Abstract
Death-associated protein kinase 1 (DAPK1) is a serine/threonine protein kinase involved in diverse fundamental cellular processes such as apoptosis and autophagy. DAPK1 isoform plays an essential role as a tumor suppressor and inhibitor of metastasis. Consequently, DAPK1 became a promising target protein for developing new anti-cancer agents. In this work, we present the rational design and complete synthetic routes of a novel series of eighteen aryl carboxamide derivatives as potential DAPK1 inhibitors. Using a custom panel of forty-five kinases, a single dose of 10 µM of the picolinamide derivative 4a was able to selectively inhibit DAPK1 kinase by 44.19%. Further investigations revealed the isonicotinamide derivative 4q as a promising DAPK1 inhibitory lead compound with an IC50 value of 1.09 µM. In an in vitro anticancer activity assay using a library of 60 cancer cell lines including blood, lung, colon, CNS, skin, ovary, renal, prostate, and breast cancers, four compounds (4d, 4e, 4o, and 4p) demonstrated high anti-proliferative activity with mean % GI ~70%. Furthermore, the most potent DAPK1 inhibitor (4q) exhibited remarkable activity against leukemia (K-562) and breast cancer (MDA-MB-468) with % GI of 72% and 75%, respectively.
1. Introduction
Death-associated protein kinase 1 (DAPK1) (a Ca2+/calmodulin dependent Ser/Thr kinase) is an essential mediator in cell death and autophagy related signals [1,2]. It consists of 1430 residues and is the largest member in the DAPK protein family, including a Ca2+/CaM autoregulatory domain, a death domain, and a serine-rich C-terminal tail whose phosphorylation activity is known to be responsible for specific forms of apoptosis [3,4]. It coordinates cell-death signaling pathways in response to various stimuli such as death receptor activation, cytokines, matrix detachment, ceramide, ischemia, and glutamate toxicity [5]. As previously discussed in various studies, DAPK1 as a stress-responsive kinase is a crucial component that transmits ER stress signals into two distinct directions, caspase activation (via regulating type I apoptotic caspase-dependent cell death) and autophagy (by controlling type II autophagic caspase-independent cell death) [5,6,7,8,9,10,11,12,13].
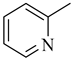
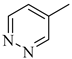
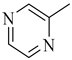
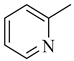
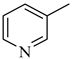
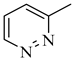
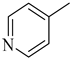
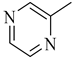
Among novel effective approaches to hinder apoptosis pathways, some fusion proteins have been reported [14]. However, a peptide-based strategy has certain potential scientific and technical cautions, such as lack of cell selectivity, instability, as well as uncertainty of the effective therapeutic concentration, which influences the peptide cargo in addition to suffering from rapid degradation after administration orally [15]. Hence, the rational drug design of small molecule inhibitors could be a unique way to overcome such drawbacks. Although DAPK1 has gained a lot of interest regarding the comprehension of its functions, only a small number of chemical scaffolds, comprehensively discussed in our recent review [4], have been found in the literature with DAPK1 inhibitory activity, i.e., aminopyridazine [16,17], imidazo [1,2-b]pyridazine [18], pyridin-3-ylmethylene-1,3-oxazol-5-one [19,20], pyrazolo [3,4-d]pyrimidinone [21,22], and 1H-pyrrolo [2,3-b]pyridine (7-azaindole) [23] (Figure 1). Inspired by the various unsolved issues of reported scaffolds, such as instability in biological systems, low potency, low selectivity profile, and/or insufficient toxicity studies, in addition to the absence of a current promising clinical candidate or an FDA-approved specific DAPK1 inhibitor, our institute has launched a project aimed at designing novel leads for DAPK1 activity modulation with potential anticancer activities.
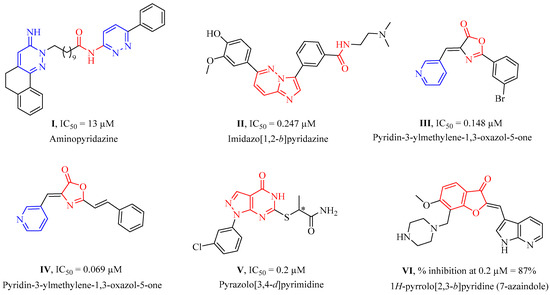
Figure 1.
Reported synthetic small molecules bearing different chemical scaffolds and their IC50 values or % inhibition against DAPK1 kinase.
In previous studies, a structure-based virtual screening strategy was followed to develop an oxazol-5-one derivative III and its related analogues as DAPK inhibitors. Further in silico studies were conducted to define the structure–activity relationship (SAR) of the developed inhibitors [19,24]. The SAR studies stated that a nitrogen containing group, such as pyridinyl moiety, is essential for DAPK activity due to the role of the N-atom in H-bond formation with the backbone NH of Val96 in the hinge region at the ATP binding site. In addition, substitution on the terminal phenyl ring was found to improve potency against DAPK compared to the unsubstituted analogues, which might be explained by the increase in electron density of the phenyl ring which allows for ring contribution in hydrophobic interactions at the ATP binding site. Regarding phenyl ring substitution, the studies reported that meta-substitution seems to be more appropriate for binding at the ATP binding site than para-substitution because of electronic effects. Additionally, substitution with electron withdrawing groups at meta-positions is more effective for activity than electron donating ones. It was also observed that the phenyl ring is located near Asp161 in a very tight area of the binding pocket, so bulky substitution would badly affect inhibitor affinity to the binding site and reduce their stability and activity, as well. Finally, docking of compound III in DAPK binding sites illustrated an observed vacancy around the phenyl group, which is considered a point of optimization for developing new DAPK inhibitors with extended substitution on the phenyl ring.
Relying on the aforementioned SAR studies, structural optimizations of the reported inhibitor III were conducted, while keeping the essential binding interactions, with the aim of producing a novel series of DAPK inhibitors (4a–r) (Figure 2). In the current work, the designed carboxamide derivatives (4a–r) were designed to retain the hinge binding interaction with the Val96 backbone by introducing different nitrogen containing groups (pyridine, pyridazine, and pyrazine). However, the principal modification was the ring opening of the central oxazole into carboxamide moiety, which has been reported in many DAPK inhibitors. The lateral phenyl group of the designed series was substituted by an electron withdrawing meta-chloro group that was reported for better hydrophobic interaction. The new designed compounds possessed a unique extension in the hydrophobic area via additional phenoxy substitutions on the terminal phenyl group in an attempt to boost molecular interaction with the enzyme pocket.
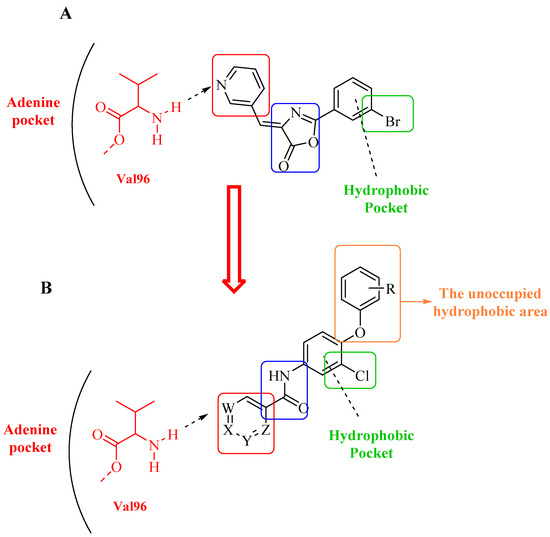
Figure 2.
Diagrammatic representation of the structural modifications of compound III for developing new DAPK inhibitor candidates: (A) molecular interactions of compound III into the DAPK active site; (B) proposed binding interaction of the new synthesized derivatives 4a–r into the DAPK kinase domain (W, X, Y, and Z could be C or N atoms).
2. Results and Discussion
2.1. Chemistry
The chemical synthesis of the target compounds 4a–r was carried out as illustrated in Scheme 1. A nucleophilic aromatic substitution reaction of the commercially available starting material 2-chloro-1-fluoro-4-nitrobenzene (1) was performed via adding the appropriate phenolic derivative and a catalytic amount of potassium carbonate to generate 2-chloro-4-nitrophenoxybenzene derivatives (2a–c). Catalytic hydrogenation of compounds 2a–c was performed using Pt/C catalyst under hydrogen atmosphere to yield compounds 3a–c. Introduction of amide functionality to intermediate 3a–c was achieved by coupling with the appropriate benzoic acid derivative using HATU to afford the final target compounds 4a–r (Table 1).
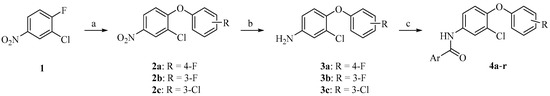
Scheme 1.
Reagents and conditions: (a) appropriate phenol derivative, K2CO3, MeCN, 85 °C, 6 h; (b) H2, Pt/C, MeOH, rt, 6 h; (c) appropriate benzoic acid derivative, DIPEA, HATU, THF, reflux, overnight.

Table 1.
Key Structure of the final target compounds 4a–r.
2.2. Biological Evaluation
2.2.1. Evaluation of Compound 4a against a Panel of 45 Kinases
In an attempt to test our design hypothesis and discover if DAPK1 is the potential conceivable kinase target of the designed candidates, a kinase selectivity assay was carried out to the first synthesized derivative (4a) against a group of forty-five different kinases. Compound 4a was evaluated at a single dose concentration of 10 μM and % inhibition was determined against each corresponding kinase (Table 2).

Table 2.
% enzyme inhibition (relative to DMSO controls) of compound 4a in a single-dose duplicate mode assay at a concentration of 10 µM against 45 kinases.
Interestingly, the preliminary data revealed a remarkable selectivity of compound 4a towards DAPK1, with mean % inhibition of 44.19%. On the other hand, the results for the other 44 kinases showed no activity for most of the kinases and low activity (less than 25%) for a few of the tested kinases (Figure 3). An additional inspection of compound 4a selectivity was carried out by evaluating its activity against other DAPK isoforms (DAPK2 and DAPK3). The obtained results showed no activity for compound 4a with DAPK2 and DAPK3 isoforms (data not shown).
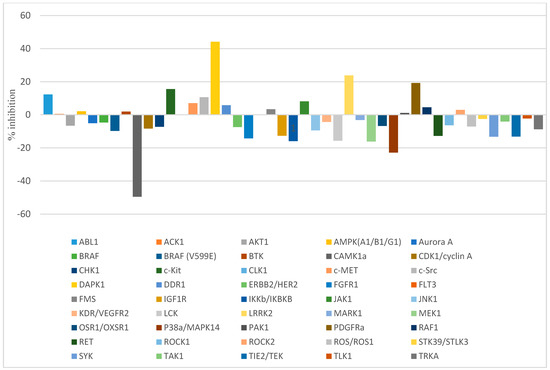
Figure 3.
A schematic chart illustrating the enzyme inhibitory activity of compound 4a (10 µM) against a panel of 45 oncogenic kinases.
2.2.2. In Vitro DAPK1 Kinase Assay and Optimization towards Lead Development
Based on the previous panel of kinase data, a series of aryl carboxamide derivatives (4b–r) were synthesized and evaluated in vitro for their inhibitory activity against DAPK1 using the ELISA technique (enzyme-linked immune sorbent assay). The enzyme inhibition assay was conducted with a 10 μM dose of the tested compounds and the mean % inhibition was measured. As illustrated in Table 3, derivatives possessing the pyridinyl carboxamide moiety (4c, 4e, 4f, 4h, 4j, 4k, 4m, 4p, and 4q) exhibited the highest inhibitory activity (59–81%) in comparison to the pyridazine or pyrazine possessing derivatives (38–61%). In addition, the attachment point of these nitrogen heterocycles (pyridine, pyridazine, and pyrazine) with the amide group did not show a remarkable effect in terms of activity. In regard to the substitution on the terminal phenoxy moiety, the overall results showed that para-substitution with the fluoro group in compounds 4a and 4h–l has a better effect on activity than the meta-substituted derivatives 4b–g, except for the pyridazine bearing derivatives 4a and 4b. Moreover, replacing the m-fluoro substitution with m-chloro in compounds 4m–r showed a slight improvement in activity, except in compounds 4d and 4o where the chloro substitution dramatically reduced activity to 37.99%. Among the tested series, compounds 4h, 4j, 4k, and 4q revealed the highest activity of 81%, 79%, 80%, and 72%, respectively.

Table 3.
DAPK1 kinase % inhibition activity for the final target compounds 4a–r at 10 µM.
2.2.3. Dose-Dependent Assay of Compounds 4h, 4j, 4k, and 4q with DAPK1 Kinase
For more clarification of the structural activity relationship of the synthesized compounds, the most active derivatives (4h, 4j, 4k ,and 4q) were subjected to a dose-dependent IC50 kinase assay. The selected candidates showed low micromolar IC50 values against DAPK1 kinase with a range of 1.09–7.26 μM. The data obtained in Table 4 revealed the impact of the attachment point of the pyridine moiety with the amide linker and, for the 4-F phenoxy derivatives, nicotinamide owing compound 4j exhibited the highest activity (IC50 = 1.7 μM). In contrast, substitution with picolinamide (4h) and isonicotinamide (4k) declined activity to 6.81 µM and 7.26 µM, respectively. Interestingly, the isonicotinamide derivative with the terminal m-chlorophenoxy moiety (4q) emerged to be the most potent inhibitor with an IC50 value of 1.09 µM. In order to determine its selectivity against DAPK1, compound 4q was evaluated at a single dose concentration (10 µM) against the other two isoforms (DAPK2/DAPK3). The two isoforms exhibited no inhibition at all by compound 4q (data not shown), indicating the high potential of compound 4q to be a promising selective DAPK1 inhibitory lead compound.

Table 4.
IC50 values of compounds 4h, 4j, 4k, and 4q against DAPK1 kinase.
2.2.4. In Vitro Anticancer Assay
The anticancer effects of all newly synthesized compounds (4a–r) were assessed through determining their anti-proliferative activities against a panel of NCI-60 human cancer cell lines belonging to nine cancer types (leukemia, non-small cell lung cancer, colon cancer, CNS cancer, melanoma, ovarian cancer, renal cancer, prostate cancer, and breast cancer). All the subjected compounds were selected for one-dose 60 cell line assays. The tested compounds showed wide spectrum activity against the different human cancer cell lines with maximum mean inhibition of 71% (Figure 4). Among the tested compounds, only compounds 4d, 4e, 4o, and 4p displayed high anti-proliferative activities (67.02%, 67.87%, 70.83%, and 69.27%, respectively). The data obtained from NCI assays are analyzed in this section.
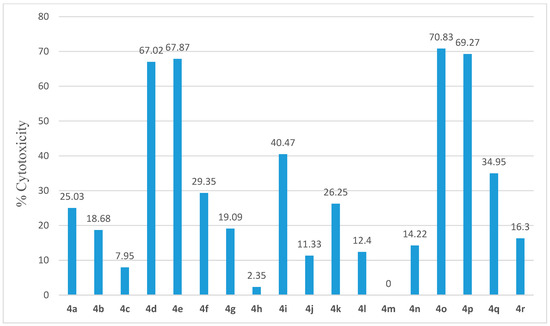
Figure 4.
Mean % inhibition of NCI-60 cell lines after treatment with a single dose (10 µM) of compounds 4a–r.
- ■
- Anti-proliferative activity against leukemia:
The efficacy of the synthesized compounds against leukemia cell lines (CCRF-CEM, HL-60(TB), K-562, MOLT-4, RPMI-8226, and SR) is illustrated in Table 5. It was observed that leukemia cells (HL-60(TB) and K-562) are the most sensitive cell lines towards treatment with the tested compounds (Figure 5). The pyrazine carboxamide derivatives 4d and 4o, with meta-(F or Cl) substitution on the terminal phenoxy group, totally inhibited the growth of the leukemia HL-60(TB) cell line with almost 100% inhibition and showed above 90% growth inhibition for leukemia K-562. Compounds 4e, 4h, and 4p exhibited satisfactory inhibitory activity against both cell lines (78–83%), while most of the tested candidates showed moderate anti-proliferative activity against leukemia (30–60%).

Table 5.
Anti-proliferative activities (% growth inhibition) of the target compounds 4a–r against leukemia cell lines.

Figure 5.
Mean % inhibition of the target compounds 4a–r against leukemia cell lines (HL-60(TB) and K-562).
- ■
- Anti-proliferative activity on non-small cell lung cancer:
The synthesized candidates were evaluated against a panel of nine NSCLCs (A549, EKVX, HOP62, HOP92, NCI-H226, NCI-H23, NCI-H322M, NCI-H460, and NCI-H522). The results are represented in Table 6. Among the tested series, compounds 4d, 4e, 4o, and 4p were the most active anti-proliferative agents against the nine NSCLCs (% GI = 55–60%).

Table 6.
Anti-proliferative activities (% growth inhibition) of the target compounds 4a–r against non-small cell lung cancer cell lines.
As depicted in Figure 6, NCI-H460 and NCI-H522 were the highest affected cell lines by the mentioned active compounds. NCI-H460 showed % GI of 77.9%, 75.34%, 84.28%, and 77.8%, while NCI- H522 growth was inhibited by 77.7%, 63.79%, 87.25%, and 72.65% upon treatment with 10 µM of 4d, 4e, 4o, and 4p, respectively. Compounds 4i and 4q elicited moderate activity with average inhibition of 40% and 36%, respectively. Meanwhile, the rest of the tested series did not exhibit significant activity towards NSCLCs (average % GI < 30%).
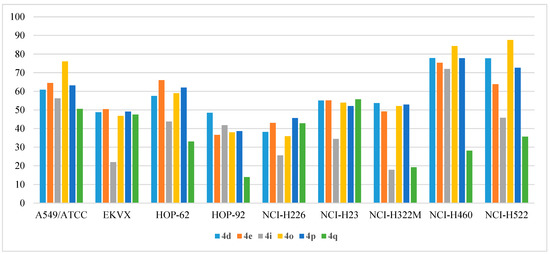
Figure 6.
Anti-proliferative activities (% GI) of compounds 4d, 4e, 4i, 4o, 4p, and 4q against NSCLCs.
- ■
- Anti-proliferative activity on colon cancers:
The target compounds were assayed against a panel of seven colon adenocarcinoma cell lines (COLO-205, HCC-2998, HCT-116, HCT-15, HT29, KM12, and SW-620) and the results are summarized in Table 7. The tested compounds revealed a wide range of inhibitory activity against the seven cell lines, with average % GI from 0% to 78%. The overall data indicated that compounds 4d, 4e, 4o, and 4p had the highest growth inhibitory activity and showed average % GI of 71.84%, 69.73%, 78.16%, and 71.11%, respectively (Figure 7). Compounds 4d and 4e significantly inhibited the growth of HOP92 adenocarcinoma with % GI of 90.87% and 87.13%, respectively. The two compounds also exhibited outstanding activity towards the COLO 205 cell line (% GI = 84.52% and 77.29%, respectively).

Table 7.
Anti-proliferative activities (% growth inhibition) of the target compounds 4a–r against colon adenocarcinoma cell lines.
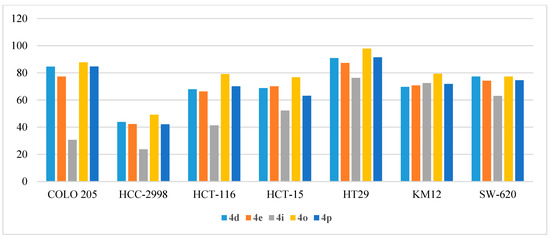
Figure 7.
Anti-proliferative activities (% GI) of compounds 4d, 4e, 4i, 4o, and 4p against colon cancer cell lines.
Moreover, compounds 4o and 4p emerged to be the most potent inhibitors of HOP92 adenocarcinoma with % GI of 97.8% and 91.4%, respectively. Furthermore, the COLO 205 cell line was also inhibited by the two compounds with % GI of 87.71% and 84.61% for the two compounds. Compound 4i moderately inhibited the growth of HCT-15, HT29, KM12, and SW-620 and exhibited % GI of 52.26%, 76.34%, 72.49%, and 63.04%, respectively (Figure 7).
- ■
- Anti-proliferative activity on CNS cancers:
The anti-proliferative activities of the target derivatives were determined against six CNS cancer cell lines (SF-268, SF-295, SF-539, SNB-19, SNB-75, and U251) and the resulting % GIs were tabulated in Table 8. As illustrated, compounds 4d, 4e, 4o, and 4p kept their ranking among the tested derivatives as the most potent anti-proliferative agents. It was observed that SF-539 and SNB-75 were the most sensitive cell lines that showed growth inhibition around 100% upon treatment with 10 µM of compounds 4d, 4o, 4p, 4e (Figure 8). In addition, the mentioned derivatives broadly inhibited the growth of the remaining four cell lines (44.50–83.53%). Compound 4q moderately inhibited the growth of SF-295 (67.42%), while there was no observable activity from the compound towards the other five cell lines.

Table 8.
Anti-proliferative activities (% growth inhibition) of the target compounds 4a–r against CNS cancer cell lines.
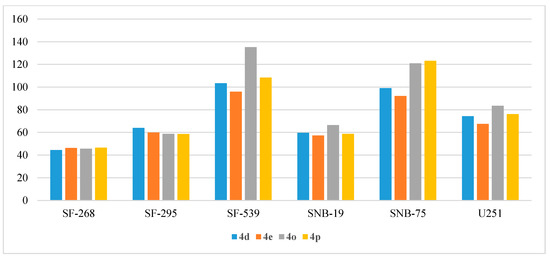
Figure 8.
Anti-proliferative activities (% GI) of compounds 4d, 4e, 4o, and 4p against CNS cancer cell lines.
- ■
- Anti-proliferative activity on melanoma:
Melanoma anti-proliferative activity of the synthesized compounds was evaluated using a panel of nine melanoma cell lines (LOX IMVI, MALME-3M, M14, MDA-MB-435, SK-MEL-2, SK-MEL-28, SK-MEL-5, UACC-257, and UACC-62) with the % GI results illustrated in Table 9. The highest potency was observed on the MDA-MB-435 cell line which was strongly inhibited by compounds 4d, 4e, 4i, 4o, and 4p (% GI = 121%, 98%, 102%, 139%, and 144%, respectively) (Figure 9). Compound 4e exhibited high potency against the SK-MEL-5 cell line with % GI of 124%, while the same cell line was highly inhibited by 4d, 4o, 4p, and 4q (% GI of 90.39%, 74.19%, 85.47%, and 73.04%, respectively) and moderately inhibited by 4a, 4f, 4i, and 4k (% GI of 69.25%, 58.84%, 50.48%, and 67.41%, respectively). Moreover, compounds 4o and 4p showed moderate activity against the M14 cell line (80.21% and 78.15%, respectively) and UACC-62 cell line (71.64% and 62.37%, respectively).

Table 9.
Anti-proliferative activities (% growth inhibition) of the target compounds 4a–r against melanoma cell lines.
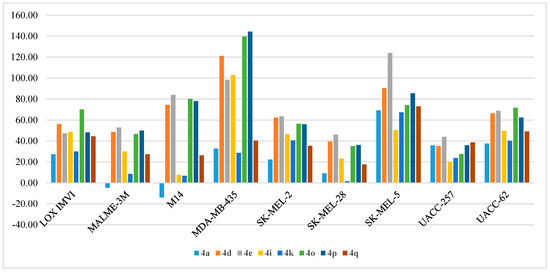
Figure 9.
Anti-proliferative activities (% GI) of compounds 4a, 4d, 4e, 4i, 4k, 4o, 4p, and 4q against Melanoma cell lines.
- ■
- Anti-proliferative activity on ovarian cancers:
The growth inhibition results of the tested compounds against a panel of six ovarian cancer cell lines are demonstrated in Table 10. Among the employed ovarian cell lines, OVCAR-3 emerged to be the most sensitive cell line, which was strongly inhibited by compounds 4d, 4e, 4o, and 4p with % GI of 117.85%, 93.03%, 107.23%, and 97.34%, respectively. The 4d, 4e, 4o, and 4p compounds also inhibited the growth of the NCI/ADR-RES cell line with % GI over 85%. OVCAR-3 and NCI/ADR-RES were moderately inhibited by compound 4i (56.36% and 59.16%, respectively). In addition, moderate growth inhibition activity was detected against three cell lines: SK-OV-3 by compounds 4e and 4p (69.98% and 65.75%, respectively); OVCAR-8 by compounds 4d, 4e, 4o, and 4p (62.49%, 65.37%, 72.05%, and 67.92%, respectively); OVCAR-4 by compounds 4d, 4e, 4f, 4o, 4p, and 4q (55.46%, 53.83%, 54.30%, 61.05%, 52.16%, 56.18%, respectively); and IGROV1 by compounds 4d, 4e, 4o, and 4p (59.49%, 58.28%, 72.96%, 57.85%, respectively) (Figure 10).

Table 10.
Anti-proliferative activities (% growth inhibition) of the target compounds 4a–r against ovarian cancer cell lines.
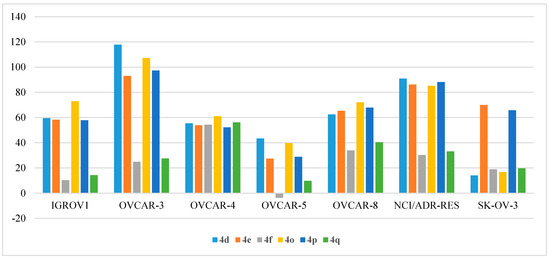
Figure 10.
Anti-proliferative activities (% GI) of compounds 4d, 4e, 4f, 4o, 4p, and 4q against ovarian cancer cell lines.
- ■
- Anti-proliferative activity on renal cancers:
The synthesized compounds 4a–r exhibited a wide range of inhibitive activity towards a panel of eight renal cancer cell lines (786-0, A498, ACHN, CAKI-1, RXF 393, SN12C, TK-10, and UO-31), as shown in Table 11. The uppermost activity was observed against the growth of the RXF 393 cell line which was totally inhibited (% GI > 100%) by compounds 4o and 4p and greatly inhibited by compounds 4d and 4e (92.63% and 86.46%, respectively). Additionally, the A498 cell line showed inhibition of 100% by compound 4p and 95.84% by compound 4e (Figure 11). Modest activity was noticed against the 786-0 cell line by compounds 4d, 4e, 4o, and 4p (% GI of 53.47%, 59.13%, 55.36%, and 60.79%, respectively), the ACHN cell line by compounds 4d, 4e, 4o, 4p, and 4q (% GI of 62.30%, 64.82%, 60.04%, 58.34%, and 52.45%, respectively), the CAKI-1 cell line by compounds 4d, 4e, 4o, and 4p (% GI of 74.84%, 67.45%, 78.37%, and 69.44%, respectively), the SN12C cell line by compounds 4d, 4e, 4o, and 4p (% GI of 54.42%, 63.63%, 55.60%, and 56.56%, respectively), and the UO-31 cell line by compounds 4d, 4e, 4o, and 4p (% GI of 56.60%, 60.28%, 62.66%, and 55.92%, respectively) (Figure 11).

Table 11.
Anti-proliferative activities (% growth inhibition) of the target compounds 4a–r against renal cancer cell lines.
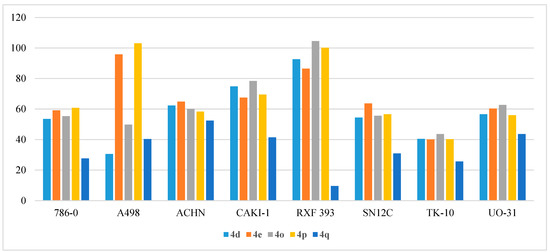
Figure 11.
Anti-proliferative activities (% GI) of compounds 4d, 4e, 4o, 4p, and 4q against renal cancer cell lines.
- ■
- Anti-proliferative activity on prostate cancers:
Two prostate cancer cell lines (PC-3 and DU-145) were used for the anti-proliferative evaluation of the target compounds 4a–r (Table 12). Among the tested series, only compounds 4d, 4e, 4o, and 4p exhibited satisfactory potency against the two cell lines. The four candidates inhibited the growth of the PC-3 cell line by % GI of 83.31%, 75.41%, 88.90%, and 85.15%, respectively. However, the DU-145 cell line was inhibited by % GI of 80.87%, 78.17%, 77.35%, and 83.88% by the four compounds, respectively (Figure 12).

Table 12.
Anti-proliferative activities (% growth inhibition) of the target compounds 4a-r against prostate cancer cell lines.
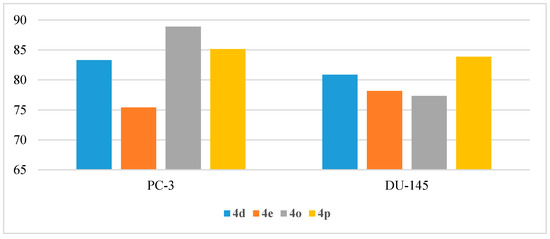
Figure 12.
Anti-proliferative activities (% GI) of compounds 4d, 4e, 4o, and 4p against prostate cancer cell lines.
- ■
- Anti-proliferative activity on breast cancers:
Finally, the target compounds were evaluated as anti-proliferative agents against six different breast cancer cell lines (MCF7, MDA-MB-231/ATCC, HS 578T, BT-549, T-47D, and MDA-MB-468) and the results were summarized. As shown in Table 13, the growth of the MDA-MB-468 cell line was completely inhibited (100–109%) by compounds 4d, 4e, 4o, and 4p, while its growth was moderately inhibited by compounds 4a, 4f, 4g, 4i, 4k, 4q, and 4r (52–76%). The MCF7 cell line was less sensitive to the tested compounds and showed maximum growth inhibition of 82.54% and 80.44% with compounds 4d and 4e, respectively. The HS 578T and T-47D cell lines were moderately inhibited by compounds 4a, 4d, 4e, 4o, and 4p (% GI 59.27–81.27%). MDA-MB-231/ATCC and BT-549 were the most resistant cell lines to the tested compounds, except for compound 4o that inhibited the growth of MDA-MB-231/ATCC by % GI of 68.29% (Figure 13).

Table 13.
Anti-proliferative activities (% growth inhibition) of the target compounds 4a–r against breast cancer cell lines.
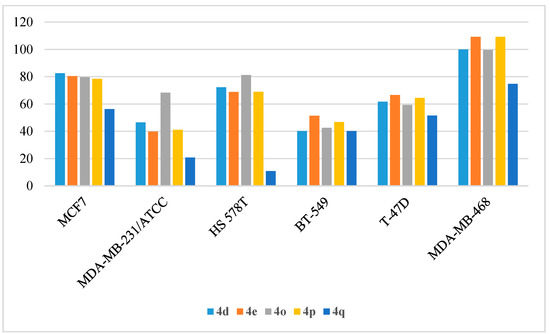
Figure 13.
Anti-proliferative activities (% GI) of compounds 4d, 4e, 4o, 4p, and 4q against breast cancer cell lines.
2.2.5. Docking Study
In an attempt to introduce a reasonable explanation for the observed DAPK1 kinase activity with respect to the inhibitors’ binding affinity, the potent inhibitor 4j was subjected to a molecular docking study as a representative example of the designed inhibitors. In this study, Molecular Operating Environment (MOE, 2014) software was used to operate the docking protocol. The X-ray crystallographic structures of DAPK1 in complex with Genistein (PDB ID: 5AUZ) were downloaded from the protein data bank (PDB). Validation of the docking protocol was achieved by re-docking of the co-crystalized ligand, Genistein, in the binding site of DAPK1. The re-docked ligand retained the same binding manner in the DAPK1 active site (docking score= −6.1511 kcal/mol) with an RMSD value of 0.8894 Å. As illustrated in Figure 14A, the iso-flavone moiety of the native ligand, Genistein, is binding in the adenine pocket via H-bonding between the hydroxyl group and Glu100 residue while the pyran ring is additionally embedded in the pocket by AreneH interaction with Val27. Furthermore, the DAPK1 hydrophobic pocket is occupied by the lateral hydroxy phenyl moiety, which exhibits H-bonding between its para-hydroxyl group and the Glu94 residue. Accordingly, the target compounds should conserve the binding mode of the native ligand, which in turn would also be an indication of their binding affinity and enzymatic activity. In Figure 14B, docking of compound 4j in the active site of DAPK1 (docking score= -5.5545 kcal/mol) illustrates that the NH of the amide linker is introduced to the adenine pocket and forms H-bonding with Glu100, while the phenoxy moiety is inserted deeply in the pocket via Arene-H interaction with Val27. On the other hand, the pyridine ring, which is substituted on the amide linker, exhibits additional H-bonding with Arg150.
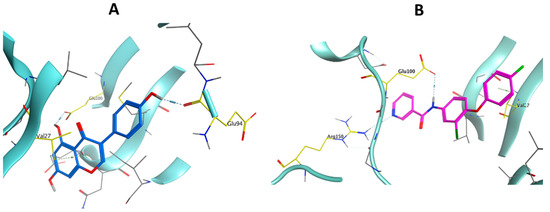
Figure 14.
3D representation of Genistein (A) and compound 4j (B) molecular interactions in the DAPK1 kinase.
3. Materials and Methods
3.1. Chemistry
General: All reactions and manipulations were performed in a nitrogen atmosphere using standard Schlenk techniques. All reaction solvents and reagents were purchased from commercial suppliers and used without further purification. The NMR spectra were obtained with a Bruker Avance 400 (400 MHz 1H and 100.6 MHz 13C NMR). 1H NMR spectra were referenced to tetramethylsilane (δ = 0.00 ppm) as an internal standard and were reported as follows: chemical shift, multiplicity (b = broad, s = singlet, d = doublet, t = triplet, dd = doublet of doublet, m = multiplet). Column chromatography was performed on Merck Silica Gel 60 (230–400 mesh) and eluting solvents for all of these chromatographic methods were noted as appropriated-mixed solvent with given volume-to-volume ratios. TLC was carried out using glass sheets pre-coated with silica gel 60 F254 purchased by Merk. For more details, see Supplementary Files
3.1.1. General Procedure of 2-Chloro-4-nitrophenoxybenzene Derivatives (2a–c)
Potassium carbonate (1.25 mmol) and the appropriate phenol (1 mmol) were added to a solution of commercially available 2-chloro-1-fluoro-4-nitrobenzene (1 mmol) in MeCN (12 mL). The reaction mixture was heated at 85 °C for 6 h. The mixture was extracted with EtOAc and water. The organic layer was dried over Na2SO4 and concentrated under reduced pressure to give the target compound.
- 2-Chloro-1-(4-fluorophenoxy)-4-nitrobenzene (2a)
Yellowish white solid, yield: 77%, mp: 76.3−92.4 °C, 1H NMR (400 MHz, CDCl3) δ 6.76 (d, J = 8.9 Hz, 1H), 7.00−7.09 (m, 4H), 7.98 (d, J = 8.9 Hz, 1H), 8.31 (s, 1H) [25,26].
- 2-Chloro-1-(3-fluorophenoxy)-4-nitrobenzene (2b)
Yellow oil, yield: 96%, 1H NMR (400 MHz, CDCl3) δ 6.80−6.87 (m, 2H), 6.95−7.00 (m, 2H), 7.40 (dd, J = 8.2 Hz, 14.6 Hz, 1H), 8.10 (dd, J = 2.7 Hz, 9.0 Hz, 1H), 8.40 (d, J = 2.7 Hz, 1H). 13C NMR (100.6 MHz, CDCl3) δ 107.68 (JC−F = 24.1 Hz), 112.49 (JC−F = 21.1 Hz), 115.27 (JC−F = 3.0 Hz), 117.91, 123.69, 125.39, 126.66, 131.27 (JC−F = 9.1 Hz), 143.29, 115.83 (JC−F = 10.1 Hz), 158.07, 163.58 (JC−F = 248.5 Hz) [27].
- 2-Chloro-1-(3-chlorophenoxy)-4-nitrobenzene (2c)
Yellow solid, yield: 90%, mp: 92.4−93.1 °C, 1H NMR (400 MHz, CDCl3) δ 6.95−6.98 (m, 2H), 7.09 (s, 1H), 7.24 (d, J = 8.0 Hz, 1H), 7.37 (t, J = 8.1 Hz, 1H), 8.09 (dd, J = 2.0 Hz, 9.0 Hz, 1H), 8.38 (d, J = 2.0 Hz, 1H). 13C NMR (100.6 MHz, CDCl3) δ 117.84, 117.91, 120.22, 123.71, 125.36, 125.71, 126.67, 131.15, 135.73, 143.28, 155.41, 158.07 [28].
3.1.2. General Procedure of 3-Choro-4-phenoxyaniline Derivatives (3a–c)
Pt/C catalyst was added to a solution of the appropriate 2-chloro-4-nitrophenoxybenzene derivative (0.1 mmol) in MeOH (15 mL) under hydrogen atmosphere. The mixture was stirred at room temperature for 6 h. The reaction mixture was filtered with celite and washed with MeOH. The filtrate was evaporated in vacuo. The residue was purified by column chromatography to give the target compound.
- 3-Chloro-4-(4-fluorophenoxy) aniline (3a)
Brown oil, yield: 65%, 1H NMR (400 MHz, DMSO-d6) δ 5.34 (s, 2H), 6.55 (d, J = 8.8 Hz, 1H), 6.72 (s, 1H), 6.80–6.84 (m, 2H), 6.90 (d, J = 8.6 Hz, 1H), 7.13 (t, J = 8.2 Hz, 2H) [29].
- 3-Chloro-4-(3-fluorophenoxy) aniline (3b)
Yellow oil, yield: 78%, 1H NMR (400 MHz, DMSO-d6) δ 5.40 (s, 2H), 6.57 (d, J = 8.6 Hz, 1H), 6.63 (d, J = 8.4 Hz, 2H), 6.73 (s, 1H), 6.86 (t, J = 8.5 Hz, 1H), 6.96 (d, J = 8.8 Hz, 1H), 7.33 (t, J = 8.0 Hz, 1H) [27,30].
- 3-Chloro-4-(3-chlorophenoxy) aniline (3c)
Brown oil, yield: 60%, 1H NMR (400 MHz, DMSO-d6) δ 5.41 (s, 2H), 6.58 (dd, J = 2.6 Hz, 8.7 Hz, 1H), 6.73 (d, J = 2.6 Hz, 1H), 6.77–6.82 (m, 2H), 6.96 (d, J = 8.6 Hz, 2H), 7.09 (dd, J = 1.0 Hz, 8.0 Hz, 1H), 7.34 (t, J = 8.1 Hz, 1H) [28,30].
3.1.3. General Procedure of Target Compounds 4a–r
The appropriate 3-chloro-4-phenoxyaniline derivative (0.1 mmol) was dissolved in THF (12 mL), followed by adding DIPEA (0.27 mmol), HATU (0.11 mmol), and the appropriate benzoic acid derivative (0.1 mmol). The reaction mixture was refluxed overnight. The mixture was extracted with EtOAc, water, and brine. The organic layer was dried over Na2SO4 and concentrated under reduced pressure. The residue was purified by column chromatography to give the target compound.
- N-(3-chloro-4-(4-fluorophenoxy) phenyl) picolinamide (4a)
Brown solid, mp: 95.6–97.0 °C, 1H NMR (400 MHz, CDCl3) δ 6.91–6.95 (m, 2H), 7.01 (dd, J = 8.8 Hz, 17.2 Hz, 3H), 7.49–7.52 (m, 1H), 7.60 (dd, J = 2.4 Hz, 8.8 Hz, 1H), 7.92 (t, J = 7.6 Hz, 1H), 8.03 (d, J = 2.4 Hz, 1H), 8.29 (d, J = 7.8 Hz, 1H), 8.61 (d, J = 4.6 Hz, 1H), 10.04 (s, 1H). 13C NMR (100.6 MHz, CDCl3) δ 116.30 (JC−F = 23.1 Hz), 118.91, 119.16 (JC−F = 31.2 Hz), 121.03, 121.97, 122.48, 126.22, 126.72, 134.55, 137.82, 148.05, 148.89, 149.37, 153.14, 158.69 (JC−F = 241.4 Hz), 162.02. HRMS (ESI) m/z calculated for C17H11ClFN3NaO2 [M+Na]+: 366.0416, found: 366.0408.
- N-(3-chloro-4-(4-fluorophenoxy) phenyl) pyridazine-3-carboxamide (4b)
White solid, mp: 134.1–134.9 °C, 1H NMR (400 MHz, CDCl3) δ 6.95–7.06 (m, 5H), 7.57 (d, J = 8.8 Hz, 1H), 7.77 (dd, J = 5.4 Hz, 7.9 Hz, 1H), 8.08 (s, 1H), 8.43 (d, J = 8.3 Hz, 1H), 9.36 (d, J = 4.4 Hz, 1H), 10.11 (s, 1H). 13C NMR (100.6 MHz, CDCl3) δ 116.37 (JC−F = 23.1 Hz), 119.15, 119.40 (JC−F = 34.2 Hz), 120.86, 122.32, 125.90, 126.23, 128.09, 133.80, 149.54, 157.27, 152.92, 153.17, 158.80 (JC−F = 241.4 Hz), 160.15. HRMS (ESI) m/z calculated for C17H11ClFN3NaO2 [M+Na]+: 366.0416, found: 366.0420.
- N-(3-chloro-4-(4-fluorophenoxy) phenyl) pyrazine-2-carboxamide (4c)
White solid, mp: 155.5–156.0 °C, 1H NMR (400 MHz, CDCl3) δ 6.92–7.05 (m, 5H), 7.59 (dd, J = 2.4 Hz, 8.8 Hz, 1H), 7.99 (d, J = 2.4 Hz, 1H), 8.60 (s, 1H), 8.84 (d, J = 2.2 Hz, 1H), 9.51 (s, 1H), 9.67 (s, 1H). 13C NMR (100.6 MHz, CDCl3) δ 116.35 (JC−F = 23.1 Hz), 119.10, 119.33 (JC−F = 27.2 Hz), 120.86, 122.18, 126.20, 133.85, 142.40, 143.98, 144.72, 147.81, 149.45, 152.92, 158.79 (JC−F = 242.4 Hz), 160.67. HRMS (ESI) m/z calculated for C18H13ClFN2O2 [M+H]+: 343.0644, found: 343.0638.
- N-(3-chloro-4-(4-fluorophenoxy) phenyl) nicotinamide (4d)
White solid, mp: 112.0–113.2 °C, 1H NMR (400 MHz, CDCl3) δ 6.89–6.93 (m, 3H), 7.02 (t, J = 9.1 Hz, 2H), 7.38–7.41 (m, 1H), 7.46 (dd, J = 2.5 Hz, 8.8 Hz, 1H), 7.83 (d, J = 2.4 Hz, 1H), 8.18 (dt, J = 1.8 Hz, 8.0 Hz, 1H), 8.71 (dd, J = 1.5 Hz, 4.8 Hz, 1H), 9.05 (s, 1H). 13C NMR (100.6 MHz, CDCl3) δ 116.38 (JC−F = 23.1 Hz), 118.85, 119.26 (JC−F = 8.0 Hz), 120.49 (JC−F = 12.1 Hz), 123.15, 123.81, 125.91, 130.43, 130.04, 135.57, 147.91, 149.70, 152.52, 152.77, 158.82 (JC−F = 241.4 Hz), 164.20. HRMS (ESI) m/z calculated for C17H12ClFN3O2 [M+H]+: 344.0597, found: 343.0600.
- N-(3-chloro-4-(4-fluorophenoxy) phenyl) isonicotinamide (4e)
White solid, mp: 57.0–58.5 °C, 1H NMR (400 MHz, CDCl3) δ 6.90–6.93 (m, 3H), 7.00–7.04 (m, 2H), 7.46 (dd, J = 2.5 Hz, 8.8 Hz, 1H), 7.68–7.70 (m, 2H), 7.83 (d, J = 2.5 Hz, 1H), 8.70–8.71 (m, 3H). 13C NMR (100.6 MHz, CDCl3) δ 116.41 (JC−F = 24.1 Hz), 118.82, 119.35 (JC−F = 9.1 Hz), 120.43 (JC−F = 10.1 Hz), 123.12, 125.91, 133.74, 141.74, 149.92, 150.58, 152.68, 158.87 (JC−F = 241.4 Hz), 164.10. HRMS (ESI) m/z calculated for C18H13ClFN2O2 [M+H]+: 343.0644, found: 343.0640.
- N-(3-chloro-4-(4-fluorophenoxy) phenyl) pyridazine-4-carboxamide (4f)
White solid, mp: 164.2–165.2 °C, 1H NMR (400 MHz, DMSO-d6) δ 7.02 (s, 2H), 7.16–7.23 (m, 3H), 7.69–7.79 (m, 1H), 8.11 (d, J = 6.1 Hz, 2H), 9.52 (d, J = 4.6 Hz, 1H), 9.65 (s, 1H), 10.89 (s, 1H). 13C NMR (100.6 MHz, DMSO-d6) δ 117.07 (JC−F = 23.1 Hz), 119.40 (JC−F = 8.0 Hz), 121.19, 121.88, 122.61 (JC−F = 11.1 Hz), 124.90, 125.01, 132.39, 135.97, 148.45, 149.34, 152.62, 153.35, 158.46 (JC−F = 225.3 Hz), 162.98.
- N-(3-chloro-4-(3-fluorophenoxy) phenyl) picolinamide (4g)
White solid, mp: 79.0–80.0 °C, 1H NMR (400 MHz, CDCl3) δ 6.65 (dt, J = 2.4 Hz, 10.2 Hz, 1H), 6.72 (dd, J = 2.2 Hz, 8.3 Hz, 1H), 6.75–6.80 (m, 1H), 7.09 (d, J = 8.8 Hz, 1H), 7.23–7.28 (m, 1H), 7.48–7.51 (m, 1H), 7.64 (dd, J = 2.6 Hz, 8.8 Hz, 1H), 7.92 (td, J = 1.7 Hz, 7.7 Hz, 1H), 8.05 (d, J = 2.6 Hz, 1H), 8.29 (d, J = 7.8 Hz, 1H), 8.61 (dt, J = 0.64 Hz, 4.12 Hz, 1H), 10.08 (s, 1H). 13C NMR (100.6 MHz, CDCl3) δ 104.73 (JC−F = 25.2 Hz), 109.76 (JC−F = 21.1 Hz), 112.49 (JC−F = 3.0 Hz), 119.36, 121.91, 122.44, 122.50, 126.77, 127.05, 130.53 (JC−F = 10.1 Hz), 135.34, 137.82, 147.49, 148.07, 149.30, 158.83 (JC−F = 10.1 Hz), 162.07, 163.55 (JC−F = 246.5 Hz). HRMS (ESI) m/z calculated for C17H12ClFN3O2 [M+H]+: 344.0597, found: 344.0601.
- N-(3-chloro-4-(3-fluorophenoxy) phenyl) pyridazine-3-carboxamide (4h)
White solid, mp: 130.3–131.2 °C, 1H NMR (400 MHz, CDCl3) δ 6.65–6.82 (m, 3H), 7.12 (d, J = 8.8 Hz, 1H), 7.27 (dd, J = 8.0 Hz, 15.0 Hz, 1H), 7.62 (dd, J = 1.8 Hz, 8.6 Hz, 1H), 7.75–7.79 (m, 1H), 8.10 (d, J = 1.8 Hz, 1H), 8.44 (d, J = 8.3 Hz, 1H), 9.37 (d, J = 4.3 Hz, 1H), 10.16 (s, 1H). 13C NMR (100.6 MHz, CDCl3) δ 104.90 (JC−F = 25.2 Hz), 109.93 (JC−F = 21.1 Hz), 112.64 (JC−F = 3.0 Hz), 119.62, 120.33, 121.61, 122.33 (JC−F = 9.1 Hz), 125.91, 127.13, 128.09, 130.55 (JC−F = 10.1 Hz), 134.60, 148.13, 152.22, 153.19, 160.19, 160.5 (JC−F = 366.2 Hz).
- N-(3-chloro-4-(3-fluorophenoxy) phenyl) pyrazine-2-carboxamide (4i)
White solid, mp: 156.6–157.2 °C, 1H NMR (400 MHz, DMSO-d6) δ 6.75 (d, J = 8.4 Hz, 1H), 6.83 (d, J = 10.3 Hz, 1H), 6.96 (t, J = 8.2 Hz, 1H), 7.30 (d, J = 8.9 Hz, 1H), 7.41 (dd, J = 8.2 Hz, 15.4 Hz, 1H), 7.95–7.97 (m, 1H), 8.28 (d, J = 2.0 Hz, 1H), 8.85 (s, 1H), 8.97 (d, J = 2.1 Hz, 1H), 9.33 (s, 1H), 11.05 (s, 1H). 13C NMR (100.6 MHz, DMSO-d6) δ 104.75 (JC−F = 25.2 Hz), 110.26 (JC−F = 21.1 Hz), 112.87, 120.26, 121.45, 122.87 (JC−F = 31.2 Hz), 125.56, 131.81 (JC−F = 10.1 Hz), 136.70, 143.74, 144.63, 145.21, 146.89, 148.37, 158.88 (JC−F = 11.1 Hz), 162.45, 163.38 (JC−F = 244.5 Hz). HRMS (ESI) m/z calculated for C17H11ClFN3NaO2 [M+Na]+: 366.0416, found: 366.0415.
- N-(3-chloro-4-(3-fluorophenoxy) phenyl) nicotinamide (4j)
White solid, mp: 117.3–118.2 °C, 1H NMR (400 MHz, MeOD) δ 6.63 (dt, J = 2.3 Hz, 10.4 Hz, 1H), 6.69 (dd, J = 2.3 Hz, 8.4 Hz, 1H), 6.81 (td, J = 1.9 Hz, 8.4 Hz, 1H), 7.12 (d, J = 8.8 Hz, 1H), 7.27–7.32 (m, 1H), 7.55–7.59 (m, 1H), 7.65 (dd, J = 2.5 Hz, 8.8 Hz, 1H), 8.02 (d, J = 2.5 Hz, 1H), 8.34 (dt, J = 1.8 Hz, 8.1 Hz, 1H), 8.72 (dd, J = 1.5 Hz, 5.0 Hz, 1H), 9.09 (d, J = 1.5 Hz, 1H). HRMS (ESI) m/z calculated for C18H13ClFN2O2 [M+H]+: 343.0644, found: 343.0648.
- N-(3-chloro-4-(3-fluorophenoxy) phenyl) isonicotinamide (4k)
White solid, mp: 49.8–51.0 °C, 1H NMR (400 MHz, CDCl3) δ 6.63 (d, J = 10.1 Hz, 1H), 6.70 (d, J = 8.2 Hz, 1H), 6.75–6.81 (m, 1H), 7.04 (t, J = 8.7 Hz, 1H), 7.26 (dd, J = 8.2 Hz, 14.9 Hz, 1H), 7.52–7.62 (m, 1H), 7.70 (d, J = 4.8 Hz, 2H), 7.88 (d, J = 1.8 Hz, 1H), 8.71 (d, J = 5.0 Hz, 2H), 8.81 (s, 1H). 13C NMR (100.6 MHz, CDCl3) δ 104.95 (JC−F = 24.1 Hz), 110.07 (JC−F = 21.1 Hz), 112.73, 120.24 (JC−F = 25.2 Hz), 121.11, 122.10, 122.49, 123.02, 126.88, 130.61 (JC−F = 10.1 Hz), 134.71, 148.43, 150.57, 158.43 (JC−F = 10.1 Hz), 163.21 (JC−F = 187.1 Hz), 164.74. HRMS (ESI) m/z calculated for C18H13ClFN2O2 [M+H]+: 343.0644, found: 343.0639.
- N-(3-chloro-4-(3-fluorophenoxy) phenyl) pyridazine-4-carboxamide (4l)
Yellow solid, mp: 141.3–142.7 °C, 1H NMR (400 MHz, CDCl3) δ 6.64 (d, J = 10.1 Hz, 1H), 6.72 (d, J = 8.4 Hz, 1H), 6.80 (t, J = 8.1 Hz, 1H), 7.06 (d, J = 8.7 Hz, 1H), 7.27 (dd, J = 8.0 Hz, 15.0 Hz, 1H), 7.58–7.68 (m, 1H), 7.95–8.02 (m, 2H), 9.30 (d, J = 5.0 Hz, 1H), 9.59 (s, 1H), 9.82 (s, 1H). 13C NMR (100.6 MHz, CDCl3) δ 105.04 (JC−F = 25.2 Hz), 110.15 (JC−F = 21.1 Hz), 112.84 (JC−F = 3.0 Hz), 120.07, 120.56, 122.05, 123.22, 125.23, 126.87, 130.63 (JC−F = 10.1 Hz), 133.09, 134.57, 148.67, 151.96, 158.29, 162.13, 163.52 (JC−F = 247.5 Hz). HRMS (ESI) m/z calculated for C17H12ClFN3O2 [M+H]+: 344.0597, found: 344.0596.
- N-(3-chloro-4-(3-chlorophenoxy) phenyl) picolinamide (4m)
Brown solid, mp: 92.7–93.5 °C, 1H NMR (400 MHz, CDCl3) δ 6.82 (dd, J = 1.9 Hz, 8.3 Hz, 1H), 6.92 (t, J = 2.1 Hz, 1H), 7.03–7.08 (m, 2H), 7.22 (t, J = 8.1 Hz, 1H), 7.47–7.50 (m, 1H), 7.63 (dd, J = 2.5 Hz, 8.8 Hz, 1H), 7.90 (td, J = 1.6 Hz, 7.7 Hz, 1H), 8.05 (d, J = 2.5 Hz, 1H), 8.28 (d, J = 7.8 Hz, 1H), 8.59 (d, J = 4.5 Hz, 1H), 10.07. 13C NMR (100.6 MHz, CDCl3) δ 115.18, 119.38, 121.91, 122.35, 122.47, 123.09, 126.76, 126.99, 130.52, 135.11, 135.35, 137.80, 147.43, 148.06, 149.27, 158.27, 162.06. HRMS (ESI) m/z calculated for C18H13Cl2N2O2 [M+H]+: 359.0349, found: 359.0346.
- N-(3-chloro-4-(3-chlorophenoxy) phenyl) pyridazine-3-carboxamide (4n)
Light brown solid, mp: 138.0–139.6 °C, 1H NMR (400 MHz, DMSO-d6) δ 6.90 (dd, J = 1.9 Hz, 8.2 Hz, 1H), 7.01 (t, J = 2.0 Hz, 1H), 7.18 (d, J = 8.0 Hz, 1H), 7.31 (d, J = 8.9 Hz, 1H), 7.40 (t, J = 8.1 Hz, 1H), 7.99–8.02 (m, 2H), 8.32–8.36 (m, 2H), 9.50 (dd, J = 1.4 Hz, 4.9 Hz, 1H), 11.40 (s, 1H). 13C NMR (100.6 MHz, DMSO-d6) δ 115.63, 116.98, 121.63, 122.86, 123.12, 123.46, 125.59, 126.67, 129.19, 131.96, 134.50, 136.81, 146.78, 153.40, 154.13, 158.49, 162.23. HRMS (ESI) m/z calculated for C17H11Cl2N3NaO2 [M+Na]+: 382.0121, found: 382.0123.
- N-(3-chloro-4-(3-chlorophenoxy) phenyl) pyrazine-2-carboxamide (4o)
Light brown solid, mp: 153.2–154.6 °C, 1H NMR (400 MHz, DMSO-d6) δ 6.90 (d, J = 8.0 Hz, 1H), 7.00 (s, 1H), 7.19 (d, J = 8.1 Hz, 1H), 7.30 (d, J = 8.8 Hz, 1H), 7.40 (t, J = 8.0 Hz, 1H), 7.96 (dd, J = 2.0 Hz, 8.8 Hz, 1H), 8.28 (d, J = 1.8 Hz, 1H), 8.84 (s, 1H), 8.96 (s, 1H), 9.32 (s, 1H), 11.05 (s, 1H). 13C NMR (100.6 MHz, DMSO-d6) δ 115.61, 116.98, 121.47, 122.72, 123.08, 123.45, 125.59, 131.94, 134.50, 136.76, 143.73, 144.63, 145.17, 146.75, 148.37, 158.47, 162.43. HRMS (ESI) m/z calculated for C17H12Cl2N3O2 [M+H]+: 360.0301, found: 360.0304.
- N-(3-chloro-4-(3-chlorophenoxy) phenyl) nicotinamide (4p)
White solid, mp: 141.0–141.5 °C, 1H NMR (400 MHz, DMSO-d6) δ 6.89 (dd, J = 1.6 Hz, 8.4 Hz, 1H), 6.99 (s, 1H), 7.18 (d, J = 9.0 Hz, 1H), 7.30 (d, J = 8.8 Hz, 1H), 7.40 (t, J = 8.2 Hz, 1H), 7.58–7.61 (m, 1H), 7.77 (dd, J = 2.4 Hz, 8.9 Hz, 1H), 8.14 (d, J = 2.4 Hz, 1H), 8.30 (d, J = 8.1 Hz, 1H), 8.78–8.79 (m, 1H), 9.12 (d, J = 1.6 Hz, 1H), 10.66 (s, 1H). 13C NMR (100.6 MHz, DMSO-d6) δ 115.61, 116.93, 121.10, 122.34, 123.23, 123.45, 124.04, 125.65, 130.68, 131.96, 134.51, 135.97, 137.39, 146.53, 149.16, 152.83, 158.53, 164.73. HRMS (ESI) m/z calculated for C18H13Cl2N2O2 [M+H]+: 359.0349, found: 359.0351.
- N-(3-chloro-4-(3-chlorophenoxy) phenyl) isonicotinamide (4q)
White solid, mp: 124.3–126.0 °C, 1H NMR (400 MHz, DMSO-d6) δ 6.91–7.00 (m, 2H), 7.19 (s, 1H), 7.28–7.40 (m, 2H), 7.78–7.88 (m, 3H), 8.15 (s, 1H), 8.82 (s, 2H), 10.72 (s, 1H).
- N-(3-chloro-4-(3-chlorophenoxy) phenyl) pyridazine-4-carboxamide (4r)
Yellow solid, mp: 133.2–134.0 °C, 1H NMR (400 MHz, DMSO-d6) δ 6.91 (dd, J = 2.2 Hz, 8.3 Hz, 1H), 7.01 (s, 1H), 7.20 (d, J = 8.0 Hz, 1H), 7.32 (d, J = 8.8 Hz, 1H), 7.42 (t, J = 8.2 Hz, 1H), 7.76 (dd, J = 2.4 Hz, 8.9 Hz, 1H), 8.13 (d, J = 2.3 Hz, 2H), 9.53 (d, J = 5.2 Hz, 1H), 9.67 (s, 1H), 10.92 (s, 1H). 13C NMR (100.6 MHz, DMSO-d6) δ 115.74, 117.09, 121.22, 122.49, 123.18, 123.57, 125.01, 125.65, 131.97, 132.34, 134.53, 136.74, 147.03, 149.32, 152.62, 158.38, 163.04. HRMS (ESI) m/z calculated for C17H12Cl2N3O2 [M+H]+: 360.0301, found: 360.0304.
3.2. Biological Evaluation
3.2.1. In Vitro Kinase Screening
Kinase assays were performed at the Reaction Biology Corporation (Malvern, PA) using the “HotSpot” assay platform. All the target compounds were evaluated according to the Reaction Biology Corp implemented protocol as reported on their website (http://www.reactionbiology.com/ 25 January 2019).
3.2.2. NCI Cell Line Screening
The target compounds were evaluated for their anti-proliferative activity at the National Cancer Institute (NCI), Bethesda, Maryland, USA, applying the standard protocol of the NCI (http://www.dtp.nci.nih.gov/).
3.2.3. Docking Methodology
The X-ray crystal structures of DAPK1 kinase in complex with Genistein (PDB ID: 5AUZ) were downloaded from the protein data bank (www.rcsb.org) in PDB format. The 2D structure of compound 4j was drawn using ChemDraw software. Molecular Operating Environment (MOE, 2014.0901) software was used for the molecular docking operation docking protocol. DAPK1 kinase was prepared for the molecular docking procedure by applying 3D protonation of both enzyme amino acids and the native ligand (Genistein). In addition, water from crystallization was removed from the kinase domain. The active site was isolated. The grid space was determined from the coordinates of the reference ligand (−22.780412, 2.367623, −10.682060) and expanded to cover all amino acid residues in the DAPK1 active site cavity (r = 8.383472). The docking protocol was validated by re-docking of Genistein with RMSD = 0.8894 Å and retained the same binding pose in the DAPK1 active site.
4. Conclusions
In the current study, a series of 3-chloro-4-(phenoxy)phenyl carboxamide derivatives featuring different substituents on the terminal phenoxy group, along with diverse nitrogen containing heterocycles on the carboxamide linker, has been designed and synthesized as DAPK1 inhibitors with potential anti-proliferative activity. A preliminary kinase inhibition evaluation was conducted on the first synthesized compound 4a in an attempt to explore its DAPK1 activity and selectivity. Among the 45 panel kinases, compound 4a exhibited higher activity and selectivity against DAPK1 kinase. Hence, DAPK1 inhibitory activity of the synthesized derivatives was detected; pyridinyl moiety owing derivatives 4h, 4j, 4f, and 4q displayed the highest activity and were subjected to 10-dose evaluation. The IC50 values of the mentioned compounds were determined and compound 4q emerged to be the most potent DAPK1 inhibitor (IC50 =1.09 µM). Additionally, all the target compounds were evaluated for their cytotoxic activity against a panel of 60 NCI cancer cell lines at 10 µM concentration. Out of the tested series, compounds 4d, 4e, 4o, and 4p revealed the highest anti-proliferative activities against most of the employed cell lines (67.02%, 67.87%, 70.83%, and 69.27%. Moreover, two leukemia cell lines (HL-60(TB) and K-562) exhibited the highest sensitivity towards the tested candidates.
Supplementary Materials
The following are available online at: https://www.mdpi.com/article/10.3390/ph15091050/s1.
Author Contributions
Conceptualization, A.E., A.H.E.H. and E.J.R.; methodology, S.P. and S.J.K., validation, A.E.; formal analysis, A.E. and E.M.H.A.; investigation, K.L. and E.J.R.; resources, A.E., K.L. and E.J.R.; data curation, A.E.; writing—original draft preparation, A.E., S.P. and E.M.H.A.; writing—review and editing, A.E. and E.M.H.A.; supervision, E.J.R.; project administration, A.E.; funding acquisition, E.J.R. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the KIST Institutional programs (Grant No. 2E 31624) from the Korea Institute of Science and Technology and the Creative Fusion Research Program through the Creative Allied Project funded by the National Research Council of Science & Technology (CAP-12-1-KIST). This research was supported by National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (No. 2018R1A5A2023127) and the BK21 FOUR program through NRF funded by the Ministry of Education of Korea.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data is contained within the article and supplementary material.
Conflicts of Interest
The authors have declared no conflict of interest.
References
- Deiss, L.P.; Feinstein, E.; Berissi, H.; Cohen, O.; Kimchi, A. Identification of a novel serine/threonine kinase and a novel 15-kD protein as potential mediators of the gamma interferon-induced cell death. Genes Dev. 1995, 9, 15–30. [Google Scholar] [CrossRef] [PubMed]
- Bialik, S.; Kimchi, A. The death-associated protein kinases: Structure, function, and beyond. Annu. Rev. Biochem. 2006, 75, 189–210. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Ravanan, P.; Talwar, P. Death Associated Protein Kinase 1 (DAPK1): A Regulator of Apoptosis and Autophagy. Front. Mol. Neurosci. 2016, 9, 46. [Google Scholar] [CrossRef] [PubMed]
- Farag Ahmed, K.; Roh Eun, J. Death-associated protein kinase (DAPK) family modulators: Current and future therapeutic outcomes. Med. Res. Rev. 2018, 39, 349–385. [Google Scholar] [CrossRef]
- Pelled, D.; Raveh, T.; Riebeling, C.; Fridkin, M.; Berissi, H.; Futerman, A.H.; Kimch, A. Death-associated protein (DAP) kinase plays a central role in ceramide-induced apoptosis in cultured hippocampal neurons. J. Biol. Chem. 2002, 277, 1957–1961. [Google Scholar] [CrossRef]
- Bialik, S.; Kimchi, A. DAP-kinase as a target for drug design in cancer and diseases associated with accelerated cell death. Semin. Cancer Biol. 2004, 14, 283–294. [Google Scholar] [CrossRef]
- Hainsworth, A.H.; Allsopp, R.C.; Jim, A.; Potter, J.F.; Lowe, J.; Talbot, C.J.; Prettyman, R.J. Death-associated protein kinase (DAPK1) in cerebral cortex of late-onset Alzheimer’s disease patients and aged controls. Neuropathol. Appl. Neurobiol. 2010, 36, 17–24. [Google Scholar] [CrossRef]
- Gade, P.; Manjegowda, S.B.; Nallar, S.C.; Maachani, U.B.; Cross, A.S.; Kalvakolanu, D.V. Regulation of the Death-Associated Protein Kinase 1 Expression and Autophagy via ATF6 Requires Apoptosis Signal-Regulating Kinase 1. Mol. Cell. Biol. 2014, 34, 4033–4048. [Google Scholar] [CrossRef]
- Gozuacik, D.; Bialik, S.; Raveh, T.; Mitou, G.; Shohat, G.; Sabanay, H.; Mizushima, N.; Yoshimori, T.; Kimchi, A. DAP-kinase is a mediator of endoplasmic reticulum stress-induced caspase activation and autophagic cell death. Cell Death Differ. 2008, 15, 1875–1886. [Google Scholar] [CrossRef]
- Shohat, G.; Spivak-Kroizman, T.; Cohen, O.; Bialik, S.; Shani, G.; Berrisi, H.; Eisenstein, M.; Kimchi, A. The Pro-apoptotic Function of Death-associated Protein Kinase Is Controlled by a Unique Inhibitory Autophosphorylation-based Mechanism. J. Biol. Chem. 2001, 276, 47460–47467. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Grupe, A.; Rowland, C.; Nowotny, P.; Kauwe, J.S.K.; Smemo, S.; Hinrichs, A.; Tacey, K.; Toombs, T.A.; Kwok, S.; et al. DAPK1 variants are associated with Alzheimer’s disease and allele-specific expression. Hum. Mol. Genet. 2006, 15, 2560–2568. [Google Scholar] [CrossRef] [PubMed]
- Mor, I.; Carlessi, R.; Ast, T.; Feinstein, E.; Kimchi, A. Death-associated protein kinase increases glycolytic rate through binding and activation of pyruvate kinase. Oncogene 2011, 31, 683. [Google Scholar] [CrossRef]
- Velentza, A.V.; Wainwright, M.S.; Zasadzki, M.; Mirzoev, S.; Schumacher, A.M.; Haiech, J.; Focia, P.J.; Egli, M.; Watterson, D.M. An aminopyridazine-based inhibitor of a pro-apoptotic protein kinase attenuates hypoxia-ischemia induced acute brain injury. Bioorganic Med. Chem. Lett. 2003, 13, 3465–3470. [Google Scholar] [CrossRef]
- Herce, H.D.; Deng, W.; Helma, J.; Leonhardt, H.; Cardoso, M.C. Visualization and targeted disruption of protein interactions in living cells. Nat. Commun. 2013, 4, 2660. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Wang, Y.; Zhang, X.; Zhang, W.; Guo, S.; Jin, F. Recent progress of cell-penetrating peptides as new carriers for intracellular cargo delivery. J. Control. Release 2014, 174, 126–136. [Google Scholar] [CrossRef] [PubMed]
- Watterson, D.M.; Velentza, A.V.; Zasadzki, M.; Craft, J.M.; Haiech, J.; van Eldik, L.J. Discovery of a new class of synthetic protein kinase inhibitors that suppress selective aspects of glial activation and protect against β-amyloid induced injury. J. Mol. Neurosci. 2003, 20, 411–423. [Google Scholar] [CrossRef]
- Mirzoeva, S.; Sawkar, A.; Zasadzki, M.; Guo, L.; Velentza, A.V.; Dunlap, V.; Bourguignon, J.-J.; Ramstrom, H.; Haiech, J.; van Eldik, L.J.; et al. Discovery of a 3-Amino-6-phenyl-pyridazine Derivative as a New Synthetic Antineuroinflammatory Compound. J. Med. Chem. 2002, 45, 563–566. [Google Scholar] [CrossRef]
- Wilbek, T.S.; Skovgaard, T.; Sorrell, F.J.; Knapp, S.; Berthelsen, J.; Strømgaard, K. Identification and characterization of a small-molecule inhibitor of death-associated protein kinase 1. ChemBioChem 2015, 16, 59–63. [Google Scholar] [CrossRef]
- Okamoto, M.; Takayama, K.; Shimizu, T.; Ishida, K.; Takahashi, O.; Furuya, T. Identification of Death-Associated Protein Kinases Inhibitors Using Structure-Based Virtual Screening. J. Med. Chem. 2009, 52, 7323–7327. [Google Scholar] [CrossRef]
- Al-Ghabkari, A.; Deng, J.-T.; McDonald, P.C.; Dedhar, S.; Alshehri, M.; Walsh, M.P.; MacDonald, J.A. A novel inhibitory effect of oxazol-5-one compounds on ROCKII signaling in human coronary artery vascular smooth muscle cells. Sci. Rep. 2016, 6, 32118. [Google Scholar] [CrossRef] [PubMed]
- Carlson, D.A.; Franke, A.S.; Weitzel, D.H.; Speer, B.L.; Hughes, P.F.; Hagerty, L.; Fortner, C.N.; Veal, J.M.; Barta, T.E.; Zieba, B.J.; et al. Fluorescence Linked Enzyme Chemoproteomic Strategy for Discovery of a Potent and Selective DAPK1 and ZIPK Inhibitor. ACS Chem. Biol. 2013, 8, 2715–2723. [Google Scholar] [CrossRef] [PubMed]
- MacDonald, J.A.; Sutherland, C.; Carlson, D.A.; Bhaidani, S.; Al-Ghabkari, A.; Swärd, K.; Haystead, T.A.J.; Walsh, M.P. A Small Molecule Pyrazolo[3,4-d]Pyrimidinone Inhibitor of Zipper-Interacting Protein Kinase Suppresses Calcium Sensitization of Vascular Smooth Muscle. Mol. Pharmacol. 2016, 89, 105–117. [Google Scholar] [CrossRef] [PubMed]
- Irie, T.; Sawa, M. 7-Azaindole: A Versatile Scaffold for Developing Kinase Inhibitors. Chem. Pharm. Bull. 2018, 66, 29–36. [Google Scholar] [CrossRef]
- Okamoto, M.; Takayama, K.; Shimizu, T.; Muroya, A.; Furuya, T. Structure–activity relationship of novel DAPK inhibitors identified by structure-based virtual screening. Bioorganic Med. Chem. 2010, 18, 2728–2734. [Google Scholar] [CrossRef]
- Yang, Z.-X.; Chen, H.B.; Zou, Y.-Z.; Hou, Y.; Wang, T.-P.; Liang, Y. Synthesis of 7-chloro-4-hydroxy-6-(phenoxy)-3-quinolinecarboxylic acid ethyl ester derivatives and determination of their activity as coccidiostats. Youji Huaxue 2008, 28, 432–435. [Google Scholar]
- Marafie, J.A.; Moseley, J.D. The application of stop-flow microwave technology to scaling-out SNAr reactions using a soluble organic base. Org. Biomol. Chem. 2010, 8, 2219–2227. [Google Scholar] [CrossRef]
- Hennequin, L.F.A.; Chen, H.; Zou, Y.; Zhu, J.; Wang, Y. Preparation of Quinazolines as Antitumor Agents; AstraZeneca AB: Södertälje, Sweden; AstraZeneca UK Limited: Cambridge, UK, 2003; p. 218. [Google Scholar]
- Bolea, C. Preparation of Amido Derivatives and Their Use as Positive Allosteric Modulators of Metabotropic Glutamate Receptors; Addex Pharma S.A.: Geneva, Switzerland, 2009; p. 68. [Google Scholar]
- Yang, Z.; Chen, H.; Zou, Y.; Zhu, J.; Wang, Y. Method for Synthesizing 6-aryloxy-7-chloro-4-hydroxyl-3-quinolinecarboxylate. CN 200610022419, 11 July 2007. [Google Scholar]
- Ishikawa, T.; Seto, M.; Banno, H.; Kawakita, Y.; Oorui, M.; Taniguchi, T.; Ohta, Y.; Tamura, T.; Nakayama, A.; Miki, H. Design and Synthesis of Novel Human Epidermal Growth Factor Receptor 2 (HER2)/Epidermal Growth Factor Receptor (EGFR) Dual Inhibitors Bearing a Pyrrolo [3,2-d] pyrimidine Scaffold. J. Med. Chem. 2011, 54, 8030–8050. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).