Predictiveness of the Human-CYP3A4-Transgenic Mouse Model (Cyp3aXAV) for Human Drug Exposure of CYP3A4-Metabolized Drugs
Abstract
1. Introduction
2. Results
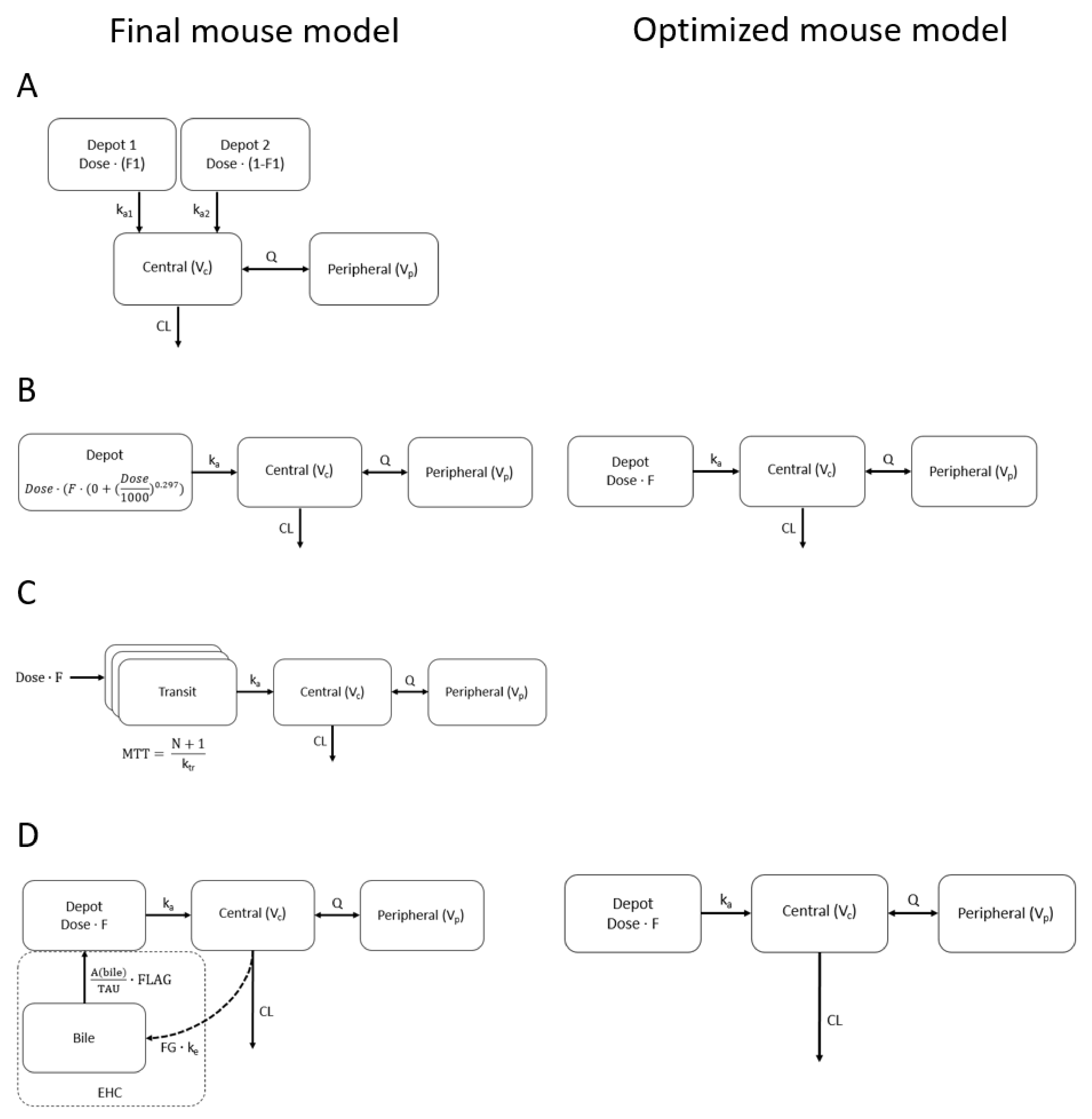
2.1. Mouse Population PK Models
2.2. Redundant Model Properties
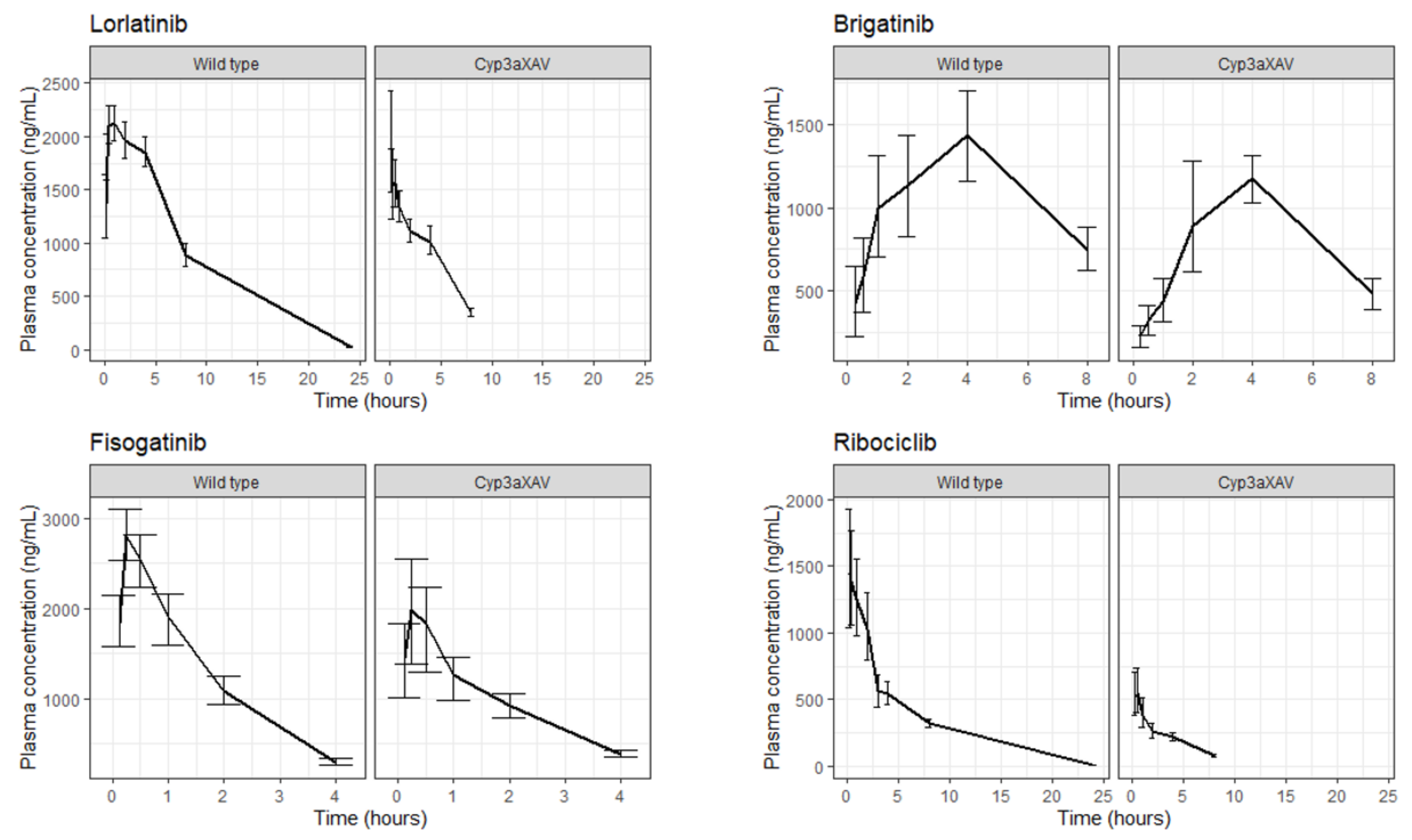
2.3. AUCinf, Cmax and Prediction Interval Comparison
3. Discussion
4. Methods
4.1. Data
4.2. Population PK Models for Cyp3aXAV and WT Mouse Strains
4.3. Extrapolation
| Lorlatinib [39,40] | Brigatinib [40,41] | Fisogatinib [26,40] | Ribociclib [40,42] | |
|---|---|---|---|---|
| Primary enzymes | CYP3A4, UGT1A4 | CYP3A4, CYP2C8 | CYP3A4 | CYP3A4, several phase-2 enzymes |
| Elimination | With feces ~41% (~9% unchanged), with urine ~48% (mostly as metabolite) | With feces, ~65% (~41% unchanged), with urine ~25% (~86% unchanged) | NA | With feces ~69% (~17% unchanged), with urine ~23% (~12% unchanged) |
| Protein binding | 66% | 91% | NA | 70% |
| Volume of distribution (L) | 390 | 307 | NA | 1090 |
| pKa | 5.71 (basic) | 8.54 (basic) | 3.79 (basic) | 8.87 (basic) |
| Water solubility (mg/mL) | 0.108 | 0.022 | 0.004 | 0.231 |
| LogP | 1.63 | 5.17 | 3.86 | 2.38 |
| Molecular mass (g/mol) | 406.4 | 584.1 | 503.4 | 434.5 |
4.4. Simulations
4.5. Comparison of Model-Derived AUCinf, Cmax and PK profiles
4.6. Software
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| CYP | cytochromes P450 |
| WT | wild-type |
| Cyp3aXAV | human CYP3A4 transgenic |
| PK | pharmacokinetic |
| ADME | absorption, distribution, metabolism and excretion |
| FIH | first-in-human |
| CL, | clearance |
| F | bioavailability |
| dOFVs | difference in objective function values |
| AUCinf | area under the plasma concentration–time curve from 0 to infinite time |
| Cmax | maximum concentration |
| EHC | enterohepatic circulation |
| RSE | relative standard error |
References
- Wong, C.H.; Siah, K.W.; Lo, A.W. Estimation of clinical trial success rates and related parameters. Biostatistics 2019, 20, 273–286. [Google Scholar] [CrossRef] [PubMed]
- Olson, H.; Betton, G.; Robinson, D.; Thomas, K.; Monro, A.; Kolaja, G.; Lilly, P.; Sanders, J.; Sipes, G.; Bracken, W.; et al. Concordance of the toxicity of pharmaceuticals in humans and in animals. Regul. Toxicol. Pharmcol. 2000, 32, 56–67. [Google Scholar] [CrossRef] [PubMed]
- Tomaszewski, J.E.; Doroshow, J.H. Preclinical Development of Molecularly Targeted Agents in Oncology. In Molecular Targeting in Oncology; Cancer Drug Discovery and Development; Kaufman, H.L., Wadler, S., Antman, K., Eds.; Humana Press: Totowa, NJ, USA, 2008; pp. 707–722. [Google Scholar] [CrossRef]
- Zhang, D.; Luo, G.; Ding, X.; Lu, C. Preclinical experimental models of drug metabolism and disposition in drug discovery and development. Acta Pharm. Sin. B 2012, 2, 549–561. [Google Scholar] [CrossRef]
- International Council for Harmonisation of Technical Requirements for Pharmaceuticals for Human Use. Safety Guidelines—S3 Toxicokinetics and Pharmacokinetics—Note for Guidance on Toxicokinetics: The Assessment of Systemic Exposure in Toxicity Studies s3a; International Council for Harmonisation of Technical Requirements for Pharmaceuticals for Human Use: Geneva, Switzerland, 1994. [Google Scholar]
- Committee for Medicinal Products for Human Use; European Medicines Agency. Guideline on Requirements for First-in-Man Clinical Trials for Potential High-Risk Medicinal Products; Committee for Medicinal Products for Human Use: Amsterdam, The Netherlands, 2007. [Google Scholar]
- US Food and Drug Administration; Center for Drug Evaluation and Research. Guidance for Industry Estimating the Maximum Safe Starting Dose in Initial Clinical Trials for Therapeutics in Adult Healthy Volunteers; US Food and Drug Administration: Silver Spring, MD, USA, 2005. [Google Scholar]
- World Health Organisation. Guidance Document for the Use of Data in Development of Chemical-Specific Adjustment Factors (CSAFs) for Interspecies Differences and Human Variability in Dose/Concentration–Response Assessment; World Health Organisation: Geneva, Switzerland, 2001. [Google Scholar]
- International Council for Harmonisation of Technical Requirements for Pharmaceuticals for Human Use. Safety Guideline—S9 Nonclinical Evaluation for Anticancer Pharmaceuticals; International Council for Harmonisation of Technical Requirements for Pharmaceuticals for Human Use: Geneva, Switzerland, 2009. [Google Scholar]
- Nelson, D.R.; Zeldin, D.C.; Hoffman, S.M.; Maltais, L.J.; Wain, H.M.; Nebert, D.W. Comparison of cytochrome P450 (CYP) genes from the mouse and human genomes, including nomenclature recommendations for genes, pseudogenes and alternative-splice variants. Pharmacogenetics 2004, 14, 1–18. [Google Scholar] [CrossRef]
- van Herwaarden, A.E.; Wagenaar, E.; van der Kruijssen, C.M.; van Waterschoot, R.A.; Smit, J.W.; Song, J.Y.; van der Valk, M.A.; van Tellingen, O.; van der Hoorn, J.W.A.; Rosing, H.; et al. Knockout of cytochrome P450 3A yields new mouse models for understanding xenobiotic metabolism. J. Clin. Investig. 2007, 117, 3583–3592. [Google Scholar] [CrossRef]
- Ma, X.; Cheung, C.; Krausz, K.W.; Shah, Y.M.; Wang, T.; Idle, J.R.; Gonzalez, F.J. A double transgenic mouse model expressing human pregnane X receptor and cytochrome P450 3A4. Drug. Metab. Dispos. Biol. Fate Chem. 2008, 36, 2506–2512. [Google Scholar] [CrossRef]
- Holmstock, N.; Gonzalez, F.J.; Baes, M.; Annaert, P.; Augustijns, P. PXR/CYP3A4-humanized mice for studying drug-drug interactions involving intestinal P-glycoprotein. Mol. Pharm. 2013, 10, 1056–1062. [Google Scholar] [CrossRef][Green Version]
- Sharma, V.; McNeill, J.H. To scale or not to scale: The principles of dose extrapolation. Br. J. Pharmacol. 2009, 157, 907–921. [Google Scholar] [CrossRef]
- Zou, P.; Yu, Y.; Zheng, N.; Yang, Y.; Paholak, H.J.; Yu, L.X.; Sun, D. Applications of human pharmacokinetic prediction in first-in-human dose estimation. AAPS J. 2012, 14, 262–281. [Google Scholar] [CrossRef]
- Thiel, C.; Schneckener, S.; Krauss, M.; Ghallab, A.; Hofmann, U.; Kanacher, T.; Zellmer, S.; Gebhardt, R.; Hengstler, j.; Kuepfer, L. A systematic evaluation of the use of physiologically based pharmacokinetic modeling for cross-species extrapolation. J. Pharm. Sci. 2015, 104, 191–206. [Google Scholar] [CrossRef]
- Wu, Q.; Peters, S.A. A Retrospective Evaluation of Allometry, Population Pharmacokinetics, and Physiologically-Based Pharmacokinetics for Pediatric Dosing Using Clearance as a Surrogate. CPT Pharmacomet. Syst. Pharmacol. 2019, 8, 220–229. [Google Scholar] [CrossRef] [PubMed]
- Damoiseaux, D.; Li, W.; Beijnen, J.H.; Schinkel, A.H.; Huitema, A.D.R.; Dorlo, T.P.C. Population pharmacokinetic modelling to support the evaluation of preclinical pharmacokinetic experiments with lorlatinib. J. Pharm. Sci. 2021, 111, 495–504. [Google Scholar] [CrossRef]
- Li, J.; Dawson, P.A. Animal models to study bile acid metabolism. Biochim. Biophys. Acta Mol. Basis Dis. 2019, 1865, 895–911. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Houk, B.; Pithavala, Y.K.; Ruiz-Garcia, A. Population pharmacokinetic model with time-varying clearance for lorlatinib using pooled data from patients with non-small cell lung cancer and healthy participants. CPT Pharmacomet. Syst. Pharmacol. 2021, 10, 148–160. [Google Scholar] [CrossRef] [PubMed]
- Gupta, N.; Wang, X.; Offman, E.; Prohn, M.; Narasimhan, N.; Kerstein, D.; Hanley, M.J.; Venkatakrishnan, K. Population Pharmacokinetics of Brigatinib in Healthy Volunteers and Patients With Cancer. Clin. Pharmacokinet. 2021, 60, 235–247. [Google Scholar] [CrossRef] [PubMed]
- Kim, R.D.; Sarker, D.; Meyer, T.; Yau, T.; Macarulla, T.; Park, J.-W.; Choo, S.P.; Hollebecque, A.; Sung, M.W.; Lim, H.-Y.; et al. First-in-Human Phase I Study of Fisogatinib (BLU-554) Validates Aberrant FGF19 Signaling as a Driver Event in Hepatocellular Carcinoma. Cancer Discov. 2019, 9, 1696–1707. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Yang, S.; Ho, Y.Y.; Ji, Y. Ribociclib Population Pharmacokinetics and Pharmacokinetic/Pharmacodynamic Analysis of Neutrophils in Cancer Patients. J. Clin. Pharmacol. 2021, 61, 1054–1068. [Google Scholar] [CrossRef]
- Perlman, R.L. Mouse models of human disease: An evolutionary perspective. Evol. Med. Public Health 2016, 2016, 170–176. [Google Scholar] [CrossRef]
- Kim, T.H.; Shin, S.; Landersdorfer, C.B.; Chi, Y.H.; Paik, S.H.; Myung, J.; Yadav, R.; Horkovics-Kovats, S.; Bulitta, J.B.; Shin, B.S. Population Pharmacokinetic Modeling of the Enterohepatic Recirculation of Fimasartan in Rats, Dogs, and Humans. AAPS J. 2015, 17, 1210–1223. [Google Scholar] [CrossRef]
- Li, W.; Sparidans, R.; El-Lari, M.; Wang, Y.; Lebre, M.C.; Beijnen, J.H.; Schinkel, A.H. P-glycoprotein (ABCB1/MDR1) limits brain accumulation and Cytochrome P450-3A (CYP3A) restricts oral availability of the novel FGFR4 inhibitor fisogatinib (BLU-554). Int. J. Pharm. 2020, 573, 118842. [Google Scholar] [CrossRef]
- Li, W.; Sparidans, R.W.; Wang, Y.; Lebre, M.C.; Beijnen, J.H.; Schinkel, A.H. Oral coadministration of elacridar and ritonavir enhances brain accumulation and oral availability of the novel ALK/ROS1 inhibitor lorlatinib. Eur. J. Pharm. Biopharm. 2019, 136, 120–130. [Google Scholar] [CrossRef]
- Li, W.; Sparidans, R.W.; Wang, Y.; Lebre, M.C.; Beijnen, J.H.; Schinkel, A.H. P-glycoprotein and breast cancer resistance protein restrict brigatinib brain accumulation and toxicity, and, alongside CYP3A, limit its oral availability. Pharmacol. Res. 2018, 137, 47–55. [Google Scholar] [CrossRef]
- Li, W.; Sparidans, R.W.; Wang, Y.; Lebre, M.C.; Wagenaar, E.; Beijnen, J.H.; Schinkel, A.H. P-glycoprotein (MDR1/ABCB1) restricts brain accumulation and cytochrome P450-3A (CYP3A) limits oral availability of the novel ALK/ROS1 inhibitor lorlatinib. Int. J. Cancer 2018, 143, 2029–2038. [Google Scholar] [CrossRef]
- Martínez-Chávez, A.; van Hoppe, S.; Rosing, H.; Lebre, M.C.; Tibben, M.; Beijnen, J.H.; Schinkel, A.H. P-glycoprotein Limits Ribociclib Brain Exposure and CYP3A4 Restricts Its Oral Bioavailability. Mol. Pharm. 2019, 16, 3842–3852. [Google Scholar] [CrossRef]
- Huwaldt, J.A. Plot Digitizer Version 2.6.9. Available online: https://sourceforge.net/projects/plotdigitizer/files/plotdigitizer/2.6.9/.2.6.9ed2020 (accessed on 6 January 2021).
- West, G.B.; Brown, J.H.; Enquist, B.J. A general model for the origin of allometric scaling laws in biology. Science 1997, 276, 122–126. [Google Scholar] [CrossRef]
- Kapetanovic, I.M.; Krishnaraj, R.; Martin-Jimenez, T.; Yuan, L.; van Breemen, R.B.; Lyubimov, A. Effects of oral dosing paradigms (gavage versus diet) on pharmacokinetics and pharmacodynamics. Chem. Biol. Interact. 2006, 164, 68–75. [Google Scholar] [CrossRef]
- US Food and Drug Administration; Center for Drug Evaluation and Research. Lobrena (lorlatinib) Product Quality Review(s). U.S. Patent 210868Orig1s000, 14 February 2017. [Google Scholar]
- US Food and Drug Administration; Center for Drug Evaluation and Research. Alunbrig (brigatinib) Chemistry Review(s). U.S. Patent 208772Orig1s000, 10 February 2017. [Google Scholar]
- US Food and Drug Administration; Center for Drug Evaluation and Research. Kisqali® (ribociclib) Chemistry Review. U.S. Patent 209092Orig1s000, 13 March 2017. [Google Scholar]
- McConnell, E.L.; Basit, A.W.; Murdan, S. Measurements of rat and mouse gastrointestinal pH, fluid and lymphoid tissue, and implications for in-vivo experiments. J. Pharm. Pharmcol. 2008, 60, 63–70. [Google Scholar] [CrossRef]
- Evans, D.F.; Pye, G.; Bramley, R.; Clark, A.G.; Dyson, T.J.; Hardcastle, J.D. Measurement of gastrointestinal pH profiles in normal ambulant human subjects. Gut 1988, 29, 1035–1041. [Google Scholar] [CrossRef]
- Committee for Medicinal Products for Human Use; European Medicines Agency. Annex I Summary of Product Characteristics. Lorviqua: EPAR—Product Information; Committee for Medicinal Products for Human Use: Amsterdam, The Netherlands, 2019. [Google Scholar]
- Wishart, D.S.; Feunang, Y.D.; Guo, A.C.; Lo, E.J.; Marcu, A.; Grant, J.R.; Sajed, T.; Johnson, D.; Li, C.; Sayeeda, Z.; et al. DrugBank 5.0: A major update to the DrugBank database for 2018. Nucleic Acids Res. 2018, 46, D1074–D1082. [Google Scholar] [CrossRef]
- Committee for Medicinal Products for Human Use; European Medicines Agency. Annex I Summary of Product Characteristics. Alunbrig: EPAR—Product Information; Committee for Medicinal Products for Human Use: Amsterdam, The Netherlands, 2018. [Google Scholar]
- Committee for Medicinal Products for Human Use; European Medicines Agency. Annex I Summary of Product Characteristics. Kisqali: EPAR—Product Information; Committee for Medicinal Products for Human Use: Amsterdam, The Netherlands, 2017. [Google Scholar]
- Keizer, R.J.; Karlsson, M.O.; Hooker, A. Modeling and Simulation Workbench for NONMEM: Tutorial on Pirana, PsN, and Xpose. CPT Pharmacomet. Syst. Pharmacol. 2013, 2, e50. [Google Scholar] [CrossRef]
- Beal, S.; Boeckmann, A.; Sheiner, L. NONMEM User Guides; University of California: San Francisco, CA, USA, 1988. [Google Scholar]

| Lorlatinib | Brigatinib | Fisogatinib | Ribociclib | |||||
|---|---|---|---|---|---|---|---|---|
| Median AUCinf (µg/mL h) | Fold Change | Median AUCinf (µg/mL h) | Fold Change | Median AUCinf (µg/mL h) | Fold Change | Median AUCinf (µg/mL h) | Fold Change | |
| Literature model | 8.2 ± 2.4 | - | 13.2 ± 7.4 | - | - | - | 20.0 ± 11.2 | - |
| Extrapolation of final mouse model | ||||||||
| Wild-type | 17.4 ± 2.4 | 2.1 | 29.8 ± 4.4 | 2.3 | 28.9 ± 4.0 | - | 21.3 ± 2.9 | 1.1 |
| Cyp3aXAV | 9.4 ± 1.3 | 1.1 | 19.2 ± 2.7 | 1.5 | 25.4 ± 3.7 | - | 6.4 ±0.9 | 0.3 |
| Extrapolation of optimized mouse model (if applicable) | ||||||||
| Wild-type | - | - | 25.5 ± 3.9 | 1.9 | 28.9 ± 4.0 | - | 20.4 ± 2.8 | 1.0 |
| Cyp3aXAV | - | - | 13.7 ± 1.9 | 1.0 | 24.4 ± 3.7 | - | 6.1 ± 0.8 | 0.3 |
| Lorlatinib | Brigatinib | Fisogatinib | Ribociclib | |||||
|---|---|---|---|---|---|---|---|---|
| Median Cmax (ng/mL) | Fold Change | Median Cmax (ng/mL) | Fold Change | Median Cmax (ng/mL) | Fold Change | Median Cmax (ng/mL) | Fold Change | |
| Literature model | 310 ± 195 | - | 615 ± 422 | - | 6404 ± 3299 | - | 1176 ± 696 | - |
| Extrapolation of final mouse model | ||||||||
| Wild-type | 632 ± 179 | 2.0 | 687 ± 124 | 1.1 | 4887 ± 829 | 0.8 | 2586 ± 390 | 2.2 |
| Cyp3aXAV | 431 ± 135 | 1.4 | 665 ± 116 | 1.1 | 2925 ± 501 | 0.5 | 1229 ± 177 | 1.0 |
| Extrapolation of optimized mouse model (if applicable) | ||||||||
| Wild-type | - | - | 499 ± 91 | 0.8 | 6693 ± 513 | 1.0 | 2189 ± 323 | 1.9 |
| Cyp3aXAV | - | - | 470 ± 85 | 0.8 | 4021 ± 278 | 0.6 | 655 ± 97 | 0.6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Damoiseaux, D.; Li, W.; Martínez-Chávez, A.; Beijnen, J.H.; Schinkel, A.H.; Huitema, A.D.R.; Dorlo, T.P.C. Predictiveness of the Human-CYP3A4-Transgenic Mouse Model (Cyp3aXAV) for Human Drug Exposure of CYP3A4-Metabolized Drugs. Pharmaceuticals 2022, 15, 860. https://doi.org/10.3390/ph15070860
Damoiseaux D, Li W, Martínez-Chávez A, Beijnen JH, Schinkel AH, Huitema ADR, Dorlo TPC. Predictiveness of the Human-CYP3A4-Transgenic Mouse Model (Cyp3aXAV) for Human Drug Exposure of CYP3A4-Metabolized Drugs. Pharmaceuticals. 2022; 15(7):860. https://doi.org/10.3390/ph15070860
Chicago/Turabian StyleDamoiseaux, David, Wenlong Li, Alejandra Martínez-Chávez, Jos H. Beijnen, Alfred H. Schinkel, Alwin D. R. Huitema, and Thomas P. C. Dorlo. 2022. "Predictiveness of the Human-CYP3A4-Transgenic Mouse Model (Cyp3aXAV) for Human Drug Exposure of CYP3A4-Metabolized Drugs" Pharmaceuticals 15, no. 7: 860. https://doi.org/10.3390/ph15070860
APA StyleDamoiseaux, D., Li, W., Martínez-Chávez, A., Beijnen, J. H., Schinkel, A. H., Huitema, A. D. R., & Dorlo, T. P. C. (2022). Predictiveness of the Human-CYP3A4-Transgenic Mouse Model (Cyp3aXAV) for Human Drug Exposure of CYP3A4-Metabolized Drugs. Pharmaceuticals, 15(7), 860. https://doi.org/10.3390/ph15070860







