Antigenic Characterization of Neuraminidase of Influenza A/H7N9 Viruses Isolated in Different Years
Abstract
:1. Introduction
2. Results
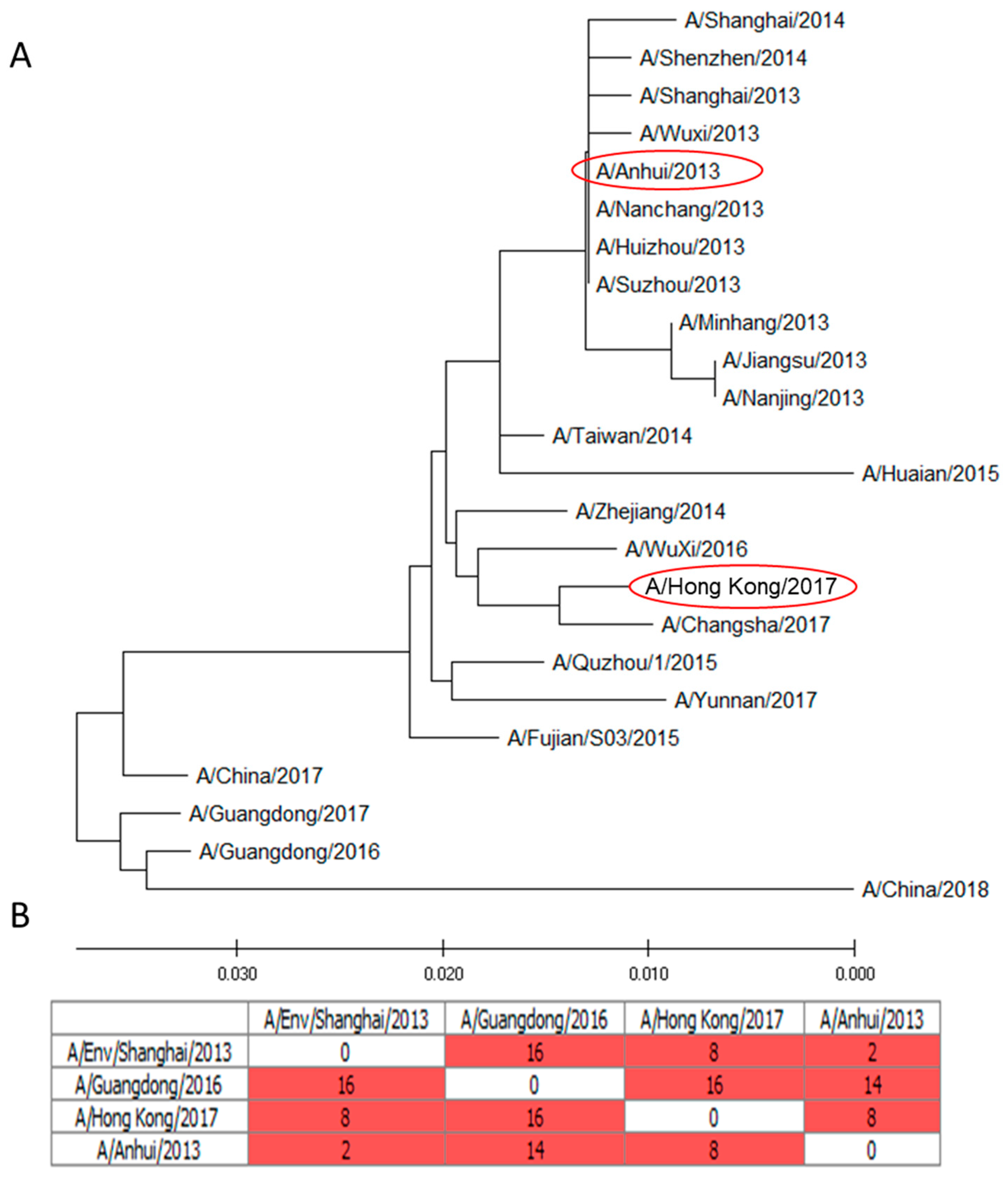
2.1. Molecular Analysis of NA A/H7N9 Viruses
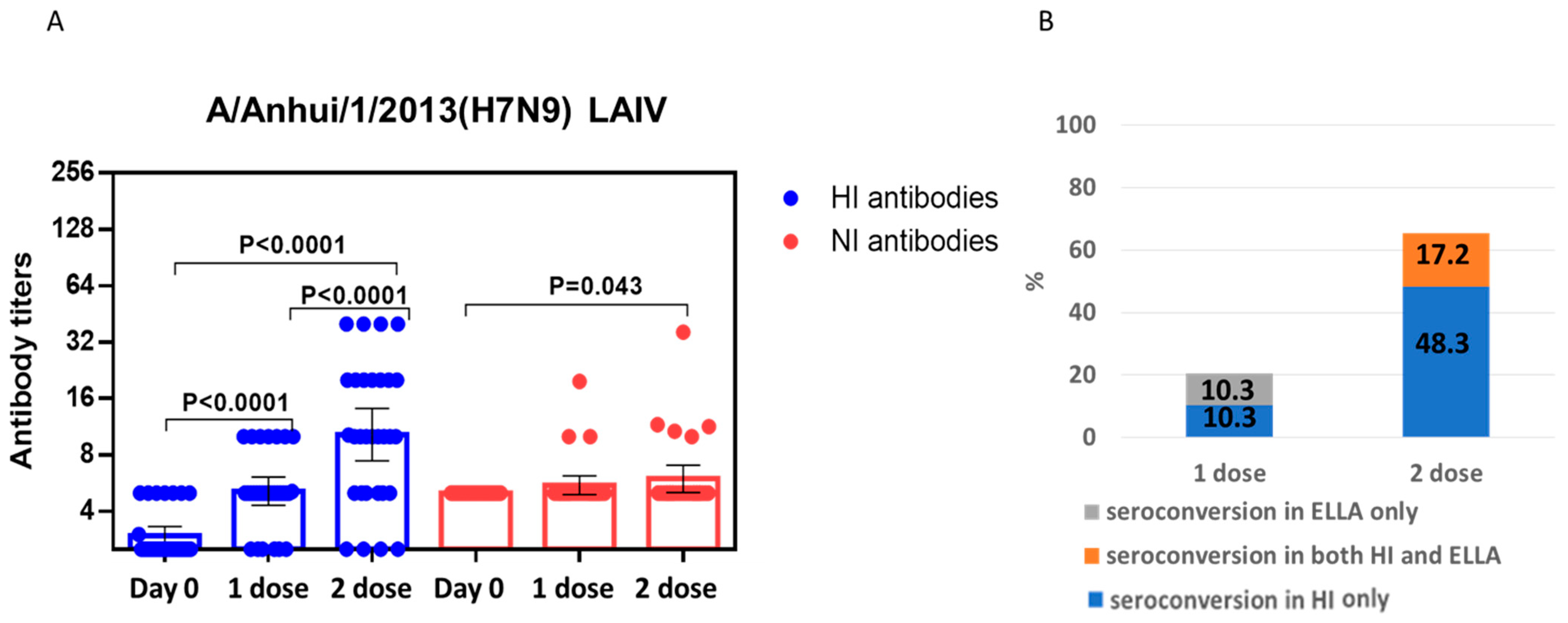
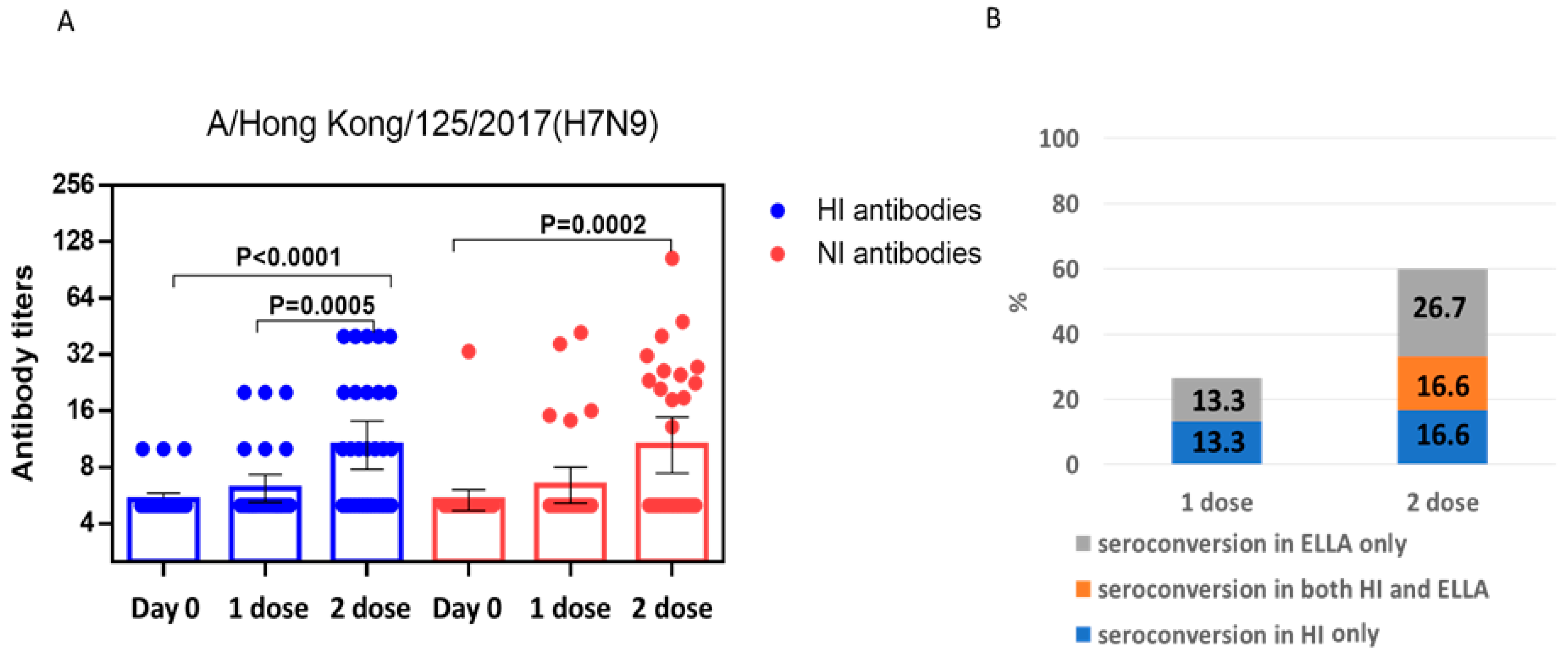
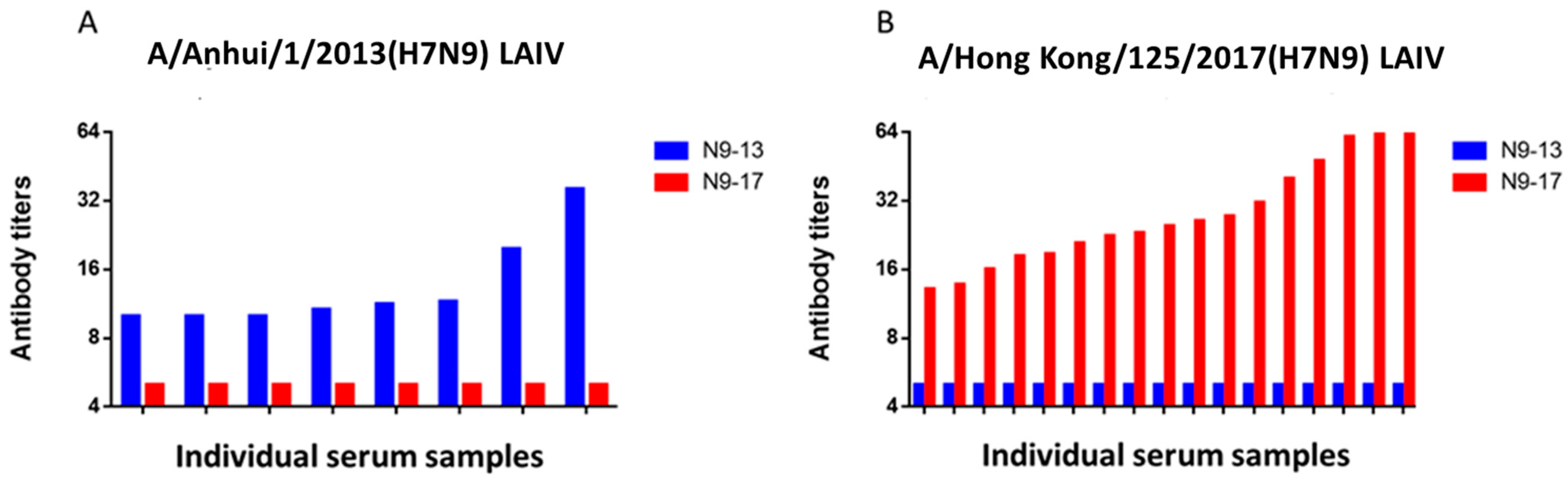
2.2. Study of the Immunogenicity of LAIV Based on A/H7N9 Viruses Isolated in 2013 and 2017
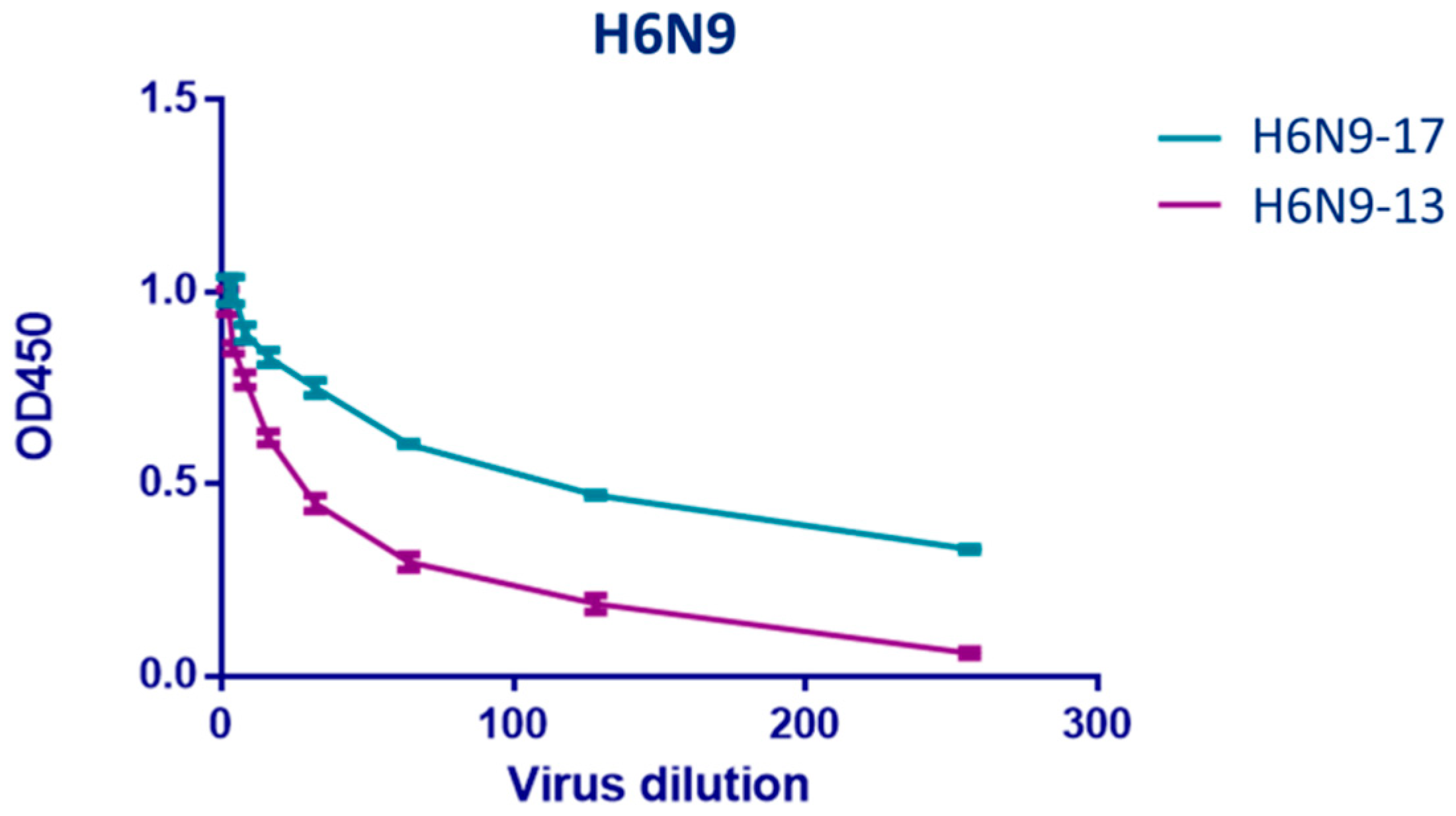
2.3. Study of Monoclonal Antibodies (mABs) to NA Subtype N9
3. Discussion
4. Materials and Methods
4.1. Ethics Statement
4.2. Viruses
4.3. Evolutionary Analysis of NA Amino Acid Sequences
4.4. Enzyme-Linked Lectin Assay (ELLA)
4.5. Hemagglutination Inhibition Test (HI)
4.6. Enzyme-Linked Immunosorbent Assay (ELISA)
4.7. Passive Immunization of Mice with Monoclonal Antibodies to N9
4.8. Statistical Analysis of Results
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- World Health Organization. WHO Risk Assessment of Human Infections with Avian Influenza A (H7N9) Virus; World Health Organization: Geneva, Switzerland, 2015. [Google Scholar]
- Kucharski, A.J.; Mills, H.L.; Donnelly, C.A.; Riley, S. Transmission potential of influenza A (H7N9) virus, China, 2013–2014. Emerg. Infect. Dis. 2015, 21, 852. [Google Scholar] [CrossRef]
- World Health Organization. Human Infection with Avian Influenza A (H7N9) Virus—China: Update; World Health Organization: Geneva, Switzerland, 2018. [Google Scholar]
- Wu, J.; Ke, C.; Lau, E.H.; Song, Y.; Cheng, K.L.; Zou, L.; Kang, M.; Song, T.; Peiris, M.; Yen, H.L. Influenza H5/H7 virus vaccination in poultry and reduction of zoonotic infections, Guangdong Province, China, 2017–2018. Emerg. Infect. Dis. 2019, 25, 116. [Google Scholar] [CrossRef] [PubMed]
- Xiang, D.; Shen, X.; Pu, Z.; Irwin, D.M.; Liao, M.; Shen, Y. Convergent evolution of human-isolated H7N9 avian influenza a viruses. J. Infect. Dis. 2018, 217, 1699–7707. [Google Scholar] [CrossRef]
- Zheng, D.; Gao, F.; Zhao, C.; Ding, Y.; Cao, Y.; Yang, T.; Xu, X.; Chen, Z. Comparative effectiveness of H7N9 vaccines in healthy individuals. Hum. Vaccines Immunother. 2019, 15, 80–90. [Google Scholar] [CrossRef] [PubMed]
- Sobhanie, M.; Matsuoka, Y.; Jegaskanda, S.; Fitzgerald, T.; Mallory, R.; Chen, Z.; Luke, C.; Treanor, J.; Subbarao, K. Evaluation of the safety and immunogenicity of a candidate pandemic live attenuated influenza vaccine (pLAIV) against influenza A (H7N9). J. Infect. Dis. 2016, 213, 922–929. [Google Scholar] [CrossRef] [PubMed]
- Rudenko, L.; Isakova-Sivak, I.; Naykhin, A.; Kiseleva, I.; Stukova, M.; Erofeeva, M.; Korenkov, D.; Matyushenko, V.; Sparrow, E.; Kieny, M.P. H7N9 live attenuated influenza vaccine in healthy adults: A randomised, double-blind, placebo-controlled, phase 1 trial. Lancet Infect. Dis. 2016, 16, 303–310. [Google Scholar] [CrossRef]
- Kiseleva, I.; Isakova-Sivak, I.; Stukova, M.; Erofeeva, M.; Donina, S.; Larionova, N.; Krutikova, E.; Bazhenova, E.; Stepanova, E.; Vasilyev, K.; et al. A phase 1 randomized placebo-controlled study to assess the safety, immunogenicity and genetic stability of a new potential pandemic H7N9 live attenuated influenza vaccine in healthy adults. Vaccines 2020, 8, 296. [Google Scholar] [CrossRef]
- Zuckerkandl, E.; Pauling, L. Evolutionary divergence and convergence in proteins. In Evolving Genes and Proteins; Academic Press: Cambridge, MA, USA, 1965; pp. 97–166. [Google Scholar]
- Webster, R.G.; Air, G.M.; Metzger, D.W.; Colman, P.M.; Varghese, J.N.; Baker, A.T.; Laver, W.G. Antigenic structure and variation in an influenza virus N9 neuraminidase. J. Virol. 1987, 61, 2910–2916. [Google Scholar] [CrossRef]
- Rajendran, M.; Nachbagauer, R.; Ermler, M.E.; Bunduc, P.; Amanat, F.; Izikson, R.; Cox, M.; Palese, P.; Eichelberger, M.; Krammer, F. Analysis of anti-influenza virus neuraminidase antibodies in children, adults, and the elderly by ELISA and enzyme inhibition: Evidence for original antigenic sin. MBio 2017, 8, e02281-16. [Google Scholar] [CrossRef]
- Starick, E.; Werner, O.; Schirrmeier, H.; Köllner, B.; Riebe, R.; Mundt, E. Establishment of a competitive ELISA (cELISA) system for the detection of influenza A virus nucleoprotein antibodies and its application to field sera from different species. J. Vet. Med. Ser. B 2006, 53, 370–375. [Google Scholar] [CrossRef]
- Aydillo, T.; Escalera, A.; Strohmeier, S.; Aslam, S.; Sanchez-Cespedes, J.; Ayllon, J.; Roca-Oporto, C.; Perez-Romero, P.; Montejo, M.; Gavalda, J.; et al. Pre-existing hemagglutinin stalk antibodies correlate with protection of lower respiratory symptoms in flu-infected transplant patients. Cell Rep. Med. 2020, 1, 100130. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Shi, J.; Deng, G.; Guo, J.; Zeng, X.; He, X.; Kong, H.; Gu, C.; Li, X.; Liu, J.; et al. H7N9 influenza viruses are transmissible in ferrets by respiratory droplet. Science 2013, 341, 410–414. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.W.; Kuo, S.M.; Yang, S.L.; Gong, Y.N.; Hsiao, M.R.; Liu, Y.C.; Shih, S.R.; Tsao, K.C. Genomic signatures for avian H7N9 viruses adapting to humans. PLoS ONE 2016, 11, e0148432. [Google Scholar] [CrossRef]
- Marjuki, H.; Mishin, V.P.; Chesnokov, A.P.; De La Cruz, J.A.; Davis, C.T.; Villanueva, J.M.; Fry, A.M.; Gubareva, L.V. Neuraminidase mutations conferring resistance to oseltamivir in influenza A (H7N9) viruses. J. Virol. 2015, 89, 5419–5426. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Hu, J.; Jin, X.; Li, X.; Wang, J.; Zhang, M.; Chen, J.; Xie, S.; Qi, W.; Liao, M.; et al. Accelerated evolution of H7N9 subtype influenza virus under vaccination pressure. Virol. Sin. 2021, 36, 1124–1132. [Google Scholar] [CrossRef] [PubMed]
- Ning, T.; Nie, J.; Huang, W.; Li, C.; Li, X.; Liu, Q.; Zhao, H.; Wang, Y. Antigenic drift of influenza A (H7N9) virus hemagglutinin. J. Infect. Dis. 2019, 219, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Varghese, J.N.; Colman, P.M.; Van Donkelaar, A.; Blick, T.J.; Sahasrabudhe, A.; McKimm-Breschkin, J.L. Structural evidence for a second sialic acid binding site in avian influenza virus neuraminidases. Proc. Natl. Acad. Sci. USA 1997, 94, 11808–11812. [Google Scholar] [CrossRef] [PubMed]
- Uhlendorff, J.; Matrosovich, T.; Klenk, H.D.; Matrosovich, M. Functional significance of the hemadsorption activity of influenza virus neuraminidase and its alteration in pandemic viruses. Arch. Virol. 2009, 154, 945–957. [Google Scholar] [CrossRef]
- Desheva, Y.; Sychev, I.; Smolonogina, T.; Rekstin, A.; Ilyushina, N.; Lugovtsev, V.; Samsonova, A.; Go, A.; Lerner, A. Anti-neuraminidase antibodies against pandemic A/H1N1 influenza viruses in healthy and influenza-infected individuals. PLoS ONE 2018, 13, e0196771. [Google Scholar] [CrossRef]
- Eichelberger, M.C.; Monto, A.S. Neuraminidase, the forgotten surface antigen, emerges as an influenza vaccine target for broadened protection. J. Infect. Dis. 2019, 219 (Suppl. S1), S75–S80. [Google Scholar] [CrossRef] [Green Version]
- Sultana, I.; Yang, K.; Getie-Kebtie, M.; Couzens, L.; Markoff, L.; Alterman, M.; Eichelberger, M.C. Stability of neuraminidase in inactivated influenza vaccines. Vaccine 2014, 32, 2225–2230. [Google Scholar] [CrossRef] [PubMed]
- Monto, A.; Kendal, A. Effect of neuraminidase antibody on Hong Kong influenza. Lancet 1973, 301, 623–625. [Google Scholar] [CrossRef]
- Dunand, C.J.; Leon, P.E.; Kaur, K.; Tan, G.S.; Zheng, N.Y.; Andrews, S.; Huang, M.; Qu, X.; Huang, Y.; Salgado-Ferrer, M.; et al. Preexisting human antibodies neutralize recently emerged H7N9 influenza strains. J. Clin. Investig. 2015, 125, 1255–1268. [Google Scholar] [CrossRef] [PubMed]
- Wan, H.; Qi, L.; Gao, J.; Couzens, L.K.; Jiang, L.; Gao, Y.; Sheng, Z.M.; Fong, S.; Hahn, M.; Khurana, S.; et al. Comparison of the efficacy of N9 neuraminidase-specific monoclonal antibodies against influenza A (H7N9) virus infection. J. Virol. 2018, 92, e01588-17. [Google Scholar] [CrossRef] [PubMed]
- Wareing, M.D.; Marsh, G.A.; Tannock, G.A. Preparation and characterisation of attenuated cold-adapted influenza A reassortants derived from the A/Leningrad/134/17/57 donor strain. Vaccine 2002, 20, 2082–2090. [Google Scholar] [CrossRef]
- Hoffmann, E.; Neumann, G.; Kawaoka, Y.; Hobom, G.; Webster, R.G. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc. Natl. Acad. Sci. USA 2000, 97, 6108–6113. [Google Scholar] [CrossRef] [PubMed]
- Okonechnikov, K.; Golosova, O.; Fursov, M.; Ugene Team. Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evolut. 1987, 4, 406–425. [Google Scholar]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evolut. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Desheva, J.A.; Lu, X.H.; Rekstin, A.R.; Rudenko, L.G.; Swayne, D.E.; Cox, N.J.; Katz, J.M.; Klimov, A.I. Characterization of an influenza A H5N2 reassortant as a candidate for live-attenuated and inactivated vaccines against highly pathogenic H5N1 viruses with pandemic potential. Vaccine 2006, 24, 6859–6866. [Google Scholar] [CrossRef]
- Hsu, W.T.; Chang, C.Y.; Tsai, C.H.; Wei, S.C.; Lo, H.R.; Lamis, R.J.; Chang, H.W.; Chao, Y.C. PEDV Infection Generates Conformation-Specific Antibodies That Can Be Effectively Detected by a Cell-Based ELISA. Viruses 2021, 13, 303. [Google Scholar] [CrossRef] [PubMed]
- Lamis, R.J.; Chiueh, T.S.; Tsai, C.H.; Lo, H.R.; Wei, S.C.; Chao, Y.C. Identification and Quantification of Anti-Gp. Mur Antibodies in Human Serum Using an Insect-Cell-Based System. Diagnostics 2021, 11, 966. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.C.; Hsu, W.T.; Chiu, C.H.; Chang, F.Y.; Lo, H.R.; Liao, C.Y.; Yang, H.I.; Chou, Y.C.; Tsai, C.H.; Chao, Y.C. An Integrated Platform for Serological Detection and Vaccination of COVID-19. Front. Immunol. 2021, 12, 771011. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Desheva, Y.; Losev, I.; Petkova, N.; Kudar, P.; Donina, S.; Mamontov, A.; Tsai, C.-H.; Chao, Y.-C. Antigenic Characterization of Neuraminidase of Influenza A/H7N9 Viruses Isolated in Different Years. Pharmaceuticals 2022, 15, 1127. https://doi.org/10.3390/ph15091127
Desheva Y, Losev I, Petkova N, Kudar P, Donina S, Mamontov A, Tsai C-H, Chao Y-C. Antigenic Characterization of Neuraminidase of Influenza A/H7N9 Viruses Isolated in Different Years. Pharmaceuticals. 2022; 15(9):1127. https://doi.org/10.3390/ph15091127
Chicago/Turabian StyleDesheva, Yulia, Igor Losev, Nadezhda Petkova, Polina Kudar, Svetlana Donina, Andrey Mamontov, Chih-Hsuan Tsai, and Yu-Chan Chao. 2022. "Antigenic Characterization of Neuraminidase of Influenza A/H7N9 Viruses Isolated in Different Years" Pharmaceuticals 15, no. 9: 1127. https://doi.org/10.3390/ph15091127
APA StyleDesheva, Y., Losev, I., Petkova, N., Kudar, P., Donina, S., Mamontov, A., Tsai, C.-H., & Chao, Y.-C. (2022). Antigenic Characterization of Neuraminidase of Influenza A/H7N9 Viruses Isolated in Different Years. Pharmaceuticals, 15(9), 1127. https://doi.org/10.3390/ph15091127






