | 0.05 |
| NATT3_09 | TRTYRNGQKR | 0.889 | −6.91 | 0.447 | 0.078 | 19.44 | 0 | 0 | 0.003 | 0.005 | 0.195 | 0.685 | 0.011 | 0 | 0.016 | 0.125 | 0.088 |
| NATT3_10 | RTYRNGQKRT | 0.91 | −6.544 | 0.439 | 0.075 | 19.89 | 0 | 0 | 0.003 | 0.005 | 0.128 | 0.688 | 0.011 | 0 | 0.012 | 0.122 | 0.076 |
| NATT4_01 | LYVAKNKYGLGKL | 0.989 | −7.356 | 0.623 | 0.024 | 18.21 | 0 | 0 | 0.056 | 0.001 | 0.578 | 0.864 | 0.022 | 0.001 | 0.007 | 0.08 | 0.132 |
| NATT4_02 | KACRDLYVAK | 0.986 | −7.455 | 0.584 | 0.039 | 9.261 | 0 | 0 | 0.003 | 0 | 1.107 | 0.79 | 0.031 | 0.008 | 0.064 | 0.069 | 0.162 |
| NATT4_03 | KITNVRYNMK | 0.95 | −6.477 | 0.549 | 0.04 | 11.28 | 0 | 0 | 0.002 | 0 | 0.986 | 0.643 | 0.017 | 0.002 | 0.007 | 0.261 | 0.069 |
| NATT4_04 | IPFTGRLTRK | 0.998 | −7.063 | 0.47 | 0.028 | 16.47 | 0 | 0 | 0.006 | 0 | 1.218 | 0.747 | 0.03 | 0.004 | 0.004 | 0.4 | 0.088 |
| NATT4_05 | PFTGRLTRKY | 0.998 | −6.943 | 0.488 | 0.024 | 26.01 | 0 | 0 | 0.003 | 0 | 1.152 | 0.761 | 0.041 | 0.003 | 0.005 | 0.027 | 0.055 |
| NATT4_06 | FTGRLTRKYS | 0.984 | −7.18 | 0.426 | 0.042 | 16.19 | 0 | 0 | 0.004 | 0 | 0.879 | 0.782 | 0.029 | 0.002 | 0.006 | 0.028 | 0.058 |
| NATT4_07 | TGRLTRKYSN | 0.923 | −7.407 | 0.452 | 0.069 | 16.24 | 0 | 0 | 0 | 0 | 0.624 | 0.698 | 0.017 | 0.001 | 0.011 | 0.054 | 0.097 |
| NATT4_08 | GRLTRKYSNG | 0.877 | −7.430 | 0.5 | 0.056 | 18.76 | 0 | 0 | 0 | 0 | 0.6 | 0.766 | 0.022 | 0.001 | 0.018 | 0.048 | 0.109 |
| NATT4_09 | RLTRKYSNGK | 0.921 | −7.018 | 0.482 | 0.055 | 17.18 | 0 | 0 | 0 | 0 | 0.638 | 0.722 | 0.022 | 0.001 | 0.017 | 0.104 | 0.124 |
| NATT4_10 | KNKYGLGKLHQS | 0.832 | −6.901 | 0.517 | 0.056 | 20.70 | 0 | 0 | 0.012 | 0 | 0.830 | 0.873 | 0.022 | 0 | 0.033 | 0.027 | 0.213 |
| NATT4_11 | KANIPFTGRLTRK | 0.999 | −6.613 | 0.425 | 0.026 | 20.34 | 0 | 0 | 0.001 | 0 | 0.557 | 0.727 | 0.009 | 0.001 | 0.004 | 0.058 | 0.066 |
| NATT4_12 | GRLTRKYSNGKVT | 0.991 | −7.574 | 0.384 | 0.041 | 22.11 | 0 | 0 | 0 | 0 | 0.113 | 0.798 | 0.008 | 0 | 0.006 | 0.036 | 0.068 |
| NATT4_13 | KVTSSSVKGIYKK | 1 | −7.250 | 0.363 | 0.02 | 18.23 | 0 | 0 | 0.003 | 0 | 0.463 | 0.918 | 0.002 | 0.001 | 0.009 | 0.042 | 0.103 |
| NATT4_14 | VTSSSVKGIYKKV | 1 | −7.499 | 0.417 | 0.025 | 16.82 | 0 | 0 | 0.005 | 0.001 | 0.544 | 0.912 | 0.002 | 0.002 | 0.004 | 0.038 | 0.065 |
| NATT4_15 | VKGIYKKVQVGEI | 0.999 | −7.261 | 0.592 | 0.019 | 20.76 | 0 | 0 | 0.008 | 0.001 | 0.712 | 0.912 | 0.003 | 0.001 | 0.003 | 0.097 | 0.085 |
| NATTP_01 | LGQALIPRCRKMP | 0.987 | −6.502 | 0.49 | 0.013 | 19.82 | 0 | 0 | 0.002 | 0 | 1.006 | 0.578 | 0.012 | 0.003 | 0.01 | 0.037 | 0.162 |
| NATTP_02 | RCRKMPGVKM | 0.985 | −6.729 | 0.483 | 0.036 | 15.67 | 0 | 0.001 | 0.001 | 0 | 1.084 | 0.773 | 0.033 | 0.003 | 0.036 | 0.036 | 0.178 |
| NATTP_03 | QALIPRCRKMPGV | 0.993 | −6.490 | 0.483 | 0.014 | 19.08 | 0 | 0 | 0.001 | 0 | 0.974 | 0.606 | 0.01 | 0.005 | 0.009 | 0.034 | 0.156 |
| NATTP_04 | ALIPRCRKMPGVK | 0.995 | −6.759 | 0.488 | 0.011 | 20.18 | 0 | 0 | 0.002 | 0 | 1.078 | 0.7 | 0.015 | 0.004 | 0.011 | 0.033 | 0.159 |
| NATTP_05 | LIPRCRKMPGVKM | 0.995 | −6.721 | 0.467 | 0.009 | 22.53 | 0 | 0 | 0.004 | 0 | 1.119 | 0.742 | 0.014 | 0.003 | 0.013 | 0.038 | 0.153 |
| Inference/Reference Range | HIA > 0.3: HIA positive HIA < 0.3: HIA negative | Optimal: higher than −5.15 | Optimal: 0.04–20 L/Kg | ≥0.1: BBB positive and <0.1: BBB negative | PPB < 90%: optimal PPB > 90%: low therapeutic index | >0.5: inhibitor <0.5: non inhibitor | >0.5: substrate <0.5: non substrate | >0.5: inhibitor <0.5: non-inhibitor | >0.5: substrate <0.5: non-substrate | High: >15 mL/min/kg Moderate: 5–15 mL/min/kg Low: <5 mL/min/kg | Long half-life: >3 h Short half-life: <3 h | >0.5: blocker <0.5: non-blocker | >0.5: hepatotoxic <0.5: non-hepatotoxic | >0.5: positive <0.5: negative | >0.5: carcinogen <0.5: non-carcinogen | >0.5: sensitizer <0.5: non-sensitizer | |
| Oral Drugs | Peptides | |||||
|---|---|---|---|---|---|---|
| Molecular Properties | Lipinski, 2001 and Veber, 2002 | Doak et al., 2014 | Santos et al., 2016 * | Diaz-Eufracio et al., 2018 ** | De Oliveira et al., 2021 # | Our Study |
| MW | ≤500 | ≤1.000 | ≤700 | 27.03 ≤ MW ≤ 5036.65 | 331.48 ≤ MW ≤ 3750.51 | 965.08 ≤ MW ≤ 2704.06 |
| LogP | ≤5 | −2 ≤ LogP ≤ 10 | ≤7.5 | −17.87 ≤ LogP ≤39.89 | −42.12 ≤ LogP ≤ 2.97 | −7.387 ≤ LogP ≤ 0.562 |
| tPSA | ≤140 | ≤250 | ≤200 | ≤2064.83 | 101.29 ≤ tPSA ≤1782.83 | 405.88 ≤ tPSA ≤ 1288.48 |
| Fsp3 | − | − | ≤0.55 | − | 0.37 ≤ Fsp3 ≤ 0.84 | 0.45 ≤ Fsp3 ≤ 0.80 |
| NRB | ≤10 | ≤20 | ≤20 | ≤209 | 9 ≤ NRB ≤ 137 | 37 ≤ NRB ≤ 117 |
| HBA | ≤10 | ≤15 | ≤10 | ≤71 | 5 ≤ HBA ≤ 55 | 25 ≤ HBA ≤ 75 |
| NAR | − | − | − | − | ≤10 | ≤5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
De Cena, G.L.; Scavassa, B.V.; Conceição, K. In Silico Prediction of Anti-Infective and Cell-Penetrating Peptides from Thalassophryne nattereri Natterin Toxins. Pharmaceuticals 2022, 15, 1141. https://doi.org/10.3390/ph15091141
De Cena GL, Scavassa BV, Conceição K. In Silico Prediction of Anti-Infective and Cell-Penetrating Peptides from Thalassophryne nattereri Natterin Toxins. Pharmaceuticals. 2022; 15(9):1141. https://doi.org/10.3390/ph15091141
Chicago/Turabian StyleDe Cena, Gabrielle Lupeti, Bruna Vitória Scavassa, and Katia Conceição. 2022. "In Silico Prediction of Anti-Infective and Cell-Penetrating Peptides from Thalassophryne nattereri Natterin Toxins" Pharmaceuticals 15, no. 9: 1141. https://doi.org/10.3390/ph15091141
APA StyleDe Cena, G. L., Scavassa, B. V., & Conceição, K. (2022). In Silico Prediction of Anti-Infective and Cell-Penetrating Peptides from Thalassophryne nattereri Natterin Toxins. Pharmaceuticals, 15(9), 1141. https://doi.org/10.3390/ph15091141






