Synthesis and Biological Evaluation of Imadazo[1,2-a]pyrazines as Anticancer and Antiviral Agents through Inhibition of CDK9 and Human Coronavirus
Abstract
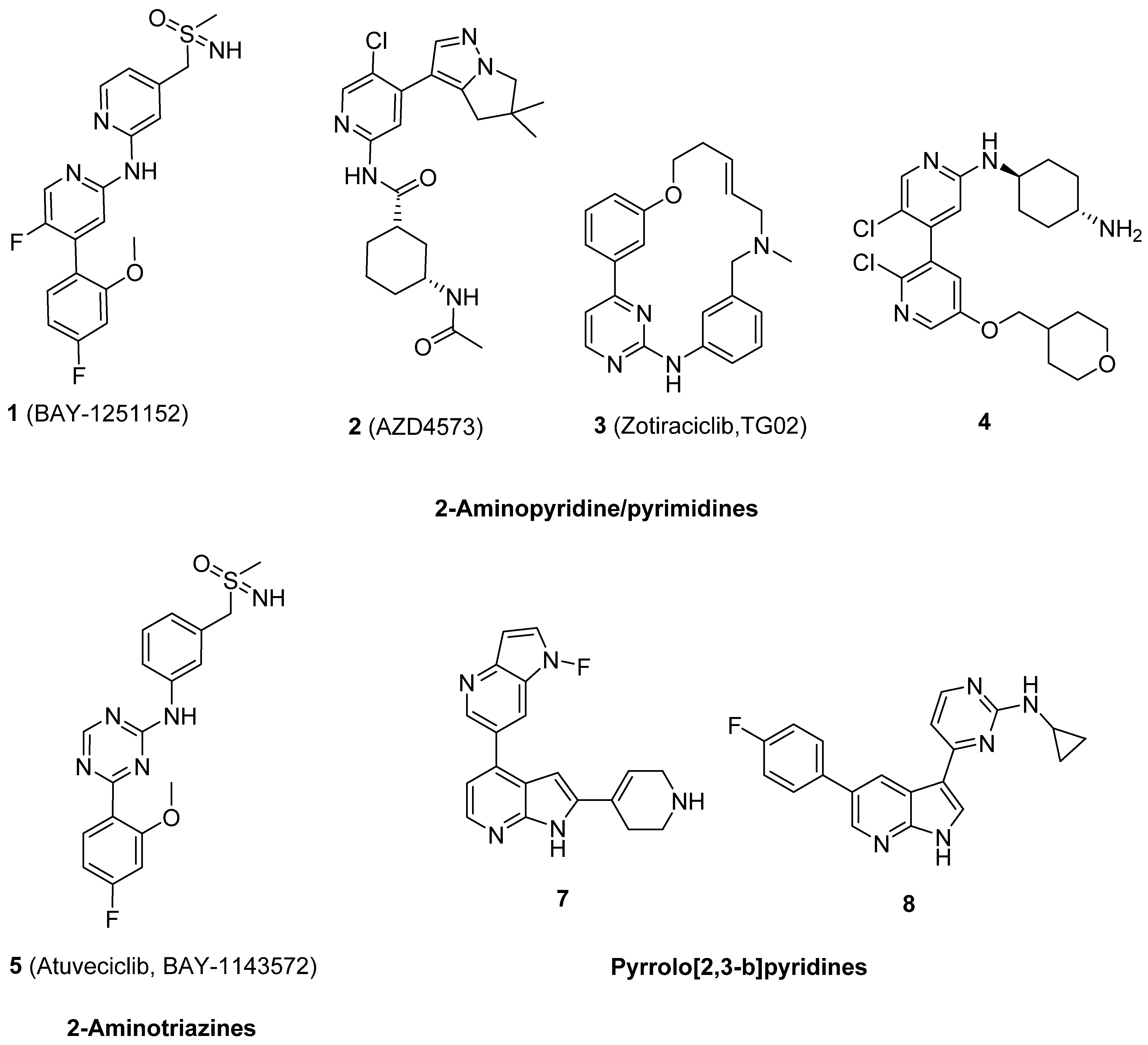
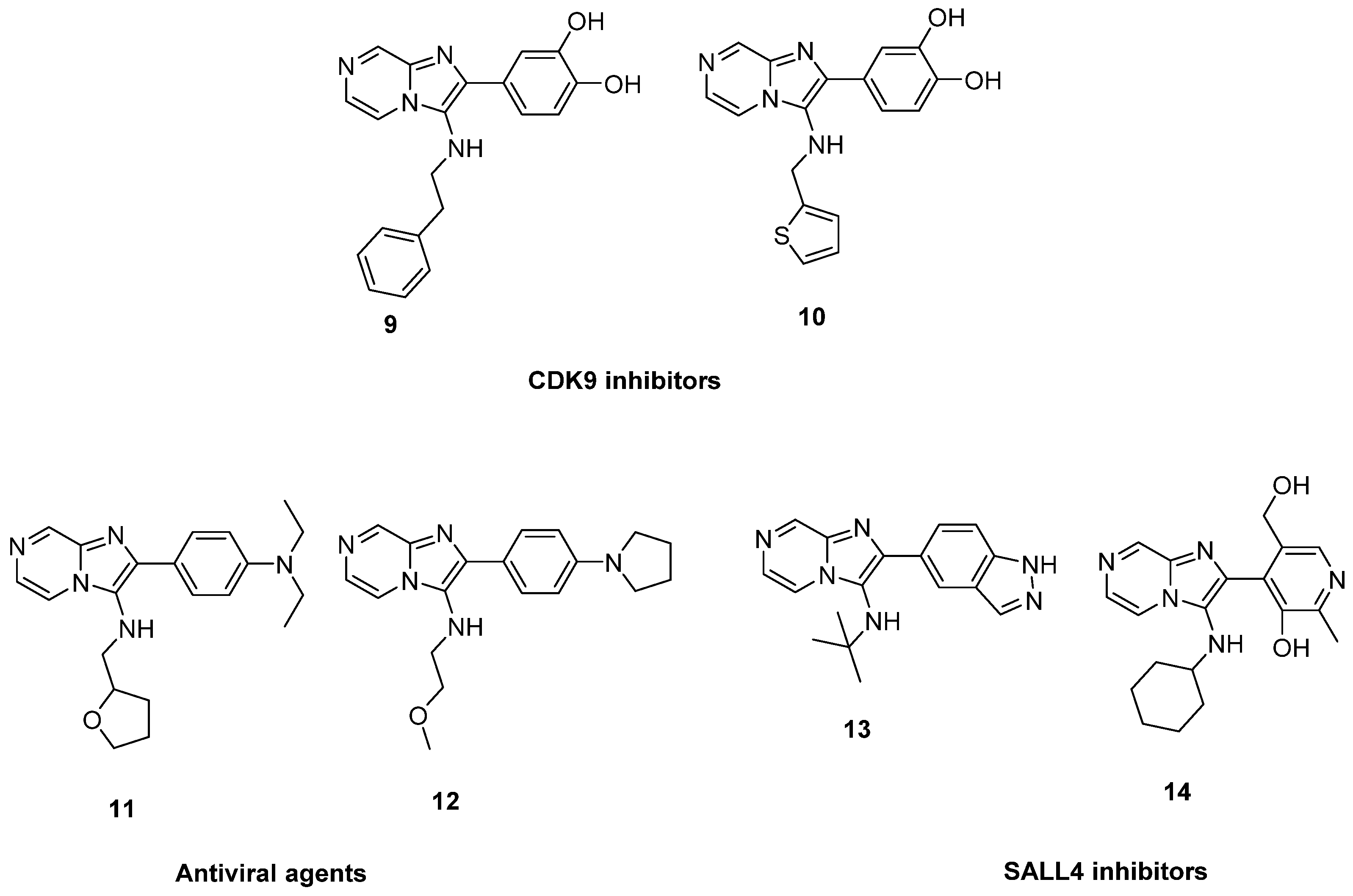
1. Introduction
2. Results and Discussion
2.1. Chemistry
2.2. Anti-Cancer Activity
2.2.1. CDK9 Inhibitory Activity
2.2.2. Cytotoxicity Activity
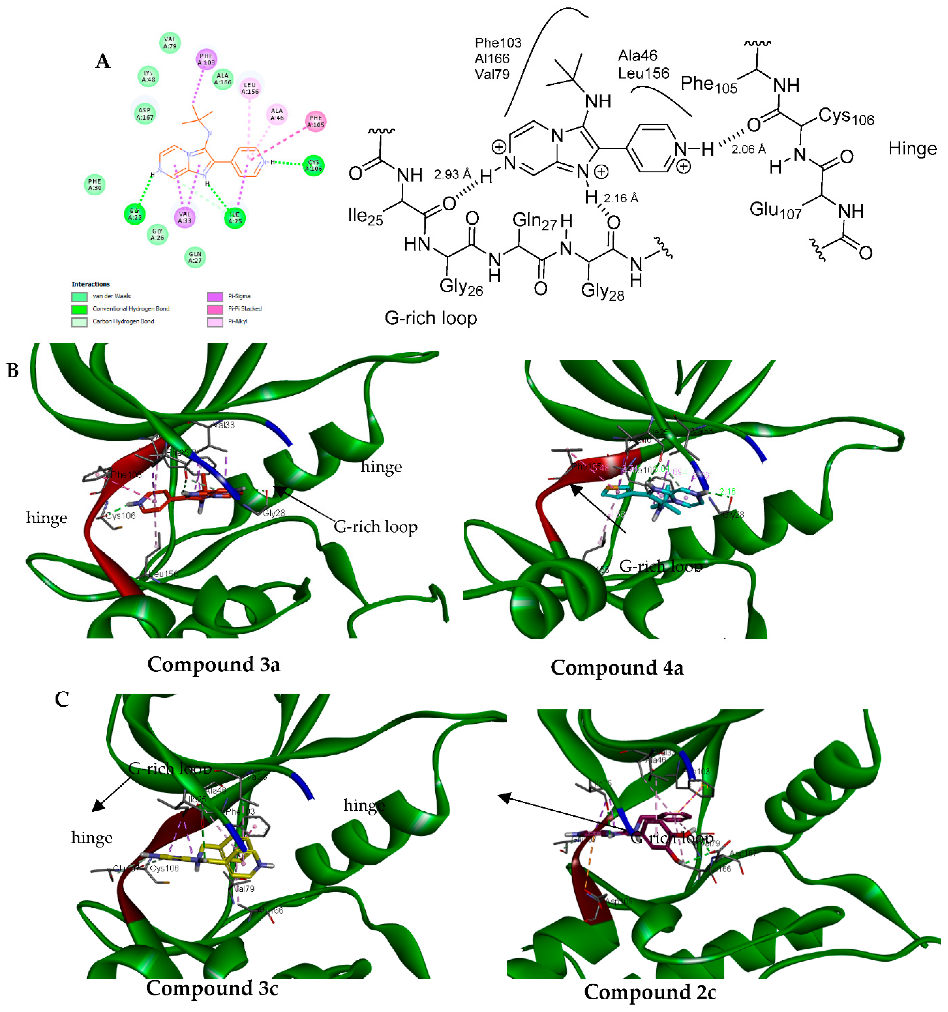
2.2.3. Molecular Docking Study into CDK9 Active Site
2.3. Antiviral Activity
2.3.1. Anti-Coronaviral Inhibitory Activity
2.3.2. Anti-Coronavirus Target Identification and Docking Studies
2.4. In Silico Predication of the Compound’s Physiochemical and Pharmacokinetic Properties
2.4.1. Physiochemical and Drug Likeness Properties
2.4.2. Pharmacokinetic and Toxicity Studies
3. Materials and Methods
3.1. Chemistry
3.2. In Vitro CDK9 Assay
3.3. MTT Cytotoxicity Assay
3.4. Antiviral Assay
3.5. Docking Studies
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Malumbres, M.; Barbacid, M. Cell cycle, CDKs and cancer: A changing paradigm. Nat. Cancer 2009, 9, 153–166. [Google Scholar] [CrossRef] [PubMed]
- Yik, J.; Chen, R.; Nishimura, R.; Jennings, J.L.; Link, A.J.; Zhou, Q. Inhibition of P-TEFb (CDK9/Cyclin T) Kinase and RNA Polymerase II Transcription by the Coordinated Actions of HEXIM1 and 7SK snRNA. Mol. Cell 2003, 12, 971–982. [Google Scholar] [CrossRef]
- Baumli, S.; Lolli, G.; Lowe, E.D.; Troiani, S.; Rusconi, L.; Bullock, A.N.; Debreczeni, J.; Knapp, S.; Johnson, L.N. The structure of P-TEFb (CDK9/cyclin T1), its complex with flavopiridol and regulation by phosphorylation. EMBO J. 2008, 27, 1907–1918. [Google Scholar] [CrossRef] [PubMed]
- Baumli, S.; Furst, R. Perspective of Cyclin-dependent kinase 9 (CDK9) as a Drug Target. Curr. Pharm. Des. 2012, 18, 2883–2890. [Google Scholar] [CrossRef]
- Franco, L.C.; Morales, F.; Boffo, S.; Giordano, A. CDK9: A key player in cancer and other diseases. J. Cell. Biochem. 2017, 119, 1273–1284. [Google Scholar] [CrossRef]
- Wu, M.; Li, C.; Zhu, X. FLT3 inhibitors in acute myeloid leukemia. J. Hematol. Oncol. 2018, 11, 133. [Google Scholar] [CrossRef]
- Schlafstein, A.J.; Withers, A.E.; Rudra, S.; Danelia, D.; Switchenko, J.M.; Mister, D.; Harari, S.; Zhang, H.; Daddacha, W.; Ehdaivand, S.; et al. CDK9 Expression Shows Role as a Potential Prognostic Biomarker in Breast Cancer Patients Who Fail to Achieve Pathologic Complete Response after Neoadjuvant Chemotherapy. Int. J. Breast Cancer 2018, 2018, 6945129. [Google Scholar] [CrossRef]
- Rahaman, M.H.; Lam, F.; Zhong, L.; Teo, T.; Adams, J.; Yu, M.; Milne, R.W.; Pepper, C.; Lokman, N.A.; Ricciardelli, C.; et al. Targeting CDK9 for treatment of colorectal cancer. Mol. Oncol. 2019, 13, 2178–2193. [Google Scholar] [CrossRef]
- Morales, F.; Giordano, A. Overview of CDK9 as a target in cancer research. Cell Cycle 2016, 15, 519–527. [Google Scholar] [CrossRef]
- Glaser, S.P.; Lee, E.F.; Trounson, E.; Bouillet, P.; Wei, A.; Fairlie, W.D.; Izon, D.J.; Zuber, J.; Rappaport, A.R.; Herold, M.J.; et al. Anti-apoptotic Mcl-1 is essential for the development and sustained growth of acute myeloid leukemia. Genes Dev. 2012, 26, 120–125. [Google Scholar] [CrossRef]
- Narita, T.; Ishida, T.; Ito, A.; Masaki, A.; Kinoshita, S.; Suzuki, S.; Takino, H.; Yoshida, T.; Ri, M.; Kusumoto, S.; et al. Cyclin-dependent kinase 9 is a novel specific molecular target in adult T-cell leukemia/lymphoma. Blood 2017, 130, 1114–1124. [Google Scholar] [CrossRef] [PubMed]
- Senderowicz, A.M. Flavopiridol: The First Cyclin-Dependent Kinase Inhibitor in Human Clinical Trials. Investig. New Drugs 1999, 17, 313–320. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.K.; LaPlant, B.; Chng, W.J.; Zonder, J.; Callander, N.; Fonseca, R.; Fruth, B.; Roy, V.; Erlichman, C.; Stewart, A.K. Dinaciclib, a novel CDK inhibitor, demonstrates encouraging single-agent activity in patients with relapsed multiple myeloma. Blood 2015, 125, 443–448. [Google Scholar] [CrossRef] [PubMed]
- Benson, C.; White, J.; De Bono, J.; O’Donnell, A.; Raynaud, F.; Cruickshank, C.; McGrath, H.; Walton, M.; Workman, P.; Kaye, S.; et al. A phase I trial of the selective oral cyclin-dependent kinase inhibitor seliciclib (CYC202; R-Roscovitine), administered twice daily for 7 days every 21 days. Br. J. Cancer 2006, 96, 29–37. [Google Scholar] [CrossRef]
- Tong, W.-G.; Chen, R.; Plunkett, W.; Siegel, D.; Sinha, R.; Harvey, R.D.; Badros, A.Z.; Popplewell, L.; Coutre, S.; Fox, J.A.; et al. Phase I and Pharmacologic Study of SNS-032, a Potent and Selective Cdk2, 7, and 9 Inhibitor, in Patients with Advanced Chronic Lymphocytic Leukemia and Multiple Myeloma. J. Clin. Oncol. 2010, 28, 3015–3022. [Google Scholar] [CrossRef] [PubMed]
- van der Biessen, D.A.; Burger, H.; de Bruijn, P.; Lamers, C.H.; Naus, N.; Loferer, H.; Wiemer, E.A.; Mathijssen, R.H.; de Jonge, M.J. Phase I Study of RGB-286638, A Novel, Multitargeted Cyclin-Dependent Kinase Inhibitor in Patients with Solid Tumors. Clin. Cancer Res. 2014, 20, 4776–4783. [Google Scholar] [CrossRef] [PubMed]
- Le Tourneau, C.; Faivre, S.; Laurence, V.; Delbaldo, C.; Vera, K.; Girre, V.; Chiao, J.; Armour, S.; Frame, S.; Green, S.R.; et al. Phase I evaluation of seliciclib (R-roscovitine), a novel oral cyclin-dependent kinase inhibitor, in patients with advanced malignancies. Eur. J. Cancer 2010, 46, 3243–3250. [Google Scholar] [CrossRef]
- Walsby, E.; Pratt, G.; Shao, H.; Abbas, A.Y.; Fischer, P.M.; Bradshaw, T.D.; Brennan, P.; Fegan, C.; Wang, S.; Pepper, C. A novel Cdk9 inhibitor preferentially targets tumor cells and synergizes with fludarabine. Oncotarget 2013, 5, 375–385. [Google Scholar] [CrossRef]
- Zhai, S.; Senderowicz, A.M.; Sausville, E.A.; Figg, W.D. Flavopiridol, a Novel Cyclin-Dependent Kinase Inhibitor, in Clinical Development. Ann. Pharmacother. 2002, 36, 905–911. [Google Scholar] [CrossRef]
- McInnes, C. Progress in the evaluation of CDK inhibitors as anti-tumor agents. Drug Discov. Today 2008, 13, 875–881. [Google Scholar] [CrossRef]
- Cidado, J.; Proia, T.; Boiko, S.; Martin, M.S.; Criscione, S.; Ferguson, D.; Shao, W.; Drew, L. Abstract 310: AZD4573, a novel CDK9 inhibitor, rapidly induces cell death in hematological tumor models through depletion of Mcl1. Cancer Res. 2018, 78, 310. [Google Scholar] [CrossRef]
- Byrne, M.; Frattini, M.G.; Ottmann, O.G.; Mantzaris, I.; Wermke, M.; Lee, D.J.; Morillo, D.; Scholz, A.; Ince, S.; Valencia, R.; et al. Phase I Study of the PTEFb Inhibitor BAY 1251152 in Patients with Acute Myelogenous Leukemia. Blood 2018, 132, 4055. [Google Scholar] [CrossRef]
- Parrott, T.; Weller, M.; Estok, T.M.; Le Rhun, E. P08.32 TG02, an oral CDK inhibitor, demonstrates activity in glioma models: EORTC Brain Tumor Group Conducts Phase 1b study (STEAM/EORTC 1608). Neuro-Oncology 2016, 18, iv48. [Google Scholar] [CrossRef]
- Goh, K.C.; Novotny-Diermayr, V.; Hart, S.; Ong, L.C.; Loh, Y.K.; Cheong, A.; Tan, Y.C.; Hu, C.; Jayaraman, R.; William, A.D.; et al. TG02, a novel oral multi-kinase inhibitor of CDKs, JAK2 and FLT3 with potent anti-leukemic properties. Leukemia 2011, 26, 236–243. [Google Scholar] [CrossRef]
- Rule, S.; Kater, A.P.; Brümmendorf, T.H.; Fegan, C.; Kaiser, M.; Radford, J.A.; Stilgenbauer, S.; Kayser, S.; Dyer, M.J.; Brossart, P.; et al. A phase 1, open-label, multicenter, non-randomized study to assess the safety, tolerability, pharmacokinetics, and preliminary antitumor activity of AZD4573, a potent and selective CDK9 inhibitor, in subjects with relapsed or refractory hematological malignancies. J. Clin. Oncol. 2018, 36, TPS7588. [Google Scholar] [CrossRef]
- Alsfouk, A. Small molecule inhibitors of cyclin-dependent kinase 9 for cancer therapy. J. Enzym. Inhib. Med. Chem. 2021, 36, 693–706. [Google Scholar] [CrossRef]
- Kim, I.K.; Nam, K.Y.; Kim, S.Y.; Park, S.J. Composition for Prevention and Treatment of Cancer Including CDK9 Inhibitor as Active Ingredient, University of Ulsan Foundation for Industry Cooperation. Patent KR1020180106188, 2019. [Google Scholar]
- Cheol-Gyu, H.; Jeong-Hyeok, Y. Novel Imidazole Pyrazine Derivative Compound, a Method for Preparing the Same, and a Pharmaceutical Composition for Antiviral Treatment Containing the Same as an Active Ingredient, Sihwa Industriy. Patent 1020110097448, 2011. [Google Scholar]
- Qi, J.; Varca, A.; Li, C. Small Molecule Inhibition of Transcription Factor SALL4 and Uses Therof, Dana-Farber Cancer Institute. U.S. Patent Application No. 16/753,536, 4 October 2017. [Google Scholar]
- Zheng, J. SARS-CoV-2: An Emerging Coronavirus That Causes a Global Threat. Int. J. Biol. Sci. 2020, 16, 1678–1685. [Google Scholar] [CrossRef]
- Malik, Y.A. Properties of Coronavirus and SARS-CoV-2. Malays. J. Pathol. 2020, 42, 3–11. [Google Scholar]
- Wang, J.; Dean, D.C.; Hornicek, F.J.; Shi, H.; Duan, Z. Cyclin-dependent kinase 9 (CDK9) is a novel prognostic marker and therapeutic target in ovarian cancer. FASEB J. 2019, 33, 5990–6000. [Google Scholar] [CrossRef]
- Ma, H.; Seebacher, N.A.; Hornicek, F.J.; Duan, Z. Cyclin-dependent kinase 9 (CDK9) is a novel prognostic marker and therapeutic target in osteosarcoma. eBioMedicine 2018, 39, 182–193. [Google Scholar] [CrossRef] [PubMed]
- Kretz, A.-L.; Schaum, M.; Richter, J.; Kitzig, E.F.; Engler, C.C.; Leithäuser, F.; Henne-Bruns, D.; Knippschild, U.; Lemke, J. CDK9 is a prognostic marker and therapeutic target in pancreatic cancer. Tumor Biol. 2017, 39, 1010428317694304. [Google Scholar] [CrossRef] [PubMed]
- Gojo, I.; Sadowska, M.; Walker, A.; Feldman, E.J.; Iyer, S.P.; Baer, M.R.; Sausville, E.A.; Lapidus, R.G.; Zhang, D.; Zhu, Y.; et al. Clinical and laboratory studies of the novel cyclin-dependent kinase inhibitor dinaciclib (SCH 727965) in acute leukemias. Cancer Chemother. Pharmacol. 2013, 72, 897–908. [Google Scholar] [CrossRef] [PubMed]
- Johnson, A.J.; Yeh, Y.-Y.; Smith, L.L.; Wagner, A.J.; Hessler, J.; Gupta, S.; Flynn, J.; Jones, J.; Zhang, X.; Bannerji, R.; et al. The novel cyclin-dependent kinase inhibitor dinaciclib (SCH727965) promotes apoptosis and abrogates microenvironmental cytokine protection in chronic lymphocytic leukemia cells. Leukemia 2012, 26, 2554–2557. [Google Scholar] [CrossRef][Green Version]
- Daina, A.; Michielin, O.; Zoete, V. SwissTargetPrediction: Updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 2019, 47, W357–W364. [Google Scholar] [CrossRef]
- V’Kovski, P.; Kratzel, A.; Steiner, S.; Stalder, H.; Thiel, V. Coronavirus biology and replication: Implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021, 19, 155–170. [Google Scholar] [CrossRef]
- Li, Y.C.; Bai, W.Z.; Hashikawa, T. The neuroinvasive potential of SARS-CoV2 may play a role in the respiratory failure of COVID-19 patients. J. Med. Virol. 2020, 92, 552–555. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef]
- Dong, J.; Wang, N.-N.; Yao, Z.-J.; Zhang, L.; Cheng, Y.; Ouyang, D.; Lu, A.-P.; Cao, D.-S. ADMETlab: A platform for systematic ADMET evaluation based on a comprehensively collected ADMET database. J. Cheminform. 2018, 10, 29. [Google Scholar] [CrossRef]
- Choi, H.J.; Song, J.H.; Park, K.S.; Kwon, D.H. Inhibitory effects of quercetin 3-rhamnoside on influenza A virus replication. Eur. J. Pharm. Sci. 2009, 37, 329–333. [Google Scholar] [CrossRef]
- Pauwels, R.; Balzarini, J.; Baba, M.; Snoeck, R.; Schols, D.; Herdewijn, P.; Desmyter, J.; De Clercq, E. Rapid and automated tetrazolium-based colorimetric assay for the detection of anti-HIV compounds. J. Virol. Methods 1988, 20, 309–321. [Google Scholar] [CrossRef]



| Compound | CDK9 Inhibitory Activity IC50 (µM) |
|---|---|
| 1a | 1.67 ± 0.025 |
| 1b | 0.25 ± 0.004 |
| 1c | 1.01 ± 0.017 |
| 1d | 1.04 ± 0.019 |
| 2a | 1.14 ± 0.019 |
| 2b | 0.89 ± 0.016 |
| 2c | 0.31 ± 0.006 |
| 2d | 2.03 ± 0.04 |
| 3a | 0.26 ± 0.004 |
| 3b | 0.41 ± 0.007 |
| 3c | 0.16 ± 0.003 |
| 3d | 0.8 ± 0.014 |
| 4a | 0.73 ± 0.011 |
| 4b | 1.32 ± 0.006 |
| 4c | 0.71 ± 0.012 |
| 4d | 3.16 ± 0.058 |
| Dinaciclib | 0.08 ± 0.002 |
| Compound | IC50 (µM) | ||
|---|---|---|---|
| MCF7 | HCT116 | K652 | |
| 1a | 405.48 ± 5.05 | 596.96 ± 8.25 | 309.74 ± 4.49 |
| 1b | 3.01 ± 0.04 | 1.43 ± 0.02 | 50.27 ± 0.8 |
| 1c | 98.95 ± 1.39 | 59.3 ± 5.05 | 88.62 ± 8.25 |
| 1d | 243.71 ± 3.62 | 75.23 ± 1.24 | 77.44 ± 1.34 |
| 2a | 76.75 ± 1.08 | 35.86 ± 0.56 | 22.79 ± 0.37 |
| 2b | 119.3 ± 1.82 | 105.43 ± 1.78 | 54.25 ± 0.96 |
| 2c | 39.71 ± 0.62 | 59.87 ± 1.04 | 88.62 ± 1.09 |
| 2d | 80.37 ± 1.31 | 161.61 ± 2.41 | 136.35 ± 2.59 |
| 3a | 4.3 ± 0.5 | 2.69 ± 0.04 | 46 ± 0.67 |
| 3b | 31.7 ± 0.44 | 8.96 ± 0.14 | 67.83 ± 1.08 |
| 3c | 8.59 ± 0.12 | 7.96 ± 0.12 | 4.45 ± 0.06 |
| 3d | 117.96 ± 1.17 | 27.91 ± 1.88 | 46.56 ± 0.48 |
| 4a | 25.37 ± 0.32 | 61.31 ± 0.12 | 12.77 ± 0.06 |
| 4b | 180.96 ± 2.63 | 27.91 ± 0.43 | 46.58 ± 0.76 |
| 4c | 55.81 ± 0.8 | 80.61 ± 1.26 | 69.19 ± 3.89 |
| 4d | 141.13 ± 2.13 | 115.39 ± 1.93 | 87.16 ± 1.53 |
| Staurosporine | 18.41 ± 0.4 | 10.86 ± 0.26 | 22.08 ± 0.56 |
| SI | ||||
|---|---|---|---|---|
| Compound | FHC IC50 (µM) | MCF7 | HCT116 | K652 |
| 1b | 122.51 ± 19.1 | 40.70 | 85.67 | 2.40 |
| 3a | 195.78 ± 2.79 | 45.53 | 72.78 | 4.25 |
| 3c | 97.19 ± 1.58 | 11.31 | 12.21 | 21.84 |
| 4a | 76.07 ± 1.11 | 3.04 | 1.2 | 6.3 |
| Staurosporine | 38.19 ± 0.97 | 2.07 | 3.5 | 1.7 |
| Compound | Docking Score (Kcal/mol) | Interaction Residue (Type of Interaction) | Bond Length (Å) |
|---|---|---|---|
| 3a | −8.6 | Cys106 (HB) | 2.06 |
| Ile25 (HB) | 2.93 | ||
| Gly28 (HB) | 2.16 | ||
| Phe105 (Pi-Pi stacked) | 5.29 | ||
| Phe103 (Pi-sigma) | 3.82 | ||
| Val33 (Pi-sigma) | 3.54 and 3.71 | ||
| Leu156 (Pi-alkyl) | 4.60 | ||
| Ala46 (Pi-alkyl) | 4.51 | ||
| 4a | −8.7 | Ile25 (HB) | 3.01 |
| Gly28 (HB) | 2.16 | ||
| Phe103 (Pi-Pi stacked) | 3.69 | ||
| Phe105 (Pi-Pi stacked) | 4.16 | ||
| Ala46 (pi-alkyl) | 3.51 | ||
| Ala166 (pi-alkyl) | 5.43 | ||
| Leu156 (pi-alkyl) | 3.69 | ||
| 3c | −8.3 | Ile25 (HB) | 3.09 |
| Phe103 (PI-Pi stacked) | 4.22 | ||
| Ala46 (pi-alkyl) | 4.52 | ||
| Ala166 (pi-alkyl) | 4.81 | ||
| Val79 (pi-alkyl) | 5.31 | ||
| Val33 (pi-sigma) | 4.93 | ||
| 2c | −8.5 | Asp167 (HB) | 2.62 and 2.47 |
| Ile25 (HB) | 2.59 | ||
| Asp109 (Pi-anion) | 4.98 | ||
| Val33 (pi-alkyl) | 4.66 | ||
| Phe103 (Pi-pi stacked) | 4.41 | ||
| Ala166 (pi-alkyl) | 4.75 | ||
| Val79 (pi-alkyl) | 5.46 | ||
| Ala56 (pi-alkyl) | 4.71 |
| Compound | CC50 (µM) | IC50 (µM) | SI |
|---|---|---|---|
| 1b | 299.32 | 145.92 | 2.051 |
| 3a | 940.19 | 393.66 | 2.39 |
| 3b | 406.86 | 56.96 | 7.14 |
| 3c | 471.51 | 379.45 | 1.24 |
| Ribavirin | 160.47 | 113.81 | 1.4 |
| Docking Score (Kcal/mol) | Interaction Residue (Type of Interaction) | Bond Length (Å) |
|---|---|---|
| −7.6 | His164 (HB) | 2.96 |
| Cys44 (HB) | 2.91 | |
| Cys145 (pi-sulfur) | 5.11, 5.13 | |
| Met165 (pi-sulfur) | 2.91 | |
| Met49 (Pi-alkyl) | 5.16 | |
| His41 (Pi-Pi stacked) | 4.38, 5.45 |
| Compound | M. W | Clog P | tPSA (Å2) | Solubility Log S | HBA | HBD | Lipinski |
|---|---|---|---|---|---|---|---|
| 1b | 292.38 | 3.24 | 42.22 | −4.78 | 2 | 1 | Yes, 0 violation |
| 3c | 301.35 | 2.68 | 55.11 | −4.07 | 3 | 1 | Yes, 0 violation |
| 4a | 272.37 | 2.82 | 70.46 | −3.98 | 2 | 1 | Yes, 0 violation |
| Compound | 1b | 3c | 4a | ||
|---|---|---|---|---|---|
| Property | Test | Recommended | |||
| Absorption | Papp (Caco-2 permeability) | >−5.15 | −4.595 | −4.86 | −4.472 |
| Pgp-inhibitor | No | No | No | ||
| Pgp-substrate | No | No | No | ||
| HIA (human intestinal absorption) | + | + | + | ||
| Bioavailability score | 0.55 | 0.55 | 0.55 | 0.55 | |
| Distribution | PPB (Plasma protein binding) | <90% | 82.523 | 87.219 | 72.632 |
| BBB (Blood–brain barrier) | + | + | + | ||
| VD (volume of distribution) | 0.04–20 L/kg | 0.698 | 0.542 | 0.29 | |
| Metabolism | CYP3A4 inhibitor | Yes | Yes | Yes | |
| CYP3A4-substrate | No | No | No | ||
| Excretion | T1/2 (Half live) | >0.5 h | 2.014 | 1.774 | 1.918 |
| Clearance | <15 mL/min/kg | 2.063 | 1.952 | 1.489 | |
| Toxicity | hERG blocker | Yes | Yes | No | |
| Ames mutagenicity | + | + | + | ||
| Skin sensitization | No | No | No | ||
| LD50 of acute toxicity | >500 mg/kg | 2.59 | 2.596 | 2.59 | |
| DILI (drug induce liver injury) | + | + | + | ||
| FDAMDD (maximum recommended daily dose) | + | + | + |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alsfouk, A.A.; Alshibl, H.M.; Alsfouk, B.A.; Altwaijry, N.A.; Al-Abdullah, E.S. Synthesis and Biological Evaluation of Imadazo[1,2-a]pyrazines as Anticancer and Antiviral Agents through Inhibition of CDK9 and Human Coronavirus. Pharmaceuticals 2022, 15, 859. https://doi.org/10.3390/ph15070859
Alsfouk AA, Alshibl HM, Alsfouk BA, Altwaijry NA, Al-Abdullah ES. Synthesis and Biological Evaluation of Imadazo[1,2-a]pyrazines as Anticancer and Antiviral Agents through Inhibition of CDK9 and Human Coronavirus. Pharmaceuticals. 2022; 15(7):859. https://doi.org/10.3390/ph15070859
Chicago/Turabian StyleAlsfouk, Aisha A., Hanan M. Alshibl, Bshra A. Alsfouk, Najla A. Altwaijry, and Ebtehal S. Al-Abdullah. 2022. "Synthesis and Biological Evaluation of Imadazo[1,2-a]pyrazines as Anticancer and Antiviral Agents through Inhibition of CDK9 and Human Coronavirus" Pharmaceuticals 15, no. 7: 859. https://doi.org/10.3390/ph15070859
APA StyleAlsfouk, A. A., Alshibl, H. M., Alsfouk, B. A., Altwaijry, N. A., & Al-Abdullah, E. S. (2022). Synthesis and Biological Evaluation of Imadazo[1,2-a]pyrazines as Anticancer and Antiviral Agents through Inhibition of CDK9 and Human Coronavirus. Pharmaceuticals, 15(7), 859. https://doi.org/10.3390/ph15070859







