Mapping Molecular Networks within Clitoria ternatea Linn. against LPS-Induced Neuroinflammation in Microglial Cells, with Molecular Docking and In Vivo Toxicity Assessment in Zebrafish
Abstract
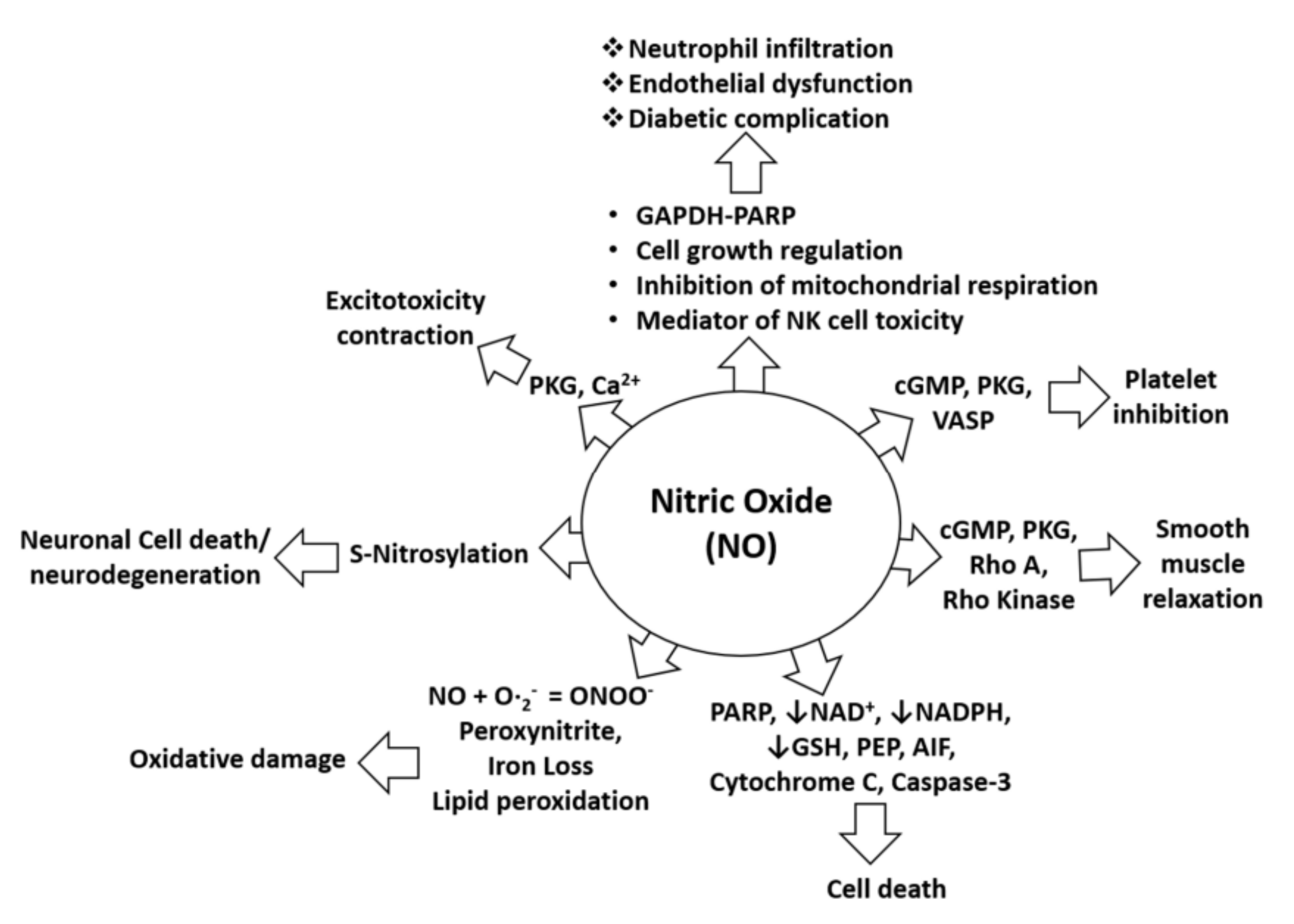
:1. Introduction
2. Results and Discussion
2.1. Biological Evaluation
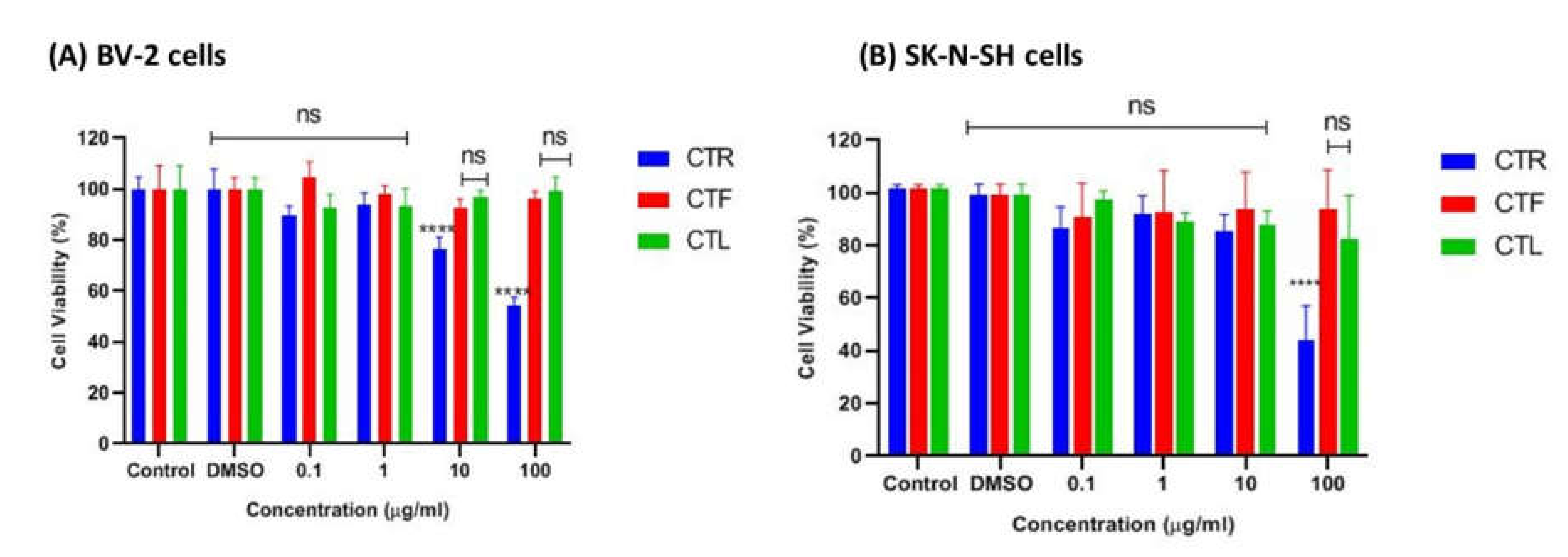
2.1.1. Effects of CT Extracts on Cell Viability Using Cultured BV-2 Microglial and SK-N-SH Neuroblastoma Cells
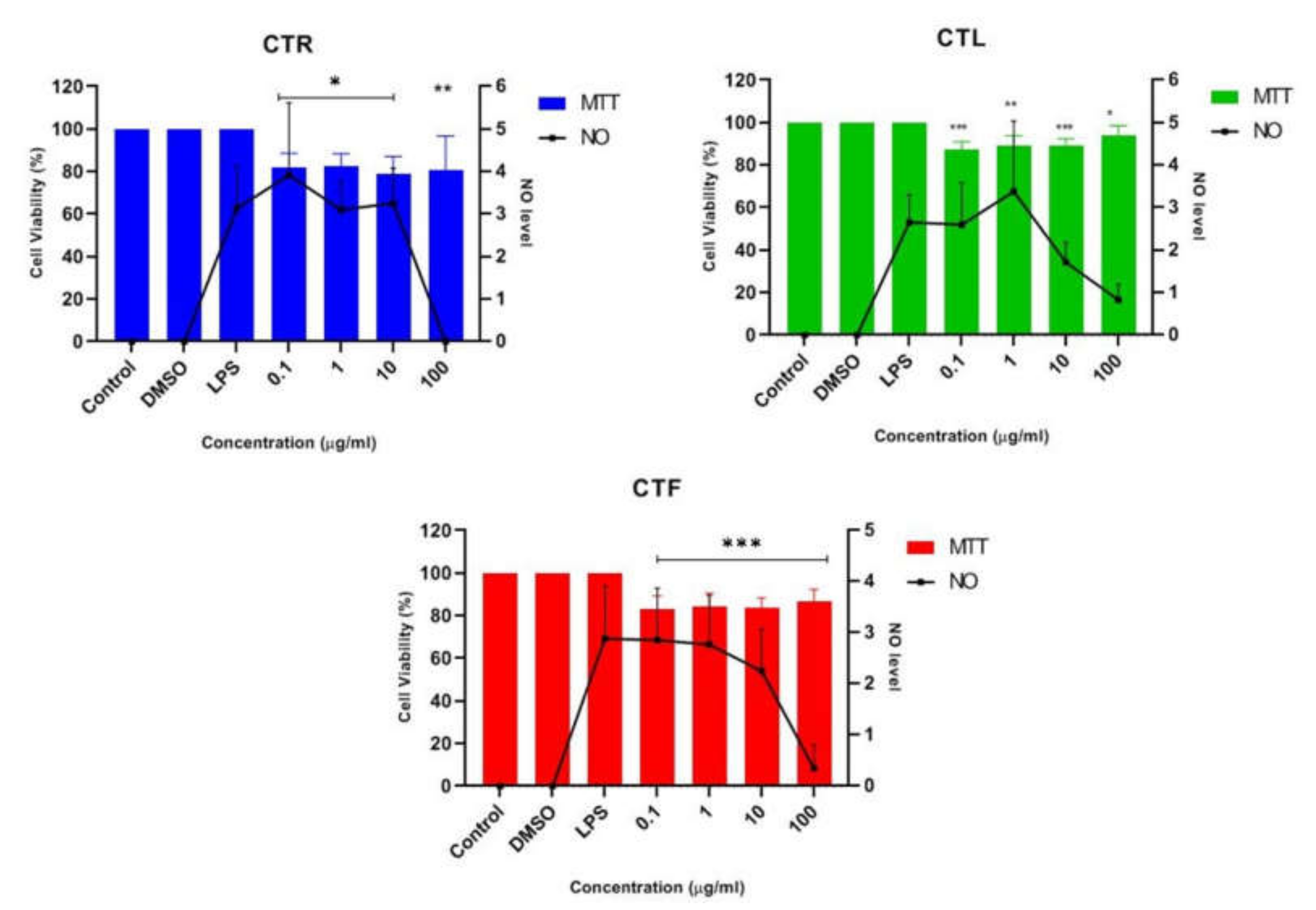
2.1.2. Inhibitory Effects of CT Extracts on NO Secretion in LPS-Induced BV-2 Cells
2.1.3. Effects of CTF Fractions on Cell Viability Using Cultured BV-2 Microglial and SK-N-SH Human Neuroblastoma Cells
2.1.4. Inhibition Effects of CTF Fractions on NO Secretion in LPS-Induced BV-2 Cells
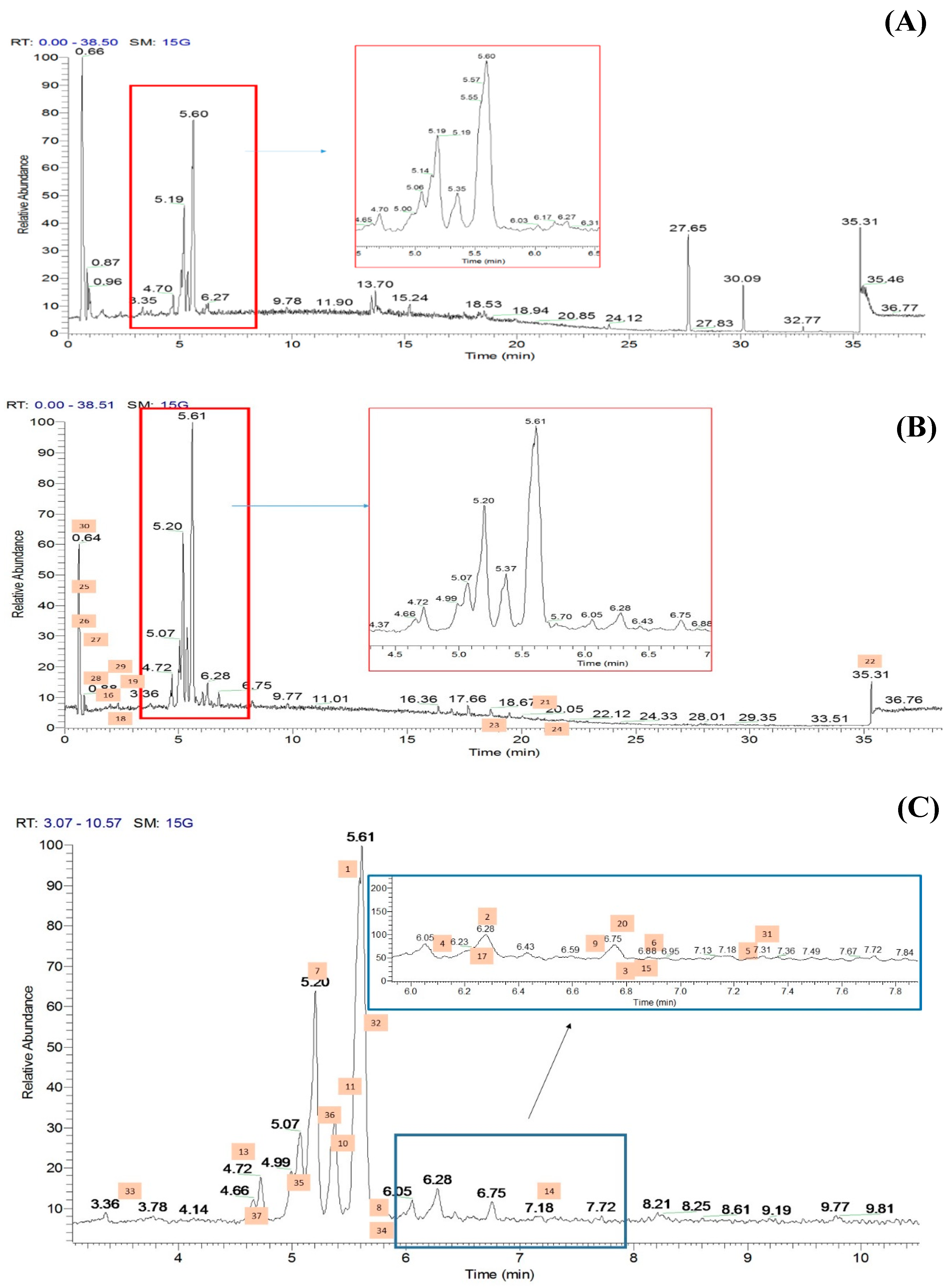
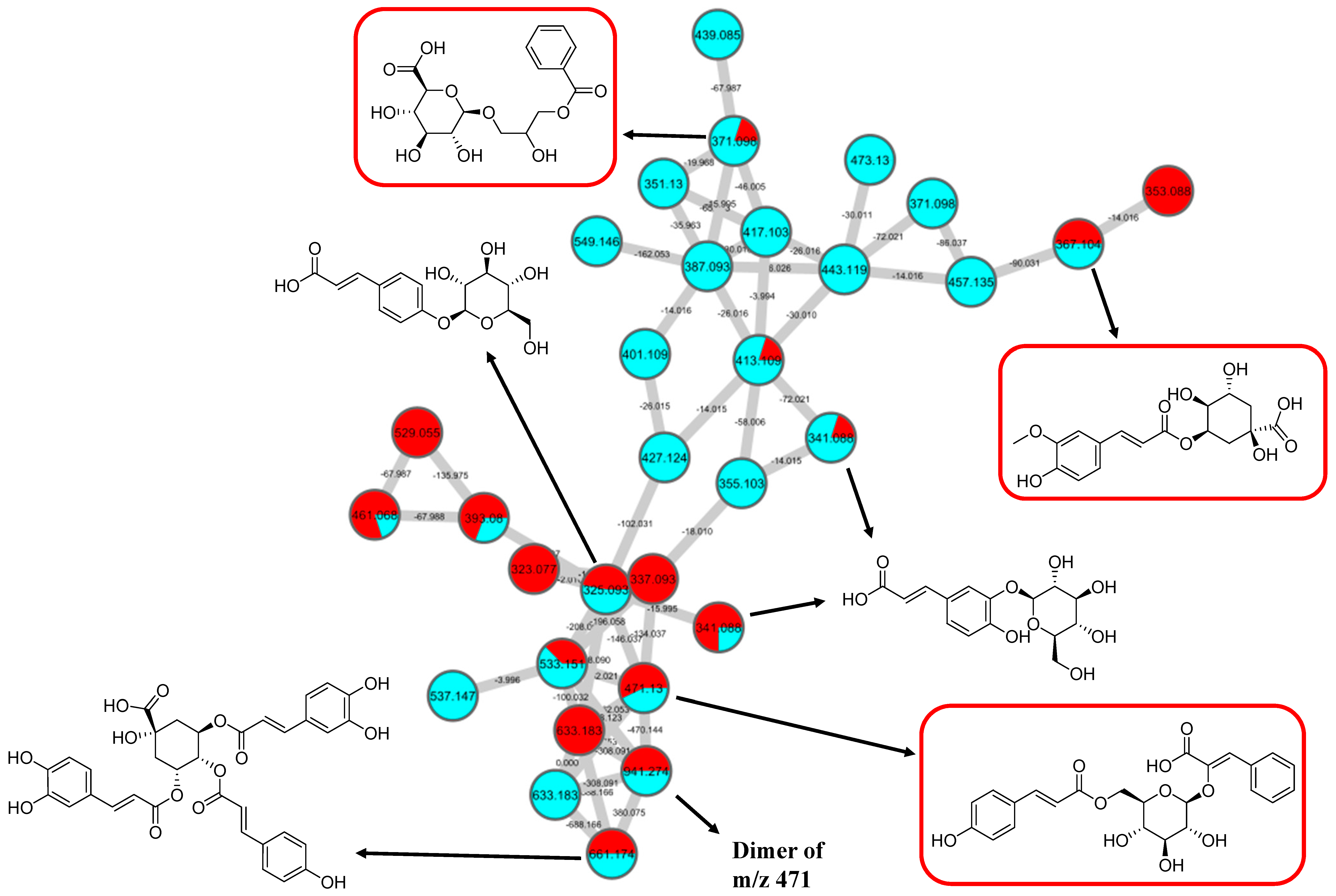
2.2. Metabolic Profiling of Ethyl Acetate Fraction of CTF (CTF_EA) by the Untargeted, Tandem-Mass-Spectrometry-Based (UHPLC–MS/MS) Molecular Networking
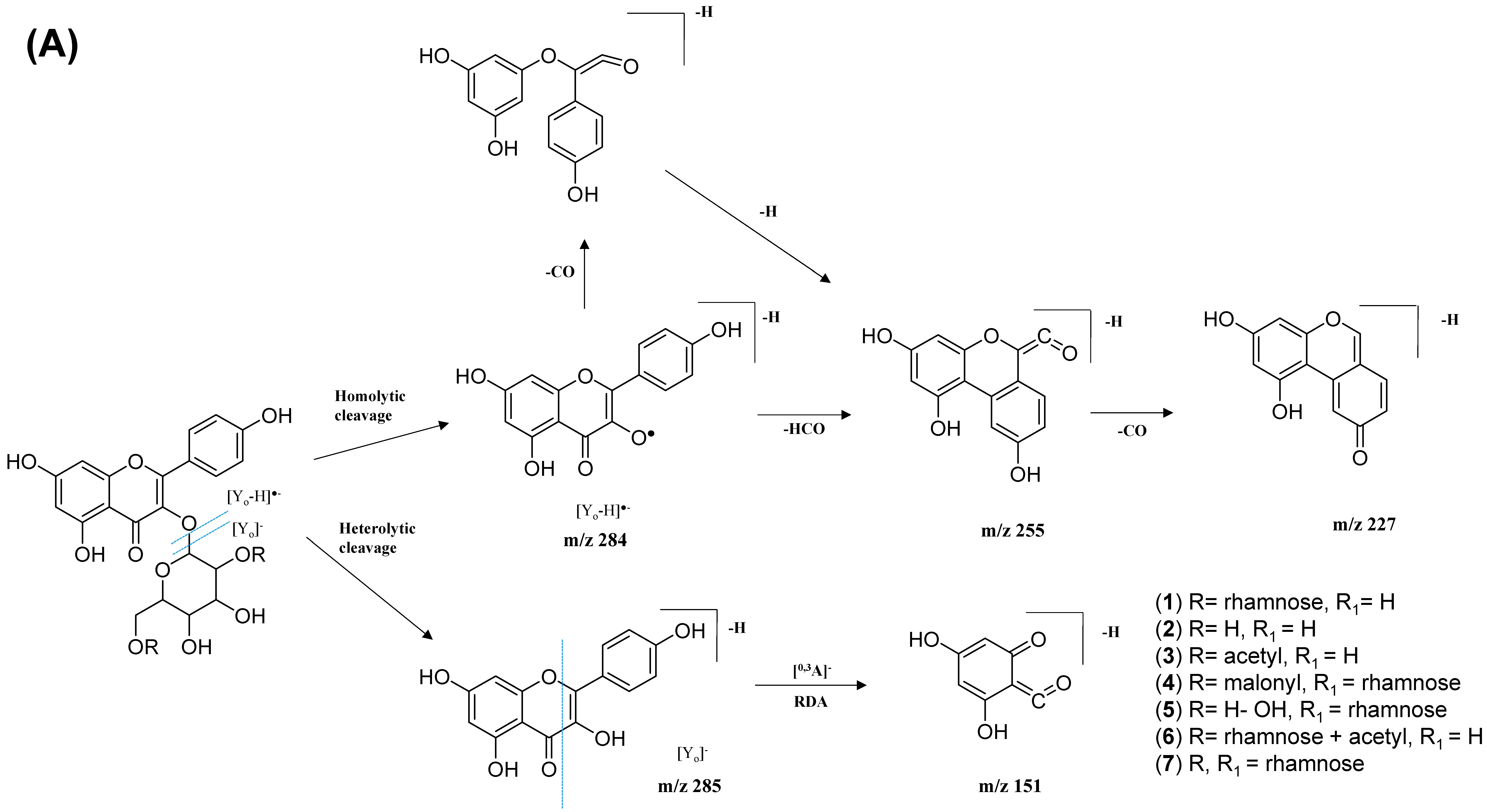
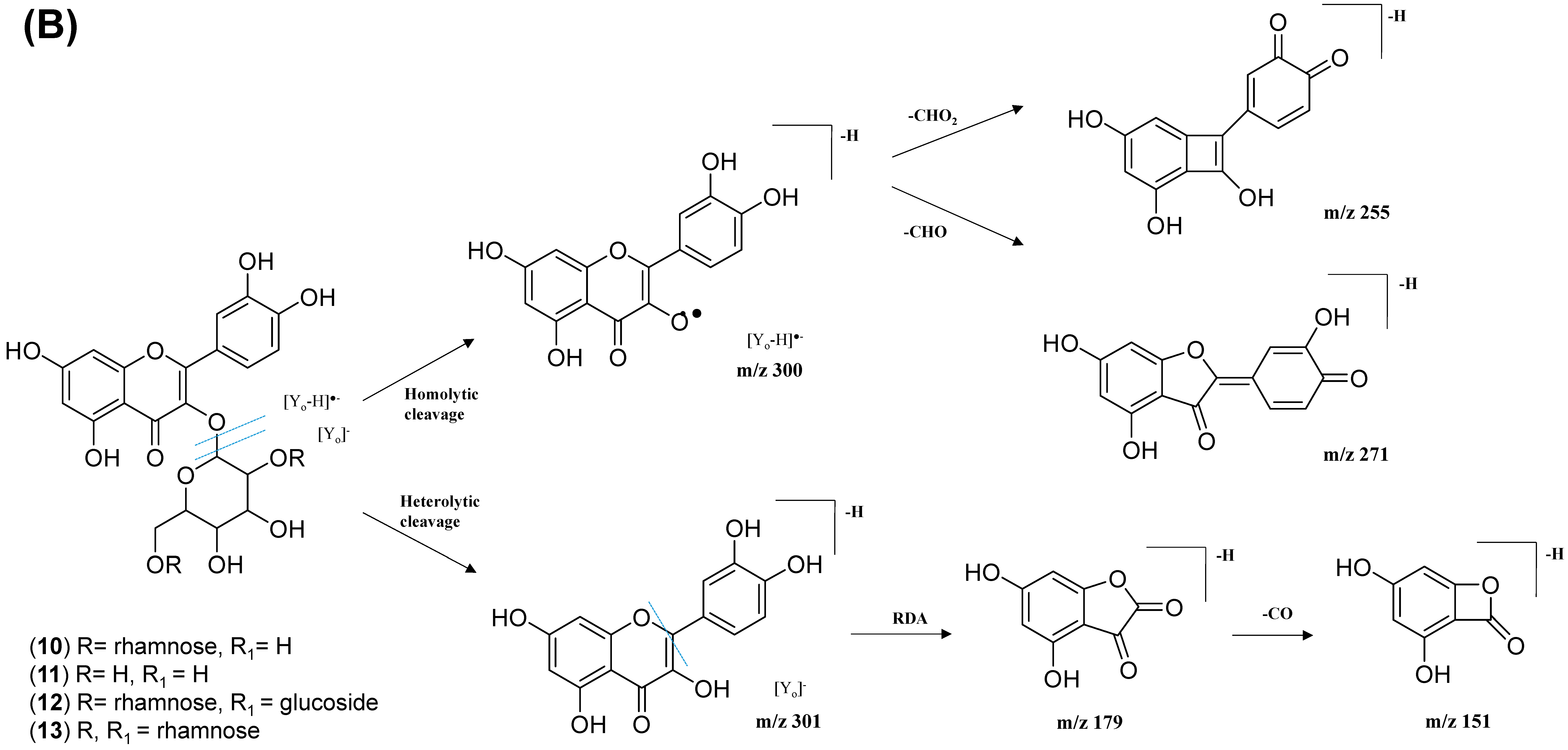
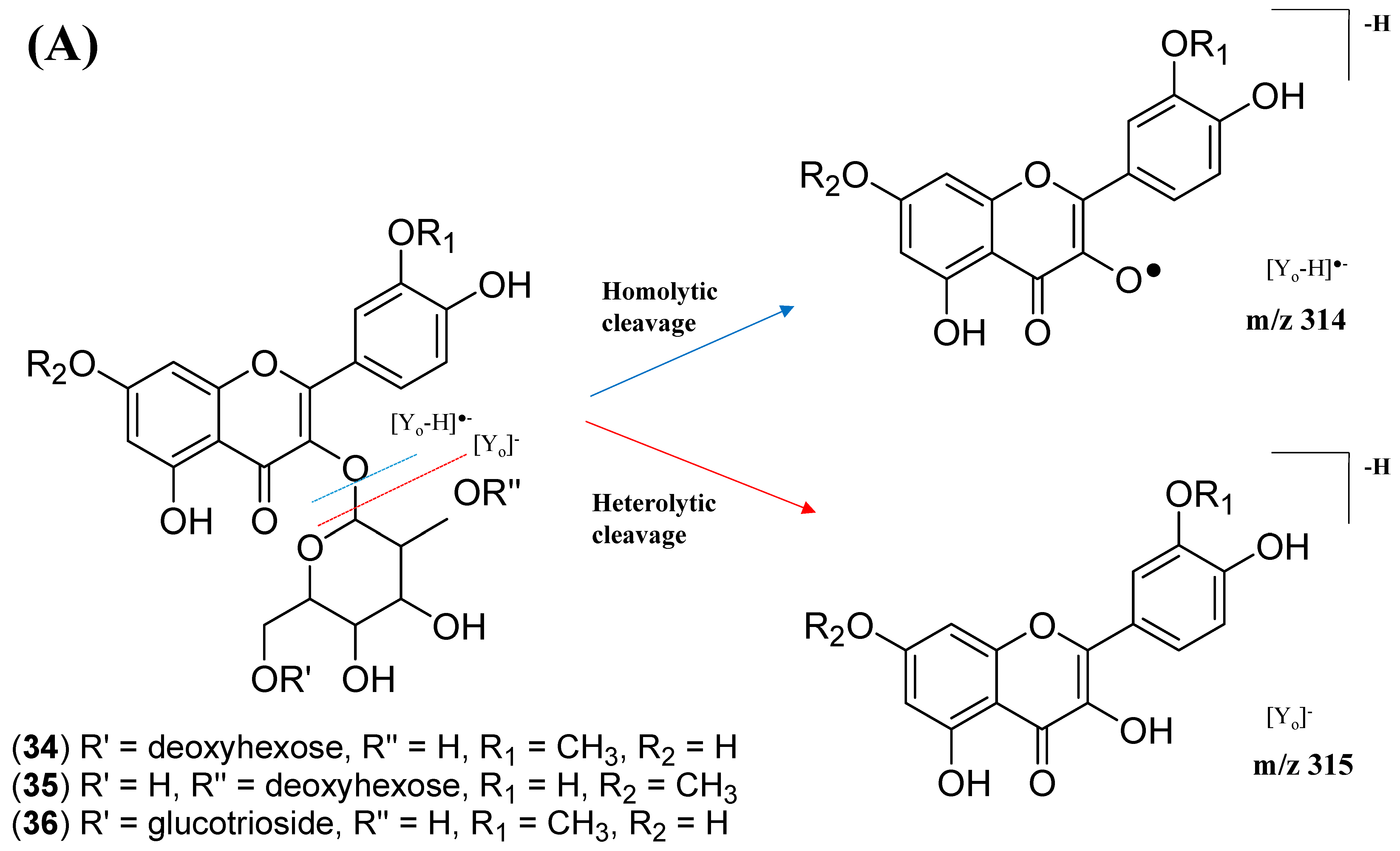
2.2.1. Flavonol-3-O-glycoside
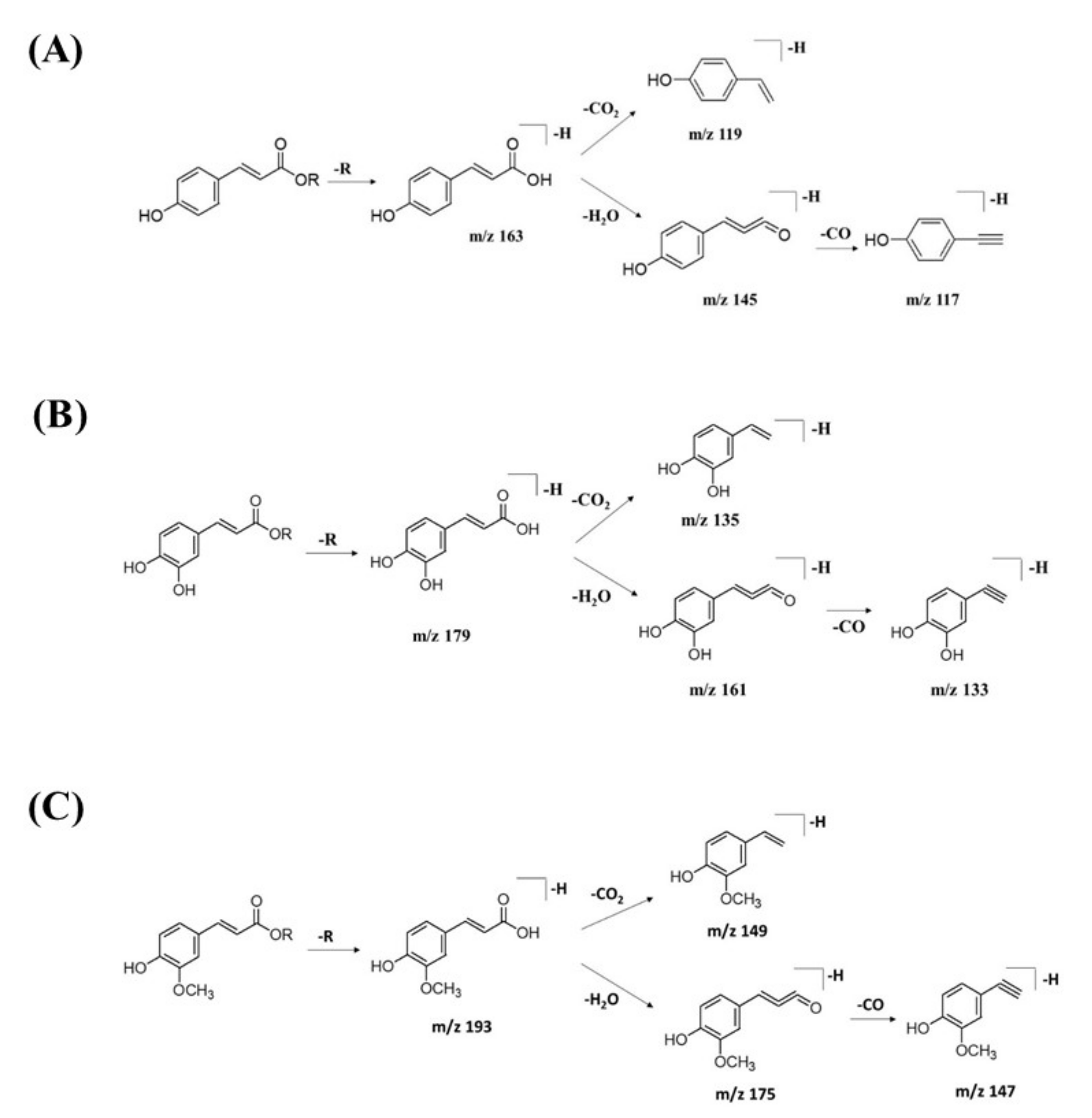
2.2.2. Hydrocinnamic Acids and Derivatives
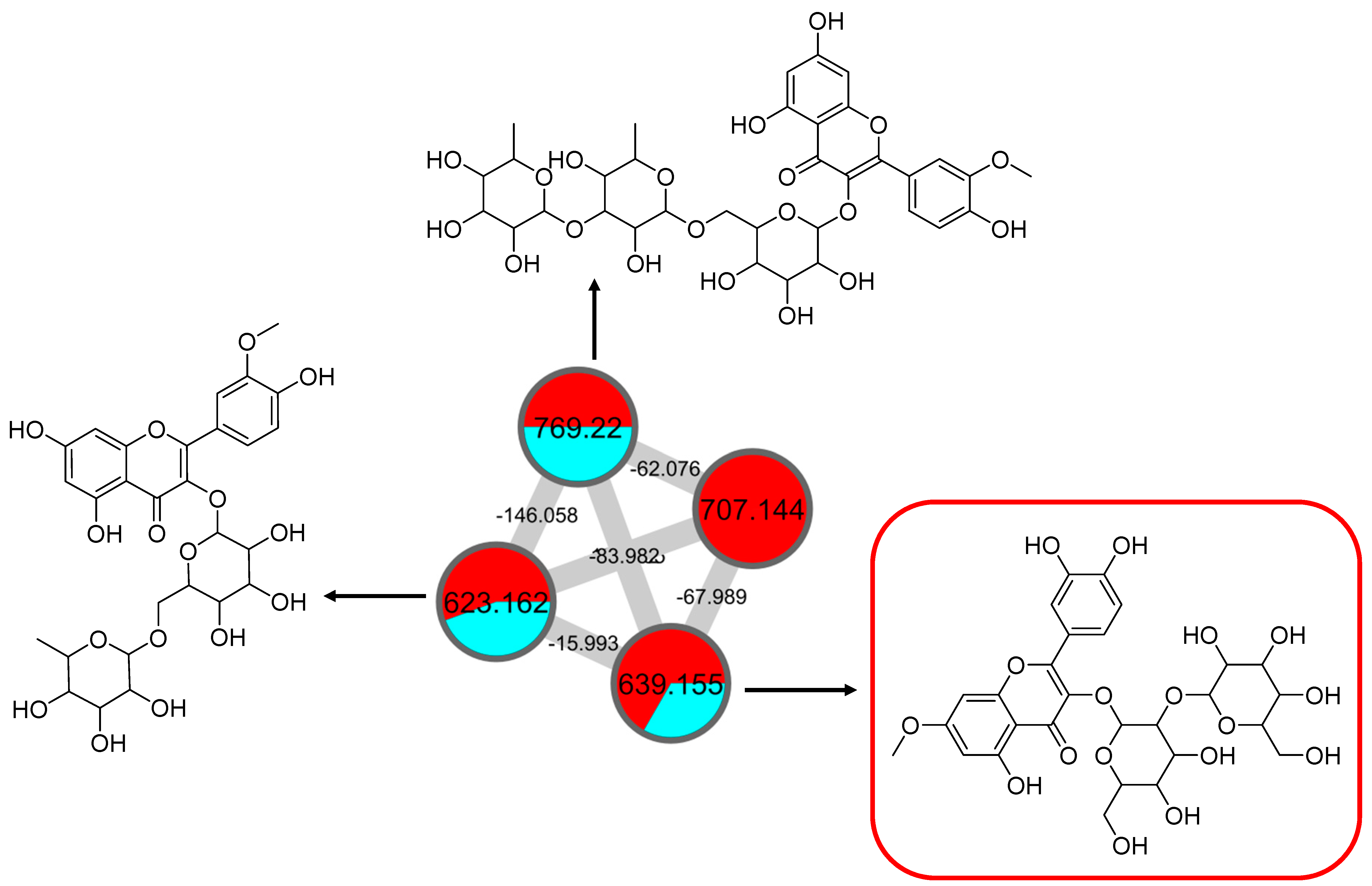
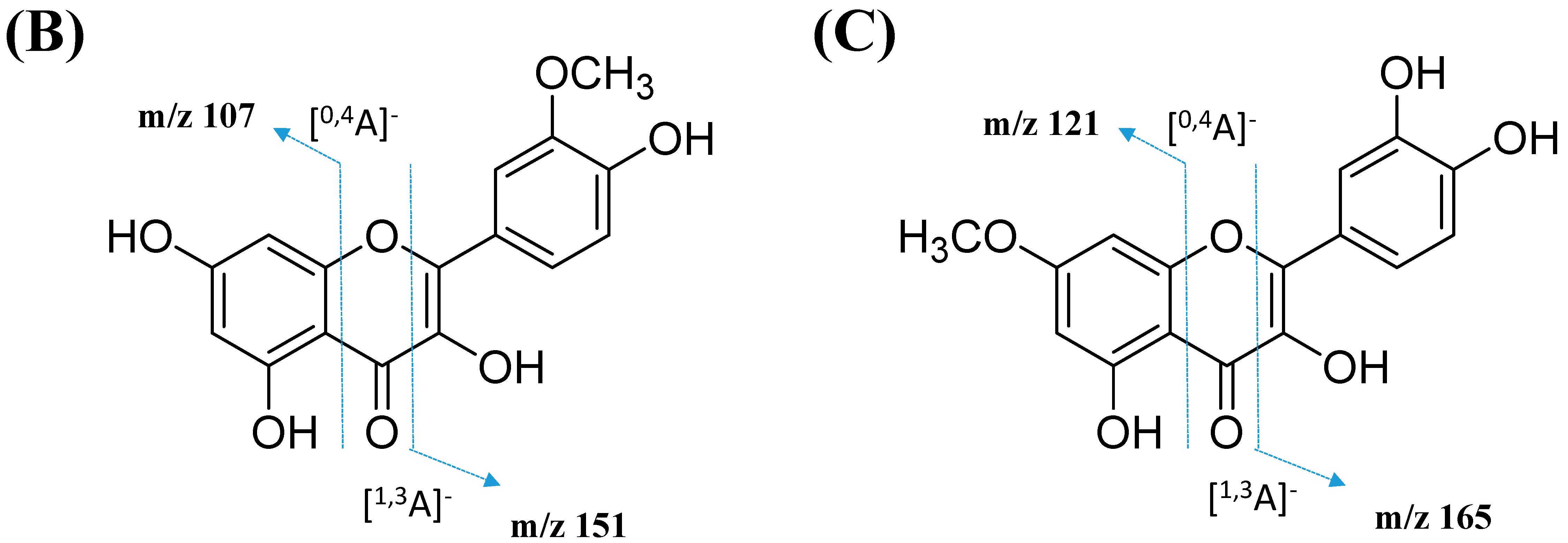
2.2.3. Mono-Methoxylflavonol 3-O-glycoside
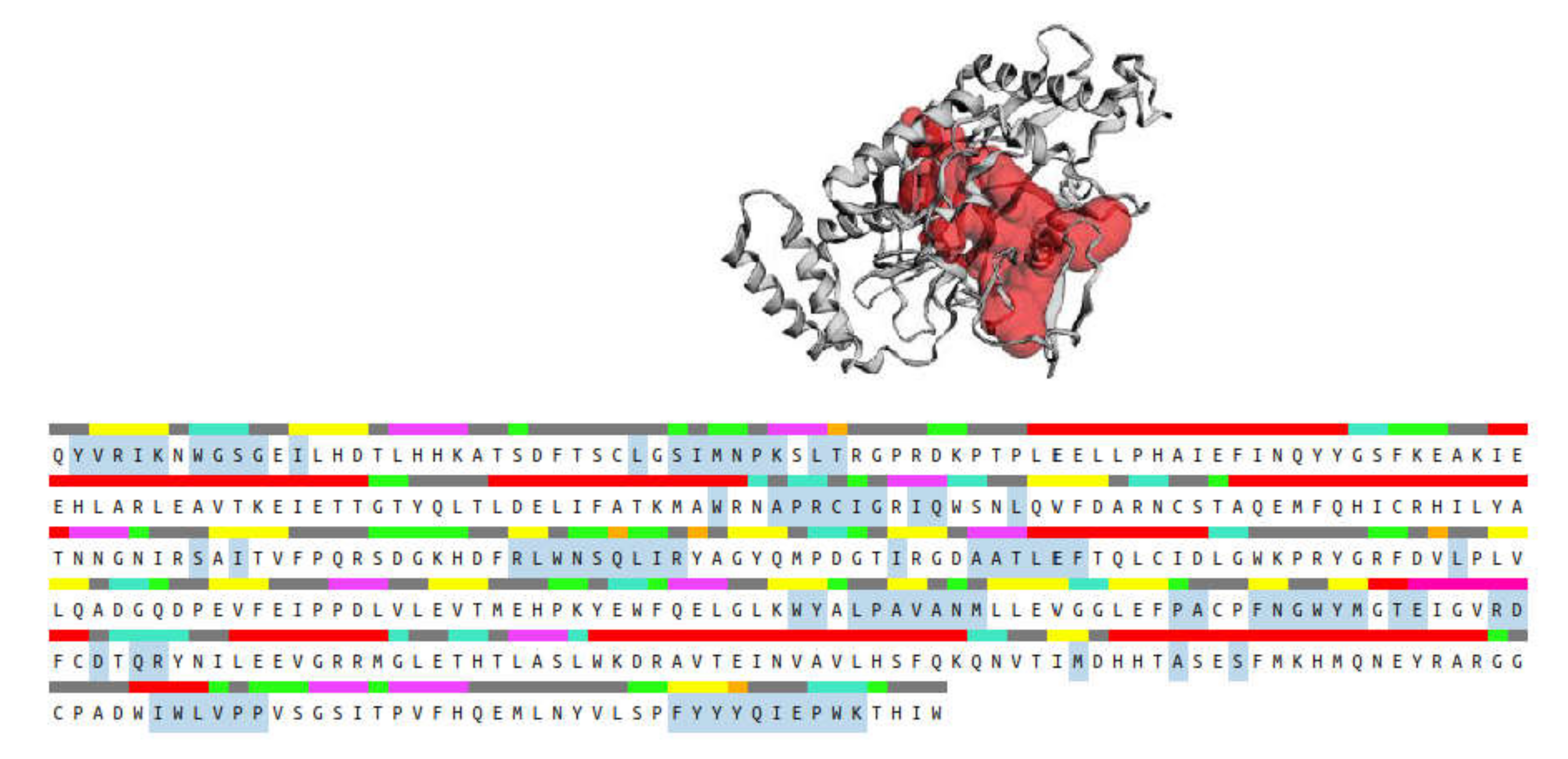
2.3. Molecular Docking
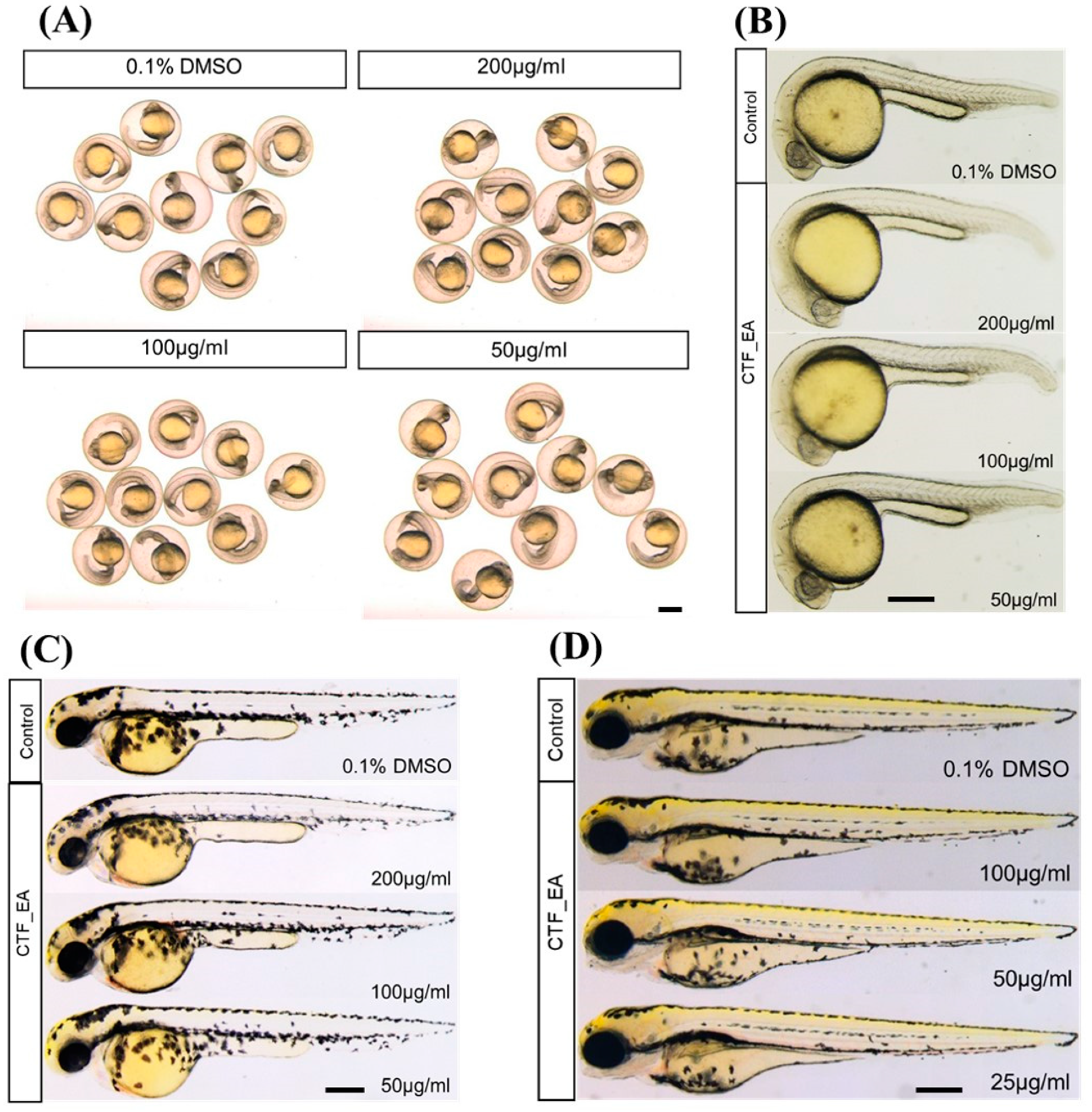
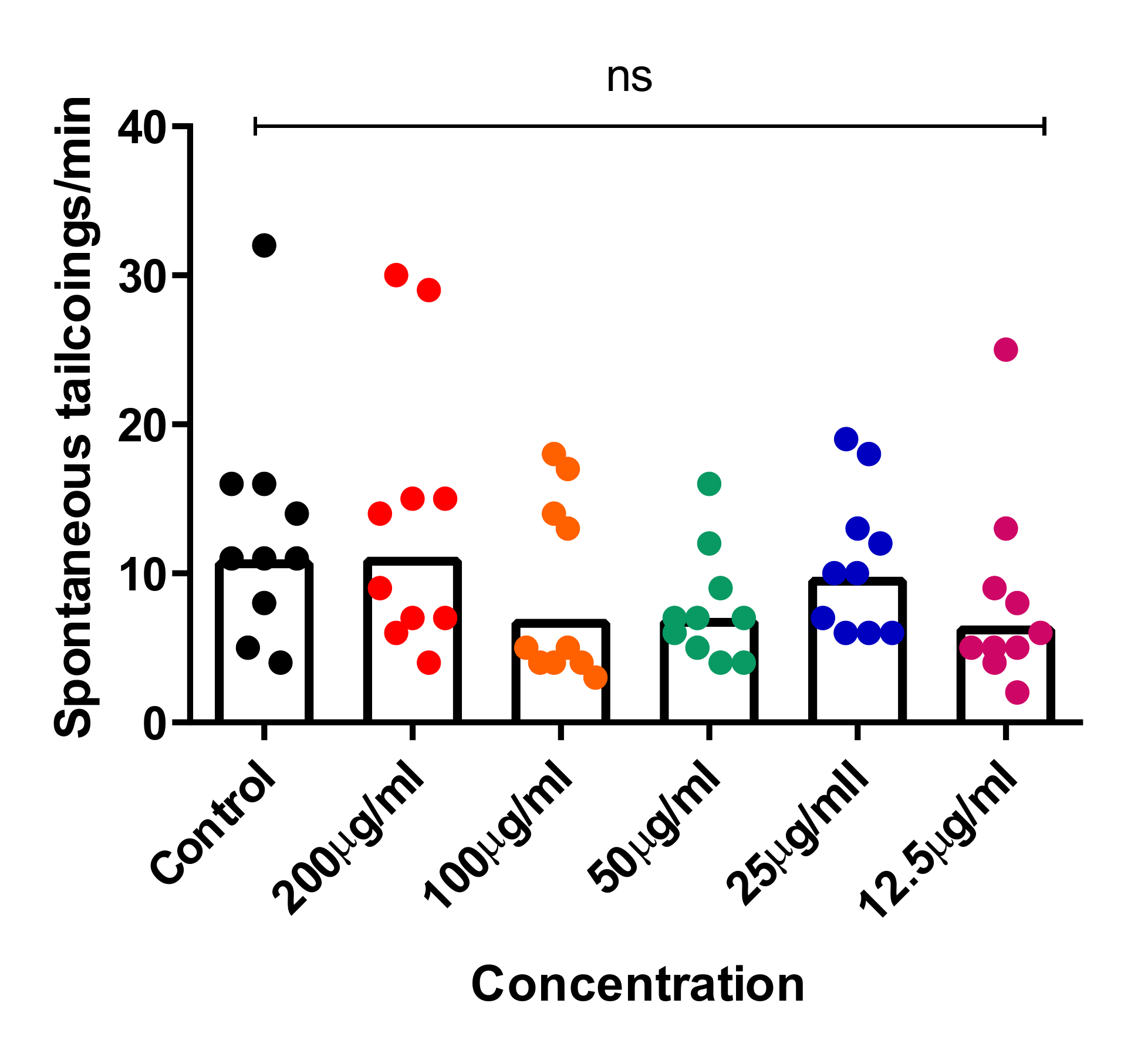
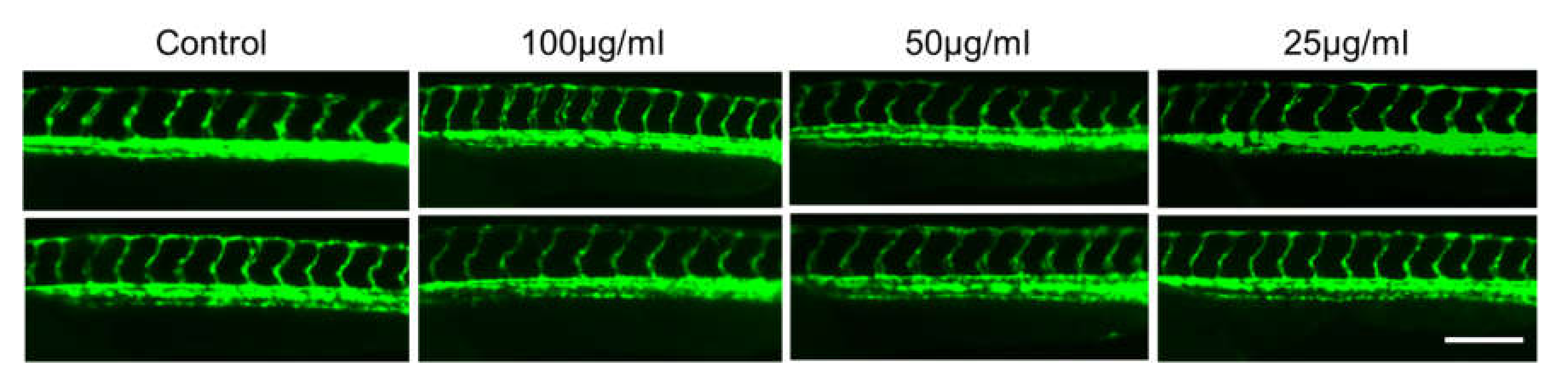
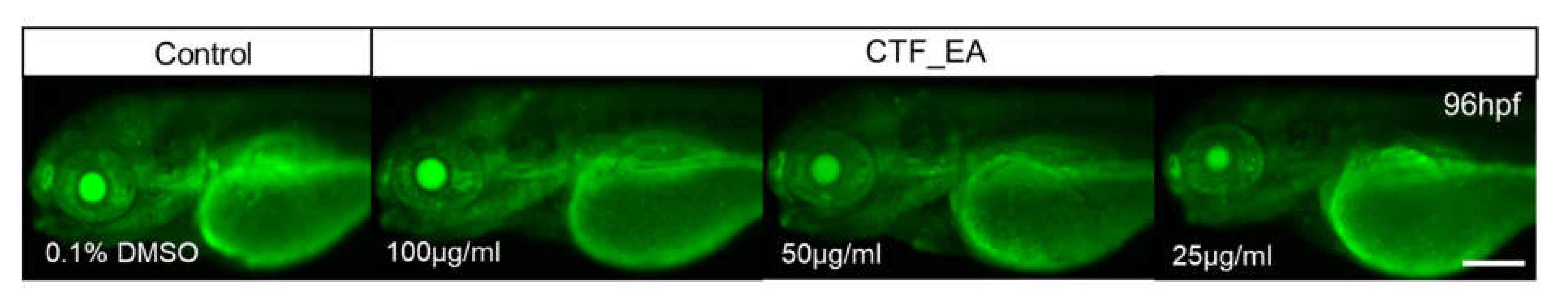
2.4. In Vivo Toxicity Test in Zebrafish Embryos
3. Material and Methods
3.1. Reagents and Materials
3.2. Plant Material
3.3. Extraction and Successive Fractionation of CT
3.4. Biological Evaluation
3.4.1. Cell Culture
3.4.2. Cell Viability Determination (MTT Assay)
3.4.3. Nitric Oxide Inhibitory Assay (Griess Assay)
3.4.4. Statistical Analysis
3.5. Ultra-High-Performance Liquid Chromatography–Tandem Mass Spectrometry (UHPLC–MS/MS) Analysis
3.6. Generation of Molecular Networks
3.7. In Silico Molecular Docking Studies
3.8. In Vivo Toxicity in Zebrafish Embryos
3.8.1. Zebrafish Husbandry and Embryo Collection
3.8.2. Acute Toxicity Testing on Zebrafish Embryos
3.8.3. Tail-Coiling Movements in Zebrafish Embryos
3.8.4. Evaluation of Adverse Effect on Development of Blood Vessels
3.8.5. Evaluation of Cell Death and Apoptosis
3.8.6. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bloom, D.E.; Cafiero, E.T.; Jané-Llopis, E.; Abrahams-Gessel, S.; Bloom, L.R.; Fathima, S.; Feigl, A.B.; Gaziano, T.; Mowafi, M.; Pandya, A.; et al. The global economic burden of noncommunicable diseases. In PGDA Working Papers 8712; Program on the Global Demography of Aging; World Economic Forum: Geneva, Switzerland, 2012. [Google Scholar]
- Nasef, N.A.; Mehta, S.; Ferguson, L.R. Susceptibility to chronic inflammation: An update. Arch. Toxicol. 2017, 91, 1131–1141. [Google Scholar] [CrossRef] [PubMed]
- Maccioni, R.B.; Rojo, L.E.; Fernández, J.A.; Kuljis, R.O. The role of neuroimmunomodulation in Alzheimer’s disease. Ann. N. Y. Acad. Sci. 2009, 1153, 240–246. [Google Scholar] [CrossRef] [PubMed]
- Guzman-Martinez, L.; Maccioni, R.B.; Andrade, V.; Navarrete, L.P.; Pastor, M.G.; Ramos-Escobar, N. Neuroinflammation as a common feature of neurodegenerative disorders. Front. Pharmacol. 2019, 10, 1008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matthews, K.A.; Xu, W.; Gaglioti, A.H.; Holt, J.B.; Croft, J.B.; Mack, D.; McGuire, L.C. Racial and ethnic estimates of Alzheimer’s disease and related dementias in the United States (2015–2060) in adults aged ≥65 years. Alzheimer’s Dement. 2019, 15, 17–24. [Google Scholar] [CrossRef]
- Alzheimer’s Association. Alzheimer’s disease facts and figures. Alzheimer’s Dement. 2019, 15, 321–387. [Google Scholar]
- Calsolaro, V.; Edison, P. Neuroinflammation in Alzheimer’s disease: Current evidence and future directions. Alzheimer’s Dement. 2016, 12, 719–732. [Google Scholar] [CrossRef] [PubMed]
- Heneka, M.T.; Carson, M.J.; El Khoury, J.; Landreth, G.E.; Brosseron, F.; Feinstein, D.L.; Jacobs, A.H.; Wyss-Coray, T.; Vitorica, J.; Ransohoff, R.M.; et al. Neuroinflammation in Alzheimer’s disease. Lancet Neurol. 2015, 14, 388–405. [Google Scholar] [CrossRef] [Green Version]
- Obulesu, M.; Jhansilakshmi, M. Neuroinflammation in Alzheimer’s disease: An understanding of physiology and pathology. Int. J. Neurosci. 2014, 124, 227–235. [Google Scholar] [CrossRef]
- Jonsson, T.; Stefansson, H.; Steinberg, S.; Jonsdottir, I.; Jonsson, P.V.; Snaedal, J.; Bjornsson, S.; Huttenlocher, J.; Levey, A.I.; Lah, J.J.; et al. Variant of TREM2 associated with the risk of Alzheimer’s disease. N. Engl. J. Med. 2013, 368, 107–116. [Google Scholar] [CrossRef] [Green Version]
- Rohn, T.T. The triggering receptor expressed on myeloid cells 2: “TREM-ming” the inflammatory component associated with Alzheimer’s disease. Oxidative Med. Cell. Longev. 2013, 2013, 860959–860967. [Google Scholar] [CrossRef] [Green Version]
- DiSabato, D.J.; Quan, N.; Godbout, J.P. Neuroinflammation: The devil is in the details. J. Neurochem. 2016, 139 (Suppl. S2), 136–153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Norden, D.M.; Trojanowski, P.J.; Villanueva, E.; Navarro, E.; Godbout, J.P. Sequential activation of microglia and astrocyte cytokine expression precedes increased iba-1 or GFAP immunoreactivity following systemic immune challenge. Glia 2016, 64, 300–316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luiking, Y.C.; Engelen, M.P.; Deutz, N.E. Regulation of nitric oxide production in health and disease. Curr. Opin. Clin. Nutr. Metab. Care 2010, 13, 97–104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Forstermann, U.; Sessa, W.C. Nitric oxide synthases: Regulation and function. Eur. Heart J. 2012, 33, 829–837. [Google Scholar] [CrossRef] [Green Version]
- Nakamura, T.; Prikhodko, O.A.; Pirie, E.; Nagar, S.; Akhtar, M.W.; Oh, C.K.; McKercher, S.R.; Ambasudhan, R.; Okamoto, S.; Lipton, S.A. Aberrant protein S-nitrosylation contributes to the pathophysiology of neurodegenerative diseases. Neurobiol. Dis. 2015, 84, 99–108. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.N.; Sun, J.; Yang, X.D.; Xiao, K.; Lv, Y.; Zhang, B.Y.; Zhou, W.; Chen, C.; Gao, C.; Shi, Q.; et al. The brain NO levels and NOS activities ascended in the early and middle stages and descended in the terminal stage in scrapie-infected animal models. Mol. Neurobiol. 2017, 54, 1786–1796. [Google Scholar] [CrossRef]
- Bourgognon, J.M.; Spiers, J.G.; Scheiblich, H.; Antonov, A.; Bradley, S.J.; Tobin, A.B.; Steinert, J.R. Alterations in neuronal metabolism contribute to the pathogenesis of prion disease. Cell Death Differ. 2018, 25, 1408–1425. [Google Scholar] [CrossRef]
- Nakamura, T.; Lipton, S.A. Protein S-nitrosylation as a therapeutic target for neurodegenerative diseases. Trends Pharmacol. Sci. 2016, 37, 73–84. [Google Scholar] [CrossRef] [Green Version]
- Nakamura, T.; Lipton, S.A. Redox modulation by S-nitrosylation contributes to protein misfolding, mitochondrial dynamics, and neuronal synaptic damage in neurodegenerative diseases. Cell Death Differ. 2011, 18, 1478–1486. [Google Scholar] [CrossRef] [Green Version]
- Wijasa, T.S.; Sylvester, M.; Brocke-Ahmadinejad, N.; Schwartz, S.; Santarelli, F.; Gieselmann, V.; Klockgether, T.; Brosseron, F.; Heneka, M.T. Quantitative proteomics of synaptosome S-nitrosylation in Alzheimer’s disease. J. Neurochem. 2019, 152, 710–726. [Google Scholar] [CrossRef] [Green Version]
- Guivernau, B.; Bonet, J.; Valls-Comamala, V.; Bosch-Morató, M.; Godoy, J.A.; Inestrosa, N.C.; Perálvarez-Marín, A.; Fernández-Busquets, X.; Andreu, D.; Oliva, B.; et al. Amyloid-β peptide nitrotyrosination stabilizes oligomers and enhances NMDAR-mediated toxicity. J. Neurosci. 2016, 6, 11693–11703. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Broom, L.; Marinova-Mutafchieva, L.; Sadeghian, M.; Davis, J.B.; Medhurst, A.D.; Dexter, D.T. Neuroprotection by the selective iNOS inhibitor GW274150 in a model of Parkinson disease. Free Radic. Biol. Med. 2011, 50, 633–640. [Google Scholar] [CrossRef] [PubMed]
- Neufeld, A.H.; Sawada, A.; Becker, B. Inhibition of nitric-oxide synthase 2 by aminoguanidine provides neuroprotection of retinal ganglion cells in a rat model of chronic glaucoma. Proc. Natl. Acad. Sci. USA 1999, 96, 9944–9948. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sil, S.; Ghosh, T.; Ghosh, R.; Gupta, P. Nitric oxide synthase inhibitor, aminoguanidine reduces intracerebroventricular colchicine induced neurodegeneration, memory impairments and changes of systemic immune responses in rats. J. Neuroimmunol. 2016, 303, 51–61. [Google Scholar] [CrossRef]
- Subedi, L.; Gaire, B.P.; Parveen, A.; Kim, S.-Y. Nitric Oxide as a Target for Phytochemicals in Anti-Neuroinflammatory Prevention Therapy. Int. J. Mol. Sci. 2021, 22, 4771. [Google Scholar] [CrossRef]
- Singh, A.K.; Rai, S.N.; Maurya, A.; Mishra, G.; Awasthi, R.; Shakya, A.; Chellappan, D.K.; Dua, K.; Vamanu, E.; Chaudhary, S.K.; et al. Therapeutic Potential of Phytoconstituents in Management of Alzheimer’s Disease. Evid. Based Complement. Altern. Med. 2021, 2021, 5578574–5578591. [Google Scholar] [CrossRef]
- Mukherjee, P.K.; Kumar, V.; Kumar, N.S.; Heinrich, M. The Ayurvedic medicine Clitoria ternatea—From traditional use to scientific assessment. J. Ethnopharmacol. 2008, 120, 291–301. [Google Scholar] [CrossRef]
- Jain, N.N.; Ohal, C.C.; Shroff, S.K.; Bhutada, R.H.; Somani, R.S.; Kasture, V.S.; Kasture, S.B. Clitoria ternatea and the CNS. Pharmacol. Biochem. Behav. 2003, 75, 529–536. [Google Scholar] [CrossRef]
- Jeyaraj, E.J.; Lim, Y.Y.; Choo, W.S. Extraction methods of butterfly pea (Clitoria ternatea) flower and biological activities of its phytochemicals. J. Food Sci. Technol. 2021, 58, 2054–2067. [Google Scholar] [CrossRef]
- Srichaikul, B. Ultrasonication extraction, bioactivity, antioxidant activity, total flavonoid, total phenolic and antioxidant of Clitoria ternatea linn flower extract for anti-aging drinks. Pharmacogn. Mag. 2018, 14, 322. [Google Scholar] [CrossRef]
- Chandra, S. Evaluation of Methanolic Extract of Clitoria ternatea Hepatoprotective & Nephroprotective Activity in Rats. J. Drug Deliv. Ther. 2019, 9, 313–319. [Google Scholar]
- Nithianantham, K.; Ping, K.Y.; Latha, L.Y.; Jothy, S.L.; Darah, I.; Chen, Y.; Chew, A.L.; Sasidharan, S. Evaluation of hepatoprotective effect of methanolic extract of Clitoria ternatea (Linn.) flower against acetaminophen-induced liver damage. Asian Pac. J. Trop. Dis. 2013, 3, 314–319. [Google Scholar] [CrossRef]
- Sarumathy, K.; Rajan, M.D.; Vijay, T.; Jayakanthi, J. Evaluation of phytoconstituents, nephro-protective and antioxidant activities of Clitoria ternatea. J. Appl. Pharm. Sci. 2011, 1, 164–172. [Google Scholar]
- Olivon, F.; Roussi, F.; Litaudon, M.; Touboul, D. Optimized experimental workflow for tandem mass spectrometry molecular networking in metabolomics. Anal. Bioanal. Chem. 2017, 409, 5767–5778. [Google Scholar] [CrossRef]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Bandeira, N. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [Green Version]
- Aron, A.T.; Gentry, E.C.; McPhail, K.L.; Nothias, L.F.; Nothias-Esposito, M.; Bouslimani, A.; Dorrestein, P.C. Reproducible molecular networking of untargeted mass spectrometry data using GNPS. Nat. Protoc. 2020, 15, 1954–1991. [Google Scholar] [CrossRef]
- Nallathamby, N.; Malek, S.N.A.; Vidyadaran, S.; Phan, C.W.; Sabaratnam, V. Lipids in an Ethyl Acetate Fraction of Caterpillar Medicinal Mushroom, Cordyceps militaris (Ascomycetes), Reduce Nitric Oxide Production in BV2 Cells via NRF2 and NF-κB Pathways. Int. J. Med. Mushrooms 2020, 22, 1215–1223. [Google Scholar] [CrossRef]
- Mohd Faudzi, S.M.; Abdullah, M.A.; Abdull Manap, M.R.; Ismail, A.Z.; Rullah, K.; Mohd Aluwi, M.F.F.; Ramli, A.N.M.; Abas, F.; Lajis, N.H. Inhibition of nitric oxide and prostaglandin E2 production by pyrrolylated-chalcones: Synthesis, biological activity, crystal structure analysis, and molecular docking studies. Bioorg. Chem. 2020, 94, 103376. [Google Scholar] [CrossRef]
- Leitner, S.; Golderer, G.; Winkler, C.; Fuchs, D.; Werner-Felmayer, G.; Werner, E.R. Interferon-γ-induced growth inhibition of neuroblastoma cells is independent of induction of nitric oxide synthase and indoleamine 2,3-dioxygenase. Pteridines 2004, 15, 91–96. [Google Scholar] [CrossRef]
- Spencer, P.S.; Lein, P. Neurotoxicity. In Encyclopedia of Toxicology; Wexler, P., Ed.; Academic Press: Cambridge, MA, USA, 2014; pp. 489–500. [Google Scholar]
- Calabrese, E.J.; Mattson, M.P. How does hormesis impact biology, toxicology, and medicine? NPJ Aging Mech. 2017, 3, 13. [Google Scholar] [CrossRef] [Green Version]
- Karakaş, D.; Ari, F.; Ulukaya, E. The MTT viability assay yields strikingly false-positive viabilities although the cells are killed by some plant extracts. Turk. J. Biol. 2017, 41, 919–925. [Google Scholar] [CrossRef] [PubMed]
- Calabrese, E.J. Hormesis: Path and progression to significance. Int. J. Mol. Sci. 2018, 19, 2871. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calabrese, E.J.; Mattson, M.P.; Dhawan, G.; Kapoor, R.; Calabrese, V.; Giordano, J. Chapter Ten—Hormesis: A potential strategic approach to the treatment of neurodegenerative disease. In International Review of Neurobiology; Söderbom, G., Esterline, R., Oscarsson, J., Mattson, M.P., Eds.; Academic Press: Cambridge, MA, USA, 2020; pp. 271–301. [Google Scholar]
- Ablajan, K.; Abliz, Z.; Shang, X.Y.; He, J.M.; Zhang, R.P.; Shi, J.G. Structural characterization of flavonol 3,7-di-O-glycosides and determination of the glycosylation position by using negative ion electrospray ionization tandem mass spectrometry. J. Mass Spectrom. 2006, 41, 352–360. [Google Scholar] [CrossRef] [PubMed]
- Justesen, U. Collision-induced fragmentation of deprotonated methoxylated flavonoids, obtained by electrospray ionization mass spectrometry. J. Mass Spectrom. 2001, 36, 169–178. [Google Scholar] [CrossRef] [PubMed]
- Leelananda, S.P.; Lindert, S. Computational methods in drug discovery. Beilstein J. Org. Chem. 2016, 12, 2694–2718. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yusuf, D.; Davis, A.M.; Kleywegt, G.J.; Schmitt, S. An alternative method for the evaluation of docking performance: RSR vs. RMSD. J. Chem. Inf. Model. 2008, 48, 1411–1422. [Google Scholar] [CrossRef] [PubMed]
- Ekor, M. The growing use of herbal medicines: Issues relating to adverse reactions and challenges in monitoring safety. Front. Pharmacol. 2014, 4, 177. [Google Scholar] [CrossRef] [Green Version]
- Garcia, C.R.; Noyes, P.D.; Tanguay, R.L. Advancements in zebrafish applications for 21st century toxicology. Pharmacol. Ther. 2016, 16, 11–21. [Google Scholar] [CrossRef] [Green Version]
- Williams, C.H.; Hong, C.C. Zebrafish small molecule screens: Taking the phenotypic plunge. Comput. Struct. Biotechnol. J. 2016, 14, 350–356. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Willett, C.; Fremgen, T. Zebrafish: An Animal Model for Toxicological Studies. Curr. Protoc. Toxicol. 2003, 17, 1–18. [Google Scholar] [CrossRef]
- Zindler, F.; Beedgen, F.; Brandt, D.; Steiner, M.; Stengel, D.; Baumann, L.; Braunbeck, T. Analysis of tail coiling activity of zebrafish (Danio rerio) embryos allows for the differentiation of neurotoxicants with different modes of action. Ecotoxicol. Environ. Saf. 2019, 186, 109754. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira, A.A.S.; Brigante, T.A.V.; Oliveira, D.P. Tail Coiling Assay in Zebrafish (Danio rerio) Embryos: Stage of Development, Promising Positive Control Candidates, and Selection of an Appropriate Organic Solvent for Screening of Developmental Neurotoxicity (DNT). Water 2021, 13, 119. [Google Scholar] [CrossRef]
- Ogungbemi, A.O.; Teixido, E.; Massei, R.; Scholz, S.; Küster, E. Automated measurement of the spontaneous tail coiling of zebrafish embryos as a sensitive behavior endpoint using a workflow in KNIME. MethodsX 2021, 8, 101330. [Google Scholar] [CrossRef] [PubMed]
- Gore, A.V.; Monzo, K.; Cha, Y.R.; Pan, W.; Weinstein, B.M. Vascular development in the zebrafish. Cold Spring Harb. Perspect. Med. 2012, 2, a006684. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Chen, X.; Gueydan, C.; Han, J. Plasma membrane changes during programmed cell deaths. Cell Res. 2018, 28, 9–21. [Google Scholar] [CrossRef]
- Mehmood, A.; Ishaq, M.; Zhao, L.; Yaqoob, S.; Safdar, B.; Nadeem, M.; Wang, C. Impact of ultrasound and conventional extraction techniques on bioactive compounds and biological activities of blue butterfly pea flower (Clitoria ternatea L.). Ultrason. Sonochem. 2019, 51, 12–19. [Google Scholar] [CrossRef] [PubMed]
- Zolkiffly, S.Z.I.; Stanslas, J.; Abdul Hamid, H.; Mehat, M.Z. Ficus deltoidea: Potential inhibitor of pro-inflammatory mediators in lipopolysaccharide-induced activation of microglial cells. J. Ethnopharmacol. 2021, 279, 114309–114320. [Google Scholar] [CrossRef]
- Norazhar, A.I.; Lee, S.Y.; Mohd Faudzi, S.M.; Shaari, K. Metabolite profiling of Christia vespertilionis leaf metabolome via molecular network approach. Appl. Sci. 2021, 11, 3526. [Google Scholar] [CrossRef]
- Garcin, E.D.; Arvai, A.S.; Rosenfeld, R.J.; Kroeger, M.D.; Crane, B.R.; Andersson, G.; Andrews, G.; Hamley, P.J.; Mallinder, P.R.; Nicholls, D.J.; et al. Anchored plasticity opens doors for selective inhibitor design in nitric oxide synthase. Nat. Chem. Biol. 2008, 4, 700–707. [Google Scholar] [CrossRef] [Green Version]
- Tian, W.; Chen, C.; Lei, X.; Zhao, J.; Liang, J. CASTp 3.0: Computed atlas of surface topography of proteins. Nucleic Acids Res. 2018, 46, W363–W367. [Google Scholar] [CrossRef] [Green Version]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdullah, M.A.; Lee, Y.R.; Mastuki, S.N.; Leong, S.W.; Wan Ibrahim, W.N.; Mohammad Latif, M.A.; Ramli, A.N.M.; Mohd Aluwi, M.F.F.; Mohd Faudzi, S.M.; Kim, C.H. Development of diarylpentadienone analogues as alpha-glucosidase inhibitor: Synthesis, in vitro biological and in vivo toxicity evaluations, and molecular docking analysis. Bioorg. Chem. 2020, 104, 104277–104291. [Google Scholar] [CrossRef] [PubMed]
- Abu Bakar, N.; Mohd Sata, N.S.; Ramlan, N.F.; Wan Ibrahim, W.N.; Zulkifli, S.Z.; Che Abdullah, C.A.; Ahmad, S.; Amal, M.N. Evaluation of the neurotoxic effects of chronic embryonic exposure with inorganic mercury on motor and anxiety-like responses in zebrafish (Danio rerio) larvae. Neurotoxicol. Teratol. 2017, 59, 53–61. [Google Scholar] [CrossRef] [PubMed]


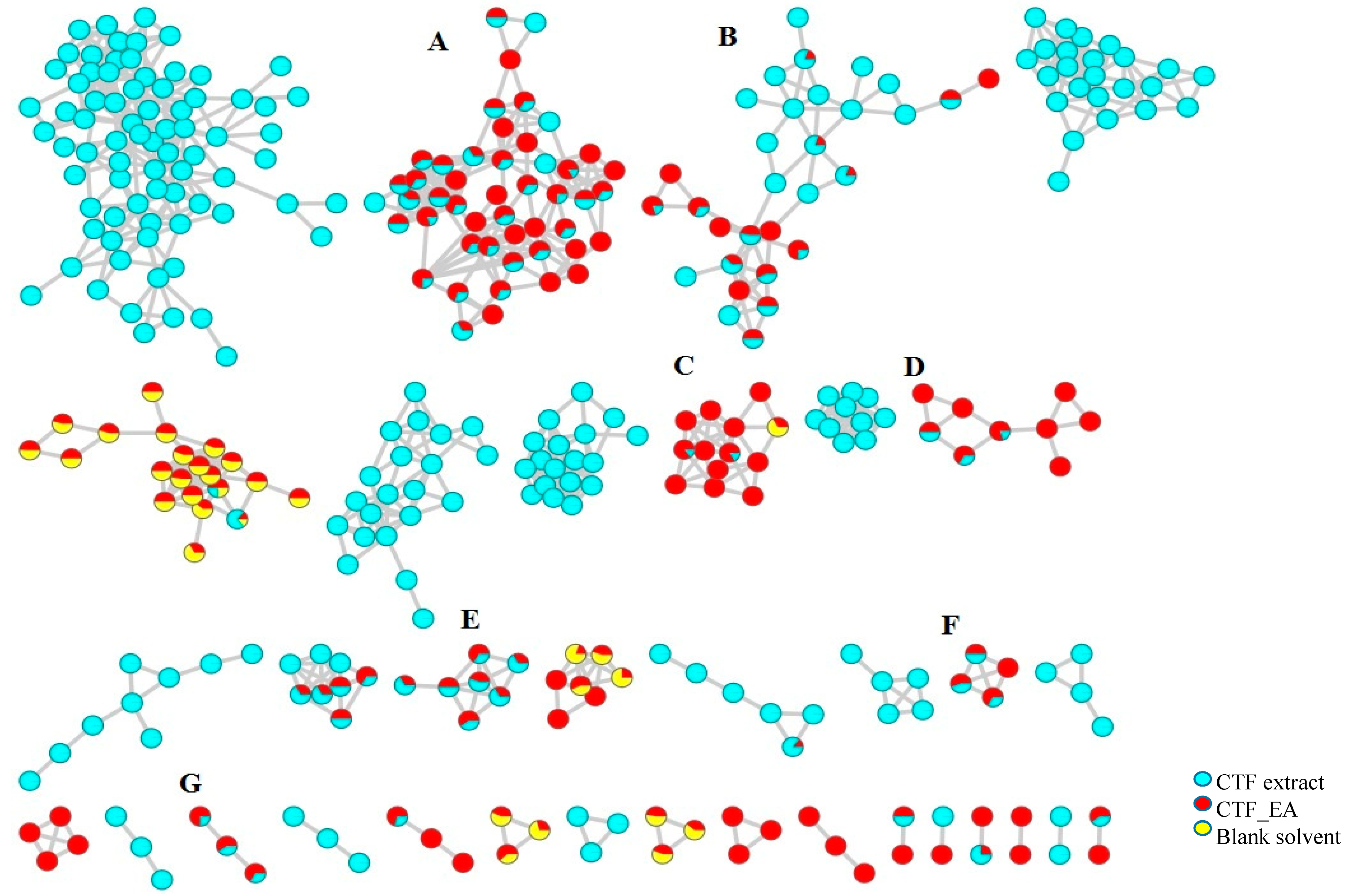
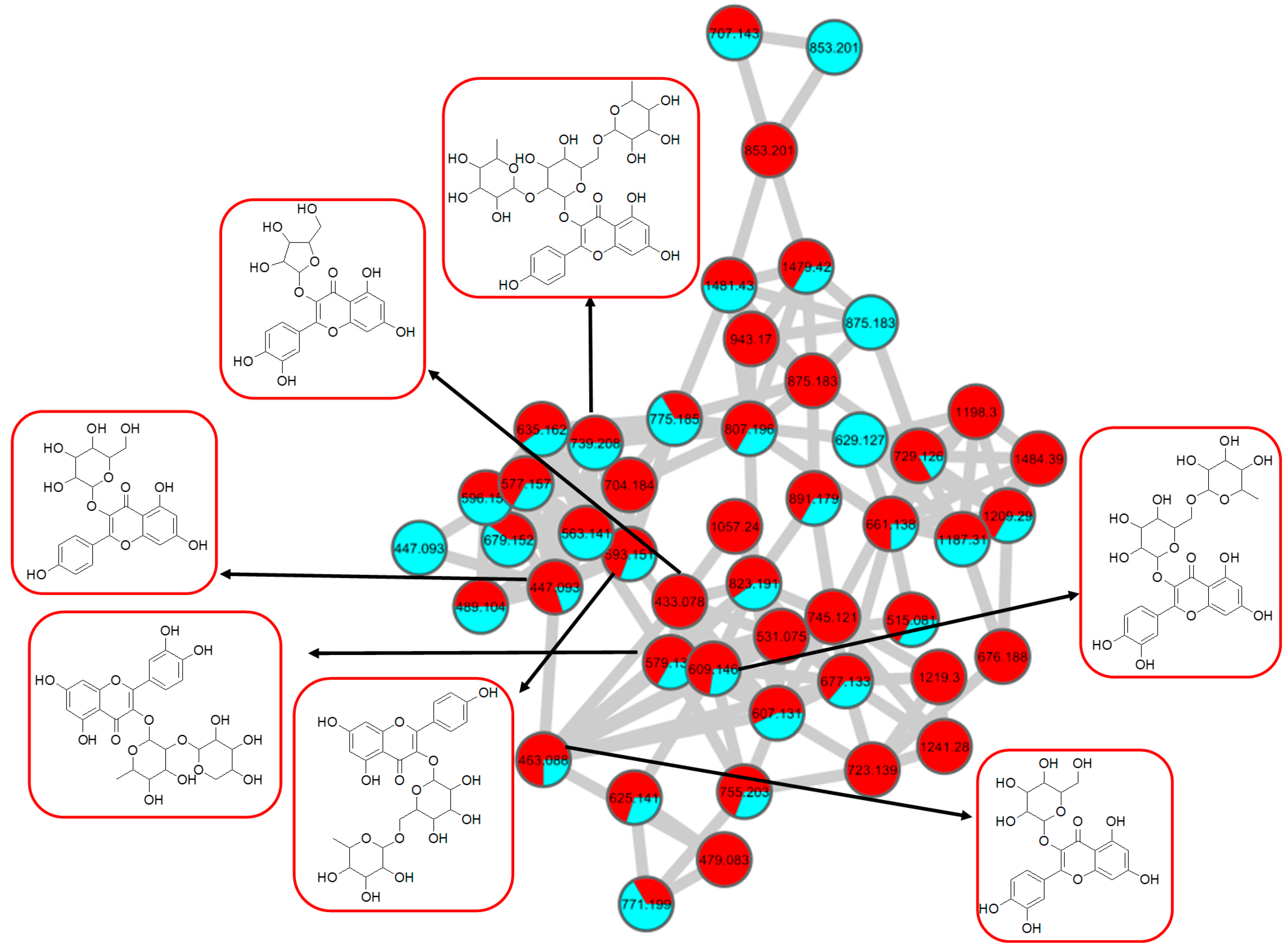

| Peak No. | Putative Identification | Molecular Formula | RT (min) | Precursor Ion (m/z) | Ion Type | Main Fragments (m/z) | Wavelength (nm) | References |
|---|---|---|---|---|---|---|---|---|
| Cluster A: Flavonol 3-O-glycosides | ||||||||
| 1. | Kaempferol-3-O-rutinoside | C27H30O15 | 5.59 | 593.1503 | [M-H]− | 284, 285, 255, 227, 151 | 208, 220, 266, 350 | GNPS |
| 2. | Kaempferol-3-O-glucoside | C21H20O11 | 6.28 | 447.1345 | [M-H]− | 284, 285, 255, 227, 151 | 218, 266, 294, 344 | GNPS |
| 3. | Kaempferol 3-(6″-acetyl-glucoside) | C23H22O12 | 6.80 | 489.1045 | [M-H]− | 285, 284, 255, 227 | 220, 272, 294 | Metabolomics workbench |
| 4. | Kaempferol 3-(6G-malonyl-neohesperidoside) | C30H32O18 | 6.16 | 679.1527 | [M-H]− | 635, 285, 284, 255, 227 | 220, 268, 298, 314, 344 | Metabolomics workbench |
| 5. | Kaempferol-3-O-α-L-rhamnosyl-(1->2)-O-L-rhamnoside | C27H30O14 | 7.28 | 577.269 | [M-H]− | 285, 284, 255, 227 | 220, 268, 298 | Metabolomics workbench |
| 6. | Kaempferol 3-O-(4″-O-acetyl)-rutinoside | C29H32O16 | 6.89 | 635.162 | [M-H]− | 284, 285, 255, 227, 151 | 220, 268, 298, 368 | https://mona.fiehnlab.ucdavis.edu/ (accessed on 13 August 2021) |
| 7. | Kaempferol-3-O-(2-rhamnosyl)-rutinoside | C33H40O19 | 5.20 | 739.1042 | [M-H]− | 284, 285, 255, 227, 151 | 198, 266, 350 | GNPS |
| 8. | Avicularin | C20H18O11 | 5.82 | 433.0780 | [M-H]− | 301, 300, 271, 255, 151 | 220,268,312 | GNPS |
| 9. | Quercetin-3-O-deoxyhexosyl- (1–2) pentoside | C26H28O15 | 5.70 | 579.1364 | [M-H]− | 301, 300, 271, 255, 151 | 218, 266, 350 | GNPS |
| 10. | Rutin | C27H30O16 | 5.40 | 609.1461 | [M-H]− | 301, 300, 271, 255, 151 | 206, 258, 354 | GNPS |
| 11. | Isoquercetin | C21H20O12 | 5.51 | 463.0887 | [M-H]− | 301, 300, 271, 255, 151 | 206, 266, 350 | GNPS |
| 12. | Quercetin 3-(2G-glucosyl-rutinoside) | C33H40O21 | 5.84 | 771.1786 | [M-H]− | 609, 463, 301, 300, 271, 255, 151 | 220, 268, 314 | Pubchem |
| 13. | Manghaslin | C33H40O20 | 4.72 | 755.2035 | [M-H]− | 301, 300, 271, 255, 151 | 206, 256, 354 | Metabolomics workbench |
| Cluster B: Hydrocinnamic acids and derivatives | ||||||||
| 14 | 3-Phenyl-2-[(2S,3R,4S,5S,6R)-3,4,5-trihydroxy-6-[[(E)-3-(4-hydroxy-phenyl)prop-2-enoyl]oxymethyl]-oxan-2-yl]oxyprop-2-enoic acid | C24H24O10 | 7.19 | 471.1300 | [M-H]− | 307, 163, 145, 119 | 220, 296, 368 | GNPS |
| 15 | Dimer 3-phenyl-2-[(2S,3R,4S,5S,6R)-3,4,5-trihydroxy-6-[[(E)-3-(4-hydroxy-phenyl)prop-2-enoyl]oxymethyl]-oxan-2-yl]oxyprop-2-enoic acid | 6.84 | 941.2737 | [M2-H]− | 779, 471, 163, 145 | 220, 272, 296, 368 | Putative annotation | |
| 16 | 3-(benzoyloxy)-2-hydroxypropyl β-D-glucopyranosiduronic acid | C16H20O10 | 2.04 | 371.0984 | [M-H]− | 370, 304, 174, 163, 146, 119 | 194, 298, 368 | GNPS |
| 17 | Feruloylquinic acid isomer | C17H20O9 | 6.23 | 367.1036 | [M-H]− | 303, 254, 193, 175, 160, 149, 134 | 218, 266, 346 | GNPS |
| 18 | Caffeic acid O-glucoside | C15H18O9 | 2.24 | 341.0877 | [M-H]− | 179, 135 | 214, 292, 368 | https://mona.fiehnlab.ucdavis.edu/ (accessed on 25 August 2021) |
| 19 | p-Coumaric acid 4-O-glucoside | C15H18O8 | 3.28 | 325.1843 | [M-H]− | 163, 145, 119 | 214, 290, 368 | (1) https://mona.fiehnlab.ucdavis.edu/ (accessed on 25 August 2021) (2) Metabolomics workbench |
| 20 | 3,5-Di-O-caffeoyl-4-O-coumaroylquinic acid | C34H30O14 | 6.78 | 661.1782 | [M-H]− | 205,163, 145, 119 | 220, 292, 368 | Metabolomics workbench (accessed on 25 August 2021) |
| Cluster C: Glycerophospholipid | ||||||||
| 21 | Lysophosphatidylmyoinositol | C27H53O12P | 21.89 | 599.3205 | [M-H]− | 283, 241, 152 | 224 | GNPS |
| 22 | Dipalmitoylphosphatidylglycerol | C38H75O10P | 35.31 | 721.3657 | [M-H]− | 255 | 220 | GNPS |
| 23 | Phosphatidylinositol lyso 16:0 | C25H49O12P | 18.61 | 571.2889 | [M-H]− | 255, 241, 152 | 224 | https://mona.fiehnlab.ucdavis.edu/ (accessed on 2 September 2021) |
| 24 | 1,2-Dioctanoyl-sn-glycero-3-phospho-1D-myo-inositol | C25H47O13P | 20.18 | 585.3047 | [M-H]− | 269, 241, 152 | 224 | Pubchem |
| Cluster D: Amino acids | ||||||||
| 25 | N-Fructosyl pyroglutamate | C11H17NO8 | 0.66 | 290.0803 | [M-H]− | 200. 128 | 196, 264, 370 | (1) https://mona.fiehnlab.ucdavis.edu/ (accessed on 7 September 2021)(2) Metabolomics workbench |
| 26 | Diglucoside pyroglutamate | C17H27NO13 | 0.72 | 470.1507 | [M-H]− | 128 | 266 | Putative annotation (accessed on 7 September 2021) |
| 27 | Fructosylvaline | C11H21NO7 | 0.74 | 278.1246 | [M-H]− | 214, 128, 116 | 256, 266 | Pubchem |
| 28 | Agropinic acid | C11H19NO8 | 0.86 | 292.8916 | [M-H]− | 274, 128 | 204, 260 | Pubchem |
| Cluster E: Carbohydrates | ||||||||
| 29 | Sucrose | C12H22O11 | 2.24 | 341.0877 | [M-H]− | 179, 135 | 214, 292, 368 | GNPS |
| 30 | Sucrose adduct chloride | C12H22O11 | 0.64 | 377.0854 | [M+Cl]− | 341, 215, 179, 89, 59 | 194, 266, 370 | Literature |
| 31 | 6-epi-7-Isocucurbic acid glucoside | C18H30O8 | 7.33 | 373.1871 | [M-H]− | 174, 119, 113, 101, 89, 71, 59 | 220, 268, 298, 368 | Pubchem |
| 32 | (2R,3R,4S,5S,6R)-2-[(3S,4S,5R)-3,4-Dihydroxy-2,5-bis(hydroxymethyl)-oxolan-2-yl]oxy-3,4,5-trihydroxy-6-(hydroxymethyl)oxan-2-yl] 3-methylbutanoate | C17H30O13 | 5.62 | 441.1745 | [M-H]− | 330, 139, 119, 113, 101, 89, 71, 59 | 208,266, 350 | Pubchem |
| 33 | Methyl 2-[(1R)-2-[(Z)-pent-2-enyl]-3-[(2R,3R,4S,5S,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)oxan-2-yl]oxy-cyclopentyl]acetate | C19H32O8 | 3.71 | 387.1152 | [M-H]− | 352, 274, 163, 113, 101, 89, 71, 59 | 216, 272, 298 | Pubchem |
| Cluster F: Mono-methoxylflavonol 3-O-glycoside | ||||||||
| 34 | Isorhamnetin-3-galactoside-6’’-rhamnoside | C28H32O16 | 5.70 | 623.1410 | [M-H]− | 315, 314, 299, 271, 151 | 218, 266, 350 | GNPS |
| 35 | Rhamnetin-3-O -gentiobioside | C28H32O17 | 5.03 | 639.2764 | [M-H]− | 315, 314, 299, 271, 255, 165, 121 | 204, 258, 354 | GNPS |
| 36 | 3-((6-(((3,5-Dihydroxy-6-methyl-4-((3,4,5-trihydroxy-6-methyl-tetrahydro-2H-pyran-2-yl)oxy)-tetrahydro-2H-pyran-2-yl)oxy)-methyl)-3,4,5-trihydroxytetrahydro-2H-pyran-2-yl)oxy)-5,7-dihydroxy-2-(4-hydroxy-3-methoxyphenyl)-4H-chromen-4-one | C34H42O20 | 5.32 | 769.2203 | [M-H]− | 605, 314, 299, 271, 151 | 206, 258, 354 | Putative annotation |
| Cluster G: Saccharolipid | ||||||||
| 37 | 1-O-[(2E)-6-[[3,4-bis-O-[(2E)-6-hydroxy-2,6-dimethyl-1-oxo-2,7-octadien-1-yl]- β-D-glucopyranosyl]-oxy]-2,6-dimethyl-1-oxo-2,7-octadien-1-yl] β-D-Glucopyranose, | C42H64O17 | 4.69 | 885.1616 | [M+HCOO-]− | 839, 793, 491, 399, 356, 303 | 206, 256, 354 | https://mona.fiehnlab.ucdavis.edu/ (accessed on 25 September 2021) |
| Enzyme | Binding Energy (kcal/mol) | |||
|---|---|---|---|---|
| Co-Crystallized Ligand | Compound 1 | Compound 7 | Compound 10 | |
| P38 | −8.85 | −7.82 | −2.29 | −6.79 |
| ERK-2 | −7.55 | −7.08 | −6.04 | −5.78 |
| iNOS | −7.64 | −10.11 | −8.78 | −8.33 |
| JNK | −9.24 | −7.95 | −7.79 | −6.69 |
| COX-2 | −10.55 | −4.98 | −4.59 | −3.47 |
| Compound | Binding Energy (kcal/mol) | Interactions | |||
|---|---|---|---|---|---|
| Hydrogen Bond | Hydrophobic | ||||
| π–Alkyl | π–Sigma | π–π Stacked | |||
| 1 | −10.11 | Thr184, 3 Arg193, Cys194, Gly196, Pro344, Asn364 | 2 Cys194, Leu203 | Ala191 | 2 Trp188, 2 Phe363 |
| 7 | −8.78 | Arg193, Cys194, Trp366, 3 Glu371, Asp376 | Ala191, Arg193 | Val346 | |
| 10 | −8.33 | Arg193, Pro344, Ala345, Asn364, Tyr367, Tyr483 | Ala191, Arg193, Cys194, Met149 | Ala191, Cys194 | Trp188, 2 Phe363 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mat Zian, N.F.A.; Swain, P.; Mohd Faudzi, S.M.; Zakaria, N.; Wan Ibrahim, W.N.; Abu Bakar, N.; Shaari, K.; Stanslas, J.; Choi, T.-I.; Kim, C.-H. Mapping Molecular Networks within Clitoria ternatea Linn. against LPS-Induced Neuroinflammation in Microglial Cells, with Molecular Docking and In Vivo Toxicity Assessment in Zebrafish. Pharmaceuticals 2022, 15, 467. https://doi.org/10.3390/ph15040467
Mat Zian NFA, Swain P, Mohd Faudzi SM, Zakaria N, Wan Ibrahim WN, Abu Bakar N, Shaari K, Stanslas J, Choi T-I, Kim C-H. Mapping Molecular Networks within Clitoria ternatea Linn. against LPS-Induced Neuroinflammation in Microglial Cells, with Molecular Docking and In Vivo Toxicity Assessment in Zebrafish. Pharmaceuticals. 2022; 15(4):467. https://doi.org/10.3390/ph15040467
Chicago/Turabian StyleMat Zian, Nurul Farah Adni, Puspanjali Swain, Siti Munirah Mohd Faudzi, Norzalina Zakaria, Wan Norhamidah Wan Ibrahim, Noraini Abu Bakar, Khozirah Shaari, Johnson Stanslas, Tae-Ik Choi, and Cheol-Hee Kim. 2022. "Mapping Molecular Networks within Clitoria ternatea Linn. against LPS-Induced Neuroinflammation in Microglial Cells, with Molecular Docking and In Vivo Toxicity Assessment in Zebrafish" Pharmaceuticals 15, no. 4: 467. https://doi.org/10.3390/ph15040467
APA StyleMat Zian, N. F. A., Swain, P., Mohd Faudzi, S. M., Zakaria, N., Wan Ibrahim, W. N., Abu Bakar, N., Shaari, K., Stanslas, J., Choi, T.-I., & Kim, C.-H. (2022). Mapping Molecular Networks within Clitoria ternatea Linn. against LPS-Induced Neuroinflammation in Microglial Cells, with Molecular Docking and In Vivo Toxicity Assessment in Zebrafish. Pharmaceuticals, 15(4), 467. https://doi.org/10.3390/ph15040467








