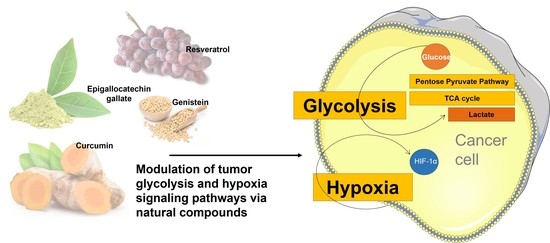
Phytochemicals as Regulators of Tumor Glycolysis and Hypoxia Signaling Pathways: Evidence from In Vitro Studies
Abstract
1. Introduction
2. Phytochemicals Targeting Glycolysis and Hypoxia Signaling Pathways—Structural Classification and Natural Sources
3. Cell Culture-Based Metabolomics to Investigate Plant-Induced Metabolic Changes
4. Aerobic Glycolysis as a Metabolic Rewiring to Sustain Cell Growth
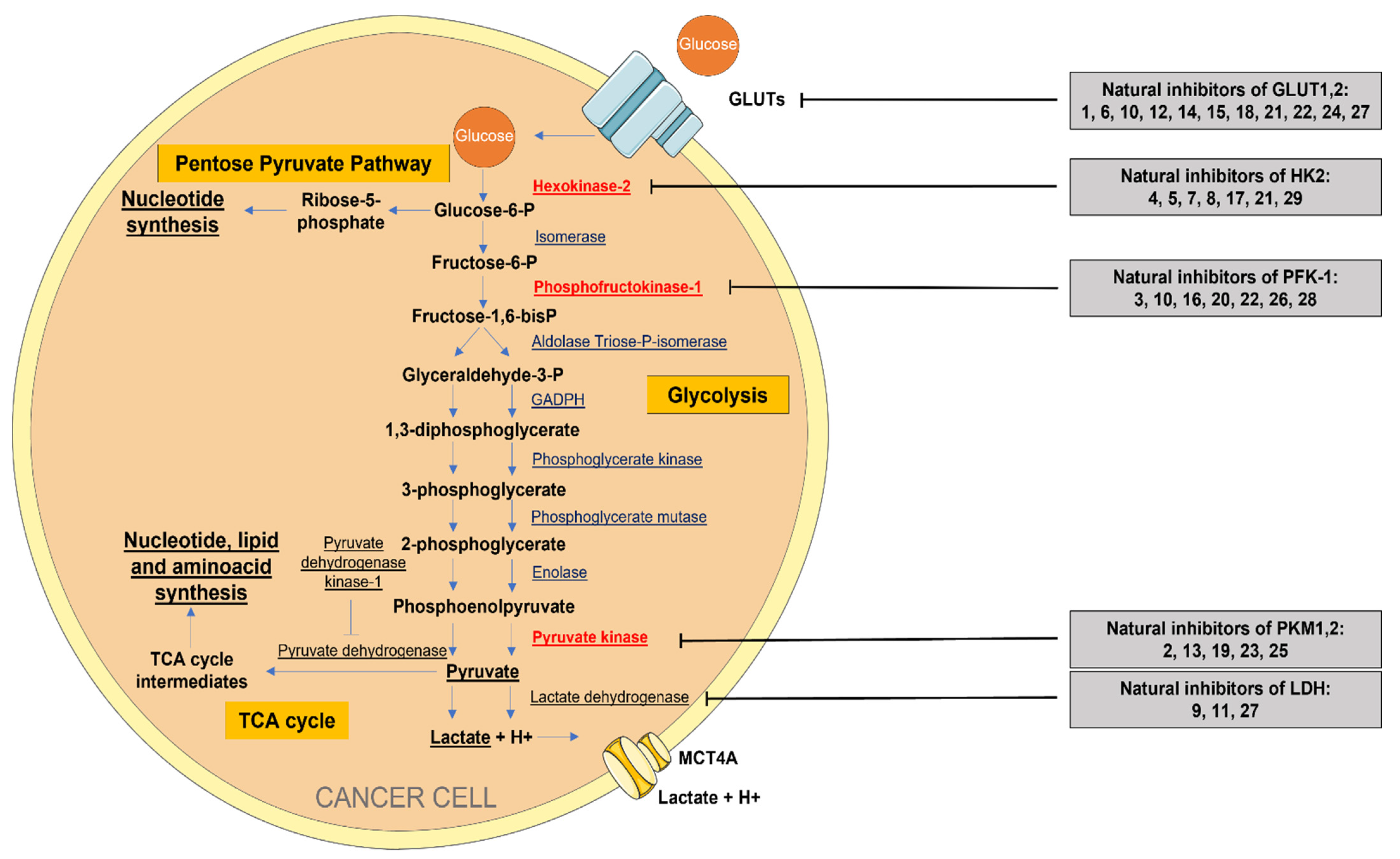
5. Natural Regulators of Aerobic Glycolysis
5.1. Inhibitors of Glucose Transporters (GLUTs)
5.2. Inhibitors of Sodium-Dependent Glucose Cotransporter (SGLT)
5.3. Inhibitors of Hexokinase 2 (HK2)
5.4. Inhibitors of Phosphofructokinase 1 (PFK-1)
5.5. Inhibitors of Pyruvate Kinase (PKM)
5.6. Inhibitors of Lactate Dehydrogenase (LDH)
6. Tumor Hypoxia and Their Natural Regulators
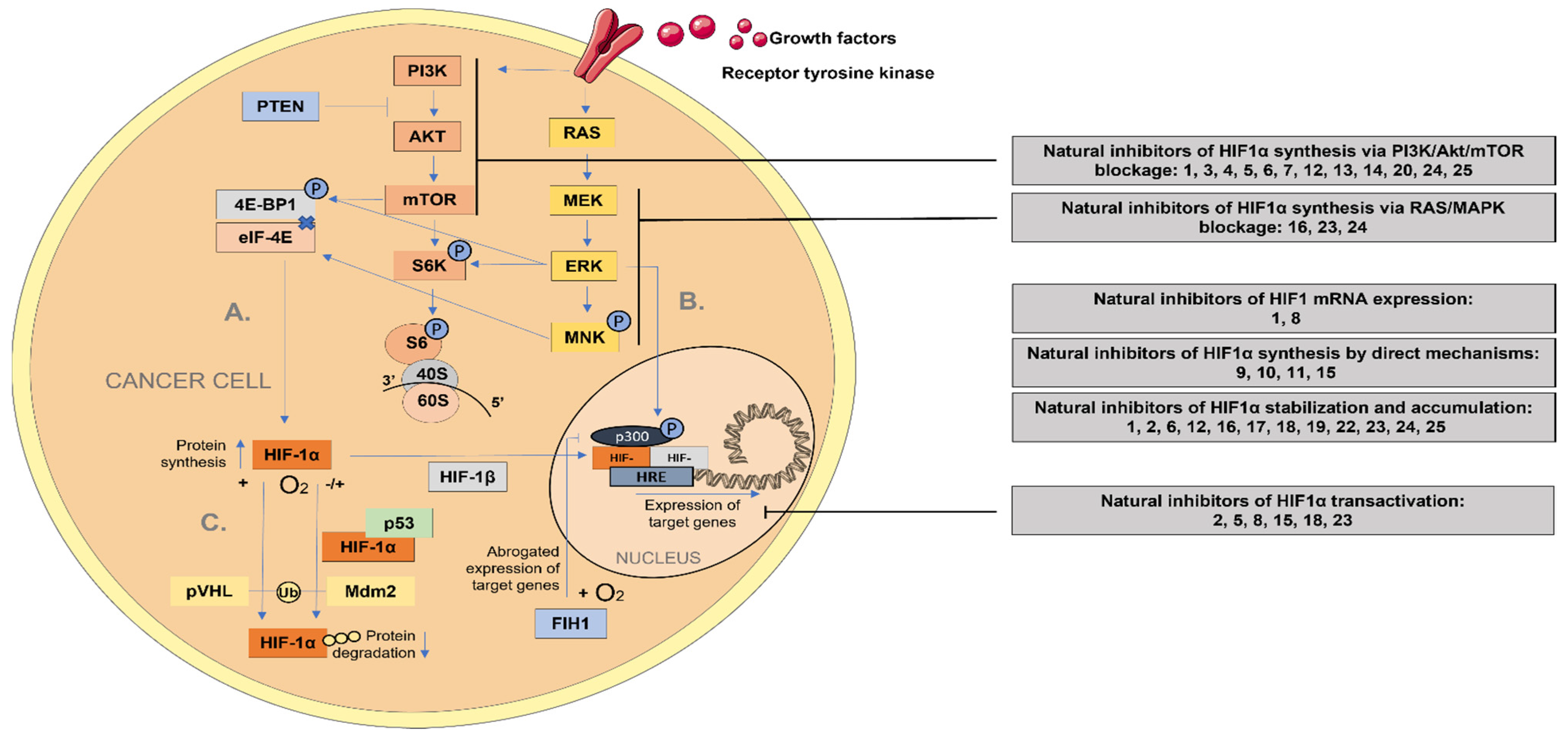
6.1. Hypoxia-Inducible Factor 1-alpha (HIF-1α) Regulating Signaling Pathway
6.2. Natural Inhibitors of Phosphoinositide 3-Kinase/Protein Kinase B/Mammalian Target of Rapamycin (PI3K/Akt/mTOR)
6.3. Natural Inhibitors of Ras/Raf/MAPK Signaling Pathway
7. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lin, X.; Xiao, Z.; Chen, T.; Liang, S.H.; Guo, H. Glucose Metabolism on Tumor Plasticity, Diagnosis and Treatment. Front. Oncol. 2020, 10, 317. [Google Scholar] [CrossRef] [PubMed]
- Masoud, G.N.; Li, W. HIF-1α pathway: Role, regulation and intervention for cancer therapy. Acta Pharm. Sin. B 2015, 5, 378–389. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Menendez, P.; Hevia, D.; Rodriguez-Garcia, A.; Mayo, J.C.; Sainz, R.M. Regulation of GLUT Transporters by Flavonoids in Androgen-Sensitive and -Insensitive Prostate Cancer Cells. Endocrinology 2014, 155, 3238–3250. [Google Scholar] [CrossRef] [PubMed]
- Fang, J.; Bao, Y.Y.; Zhou, S.H.; Fan, J. Apigenin inhibits the proliferation of adenoid cystic carcinoma via suppression of glucose transporter-1. Mol. Med. Rep. 2015, 12, 6461–6466. [Google Scholar] [CrossRef]
- Korga, A.; Ostrowska, M.; Jozefczyk, A.; Iwan, M.; Wojcik, R.; Zgorka, G.; Herbet, M.; Vilarrubla, G.G.; Dudka, J. Apigenin and hesperidin augment the toxic effect of doxorubicin against HepG2 cells. BMC Pharmacol. Toxicol. 2019, 20, 1–13. [Google Scholar] [CrossRef]
- Shi, Y.; Lian, K.; Jia, J. Apigenin Suppresses the Warburg Effect and Stem-like Properties in SOSP-9607 Cells by Inactivating the PI3K/Akt/mTOR Signaling Pathway. Evid.-Based Complement. Altern. Med. 2022, 2022, 1–10. [Google Scholar] [CrossRef]
- Shan, S.; Shi, J.; Yang, P.; Jia, B.; Wu, H.; Zhang, X.; Li, Z. Apigenin Restrains Colon Cancer Cell Proliferation via Targeted Blocking of Pyruvate Kinase M2-Dependent Glycolysis. J. Agric. Food Chem. 2017, 65, 8136–8144. [Google Scholar] [CrossRef]
- Fang, J.; Zhou, Q.; Liu, L.Z.; Xia, C.; Hu, X.; Shi, X.; Jiang, B.H. Apigenin inhibits tumor angiogenesis through decreasing HIF-1α and VEGF expression. Carcinogenesis 2007, 28, 858–864. [Google Scholar] [CrossRef]
- Yang, J.; Pi, C.; Wang, G. Inhibition of PI3K/Akt/mTOR pathway by apigenin induces apoptosis and autophagy in hepatocellular carcinoma cells. Biomed. Pharmacother. 2018, 103, 699–707. [Google Scholar] [CrossRef]
- Wang, M.; Firrman, J.; Liu, L.S.; Yam, K. A review on flavonoid apigenin: Dietary intake, ADME, antimicrobial effects, and interactions with human gut microbiota. BioMed Res. Int. 2019, 2019, 1–45. [Google Scholar] [CrossRef]
- Salehi, B.; Venditti, A.; Sharifi-Rad, M.; Kręgiel, D.; Sharifi-Rad, J.; Durazzo, A.; Lucarini, M.; Santini, A.; Souto, E.B.; Novellino, E.; et al. The Therapeutic Potential of Apigenin. Int. J. Mol. Sci. 2019, 20, 1305. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Lu, J.J.; Ding, J. Natural Products in Cancer Therapy: Past, Present and Future. Nat. Prod. Bioprospecting 2021, 11, 5–13. [Google Scholar] [CrossRef] [PubMed]
- Roy, M.; Datta, A. Cancer Genetics and Therapeutics: Focus on Phytochemicals; Springer: Singapore, 2019; pp. 1–227. [Google Scholar]
- Salanță, L.C.; Uifălean, A.; Iuga, C.A.; Tofană, M.; Cropotova, J.; Pop, O.L.; Pop, C.R.; Rotar, M.A.; Bautista-Ávila, M.; González, C.V. Valuable Food Molecules with Potential Benefits for Human Health. In The Health Benefits of Foods; Salanță, L.C., Ed.; IntechOpen: London, UK, 2020. [Google Scholar]
- Lichota, A.; Gwozdzinski, K. Anticancer Activity of Natural Compounds from Plant and Marine Environment. Int. J. Mol. Sci. 2018, 19, 3533. [Google Scholar] [CrossRef] [PubMed]
- Briguglio, G.; Costa, C.; Pollicino, M.; Giambò, F.; Catania, S.; Fenga, C. Polyphenols in cancer prevention: New insights (Review). Int. J. Funct. Nutr. 2020, 1, 9. [Google Scholar] [CrossRef]
- Samec, M.; Liskova, A.; Koklesova, L.; Samuel, S.M.; Zhai, K.; Buhrmann, C.; Varghese, E.; Abotaleb, M.; Qaradakhi, T.; Zulli, A.; et al. Flavonoids against the Warburg phenotype—Concepts of predictive, preventive and personalised medicine to cut the Gordian knot of cancer cell metabolism. EPMA J. 2020, 113, 377–398. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, Y.; Du, X.; Ma, H.; Yao, J. The Anti-Cancer Mechanisms of Berberine: A Review. Cancer Manag. Res. 2020, 12, 695–702. [Google Scholar] [CrossRef]
- Laghezza Masci, V.; Bernardini, S.; Modesti, L.; Ovidi, E.; Tiezzi, A. Medicinal Plants as a Source of Alkaloids. In Microorganisms for Sustainability; Kothari, R., Singh, A., Arora, N.K., Eds.; Springer: Singapore, 2019; pp. 85–113. [Google Scholar]
- Miekus, N.; Marszałek, K.; Podlacha, M.; Iqbal, A.; Puchalski, C.; Swiergiel, A.H. Health Benefits of Plant-Derived Sulfur Compounds, Glucosinolates, and Organosulfur Compounds. Molecules 2020, 25, 3804. [Google Scholar] [CrossRef]
- Yun, B.D.; Son, S.W.; Choi, S.Y.; Kuh, H.J.; Oh, T.J.; Park, J.K. Anti-Cancer Activity of Phytochemicals Targeting Hypoxia-Inducible Factor-1 Alpha. Int. J. Mol. Sci. 2021, 22, 9819. [Google Scholar] [CrossRef]
- Goncharov, N.V.; Belinskaia, D.A.; Ukolov, A.I.; Jenkins, R.O.; Avdonin, P.V. Organosulfur compounds as nutraceuticals. In Nutraceuticals. Efficacy, Safety and Toxicity, 2nd ed.; Gupta, R.C., Lall, R., Srivastava, A., Eds.; Academic Press: Cambridge, MA, USA, 2021; pp. 911–924. [Google Scholar]
- Jiang, L.; Zhao, X.; Xu, J.; Li, C.; Yu, Y.; Wang, W.; Zhu, L. The Protective Effect of Dietary Phytosterols on Cancer Risk: A Systematic Meta-Analysis. J. Oncol. 2019, 2019, 7479518. [Google Scholar] [CrossRef]
- Schneider, N.F.Z.; Cerella, C.; Simões, C.M.O.; Diederich, M. Anticancer and Immunogenic Properties of Cardiac Glycosides. Molecules 2017, 22, 1932. [Google Scholar] [CrossRef]
- Martin, L.J.; Cairns, E.A.; Heblinski, M.; Fletcher, C.; Krycer, J.R.; Arnold, J.C.; McGregor, I.S.; Bowen, M.T.; Anderson, L.L. Cannabichromene and Δ9-Tetrahydrocannabinolic Acid Identified as Lactate Dehydrogenase-A Inhibitors by in Silico and in Vitro Screening. J. Nat. Prod. 2021, 84, 1469–1477. [Google Scholar] [CrossRef] [PubMed]
- Halama, A. Metabolomics in cell culture-a strategy to study crucial metabolic pathways in cancer development and the response to treatment. Arch. Biochem. Biophys. 2014, 564, 100–109. [Google Scholar] [CrossRef] [PubMed]
- Álvarez-Sánchez, B.; Priego-Capote, F.; Luque de Castro, M.D. Metabolomics analysis II. Preparation of biological samples prior to detection. TrAC Trends Anal. Chem. 2010, 29, 120–127. [Google Scholar] [CrossRef]
- Balcerczyk, A.; Damblon, C.; Elena-Herrmann, B.; Panthu, B.; Rautureau, G.J.P. Metabolomic Approaches to Study Chemical Exposure-Related Metabolism Alterations in Mammalian Cell Cultures. Int. J. Mol. Sci. 2020, 21, 6843. [Google Scholar] [CrossRef]
- Valdés, A.; García-Cañas, V.; Simó, C.; Ibáñez, C.; Micol, V.; Ferragut, J.A.; Cifuentes, A. Comprehensive foodomics study on the mechanisms operating at various molecular levels in cancer cells in response to individual rosemary polyphenols. Anal. Chem. 2014, 86, 9807–9815. [Google Scholar] [CrossRef]
- Ibáñez, C.; Simó, C.; García-Cañas, V.; Gómez-Martínez, Á.; Ferragut, J.A.; Cifuentes, A. CE/LC-MS multiplatform for broad metabolomic analysis of dietary polyphenols effect on colon cancer cells proliferation. Electrophoresis 2012, 33, 2328–2336. [Google Scholar] [CrossRef]
- Valdés, A.; Simó, C.; Ibáñez, C.; Rocamora-Reverte, L.; Ferragut, J.A.; García-Cañas, V.; Cifuentes, A. Effect of dietary polyphenols on K562 leukemia cells: A Foodomics approach. Electrophoresis 2012, 33, 2314–2327. [Google Scholar] [CrossRef]
- Daddiouaissa, D.; Amid, A.; Abdullah Sani, M.S.; Elnour, A.A.M. Evaluation of metabolomics behavior of human colon cancer HT29 cell lines treated with ionic liquid graviola fruit pulp extract. J. Ethnopharmacol. 2021, 270, 113813. [Google Scholar] [CrossRef]
- Uifălean, A.; Schneider, S.; Gierok, P.; Ionescu, C.; Iuga, C.A.; Lalk, M. The Impact of Soy Isoflavones on MCF-7 and MDA-MB-231 Breast Cancer Cells Using a Global Metabolomic Approach. Int. J. Mol. Sci. 2016, 17, 1443. [Google Scholar] [CrossRef]
- Fernández-Arroyo, S.; Gómez-Martínez, A.; Rocamora-Reverte, L.; Quirantes-Piné, R.; Segura-Carretero, A.; Fernández-Gutiérrez, A.; Ferragut, J.A. Application of nanoLC-ESI-TOF-MS for the metabolomic analysis of phenolic compounds from extra-virgin olive oil in treated colon-cancer cells. J. Pharm Biomed. Anal. 2012, 63, 128–134. [Google Scholar] [CrossRef]
- Massimi, M.; Tomassini, A.; Sciubba, F.; Sobolev, A.P.; Devirgiliis, L.C.; Miccheli, A. Effects of resveratrol on HepG2 cells as revealed by (1)H-NMR based metabolic profiling. Biochim. Biophys. Acta (BBA)-Gen. Subj. 2012, 1820, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Jäger, W.; Gruber, A.; Giessrigl, B.; Krupitza, G.; Szekeres, T.; Sonntag, D. Metabolomic Analysis of Resveratrol-Induced Effects in the Human Breast Cancer Cell Lines MCF-7 and MDA-MB-231. OMICS A J. Integr. Biol. 2011, 15, 9–14. [Google Scholar] [CrossRef] [PubMed]
- Bayet-Robert, M.; Morvan, D. Metabolomics Reveals Metabolic Targets and Biphasic Responses in Breast Cancer Cells Treated by Curcumin Alone and in Association with Docetaxel. PLoS ONE 2013, 8, e57971. [Google Scholar] [CrossRef] [PubMed]
- Warburg, O.; Wind, F.; Negelein, E. The Metabolism of Tumors in the Body. J. Gen. Physiol. 1927, 8, 519–530. [Google Scholar] [CrossRef] [PubMed]
- Ganapathy-Kanniappan, S.; Geschwind, J.F.H. Tumor glycolysis as a target for cancer therapy: Progress and prospects. Mol. Cancer 2013, 12, 152. [Google Scholar] [CrossRef]
- Ferreira, L.M.R. Cancer metabolism: The Warburg effect today. Exp. Mol. Pathol. 2010, 89, 372–380. [Google Scholar] [CrossRef]
- Shestov, A.A.; Liu, X.; Ser, Z.; Cluntun, A.A.; Hung, Y.P.; Huang, L.; Kim, D.; Le, A.; Yellen, G.; Albeck, J.G.; et al. Quantitative determinants of aerobic glycolysis identify flux through the enzyme GAPDH as a limiting step. eLife 2014, 3, e03342. [Google Scholar] [CrossRef]
- Slavov, N.; Budnik, B.A.; Schwab, D.; Airoldi, E.M.; van Oudenaarden, A. Constant Growth Rate Can Be Supported by Decreasing Energy Flux and Increasing Aerobic Glycolysis. Cell Rep. 2014, 7, 705–714. [Google Scholar] [CrossRef]
- DeBerardinis, R.J.; Lum, J.J.; Hatzivassiliou, G.; Thompson, C.B. The Biology of Cancer: Metabolic Reprogramming Fuels Cell Growth and Proliferation. Cell Metab. 2008, 7, 11–20. [Google Scholar] [CrossRef]
- Locasale, J.W.; Cantley, L.C. Metabolic Flux and the Regulation of Mammalian Cell Growth. Cell Metab. 2011, 14, 443–451. [Google Scholar] [CrossRef]
- Estrella, V.; Chen, T.; Lloyd, M.; Wojtkowiak, J.; Cornnell, H.H.; Ibrahim-Hashim, A.; Bailey, K.; Balagurunathan, Y.; Rothberg, J.M.; Sloane, B.F.; et al. Acidity generated by the tumor microenvironment drives local invasion. Cancer Res. 2013, 73, 1524–1535. [Google Scholar] [CrossRef] [PubMed]
- Zambrano, A.; Molt, M.; Uribe, E.; Salas, M. Glut 1 in Cancer Cells and the Inhibitory Action of Resveratrol as A Potential Therapeutic Strategy. Int. J. Mol. Sci. 2019, 20, 3374. [Google Scholar] [CrossRef] [PubMed]
- Pliszka, M.; Szablewski, L. Glucose Transporters as a Target for Anticancer Therapy. Cancers 2021, 13, 4184. [Google Scholar] [CrossRef]
- Ghanavat, M.; Shahrouzian, M.; Zayeri, Z.D.; Banihashemi, S.; Kazemi, S.M.; Saki, N. Digging deeper through glucose metabolism and its regulators in cancer and metastasis. Life Sci. 2021, 264, 118603. [Google Scholar] [CrossRef] [PubMed]
- Azevedo, C.; Correia-Branco, A.; Araújo, J.R.; Guimarães, J.T.; Keating, E.; Martel, F. The Chemopreventive Effect of the Dietary Compound Kaempferol on the MCF-7 Human Breast Cancer Cell Line Is Dependent on Inhibition of Glucose Cellular Uptake. Nutr. Cancer 2015, 67, 504–513. [Google Scholar] [CrossRef] [PubMed]
- Catanzaro, D.; Gabbia, D.; Cocetta, V.; Biagi, M.; Ragazzi, E.; Montopoli, M.; Carrara, M. Silybin counteracts doxorubicin resistance by inhibiting GLUT1 expression. Fitoterapia 2018, 124, 42–48. [Google Scholar] [CrossRef]
- Zhan, T.; Digel, M.; Küch, E.M.; Stremmel, W.; Füllekrug, J. Silybin and dehydrosilybin decrease glucose uptake by inhibiting GLUT proteins. J. Cell. Biochem. 2011, 112, 849–859. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhang, L.; Wu, Y.; Dai, Q.; Zhou, Y.; Li, Z.; Yang, L.; Guo, Q.; Lu, N. Selective anti-tumor activity of wogonin targeting the Warburg effect through stablizing p53. Pharm. Res. 2018, 135, 49–59. [Google Scholar] [CrossRef]
- Hamilton, K.E.; Rekman, J.F.; Gunnink, L.K.; Busscher, B.M.; Scott, J.L.; Tidball, A.M.; Stehouwer, N.R.; Johnecheck, G.N.; Looyenga, B.D.; Louters, L.L. Quercetin inhibits glucose transport by binding to an exofacial site on GLUT1. Biochimie 2018, 151, 107–114. [Google Scholar] [CrossRef]
- Jia, L.; Huang, S.; Yin, X.; Zan, Y.; Guo, Y.; Han, L. Quercetin suppresses the mobility of breast cancer by suppressing glycolysis through Akt-mTOR pathway mediated autophagy induction. Life Sci. 2018, 208, 123–130. [Google Scholar] [CrossRef]
- Moreira, L.; Araújo, I.; Costa, T.; Correia-Branco, A.; Faria, A.; Martel, F.; Keating, E. Quercetin and epigallocatechin gallate inhibit glucose uptake and metabolism by breast cancer cells by an estrogen receptor-independent mechanism. Exp. Cell Res. 2013, 319, 1784–1795. [Google Scholar] [CrossRef] [PubMed]
- Li, L.J.; Li, G.W.; Xie, Y. [Regulatory effects of glabridin and quercetin on energy metabolism of breast cancer cells]. Zhongguo Zhongyao Zazhi 2019, 44, 3786–3791. [Google Scholar] [PubMed]
- Wei, R.; Mao, L.; Xu, P.; Zheng, X.; Hackman, R.M.; Mackenzie, G.G.; Wang, Y. Suppressing glucose metabolism with epigallocatechin-3-gallate (EGCG) reduces breast cancer cell growth in preclinical models. Food Funct. 2018, 9, 5682–5696. [Google Scholar] [CrossRef] [PubMed]
- Torres-Villarreal, D.; Camacho, A.; Milagro, F.I.; Ortiz-Lopez, R.; de la Garza, A.L. Quercetin-3-O-glucoside Improves Glucose Tolerance in Rats and Decreases Intestinal Sugar Uptake in Caco-2 Cells. Nat. Prod. Commun. 2017, 12, 1709–1712. [Google Scholar] [CrossRef]
- Jiang, H.; Yamashita, Y.; Nakamura, A.; Croft, K.; Ashida, H. Quercetin and its metabolite isorhamnetin promote glucose uptake through different signalling pathways in myotubes. Sci. Rep. 2019, 9, 2690. [Google Scholar] [CrossRef]
- Kitagawa, M.; Ikeda, S.; Tashiro, E.; Soga, T.; Imoto, M. Metabolomic identification of the target of the filopodia protrusion inhibitor glucopiericidin A. Chem. Biol. 2010, 17, 989–998. [Google Scholar] [CrossRef]
- Dhar, D.; Raina, K.; Kant, R.; Wempe, M.F.; Serkova, N.J.; Agarwal, C.; Agarwal, R. Bitter melon juice-intake modulates glucose metabolism and lactate efflux in tumors in its efficacy against pancreatic cancer. Carcinogenesis 2019, 40, 1164–1176. [Google Scholar] [CrossRef]
- Wright, E.M. SGLT2 and cancer. Pflügers Arch. Eur. J. Physiol. 2020, 472, 1407–1414. [Google Scholar] [CrossRef]
- Tsai, K.F.; Chen, Y.L.; Chiou, T.T.Y.; Chu, T.H.; Li, L.C.; Ng, H.Y.; Lee, W.C.; Lee, C.T. Emergence of SGLT2 Inhibitors as Powerful Antioxidants in Human Diseases. Antioxidants 2021, 10, 1166. [Google Scholar] [CrossRef]
- Blaschek, W. Natural Products as Lead Compounds for Sodium Glucose Cotransporter (SGLT) Inhibitors. Planta Med. 2017, 83, 985–993. [Google Scholar] [CrossRef]
- Sato, S.; Takeo, J.; Aoyama, C.; Kawahara, H. Na+-Glucose cotransporter (SGLT) inhibitory flavonoids from the roots of Sophora flavescens. Bioorg. Med. Chem. 2007, 15, 3445–3449. [Google Scholar] [CrossRef] [PubMed]
- Shimokawa, Y.; Akao, Y.; Hirasawa, Y.; Awang, K.; Hadi, A.H.A.; Sato, S.; Aoyama, C.; Takeo, J.; Shiro, M.; Morita, H. Gneyulins A and B, stilbene trimers, and noidesols A and B, dihydroflavonol- C -glucosides, from the bark of gnetum gnemonoides. J. Nat. Prod. 2010, 73, 763–767. [Google Scholar] [CrossRef] [PubMed]
- Valdés, M.; Calzada, F.; Mendieta-Wejebe, J.E.; Merlín-Lucas, V.; Velázquez, C.; Barbosa, E. Antihyperglycemic Effects of Annona diversifolia Safford and Its Acyclic Terpenoids: α-Glucosidase and Selective SGLT1 Inhibitiors. Molecules 2020, 25, 3361. [Google Scholar] [CrossRef]
- Oranje, P.; Gouka, R.; Burggraaff, L.; Vermeer, M.; Chalet, C.; Duchateau, G.; van der Pijl, P.; Geldof, M.; de Roo, N.; Clauwaert, F.; et al. Novel natural and synthetic inhibitors of solute carriers SGLT1 and SGLT2. Pharm. Res. Perspect. 2019, 7, e00504. [Google Scholar] [CrossRef]
- Mashraqi, M.M.; Chaturvedi, N.; Alam, Q.; Alshamrani, S.; Bahnass, M.M.; Ahmad, K.; Alqosaibi, A.I.; Alnamshan, M.M.; Ahmad, S.S.; Beg, M.M.A.; et al. Biocomputational Prediction Approach Targeting FimH by Natural SGLT2 Inhibitors: A Possible Way to Overcome the Uropathogenic Effect of SGLT2 Inhibitor Drugs. Molecules 2021, 26, 582. [Google Scholar] [CrossRef]
- Roberts, D.J.; Miyamoto, S. Hexokinase II integrates energy metabolism and cellular protection: Akting on mitochondria and TORCing to autophagy. Cell Death Differ. 2015, 22, 248–257. [Google Scholar] [CrossRef] [PubMed]
- Garcia, S.N.; Guedes, R.C.; Marques, M.M. Unlocking the Potential of HK2 in Cancer Metabolism and Therapeutics. Curr. Med. Chem. 2018, 26, 7285–7322. [Google Scholar] [CrossRef] [PubMed]
- Ciscato, F.; Ferrone, L.; Masgras, I.; Laquatra, C.; Rasola, A. Hexokinase 2 in Cancer: A Prima Donna Playing Multiple Characters. Int. J. Mol. Sci. 2021, 22, 4716. [Google Scholar] [CrossRef]
- Liu, W.; Li, W.; Liu, H.; Yu, X. Xanthohumol inhibits colorectal cancer cells via downregulation of hexokinases II-mediated glycolysis. Int. J. Biol. Sci. 2019, 15, 2497–2508. [Google Scholar] [CrossRef]
- Wu, H.; Pan, L.; Gao, C.; Xu, H.; Li, Y.; Zhang, L.; Ma, L.; Meng, L.; Sun, X.; Qin, H. Quercetin Inhibits the Proliferation of Glycolysis-Addicted HCC Cells by Reducing Hexokinase 2 and Akt-mTOR Pathway. Molecules 2019, 24, 1993. [Google Scholar] [CrossRef]
- Li, W.; Gao, F.; Ma, X.; Wang, R.; Dong, X.; Wang, W. Deguelin inhibits non-small cell lung cancer via down-regulating Hexokinases II-mediated glycolysis. Oncotarget 2017, 8, 32586–32599. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Jin, J.; Yu, H.; Zhao, Z.; Ma, D.; Zhang, C.; Jiang, H. Chrysin inhibited tumor glycolysis and induced apoptosis in hepatocellular carcinoma by targeting hexokinase-2. J. Exp. Clin. Cancer Res. 2017, 36, 44. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Zhou, Y.; Dai, Q.; Qiao, C.; Zhao, L.; Hui, H.; Lu, N.; Guo, Q.L. Oroxylin A induces dissociation of hexokinase II from the mitochondria and inhibits glycolysis by SIRT3-mediated deacetylation of cyclophilin D in breast carcinoma. Cell Death Dis. 2013, 4, e601. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Han, X.; Tan, G.; Zhu, Q.; Chen, H.; Xia, Y.; Gong, J.; Wang, Z.; Wang, Y.; Yan, J. Dioscin Inhibited Glycolysis and Induced Cell Apoptosis in Colorectal Cancer via Promoting c-myc Ubiquitination and Subsequent Hexokinase-2 Suppression. Onco Targets Ther. 2020, 13, 31–44. [Google Scholar] [CrossRef]
- Ban, D.; Hua, S.; Zhang, W.; Shen, C.; Miao, X.; Liu, W. Costunolide reduces glycolysis-associated activation of hepatic stellate cells via inhibition of hexokinase-2. Cell. Mol. Biol. Lett. 2019, 24, 52. [Google Scholar] [CrossRef]
- Swargiary, G.; Mani, S. Molecular docking and simulation studies of phytocompounds derived from Centella asiatica and Andrographis paniculata against hexokinase II as mitocan agents. Mitochondrion 2021, 61, 138–146. [Google Scholar] [CrossRef]
- Bao, F.; Yang, K.; Wu, C.; Gao, S.; Wang, P.; Chen, L.; Li, H. New natural inhibitors of hexokinase 2 (HK2): Steroids from Ganoderma sinense. Fitoterapia 2018, 125, 123–129. [Google Scholar] [CrossRef]
- Sciacovelli, M.; Gaude, E.; Hilvo, M.; Frezza, C. The Metabolic Alterations of Cancer Cells. In Methods in Enzymology; Galluzzi, L., Kroemer, G., Eds.; Academic Press: Cambridge, MA, USA, 2014; pp. 1–23. [Google Scholar]
- Berg, J.M.; Tymoczko, J.L.; Stryer, L. The glycolytic pathway is tightly controlled. In Biochemistry, 5th ed.; W H Freeman: New York, NY, USA, 2002; p. 477. [Google Scholar]
- Yalcin, A.; Telang, S.; Clem, B.; Chesney, J. Regulation of glucose metabolism by 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatases in cancer. Exp. Mol. Pathol. 2009, 86, 174–179. [Google Scholar] [CrossRef]
- Lee, J.H.; Liu, R.; Li, J.; Zhang, C.; Wang, Y.; Cai, Q.; Qian, X.; Xia, Y.; Zheng, Y.; Piao, Y.; et al. Stabilization of phosphofructokinase 1 platelet isoform by AKT promotes tumorigenesis. Nat. Commun. 2017, 8, 949. [Google Scholar] [CrossRef]
- Li, S.; He, P.; Wang, Z.; Liang, M.; Liao, W.; Huang, Y.; Chi, M.; Liu, F.; Zen, N.; Su, R.; et al. RNAi-mediated knockdown of PFK1 decreases the invasive capability and metastasis of nasopharyngeal carcinoma cell line, CNE-2. Cell Cycle 2021, 20, 154–165. [Google Scholar] [CrossRef]
- Gomez, L.S.; Zancan, P.; Marcondes, M.C.; Ramos-Santos, L.; Meyer-Fernandes, J.R.; Sola-Penna, M.; Da Silva, D. Resveratrol decreases breast cancer cell viability and glucose metabolism by inhibiting 6-phosphofructo-1-kinase. Biochimie 2013, 95, 1336–1343. [Google Scholar] [CrossRef] [PubMed]
- Santoro, M.; Guido, C.; De Amicis, F.; Sisci, D.; Cione, E.; Dolce, V.; Donà, A.; Panno, M.L.; Aquila, S. Bergapten induces metabolic reprogramming in breast cancer cells. Oncol. Rep. 2016, 35, 568–576. [Google Scholar] [CrossRef] [PubMed]
- Heghes, S.C.; Vostinaru, O.; Mogosan, C.; Miere, D.; Iuga, C.A.; Filip, L. Safety Profile of Nutraceuticals Rich in Coumarins: An Update. Front. Pharmacol. 2022, 13, 48. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Wu, L.; Feng, J.; Li, J.; Liu, T.; Zhang, R.; Xu, S.; Cheng, K.; Zhou, Y.; Zhou, S.; et al. In vitro and in vivo study of epigallocatechin-3-gallate-induced apoptosis in aerobic glycolytic hepatocellular carcinoma cells involving inhibition of phosphofructokinase activity. Sci. Rep. 2016, 6, 28479. [Google Scholar] [CrossRef]
- Jeon, Y.K.; Yoo, D.R.; Jang, Y.H.; Jang, S.Y.; Nam, M.J. Sulforaphane induces apoptosis in human hepatic cancer cells through inhibition of 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase4, mediated by hypoxia inducible factor-1-dependent pathway. Biochim. Biophys. Acta (BBA)-Proteins Proteom. 2011, 1814, 1340–1348. [Google Scholar] [CrossRef]
- Ji, L.; Shen, W.; Zhang, F.; Qian, J.; Jiang, J.; Weng, L.; Tan, J.; Li, L.; Chen, Y.; Cheng, H.; et al. Worenine reverses the Warburg effect and inhibits colon cancer cell growth by negatively regulating HIF-1α. Cell. Mol. Biol. Lett. 2021, 26, 19. [Google Scholar] [CrossRef]
- Li, Y.; Xu, Q.; Yang, W.; Wu, T.; Lu, X. Oleanolic acid reduces aerobic glycolysis-associated proliferation by inhibiting yes-associated protein in gastric cancer cells. Gene 2019, 712, 143956. [Google Scholar] [CrossRef]
- Wang, T.X.; Zhang, Z.Q.; Cong, Y.; Shi, X.Y.; Liu, Y.H.; Zhao, F.L. Prosapogenin A induces apoptosis in human cancer cells in vitro via inhibition of the STAT3 signaling pathway and glycolysis. Oncol. Lett. 2013, 6, 1323–1328. [Google Scholar] [CrossRef]
- Rihan, M.; Nalla, L.V.; Dharavath, A.; Shard, A.; Kalia, K.; Khairnar, A. Pyruvate Kinase M2: A Metabolic Bug in Re-Wiring the Tumor Microenvironment. Cancer Microenviron. 2019, 12, 149–167. [Google Scholar] [CrossRef]
- Puckett, D.L.; Alquraishi, M.; Chowanadisai, W.; Bettaieb, A. The Role of PKM2 in Metabolic Reprogramming: Insights into the Regulatory Roles of Non-Coding RNAs. Int. J. Mol. Sci. 2021, 22, 1171. [Google Scholar] [CrossRef]
- Zahra, K.; Dey, T.; Ashish; Mishra, S.P.; Pandey, U. Pyruvate Kinase M2 and Cancer: The Role of PKM2 in Promoting Tumorigenesis. Front. Oncol. 2020, 10, 159. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Li, H.; Lu, Y.; Yang, P.; Li, Z. Berberine Inhibited the Proliferation of Cancer Cells by Suppressing the Activity of Tumor Pyruvate Kinase M2. Nat. Prod. Commun. 2017, 12, 1415–1418. [Google Scholar] [CrossRef]
- You, L.; Zhu, H.; Wang, C.; Wang, F.; Li, Y.; Li, Y.; Wang, Y.; He, B. Scutellarin inhibits Hela cell growth and glycolysis by inhibiting the activity of pyruvate kinase M2. Bioorganic Med. Chem. Lett. 2017, 27, 5404–5408. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Ge, Y.; Cui, J.; Yu, Y.; Liu, B. Scutellarin resensitizes oxaliplatin-resistant colorectal cancer cells to oxaliplatin treatment through inhibition of PKM2. Mol. Ther.-Oncolytics 2021, 21, 87–97. [Google Scholar] [CrossRef]
- Tang, W.; Liu, Z.L.; Mai, X.Y.; Qi, X.; Li, D.H.; Gu, Q.Q.; Li, J. Identification of Gliotoxin isolated from marine fungus as a new pyruvate kinase M2 inhibitor. Biochem. Biophys. Res. Commun. 2020, 528, 594–600. [Google Scholar] [CrossRef]
- Anjum, K.; Bi, H.; Chai, W.; Lian, X.-Y.; Zhang, Z. Antiglioma pseurotin A from marine Bacillus sp. FS8D regulating tumour metabolic enzymes. Nat. Prod. Res. 2017, 32, 1353–1356. [Google Scholar] [CrossRef]
- Chen, J.; Xie, J.; Jiang, Z.; Wang, B.; Wang, Y.; Hu, X. Shikonin and its analogs inhibit cancer cell glycolysis by targeting tumor pyruvate kinase-M2. Oncogene 2011, 30, 4297–4306. [Google Scholar] [CrossRef]
- Chai, X.X.; Le, Y.F.; Wang, J.C.; Mei, C.X.; Feng, J.F.; Zhao, H.; Wang, C.; Lu, D.Z. Carpesium abrotanoides (L.) Root as a Potential Source of Natural Anticancer Compounds: Targeting Glucose Metabolism and PKM2/HIF-1α Axis of Breast Cancer Cells. J. Food Sci. 2019, 84, 3825–3832. [Google Scholar] [CrossRef]
- Ruzzolini, J.; Peppicelli, S.; Bianchini, F.; Andreucci, E.; Urciuoli, S.; Romani, A.; Tortora, K.; Caderni, G.; Nediani, C.; Calorini, L. Cancer Glycolytic Dependence as a New Target of Olive Leaf Extract. Cancers 2020, 12, 317. [Google Scholar] [CrossRef]
- Woodford, M.R.; Chen, V.Z.; Backe, S.J.; Bratslavsky, G.; Mollapour, M. Structural and functional regulation of lactate dehydrogenase-A in cancer. Future Med. Chem. 2020, 12, 439–455. [Google Scholar] [CrossRef]
- Lin, Y.C.; Chitcharoenthum, M.; Rikihisa, Y. Effect of gossypol on spermatozoal lactate dehydrogenase-X (LDH-X) in male rats. Contraception 1987, 36, 581–592. [Google Scholar] [CrossRef]
- Wang, S.; Zhao, J.; Ren, S.; Sun, W.; Zhang, W.; Zhang, J. Wogonin affects proliferation and the energy metabolism of SGC-7901 and A549 cells. Exp. Ther. Med. 2019, 17, 911–918. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Cui, X.; Chen, Z. Screening of lactate dehydrogenase inhibitor from bioactive compounds in natural products by electrophoretically mediated microanalysis. J. Chromatogr. A 2021, 1656, 462554. [Google Scholar] [CrossRef] [PubMed]
- Bader, A.; Tuccinardi, T.; Granchi, C.; Martinelli, A.; Macchia, M.; Minutolo, F.; De Tommasi, N.; Braca, A. Phenylpropanoids and flavonoids from Phlomis kurdica as inhibitors of human lactate dehydrogenase. Phytochemistry 2015, 116, 262–268. [Google Scholar] [CrossRef]
- Manerba, M.; Vettraino, M.; Fiume, L.; Di Stefano, G.; Sartini, A.; Giacomini, E.; Buonfiglio, R.; Roberti, M.; Recanatini, M. Galloflavin (CAS 568-80-9): A Novel Inhibitor of Lactate Dehydrogenase. ChemMedChem 2012, 7, 311–317. [Google Scholar] [CrossRef]
- De Leo, M.; Peruzzi, L.; Granchi, C.; Tuccinardi, T.; Minutolo, F.; Tommasi, N.D.; Braca, A. Constituents of Polygala flavescens ssp. flavescens and Their Activity as Inhibitors of Human Lactate Dehydrogenase. J. Nat. Prod. 2017, 80, 2077–2087. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, D.; Han, S.; Wang, N.; Mo, F.; Loo, T.Y.; Shen, J.; Huang, H.; Chen, J. Bioactivity-Guided Identification and Cell Signaling Technology to Delineate the Lactate Dehydrogenase A Inhibition Effects of Spatholobus suberectus on Breast Cancer. PLoS ONE 2013, 8, e56631. [Google Scholar] [CrossRef]
- Cheng, G.; Pi, Z.; Zheng, Z.; Liu, S.; Liu, Z.; Song, F. Magnetic nanoparticles-based lactate dehydrogenase microreactor as a drug discovery tool for rapid screening inhibitors from natural products. Talanta 2020, 209, 120554. [Google Scholar] [CrossRef]
- Salas, M.; Obando, P.; Ojeda, L.; Ojeda, P.; Pérez, A.; Vargas-Uribe, M.; Rivas, C.I.; Vera, J.C.; Reyes, A.M. Resolution of the direct interaction with and inhibition of the human GLUT1 hexose transporter by resveratrol from its effect on glucose accumulation. Am. J. Physiol. Cell Physiol. 2013, 305, 90–99. [Google Scholar] [CrossRef]
- Maxwell, P.H.; Pugh, C.W.; Ratcliffe, P.J. Activation of the HIF pathway in cancer. Curr. Opin. Genet. Dev. 2001, 11, 293–299. [Google Scholar] [CrossRef]
- Kaelin, W.G., Jr.; Ratcliffe, P.J. Oxygen sensing by metazoans: The central role of the HIF hydroxylase pathway. Mol. Cell 2008, 30, 393–402. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.; Arora, R.; Kaur, P.; Singh, B.; Mannan, R.; Arora, S. Overexpression of hypoxia-inducible factor and metabolic pathways: Possible targets of cancer. Cell Biosci. 2017, 71, 62. [Google Scholar] [CrossRef] [PubMed]
- Semenza, G.L. Defining the role of hypoxia-inducible factor 1 in cancer biology and therapeutics. Oncogene 2010, 29, 625–634. [Google Scholar] [CrossRef] [PubMed]
- Laughner, E.; Taghavi, P.; Chiles, K.; Mahon, P.C.; Semenza, G.L. HER2 (neu) Signaling Increases the Rate of Hypoxia-Inducible Factor 1α (HIF-1α) Synthesis: Novel Mechanism for HIF-1-Mediated Vascular Endothelial Growth Factor Expression. Mol. Cell. Biol. 2001, 21, 3995–4004. [Google Scholar] [CrossRef]
- Zou, D.; Han, T.; Deng, H.; Shao, X.; Guo, X.; Qi, X. Hypoxia-inducible factors in cancer: An overview of major findings from meta-analyses. AME Med. J. 2017, 2, 48. [Google Scholar] [CrossRef]
- Xie, Y.; Shi, X.; Sheng, K.; Han, G.; Li, W.; Zhao, Q.; Jiang, B.; Feng, J.; Li, J.; Gu, Y. PI3K/Akt signaling transduction pathway, erythropoiesis and glycolysis in hypoxia (Review). Mol. Med. Rep. 2019, 19, 783–791. [Google Scholar] [CrossRef]
- Woo, Y.M.; Shin, Y.; Lee, E.J.; Lee, S.; Jeong, S.H.; Kong, H.K.; Park, E.Y.; Kim, H.K.; Han, J.; Chang, M.; et al. Inhibition of Aerobic Glycolysis Represses Akt/mTOR/HIF-1α Axis and Restores Tamoxifen Sensitivity in Antiestrogen-Resistant Breast Cancer Cells. PLoS ONE 2015, 10, e0132285. [Google Scholar] [CrossRef]
- Dong, Q.; Li, Q.; Duan, L.; Yin, H.; Wang, X.; Liu, Y.; Wang, B.; Li, K.; Yao, X.; Yuan, G.; et al. Biochanin A Inhibits Glioblastoma Growth via Restricting Glycolysis and Mitochondrial Oxidative Phosphorylation. Front. Oncol. 2021, 11, 652008. [Google Scholar] [CrossRef]
- Mao, L.; Chen, Q.; Gong, K.; Xu, X.; Xie, Y.; Zhang, W.; Cao, H.; Hu, T.; Hong, X.; Zhan, Y. Berberine decelerates glucose metabolism via suppression of mTOR-dependent HIF-1α protein synthesis in colon cancer cells. Oncol. Rep. 2018, 39, 2436–2442. [Google Scholar] [CrossRef]
- Wang, J.; Qi, Q.; Feng, Z.; Zhang, X.; Huang, B.; Chen, A.; Prestegarden, L.; Li, X.; Wang, J.; Wang, J.; et al. Berberine induces autophagy in glioblastoma by targeting the AMPK/mTOR/ULK1-pathway. Oncotarget 2016, 7, 66944–66958. [Google Scholar] [CrossRef]
- Siddiqui, F.A.; Prakasam, G.; Chattopadhyay, S.; Rehman, A.U.; Padder, R.A.; Ansari, M.A.; Irshad, R.; Mangalhara, K.; Bamezai, R.N.K.; Husain, M.; et al. Curcumin decreases Warburg effect in cancer cells by down-regulating pyruvate kinase M2 via mTOR-HIF1α inhibition. Sci. Rep. 2018, 8, 8323. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Fan, H.; Chen, Q.; Ma, G.; Zhu, M.; Zhang, X.; Zhang, Y.; Yu, J. Curcumin inhibits aerobic glycolysis and induces mitochondrial-mediated apoptosis through hexokinase II in human colorectal cancer cells in vitro. Anti-Cancer Drugs 2015, 26, 15–24. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Zhang, X.; Wang, Y.; Sun, Q.; Chen, M.; Liu, S.; Zou, X. Licochalcone A suppresses hexokinase 2-mediated tumor glycolysis in gastric cancer via downregulation of the Akt signaling pathway. Oncol. Rep. 2018, 39, 1181–1190. [Google Scholar] [CrossRef] [PubMed]
- Xue, L.; Zhang, W.J.; Fan, Q.X.; Wang, L.X. Licochalcone A inhibits PI3K/Akt/mTOR signaling pathway activation and promotes autophagy in breast cancer cells. Oncol. Lett. 2018, 15, 1869–1873. [Google Scholar] [CrossRef]
- Liu, Y.; Tong, L.; Luo, Y.; Li, X.; Chen, G.; Wang, Y. Resveratrol inhibits the proliferation and induces the apoptosis in ovarian cancer cells via inhibiting glycolysis and targeting AMPK/mTOR signaling pathway. J. Cell. Biochem. 2018, 119, 6162–6172. [Google Scholar] [CrossRef]
- Li, W.; Ma, X.; Li, N.; Liu, H.; Dong, Q.; Zhang, J.; Yang, C.; Liu, Y.; Liang, Q.; Zhang, S.; et al. Resveratrol inhibits Hexokinases II mediated glycolysis in non-small cell lung cancer via targeting Akt signaling pathway. Exp. Cell Res. 2016, 349, 320–327. [Google Scholar] [CrossRef]
- Yuan, J.; Peng, G.; Xiao, G.; Yang, Z.; Huang, J.; Liu, Q.; Yang, Z.; Liu, D. Xanthohumol suppresses glioblastoma via modulation of Hexokinase 2 -mediated glycolysis. J. Cancer 2020, 11, 4047–4058. [Google Scholar] [CrossRef]
- Yang, W.; Zheng, Y.; Xia, Y.; Ji, H.; Chen, X.; Guo, F.; Lyssiotis, C.A.; Aldape, K.; Cantley, L.C.; Lu, Z. ERK1/2-dependent phosphorylation and nuclear translocation of PKM2 promotes the Warburg effect. Nat. Cell Biol. 2012, 14, 1295–1304. [Google Scholar] [CrossRef]
- Papa, S.; Choy, P.M.; Bubici, C. The ERK and JNK pathways in the regulation of metabolic reprogramming. Oncogene 2018, 38, 2223–2240. [Google Scholar] [CrossRef]
- Kim, H.S.; Wannatung, T.; Lee, S.; Yang, W.K.; Chung, S.H.; Lim, J.S.; Choe, W.; Kang, I.; Kim, S.S.; Ha, J. Quercetin enhances hypoxia-mediated apoptosis via direct inhibition of AMPK activity in HCT116 colon cancer. Apoptosis 2012, 17, 938–949. [Google Scholar] [CrossRef]
- Triantafyllou, A.; Mylonis, I.; Simos, G.; Bonanou, S.; Tsakalof, A. Flavonoids induce HIF-1alpha but impair its nuclear accumulation and activity. Free Radic. Biol. Med. 2008, 44, 657–670. [Google Scholar] [CrossRef] [PubMed]
- Mylonis, I.; Lakka, A.; Tsakalof, A.; Simos, G. The dietary flavonoid kaempferol effectively inhibits HIF-1 activity and hepatoma cancer cell viability under hypoxic conditions. Biochem. Biophys. Res. Commun. 2010, 398, 74–78. [Google Scholar] [CrossRef] [PubMed]
- Ilieș, M.; Uifălean, A.; Pașca, S.; Dhople, V.M.; Lalk, M.; Iuga, C.A.; Hammer, E. From Proteomics to Personalized Medicine: The Importance of Isoflavone Dose and Estrogen Receptor Status in Breast Cancer Cells. J. Pers. Med. 2020, 10, 292. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.; Lee, H.Y.; Ahn, D.R.; Kim, S.Y.; Kim, S.; Lee, K.B.; Lee, Y.M.; Park, H.; Yang, E.G. Baicalein Induces Functional Hypoxia-Inducible Factor-1α and Angiogenesis. Mol. Pharmacol. 2008, 74, 70–81. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Chen, A.Y.; Rojanasakul, Y.; Ye, X.; Rankin, G.O.; Chen, Y.C. Dietary compounds galangin and myricetin suppress ovarian cancer cell angiogenesis. J. Funct. Foods 2015, 15, 464–475. [Google Scholar] [CrossRef]
- Chen, K.; Ye, J.; Qi, L.; Liao, Y.; Li, R.; Song, S.; Zhou, C.; Feng, R.; Zhai, W. Oridonin inhibits hypoxia-induced epithelial-mesenchymal transition and cell migration by the hypoxia-inducible factor-1α/matrix metallopeptidase-9 signal pathway in gallbladder cancer. Anti-Cancer Drugs 2019, 30, 925–932. [Google Scholar] [CrossRef]
- Fu, B.; Xue, J.; Li, Z.; Shi, X.; Jiang, B.H.; Fang, J. Chrysin inhibits expression of hypoxia-inducible factor-1alpha through reducing hypoxia-inducible factor-1alpha stability and inhibiting its protein synthesis. Mol. Cancer Ther. 2007, 6, 220–226. [Google Scholar] [CrossRef]
- Park, J.J.; Hwang, S.J.; Park, J.H.; Lee, H.J. Chlorogenic acid inhibits hypoxia-induced angiogenesis via down-regulation of the HIF-1α/AKT pathway. Cell. Oncol. 2015, 38, 111–118. [Google Scholar] [CrossRef]
- Liu, Y.; Lin, F.; Chen, Y.; Wang, R.; Liu, J.; Jin, Y.; An, R. Cryptotanshinone inhibites bladder cancer cell proliferation and promotes apoptosis via the PTEN/PI3K/AKT pathway. J. Cancer 2020, 11, 488–499. [Google Scholar] [CrossRef]
- Zhang, Q.; Tang, X.; Lu, Q.; Zhang, Z.; Rao, J.; Le, A.D. Green tea extract and (−)-epigallocatechin-3-gallate inhibit hypoxia- and serum-induced HIF-1α protein accumulation and VEGF expression in human cervical carcinoma and hepatoma cells. Mol. Cancer Ther. 2006, 5, 1227–1238. [Google Scholar] [CrossRef]
- Wang, F.; Zhang, W.; Guo, L.; Bao, W.; Jin, N.; Liu, R.; Liu, P.; Wang, Y.; Guo, Q.; Chen, B. Gambogic acid suppresses hypoxia-induced hypoxia-inducible factor-1α/vascular endothelial growth factor expression via inhibiting phosphatidylinositol 3-kinase/Akt/mammalian target protein of rapamycin pathway in multiple myeloma cells. Cancer Sci. 2014, 105, 1063–1070. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Tang, X.; Lu, Q.Y.; Zhang, Z.F.; Brown, J.; Le, A.D. Resveratrol inhibits hypoxia-induced accumulation of hypoxia-inducible factor-1α and VEGF expression in human tongue squamous cell carcinoma and hepatoma cells. Mol. Cancer Ther. 2005, 4, 1465–1474. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhao, L.; Zhu, L.T.; Wang, Y.; Pan, D.; Yao, J.; You, Q.D.; Guo, Q.L. Wogonin reverses hypoxia resistance of human colon cancer HCT116 cells via downregulation of HIF-1α and glycolysis, by inhibiting PI3K/Akt signaling pathway. Mol. Carcinog. 2014, 53, 22052. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.H.; Woo, J.K.; Jin, Q.; Kang, H.J.; Jeong, J.W.; Kim, K.W.; Hong, W.K.; Lee, H.Y. Identification of novel antiangiogenic anticancer activities of deguelin targeting hypoxia-inducible factor-1 alpha. Int. J. Cancer 2008, 122, 5–14. [Google Scholar] [CrossRef]
- Wang, J.Y.; Wang, Z.; Li, M.Y.; Zhang, Z.; Mi, C.; Zuo, H.X.; Xing, Y.; Wu, Y.L.; Lian, L.H.; Xu, G.H.; et al. Dictamnine promotes apoptosis and inhibits epithelial-mesenchymal transition, migration, invasion and proliferation by downregulating the HIF-1α and Slug signaling pathways. Chem.-Biol. Interact. 2018, 296, 134–144. [Google Scholar] [CrossRef]
- Zhu, Z.; Wang, Y.; Liu, Z.; Wang, F.; Zhao, Q. Inhibitory effects of epigallocatechin-3-gallate on cell proliferation and the expression of HIF-1α and P-gp in the human pancreatic carcinoma cell line PANC-1. Oncol. Rep. 2012, 27, 1567–1572. [Google Scholar] [CrossRef][Green Version]
- Mukund, V.; Saddala, M.S.; Farran, B.; Mannavarapu, M.; Alam, A.; Nagaraju, G.P. Molecular docking studies of angiogenesis target protein HIF-1α and genistein in breast cancer. Gene 2019, 701, 169–172. [Google Scholar] [CrossRef]
- Melstrom, L.G.; Salabat, M.R.; Ding, X.Z.; Strouch, M.J.; Grippo, P.J.; Mirzoeva, S.; Pelling, J.C.; Bentrem, D.J. Apigenin Down-Regulates the Hypoxia Response Genes: HIF-1α, GLUT-1, and VEGF in Human Pancreatic Cancer Cells. J. Surg. Res. 2011, 167, 173–181. [Google Scholar] [CrossRef]
- Shan, B.; Schaaf, C.; Schmidt, A.; Lucia, K.; Buchfelder, M.; Losa, M.; Kuhlen, D.; Kreutzer, J.; Perone, M.J.; Arzt, E.; et al. Curcumin suppresses HIF1A synthesis and VEGFA release in pituitary adenomas. J. Endocrinol. 2012, 214, 389–398. [Google Scholar] [CrossRef]
- Tan, C.; Zhang, L.; Cheng, X.; Lin, X.F.; Lu, R.R.; Bao, J.D.; Yu, H.X. Curcumin inhibits hypoxia-induced migration in K1 papillary thyroid cancer cells. Exp. Biol. Med. 2014, 240, 925–935. [Google Scholar] [CrossRef]
- Gade, S.; Gandhi, N.M. Baicalein Inhibits MCF-7 Cell Proliferation In Vitro, Induces Radiosensitivity, and Inhibits Hypoxia Inducible Factor. J. Environ. Pathol. Toxicol. Oncol. 2015, 34, 299–308. [Google Scholar] [CrossRef] [PubMed]
- Park, M.K.; Ji, J.; Haam, K.; Han, T.H.; Lim, S.; Kang, M.J.; Lim, S.S.; Ban, H.S. Licochalcone A inhibits hypoxia-inducible factor-1α accumulation by suppressing mitochondrial respiration in hypoxic cancer cells. Biomed. Pharmacother. 2021, 133, 111082. [Google Scholar] [CrossRef] [PubMed]
- Monti, E.; Marras, E.; Prini, P.; Gariboldi, M.B. Luteolin impairs hypoxia adaptation and progression in human breast and colon cancer cells. Eur. J. Pharmacol. 2020, 881, 173210. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.C.; Lee, C.F.; Huang, W.H.; Chou, T.C. Magnolol suppresses hypoxia-induced angiogenesis via inhibition of HIF-1α/VEGF signaling pathway in human bladder cancer cells. Biochem. Pharmacol. 2013, 85, 1278–1287. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Zhou, Y.; Qiao, C.; Ni, T.; Li, Z.; You, Q.; Guo, Q.; Lu, N. Oroxylin A inhibits glycolysis-dependent proliferation of human breast cancer via promoting SIRT3-mediated SOD2 transcription and HIF1α destabilization. Cell Death Dis. 2015, 64, e1714. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.H.; Lee, Y.J. Quercetin suppresses hypoxia-induced accumulation of hypoxia-inducible factor-1α (HIF-1α) through inhibiting protein synthesis. J. Cell. Biochem. 2008, 105, 546–553. [Google Scholar] [CrossRef]
- Sun, Y.; Wang, H.; Liu, M.; Lin, F.; Hua, J. Resveratrol abrogates the effects of hypoxia on cell proliferation, invasion and EMT in osteosarcoma cells through downregulation of the HIF-1α protein. Mol. Med. Rep. 2015, 11, 1975–1981. [Google Scholar] [CrossRef]
- Fu, R.; Chen, Y.; Wang, X.P.; An, T.; Tao, L.; Zhou, Y.X.; Huang, Y.J.; Chen, B.A.; Li, Z.Y.; You, Q.D.; et al. Wogonin inhibits multiple myeloma-stimulated angiogenesis via c-Myc/VHL/HIF-1α; signaling axis. Oncotarget 2015, 7, 5715–5727. [Google Scholar] [CrossRef]
- Bae, M.K.; Kim, S.H.; Jeong, J.W.; Lee, Y.M.; Kim, H.S.; Kim, S.R.; Yun, I.; Bae, S.K.; Kim, K.W. Curcumin inhibits hypoxia-induced angiogenesis via down-regulation of HIF-1. Oncol. Rep. 2006, 15, 1557–1562. [Google Scholar] [CrossRef]
- Li, S.; Li, J.; Dai, W.; Zhang, Q.; Feng, J.; Wu, L.; Liu, T.; Yu, Q.; Xu, S.; Wang, W.; et al. Genistein suppresses aerobic glycolysis and induces hepatocellular carcinoma cell death. Br. J. Cancer 2017, 117, 1518–1528. [Google Scholar] [CrossRef]

| Natural Compound/Extract | In Vitro Model | Study Design | Analytical Methods | Effect | Altered Pathways | Ref |
|---|---|---|---|---|---|---|
| Rosmary extract containing 256 µg/mL carnosic acid and 37.1 µg/mL carnosol Standard carnosic acid | Colorectal adenocarcinoma cells (HT-29) | 9.9 µg/mL standard carnosic acid for 48 h | CE-TOF MS and HILIC/UHPLC-TOF MS | ↑ GSH ↓ GSSG ↑ GSH/GSSG ratio ↓ N-acetylputrescine | glutathione metabolism, polyamine metabolism | [29] |
| Rosemary extract containing 226.39 µg/mg carnosol and 51.55 µg/mg carnosic acid | Colorectal adenocarcinoma cells (HT-29) | 10 µM rosemary phenols | CE-ESI-TOF MS HILIC/UPLC-ESI-TOF MS | ↓ GSH/GSSG ratio | glutathione metabolism, polyamine metabolism, urea cycle and metabolism of amino groups | [30] |
| Five rosemary phenolic extracts | Chronic myelogenous leukemia cells (K-562) and daunomycin-resistant chronic myelogenous leukemia cells (K-562/R) | 5, 10 µM | CE-TOF MS and UPLC-TOF MS | ↑ methionine ↑ leucine ↑ glutamine ↑ tyrosine ↓ lysine | aminoacyl-tRNA biosynthesis, glutathione metabolism, arginine, proline metabolism, nitrogen metabolism, urea cycle | [31] |
| Ionic liquid-Graviola fruit pulp extract | Colorectal adenocarcinoma cells (HT-29) | 10.56 μg/mL for 48 h | GC-TOF MS | ↑ thiocyanic acid ↓ lactic acid ↓ L-alanine ↓ tricosadiynoic acid | amino acid metabolism, aerobic glycolysis, urea cycle, ketone bodies metabolism | [32] |
| Soy seed extract (SSE), standard genistein (Gen), standard daidzein (Dai) | Breast adenocarcinoma cells (MCF7, MDA-MB-231) | For MCF7 cells: 23 μM Gen, 52 μM Dai, 166 μg/mL SSE; For MDA-MB-231 cells: 11 μM Gen, 36 μM Dai, 26 μg/mL SSE | 1H-NMR GC-MS | ↓ glucose uptake ↓ glutamine uptake | glucose transport, glycolysis, protein biosynthesis | [33] |
| Extra virgin olive oil extract | Colorectal adenocarcinoma cells (HT-29, SW480) | 0.01–0.1% | nanoLC-ESI-TOF MS | cell cycle, metabolism of polyphenols | [34] | |
| Resveratrol | Hepatocellular carcinoma cells (Hep G2) | 40 μM | 1H-NMR | ↓ use of glucose and amino acids ↓ lactate release ↑ succinate use | glycolytic activity, energy production | [35] |
| Breast adenocarcinoma cells (MCF7, MDA-MB-231) | 5–100 mM for 72 h | LC-MS (Targeted approach) | ↑ amino acids levels, ↑ serotonin, kynurenine, and putrescine synthesis | biogenic amine metabolism, arachidonic acid pathway | [36] | |
| Curcumin | Breast adenocarcinoma cells (MCF7, MDA-MB-231) | 0.5, 2.5, 10, 25, and 50 mg/L for 24 h for dose-dependent effect analysis; 10 mg/L for 24, 48, 72, and 96 h for the analysis of the time-dependent response | 1H-NMR | -biphasic effect (≤28 μM, ↑ total GSH; ≥70 μM ↓ total GSH) -at high doses: accumulation of polyunsaturated and total free fatty acids, decrease in glycerophospho-ethanolamine and choline | glutathione metabolism, lipid metabolism | [37] |
| Natural Compound | In Vitro Models | Study Design | Ref |
|---|---|---|---|
| Inhibitors of GLUT transporters | |||
| Genistein Phloretin Apigenin Daidzein | Prostate carcinoma cells (LNCaP) Normal prostatic epithelial cells immortalized with simian virus 40 (PNT1A) | Cell proliferation assay (Hoechst assay—48 h): 0 μM–100 μM for genistein and phloretin, 0 μM–50 μM for apigenin, 0 μM–140 μM for daidzein Glucose uptake (20 min, 1 h, 24 h) at IC50 Western blot for GLUT1,4 protein levels (6, 24, 48 h) at IC50 Immunocytochemistry for GLUT1,4 subcellular location (24, 48 h) at IC50 Molecular docking study (Target XylE 0 E. coli homolog of GLUT1-4) | [3] |
| Silybin | Colorectal adenocarcinoma cells (LoVo) Doxorubicin-resistant colorectal adenocarcinoma cells (LoVo-DOX) | Cell viability assay (MTT assay, 24, 48, 72 h) at 5, 10, 50 μM Western blot for GLUT1 protein levels (48 h) at 10, 50 μM | [50] |
| Wogonin | Colorectal carcinoma cells with different p53 expressions (HCT116, HT-29), normal colon epithelial cells (NCM460); Hepatocellular carcinoma cells with different p53 expressions (Hep G2, SMMC-7721), normal hepatic cells (L02); Ovarian cancer cells with different p53 expressions (A2780, SK-OV-3), normal ovarian cells (IOSE-80) | Cell viability assay (MTT assay, 24 h) Measurement of intracellular ROS Measurement of glucose uptake level and lactate generation ATP assessment Western blot for the protein expression and real-time PCR for the mRNA level of p53, TIGAR, HK2, LDH-A, PDK, GLUT1, and PGM In vivo tumor growth | [52] |
| Resveratrol | Acute promyelocytic leukemia cells (HL-60) and histiocytic lymphoma cells (U-937) | Glucose uptake at 0.1–100 μM resveratrol | [115] |
| Glucopiericidin A | Epidermoid carcinoma (A-431) | CE-MS metabolomics, cells grown in serum-reduced media for 18 h and treated for 30 min CE-MS [13C]-glucose labeling study Glucose uptake | [60] |
| Bitter melon juice (MBJ, derived from Momordica charantia fruits) | Pancreatic cancer cells (PANC-1, BxPC3) | Cell growth and viability assay (Trypan blue exclusion assay, 72, 96, 120, 144 h) NMR metabolomics analysis with 2% BMJ Immunofluorescence analysis of glucose and lactate transporter expression status after treatment with 2% BMJ | [61] |
| Inhibitors of hexokinase 2 | |||
| Xanthohumol | Colorectal carcinoma cells (FHC, CCD841 CoN, HT-29, SW480, LoVo, HCT116, and SW620) | Cell viability assay (MTS assay, 24, 48, 72 h) at 2, 4, 8 μM Glucose uptake and lactate production measurement Immunoblotting for HK2 expression In vivo tumor growth | [73] |
| Quercetin | Hepatocellular carcinoma cells (SMMC-7721, BEL-7402) Normal hepatic cells (LO2) | Cell proliferation assay (MTS assay, 24 h) at 12.5, 25, 50 µM Glucose uptake and lactate production assays Western blot assay for HK2 protein expression Western blot for the expression of p-Akt/Akt, p-mTOR/mTOR In vivo tumor growth and IHC staining | [74] |
| Deguelin | Non-small cell lung cancer cells (H460, H1650, H1299, H520, HCC827, H1975, and H358) | Cell viability assay (MTS assay, 24, 48, 72 h) at 1, 2, 5 μM Western blot assay for HK2 protein expression Glucose uptake and lactate production measurement In vivo tumor growth | [75] |
| Chrysin | Hepatocellular carcinoma cells (Hep G2, Hep3B, Huh-7, HCC-LM3, BEL-7402, and SMMC-7721) Normal hepatic cells (LO2) | Cell viability assay (Cell Titer-Glo assay, 0, 24, 48, 72 h) at 15, 30, 60 μM Western blot assay for HK2 protein expression Apoptosis assays Glucose uptake and lactate production measurement at 15, 30, and 60 μM In vivo tumor growth assay and IHC staining | [76] |
| Orxoylin A | Breast adenocarcinoma cells (MDA-MB-231, MCF7) | Cell viability assay (MTT assay, 48 h) at 0–250 μM Glucose uptake and lactate production measurement Western blot assays for PFKFB1/4, PFKFB2, PKM2, LDH and GLUT1 (at 150 μM) Western blot analysis for HK2 expression (at 100, 150, and 200 μM) | [77] |
| Dioscin | Colorectal carcinoma cells (HCT116, HT-29, DLD1, and SW620) | Cell viability assay (Cell Titer-Glo assay, 24, 48, 72 h) at 0, 1, 2, 5 µM Tumor glycolysis measurement Western blot analysis for HK2 protein expression Apoptosis assays In vivo tumor growth and IHC staining | [78] |
| Costunolide | Primary hepatic stellate cells | Cell viability assay (MTT assay, 24 h) at 10, 20, and 30 μM Glucose uptake and intracellular lactate Measurement of hexokinase activity Western blot assay for HK2 protein expression | [79] |
| (22E,24R)-6β-methoxyergosta-7,9(11),22-triene- 3β,5α-diol | Pancreatic ductal adenocarcinoma cells (SW1990) African green monkey kidney cells (Vero cells) | In vitro HK2 enzyme inhibition assay Molecular docking to human HK2 Microscale thermophoresis assay for the binding affinity of Ganoderma sinense potential ligands to target Cytotoxicity assay (Cell Counting Kit -8-48 h) to 0–100 μM | [81] |
| Inhibitors of phosphofructokinase 1 (PFK-1) | |||
| Resveratrol | Breast adenocarcinoma cells (MCF7) | Cell viability assay (MTT assay, 24 h) at 0, 1, 5, 15, 50, and 100 µM Glucose consumption and lactate production assays LDH leakage, ATP content, and PFK activity assay Radioassay for PFK activity and PFK intrinsic fluorescence measurement | [87] |
| Bergapten | Breast cancer cells (MCF7, ZR75-1) | Western blot for PFK-1 expression (6 and 16 h, at 20 and 50 µM) Glucose and LDH assays Triglyceride and lipase activity assay G6PDH activity and ATP assay Isocitrate dehydrogenase and malic enzyme activity assay | [88] |
| Epigallocatechin-3-gallate | Hepatocellular carcinoma cells (HCC-LM3, Huh-7, Hep G2, Hep3B, SMMC-7721) Normal hepatic cells (LO2) | Glucose uptake, lactate production (25, 50, 100 μM) RT-PCR analysis for the expression of glycolysis-associated genes Western blot assay for PKF protein expression PFK activity (25, 50, 100, and 200 μM) Cell proliferation assay (CCK assay -8; 24 h; 25, 50, 100, 200 and 400 μM) Animal experiments | [90] |
| Sulforaphane | Hepatocellular carcinoma cells (Huh-7, SNU-449, and NCTC cells) | Cell proliferation assay (Real-time cell electronic sensing) Apoptosis assays, Cell cycle analysis Protein analysis (MS-based protein analysis and RT-PCR validation) PFK expression validation through immunoblot and RNAi technique | [91] |
| Worenin | Colorectal carcinoma cells (HCT-116, SW-620) Normal colon cells FHC (CRL-1831) | Cell viability assay (MTT assay, 24 h at 0, 1, 5, 10, 20, 40 or 80 μM) Colony formation test and cell cycle analysis Glucose consumption and lactate production assays Western blot for GLUT3, HK2, PKM2, and LDHA protein expressions RT-PCR analysis for GLUT3, HK2, PKM2, and LDHA mRNA expressions Western blot and immunofluorescence staining for nuclear HIF-1α expression | [92] |
| Oleanolic acid | Gastric cancer cells (MKN-45, SGC-7901) Normal gastric mucosal epithelium cells (GES-1) | Cell viability assay (MTT assay) and proliferation (BrdU incorporation) assays (24 h, at 0, 10, 20, 30 µM for the cancerous cells, and up to 80 µM for the normal cells) Glucose consumption and lactate production assays Western blot assay for HK2, PFK-1, HIF-1α, and YAP protein expression RT-PCR assay for HIF-1α, HK2, and PFK-1 mRNA expression | [93] |
| Prosapogenin A | Cervical adenocarcinoma cells (HeLa) Hepatocellular carcinoma cells (Hep G2) Breast adenocarcinoma cells (MCF7) Non-cancer cells (7701, 293) | Cell viability assay (MTT assay, 24, 48, and 72 h) at 10 μM Cell apoptosis and cell cycle analysis Western blot assay to detect the STAT3 mRNA protein level RT-PCR to detect the STAT3, GLUT1, HK, and PFKL mRNA level (5 µM, 48 h) | [94] |
| Inhibitors of pyruvate kinase (PKM) | |||
| Berberine | Colorectal cancer cells (HCT-116) Cervical adenocarcinoma cells (HeLa) | Cell viability assay (MTT assay, 24 h) at 50–300 μM Western blot assay for PKM protein expression Pyruvate kinase assay (100–300 μM) | [98] |
| Scutellarin | Cervical adenocarcinoma cells (HeLa) | Biotinylated scutellareins as probes for target identification in HeLa lysate | [99] |
| Colorectal adenocarcinoma cells (SW480, HT-29) Oxaliplatin-resistant colorectal cancer cells (OR-SW480, OR-HT-29) | Cell viability assay (MTT assay, 24, 48 h) at 0–40 μM Western blot analysis for PKM protein expression Glucose, lactate, and ATP assays | [100] | |
| Gliotoxin | Brain cancer (glioblastoma) cells (U87, U251) Leukemia cells (HL-60, K-562) Non-small cell lung cancer cells (H1975) Prostate adenocarcinoma cells (PC-3) Colorectal cancer cells (HCT-116) Cervical adenocarcinoma cells (HeLa) | PK activity assay Cell viability assay (MTT assay, 72 h)—all cell lines Western blot assay (0.1–0.5, 48 h U87) Cellular thermal shift assay (CETSA assay 6 h, U87) Glucose and lactic acid assays (U87, U251) | [101] |
| Pseurotin A | Brain cancer (glioblastoma) cells (U87-MG, C6, U251, and SHG-44) | Cell growth inhibition assay (sulforhodamine B assay) Western blot assay for PKM protein expression | [102] |
| Shikonin | Breast adenocarcinoma cells (MCF7) Lung adenocarcinoma cells (A549) Cervical adenocarcinoma cells (HeLa) | Gel electrophoresis protein separation and MS detection PKM and LDH activity assay (20–200 μM and 40 μM) Glucose and lactate assays (2.5–20 μM) Cell viability assay (trypan blue assay, 6 h) at 10 μM | [103] |
| Olive leaf extract enriched in oleuropein | Melanoma cells (A375) | Cell viability assay (MTT assay, 24, 48, 72 h) at 0–800 µM Cell cycle analysis and wound healing assay RT-PCR and western blot for PKM2, GLUT1, and MCT4 expressions | [105] |
| Inhibitors of lactate dehydrogenase (LDH) | |||
| Galloflavin | Hepatoma carcinoma cells (PLC/PRF/5) | Virtual screening (LDH-A) Assay on purified human LDH-A and LDH-B Cell proliferation assay (neutral red assay, 72 h) Mechanism of cell death (apotox-glotm Triplex Assay, 24 h) Inhibition of lactate production | [111] |
| Epigallocatechin (Spatholobus suberectus Dunn. Subfractionation) | Breast adenocarcinoma cells (MCF7, MDA-MB-231) | In vitro LDH-A activity assay (screening of subfractions) RT-PCR assay for LDH-A mRNA level Western blot assay for LDH-A protein expression In vivo tumor growth | [113] |
| Wogonin | Gastric cancer cells (SGC-7901) and lung adenocarcinoma cells (A549) | Cell viability assay (MTT assay, 48 h) at 5, 10, 15, 20, 25 and 30 µg/mL Trypan blue exclusion assay and cell morphological assessment (48 h, at 15 µg/mL) Enzyme activity assays (48 h at 15 µg/mL) | [108] |
| (−)-epicatechin-3-O-gallate, ω-hydroxyemodin, emodin-1-O-gluciside, lunatin, chrysophanic acid, rhein-O-(acetyl)-glucoside, aloe-emodin, rhein and emodin | LDH functionalized magnetic nanoparticles study | [114] | |
| Natural Compound | In Vitro Models | Study Design | Ref |
|---|---|---|---|
| Inhibitors of HIF-1α synthesis | |||
| By targeting the upstream pathways | |||
| Apigenin | Hepatocellular carcinoma cells (Hep G2) | 0, 10, 20, and 40 μM for 12 h | [9] |
| Biochanin A | Glioblastoma multiforme cells (U251) | 0, 50, and 100 μM for 48 h | [124] |
| Berberine | Colorectal carcinoma cells (HCT116, KM12C) | 0–100 μM for 24 h for HCT116 cells or 15 h for KM12C cells | [125] |
| Chrysin | Prostate cancer cells (DU145) | serum-starved cells stimulated with insulin (200 nmol/L) for 6 h, followed by 30 μmol/L chrysin for 30 min | [143] |
| Chlorogenic acid | Lung cancer cells (A549) | 2 μM or 10 μM for 16 h, followed by exposure to 200 μM cobalt chloride for 6 h | [144] |
| Cryptotanshinone | Bladder carcinoma cells (5637, T24) | 0, 20, 40, and 80 μM for 48 h | [145] |
| Epigallocatechin-3-gallate (EGCT) and green tea extract (GTE) | Cervical adenocarcinoma cells (HeLa) and hepatocellular carcinoma (Hep G2) cells | 10–80 μg/mL GTE, 10–100 μM EGCG, under hypoxic and normoxic conditions for 16 h | [146] |
| Gambogic acid | Multiple myeloma cells (U266) | 0.2 μM for 4 h under hypoxic conditions | [147] |
| Quercetin | Colorectal carcinoma cells (HCT116), prostate cancer cells (DU145), cervical adenocarcinoma cells (HeLa S3) | 0, 50, and 100 μM for 12 h | [136] |
| Resveratrol | Tongue squamous cell carcinoma (SCC-9) and hepatocellular carcinoma cells (Hep G2) | 5, 50, and 100 μM under hypoxic and normoxic conditions for 1 h or 16 h | [148] |
| Wogonin | Colorectal carcinoma cells (HCT116) | 20, 40, 60, 80, and 100 mM for 24 h | [149] |
| By direct mechanisms | |||
| Deguelin | Non-small cell lung cancer cells (H1299, A549), prostate adenocarcinoma cells (PC-3), gastric cancer cells (MKN-45), breast adenocarcinoma cells (MCF7), renal carcinoma cells (786-0) | 100 nM for 6 h, under hypoxic and normoxic conditions | [150] |
| Dictamnine | Colorectal carcinoma cells (HCT116), cervical adenocarcinoma cells (HeLa), hepatic adenocarcinoma cells (SK-Hep1), lung carcinoma cells (A549) | 0, 10, 30, and 100 μM for 12 h, under hypoxic and normoxic conditions | [151] |
| Epigallocatechin-3-gallate (EGCG) | Pancreatic cancer cells (PANC-1) | 0, 20, 40, and 80 µg/mL EGCG under hypoxic conditions for 24 h, no EGCG under normoxic conditions | [152] |
| Genistein | Breast cancer cells (MDA-MB-231, T-47D) | for MDA-MB-231 cells—100 µM for 24 h; for T-47D cells—50 µM for 24 h | [153] |
| Inhibitors of HIF-1α mRNA expression | |||
| Apigenin | Pancreatic cancer cells (S2-013, CD18) | 0-50 µM for 24 h, under hypoxic and normoxic conditions | [154] |
| Curcumin | Rodent (AtT20, GH3) and human pituitary tumor cells | 0, 10, 20, and 30 μM for 30 min, followed by 125 or 250 μM cobalt chloride exposure for 3 h | [155] |
| Papillary thyroid cancer cells (K1 PTC) | 12.5, 25, and 50 mmol/L for 1 h, followed by exposure to hypoxia for an additional 12 h | [156] | |
| Inhibitors of protein stabilization and accumulation | |||
| Apigenin | Prostate adenocarcinoma cells (PC-3, DU145, and LNCaP), ovarian cancer cells (OVCAR-3), colon cancer cells (HCT-8), and breast adenocarcinoma cells (MCF7) | 0, 20, and 40 μM for 1 h—depending on the experiment purpose | [8] |
| Baicalein | Breast adenocarcinoma cells (MCF7) | 50 µM under hypoxic conditions or with 150 µM cobalt chloride for 8 h | [157] |
| Chrysin | Prostate cancer cells (DU145) | serum-starved cells stimulated with 200 nmol/L insulin for 6 h, followed by 30 μmol/L chrysin for 30 min | [143] |
| Epigallocatechin-3-gallate (EGCG) and green tea extract (GTE) | Cervical adenocarcinoma cells (HeLa) and hepatocellular carcinoma (Hep G2) cells | 10–80 μg/mL GTE, 10–100 μM EGCG, under hypoxic and normoxic conditions for 16 h | [146] |
| Kaempferol | Hepatocellular carcinoma cells (Huh-7) | 0, 1, 5, 10, and 50 μM for 4 h under hypoxic conditions (1% O2) | [138] |
| Licochalcone A | Colorectal carcinoma cells (HCT116), Non-small cell lung cancer cells (H1299), and bronchoalveolar carcinoma cells (H322) | For HCT116 cells: 5–20 μM, for 6 h (or 2–6 h) under hypoxic conditions | [158] |
| Luteolin | Colorectal carcinoma cells (HCT116), breast adenocarcinoma cells (MDA-MB-231) | 0, 10, 25, and 50 μM for 48 h in the presence of 100 μM cobalt chloride for the last 24 h | [159] |
| Magnolol | Bladder cancer cells (T24) | 0, 1, 5, and 10 µM for 8 h under normoxic or hypoxic conditions | [160] |
| Oroxylin A | Breast adenocarcinoma cells (MDA-MB-231) | 50, 100, and 200 μM for 10 h, under hypoxic conditions | [161] |
| Quercetin | Prostate carcinoma cells (LNCaP), colon cancer cells (CX-1), and breast adenocarcinoma cells (SkBr3) | 10–100 mM for 1, 2, 4, or 8 h, under normoxic or hypoxic conditions—depending on the experiment purpose | [162] |
| Resveratrol | Human osteosarcoma cells (Saos-2) | 50 μM for 24 h | [163] |
| Tongue squamous cell carcinoma (SCC-9) and hepatocellular carcinoma cells (Hep G2) | 5, 50, 100 μM for 1 or 16 h, under hypoxic and normoxic conditions | [148] | |
| Wogonin | Multiple myeloma cells (RPMI 8226, U266) | 0, 20, 40, 80 μM for 24 h, under hypoxic and normoxic conditions | [164] |
| Inhibitors of transcriptional activity | |||
| Baicalein | Breast adenocarcinoma cells (MCF7) | 50 µM under hypoxic conditions or with 150 µM cobalt chloride for 8 h | [157] |
| Chlorogenic acid | Lung cancer cells (A549) | 2, 10 μM for 16 h, followed by exposure to 200 μM cobalt chloride for 6 h | [144] |
| Curcumin | Hepatocellular carcinoma cells (Hep G2) | 0, 25 μM, and 50 μM for 6 h under hypoxic conditions | [165] |
| Genistein | Hepatocellular carcinoma cells (HCC-LM3, SMMC-7721, Hep3B, BEL-7402, and Huh-7), normal hepatic cells (LO2) | for HCC-LM3 cells: 60 μM for 24 h | [166] |
| Luteolin | Colorectal carcinoma cells (HCT116), breast adenocarcinoma cells (MDA-MB-231) | 0, 10, 25, and 50 μM for 48 h with 100 μM cobalt chloride for the last 24 h | [159] |
| Quercetin | Colorectal carcinoma cells (HCT116), prostate cancer cells (DU145), cervical adenocarcinoma cells (HeLa S3) | 0, 50, and 100 μM for 12 h | [136] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pralea, I.-E.; Petrache, A.-M.; Tigu, A.B.; Gulei, D.; Moldovan, R.-C.; Ilieș, M.; Nicoară, R.; Hegheș, S.-C.; Uifălean, A.; Iuga, C.-A. Phytochemicals as Regulators of Tumor Glycolysis and Hypoxia Signaling Pathways: Evidence from In Vitro Studies. Pharmaceuticals 2022, 15, 808. https://doi.org/10.3390/ph15070808
Pralea I-E, Petrache A-M, Tigu AB, Gulei D, Moldovan R-C, Ilieș M, Nicoară R, Hegheș S-C, Uifălean A, Iuga C-A. Phytochemicals as Regulators of Tumor Glycolysis and Hypoxia Signaling Pathways: Evidence from In Vitro Studies. Pharmaceuticals. 2022; 15(7):808. https://doi.org/10.3390/ph15070808
Chicago/Turabian StylePralea, Ioana-Ecaterina, Alina-Maria Petrache, Adrian Bogdan Tigu, Diana Gulei, Radu-Cristian Moldovan, Maria Ilieș, Raul Nicoară, Simona-Codruța Hegheș, Alina Uifălean, and Cristina-Adela Iuga. 2022. "Phytochemicals as Regulators of Tumor Glycolysis and Hypoxia Signaling Pathways: Evidence from In Vitro Studies" Pharmaceuticals 15, no. 7: 808. https://doi.org/10.3390/ph15070808
APA StylePralea, I.-E., Petrache, A.-M., Tigu, A. B., Gulei, D., Moldovan, R.-C., Ilieș, M., Nicoară, R., Hegheș, S.-C., Uifălean, A., & Iuga, C.-A. (2022). Phytochemicals as Regulators of Tumor Glycolysis and Hypoxia Signaling Pathways: Evidence from In Vitro Studies. Pharmaceuticals, 15(7), 808. https://doi.org/10.3390/ph15070808